Abstract
Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base-pairing manually for any given target structure. Here, we report a general, top-down strategy to design nearly arbitrary DNA architectures autonomously based only on target shape. Objects are represented as closed surfaces rendered as polyhedral networks of parallel DNA duplexes, which enables complete DNA scaffold routing with a spanning tree algorithm. The asymmetric polymerase chain reaction was applied to produce stable, monodisperse assemblies with custom scaffold length and sequence that are verified structurally in 3D to be high fidelity using single-particle cryo-electron microscopy. Their long-term stability in serum and low-salt buffer confirms their utility for biological as well as nonbiological applications.
DNA nanotechnology offers the ability to synthesize highly structured nanometer-scale assemblies that in principle could rival the geometric complexity found in natural protein and nucleic acid assemblies. The past decade has witnessed dramatic growth in the diversity of structured DNA assemblies that can be programmed from the bottom-up to self-assemble into target shapes using complementary Watson-Crick base pairing (1–7). Scaffolded DNA origami is a particularly powerful means of synthesizing structured DNA assemblies, offering full control over both molecular weight and intricate nanometer-scale structure, with near quantitative yield of the programmed product that relies on a single-stranded DNA template (2, 5, 8, 9). Wireframe topologies based on the scaffolding principle have further demonstrated highly versatile control over 2D and 3D spatial architecture (10–13).
Similar to the challenge of structure-based protein sequence design, which seeks to infer the amino acid sequence needed to fold a target protein structure of interest (14, 15), achieving a general strategy for structure-based design of synthetic DNA assemblies represents a major challenge as well as opportunity for nanotechnology. While numerous computational design tools exist to aid in the bottom-up, manual programming of scaffolded DNA origami, which requires complex scaffold routing and staple design to realize a target geometry based on Watson-Crick base complementarity, only one approach offers a solution to the inverse problem of sequence design based on specification of target geometry (13). However, this approach is only semi-automated and relies on single duplex DNA arms and multi-way-junctions to represent polyhedral geometries, which may result in compliant and unstable assemblies that are unsuitable for many applications. Moreover, programmed geometries must be topologically equivalent to a sphere, significantly limiting its scope.
As an alternative, here we introduce the fully automatic inverse design procedure DAEDALUS (DNA Origami Sequence Design Algorithm for User-defined Structures) that programs arbitrary wireframe DNA assemblies based on an input wireframe mesh without reliance on user feedback or limitation to spherical topologies. We apply our procedure to design 35 Platonic, Archimedean, Johnson, and Catalan solids, six asymmetric structures specified using surface geometry alone, as well as four polyhedra with nonspherical topologies. Designed sequences are used to synthesize icosahedral, tetrahedral, cuboctahedral, octahedral, and reinforced hexahedral structures using the asymmetric PCR (aPCR) for facile production of single-stranded scaffolds of custom length and sequence. Programmed objects are confirmed using cryo-electron microscopy (cryo-EM), folding, and stability assays, to be both high fidelity structurally as well as stable under low-salt buffer conditions important to biological as well as in vitro applications. These results demonstrate the broad applicability of our design and synthesis strategy for numerous potential applications in biomolecular science and nanotechnology including nanoparticle (NP) delivery (16, 17), photonics (18, 19), inorganic NP synthesis (20, 21), memory storage (22–24), and single-particle cryo-EM analysis (3, 25–27), among others (7, 28, 29). The ability to synthesize nearly arbitrary geometric shapes that are automatically rendered from the top-down should enable the broad participation of nonexperts in this powerful molecular design paradigm.
Top-down Automatic Sequence Design
To enable the fully automatic and robust inverse design of programmed DNA assemblies, we chose to render arbitrary geometries as node-edge networks based on the DX-based wireframe motif in which inter-connected edges consist of two duplexes joined using antiparallel (DX) crossovers (Fig. 1) (3, 10–12, 30). This strategy offered application of our procedure to any closed geometric surface including nonspherical topologies such as a torus, provided that it can be rendered using polyhedral surface meshes. Using this approach, the spatial coordinates of all vertices, the edge connectivities between vertices, and the faces to which vertices belong fully specify the target object (31). Standard polyhedron file formats containing this information are converted into this set of arrays, providing input to our scaffold routing and staple design procedure (Fig. 1, step i, and Fig. 2A). Programmed edges are required to consist of multiples of 10.5 bp rounded to the nearest nucleotide, as commonly assumed in DNA origami design to satisfy the natural helicity of B-form DNA. Obeying the natural geometry of DNA ensures that no over- or underwinding in duplexes occurs (32, 33), which may otherwise result in shape distortions that force deviation from the target geometry and would require iterative, ad hoc adjustment of edge lengths and sequence design (13). Notably, our algorithm has no theoretical limitation on the length of scaffold that can be used to program the target DNA origami object.
Fig. 1. Top-down sequence design procedure for scaffolded DNA origami nanoparticles of arbitrary shape.
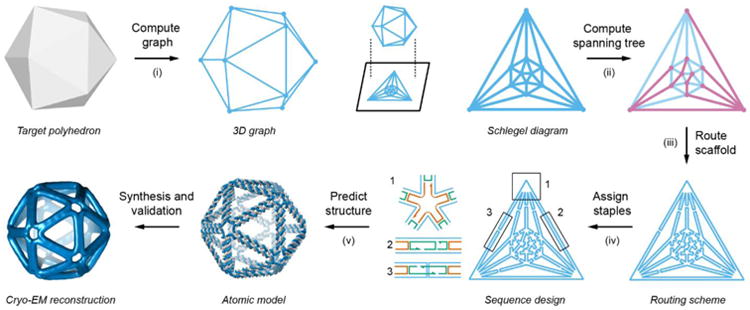
Specification of the arbitrary target geometry is based on a continuous, closed surface that is discretized using polyhedra. This discrete representation is used (step i) to compute the corresponding 3D graph and (step ii) spanning tree. The spanning tree is used (step iii) to route the single-stranded DNA scaffold throughout the entire origami object automatically, which then enables (step iv) the assignment of complementary staple strands. Finally, (step v) a 3D atomic-level structural model is generated assuming canonical B-form DNA geometry, which is validated using 3D cryo-EM reconstruction.
Fig. 2. Fully automatic sequence design of 45 diverse scaffolded DNA origami nanoparticles.
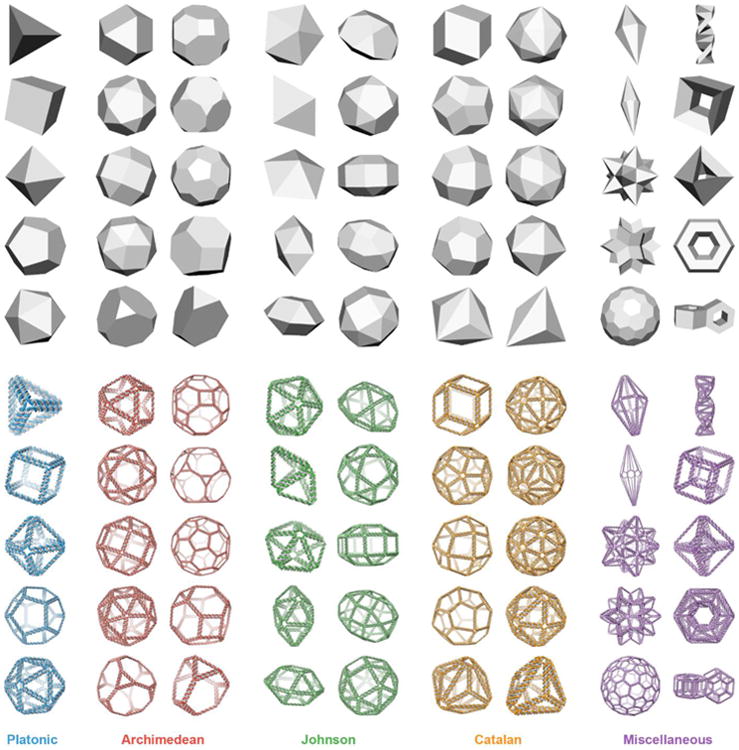
(Top) Face-shaded 3D representations of geometric models used as input to the algorithm. (Bottom) 3D atomic models of DNA-rendered nanoparticles for (blue) Platonic, (red) Archimedean, (green) Johnson, (orange) Catalan, and (violet) miscellaneous polyhedra generated using the automatic scaffold routing and sequence design procedure (particles are not shown to scale). Miscellaneous polyhedra include (first column) heptagonal bipyramid; enneagonal trapezohedron; small stellated dodecahedron, a type of Kepler-Poinsot solid; rhombic hexecontahedron, a type of zonohedron; Goldberg polyhedron G(2,1) with symmetry of Papillomaviridae; (second column) double helix; nested cube; nested octahedron; torus; and double torus. Platonic, Archimedean, and Johnson solids each have 52-bp edge length, Catalan solids and the first column of miscellaneous polyhedra have minimum 42-bp edge length, and the second column of miscellaneous polyhedra have minimum 31-bp edge length. 30 of the 45 structures shown have scaffolds smaller than the 7,249-nt M13mp18 whereas 15 have scaffold lengths that exceed it (Table S2).
Representing the target geometry as a polyhedral mesh that satisfies the preceding design criteria guarantees that a single-stranded scaffold can be routed uniquely throughout the entire object with an Eulerian circuit, without modifications to the target geometry. From the mesh, the graph of the target structure is computed, containing the vertex, edge, and face information (Fig. 1, step i). Scaffold routing is then assigned using a spanning tree (Fig. 1, step ii) generated with Prim's algorithm (Fig. S1) (34). Each of the edges that is a member of the spanning tree is assigned no scaffold crossover, whereas each remaining edge is assigned one scaffold crossover. For a given graph, every spanning tree therefore corresponds to a unique scaffold routing, where by default Prim's algorithm generates a maximally-branching spanning tree (Fig. S1 to S2) that has been suggested for folding 2D nets to self-assemble more reliably than linear trees (35). The use of DX arms to represent edges ensures that a solution to the scaffold routing problem is obtained efficiently in solution time that scales as E log V, where E and V are the number of edges and vertices, respectively (34). To complete the scaffold routing, a scaffold crossover is placed at the center of each edge that is not part of the preceding spanning tree. From the scaffold crossover positions and spanning tree, the circuit for the scaffold routing is determined, specifying the order in which the scaffold visits each vertex and crossover while ensuring that the scaffold does not intersect itself at vertices (Fig. 1, step iii) (36). A linear scaffold nick position is set to ensure that it is non-coincident with crossovers and other nicks, with the polarity of its routing chosen to be counter-clockwise around each face because of the preference of the major groove to orient inwards at vertices (37). Thus, use of our spanning tree approach enables fully automatic conversion of the input polyhedral geometry to full scaffold routing based on the single circuit that traverses each duplex once.
With the scaffold routing determined, staple strands are assigned automatically by using distinct rules for vertex versus edge staples enabling the assignment of staple strand sequences assuming Watson-Crick base complementarity (Fig. 1, step iv) (31). Vertex staples hybridize to the scaffold in the 10–11 bp closest to vertices, comprising 52- and 78-nt staples, the numbers of which are determined by the degree of the vertex (31). Edge staples occupy the intermediate regions, spanning across scaffold crossovers to help rigidify each edge by creating crossovers every 10, 11, or 21 bp (31). Finally, the positions and orientations of each nucleotide are modeled to predict the 3D structure of the NP (Fig. 1, step v, and Fig. S3) (31). Critically, in contrast with previous tile-based approaches that used this DX-motif to synthesize NPs of diverse form (3), the use of a single-stranded DNA scaffold that is routed throughout the entire object in our strategy offers near quantitative yield of the final product in its self-assembly, no dependence on relative multi-arm junction tile concentrations, and full control over DNA sequence. This final feature is essential to biomolecular applications that use spatially specific asymmetric sequence programming for protein or RNA scaffolding as well as other chemical functionalization.
To test the generality and robustness of our design procedure to be applied to diverse polyhedral geometries, we first applied it to design Platonic solids that have equal edge lengths, angles, and vertex-degree, followed by geometries of increasing complexity including Archimedean solids with unequal vertex angles, Johnson solids that include heterogeneity in vertex degree, and Catalan solids that have unequal edge lengths (Fig. 2). Applicability to asymmetric and non-convex objects specified using surface geometry alone (13) was also confirmed (Fig. 2 and Fig. S4), in addition to nonspherical topologies including a nested cube, a nested octahedron, and tori that have not been previously realized experimentally (3, 12) and cannot be solved computationally using existing procedures (13). 15 of these structures required scaffolds longer than the 7,249-nt M13mp18 (38), for which random sequences of appropriate length were generated (Fig. 2, bottom). Taken together, these examples illustrate the broad ability of our procedure to automatically generate complex scaffold and staple routings for diverse geometries based on top-down geometric specification alone (Fig. 2).
Custom Scaffold Production with Asymmetric PCR
To investigate the synthetic yield and homogeneity of self-assembled objects programmed with our computationally-generated scaffold and staple designs, we used the aPCR (39) to generate object-specific scaffolds for folding (Figs. S6 to S7) (31). These custom scaffolds minimize excess single-stranded DNA in the final structure, which may result in nonspecific object aggregation or otherwise interfere with folding as well as downstream chemical functionalization (Fig. 3 and Fig. S8) (2, 40, 41). Monodispersity of multiple custom linear short scaffold strands synthesized ranging from 450 to 3,400 nucleotides were first confirmed using gel electrophoresis of aPCR products based either on the M13pm18 ssDNA plasmid or dsDNA fragments as templates (Fig. 3). Custom scaffolds were used to fold tetrahedra of 31-, 42-, 52-, 63-, and 73-bp edge lengths in addition to an octahedron, two pentagonal bipyramids (42- and 52-bp edge lengths), a cube, a reinforced cube, an icosahedron, and a cuboctahedron (Fig. 4 and Figs. S8 to S18) (31). Folded objects were analyzed using single-particle AFM following purification, which confirmed their folding yields of up to 90% (Table S3) and particle homogeneity that is characteristic of scaffolded DNA origami objects (Fig. 2 and Figs. S12 to 18). Importantly, application of our aPCR approach offers folded sample purity that is similar to existing synthesis strategies that utilize restriction enzymes to generate subfragment scaffolds (Fig. 3 and Figs. S6 to S7) (42), yet without dependence on restriction sites and with higher synthetic yield. Redesign of vertex staple nicks to be positioned at crossovers instead of interior segments of duplexes also resulted in increased folding stability (Fig. S19) (31). Thus, diverse polyhedral origami objects from 200 kDa to 1 MDa programmed with our top-down, inverse sequence design procedure self-assembled robustly by using scaffolds of custom length and sequence, which may be natural or synthetic.
Fig. 3. aPCR strategy to synthesize custom single-stranded DNA scaffolds.
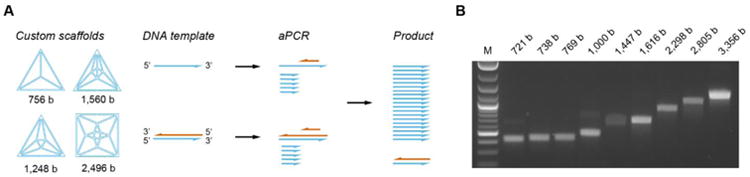
(A) Single-stranded DNA (ssDNA) scaffolds of custom length and sequence for each target structure are amplified using either a single- or double-stranded DNA template mixed with appropriate primer pairs consisting of 50× sense primer and 1× anti-sense primer concentration relative to the scaffold concentration. (B) Amplified ssDNA products are purified and analyzed using agarose gel electrophoresis.
Fig. 4. Folding and 2D structural characterization of scaffolded DNA origami nanoparticles.
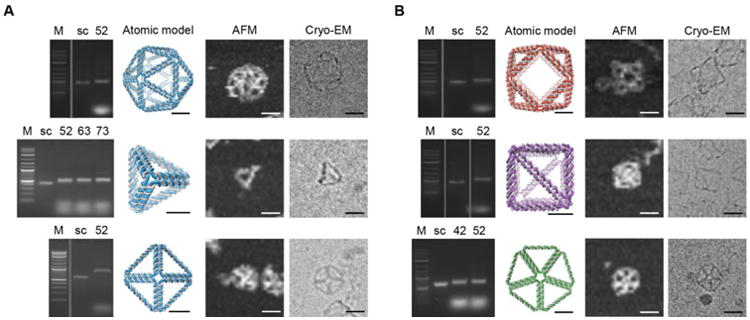
(A) Characterization of folding for five platonic solids (52-bp, 63-bp, and 73-bp edge-length tetrahedra; 52-bp edge-length octahedron; 52-bp edge-length icosahedron) using AGE, AFM and cryo-EM. (B) Characterization of folding for one Archimedean solid (52-bp edge-length cuboctahedron), one miscellaneous solid (reinforced cube with 52- and 73-bp edge lengths), and one Johnson solid (42- and 52-bp edge-length pentagonal bipyramid); using AGE, AFM and cryo-EM. M: DNA marker; sc: custom ssDNA scaffold. Scale bars are 20 nm for AFM and cryo-EM and 10 nm for atomic models.
Cryo-EM 3D Reconstruction
Next, we evaluated the structural fidelity of programmed origami objects using single-particle cryo-EM and 3D reconstruction (Fig. 5 and Figs. S20 to S27) (31). Cryo-EM imaging confirmed the abundance of well-folded single DNA NPs of expected sizes and shapes, which were used to generate 3D density maps (Fig. 5). All maps were validated by matching class averages, map projections, and tilted-pair images (Figs. S28 to S33) (31). 3D atomic structures of programmed DNA origami objects were predicted using a rigid duplex model in which each edge is composed of two parallel duplexes with the central axis of each edge meeting at a single point that is specified in the input graph (31). Quantitative comparison of model predictions with cryo-EM reconstructions revealed favorable agreement, with correlations ranging from 0.73 for the tetrahedron to 0.92 for the cuboctahedron, and resolutions comparable to previous wireframe DX designs based on tile assembly that produces polydisperse samples (Fig. 5A and Fig. S28) (3). Interestingly, the tetrahedron exhibited outward bowing of its edges (Fig. 5B and Fig. S29), which has been previously observed for single-duplex edges (43), and may be attributable to its smaller acute interior angle compared with other designed objects, which may result in steric overlap of adjoining DNA duplexes at vertices. Structure-based molecular modeling is of interest to test possible redesigns of the tetrahedral object that may improve agreement with the target, input geometry. Comparison of the cuboctahedron reconstruction with two competing models for vertex geometry suggested that duplex overlap is preferred over backbone stretching in determining overall equilibrium shape (Fig. 5D and Figs. S3 and S30).
Fig. 5. 3D structural characterization of scaffolded DNA origami nanoparticles using cryo-EM reconstruction and comparison with model predictions.

(A) Programmed edges of the 52-bp edge-length icosahedron are straight and vertices are rotationally symmetric, as designed. Cryo-EM resolution is 2.0 nm and correlation with model is 0.85 (54). (B) Edges of the 63-bp edge-length tetrahedron reveal significant outward bowing (arrow) that is attributable to its acute interior angles that might result in steric hindrance. Cryo-EM resolution is 1.8–2.2 nm and correlation with model is 0.72. (C) 15° right-handed twist is visible at each vertex (arrow) of the 52-bp edge-length octahedron, which suggests that the structure folds as prescribed rather than “inside-out.” Cryo-EM resolution is 2.5 nm and correlation with model is 0.89. (D) 52-bp edge-length cuboctahedron has unequal angles between edges that meet at vertices (arrows), which supports a rigid-duplex model in which phosphate backbone stretch is minimized (31). Cryo-EM resolution is 2.9 nm and correlation with model is 0.92. (E) The addition of 73-bp reinforcing struts to a simple cube of 52-bp edge-length increases its structural homogeneity to produce a 3D reconstruction with 915 particles. With the reinforcement, the particles maintain right-angled vertices (upper arrow). The diagonal edges form a tetrahedral symmetry that exhibits outward bowing (lower arrow). Cryo-EM resolution is 2.7 nm and correlation with model is 0.72. (F) 3D reconstruction of a nested cube within a cube that has nonspherical topology. The 73-bp edge-length outer cube is connected to a 32-bp edge-length inner cube by eight 31-bp edge-length diagonals. Cryo-EM resolution is 4.0–4.5 nm and correlation with model is 0.74. Scale bars are 5 nm.
Importantly, cryo-EM reconstructions suggested that origami objects assembled as designed instead of “inside-out” while satisfying programmed Watson-Crick base pairing from sequence design (31). This result reaffirms the suitability of our sequence design algorithm to choose to point the major groove inwards at vertices, which was based on the previous observation that DNA origami folds in this manner (37). These two variants are distinguished experimentally by the designed asymmetry that places a 1-bp overhang on the 5′ end of the scaffold on each edge to keep the scaffold and staple crossovers perpendicular to the helical axis. While 3′ end overhangs would have also achieved this, our choice of 5′ end overhangs anticipates a right-handed twist of the central hole of each vertex when the structures fold as prescribed, which is supported experimentally for the octahedron in which the chirality of the vertex twist is apparent (Fig. 5C and Fig. S31). Notwithstanding, the extent to which the edges twist is not predicted by a simple geometric model that assumes edges meet straight-on at vertices, deviating from the approximately 15° right-handed twist observed experimentally.
Although individual DX-based polyhedral edges are expected to be more structurally rigid than single-duplex edges employed previously (13), some surfaced-rendered polyhedral objects may still be expected to be structurally compliant because of their overall geometry, as observed in the case of the simple cube with 52-bp edges that was initially observed using AFM and cryo-EM to fold into heterogeneous single particles (Fig. 2 and Figs. S34 to S36). To test the ability of our algorithm to reprogram such flexible objects to render them rigid for applications including single-particle cryo-EM at subnanometer resolution that requires highly homogeneous structural populations for reconstruction, we applied it to redesign a new, reinforced cube (Fig. 5E and Fig. S32). We hypothesized that the observed heterogeneity in the simple cube resided in its vertex flexibilities, in which right-angle vertices can change in concert while maintaining constant edge lengths in shearing modes. To eliminate this compliant mode of deformation we sought to introduce cross-bars on each face. Reinforced structures folded into highly homogeneous hexahedral objects that enabled single-particle cryo-EM imaging and reconstruction that agreed with 3D atomic model predictions (Fig. 5E and Fig. S32). Surprisingly, constraining edge lengths to be multiples of 10.5 bp did not prove restrictive in achieving this target redesign, which required introduction of 73-bp cross-bars across each face (Fig. 5E and Fig. S32). While in principle this agreement may be attributable to the overall symmetry of the object, further bowing of edges may equally have been anticipated based on steric overlap or duplex crowding at vertices. Given that uniform triangular polyhedra or other simple rules do not apply generally to the ultimate aim of rendering mechanically rigid surface geometries, generalizing this preceding redesign strategy from the hexahedron to other objects represents an important future challenge and demonstrates the versatility of our top-down sequence design procedure.
To test the ability of our algorithm to design scaffolded DNA origami objects of nonspherical topology and with internal structure that have not been previously realized experimentally (3, 12) and cannot be designed using existing top-down computational procedures (13), we synthesized the nested cube (Fig. 2B) and reconstructed its 3D structure using cryo-EM (Fig. 5F and Fig. S33). While the folding yield and cryo-EM resolution of this object were somewhat lower than for other objects (31), possibly due to the flexibility that is intrinsic to cube-like geometries, noted above, the ability to program internal structure significantly broadens the scope for synthesizing such complex scaffolded origami objects (44) in a one-step folding reaction.
Folding and Stability Characterization
An important limitation of DNA origami for biological as well as in vitro applications has been the requirement of high concentrations of either magnesium or monovalent cations for their folding and stability (41, 45), which was recently shown to be alleviated by the use of single-duplex edge meshworks that fold and are stable in physiological buffer and salt conditions (13). Investigation of the folding properties of DX-based objects synthesized here revealed that objects fold effectively in cation concentrations as low as 4 mM Mg2+ and 500 mM Na+ (Fig. 6 and Figs. S37 to S48), as well as in PBS alone (Figs. S49 to S53). While these results mirror those for wireframe structures employing single-duplex edges (13), our use of DX-arms and multiway junctions here results in structurally stable, rigid assemblies that are crucial to most applications (10, 11). To test the utility of our objects for cellular assays, post-folding in TAE-Mg2+ particles were transferred to PBS and Dulbecco's Modified Eagle's Medium (DMEM) containing 0 to 10% FBS, where they were found to be stable for at least 6 hours (Fig. 7 and Figs. S54 to S59). When transferred to salt-free solution post-folding, however, particles were observed to be unstable, confirming the well-known importance of a minimal amount of added salt for their longer-term stability (Fig. 7 and Fig. S58). Interestingly, folding of the 52-bp tetrahedron using the full M13 scaffold instead of its custom scaffold also required higher magnesium concentrations to achieve folding, reaffirming the importance of the use of scaffolds that eliminate excess in single-stranded DNA (Fig. S8).
Fig. 6. Characterization of scaffolded DNA origami nanoparticle folding in variable added salt.
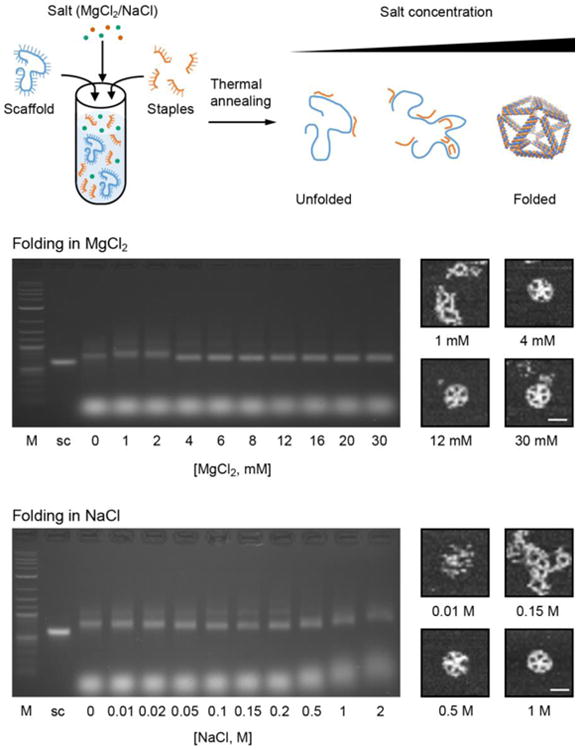
Characterization of folding of the 52-bp edge-length pentagonal bipyramid in increasing magnesium chloride (MgCl2) and sodium chloride (NaCl) concentration using 2% AGE and AFM imaging. Critical concentrations for folding are 4 mM MgCl2 and 500 mM NaCl in TRIS-acetate pH 8.0. M: DNA marker; sc: custom ssDNA scaffold. Scale bars are 30 nm.
Fig. 7. Characterization of scaffolded DNA origami nanoparticle stability in physiological buffer and serum.
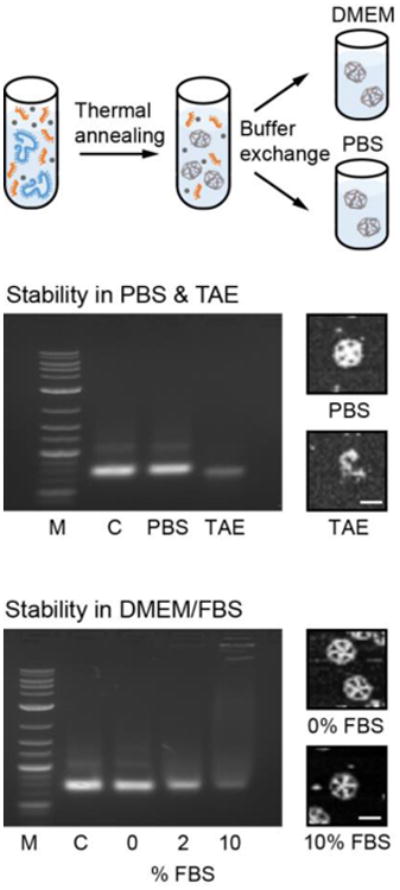
AGE and AFM structural characterization of the 52-bp edge-length pentagonal bipyramid after 6 hours in PBS, TAE (without added NaCl or MgCl2), and DMEM buffer with increasing concentration of FBS (0, 2, and 10%) after folding in TAE-Mg2+ buffer (12 mM MgCl2) followed by buffer exchange. Stability is observed for structures in PBS buffer but not in TAE due to the absence of salt, which demonstrates the importance of a minimal salt concentration for stability. AFM imaging reveals the presence of intact objects after 6 hours in DMEM media in the presence of 2 to 10% FBS despite partial degradation is observed in AGE. Scale bars are 30 nm.
Outlook
Structure-based, rational design of macromolecular assemblies including both nucleic acids and proteins is a long-standing aim of nanotechnology and biological engineering. Unlike proteins, which contain a myriad of specific and non-specific inter-residue interactions that determine their local and global folds, and RNA, which exhibits promiscuity in base-pairing and secondary structure, synthetic DNA assemblies are well established to be highly programmable using Watson-Crick base pairing alone (2, 46). In particular, wireframe polyhedral geometries offer the powerful ability to program nearly arbitrary 3D geometries on the nanometer scale, limited only by current size constraints imposed by single-stranded scaffold lengths. This important and versatile class of topologies therefore has broad potential for programming complex nanoscale geometries including biomimetic systems inspired by viruses, photosynthetic systems, as well as other natural highly evolved macromolecular assemblies. Achieving full automation of inverse sequence design using this versatile wireframe approach has the potential to realize the original vision of Ned Seeman to program nanoscale materials with full 3D control over positioning of all atomic-level groups (11, 46). As a major advance in this direction, we developed a top-down, geometry-driven sequence design procedure that uses a spanning tree algorithm to determine scaffold crossover positions. This enables efficient and unique routing of the single-stranded scaffold throughout any target origami object of arbitrary polyhedral shape and molecular weight, as well as automated staple assignment to enable custom synthesis of programmed origami objects of near quantitative yield and high fidelity atomic-level structure. Asymmetric PCR is demonstrated to provide full control over scaffold sequence and length, verified between 449 to 3356 nt, and use of the DX-based design further confers folding capacity and stability under diverse conditions including cell-compatible buffers. Combined, our strategy to realize the top-down design of nanoscale DNA assemblies offers full control over both 3D structure and local sequence which, together with our broadly usable software and experimental protocols, provides a versatile approach to the design of functionalized DNA objects of nearly arbitrary shape for photonic applications (18, 47, 48) including self-assembled super-lattices (49), cellular delivery (17, 50), memory storage devices (22-24), and single-particle cryo-EM reconstruction assays (3, 25-27) for proteins and RNAs that are not otherwise amenable to crystallography or NMR. Possible future generalizations of our approach may include rendering polyhedral networks with edges of arbitrary cross-section (4) for applications requiring further enhanced NP rigidity and closed surface topologies, such as inorganic nanoparticle synthesis (20, 21) and hybrid nanoscale materials (51).
Materials and Methods
Top-down sequence design
DAEDALUS is available for use as open source software (31) and online at http://daedalus-dna-origami.org. Additional details can be found in Supplementary Materials (31) and in documentation provided with the software.
Single stranded DNA scaffold amplification using aPCR
aPCR was performed using a sense primer concentration of 1 μM, an antisense primer concentration of 20 nM (Table S2), 30 ng of M13mp18 ssDNA template or 10 ng of dsDNA gBlocks, 200 μM of dNTPs mix in a final volume of 50 μL and 1 unit of Accustart Taq DNA polymerase HiFi. The aPCR program used is as follows: 94°C, 1 min for the initial denaturation; followed by 30–40 cycles of 94°C, 20 sec; 55–59°C, 30 sec; 68°C, 1 min per kb to amplify. PCR products were run through 1% low melting temperature agarose gel and were extracted and purified using Zymoclean Gel DNA recovery kit.
DNA origami assembly and purification
In a one pot reaction, 5 to 40 nM scaffold strand was mixed with 50 to 800 nM of a staple strands mix in a Tris-Acetate EDTA-MgCl2 buffer (40 mM Tris, 20 mM acetic acid, 2 mM EDTA, 12 mM MgCl2, pH 8.0). Annealing was performed in a thermal cycler with the following program: 95°C for 5 min, 80–75°C at 1°C per 5 min, 75–30°C at 1°C per 15 min, and 30–25°C at 1°C per 10 min. Fresh DNA origami solutions were purified using Amicon Ultra-0.5 mL centrifugal filter (MWCO 100 kDa) to remove excess of staple strands and stored at 4°C.
Stability experiments
After buffer exchange using Amicon Ultra-0.5 (MWCO 100 kDa DNA origami stability was evaluated for 6h in TAE, PBS, or DMEM buffer complemented with FBS.
Agarose gel electrophoresis
Samples were loaded in 2% agarose gel in Tris-Acetate EDTA buffer supplemented with 12 mM MgCl2 and pre-stained with EtBr. Gels were run on a BioRad electrophoresis unit at 4°C for 3–4 h under a constant voltage of 70 V. Gels were imaged using a Gene flash gel imager (Syngene, Inc.), and yield was estimated by analyzing the band intensity with the Gel Analyzer program in the ImageJ software (52).
Atomic force microscopy (AFM)
Samples for AFM were annealed at a concentration of 5 nM in TAE-Mg2+ (12 mM) buffer and subsequently purified before AFM imaging. 1.5 μL of the samples were deposited onto freshly peeled mica (Ted Pella, Inc.) and 10 μL of 1× TAE-Mg2+ buffer were added to enlarge the solution drop to cover the whole mica surface. 1.5 μL NiCl2 (100 mM) were added to the drop immediately. After leaving about 30 seconds for adsorption to the mica surface, 70 μL of 1× TAE-Mg2+ buffer were added to the samples and an extra 40 μL of the same buffer were deposited on the AFM tip. The samples were scanned in “ScanAssyst mode in fluid” using an AFM (Dimension FastScan, Bruker Corporation, Inc.) with SCANASSYST-FLUID+ tips.
Cryo-EM
15-25 tubes of samples were freshly annealed at a scaffold concentration of 20 nM in 50 μL TAE-Mg2+ buffer, pooled, purified, and concentrated using an Amicon column Ultra-0.5 mL centrifugal filter (MWCO 100 kDa) to a final volume of 20–30 μL. 2 μL of the freshly concentrated DNA nanostructure solution were applied onto the glow-discharged 200-mesh Quantifoil grid, blotted for 1.5 sec and rapidly frozen in liquid ethane using a Vitrobot Mark IV (FEI). All grids were screened on a JEM2200FS cryo-electron microscope (JEOL) operated at 200 kV with in-column energy filter with a slit of 20 eV. Micrographs of the icosahedron, tetrahedron, cuboctahedron, and octahedron were recorded with a direct detection device (DDD) (DE-20 4k×5k camera, Direct Electron, LP) operating in movie mode at a recording rate of 25 raw frames per second at 25,000× microscope magnification (corresponding to a calibrated sampling of 2.51 Å per pixel) and a dose rate of ∼21 electrons/sec/Å2 with a total exposure time of 3 sec. Micrographs of the reinforced cube and the nested cube were recorded on a 4k×4k CCD camera (Gatan, Inc.) at 40,000× microscope magnification (corresponding to a calibrated sampling of 2.95 Å per pixel) and a dose rate of ∼20 electrons/sec/Å2 with a total exposure time of 3 sec. A total of 180 images for the icosahedron, 91 images for the tetrahedron, 101 images for the cuboctahedron, 100 images for the octahedron, 36 images for the reinforced cube, and 152 images for the nested cube were collected with a defocus range of ∼1.5–4 μm.
Single-particle image processing and 3D reconstruction
Particle images recorded on DE-20 detector were motion corrected and radiation damage compensated (24). The motion correction was done with running averages of three consecutive frames using the DE_process_frames.py script (Direct Electron, LP). Single-particle image processing and 3D reconstruction was performed using the image processing software package EMAN2 (53). EMAN2 was used for initial micrograph evaluation, particle picking, contrast transfer function correction, 2D reference free class averaging, initial model building, and 3D refinement. All initial models for the DNA origami objects were built from 2D reference-free class averages. 3,650 particles for the icosahedron, 2,183 particles for the tetrahedron, 1,758 particles for the cuboctahedron, 2,678 particles for the octahedron, 915 particles for the reinforced cube, and 2,008 particles for the nested cube were used for final refinement, applying icosahedral, tetrahedral, octahedral, octahedral, tetrahedral, and octahedral symmetries, respectively. Resolutions for the final maps were estimated using the 0.143 criterion of the Fourier shell correlation (FSC) curve without any mask. A Gaussian low-pass filter was applied to the final 3D maps displayed in the UCSF Chimera software package (54). Correlation of each map with its corresponding atomic model is calculated by the UCSF Chimera fitmap function.
Tilt-pair validation for the cryo-EM map (55) was performed by collecting data at two goniometer angles, 0° and -10°, for each region of the grid. The test was performed using the e2tiltvalidate.py program in EMAN2. Additional details on the tilt-pair validation are provided in Supplementary Table S3.
qPCR thermal analysis
qPCR analyses were performed in a Roche LightCycler® 480. The scaffold concentration used for the tetrahedron folding analysis was 80 nM and the concentrations of each strand were adjusted to 1 μM for the three-way junction model. Samples were complemented with 1× final concentration of SYBR Green in a final volume of 20 μL. Fluorescence curves obtained were analyzed using first-order derivatives to identify transition temperatures.
Supplementary Material
Fig. 0. DNA nanoparticle design, synthesis, and characterization.
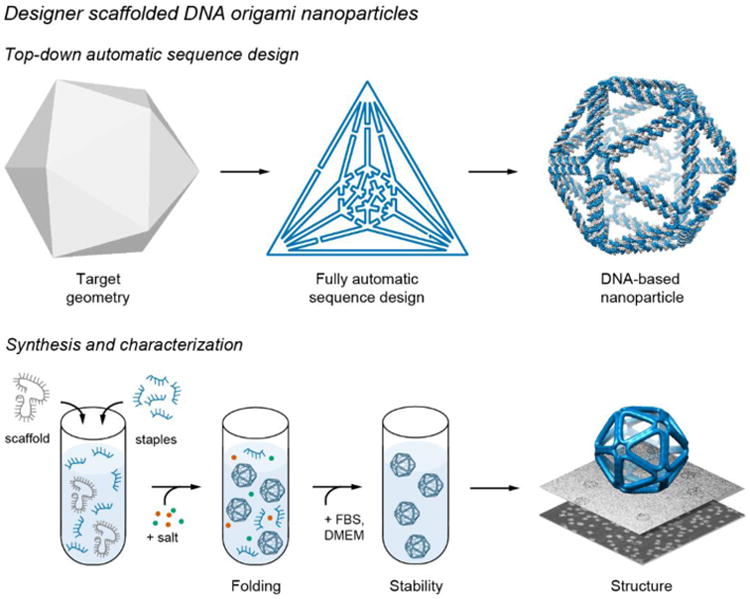
(Top) Top-down geometric specification of the target geometry is followed by fully automatic sequence design and 3D atomic-level structure prediction. (Bottom) The asymmetric PCR is used to synthesize object-specific single-stranded DNA scaffold for folding. Nanoparticle stability is characterized in cellular media with serum and nanoparticle 3D structure is characterized using single-particle cryo-electron microscopy.
Acknowledgments
Funding from the Office of Naval Research (ONR) N000141410609, the Human Frontier Science Program (HFSP) RGP0029/2015, and National Science Foundation (NSF-EAGER) CCF-1547999 to M.B.; the National Science Foundation (NSF CMMI) 1334109 to M.B. and H.Y.; and the National Institutes of Health (NIH) P41GM103832 and P50GM103297 to W.C. is gratefully acknowledged. Patent applications have been filed by the MIT Technology & Licensing Office on behalf of M.B., R.V., and S.R. related to this work. M.B. is scientific founder of Structured DNA Technologies LLC. K. Pan is kindly acknowledged for his technical assistance with atomic-level rendering of origami models and with website construction; C. Hill for his technical assistance with website construction; V. Kaushik, A. Leed, and M. Adendorff for their technical assistance with qPCR; and S. Ludtke for his advice on cryo-EM image processing.
References and Notes
- 1.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 4.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 6.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones MR, Seeman NC, Mirkin CA. Programmable materials and the nature of the DNA bond. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 8.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 9.Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H. A primer to scaffolded DNA origami. Nat Methods. 2011;8:221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 11.Rothemund PW. Nanotechnology: Science and Computation. Springer; 2006. Scaffolded DNA origami: From generalized multicrossovers to polygonal networks; pp. 3–21. [Google Scholar]
- 12.Zhang F, Jiang S, Wu S, Li Y, Mao C, Liu Y, Yan H. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat Nanotechnol. 2015;10:779–784. doi: 10.1038/nnano.2015.162. [DOI] [PubMed] [Google Scholar]
- 13.Benson E, Mohammed A, Gardell J, Masich S, Czeizler E, Orponen P, Högberg B. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair JC, Davies KM, Vénien-Bryan C, Noble MEM. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat Nanotechnol. 2011;6:558–562. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 15.Gradišar H, Božič S, Doles T, Vengust D, Hafner-Bratkovič I, Mertelj A, Webb B, Šali A, Klavžar S, Jerala R. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat Chem Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat Commun. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 17.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 18.Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller EM, Högele A, Simmel FC, Govorov AO, Liedl T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 19.Acuna GP, Möller FM, Holzmeister P, Beater S, Lalkens B, Tinnefeld P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science. 2012;338:506–510. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Boulais E, Hakobyan Y, Wang WL, Guan A, Bathe M, Yin P. Casting inorganic structures with DNA molds. Science. 2014;346:1258361. doi: 10.1126/science.1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmi S, Ziegler C, Kauert DJ, Seidel R. Shape-controlled synthesis of gold nanostructures using DNA origami molds. Nano Lett. 2014;14:6693–6698. doi: 10.1021/nl503441v. [DOI] [PubMed] [Google Scholar]
- 22.Zhirnov V, Zadegan RM, Sandhu GS, Church GM, Hughes WL. Nucleic acid memory. Nat Mater. 2016;15:366–370. doi: 10.1038/nmat4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. 2012;337:1628–1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 24.Goldman N, Bertone P, Chen S, Dessimoz C, LeProust EM, Sipos B, Birney E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013;494:77–80. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai X, Martin TG, Scheres SHW, Dietz H. Cryo-EM structure of a 3D DNA-origami object. Proc Natl Acad Sci. 2012;109:20012–20017. doi: 10.1073/pnas.1215713109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, Chen DH, Liu X, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat Commun. 2014;5:4808. doi: 10.1038/ncomms5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irobalieva RN, Fogg JM, Catanese DJ, Sutthibutpong T, Chen M, Barker AK, Ludtke SJ, Harris SA, Schmid MF, Chiu W, Zechiedrich L. Structural diversity of supercoiled DNA. Nat Commun. 2015;6:8440. doi: 10.1038/ncomms9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan Y, Bathe M. Designer nucleic acids to probe and program the cell. Trends Cell Biol. 2012;22:624–633. doi: 10.1016/j.tcb.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry (Mosc) 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 31.See supplementary materials.
- 32.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DN, Kilchherr F, Dietz H, Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012;40:2862–2868. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prim RC. Shortest connection networks and some generalizations. Bell Syst Tech J. 1957;36:1389–1401. [Google Scholar]
- 35.Pandey S, Ewing M, Kunas A, Nguyen N, Gracias DH, Menon G. Algorithmic design of self-folding polyhedra. Proc Natl Acad Sci. 2011;108:19885–19890. doi: 10.1073/pnas.1110857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bent SW, Manber U. On non-intersecting Eulerian circuits. Discrete Appl Math. 1987;18:87–94. [Google Scholar]
- 37.He Y, Su M, Fang P, Zhang C, Ribbe AE, Jiang W, Mao C. On the chirality of self-assembled DNA octahedra. Angew Chem. 2010;122:760–763. doi: 10.1002/anie.200904513. [DOI] [PubMed] [Google Scholar]
- 38.Marchi AN, Saaem I, Vogen BN, Brown S, LaBean TH. Toward larger DNA origami. Nano Lett. 2014;14:5740–5747. doi: 10.1021/nl502626s. [DOI] [PubMed] [Google Scholar]
- 39.Wooddell CI, Burgess RR. Use of asymmetric PCR to generate long primers and single-stranded DNA for incorporating cross-linking analogs into specific sites in a DNA probe. Genome Res. 1996;6:886–892. doi: 10.1101/gr.6.9.886. [DOI] [PubMed] [Google Scholar]
- 40.Dunn KE, Dannenberg F, Ouldridge TE, Kwiatkowska M, Turberfield AJ, Bath J. Guiding the folding pathway of DNA origami. Nature. 2015;525:82–86. doi: 10.1038/nature14860. [DOI] [PubMed] [Google Scholar]
- 41.Sobczak JPJ, Martin TG, Gerling T, Dietz H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science. 2012;338:1458–1461. doi: 10.1126/science.1229919. [DOI] [PubMed] [Google Scholar]
- 42.Said H, Schüller VJ, Eber FJ, Wege C, Liedl T, Richert C. M1.3--a small scaffold for DNA origami. Nanoscale. 2013;5:284–290. doi: 10.1039/c2nr32393a. [DOI] [PubMed] [Google Scholar]
- 43.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Tian C, Yu J, Li Y, Jiang W, Mao C. Self-assembly of responsive multilayered DNA nanocages. J Am Chem Soc. 2015;137:1730–1733. doi: 10.1021/ja5101307. [DOI] [PubMed] [Google Scholar]
- 45.Martin TG, Dietz H. Magnesium-free self-assembly of multi-layer DNA objects. Nat Commun. 2012;3:1103. doi: 10.1038/ncomms2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeman NC, Kallenbach NR. Design of immobile nucleic acid junctions. Biophys J. 1983;44:201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan K, Boulais E, Yang L, Bathe M. Structure-based model for light-harvesting properties of nucleic acid nanostructures. Nucleic Acids Res. 2014;42:2159–2170. doi: 10.1093/nar/gkt1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dutta PK, Varghese R, Nangreave J, Lin S, Yan H, Liu Y. DNA-directed artificial light-harvesting antenna. J Am Chem Soc. 2011;133:11985–11993. doi: 10.1021/ja1115138. [DOI] [PubMed] [Google Scholar]
- 49.Liu W, Tagawa M, Xin HL, Wang T, Emamy H, Li H, Yager KG, Starr FW, Tkachenko AV, Gang O. Diamond family of nanoparticle superlattices. Science. 2016;351:582–586. doi: 10.1126/science.aad2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surana S, Shenoy AR, Krishnan Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat Nanotechnol. 2015;10:741–747. doi: 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Y, Zhang Y, Wang T, Xin HL, Li H, Gang O. Lattice engineering through nanoparticle-DNA frameworks. Nat Mater. 2016 doi: 10.1038/nmat4571. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellot G, McClintock MA, Lin C, Shih WM. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat Methods. 2011;8:192–194. doi: 10.1038/nmeth0311-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Ellis-Monaghan JA, McDowell A, Moffatt I, Pangborn G. DNA origami and the complexity of Eulerian circuits with turning costs. Nat Comput. 2014;14:1–13. [Google Scholar]
- 57.Kallenbach NR, Ma RI, Seeman NC. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 58.Pan K, Kim DN, Zhang F, Adendorff MR, Yan H, Bathe M. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nat Commun. 2014;5:5578. doi: 10.1038/ncomms6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 61.Mei Q, Wei X, Su F, Liu Y, Youngbull C, Johnson R, Lindsay S, Yan H, Meldrum D. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 2011;11:1477–1482. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray SC, Flanagan J, Popova OB, Chiu W, Ludtke SJ, Serysheva II. Validation of cryo-EM structure of IP3R1 channel. Structure. 2013;21:900–909. doi: 10.1016/j.str.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


