Summary
Protein synthesis continues in platelets and maturing reticulocytes, although these blood cells lack nuclei and do not make new mRNA or ribosomes. Here, we analyze translation in primary human cells from both anucleate lineages by ribosome profiling and uncover dramatic accumulation of post-termination, unrecycled ribosomes in the 3´UTRs of mRNAs. We demonstrate that these ribosomes accumulate as a result of the natural loss of the ribosome recycling factor ABCE1 during terminal differentiation. Induction of ribosome rescue factors PELO and HBS1L is required to support protein synthesis when ABCE1 levels fall. including for hemoglobin production during blood cell development. Our observations suggest that this distinctive loss of ABCE1 in anucleate blood lineages could sensitize them to defects in ribosome homeostasis, perhaps explaining in part why genetic defects in the fundamental process of ribosome production (“ribosomopathies”) often affect hematopoiesis specifically.
Graphical Abstract
Introduction
In mammals, immature red blood cells (reticulocytes) and platelets lack nuclei and therefore cannot transcribe new RNA. Despite this, retained ribosomes and pre-synthesized mRNAs permit continued protein synthesis during terminal differentiation in these cells (Heynen, 1990; Ji et al., 2011). Reticulocytes are highly translationally active and circulate for 2–3 days before completely losing ribosomes, organelles, and mRNA; these latter events define their transition to mature erythrocytes (Heynen, 1990; Wiczling and Krzyzanski, 2008). Translation by reticulocytes is prolific – as much as one third of the final hemoglobin content of an erythrocyte is synthesized after enucleation (Heynen and Verwilghen, 1982; Heynen, 1990). Similarly, nascent platelets are released from the bone marrow containing ribosomes, organelles, and mRNA derived from the precursor megakaryocyte (Bray et al., 2013; Kissopoulou et al., 2013; Rowley et al., 2011). Platelet mRNA and ribosomes are stable for a period of at least several days (Ault, 1993; Ault et al., 1992) and during this time they direct protein synthesis (Booyse and Rafelson, 1967; Weyrich et al., 1998).
Ribosome homeostasis, which refers broadly to the regulation of ribosome levels in cells, plays an essential role in determining which mRNAs are translated (Ludwig et al., 2014; Signer et al., 2014). Haploinsufficiencies of specific ribosomal proteins and defects in ribosome biogenesis perturb ribosome homeostasis and disproportionately result in hematopoietic dysfunction (Barlow et al., 2010; Buchanan et al., 1981; Draptchinskaia et al., 1999; Ebert et al., 2008; Neuwirtova et al., 2013; Ricciardi et al., 2015). Additionally, disrupted ribosome biogenesis increases free ribosomal proteins, which can activate p53 (Zhang and Lu, 2009), further modulating these phenotypes (Barlow et al., 2010). It is not well understood why these defects lead to hematopoietic tissue-specific phenotypes. Moreover, mechanisms that regulate ribosome availability in anucleate blood lineages have not been identified, though autophagy (Kundu et al., 2008), ubiquitin-dependent degradation (Etlinger and Goldberg, 1977; Wefes et al., 1995), as well as specific ribonucleases (Pisareva et al., 2015; Valentine et al., 1974) have been proposed to degrade ribosomes in reticulocytes. Delayed ribosome clearance is associated with abnormal erythroid maturation (Kundu et al., 2008; Valentine et al., 1974) and platelet dysfunction in humans (Fletcher et al., 2015; Marconi et al., 2016).
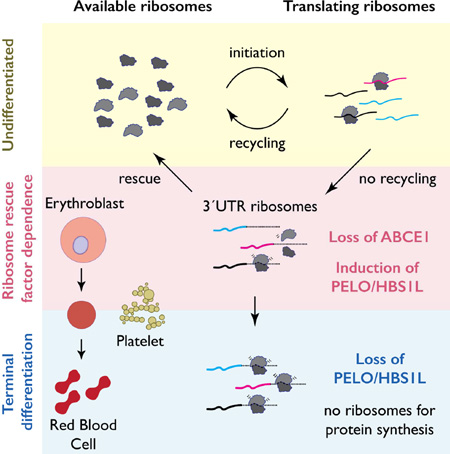
We show here that the active dissociation of the ribosomal subunits, a distinct step in the translation cycle termed ribosome recycling (Pisarev et al., 2010; Shoemaker et al., 2010), regulates ribosome homeostasis in reticulocytes and platelets. Our data suggest that loss of the ribosome recycling factor ABCE1 (Rli1p in S. cerevisiae), limits ribosome availability during terminal differentiation of these anucleate blood lineages leading to the accumulation of post-termination, unrecycled ribosomes in the 3´UTRs of mRNAs. These 3´UTR ribosomes are cleared by the induction of ribosome rescue factors PELO and HBS1L (Dom34p and Hbs1p in S. cerevisiae), which return ribosomes to the active cellular pool. These factors act as a heteroduplex to release stalled and unrecycled ribosomes (Guydosh and Green, 2014) and compensate for RLI1 defects in yeast (Young et al., 2015). We propose that enhanced ribosome rescue compensates for the loss of ABCE1-mediated recycling in anucleate blood lineages. Our results delineate the functional significance of this regulatory switch by demonstrating that hemoglobin translation depends on PELO/HBS1L expression. This work may help rationalize the sensitivity of erythroid cell lineages to genetic diseases caused by mutations in components of the translational machinery.
Results
Anucleate cells accumulate 3´UTR ribosomes
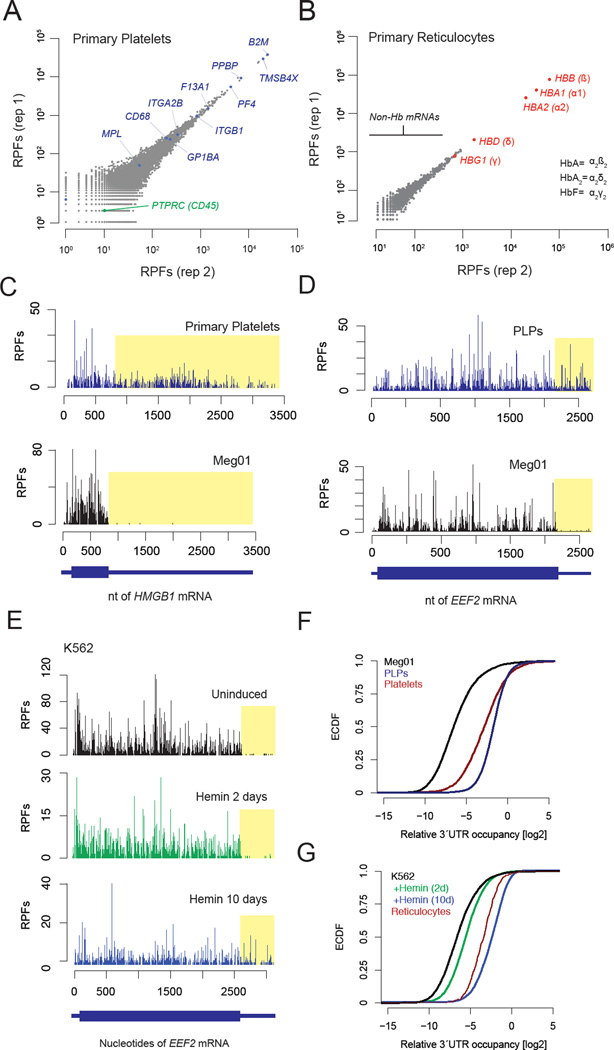
We analyzed translation in primary human anucleate blood lineages by ribosome profiling, which relies on the deep sequencing of ribosome-protected fragments (RPFs) (Ingolia et al., 2011; Ingolia et al., 2009). Analysis of these data revealed ~6,700 ribosome-occupied mRNAs in platelets using a cut-off of 0.3 reads per kilobase of coding sequence per million mapped reads (Figure 1A), consistent with the high complexity of the platelet transcriptome (Bray et al., 2013; Gnatenko et al., 2003; Kissopoulou et al., 2013; Rowley et al., 2011) and proteome (Burkhart et al., 2012; Kim et al., 2014; Martens et al., 2005; Vaudel et al., 2012; Wilhelm et al., 2014). Reticulocyte ribosomes predominantly occupied adult hemoglobin (Hb) mRNAs (Figure 1B), consistent with the fact that Hb transcripts make up the majority of the reticulocyte transcriptome (Clissold et al., 1977) and mostly hemoglobin protein (Denton et al., 1975) is made at this stage of differentiation. We also detected ribosomes occupying ~400 additional mRNAs in reticulocytes, indicating that many non-Hb mRNAs are also translated during this stage of development.
Fig. 1. Platelet and reticulocyte ribosome profiling show abundant 3´UTR RPFs.
A. Scatter plots of RPF counts per mRNA coding sequence for replicates from a single healthy human donor of primary platelets. Known platelet proteins are highlighted in blue. PTPRC (CD45) is a white blood cell marker (green). B. Replicates of RPF counts from CD71+ primary human reticulocytes. C. Ribosome profile of the HMGB1 mRNA in platelets (upper panel) and Meg01 cells (lower panel). D. Ribosome profile of the EEF2 mRNA from in vitro platelet-like particles (PLPs) (upper panel) and precursor Meg01 cells (lower panel). E. Accumulation of 3´UTR RPFs during erythroid-like differentiation. F and G. Cumulative histograms of relative 3´UTR occupancy for (F) primary human platelets, and cell culture models, and (G) primary reticulocytes and cell culture models.
Surprisingly, in primary human platelets and reticulocytes, we observed substantial ribosome occupancy on the 3´UTRs (Figure 1C, 1F–G). While these regions are devoid of ribosome footprints in other cell types, we quantified an approximately 30-fold increase in RPFs in platelets and reticulocytes compared to nucleated peripheral blood mononuclear cells (PBMCs) (Figure 1F–G, Figure S2A), neutrophils (Guo et al., 2010), or macrophages (Su et al., 2015). The increased density of 3´UTR RPFs is a global rather than a transcript-specific effect. In platelets, 3´UTR RPFs represented ~30% of all RPFs mapped to mRNA transcripts, while in reticulocytes (which have a far less complex transcriptome) 4% of all RPFs were mapped to 3´UTRs.
To explore the mechanisms underlying the 3´UTR ribosome accumulation in primary human cells, we tested whether these effects could be recapitulated by in vitro systems of blood cell differentiation. We purified in vitro platelet-like particles (PLPs) from megakaryoblast (Meg01) precursor cells that generate these particles in tissue culture (Clarke et al., 2003; Takeuchi et al., 1991; Takeuchi et al., 1998) and showed that they are translationally active (Figure S1F–J; See Supplemental Methods). Ribosome profiling in PLPs showed that, like primary human platelets, PLPs display increased 3´UTR RPF density relative to precursor Meg01 cells (Figure 1D, F). Next, we induced K562 cells towards erythroid differentiation by supplementing the growth media with hemin (Rutherford et al., 1979). These cells also exhibited a global, differentiation-dependent increase in 3´UTR RPF density (Figure 1E), consistent with observations from primary reticulocytes (Figure 1G). Together, these data establish that the striking increase in 3´UTR RPF density we observed in primary platelets and reticulocytes also occurs in cell models for these lineages.
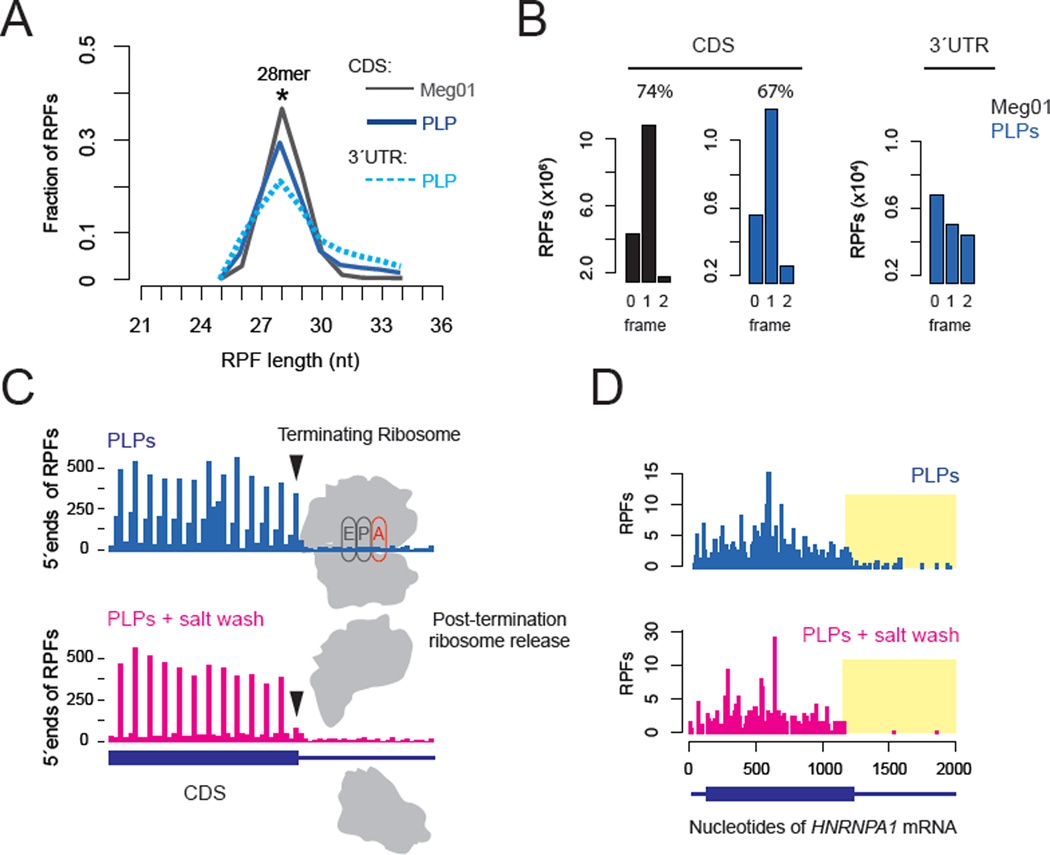
We next set out to verify that these 3’UTR RPFs are derived from bona fide ribosomes and test whether these ribosomes are actively engaged in translation. As previous work has shown that genetic reduction of ribosome rescue and recycling factors in yeast generates an accumulation of 3´UTR ribosomes (Young et al., 2015) we focused on the possibility that a failure of ribosome recycling may explain the abundance of 3´UTR RPFs observed in platelets and reticulocytes. The distribution of fragment sizes that mapped to 3´UTRs was nearly indistinguishable from those that mapped to the CDS (Figure 2A), suggesting that they are authentic RPFs, and not the result of non-ribosomal ribonucleoprotein (RNP) complexes protecting mRNA fragments from nuclease digestion (Ingolia et al., 2014). The 3´UTR RPFs did not preferentially occupy a single open reading frame (ORF) as would be anticipated from stop codon read-through (Figure 2B) (Dunn et al., 2013), however. We thus tested whether 3´UTR ribosomes are associated with a nascent polypeptide chain (indicative of active translation) by treating lysates with high salt concentrations to release vacant ribosomes lacking a nascent polypeptide (Blobel and Sabatini, 1971). This treatment released ~80% of ribosomes with a stop codon in the A site (Figure 2C, black arrows), consistent with the expectation that these ribosomes have terminated translation and released their nascent polypeptide. Critically, ribosomes in the 3´UTR were similarly released by the high salt wash, indicating that they also lack a nascent polypeptide (Figure 2D, Figure S2B); by contrast, ribosomes within the ORF were not affected. Based on these data (and additional genetic experiments below), we conclude that a majority of ribosomes in the 3´UTR are post-termination, unrecycled ribosomes not actively translating.
Fig. 2. 3´UTR RPFs result from post-termination ribosomes.
A. RPF size distributions of 3´UTR footprints relative to CDSes. B. Reading frame analysis of RPFs mapped to coding sequences (CDS) or 3´UTRs. C. 5´ends of RPFs around the stop codon. Arrows indicate ribosomes with a stop codon in the A site, and are dissociated a high salt wash (1M KCl). D. Ribosome profile for the HNRNPA1 mRNA, with 3´UTR regions highlighted in yellow.
Dynamic regulation of ribosome recycling and ribosome rescue
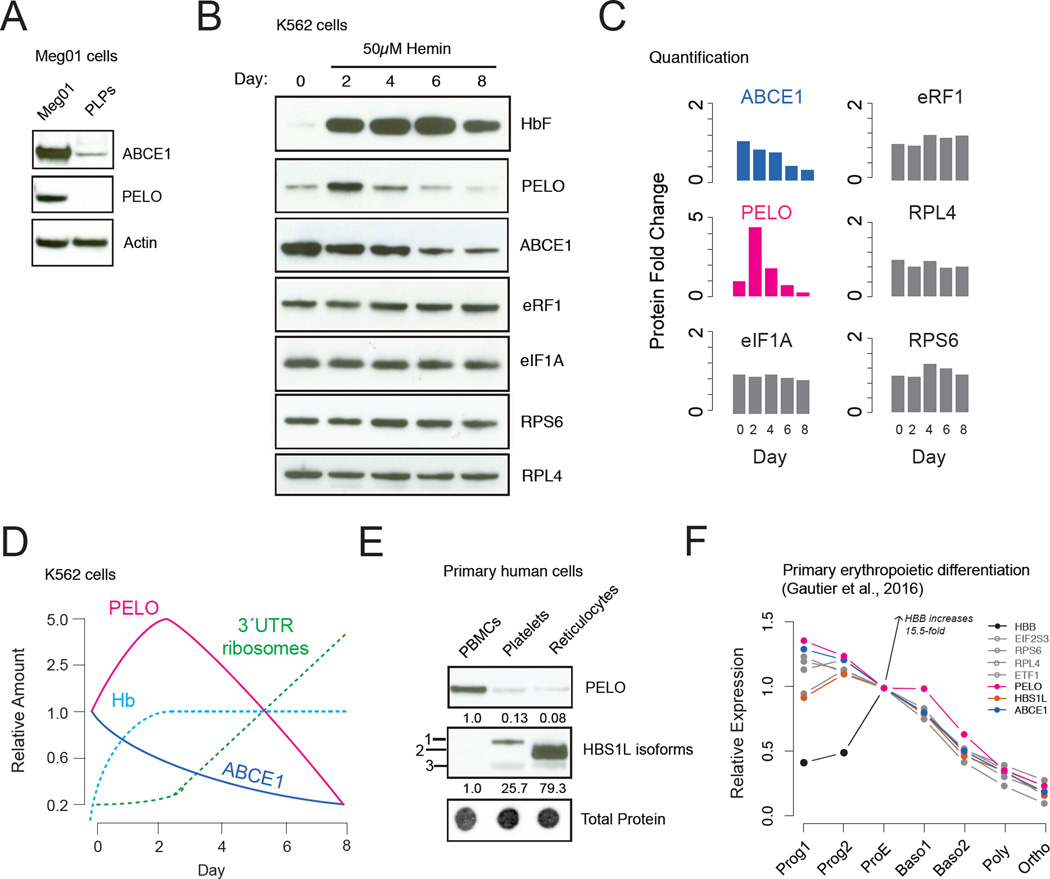
The accumulation of vacant ribosomes downstream of the stop codon is consistent with a recycling defect. Indeed, we found that the levels of the canonical ribosome recycling factor ABCE1 (Pisarev et al., 2010) are strongly reduced in platelets compared to precursor megakaryoblast cells (Meg01) and in reticulocytes when compared to undifferentiated K562 cells (Figure S2C). Moreover, declining ABCE1 levels have been observed in proteomic analyses of erythroid differentiation from primary human cells (Gautier et al., 2016; Prenni et al., 2012), although the consequences of this decline were not explored. Interestingly, ABCE1 is also depleted in PLPs (Figure 3A) and in K562 cells induced toward erythroid differentiation (Figure 3B–C), where RNA levels also decline (Figure S4). Natural reduction of ABCE1 in primary anucleate lineages thus occurs our in vitro models, which also show 3´UTR ribosome accumulation, while levels of other translation factors, including the initiation factor eIF1A, the termination factor eRF1, and core ribosomal proteins RPS6 or RPL4 do not change (Figure 3B–C). In reticulocytes, a likely explanation for the loss of ABCE1 is that this protein contains an obligate FeS cluster (Barthelme et al., 2007) (Figure S2F–G) and natural clearance of mitochondria during maturation (Kundu et al., 2008; Novak et al., 2010; Schweers et al., 2007) will inevitably impair FeS cluster biogenesis. Consistent with this model, we see clear evidence for loss of mitochondrially encoded transcripts in our RNA-Seq data (Figure S2F).
Fig. 3. Dynamic regulation of the ribosome recycling machinery.
A. PELO and ABCE1 are present in Meg01 cells but absent in PLPs. B. Translation factor expression in differentiating K562 cells. C. Quantification of western blots in (B), normalized to day 0. D. Schematic of recycling/rescue factor changes during K562 cell differentiation in relation to hemoglobin and 3´UTR ribosome accumulation. E. Differential expression of PELO and HBS1L isoforms in primary blood cells (PBMCs: peripheral blood mononuclear cells) with band intensities normalized to total protein. F. Quantitative mass spectrometry data from Gautier et al., 2016 showing relative protein levels during erythropoietic differentiation of primary CD34+ cells.
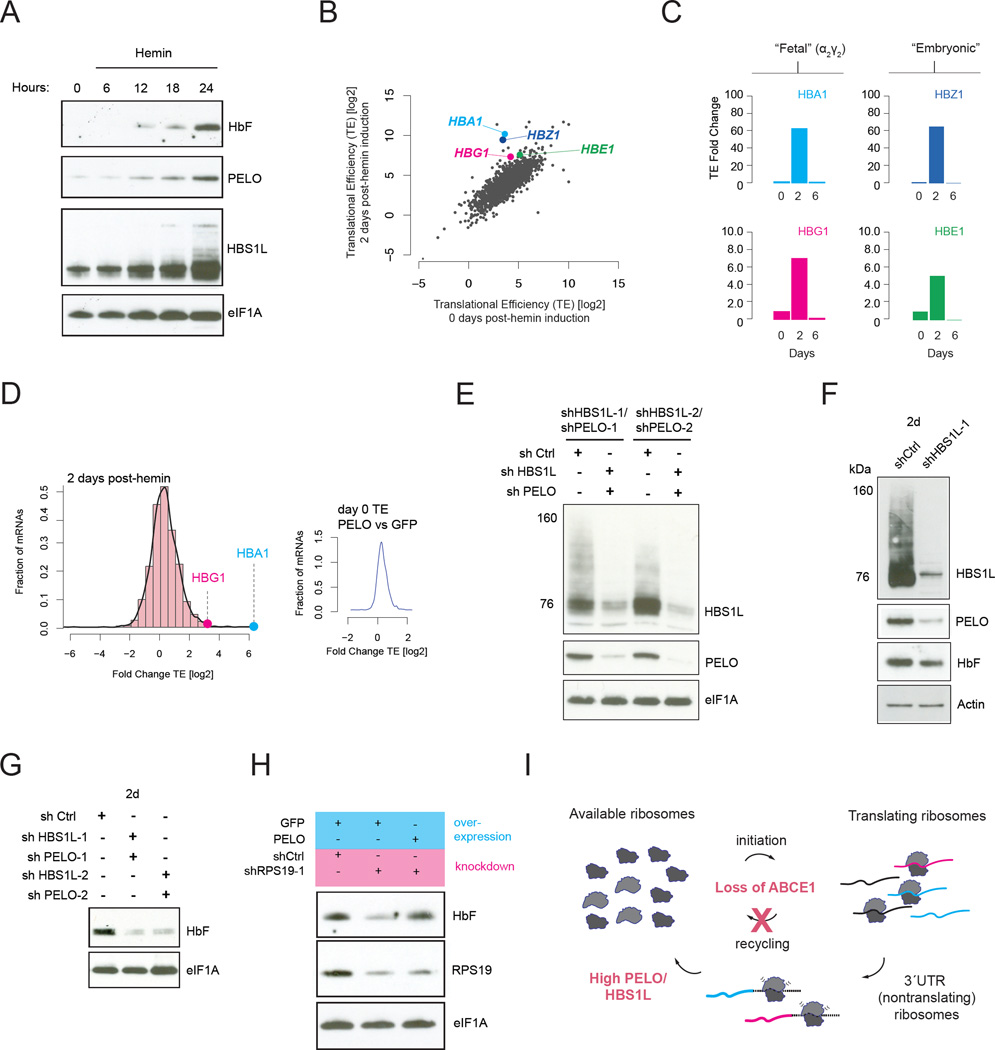
Reduced ABCE1 levels suggest a plausible mechanistic explanation for 3´UTR ribosome occupancy. However, while ABCE1 levels begin to decline immediately after K562 cells are induced to differentiate, 3’ UTR ribosome accumulation was modest 2 days after induction (Figure 1E, G). These observations suggest a compensatory response might allow these cells to remove post-termination ribosomes from the 3´UTRs despite reduced levels of ABCE1. To test this hypothesis, we measured the expression of the ribosome rescue factors PELO and HBS1L, which act together to release stalled or unrecycled ribosomes that are not engaged in elongation and make them available for translation (Guydosh and Green, 2014; Shoemaker et al., 2010). In primary platelets and reticulocytes, PELO levels were strongly diminished (Figure 3E) and this down regulation was recapitulated in PLPs (Figure 3A) and fully differentiated K562 cells (Figure 3B). Both recycling and rescue factors are attenuated in fully mature platelets and reticulocytes, consistent with the observed abundance of 3´UTR ribosomes in these lineages. Strikingly however, PELO was initially up-regulated ~5-fold in the 2 days following hemin induction in K562 cells (Figure 3B–D) and more modestly in published quantitative mass spectrometry data from erythropoietic differentiation of primary CD34+ cells (Fig. 3F) (Gautier et al., 2016).
Our data reveal dynamic regulation of ribosome recycling and rescue factors in anucleate hematopoietic lineages. The delay between the decline in ABCE1 levels and the accumulation of 3´UTR ribosomes leads to a model wherein transient activation of the ribosome rescue pathway compensates for the natural decline of ribosome recycling. to permit ongoing translation of critical mRNAs during terminal differentiation (Figure 3D). Subsequent, combined loss of both ABCE1 and PELO late in differentiation, when less translation is required, may allow the dramatic accumulation of unrecycled ribosomes observed by ribosome profiling.
PELO compensates for the loss of ABCE1
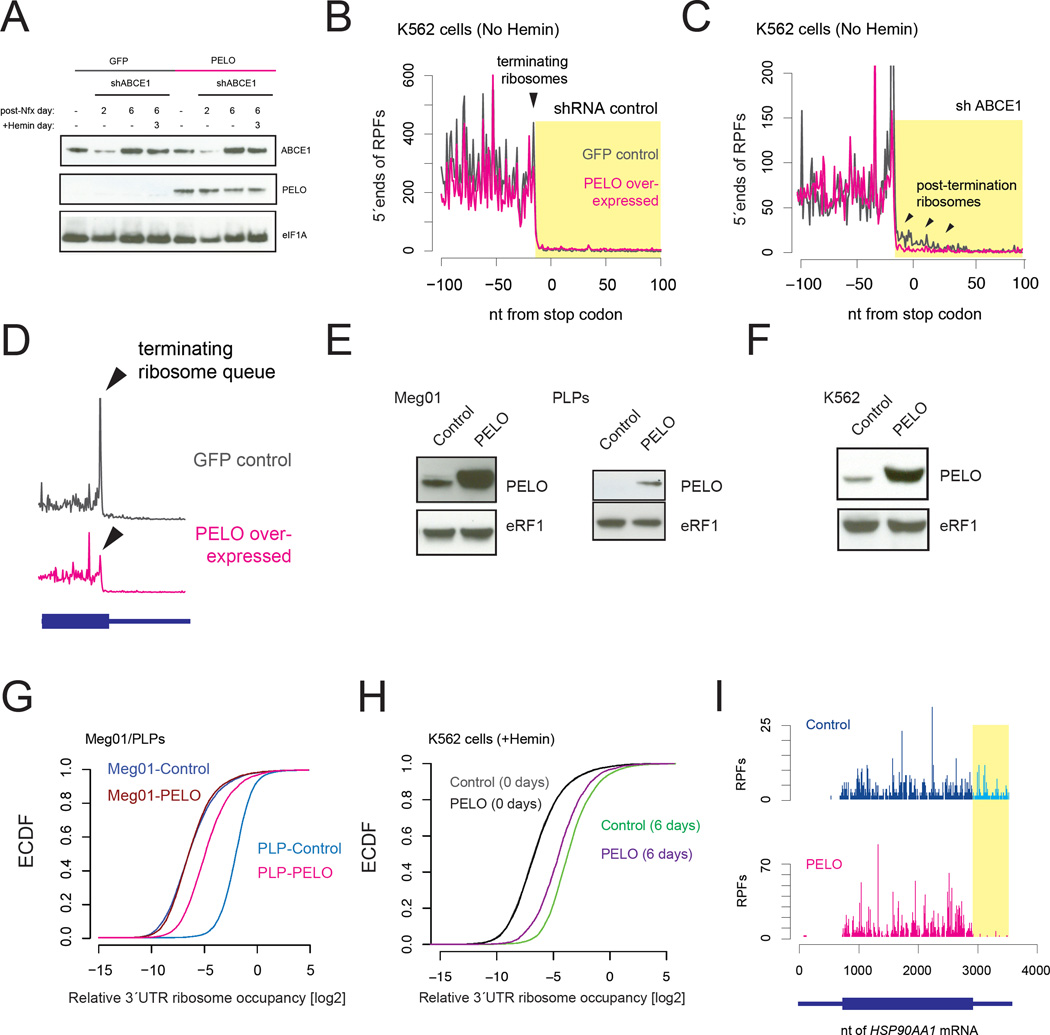
We set out to directly test our model by manipulating the levels of ribosome recycling/rescue factors in vitro. Indeed, shRNA knock-down of ABCE1 undifferentiated K562 cells (Figure 4A), which normally do not accumulate 3´UTR ribosomes, was sufficient to drive their accumulation (Figure 4B–C, black traces). Further, these accumulated 3´UTR ribosomes could be “rescued” by over expression of PELO (Figure 4B–C, pink traces; Figure 4F), further validating that 3´UTR RPFs in K562 cells and PLPs represent authentic ribosomes. As anticipated, ABCE1 knock down also led to a build-up of terminated, unrecycled ribosomes at the stop codon (Figure 4D black trace) that was cleared by over expression of PELO (Figure 4D, pink trace). Importantly, over expression of PELO did not alter global translation patterns or RNA levels in the presence of normal levels of ABCE1 (Figure S3G–H, Days 0–2). These data establish that loss of ABCE1 is sufficient to cause the accumulation of post-termination 3´UTR ribosomes in mammalian cells and that increased expression of PELO can at least in part suppress this effect.
Fig. 4. Ribosome rescue by PELO/HBS1L compensates for declining ABCE1 levels.
A. shRNA knock down of ABCE1 in K562 cells overexpressing GFP or PELO. B and C. Profile of 28–32nt RPFs around stop codons. ABCE1 knockdown leads to the accumulation of post-termination ribosomes, indicated by black arrows, either at the stop codon (in B) or in the 3´ UTR (in C). PELO overexpression suppresses this effect. D. ABCE1 knock down leads to a queue of terminating ribosomes on stop codons (gray trace) and over expression of PELO prevents this effect. E and F. Transgenic over expression of PELO increases protein levels relative to eRF1 (control). Restoration of PELO releases 3´UTR ribosomes in PLPs (G) and 6 day differentiated K562 cells (H). I. ribosome profile of the HSP90AA1 mRNA.
To test whether PELO can also rescue the 3´UTR ribosomes that accumulate physiologically during differentiation, we stably over expressed PELO (or GFP as our control) in Meg01 or K562 cells (Figure 4E–F) and performed ribosome profiling on PLPs and hemin-differentiated K562 cells. As anticipated, overexpression of PELO suppressed 3´UTR ribosomes strongly in PLPs (Figure 4G and 4I; Figure S3C–E) and more modestly in day 6 differentiated K562 cells, when endogenous levels of PELO were low (Figure 4H, Figure S3F). We conclude that the 3´UTR ribosomes that accumulate on platelet and reticulocyte mRNAs are true substrates for ribosome rescue by PELO.
PELO/HBS1L regulate ribosome homeostasis
We propose that induction of the ribosome rescue factor PELO during platelet and reticulocyte maturation plays a critical role in rescuing ribosomes to maintain translational homeostasis and support the synthesis of critical proteins as ABCE1 levels decline. Reticulocytes synthesize large quantities of hemoglobin subunit proteins, as do K562 cells differentiating in response to hemin treatment (Rutherford et al., 1979), and we observed that the induction of PELO and HBS1L coincides closely with hemoglobin production (Figure 5A and 3B). We note that K562 cells primarily synthesize embryonic and fetal hemoglobin tetramers (α2γ2 and α2ε2) (Rutherford et al., 1979) rather than the adult hemoglobin (α2β2) that is made in primary reticulocytes from adults. Through analysis of our ribosome profiling and RNA-Seq data, we see that hemoglobin accumulation is primarily driven by increased translation (Figure 5B). We calculated the translational efficiency (TE) as the ratio of RPFs relative to mRNA abundance and saw that TE of the individual alpha and gamma mRNAs encoding the HbF (α2γ2) tetramer rises dramatically in the first 2 days of differentiation (~75-fold for α-globin mRNA and ~8-fold for γ-globin mRNA) and drops off rapidly thereafter (Figure 5B–C, Figure S4). The time course of hemoglobin production closely matches that of PELO expression (Figure 3B, Figure 5A), suggesting that PELO/HBS1L induction may be critical in providing available ribosome subunits for its translation.
Fig. 5. PELO/HBS1L regulate ribosome homeostasis.
A. Hemoglobin, PELO, and HBS1L expression during the first 24 hours of hemin induction. B. Global translational efficiency (TE) changes after 2 days of hemin induction. C. TE changes for mRNAs encoding hemoglobin subunits measured two days post-induction. D. Histogram of TE changes for all mRNAs after 2 days of hemin treatment. Inset: TE ratio (PELO vs GFP) prior to Hemin induction. E. shRNA knockdown of PELO and HBS1L with two different shRNA hairpin sequences each. F and G. Hemoglobin expression after hemin induction is reduced by knock-down of HBS1L, enhanced by knock-down of PELO. H. RPS19 knockdown reduces hemin-induced hemoglobin accumulation, suppressed by overexpression of PELO. I. Model showing the loss of ABCE1 in differentiating K562 cells accompanied by induction of PELO/HBS1L to maintain ribosome homeostasis.
PELO/HBS1L is required for hemin-induced HbF accumulation
In light of the tightly coincidence between PELO induction and hemoglobin expression, we next asked whether PELO/HBS1L induction is required to promote hemoglobin accumulation. Defects in ribosome rescue (Figure 5E), either by knock-down of HBS1L alone (Figure 5F) or with PELO (Figure 5G), abrogated hemoglobin production. Levels of control proteins eIF1A or actin remained unchanged, indicating that these changes do not reflect a loss of cell viability. These data instead argue that PELO/HBS1L induction is required for translation of a group of mRNAs that includes hemoglobin in K562 cells, where they compensate for the natural loss of ABCE1 to maintain ribosome availability. We propose that this release of unrecycled ribosomes by PELO/HBS1L allows for the translation of critical mRNAs, such as those encoding hemoglobin (Lodish, 1974). As hemoglobin mRNAs are well known to have relatively high initiation rates (Lodish, 1971), and should be kinetically favored in competitions with other mRNAs, an increase in ribosomes available for initiation could differentially enhance translation of hemoglobin transcripts.
Ribosome rescue and disorders of ribosome homeostasis
Our model for the regulation of ribosome homeostasis in anucleate hematopoietic cells raised the question of how recycling and rescue factors interact with other perturbations in ribosome availability in these lineages. “Ribosomopathies” are a group of heterogeneous disorders that result from perturbations in ribosome homeostasis and often manifest in hematopoietic dysfunction (in addition to defects in other tissues) (Narla and Ebert, 2010). Heterozygous loss of several specific core ribosomal protein genes, including RPS19, leads to Diamond-Blackfan Anemia (DBA) (Draptchinskaia et al., 1999), a disorder associated with reduced erythroid progenitor populations (Nathan et al., 1978) that causes congenital hypoplastic anemia (Diamond and Blackfan, 1938). The pathogenesis of RPS19-deficient DBA is complex, but involves a defect in 40S ribosome biogenesis (Flygare et al., 2007) that leads to impaired survival of early erythroid progenitors (Flygare et al., 2005).
To examine the potential relevance of our observations to DBA, we depleted RPS19 in K562 cells by shRNA knockdown (Figure 5H, Figure S5), which should reduce ribosome production by restricting ribosome proteins available for assembly. While specific stages of erythroid development lack reliable matched counterparts in K562 differentiation, the early phase of erythrogenesis resembles an early, 24-hour timepoint in K562 cells since they still have nuclei and have not yet lost all ribosome recycling/ribosome rescue or arrested protein synthesis (as in Figure 3D). Hemoglobin accumulation was strongly abrogated by RPS19 depletion (Figure 5H, lanes 1 and 2). Strikingly, transgenic over expression of PELO/HBS1L restored hemoglobin levels (Figure 5H, lane 3; See also Figure S10B). PELO/HBS1L can thus counteract general defects in ribosome availability, resulting from defects in ribosome production as well as recycling.
Discussion
Here, we reveal a surprising accumulation of ribosomes on the 3´UTRs of mRNA transcripts in primary platelets and reticulocytes, and argue that they are driven by cell type specific regulation of the core ribosome recycling and rescue machinery present in all cells (Figure 5I). These distinctive features of anucleate blood cell lineages are recapitulated in cell culture models. We found that the natural loss of ABCE1-mediated ribosome recycling during terminal differentiation causes vacant, un-recycled ribosomes to enter the 3´UTR of platelet and reticulocyte mRNAs. These 3´UTR RPFs can be released by overexpression of ribosome rescue factors PELO and HBS1L.
We further show that these ribosome rescue factors, PELO and HBS1L, along with the recycling factor ABCE1, are dynamically regulated during erythroid and platelet maturation. We propose that the up-regulation of PELO/HBS1L is critical for maintaining the pool of available ribosomes for translation when ABCE1 activity declines during the differentiation of anucleate blood cell lineages. The events that trigger ABCE1 loss are unknown, but we speculate that its obligate FeS cluster (Barthelme et al., 2007) can no longer by synthesized after mitochondrial break down during reticulocyte maturation.
We have identified the regulation of ribosome availability in platelets and reticulocytes as a critical role for mammalian ribosome rescue factors, distinct from their known role in mRNA surveillance. Our results also point to a broader potential for rescue factors in mitigating the effects of ribosome shortage resulting from impaired ribosome biogenesis as well as physiological loss of ABCE1. Natural levels of ribosome rescue factors could modulate the sensitivity of cells to the availability of ribosomes, and PELO/HBS1L levels may increase to meet transient cellular demands for ribosomes, including those posed by declining levels of ABCE1 or ribosomal protein haploinsufficiency (Bhattacharya et al., 2010; Flygare et al., 2007). Intriguingly, genomic variants near the mammalian HBS1L-MYB locus are associated with multiple hematologic traits (Menzel et al., 2007). Whether these polymorphic variants affect the regulation of PELO/HBS1L that we observed in platelets and erythroid cells, in addition to their well-documented effects on MYB expression (Stadhouders et al., 2014), remains an exciting area of future investigation.
Experimental Procedures
Primary platelet and reticulocytes
Leukoreduced plateletpheresis samples were obtained from individual healthy donors in accordance with IRB #00001463 (Johns Hopkins University). Platelet samples were prepared from single donor apheresis samples to achieve <1×106 WBCs per unit of >1×1011 platelets (or a WBC frequency of 1×10−6).
Ribosome profiling
Ribosome profiling was carried out as previously described (Ingolia et al., 2012). For high salt washes, lysates were brought to 1M KCl (final concentration) for 30 minutes on ice and exchanged using a desalting column (Zeba) prior to footprinting.
Meg01/PLP cell culture experiments
Meg01 cells were obtained from ATCC and cultured in RPMI 1640 (10% FBS). PLPs were purified from Meg01 culture supernatants by 5µm filtration and centrifugation at 3,000×g for 10 minutes (Figure S3A–C; See Supplemental Methods).
K562 differentiation experiments
K562 cells were obtained from ATCC and cultured in RPMI 1640 (10% FBS) supplemented with 2mM Glutamine. Hemin (Sigma) was dissolved in 20mM NaOH and sterile filtered through a 0.22µm filter (Millipore) before adding to cells.
Supplementary Material
Acknowledgments
We thank S. Eacker for help with lentiviral preparations. This work was supported by an AHA fellowship (E.W.M.), MSTP funding (E.W.M.), the Searle Scholars Program 11-SSP-229 (N.T.I.), an NIH training grant (J.W), and HHMI (R.G.). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.W.M., R.G., and N.T.I designed the experiments and interpreted results; E.W.M performed the experiments; E.W.M & J.W performed initial hemin experiments; E.W.M., R.G., and N.T.I. wrote the paper with input from all authors.
Accession Numbers
Data has been deposited in the NCBI GEO database under the accession number GSE85864.
References
- Ault KA. Flow cytometric measurement of platelet function and reticulated platelets. Ann. N. Y. Acad. Sci. 1993;677:293–308. doi: 10.1111/j.1749-6632.1993.tb38785.x. [DOI] [PubMed] [Google Scholar]
- Ault KA, Rinder HM, Mitchell J, Carmody MB, Vary CP, Hillman RS. The significance of platelets with increased RNA content (reticulated platelets). A measure of the rate of thrombopoiesis. Am. J. Clin. Pathol. 1992;98:637–646. doi: 10.1093/ajcp/98.6.637. [DOI] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q-syndrome. Nat. Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Scheele U, Dinkelaker S, Janoschka A, Macmillan F, Albers SV, Driessen AJ, Stagni MS, Bill E, Meyer-Klaucke W, Schunemann V, Tampe R. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 2007;282:14598–14607. doi: 10.1074/jbc.M700825200. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol. Cell. Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U. S. A. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booyse F, Rafelson ME. In vitro incorporation of amino-acids into the contractile protein of human blood platelets. Nature. 1967;215:283–284. doi: 10.1038/215283a0. [DOI] [PubMed] [Google Scholar]
- Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, Kupper J, Jing Y, Londin E, Loher P, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14 doi: 10.1186/1471-2164-14-1. 1-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan GR, Alter BP, Holtkamp CA, Walsh EG. Platelet number and function in Diamond-Blackfan anemia. Pediatrics. 1981;68:238–241. [PubMed] [Google Scholar]
- Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120:e73–e82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- Clarke MC, Savill J, Jones DB, Noble BS, Brown SB. Compartmentalized megakaryocyte death generates functional platelets committed to caspase-independent death. J. Cell Biol. 2003;160:577–587. doi: 10.1083/jcb.200210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold PM, Arnstein HR, Chesterton CJ. Quantitation of globin mRNA levels during erythroid development in the rabbit and discovery of a new β-related species in immature erythroblasts. Cell. 1977;11:353–361. doi: 10.1016/0092-8674(77)90052-6. [DOI] [PubMed] [Google Scholar]
- Denton MJ, Spencer N, Arnstein H. Biochemical and enzymic changes during erythrocyte differentiation. The significance of the final cell division. Biochem. J. 1975;146:205–211. doi: 10.1042/bj1460205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LK, Blackfan K. Hypoplastic anemia. Am. J. Dis. Child. 1938;56:464–467. (1938) [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etlinger JD, Goldberg AL. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. U. S. A. 1977;74:54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SJ, Johnson B, Lowe GC, Bem D, Drake S, Lordkipanidzé M, Guiú IS, Dawood B, Rivera J, Simpson MA. SLFN14 mutations underlie thrombocytopenia with excessive bleeding and platelet secretion defects. J. Clin. Invest. 2015;125:3600–3605. doi: 10.1172/JCI80347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J, Kiefer T, Miyake K, Utsugisawa T, Hamaguchi I, Da Costa L, Richter J, Davey EJ, Matsson H, Dahl N, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- Gautier E, Ducamp S, Leduc M, Salnot V, Guillonneau F, Dussiot M, Hale J, Giarratana M, Raimbault A, Douay L. Comprehensive Proteomic Analysis of Human Erythropoiesis. Cell Reports. 2016;16:1470–1484. doi: 10.1016/j.celrep.2016.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatenko DV, Dunn JJ, McCorkle SR, Weissmann D, Perrotta PL, Bahou WF. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heynen M, Verwilghen R. A quantitative ultrastructural study of normal rat erythroblasts and reticulocytes. Cell Tissue Res. 1982;224:397–408. doi: 10.1007/BF00216882. [DOI] [PubMed] [Google Scholar]
- Heynen M. Ultrastructural Changes during Erythroid Development. In: Harris JR, editor. Blood Cell Biochemistry. 1990. pp. 1–26. [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Murata-Hori M, Lodish HF. Formation of mammalian erythrocytes: chromatin condensation and enucleation. Trends Cell Biol. 2011;21:409–415. doi: 10.1016/j.tcb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissopoulou A, Jonasson J, Lindahl TL, Osman A. Next generation sequencing analysis of human platelet PolyA mRNAs and rRNA-depleted total RNA. 2013 doi: 10.1371/journal.pone.0081809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- Lodish HF. Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J. Biol. Chem. 1971;246:7131–7138. [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014 doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi C, Di Buduo CA, Barozzi S, Palombo F, Pardini S, Zaninetti C, Pippucci T, Noris P, Balduini A, Seri M, Pecci A. SLFN14-related thrombocytopenia: identification within a large series of patients with inherited thrombocytopenia. Thromb. Haemost. 2016;115:1076–1079. doi: 10.1160/TH15-11-0884. [DOI] [PubMed] [Google Scholar]
- Martens L, Van Damme P, Van Damme J, Staes A, Timmerman E, Ghesquière B, Thomas GR, Vandekerckhove J, Gevaert K. The human platelet proteome mapped by peptide-centric proteomics: A functional protein profile. Proteomics. 2005;5:3193–3204. doi: 10.1002/pmic.200401142. [DOI] [PubMed] [Google Scholar]
- Menzel S, Jiang J, Silver N, Gallagher J, Cunningham J, Surdulescu G, Lathrop M, Farrall M, Spector TD, Thein SL. The HBS1L-MYB intergenic region on chromosome 6q23.3 influences erythrocyte, platelet, and monocyte counts in humans. Blood. 2007;110:3624–3626. doi: 10.1182/blood-2007-05-093419. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DG, Hillman DG, Chess L, Alter BP, Clarke BJ, Breard J, Housman DE. Normal erythropoietic helper T cells in congenital hypoplastic (Diamond-Blackfan) anemia. N. Engl. J. Med. 1978;298:1049–1051. doi: 10.1056/NEJM197805112981903. [DOI] [PubMed] [Google Scholar]
- Neuwirtova R, Fuchs O, Holicka M, Vostry M, Kostecka A, Hajkova H, Jonasova A, Cermak J, Cmejla R, Pospisilova D. Transcription factors Fli1 and EKLF in the differentiation of megakaryocytic and erythroid progenitor in 5q-syndrome and in Diamond–Blackfan anemia. Ann. Hematol. 2013;92:11–18. doi: 10.1007/s00277-012-1568-1. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Muslimov IA, Tcherepanov A, Pisarev AV. Characterization of Novel Ribosome-Associated Endoribonuclease SLFN14 from Rabbit Reticulocytes. Biochemistry (N. Y.) 2015 doi: 10.1021/acs.biochem.5b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenni JE, Vidal M, Olver CS. Preliminary characterization of the murine membrane reticulocyte proteome. Blood Cells, Molecules, and Diseases. 2012;49:74–82. doi: 10.1016/j.bcmd.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Ricciardi S, Miluzio A, Brina D, Clarke K, Bonomo M, Aiolfi R, Guidotti L, Falciani F, Biffo S. eIF6 is a novel regulator of ROS-dependent megakaryocyte maturation. Journal of Thrombosis and Haemostasis. 2015 doi: 10.1111/jth.13150. [DOI] [PubMed] [Google Scholar]
- Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford T, Clegg J, Weatherall D. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. 1979 doi: 10.1038/280164a0. [DOI] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Rätsch G, Rice CM, Ivashkiv LB. Interferon-[gamma] regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Ogura M, Saito H, Satoh M, Takeuchi M. Production of platelet-like particles by a human megakaryoblastic leukemia cell line (MEG-01) Exp. Cell Res. 1991;193:223–226. doi: 10.1016/0014-4827(91)90560-h. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Satoh M, Kuno H, Yoshida T, Kondo H, Takeuchi M. Platelet-like particle formation in the human megakaryoblastic leukaemia cell lines, MEG-01 and MEG-01s. Br. J. Haematol. 1998;100:436–444. doi: 10.1046/j.1365-2141.1998.00576.x. [DOI] [PubMed] [Google Scholar]
- Valentine WN, Fink K, Paglia DE, Harris SR, Adams WS. Hereditary hemolytic anemia with human erythrocyte pyrimidine 5'-nucleotidase deficiency. J. Clin. Invest. 1974;54:866–879. doi: 10.1172/JCI107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudel M, Burkhart JM, Radau S, Zahedi RP, Martens L, Sickmann A. Integral quantification accuracy estimation for reporter ion-based quantitative proteomics (iQuARI) Journal of Proteome Research. 2012;11:5072–5080. doi: 10.1021/pr300247u. [DOI] [PubMed] [Google Scholar]
- Wefes I, Mastrandrea LD, Haldeman M, Koury ST, Tamburlin J, Pickart CM, Finley D. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4982–4986. doi: 10.1073/pnas.92.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyrich AS, Dixon DA, Pabla R, Elstad MR, McIntyre TM, Prescott SM, Zimmerman GA. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiczling P, Krzyzanski W. Flow cytometric assessment of homeostatic aging of reticulocytes in rats. Exp. Hematol. 2008;36:119–127. doi: 10.1016/j.exphem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3′ UTRs In Vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.