Abstract
Because of the methodological complexity of network meta-analyses (NMAs), NMAs may be more vulnerable to methodological risks than conventional pair-wise meta-analysis. Our study aims to investigate epidemiology characteristics, conduction of literature search, methodological quality and reporting of statistical analysis process in the field of cancer based on PRISMA extension statement and modified AMSTAR checklist. We identified and included 102 NMAs in the field of cancer. 61 NMAs were conducted using a Bayesian framework. Of them, more than half of NMAs did not report assessment of convergence (60.66%). Inconsistency was assessed in 27.87% of NMAs. Assessment of heterogeneity in traditional meta-analyses was more common (42.62%) than in NMAs (6.56%). Most of NMAs did not report assessment of similarity (86.89%) and did not used GRADE tool to assess quality of evidence (95.08%). 43 NMAs were adjusted indirect comparisons, the methods used were described in 53.49% NMAs. Only 4.65% NMAs described the details of handling of multi group trials and 6.98% described the methods of similarity assessment. The median total AMSTAR-score was 8.00 (IQR: 6.00–8.25). Methodological quality and reporting of statistical analysis did not substantially differ by selected general characteristics. Overall, the quality of NMAs in the field of cancer was generally acceptable.
Cancer is a leading cause of death worldwide and the total number of global cancer deaths is projected to increase by 45% from 7.9 million in 2007 to 11.5 million in 20301. For patients suffering from cancer, healthcare interventions aim to cure or considerably prolong the life of patients and to ensure the best possible quality of life for cancer survivors1. Treatment decisions should be based on evidence of the existing most effective treatment given available resources. High quality systematic reviews/meta-analyses of randomized controlled trials (RCTs) can provide the most valid evidence2. However, conventional meta-analysis becomes inadequate when there are no head-to-head trials comparing alternative interventions, or when more than two interventions need to be compared simultaneously3. For example, although there are trials directly comparing each of the newer antineoplastic agents with the current standard treatment (or placebo) for patients with neoplasm, there are no trials that directly compared different newer antineoplastic agents. Another example is a lack of direct comparison of 19 different chemotherapy regimens that are currently available for the treatment of advanced pancreatic cancer4.
Network meta-analyses (NMAs), as a generalization of pairwise meta-analysis, is becoming increasingly popular5,6,7,8. In the absence of or insufficient head-to-head comparisons of competing interventions of interest, NMAs using indirect treatment comparison analyses can provide useful evidence to inform health-care decision making. Even when evidence from direct comparisons are available, combining them with indirect estimates in a mixed treatment comparison may yield more refined estimates8,9. Formally, NMAs can be defined as a statistical combination of all available evidence for an outcome from several studies across multiple treatment to generate estimates of pairwise comparisons of each intervention to every other intervention within a network10. It has been considered that NMAs would be the next generation evidence synthesis toolkit which, when properly applied, could serve decision-making better than the conventional pair-wise meta-analysis11. However, NMAs are subject to similar methodological risks as standard pairwise systematic reviews. Because of its methodological complexity, it is probable that NMAs may be more vulnerable to such risks12. Therefore, it is important to assess the quality of published NMAs before their results are implemented into clinical or public health practice.
Previous studies have examined methodological problems in published indirect comparisons and NMAs, especially regarding reporting quality of statistical analysis12,13,14,15. It was concluded that the key methodological components of the NMAs process were often inadequately reported in published NMAs12. Currently, there are 30 tools available to assess the methodological quality of systematic reviews or meta-analyses16. To the best of our knowledge, no standard tool has been developed currently to assess the methodological quality of NMAs. AMSTAR (a measurement tool to assess the methodological quality of systematic reviews) tool is probably the most commonly used quality assessment tool for systematic reviews, which has been proven with good reliability, validity, and responsibility17,18,19.
The objective of this study is to conduct a methodological review of published NMAs in the field of cancer, summarise their characteristics, methodological quality, and reporting of key statistical analysis process. We also aim to compare the methodological quality and reporting of statistical analysis by selected general characteristics.
Results
Search results
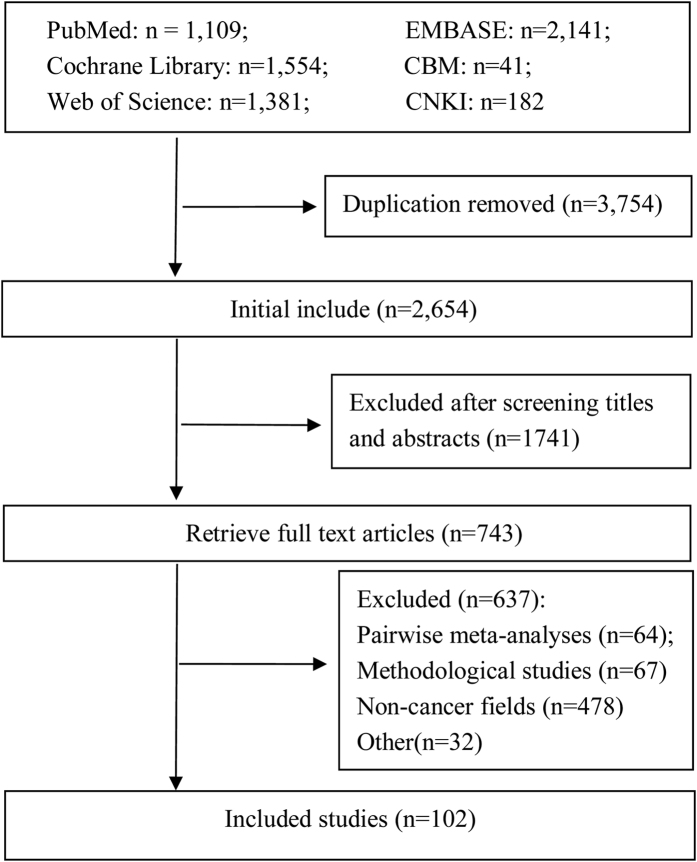
Initial literature search retrieved 6,408 citations. Of them, 3,754 citations were duplicates, so 2,654 citations were sent for further screening. Based on titles and abstracts, 1,741 citations were excluded. Then 637 articles were excluded based on reading full-texts, for reasons including: traditional pair-wise meta analysis (n = 64), methodological studies (n = 67), NMAs not related to cancer (n = 478), abstracts/letters/editorials/correspondences (n = 26), cost-effectiveness reviews (n = 6). Finally, 102 NMAs in the field of cancer were included (Fig. 1), including 92 published in English and 10 in Chinese. A list of included NMAs could be found in Appendix 1.
Figure 1. The details of literature selection.
General characteristics of included NMAs
The first NMA in the field of cancer was published in 200620. The number of published NMAs increased slowly until 2010, and then increased quickly. 43.14% (44/102) of the included NMAs were published since 2014 (Fig. 2). 98 NMAs involved 24 kinds of cancer, although 4 NMAs did not specify types of cancer. Non-small cell lung cancer (19/102, 18.63%) and breast cancer (12/102, 11.76%) were the most or secondly common type of cancer studies in the included NMAs (Fig. 3). NMAs were often performed by researchers based in China (29/102, 28.43%), UK (24/102, 23.53%), and USA (11/102, 10.78%) (Fig. 4). 99 NMAs were published in 60 different journals and 3 NMAs were doctorate dissertations. 85.30% (87/102) of NMAs were indexed by Science Citation Index (SCI) and 31.37% (32/102) were published in journals with high impact factors. According to 56 NMAs (54.90%) that reported dates of manuscript reception and acceptation, the median publishing period was 101 days (IQR: 47–187 days). According to 60 NMAs (58.82%) with information on funding source, 46 NMAs (45.10%) received funding support. The median number of interventions assessed per NMA was five (IQR: 3–9). The median number of trials included per NMAs was 12 (IQR: 7–23), and the median number of patients included in NMAs was 3,605 (IQR: 1,950–7,564). The main characteristics of the included NMAs were shown in Table 1. A more detailed characteristics and reporting of statistical analysis process could be found in Appendix 2.
Figure 2. The trend of year of publications.
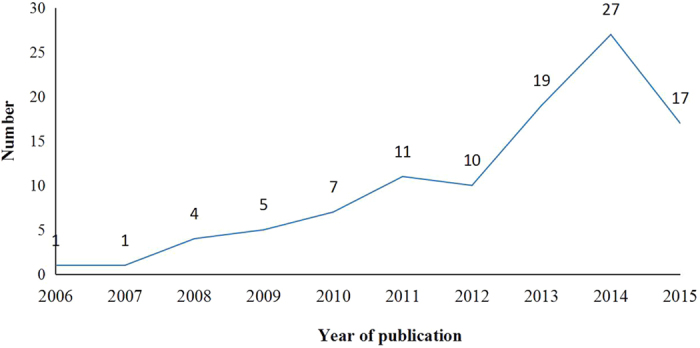
Figure 3. Categories of disease of included NMAs.
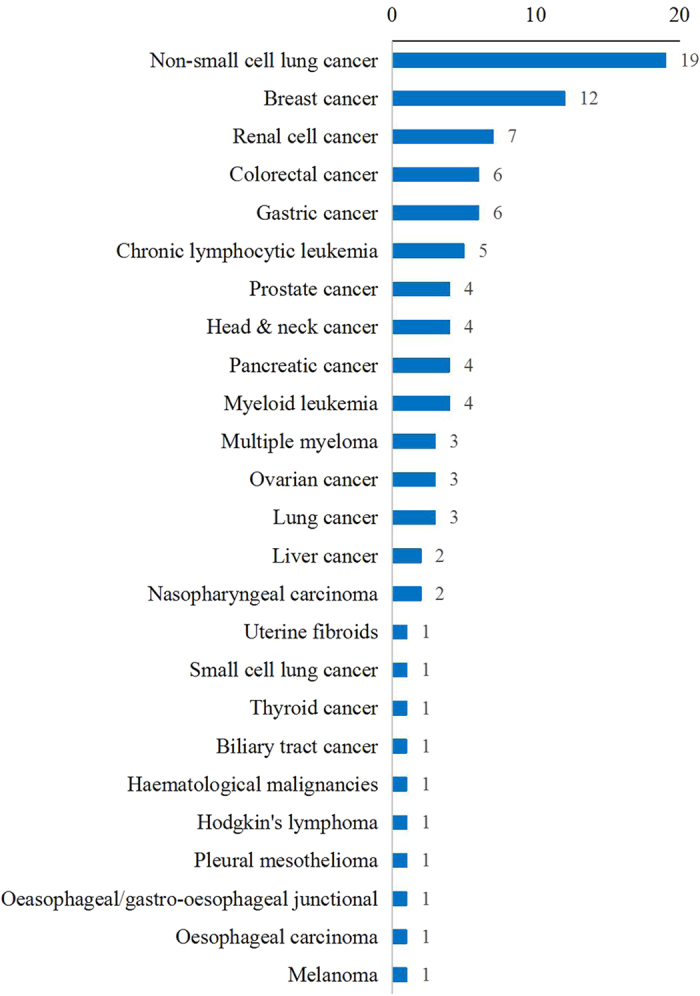
Figure 4. Countries of included NMAs.
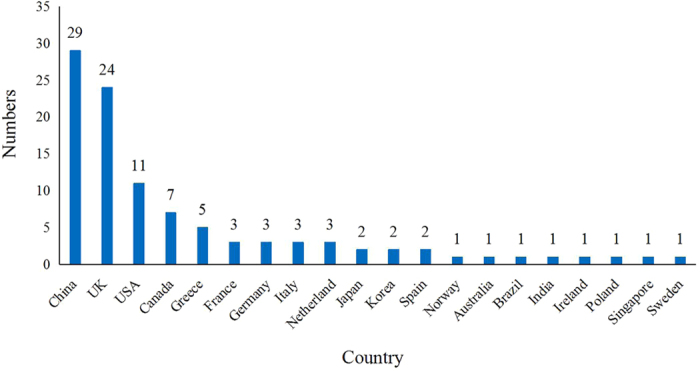
Table 1. Characteristics of the included NMA.
| Category | Frequency | Proportion [%(95% CI)] | |
|---|---|---|---|
| Language (n = 102) | English | 92 | 90.20 (82.70–94.60) |
| Chinese | 10 | 9.80 (5.40–17.30) | |
| Journal quality (n = 102) | SCI | 87 | 85.30 (77.00–90.90) |
| Non-SCI | 15 | 14.70 (9.10–23.00) | |
| CSCD | 3 | 2.90 (1.00–8.70) | |
| Non-CSCD | 7 | 6.90 (3.30–13.70) | |
| Impact factor (n = 87) | Median (IQR) | 3.94 (2.721–5.635) | |
| 0.0–2.0 | 6 | 6.90 (3.10–14.50) | |
| 2.0–5.0 | 49 | 56.30 (45.80–66.30) | |
| 5.0–10.0 | 22 | 25.30 (17.30–35.40) | |
| ≥10.0 | 10 | 11.50 (6.30–20.10) | |
| Publishing period (n = 56) | Median (IQR) | 101 (47–187) day | |
| 1–30 days | 12 | 21.40 (12.60–34.10) | |
| 30–60 days | 10 | 17.90 (9.90–30.10) | |
| 60–90 days | 4 | 7.10 (2.70–17.50) | |
| 90–150 days | 12 | 21.40 (12.60–34.10) | |
| ≥150 days | 19 | 33.90 (22.80–47.20) | |
| Funding source (n = 60) | Unfunded (n = 14) | 14 | 23.30 (14.30–35.60) |
| Industry-supported (n = 20) | 20 | 33.30 (22.60–46.10) | |
| Government-supported (n = 23) | 23 | 38.30 (27.00–51.10) | |
| Industry + Government (n = 3) | 3 | 5.00 (1.60–14.40) | |
| Number of author | Median (IQR) | 6 (5–8) | |
| Type of included study (n = 102) | RCT | 100 | 98.00 (92.50–99.50) |
| Meta-analysis | 2 | 2.00 (0.50–7.50) | |
| Number of included trial | Median (IQR) | 12 (7–23) | |
| Intervention | Median (IQR) | 5 (3–9) | |
| Sample size (n = 102) | Median (IQR) | 3605 (1950–7564) | |
| Not reported | 7 | 6.90 (3.30–13.70) | |
| Outcome (n = 102) | Median (IQR) | 3 (2–5) | |
| Dichotomous | 73 | 71.60 (62.10–79.50) | |
| Continue | 5 | 4.90 (2.10–11.20) | |
| Survival time | 79 | 77.50 (68.30–84.50) | |
| Number of study arm (n = 102) | Two arms | 50 | 49.00 (39.50–58.60) |
| Three arms | 7 | 6.90 (3.30–13.70) | |
| Two + Three arms/more | 36 | 35.30 (26.70–45.00) |
Note: CI, confidence interval; NMA, network meta-analysis; SCI, Science Citation Index; CSCD, Chinese Science Citation Database; IQR, interquartile range.
Reporting of literature search
Thirteen NMAs did not report any information on literature search, whereas one NMAs was conducted based on previous meta-analyses without additional searching. 98.90% (88/89) NMAs searched only English databases. The median number of Chinese databases searched was 5 (IQR: 3–6), and it was 3 (IQR: 3–4) for English databases. 22.50% (20/89) NMAs reported the search strategy, and the median number of search strategies reported was 2 (IQR: 1–3). 27.00% (24/89) NMAs searched previous published meta-analyses as a supplemental literature search. Other supplemental literature search methods included reference list checking, clinical trial registration platform, conference abstracts or web sites, and google engine (Table 2). PubMed/MEDLINE was the most common single database searched, and it was often combined with a search of Cochrane Library. The details of databases searched were showed in Table 3.
Table 2. Reporting information of literature search.
| Category | Frequency | Proportion [%(95% CI)] | |
|---|---|---|---|
| Reported database searched (n = 102) | Yes | 89 | 87.30 (79.30–92.50) |
| Language of database searched (n = 89)* | English | 88 | 98.90 (92.50–99.80) |
| Chinese | 10 | 11.20 (6.20–19.60) | |
| Chinese + English | 9 | 10.10 (5.30–18.30) | |
| Number of Chinese database searched | Median (IQR) | 5(3–6) | |
| Number of English database searched | Median (IQR) | 3(3–4) | |
| Number of search strategy | Median (IQR) | 2(1–3) | |
| Reported search strategy (n = 89) | Yes | 20 | 22.50 (15.00–32.30) |
| Presented search strategy (n = 20) | Manuscript | 9 | 45.00 (25.30–66.40) |
| Supplement | 5 | 25.00 (10.80–47.80) | |
| Online supplement | 6 | 30.00 (14.10–52.70) | |
| Searched previous published meta analysis (n = 89) | Yes | 24 | 27.00 (18.80–37.10) |
| Supplemental literature search (n = 89) | Yes | 75 | 84.30 (75.20–90.50) |
| Supplemental literature search method (n = 75) | Reference lists checking | 42 | 56.00 (44.70–66.80) |
| Clinical trial registration platform | 27 | 36.00 (26.00–47.40) | |
| Conference abstracts or Web sites | 43 | 57.30 (46.00–68.00) | |
| Google engine | 17 | 22.70 (14.60–33.50) | |
*13 studies did not search databases. CI, confidence interval; IQR, interquartile range.
Table 3. Information of databases searched (n = 89).
| Category | Frequency | Proportion [%(95% CI)] | |
|---|---|---|---|
| Name of database | PubMed/MEDLINE | 84 | 94.40 (87.20–97.60) |
| EMBASE | 69 | 77.50 (67.70–85.00) | |
| The Cochrane Library | 71 | 79.80 (70.20–86.90) | |
| ISI Web of Knowledge | 17 | 19.10 (12.20–28.60) | |
| CNKI | 9 | 10.10 (5.30–18.30) | |
| CBM | 9 | 10.10 (5.30–18.30) | |
| Wanfang | 7 | 7.90 (3.80–15.60) | |
| VIP | 8 | 9.00 (4.60–17.00) | |
| CSCD | 7 | 7.90 (3.80–15.60) | |
| Others | 35 | 39.30 (29.80–49.80) | |
| Common combination of database | PubMed/MEDLINE + EMBASE | 65 | 73.00 (62.90–81.20) |
| PubMed/MEDLINE + Cochrane | 69 | 77.50 (67.70–85.00) | |
| EMBASE + Cochrane | 60 | 67.40 (57.00–76.30) | |
| PubMed/MEDLINE + EMBASE + Cochrane | 56 | 62.90 (52.50–72.30) | |
| PubMed/MEDLINE + ISI Web of Knowledge | 17 | 19.10 (12.20–28.60) | |
| PubMed/MEDLINE + EMBASE + Cochrane + ISI Web of Knowledge | 14 | 15.70 (9.50–24.80) | |
| PubMed/MEDLINE + EMBASE + CBM | 8 | 9.00 (4.60–17.00) | |
| PubMed/MEDLINE + CNKI | 7 | 7.90 (3.80–15.60) |
Reporting of statistical analysis processes
Sixty-one (59.80%) NMAs were conducted using a Bayesian framework (2 reviews are adjusted indirect comparisons). 43 reviews were adjusted indirect comparisons (2 adjusted indirect comparisons use Bayesian framework).
For NMAs using a Bayesian framework, more than half also included traditional meta-analyses (42/61, 68.85%). The majority of NMAs reported summary measures (57/61, 93.44%). 75.41% (46/61) NMAs reported the model used. Of the 24 (24/61, 39.34%) NMAs that tested model fit, the most common method was the use of deviance information criterion (15/24, 62.50%). The majority of NMAs (40/61, 65.57%) did not make their code available to journal readers. 16 (26.23%) NMAs provided the model source cited, however, it was unclear for the details of model used. 91.80% (56/61) did not report whether there was an adjustment for multiple arms. Half of NMAs (31/61, 50.82%) specified the prior distributions used, and the most common prior used was non-informative prior (18/31, 58.06%). Only 4 (6.56%) NMAs performed pre-specified sensitivity analyses. More than half of NMAs did not report assessment of convergence (37/61, 60.66%) and sensitivity analyses performed (37/61, 60.66%). Inconsistency was assessed in 27.87% of NMAs. Assessment of heterogeneity in traditional meta-analyses was more common (26/61, 42.62%) than in NMAs (4/61, 6.56%). Most of the included NMAs did not report assessment of similarity (53/61, 86.89%), publication or reporting bias (60/61, 98.36%), subgroup analyses or meta-regression performed (49/61, 80.33%), and whether GRADE tool was used to assess quality of evidence (58/61, 95.08%). NMAs published in journals with higher impact factors more often provided model code (57.69% versus 23.08%, p = 0.012) and assessed the heterogeneity of NMAs (0% versus 15.38%, p = 0.039). Based on the median division of the number of included NMAs, we chose December 31st 2013 as cut-off point. NMAs published prior to December 31st 2013 more often reported models used (89.66% versus 62.50%, p = 0.015), model code used (48.28% versus 21.88%, p = 0.032), and assessment of heterogeneity of NMAs (13.79% versus 0%, p = 0.031). Other results did not differ by journal impact factor or year of publication. The more details of statistical reporting in Bayesian NMAs was showed in Table 4.
Table 4. Statistical Reporting in Bayesian analysis [n/N(%)].
| Items | All studies (n = 61) | Journal impact factor* | Year of publication | ||||
|---|---|---|---|---|---|---|---|
| Low (<5.00)(n = 26) | High (≥5.00)(n = 26) | P-value | Older studies(~2013) (n = 29) | Recent studies(2014~2015) (n = 32) | P-value | ||
| Was traditional meta-analysis conducted? (Yes) | 42/61(68.85) | 15/26(57.69) | 20/26(76.92) | 0.143 | 18/29(62.07) | 24/32(93.10) | 0.280 |
| Were summary measures reported? (Yes) | 57/61(93.44) | 24/26(92.31) | 25/26(96.15) | 0.556 | 27/29(93.10) | 30/32(93.75) | 0.920 |
| Planned methods of analysis: | |||||||
| What was the model used? | |||||||
| Fixed-effect | 7/46(15.22) | 4/20(20.00) | 2/24(8.33) | 0.267 | 6/26(23.08) | 1/20(5.00) | 0.094 |
| Random-effects | 11/46(23.91) | 4/20(20.00) | 6/24(25.00) | 0.697 | 6/26(23.08) | 5/20(25.00) | 0.881 |
| Fixed- and random-effects | 18/46(39.13) | 12/20(60.00) | 6/24(25.00) | 0.020 | 7/26(26.92) | 11/20(55.00) | 0.056 |
| Other models | 10/46(21.74) | 0/20(0.00) | 10/24(41.67) | 0.001 | 7/26(26.92) | 3/20(15.00) | 0.336 |
| Nor reported | 15/61(24.59) | 6/26(23.08) | 2/26(7.69) | 0.128 | 3/29(10.34) | 12/32(37.50) | 0.015 |
| How to present the model code? | |||||||
| Supplement | 5/21(23.81) | 2/6(33.33) | 3/15(20.00) | 0.527 | 2/14(14.29) | 3/7(42.86) | 0.157 |
| Reference | 16/21(76.19) | 4/6(66.67) | 12/15(80.00) | 0.527 | 12/14(85.71) | 4/7(57.14) | 0.157 |
| Not provided | 40/61(65.57) | 20/26(76.92) | 11/26(42.31) | 0.012 | 15/29(51.72) | 25/32(78.13) | 0.032 |
| How to assess the model fit? | |||||||
| Residual deviance | 2/24(8.33) | 1/14(7.14) | 1/10(10.00) | 0.807 | 1/9(11.11) | 1/15(6.67) | 0.709 |
| Deviance information criterion | 15/24(62.50) | 8/14(57.14) | 7/10(70.00) | 0.530 | 6/9(66.67) | 9/15(60.00) | 0.749 |
| Residual deviance+Deviance information criterion | 7/24(29.17) | 5/14(35.71) | 2/10(20.00) | 0.414 | 2/9(22.22) | 5/15(33.33) | 0.570 |
| Not reported | 37/61(60.66) | 12/26(46.15) | 16/26(61.54) | 0.270 | 20/29(68.97) | 17/32(53.13) | 0.210 |
| Was handling of multigroup trials reported? (Yes) | 5/61(8.20) | 1/26(3.85) | 3/26(11.54) | 0.303 | 3/29(10.34) | 2/32(6.25) | 0.564 |
| Were the prior distributions reported? | |||||||
| Noninformative prior | 18/31(58.06) | 9/13(69.23) | 10/17(58.82) | 0.564 | 8/16(50.00) | 10/15(66.67) | 0.355 |
| Informative prior | 3/31(9.68) | 1/13(7.69) | 2/17(11.76) | 0.717 | 1/16(6.25) | 2/15(13.33) | 0.512 |
| Vague prior | 10/31(32.26) | 4/13(30.77) | 6/17(35.29) | 0.798 | 7/16(43.75) | 3/15(20.00) | 0.164 |
| Not specified | 30/61(49.18) | 13/26(50.00) | 9/26(34.62) | 0.266 | 13/29(44.83) | 17/32(53.13) | 0.521 |
| Sensitivity analysis based on priors | 4/61(6.56) | 2/26(7.69) | 2/26(7.69) | 1.000 | 2/29(6.90) | 2/32(6.25) | 0.920 |
| Was the convergence assessed? | |||||||
| Not reported | 37/61(60.66) | 15/26(57.69) | 14/26(53.85) | 0.782 | 17/29(58.62) | 20/32(62.50) | 0.759 |
| Gelman Rubin statistic | 21/24(87.50) | 11/11(100.00) | 10/12(83.33) | 0.166 | 9/12(75.00) | 12/12(100.00) | 0.070 |
| Visual plot inspection | 13/24(54.17) | 5/11(45.45) | 7/12(58.33) | 0.546 | 7/12(58.33) | 6/12(50.00) | 0.688 |
| Observation of chain mix | 2/24(8.33) | 2/11(18.18) | 0/12(0.00) | 0.131 | 1/12(8.33) | 1/12(8.33) | 1.000 |
| Was the heterogeneity in NMA assessed? | |||||||
| Precision (Tau2) | 4/61(6.56) | 0/26(0.00) | 4/26(15.38) | 0.039 | 4/29(13.79) | 0/32(0.00) | 0.031 |
| Not reported | 57/61(93.44) | 26/26(100.00) | 22/26(84.62) | 0.039 | 25/29(86.21) | 32/32(100.00) | 0.031 |
| Was the similarity in NMA assessed? | |||||||
| Not reported | 53/61(86.89) | 22/26(84.62) | 22/26(84.62) | 1.000 | 23/29(79.31) | 30/32(93.75) | 0.098 |
| Comparing patients’ or trials’ characteristics | 5/8(62.50) | 2/4(50.00) | 3/4(75.00) | 0.495 | 4/6(66.67) | 1/2(50.00) | 0.693 |
| Investigating potential effect modifying covariates | 3/8(37.50) | 2/4(50.00) | 1/4(25.00) | 0.495 | 2/6(33.33) | 1/2(50.00) | 0.693 |
| Were the inconsistency assessed? | |||||||
| Not reported | 44/61(72.13) | 20/26(76.92) | 16/26(61.54) | 0.234 | 20/29(68.97) | 24/32(75.00) | 0.603 |
| Inconsistency/incoherence factors | 4/17(23.53) | 1/6(16.67) | 2/10(20.00) | 0.873 | 3/9(33.33) | 1/8(12.50) | 0.327 |
| Hypothesis test | 8/17(47.06) | 2/6(33.33) | 6/10(60.00) | 0.317 | 4/9(44.44) | 4/8(50.00) | 0.824 |
| Node-splitting analysis | 5/17(29.41) | 3/6(50.00) | 2/10(20.00) | 0.225 | 2/9(22.22) | 3/8(37.50) | 0.503 |
| Was the publication or reporting bias assessed? | |||||||
| Comparison-adjusted funnel plot | 1/61(1.64) | 0/26(0.00) | 1/26(3.85) | 0.317 | 0/29(0.00) | 1/32(3.13) | 0.341 |
| Not reported | 60/61(98.36) | 26/26(100.00) | 25/26(96.15) | 0.317 | 29/29(100.00) | 31/32(96.87) | 0.341 |
| Additional analyses: | |||||||
| Was a sensitivity analysis performed? (Yes) | 24/61(39.34) | 10/26(38.46) | 12/26(46.15) | 0.578 | 12/29(41.38) | 12/32(37.50) | 0.759 |
| Was subgroup analysis or meta-regression performed? (Yes) | 12/61(19.67) | 3/26(11.54) | 7/26(26.92) | 0.163 | 7/29(24.14) | 5/32(15.63) | 0.407 |
| Was the GRADE used? (Yes) | 3/61(4.92) | 1/26(3.85) | 2/26(7.69) | 0.556 | 2/29(6.90) | 1/32(3.13) | 0.500 |
*9 studies published in journals with no associated impact factor.
For adjusted indirect comparisons, the majority of NMAs (42/43, 97.67%) also conducted traditional meta-analyses and 53.49% (23/43) adjusted indirect comparisons were performed using methods described by Bucher21. 58.14% (25/43) assessed the heterogeneity of direct comparisons, but none of NMAs assessed the heterogeneity of indirect comparisons. Only two (4.65%) NMAs described the details of handling of multi group trials and three (6.98%) described the methods of similarity assessment. Most of NMAs did not report whether sensitivity analyses were performed (38/43, 88.37%) and whether subgroup analyses or meta-regression were performed (34/43, 79.07%). These results did not differ by journal quality or year of publication. The details of statistical reporting for adjusted indirect comparisons was showed in Table 5.
Table 5. Statistical Reporting in adjusted indirect comparisons [n/N(%)].
| Items | n/N | Journal impact factor (n = 37)* | Year of publication (n = 43)# | ||||
|---|---|---|---|---|---|---|---|
| Low (<5.00)(n = 29) | High (≥5.00)(n = 8) | P-value | Older studies(~2013) (n = 31) | Recent studies(2014~2015) (n = 12) | P-value | ||
| Was traditional meta-analysis conducted? (Yes) | 42/43(97.67) | 28/29(96.55) | 8/8(100.00) | 0.599 | 30/31(96.77) | 12/12(100.00) | 0.534 |
| Were summary measures reported? (Yes) | 41/43(97.62) | 26/29(89.66) | 8/8(100.00) | 0.976 | 29/31(93.55) | 12/12(100.00) | 0.373 |
| Planned methods of analysis in adjusted indirect comparisons: | |||||||
| What was the method used? | |||||||
| Bucher | 23/28(82.14) | 17/20(85.00) | 2/4(50.00) | 0.140 | 15/20(75.00) | 8/8(100.00) | 0.240 |
| Other methods | 5/28(17.86) | 3/20(15.00) | 2/4(50.00) | 0.123 | 5/20(25.00) | 0/8(0.00) | 0.125 |
| Not reported | 15/43(34.88) | 9/29(31.03) | 4/8(50.00) | 0.326 | 11/31(35.48) | 4/12(33.33) | 0.896 |
| Was the method presented or source cited? | |||||||
| Manuscript | 5/23(21.74) | 1/16(6.25) | 2/4(50.00) | 0.033 | 4/20(20.00) | 1/3(33.33) | 0.610 |
| Reference | 18/23(78.26) | 15/16(93.75) | 2/4(50.00) | 0.033 | 16/20(80.00) | 2/3(66.67) | 0.610 |
| Not provided | 20/43(46.51) | 13/29(44.83) | 4/8(50.00) | 0.798 | 11/31(35.48) | 9/12(75.00) | 0.021 |
| Was handling of multigroup trials reported? (Yes) | 2/43(4.65) | 0/29(0.00) | 1/8(12.50) | 0.057 | 2/31(6.45) | 0/12(0.00) | 0.373 |
| Was the similarity assessed? | 3/43(6.98) | 3/29(10.34) | 0/8(0.00) | 0.349 | 3/31(9.68) | 0/12(0.00) | 0.269 |
| Was the inconsistency assessed? | |||||||
| Not reported | 36/43(83.72) | 25/29(86.21) | 5/8(62.50) | 0.150 | 26/31(83.87) | 10/12(83.33) | 0.970 |
| Not application | 4/7(57.14) | 4/4(100.00) | 0/3(0.00) | 0.050 | 3/5(60.00) | 1/2(50.00) | 0.810 |
| Hypothesis test | 3/7(42.86) | 0/4(0.00) | 3/3(100.00) | 0.050 | 2/5(40.00) | 1/2(50.00) | 0.810 |
| Additional analyses: | |||||||
| Was a sensitivity analysis performed? | 5/43(11.63) | 1/29(3.45) | 3/8(37.50) | 0.007 | 3/31(9.68) | 2/12(16.67) | 0.526 |
| Was subgroup analysis or meta-regression performed? | 9/43(20.93) | 8/29(27.59) | 1/8(12.50) | 0.385 | 6/31(19.35) | 3/12(25.00) | 0.827 |
*6 studies published in journals with no associated impact factor.
#Based on the median division of number of included NMAs, December 31st 2013 is the cut-off point.
Methodological quality assessment
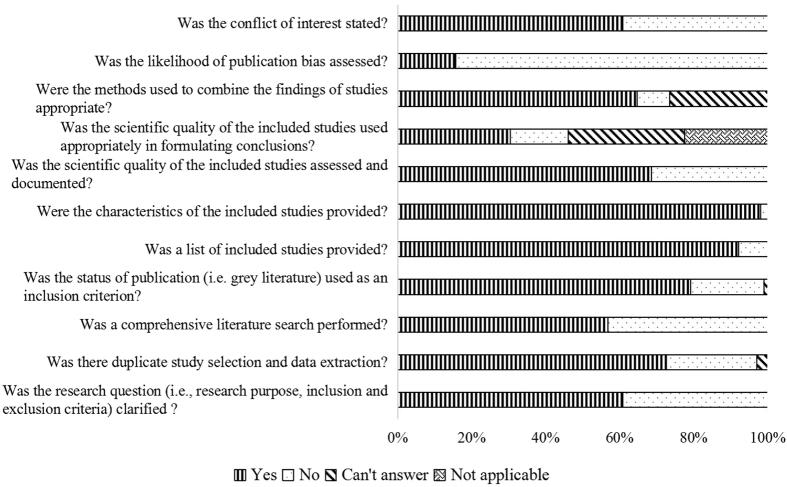
The results of methodological quality assessment based on modified AMSTAR checklist were presented in Fig. 5. The median total score was 8.00 (IQR: 6.00–8.25). Approximately half of the included NMAs did not perform a comprehensive literature search (Item 3, 42.31%). More than half of NMAs (69.61%) did not consider the scientific quality of the included studies in formulating conclusions, and 84.31% NMAs did not assess the likelihood of publication bias.
Figure 5. The results of methodological quality assessment.
Table 6 presented the results of stratified analyses of methodological quality assessment. NMAs published in journals with higher impact factors more often performed a comprehensive literature search (78.13% versus 45.45%, p = 0.002), reported appropriate methods used to combine the findings of studies (81.25% versus 58.18%, p = 0.019), and assessed the likelihood of publication bias (25.00% versus 5.45%, p = 0.017). NMAs published after December 31st 2013 more often assessed the scientific quality of the included studies (86.36% versus 55.17%, p = 0.001) and considered the scientific quality in formulating conclusions (43.18% versus 20.69%, p = 0.015). Most of these items did not differ between funding support and non-funding support. NMAs published in China more often reported two independent reviewers for study selection and data extraction (89.66% versus 65.75%, p = 0.015), assessed the scientific quality of the included studies (86.21% versus 61.64%, p = 0.016) and considered the scientific quality in formulating conclusions (68.97% versus 15.07%, p = 0.000). Moreover, Bayesian NMAs more often reported two independent reviewers for study selection and data extraction (81.97% versus 60.47%, p = 0.015), performed a comprehensive literature search (70.49% versus 39.53%, p = 0.002), considered the status of publication (i.e. grey literature) used as an inclusion criterion (86.89% versus 67.44%, p = 0.017), and assessed the scientific quality of the included studies (78.69% versus 55.81%, p = 0.013).
Table 6. Subgroup analyses of methodological quality assessment (n/%).
| Items (Yes) | All NMAs (n = 102) | Journal impact factor* | Year of publication# | Funding source | Country of corresponding author | Type of NMAs | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low (<5.00) (n = 55) vs. High (≥5.00) (n = 32) | P-value | Older (n = 58) vs. Recent (n = 44) | P-value | Funding (n = 46) vs. Non-funding (n = 56) | P-value | China (n = 29) vs. Others (n = 73) | P-value | Bayesian NMAs (n = 61) vs. Adjusted indirect comparisons (n = 43)& | P-value | ||
| Was the research question (i.e., research purpose, inclusion and exclusion criteria) clarified? | 62/60.78 | 31/56.36 vs. 20/62.50 | 0.568 | 33/56.90 vs. 29/65.91 | 0.358 | 26/56.52 vs. 36/64.29 | 0.426 | 20/68.97 vs. 42/57.53 | 0.288 | 38/62.30 vs. 25/58.14 | 0.671 |
| Was there duplicate study selection and data extraction? | 74/72.55 | 38/69.09 vs. 24/75.00 | 0.547 | 41/70.69 vs. 33/75.00 | 0.631 | 32/69.57 vs. 42/75.00 | 0.543 | 26/89.66 vs. 48/65.75 | 0.015 | 50/81.97 vs. 26/60.47 | 0.015 |
| Was a comprehensive literature search performed? | 58/56.86 | 25/45.45 vs. 25/78.13 | 0.002 | 31/53.45 vs. 27/61.36 | 0.426 | 26/56.52 vs. 32/57.14 | 0.950 | 16/55.17 vs. 42/57.53 | 0.829 | 43/70.49 vs. 17/39.53 | 0.002 |
| Was the status of publication (i.e. grey literature) used as an inclusion criterion? | 81/79.41 | 43/78.18 vs. 26/81.25 | 0.728 | 46/79.31 vs. 35/79.55 | 0.977 | 35/76.09 vs. 46/82.14 | 0.454 | 26/89.66 vs. 55/75.34 | 0.109 | 53/86.89 vs. 29/67.44 | 0.017 |
| Was a list of included studies provided? | 94/92.16 | 50/90.91 vs. 29/90.63 | 0.965 | 53/91.38 vs. 41/93.18 | 0.739 | 44/95.65 vs. 50/89.29 | 0.236 | 28/96.55 vs. 66/90.41 | 0.300 | 56/91.80 vs. 40/93.02 | 0.819 |
| Were the characteristics of the included studies provided? | 100/98.04 | 54/98.18 vs. 31/96.88 | 0.715 | 58/100.00 vs. 42/95.45 | 0.103 | 45/97.83 vs. 55/98.21 | 0.889 | 29/100.00 vs. 71/97.26 | 0.370 | 59/96.72 vs. 43/100.00 | 0.233 |
| Was the scientific quality of the included studies assessed and documented? | 70/68.63 | 34/61.82 vs. 25/78.13 | 0.098 | 32/55.17 vs. 38/86.36 | 0.001 | 29/63.04 vs. 41/73.21 | 0.273 | 25/86.21 vs. 45/61.64 | 0.016 | 48/78.69 vs. 24/55.81 | 0.013 |
| Was the scientific quality of the included studies used appropriately in formulating conclusions? | 31/30.39 | 11/20.00 vs. 8/25.00 | 0.584 | 12/20.69 vs. 19/43.18 | 0.015 | 9/19.57 vs. 22/39.29 | 0.032 | 20/68.97 vs. 11/15.07 | 0.000 | 21/34.43 vs. 10/23.26 | 0.222 |
| Were the methods used to combine the findings of studies appropriate? | 66/64.71 | 32/58.18 vs. 26/81.25 | 0.019 | 37/63.79 vs. 29/65.91 | 0.826 | 28/60.87 vs. 38/67.86 | 0.465 | 18/62.07 vs. 48/65.75 | 0.727 | 39/63.93 vs. 29/67.44 | 0.713 |
| Was the likelihood of publication bias assessed? | 16/15.69 | 3/5.45 vs. 8/25.00 | 0.017 | 10/17.24 vs. 6/13.64 | 0.622 | 5/10.87 vs. 11/19.64 | 0.228 | 6/20.69 vs. 10/13.70 | 0.383 | 8/13.11 vs. 9/20.93 | 0.291 |
| Was the conflict of interest stated? | 62/60.78 | 38/69.09 vs. 22/68.75 | 0.974 | 37/63.79 vs. 25/56.82 | 0.477 | 29/63.04 vs. 33/58.93 | 0.673 | 11/37.93 vs. 51/69.86 | 0.001 | 35/57.38 vs. 27/62.79 | 0.581 |
*6 studies published in journals with no associated impact factor.
#Based on the median division of number of included NMAs, December 31st 2013 is the cut-off point.
&2 adjusted indirect comparisons also were conducted using Bayesian framework.
Table 7 presented the association of total AMSTAR-score and selected general characteristics. Although the AMSTAR-score of NMAs published in China was higher than NMAs published in others (p = 0.023), there were no significant differences between AMSTAR-score and different countries (p = 0.465). The differences were not significant between AMSTAR-score and other selected general characteristics.
Table 7. The association of total AMSTAR-score and selected general characteristics [median (IQR)].
| Category | Overall score | AMSTAR-score | Statistic | P-value |
|---|---|---|---|---|
| 8.00 (6.00, 8.25) | ||||
| Year of publication | 1.194 | 0.232 | ||
| 2006~2013 (n = 58) | 7.00(5.00, 9.00) | |||
| 2014~2015 (n = 44) | 8.00(6.25, 8.00) | |||
| Country of corresponding author I | 2.557 | 0.465 | ||
| East Asia (n = 34) | 8.00(7.00, 9.00) | |||
| Europe (n = 47) | 7.00(5.00, 8.00) | |||
| North America (n = 18) | 7.00(5.00, 8.25) | |||
| Others (n = 3) | 7.00(6.00, 7.00) | |||
| Country of corresponding author II | 2.272 | 0.023 | ||
| China (n = 29) | 8.00(7.00, 9.00) | |||
| Others (n = 73) | 7.00(5.00, 8.00) | |||
| Impact factors of SCI | 5.503 | 0.239 | ||
| Not SCI (n = 15) | 8.00(6.00, 8.00) | |||
| 0.0–2.0 (n = 6) | 5.00(2.00, 9.00) | |||
| 2.0–5.0 (n = 49) | 7.00(5.00, 8.00) | |||
| 5.0–10.0 (n = 22) | 8.00(6.00, 9.00) | |||
| ≥10.0 (n = 10) | 7.00(7.00, 9.00) | |||
| Funding source | 1.512 | 0.469 | ||
| Not report (n = 42) | 8.00(6.00, 9.00) | |||
| Unfunded (n = 14) | 7.50(7.00, 8.00) | |||
| Funding-supported (n = 46) | 7.00(5.00, 8.25) | |||
| Method | 1.601 | 0.109 | ||
| Indirect comparison (n = 41) | 7.00(3.50, 8.00) | |||
| Bayes (n = 61) | 8.00(6.00, 9.00) | |||
| Categories of disease | 5.649 | 0.342 | ||
| Non-small cell lung cancer (n = 19) | 8.00(7.00, 9.00) | |||
| Breast cancer (n = 12) | 7.00(4.25, 8.00) | |||
| Renal cell carcinoma (n = 7) | 6.00(5.00, 8.00) | |||
| Colorectal cancer (n = 6) | 7.50(6.00, 10.25) | |||
| Gastric cancer (n = 6) | 8.00(7.00, 9.00) | |||
| Others (n = 52) | 7.50(5.25, 8.75) |
Discussion
We identified 102 NMAs involving 24 kinds of cancer. Methodological quality and statistical reporting were assessed based on PRISMA extension statement and modified AMSTAR checklist. In addition, we also assessed the conduct of literature search in the included NMAs. Some key methodological components including the literature search and statistical analysis were missing or inadequate in most of included NMAs, such as only 22.50% of NMAs reported search strategy, 6.56% assessed the heterogeneity in NMAs. Methodological quality and reporting of statistical analysis did not substantially differ by selected general characteristics of NMAs.
NMAs could provide useful evidence on relative effectiveness of different interventions for decision-making when there are no or insufficient direct comparison trials11. Methodological quality of NMAs is a crucial point for health care decision-makers and researchers. We assessed the methodological quality of NMAs in the field of cancer based on modified AMSTAR checklist. Some methodological flaws were identified, especially regarding to literature search (Item 3), assessment of scientific quality (Item 7) and scientific quality used appropriately in formulating conclusions (Item 8), the methods used to combine the findings of studies (Item 9), and assessment of publication bias (Item 10).
NMAs aimed to rank the benefits (or harms) of interventions, based on all available RCTs. Thus, the identification of all relevant data is critical7. Most of the included NMAs (80.39%, 82/102) did not report database search strategy. For those that reported search strategy, 26.96% only searched previous published meta-analyses. It is important to search, track, and include previous systematic reviews and meta-analyses in conducting NMAs22. PubMed/MEDLINE was the most commonly used databases and the most common combination of databases was PubMed/MEDLINE and EMBASE. The majority of NMAs did not search Chinese databases. Cohen et al.’ study showed that searching Chinese databases might lead to the identification of a large amount of additional clinical evidence, and suggested that Chinese biomedical databases should be searched when performing systematic reviews23.
The assessment of scientific quality of individual studies could affect findings of NMAs24. However, 31.37% of the included NMAs did not report methods for assessing the risk of bias of individual studies in methods sections. And 69.61% did not consider the scientific quality of the included studies in formulating conclusions. Although reporting bias could have a substantial effect on the conclusions of a NMA12, most of the included NMAs (84.31%) did not report a method to assess publication bias.
The complex nature of NMA mainly reflected in the diversification of interventions and complex statistical analysis process. Homogeneity and consistency assumptions underlie NMA25. Although assessment of heterogeneity in traditional meta-analyses was common, only 4 NMAs (3.92%) assessed the heterogeneity in the entire network by heterogeneity variance parameter (Tau2). Eleven (10.78%) explicitly reported the methods of assessment of similarity. For those with Bayesian framework, 17 (27.87%) assessed the inconsistency between direct comparisons and indirect comparisons. GRADE tool was proposed to assess the quality of evidence from NMAs in 201426. However, it still was rarely used to assess quality of evidence in NMAs related to cancer.
To the best of our knowledge, this is the first review to comprehensively assess the methodological quality using a modified AMSTAR checklist, and simultaneously assess the quality of reporting of literature search and statistical analysis methods. Two recent reviews that also focused on the methodological problems of published network meta analyses12,15 covered a wide range of medical areas and some details of reporting of literature search and statistical analysis were missing. Bafeta A et al.12 included 121 NMAs to examine the methodological reporting of NMAs, the results showed that 73% did not report the electronic search strategy for each database compared with 77.5% in our study. Most of NMAs did not assess quality of evidence using GRADE tool (3% vs. 4.92%). The results of methodological reporting were similar to our study. Chambers J et al.15 also showed that there were similar methodological quality problems in their included NMAs. However, AMSTAR checklist has not been used to systematically assess the methodological quality of NMAs. Furthermore, we explored the potential factors influencing methodological quality and statistical reporting according to general characteristics of the included NMAs. There were no substantial differences by selected general characteristics of NMAs.
Our study also have some limitations. There was no standard tool to assess the methodological quality of NMAs. We slightly modified three of the 11 AMSTAR items (Item 1, Item 5, and Item 9) to assess the methodological quality of NMAs. However, there are still some problems or uncertain issues, such as the difficulty in defining type of interventions and type of comparisons for inclusion in NMAs, how to draw geometry of the network, how to handle multi group trials, how to decide whether the assessment of similarity and consistency was appropriate, and whether statistical analysis methods were appropriate for NMAs. The complex nature of statistical analysis of NMAs raised the necessity to develop a guideline about the reporting of statistical analysis of NMAs. As with other methodological studies, assessing methodological quality and reporting quality from published reports alone could be misleading. The study authors may have used adequate methods but omitted important details from published reports12, or published reports were sufficient referring to relevant reporting guidelines but not rigorous during the conduct process. For example, while we distinguished whether study selection and data extraction were performed by least two independent reviewers, we did not know whether the processes were really performed by two independent reviewers. Finally, we did not identify any eligible NMAs related to diagnostic test accuracy and animal study. We also did not include reviews based on individual patient data (IPD) due to the differences of method and statistical analysis processes between IPD and aggregated data.
Overall, the methodological quality of NMAs in the field of cancer was generally acceptable. However, some methodological flaws have been identified in published NMAs, especially regarding to literature search, assessment of scientific quality and scientific quality used appropriately in formulating conclusions, the methods used to combine findings of studies, and assessment of publication bias. Methodological quality and statistical reporting did not substantially differ by general characteristics.
Methods
Search strategy
PubMed, EMBASE, Web of Science, Science Citation Index Expanded, Social Sciences Citation Index, The Cochrane Library, Cochrane Database of Systematic Reviews, Database of Abstracts of Reviews of Effects, Health Technology Assessment Database, NHS Economic Evaluation Database, Chinese Biomedical Literature Database (CBM), and China National Knowledge Infrastructure (CNKI) were searched from inception to February 26th, 2014. The search strategy was recently reported in a published paper7. All searches were updated on 9th July, 2015.
Eligibility criteria
We included any NMAs in the field of cancer in the English and Chinese languages, regardless of interventions. NMAs were defined as meta-analyses that used network meta-analytic methods to analyze, simultaneously, three or more different interventions7, adjusted indirect comparisons were also included. If the same NMA had duplicate publications, the latest was included. We excluded methodological articles, conference abstracts, letters, editorials, correspondences, cost-effectiveness reviews, and reviews based on individual patient data.
Study selection
Literature search records were imported into ENDNOTE X6 literature management software. Two independent reviewers (LG, LL) examined the title and abstract of retrieved studies to identify potentially relevant studies according to the eligibility criteria. Then, full-text versions of all potentially relevant studies were obtained. Excluded trials and the reasons for their exclusion were listed, conflicts were resolved by a third reviewer (J-HT, or K-HY).
Data extraction and management
A standard data abstraction form was created using Microsoft Excel 2013 (Microsoft Corp, Redmond, WA, www.microsoft.com) to collect data of interest. A pilot-test was performed for literature selection and data extraction, and a “cheat sheet” with detailed definitions and examples were developed to ensure high inter-rater reliability among the reviewers.
General characteristics
The following general characteristics were collected by one reviewer (LG): first author, year of publication, country of corresponding author, journal name, publishing period (time from received to accepted), funding source (industry-supported, non-industry-supported, unfunded or not report), number of author, language of publication (English or Chinese), number and type of included original studies, sample size of included original studies, number of study arm, type of outcome (dichotomous, continue, or survival time), categories of disease, and number of interventions included in the network. We categorised journal types into Science Citation Index (SCI) or non-SCI; we also identified journals with high impact factors (IF ≥ 5.000, as reported on Journal Citation Reports 2014)27 or low impact factors (IF < 5.000). We also categorised NMAs into older studies or recent studies based on the median division of number of included NMAs.
Reporting of literature search
One reviewer (XQ) extracted following information regarding reporting of literature search: number of databases searched (Chinese, English, or both), name of databases searched, whether the search strategy was provided, whether the previous systematic reviews/meta-analyses were searched, name and number of other sources searched (e.g., reference lists checking, clinical trial registration platform, conference abstracts or web sites, Google engine).
Reporting of statistical analysis processes
We assessed the reporting and quality of statistical analysis processes in the methods sections of each NMA report according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) extension statement for NMAs28. The following questions were designed according to the statistical analysis section of PRISMA extension statement, and were extracted by two independent reviewers (LG, JZ), and conflicts were resolved by a third reviewer (J-HT, or K-HY):
Was traditional meta-analysis conducted?
Were summary measures reported?State the principal summary measures (e.g., risk ratio, odd ratio, mean difference, hazard ratio). Also describe the use of additional summary measures assessed, such as treatment rankings (e.g., treatment rankings, best, or surface under the cumulative ranking curve (SUCRA) values), shape and scale parameters for survival data29.
Planned methods of analysis, this should include: Was a Bayesian or a frequentist framework used? What was the model used? (random-effects model, fixed-effect model, others, or not report). What was the method used to undertake the indirect comparisons30?Was the model code presented or source cited in Bayesian analyses? (not provided, manuscript, online supplement, external web site, reference, or others). Was the model fit assessed? (e.g., residual deviance31, deviance information criterion31, other, not reported). Was handling of multigroup trials reported? Was selection of prior distributions in Bayesian analyses described? Was the convergence in Bayesian analyses assessed32? Was the heterogeneity in traditional meta-analysis assessed and how to handle the heterogeneity33? Was the heterogeneity in the entire network of NMA assessed and how to handle the heterogeneity34? Was the transitivity/similarity in NMA assessed35?
Were the inconsistency assessed and how to handle the inconsistency36?
Was the publication or reporting bias assessed37?
Additional analyses included: Was a sensitivity analysis performed? (e.g., excluding studies, alternative prior distributions for Bayesian analyses, alternative formulations of the treatment network). Was subgroup analysis or meta-regression performed?
Was the Grading of Recommendations Assessment, Development and Evaluation (GRADE) tool used to assess quality of evidence26?
Methodological quality assessment
There were no consensuses to assess the methodological quality of NMAs. We assessed the methodological quality of included NMAs using a modified AMSTAR checklist. This checklist included 11 items, with possible responses of “Yes” (item/question fully addressed), “No” (item/question not addressed), “Cannot answer” (not enough information to answer the question), and “Not applicable”. Two reviewers (XQ, G-QP) independently extracted data, and conflicts were resolved by a third reviewer (LG, or J-HT). The total score using AMSTAR was obtained by summing one point for each “yes” and no points for any other responses (“no”, “Cannot answer” and “Not applicable”), ranging from 0 to 11. In our study, three of the 11 items were slightly modified as follows (Appendix 3):
“Was an ‘a priori’ design provided?” was amended to “Was the research question (i.e., research purpose, inclusion and exclusion criteria) clarified?”The reason for this modification was that only a small minority of published non-Cochrane reviews reported a protocol38. Where a protocol providing this information was available, the answer to this question would be “Yes”. Where no protocol was available but detailed information about research purpose and inclusion and exclusion criteria (patients, interventions, comparators, outcome, and study design) were supplied, we also considered answer this question “Yes”.
“Was a list of studies (included and excluded) provided?” was amended to “Were a list of included studies and flow diagram provided?”The reason for this modification was that most of published systematic reviews did not provide a list of excluded studies. Where a list of included studies and flow diagram of literature selection were provided (as references, electronic link, or supplement), we considered answer this question “Yes”.
Were the methods used to combine the findings of studies appropriate?For pairwise meta-analysis, we scored “Yes” if they mentioned or described heterogeneity and reported how to handle heterogeneity. For NMA, the following factors should be taken into consideration except heterogeneity, but not be limited to: summary measures, model used, model fit, prior distributions (Bayesian analysis), convergence (Bayesian analysis), and inconsistency.
Statistical analysis
Quantitative data were summarised by medians and interquartile range (IQR), and categorical data summarised by numbers and percentages. The association between methodological quality and following characteristic variables was explored using the Mann-Whitney U test and Kruskai-Wallis test: journal impact factor, year of publication, funding source, country of corresponding author, type of NMAs, and categories of disease. Moreover, the subgroup analyses for statistical reporting were performed according to journal impact factor (high vs. low impact factors) and year of publication (older vs. recent studies). Proportion results were analysed by Chi-square test using STATA version 12.039. All tests were two sided, and P ≤ 0.05 was considered statistically significant.
Additional Information
How to cite this article: Ge, L. et al. Epidemiology Characteristics, Methodological Assessment and Reporting of Statistical Analysis of Network Meta-Analyses in the Field of Cancer. Sci. Rep. 6, 37208; doi: 10.1038/srep37208 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
No funding bodies played any role in study design, data collection and analysis, decision to publish, or preparation this manuscript.
Footnotes
Author Contributions G.L., T.J.H., and Y.K.H. planned and designed the research; S.F.J. provided methodological support/advice; L.L. and S.F.J. tested the feasibility of the study; G.L., T.J.H., L.X.X., Z.J., Q.X., and P.G.Q. extract data; G.L., T.J.H., and L.G. performed the statistical analysis; G.L. wrote the manuscript; all authors approved the final version of the manuscript.
References
- World Health Organization. Cancer. Available: http://www.who.int/topics/cancer/en/ (2015).
- Li J. L. et al. Quality of reporting of systematic reviews published in “evidence-based” Chinese journals. Syst Rev. 3, 58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C., Sturtz S. & Bender R. Indirect Comparisons and Network Meta-Analyses. Dtsch Arztebl Int. 112, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham G. K., Wells G. A., Gill S., Cameron C. & Jonker D. J. Chemotherapy regimens for advanced pancreatic cancer: a systematic review and network meta-analysis. BMC Cancer. 14, 471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani A., Higgins J. P., Geddes J. R. & Salanti G. Conceptual and technical challenges in network meta-analysis. Ann Intern Med. 159, 130e7 (2013). [DOI] [PubMed] [Google Scholar]
- Salanti G., Higgins J. P. T., Ades A. E. & Ioannidis J. P. Evaluation of networks of randomized trials. Stat Methods Med Res. 17, 279e301 (2008). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Network meta-analyses could be improved by searching more sources and by involving a librarian. J Clin Epidemiol. 67, 1001–1007 (2014). [DOI] [PubMed] [Google Scholar]
- Lee A. W. Review of mixed treatment comparisons in published systematic reviews shows marked increase since 2009. J Clin Epidemio. 67, 138–143 (2014). [DOI] [PubMed] [Google Scholar]
- Jansen J. P., Schmid C. H. & Salanti G. Directed acyclic graphs can help understand bias in indirect and mixed treatment comparisons. J Clin Epidemiol. 65, 798e807 (2012). [DOI] [PubMed] [Google Scholar]
- Lumley T. Network meta-analysis for indirect treatment comparisons. Stat Med. 21, 2313–2324 (2002). [DOI] [PubMed] [Google Scholar]
- Salanti G. Indirect and mixed-treatment comparison, network, or multiple-treatments meta-analysis: many names, many benefits, many concerns for the next generation evidence synthesis tool. Res Synth Methods. 3, 80–97 (2012). [DOI] [PubMed] [Google Scholar]
- Bafeta A., Trinquart L., Seror R. & Ravaud P. Analysis-of the-systematic reviews-process-in-reports-of-network-meta-analyses: methodological-systematic review. BMJ. 347, f3675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F. et al. Methodological problems in the use of indirect comparisons for evaluating healthcare interventions: survey of published systematic reviews. BMJ. 338, b1147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan S., Williamson P., Gamble C. & Tudur-Smith C. Indirect comparisons: a review of reporting and methodological quality. PLoS One. 5, e11054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J. D. et al. An assessment of the-methodological-quality-of published network meta-analyses: a systematic review. PLoS One. 10, e0121715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. X., Li X., Wang L., Li Y. P. & Tang X. L. Background Research for the Decision Support Tool of Evidence-Based Evaluation for Essential Medicines: 1. Overview of Quality Assessment Tools for Systematic Review of Randomized Controlled Trials. Chin J Evid-based Med. 15, 723–729 (2015). [Google Scholar]
- Shea B. J. et al. Development of AMSTAR: a measurement tool to assess the methodological quality of systematic reviews. BMC Med Res Methodol. 7, 10 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea B. J. et al. AMSTAR is a reliable and valid measurement tool to assess the methodological quality of systematic reviews. J Clin Epidemiol. 62, 1013–1020 (2009). [DOI] [PubMed] [Google Scholar]
- Shea B. J. et al. External validation of a measurement tool to assess systematic reviews (AMSTAR). PLoS One. 2, e1350 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrgiou M., Salanti G., Pavlidis N., Paraskevaidis E. & Ioannidis J. P. Survival benefits with diverse chemotherapy regimens for ovarian cancer: meta-analysis of multiple treatments. J Natl Cancer Inst. 98, 1655–1663 (2006). [DOI] [PubMed] [Google Scholar]
- Bucher H. C., Guyatt G. H., Griffith L. E. & Walter S. D. The results of direct and indirect treatment comparisons in meta-analysis of randomized controlled trials. J Clin Epidemiol. 50, 683–691 (1997). [DOI] [PubMed] [Google Scholar]
- Tian J. H., Ge L. & Li L. The PRISMA Extension Statement. Ann Intern Med. 163, 566 (2015). [DOI] [PubMed] [Google Scholar]
- Cohen J. F., Korevaar D. A., Wang J., Spijker R. & Bossuyt P. M. Should we search Chinese biomedical databases when performing systematic reviews? Syst Rev. 4, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S., Welton N. J., Marinho V., Salanti G., Higgins J. P. & Ades A. E. Estimation and adjustment of bias in randomized evidence by using mixed treatment comparison meta-analysis. J R Stat Soc. 173, 613–629 (2010). [Google Scholar]
- Donegan S., Williamson P., D’Alessandro U. & Tudur Smith C. Assessing key assumptions of network meta-analysis: a review of methods. Res Synth Methods. 4, 291–323 (2013). [DOI] [PubMed] [Google Scholar]
- Salanti G., Del Giovane C., Chaimani A., Caldwell D. M. & Higgins J. P. T. Evaluating the Quality of Evidence from a Network Meta-Analysis. PLoS ONE. 9, e99682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson Reuters. 2014 Journal Citation Reports. Available: http://thomsonreuters.com/journalcitation-reports/ (2014).
- Hutton B. et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med. 162, 777–784 (2015). [DOI] [PubMed] [Google Scholar]
- Ouwens M. J., Philips Z. & Jansen J. P. Network meta-analysis of parametric survival curves. Res Synth Methods. 1, 258–271 (2010). [DOI] [PubMed] [Google Scholar]
- Song F. J., Harvey I. & Lilford R. Adjusted indirect comparison may be less biased than direct comparison for evaluating new pharmaceutical interventions. J Clin Epidemiol. 61, 455–463 (2008). [DOI] [PubMed] [Google Scholar]
- Spiegelhalter D. J., Best N. G., Carlin B. P. & van der Linde A. Bayesian measures of model complexity and fit. J R Statist Soc B. 64, 57 (2002). [Google Scholar]
- Brooks S. P. & Gelman A. General Methods for Monitoring Convergence of Iterative Simulations. J Comput Graph Stat. 7(4), 434–455 (1998). [Google Scholar]
- Higgins J. & Green S. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.0.0 [updated February 2008]. The Cochrane Collaboration. Available: www.cochrane-handbook.org (2008).
- Cooper N., Sutton A., Morris D., Ades A. & Welton N. Addressing between study heterogeneity and inconsistency in mixed treatment comparisons: Application to stroke prevention treatments in individuals with non-rheumatic atrial fibrillation. Stat Med. 28, 1861–1881 (2009). [DOI] [PubMed] [Google Scholar]
- Glenny A. M. et al. Indirect comparisons of competing interventions. Health Technol Assess. 9, 1–134 (2005). [DOI] [PubMed] [Google Scholar]
- Veroniki A. A., Vasiliadis H. S., Higgins J. P. & Salanti G. Evaluation of inconsistency in networks of interventions. Int J Epidemiol. 42, 332–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y. et al. Comparative effectiveness of renin-angiotensin system blockers and other antihypertensive drugs in patients with diabetes: systematic review and Bayesian network meta-analysis. BMJ. 347, f6008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Tetzlaff J., Tricco A. C., Sampson M. & Altman D. G. Epidemiology-and-reporting-characteristics-of-systematic reviews. PLoS Med. 4, e78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. Stata Statistical Software. College Station: StataCorp LP (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.