Abstract
Background
Hepatitis B virus, with 8 known distinct genotypes, is one of the most serious health problems which results to liver injuries. The surface gene of Hepatitis B virus completely overlaps with the polymerase gene. Mutations in the RT gene result in changes in the overlapping hepatitis B surface antigen.
Objectives
The present study aimed to evaluate the genotypes and prevalence of mutations in a segment of S and RT gene in HBV isolates in Southern Khorasan, Iran.
Methods
This was a population-based study comprising 5,235 randomized samples for HBV screening. A nested-polymerase chain reaction (PCR) test was followed by direct sequencing, and the sequences blast with present sequences of NCBI database for genotyping. Alignment and phylogenic analysis was performed using MEGA-6 software, and mutation pattern of this segment was finally surveyed in Bioedit software.
Results
The mean age was 39.07 ± 14.04 years, with 52.2% female and 47.8% male. All isolates belonged to HBV genotype D, sub-genotype D1. The most amino acid substitutions of surface protein were Q129H (34.42%) and A168V (8.2%), other escape mutants observed in this study were P127L/T, S117G, T125M, S143L, D144E and E164D. In the RT gene, Q149K was the most frequently identified amino acid substitution (9.83%), followed by L122F (8.19%), N118D/T (6.55%), L157M (4.91%), and H124Y (3.27%).
Conclusions
This finding represents an ongoing dominancy of HBV genotype D in Eastern Iran, corresponding to other parts of Iran. There were a lot of variations in the S gene leading to an escape mutation, some of which affected the corresponding area of the RT region.
Keywords: Hepatitis B, Genotyping, Mutations, HBsAg, RT, Southern Khorasan, Iran
1. Background
Worldwide, over 240 million people are chronically infected with Hepatitis B virus (HBV) (1) which can lead to cirrhosis, end-stage liver disease and hepatocellular carcinoma (2). HBV is an enveloped, partially double stranded DNA virus, belonging to the Hepadenaviridae family and is transmitted through blood, blood products, sexual and maternal routes (3). Sequence analysis of the viral isolates indicated that the virus genome can be divided into eight separate genotypes, A to H, among which there exist about 8 to 15% dissimilarity (4). HBV genotypes possess distinguished geographical distributions, and they seem to have different biological properties, which affect the clinical outcome of HBV disease (5). The primary classification of HBV has been based on the dissimilarity in amino acid sequences in the HBsAg. This scale divides HBV into four main HBsAg serotypes: adr, adw, ayr, ayw, while additional differences expands this classification into nine serotypes (ayw 1 to 4, ayr, adw2, adw4, adrq (6, 7).
The HBV genome encodes only one viral enzyme, the HBV reverse transcriptase (RT) (8), which is an error-prone enzyme without exonuclease proofreading activity, similar to HIV-RT. Together with a high virus replication titer, this produces various HBV mutants (9). The S open reading frame (ORF), which encodes the surface protein, is completely overlapped by the polymerase (P) gene (10). Hence, mutations in the RT can result in changes in the HBsAg protein and vice versa (11).
The HBsAg ‘‘a’’ determinant (amino acids 124 - 147) is located within the major hydrophilic region (MHR, amino acids 99 - 169) of this surface protein (12) which produces the potent B-cell epitope of HBs Ag(13), and it has been proven that amino acid substitutions within this region produce escape mutants. Immune escape mutants allow for the replication of HBV in vaccinated persons (14, 15), since antibodies induced by current vaccines do not recognize critical changes in the surface antigen domain (16).
Inhibitors of RT, nucleos(t)ide analogues (NAs), such as lamivudine, adefovir, dipivoxil, entecavir, telbivudine, etc are approved for the treatment of chronic hepatitis B (CHB) (17). An important issue with NAs treatment is the selection of antiviral-resistant mutations (18). HBV mutants has the tendency of exhibiting enhanced virulence, resistance to antivirals, facilitated cell attachment or alteration of epitopes which are important in the host immune response (19).
Various studies have shown that HBV genotype D is the most predominant genotype in Iran (20, 21), although some studies reported non-D (22) or a diversity on subgenotypes of D (23). To date, to the best of our knowledge, there is no clear information regarding the genotype distribution of HBV in Southern Khorasan of Iran, and also the mutation profile of S gene is unclear.
2. Objectives
The aim of this study was firstly to determine the distribution of genotype and subgenotype of HBV among health carriers in Southern Khorasan of Iran, and secondly to assess the frequency of mutations in surface gene and concomitantly polymerase gene associated with viral escape and antiviral resistance, respectively.
3. Methods
3.1. Patients
The study was conducted between 2014 and 2015 in Birjand, the centre of South Khorasan province of Iran. It was a population-based cross-sectional study which comprised 5,235 cases with an age range between 15 and 70 years. This study was approved by the Iranian society for supporting patients with infectious diseases ethics committee, and informed consent was taken from the patients before registration. All samples were tested for HBsAg, among whom were 85 HBs-Ag positive cases of which all were treatment naive health carriers. This study was also approved by the ethics committee of the research center of Birjand University of Medical Sciences (BUMS).
3.2. Extraction of HBV DNA
Patients were referred to the hepatitis research center of BUMS, and after registration and completion of questionnaire, sera samples were collected from patients and stored at -70°C until initiation of the experiments. HBV DNA was isolated from 200 micro litters (µL) of serum samples using a viral nucleic acid extraction kit, viral gene spine (iNtRON Biotechnology) according to the manufacturers’ instruction but with some minor alterations.
3.3. Selection of Primer
Selected primers (24) corresponded to the conserved regions across the different genotypes, and also corresponded to the surrounding heterogeneous regions between them, which allowed for the distinction of HBV genotypes. It also covered the amino acid loop corresponding to a, d/y and w/r specificities and mutations in the nucleotide sequences related to hepatitis B Immunoglobulin (Ig), anti-HBs monoclonal antibody, and vaccine resistance. In the concomitantly overlapping region of RT gene, some resistance mutations of RT were also detected. This region amplified a 417-bp fragment covering a region of S gene with marked heterogeneity among genotypes, flanked by a conserved region to allow their equal amplification (Table 1).
Table 1. The Primer Sequences and Positions Used in Nested PCR for Partially S Gene Amplification with Their Positions.
| Primer | Sequence | Position |
|---|---|---|
| FHBS1 | 5’ GAG TCT AGA CTC GTG GTG GAC TTC 3’ | 244 to 267 |
| FHBS2 | 5’ CGT GGT GGA CTT CTC TCA ATT TTC 3’ | 255 to 278 |
| RHBS1 | 5’ AAA TKG CAC TAG TAA ACT GAG CCA 3’ | 648 to 671 |
| RHBS2 | 5’ GCC ARG AGA AAC GGR CTG AGG CCC 3’ | 668 to 691 |
3.4. HBsAg Gene Partial Amplification
HBs-Ag positive cases were subjected to S-gene partial amplification, a nested PCR test. The first-round of PCR was performed for 30 cycles (94°C for 30 seconds, 55°C for 20 seconds and 72°C for 30 seconds) and a final extension step at 72°C for 1 minutes in a 25 µL reaction volume containing 50 ng of extracted DNA, 1× PCR buffer, 200 Mm of each dNTPs, 1.5 mM MgCl2, 25 pmol/mL of each outer primers and 1U Taq DNA polymerase (Invitrogen, France). Three (3) µL of the first-round PCR product was then subjected for the second-round PCR under the same conditions but with the inner primers. Appropriate negative and positive controls were included in each test. After the second round, a 417 bp fragment was obtained and rounded by electrophoresis in a 2% agarose gel; observation was carried out using a gel documentation device.
3.5. Sequencing and Determination of HBV Genotype, Subgenotype, and Serotypes
The second round PCR products which contained a sharp band on agarose gel electrophoresis were subjected to direct sequencing in Bionear company, South Korea (BigDyeTM terminator cyclic sequencing was performed and then analysis was made using ABI 3730XL DNA analyser). The obtained sequences were blast with the sequences present in the gene bank database of NCBI for HBV genotyping. Nucleotide sequences were aligned with CLUSTALW program using Bioedit software (BioEdit sequence alignment editor software, department of microbiology, and North California State university). For phylogenetic analysis, the sequences of the current project were entered into MEGA-6 software (Tamura, Stecher, Peterson, Filipski, and Kumar 2013) together with the reference sequence of HBV genotype D1 and all other known genotypes. The phylogenetic tree was designed according to the maximum likelihood model.
The HBsAg serotypes were deduced from the sequence of the same S gene region used for genotyping by a newly described algorithm (25), based on the identification of amino acids at positions 122 (Lys-Arg for d-y determinants), 160 (Lys-Arg for w-r determinants), 127 (Pro-Thr-Leu/Ile for w2-w3-w4), and in the case of Arg122 Pro127 Lys160, also at positions 159 (Ala-not Ala for ayw1-ayw2 and ayw4) and 140 (not Ser-Ser for ayw2-ayw4).
3.6. Determination of Surface and Polymerase Gene Mutations
The accession number GQ183486.1 that was found to have most proximity to the Iranian HBV sequences, was selected as reference sequence. The whole RT and S gene of GQ183486.1 were separately aligned with all sequences of current project using Bioedit software by ClustalW program, so that each amino acids of ref seq put in right position. After alignment of nucleotide sequences, the session converted to amino acid and mutations were revealed as an altered amino acid.
4. Results
4.1. Baseline Characteristics of the Study Subjects
There were 85 cases with HBsAg among 5,232 randomized samples in Southern Khorasan. Among them, 61 samples had HBV-DNA with the nested PCR test (72%). The mean age was 39.07 ± 14.04 years, with 52.2% female and 47.8% male. These patients were all naive to any NAs and did not give any antiviral treatment. Among the selected population, 29.2% were vaccinated against HBV and had taken the full course of HBs Ag containing vaccine.
4.2. Genotyping
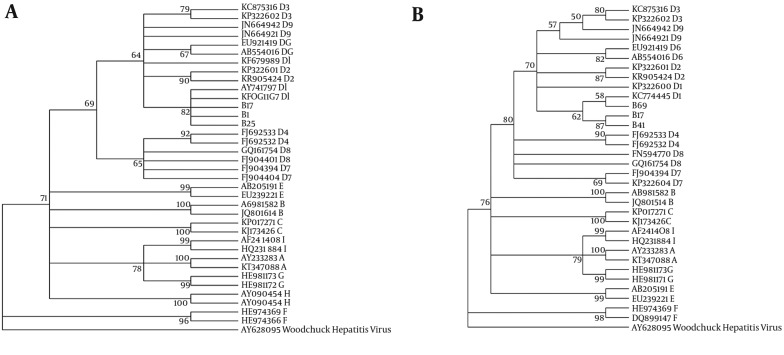
The sequences of the S gene region of 61 isolates (the accession number KX149129 and KX180836 to KX180893) were analyzed and compared with the reference sequences for all 8 known HBV genotypes from the DDBJ/EMBL/GenBank database. The results obtained from the analysis are summarized in two phylogenetic trees from S and RT gene (Figure 1). Genotype D was the only genotype detected among these isolates. For the determination of subgenotypes, the isolates were compared with the reference sequences for D1 - D9 subgenotypes, the results showed that all the isolates belonged to D1.
Figure 1. The maximum likelihood phylogenic (ML) tree of surface gene (A) and rt gene (B) sequences from current isolates in an alignment with reference isolates of all genotypes and sub genotypes of D1, constructed in MEGA-6 software. Model selection with (ML) test was used before phylogeny. Phylogeny reconstruction was done with ML test and Kimura 2-parameter model, rates among sites was gamma distributed, selected codon were 1st, 2nd, 3rd and non- coding sites. In order to test the validity of tree, the bootstrapping of 1000 replication was used. The S gene sequence of Woodchuck hepatitis virus was used to root the tree. The accession numbers, genotype and subgenotypes were given. The symbols B17, 41, 69, and 25 are sequences of current study.
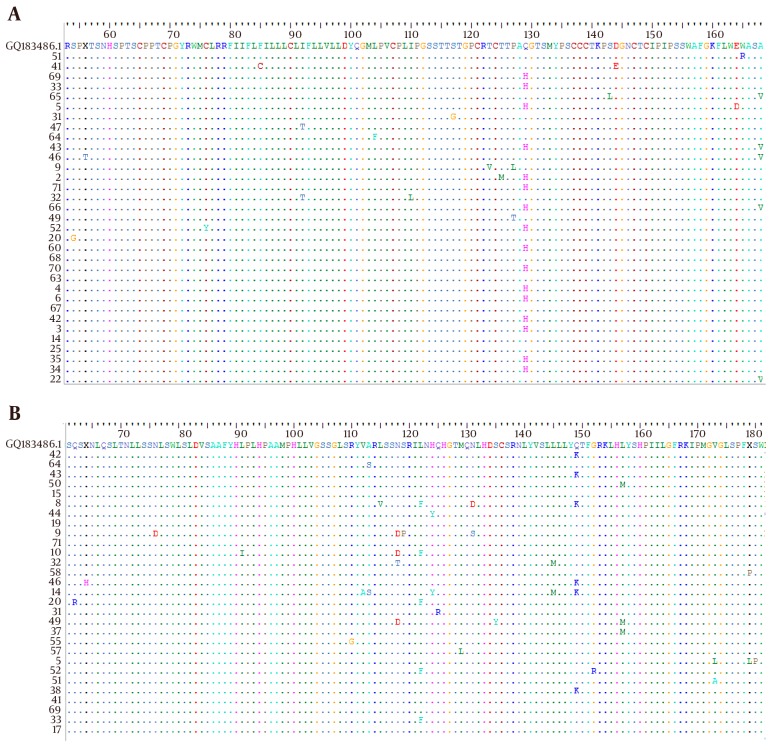
HBsAg serotypes were inferred by identifying amino acids at positions 122, 160, 127, 159 and 140 according to the alignment of the study isolates with reference HBs-Ag sequence of accession number GQ183486 (Figure 2A). The result showed that all isolates belonged to the serotype ayw2, with exception of one with ayw3 and one with ayw4.
Figure 2. The Amino Acid Sequence of HBsAg A, and RT B, from current isolates, “a” determinant aligned in Bioedite software with the reference isolate of genotype D1.
4.3. Mutations in the HBsAg
To explore the mutations affecting the antigenicity of HBsAg, MHR within the major antigenic “a” determinant was analyzed. The result of this survey in the Bioedit software showed that there existed substitutions in 35 positions in the level of nucleotide. The whole surface protein that was tested comprise of 144 amino acids which mutations have occurred in 17 positions (11.8%).
It was obvious that most of mutations were accumulated in the MHR region (13 positions of 70 amino acids, 18.57%), including “a” determinant region (in 5 positions of 23 amino acids, 21.73%). All mutations observed in this segment of S gene including escape ones were summarized in Table 2.
Table 2. Amino Acid Substitutions in the HBs- Ag with Known and Unknown Functional Effects.
| Codon /Position | aa Substitution | Frequency, % | Vaccinated frq, % | Function | References |
|---|---|---|---|---|---|
| AGT/GGT - 160 | S54G | 1 (1.63) | 100 | Unknown | |
| TGT/TAT - 227 | C76Y | 1 (1.63) | 0 | Unknown | |
| TTC/TGC - 254 | F85C | 1 (1.63) | 0 | Unknown | |
| ATC /ACC - 275 | I92T | 2 (3.27) | 0 | Unknown | |
| TTG/TTT - 312 | L104F | 1 (1.63) | 100 | Unknown | |
| ATT/CTT - 330 | I110L | 1 (1.63) | 0 | Unknown | |
| AGC/GGC - 349 | S117G | 1 (1.63) | 100 | Escape | (26) |
| ACC/GTC - 367 | T123V | 1 (1.63) | 0 | Unknown | |
| ACG/ATG -368 | T125M | 1 (1.63) | 0 | Escape | (26) |
| CCT/ACT - 379 | P127L/T | 2 (3.27) | 50 | Escape | (27) |
| CAA/CAC - 387 | Q129H | 21 (34.42) | 35.3 | Escape, detection, Ig therapy | (26) |
| TCG/TTG - 428 | S143L | 1 (1.63) | 0 | Escape | (9) |
| GAC/GAA - 432 | D144E | 1 (1.63) | 0 | Escape, detection, Ig therapy | (28) |
| GAG/GAT - 492 | E164D | 1 (1.63) | 0 | Escape | (26) |
| TGG/ CGG - 493 | W165R | 1 (1.63) | 0 | Unknown | |
| GCC/ GTC - 503 | A168V | 5 (8.2%), | 20 | Unknown |
4.4. Analysis of Patient Samples for Polymerase Mutations
As the whole S gene overlapped with Pol gene, most nucleotide substitutions in the S region contributed to a mutation in RT sequence. This study showed that most, but not all mutations of the S gene caused an amino acid change in RT. However, some changes were observed in RT without change of the S sequence. In the level of nucleotide there were 38 substitutions of 364 (10.43%), while there were 22 mutations in 122 amino acids. It means 42.1% of substitutions eventuate to a non- sense mutation. The result of this study showed that most mutations were occurred in position 110 to 157 (including 16 mutations in 47 residues, 34%); it means this region is less conserved and can select some variations. The Q149K was the most frequently identified amino acid substitution observed in these isolates (9.83%), followed by L122F (8.19%), N118D/T (6.55%), L157M (4.91%), and H124Y (3.27%). Other mutations that each one has seen only in one patient listed below:
Q62R, N76D, S78T, L91I, R110G, V112A, A113V/S, L115V, S119P, Q125R, M129L, N131D/S, S135Y, L145M, G152R, V173A/L, S180P.
5. Discussion
To the best of our knowledge, this is the first report of HBV genotyping with an acceptable sample size from this part of Iran, sharing borders with Afghanistan. Results of this study showed that genotype D, subgenotype D1 is the only genotype of HBV in this province. The study also showed that ayw2 is the most prevalent serotype of HBs Ag in this province; however, there were two exceptions, ayw3 and ayw4. This finding is compatible with those of similar studies from other parts of Iran.
Several studies in Iran have shown that genotype D is the only genotype observed in different parts of the country (29). However, genotype B was reported in a case in Kermanshah Province, which is located in western Iran (22). Studies from countries around Iran, such as Turkey (30), Pakistan (31), and Saudi Arabia (32) showed genotype D as the predominant HBV genotype. However, lower rates of other genotypes have also been reported in Eastern Azerbaijan, Northwest Iran (33). Attaullah demonstrated that genotype D (35.67%) is the predominant genotype among Afghani population, followed by genotype C (32.16%), genotype A (19.30%), and genotype B (7.02%), respectively (34). Due to the proximity of Southern Khorasan to Afghanistan, and the substantial immigration of Afghani people through this province, our results are interesting and may contribute to the social and cultural behavior, as well as the health level.
Other studies have also shown that HBV serotype ayw2 is dominant in Iran (29) but was 97.64% according to the present research figures. The serotypes ayw3 and ayw4 reported 0.2% and 0.4%, respectively by Mohebbi et al. (35), which was 1.17% (one case) for each one in the present study. The other sub genotypes of D, D2 and D3, were reported in a few studies around the Iran (35-37). Genotype other than genotype D was seen only in Western areas of Iran, genotype B was reported in two studies in Kermanshah and Khuzestan (22, 37) which may be related to neighboring countries.
HBV vaccine contains the HBsAg protein that induces immune response against MHR, the so called “a” determinant, which is involved in amino acid residues 100 - 160 of the HBs- Ag (38). The most frequently identified HBsAg amino acid substitutions in previous studies include P120T, M33I, S143L, D144E/A, G145R, E164D, W172*, and W182* which are the most commonly occurring pattern (39). The well-known G145R mutation occurs in the S-gene with overlapping rtW153Q mutation in the Pol-gene (HBsAg classical vaccine escape strain), which was absent in isolates of the current project. The Q129H was the most frequently detected escape mutant in this study (34.42%), followed by P127L/T (3.27%) and E164D, S117G/T, D144E, T125M, S143L, 1.63% for each; and their roles in the vaccine escape, Ig response and/or detection have already been established. It is noteworthy that 29.2% of these patients were vaccinated against HBV, among them, 9 cases had an escape mutation. The most prevalent escape mutation among the vaccinated was Q129H/P (6 cases), and S117G, P127L one case for each, while other escape mutations occurred just in the unvaccinated populations.
Sayan reported multiple HBV vaccine-escape mutations (S143T, D144E, G145R, E164D, I195M) (9), of which 3 of them (D144E, S143L and E164D, one case for each) were seen in the current study. Avellon reported M133T (2.2%), Q129H, M133I, F/Y134N (1.8%), F/Y134L, G145A (1.5%), and P120T (1.1%) as the most frequent mutations of S region, while G145R was found at a frequency of less than 1% (0.4%) (26), while among them just Q129H was found in the current study.
In a previous study in this estate of Iran, only P127L was present in one case and Q129H in two cases at MHR region in 25 patients. There were 16 other mutations in S gene with any patient having at least one mutation (40), which had smaller mutation frequency than the current study. Of course, part of it may be related to the small sample size. In another study in Iran, P120T/S and R122K/T were found in 4% of patients, and G145R was found only in one case (13); which none of them occurred in the current project. These mutations frequently occurred and it was suggested that this frequency may be due to the pressure of the immune system or vaccine on their selection, considering the fact that these patients did not receive NAs.
It was observed that the polymerase gene is more conserved and that not all variants of MHR were accompanied by amino acid substitutions in the polymerase. Nevertheless, most of S variants led to a change in RT region. Almost all the escape mutations that were found in this study led to a silent mutation on RT region without any change in the level of amino acid. It should be noted that some mutations occurred in RT without any amino acid substitution in the corresponding region of HBsAg, suggesting the mutual effect of some nucleotide substitution of S/RT - ORF.
The most prevalent mutations in RT region of the study isolates was Q149K (9.83%) followed by N118D/T (6.55%), L157M (4.91%) and H124Y, A113Y/S, N131D/S, L145M, V173A/L, 3.27% for each one. Almost all these mutations were reported previously and there were also many other mutations in this segment of RT gene which had been identified by earlier reports (41, 42). The well known mutation rtV173L observed in two cases (3.2%), which is a compensatory mutation and could be selected in lamivudine-resistant patients due to an enhanced replication phenotype (43). The S135Y that occurred in one case is reported to be related to lamivudine resistance (44). The rare S78T mutation also observed in one patient can lead to tenofovir resistance (45). Other mutation that observed in this study was L91I which reported as an Entecavir, lamivudine -related resistance mutation (46, 47).
It is noteworthy that mutations of RT observed in current study occurred naturally and these patients did not give any NAs. This was a limitation for this project which prevented the amplification of a bigger sequence of the RT region. Meanwhile, a lot of mutation occurred in this fairly short segment of RT which showed the importance of the determination of RT mutants before NAs prescription.
Although it has been proven that genotype D is predominant in Iran by it sharing borders with countries such Afghanistan, Pakistan and some Arab countries which are not like Iran and are not devoid of other genotypes, the rise of other genotypes is still not unexpected. Therefore, genotyping of HBV in different regions of Iran in time intervals could be very important.
Considering the fact that chronic carriers are the major reservoir of HBV infection, the selection of such naturally occurring variants in these patients could increase the transmission of these variants in the general population. Therefore, the epidemiological monitoring of naturally occurring MHR variants is essential, especially for immunotherapy and vaccination efficacy.
5.1. Conclusion
Briefly, this study showed that HBV genotype D was still predominant in Iran, as well as Southern Khorasan. There were some variations in the S gene of HBV which occurred spontaneously and or in the presence of immune system which caused escape mutation or NA resistance in RT region. Considering that some vaccinated people were infected with an escape mutant, the screening of such variants among health carriers is very important.
Acknowledgments
We would like to express sincere thanks to the vice chancellor for research at BUMS for full support of this project and the future one related to this. We also acknowledge and appreciate the kind revision and editing of this paper by Mr. Ruzegar.
Footnotes
Authors’ Contribution:Masood Ziaee get the idea and design the study. Davod Javanmard performed the expriments, analysed the data and drafting the manuscripts. Gholamreza Sharifzadeh, Mohammad Hasan Namaei and Ghodsiyeh Azarkar all had equal roles in administrative, technical, and material support.
Funding/Support:This study was both supported and funded by Birjand University of Medical Sciences (BUMS), Birjand, Iran.
References
- 1.WHO. Hepatitis B Fact sheet N°204. Geneva: world health organization; 2015. [updated July 2016]. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- 2.Simon B, Kundi M, Puchhammer E. Analysis of mutations in the S gene of hepatitis B virus strains in patients with chronic infection by online bioinformatics tools. J Clin Microbiol. 2013;51(1):163–8. doi: 10.1128/JCM.01630-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunford L, Carr MJ, Dean J, Nguyen LT, Ta Thi TH, Nguyen BT, et al. A multicentre molecular analysis of hepatitis B and blood-borne virus coinfections in Viet Nam. PLoS One. 2012;7(6):39027. doi: 10.1371/journal.pone.0039027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eftekhari Y, Kazemi Arababadi M, Hakimi H, Rezazadeh Zarandi E. Common HBV genotype in southeastern Iranian patients. Arch Iran Med. 2010;13(2):147–9. [PubMed] [Google Scholar]
- 5.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13(1):14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A, Datta S, Chandra PK, Roychowdhury S, Panda CK, Chakravarty R. Distribution of hepatitis B virus genotypes: phylogenetic analysis and virological characteristics of genotype C circulating among HBV carriers in Kolkata, Eastern India. World J Gastroenterol. 2006;12(37):5964–71. doi: 10.3748/wjg.v12.i37.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazarevic I, Cupic M, Delic D, Svirtlih NS, Simonovic J, Jovanovic T. Distribution of HBV genotypes, subgenotypes and HBsAg subtypes among chronically infected patients in Serbia. Arch Virol. 2007;152(11):2017–25. doi: 10.1007/s00705-007-1031-0. [DOI] [PubMed] [Google Scholar]
- 8.Nijhuis M, van Maarseveen NM, Boucher CA. Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. Handb Exp Pharmacol. 2009;(189):299–320. doi: 10.1007/978-3-540-79086-0_11. [DOI] [PubMed] [Google Scholar]
- 9.Sayan M, Senturk O, Akhan SC, Hulagu S, Cekmen MB. Monitoring of hepatitis B virus surface antigen escape mutations and concomitantly nucleos(t)ide analog resistance mutations in Turkish patients with chronic hepatitis B. Int J Infect Dis. 2010;14 Suppl 3:136–41. doi: 10.1016/j.ijid.2009.11.039. [DOI] [PubMed] [Google Scholar]
- 10.Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25(2):97–106. doi: 10.1016/s1386-6532(02)00049-5. [DOI] [PubMed] [Google Scholar]
- 11.Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48(1):88–98. doi: 10.1002/hep.22295. [DOI] [PubMed] [Google Scholar]
- 12.Weber B. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J Clin Virol. 2005;32(2):102–12. doi: 10.1016/j.jcv.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Moradi A, Zhand S, Ghaemi A, Javid N, Tabarraei A. Mutations in the S gene region of hepatitis B virus genotype D in Golestan Province-Iran. Virus Genes. 2012;44(3):382–7. doi: 10.1007/s11262-012-0715-z. [DOI] [PubMed] [Google Scholar]
- 14.Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, et al. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293(2):305–13. doi: 10.1006/viro.2001.1246. [DOI] [PubMed] [Google Scholar]
- 15.Sheldon J, Soriano V. Hepatitis B virus escape mutants induced by antiviral therapy. J Antimicrob Chemother. 2008;61(4):766–8. doi: 10.1093/jac/dkn014. [DOI] [PubMed] [Google Scholar]
- 16.Zuckerman JN, Zuckerman AJ. Mutations of the surface protein of hepatitis B virus. Antiviral Res. 2003;60(2):75–8. doi: 10.1016/j.antiviral.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Gines P, Angeli P, Lenz K, Moller S, Moore K, Moreu R, et al. EASL clinical practice guidelines on the management of ascites, spontaneous bacterial peritonitis, and hepatorenal syndrome in cirrhosis. J Hepatol. 2010;53(3):397–417. doi: 10.1016/j.jhep.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol. 2006;44(3):593–606. doi: 10.1016/j.jhep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Cao G. Genotypes, mutations, and viral load of hepatitis B virus and the risk of hepatocellular carcinoma: HBV properties and hepatocarcinogenesis. Hepat Mon. 2011;11(2):86–91. [PMC free article] [PubMed] [Google Scholar]
- 20.Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Adeli A, Sabahi F, Abachi M, Azizi M, et al. Complete genomic sequence and phylogenetic relatedness of hepatitis B virus isolates from Iran. J Med Virol. 2005;76(3):318–26. doi: 10.1002/jmv.20362. [DOI] [PubMed] [Google Scholar]
- 21.Mojiri A, Behzad-Behbahani A, Saberifirozi M, Ardabili M, Beheshti M, Rahsaz M, et al. Hepatitis B virus genotypes in southwest Iran: molecular, serological and clinical outcomes. World J Gastroenterol. 2008;14(10):1510–3. doi: 10.3748/wjg.14.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dokanehiifard S, Bidmeshkipour A. Study of hepatitis b virus (hbv) genotypes in kermanshah province, west of Iran. J Biol Sci. 2009;1(1):113–20. [Google Scholar]
- 23.Mohebbi SR, Amini-Bavil-Olyaee S, Zali N, Derakhshan F, Sabahi F, Zali MR. An Extremely Aberrant Subtype of Hepatitis B Virus Genotype D in Iran. Hepat Mon. 2009;9(1):73–5. [Google Scholar]
- 24.Sitnik R, Pinho JR, Bertolini DA, Bernardini AP, Da Silva LC, Carrilho FJ. Hepatitis B virus genotypes and precore and core mutants in Brazilian patients. J Clin Microbiol. 2004;42(6):2455–60. doi: 10.1128/JCM.42.6.2455-2460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purdy MA, Talekar G, Swenson P, Araujo A, Fields H. A new algorithm for deduction of hepatitis B surface antigen subtype determinants from the amino acid sequence. Intervirology. 2007;50(1):45–51. doi: 10.1159/000096312. [DOI] [PubMed] [Google Scholar]
- 26.Avellon A, Echevarria JM. Frequency of hepatitis B virus 'a' determinant variants in unselected Spanish chronic carriers. J Med Virol. 2006;78(1):24–36. doi: 10.1002/jmv.20516. [DOI] [PubMed] [Google Scholar]
- 27.Mele A, Tancredi F, Romano L, Giuseppone A, Colucci M, Sangiuolo A, et al. Effectiveness of hepatitis B vaccination in babies born to hepatitis B surface antigen-positive mothers in Italy. J Infect Dis. 2001;184(7):905–8. doi: 10.1086/323396. [DOI] [PubMed] [Google Scholar]
- 28.Awerkiew S, Daumer M, Reiser M, Wend UC, Pfister H, Kaiser R, et al. Reactivation of an occult hepatitis B virus escape mutant in an anti-HBs positive, anti-HBc negative lymphoma patient. J Clin Virol. 2007;38(1):83–6. doi: 10.1016/j.jcv.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Haghshenas MR, Arabi M, Mousavi T. Hepatitis B genotypes in iran. Mater Sociomed. 2014;26(2):129–33. doi: 10.5455/msm.2014.26.129-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunbul M, Leblebicioglu H. Distribution of hepatitis B virus genotypes in patients with chronic hepatitis B in Turkey. World J Gastroenterol. 2005;11(13):1976–80. doi: 10.3748/wjg.v11.i13.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumtaz K, Hamid S, Ahmed S, Moatter T, Mushtaq S, Khan A, et al. A study of genotypes, mutants and nucleotide sequence of hepatitis B virus in Pakistan: HBV genotypes in pakistan. Hepat Mon. 2011;11(1):14–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Abdo AA, Al-Jarallah BM, Sanai FM, Hersi AS, Al-Swat K, Azzam NA, et al. Hepatitis B genotypes: relation to clinical outcome in patients with chronic hepatitis B in Saudi Arabia. World J Gastroenterol. 2006;12(43):7019–24. doi: 10.3748/wjg.v12.i43.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammadnejad L, Farajnia S, Parivar K, Naghili B, Yousefzadeh Kheirnagsh R. Hepatitis B virus genotypes in eastern Azerbaijan, Northwest Iran. Arch Iran Med. 2012;15(7):446–8. [PubMed] [Google Scholar]
- 34.Attaullah S, Rehman S, Khan S, Ali I, Ali S, Khan SN. Prevalence of hepatitis B virus genotypes in HBsAg positive individuals of Afghanistan. Virol J. 2011;8:281. doi: 10.1186/1743-422X-8-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohebbi SR, Amini-Bavil-Olyaee S, Zali N, Noorinayer B, Derakhshan F, Chiani M, et al. Molecular epidemiology of hepatitis B virus in Iran. Clin Microbiol Infect. 2008;14(9):858–66. doi: 10.1111/j.1469-0691.2008.02053.x. [DOI] [PubMed] [Google Scholar]
- 36.Garmiri P, Rezvan H, Abolghasemi H, Allain JP. Full genome characterization of hepatitis B virus strains from blood donors in Iran. J Med Virol. 2011;83(6):948–52. doi: 10.1002/jmv.21772. [DOI] [PubMed] [Google Scholar]
- 37.Neisi N, Makvandi M, Samarbaf-Zadeh AR. A study on genotypes of hepatitis B virus among hemodialysis patients in Khuzestan province. Jundishapur J Microbiol. 2011;4(2):65–70. [Google Scholar]
- 38.Al Baqlani SA, Sy BT, Ratsch BA, Al Naamani K, Al Awaidy S, Busaidy SA, et al. Molecular epidemiology and genotyping of hepatitis B virus of HBsAg-positive patients in Oman. PLoS One. 2014;9(5):97759. doi: 10.1371/journal.pone.0097759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheldon J, Rodes B, Zoulim F, Bartholomeusz A, Soriano V. Mutations affecting the replication capacity of the hepatitis B virus. J Viral Hepat. 2006;13(7):427–34. doi: 10.1111/j.1365-2893.2005.00713.x. [DOI] [PubMed] [Google Scholar]
- 40.Ghaziasadi A, Ziaee M, Norouzi M, Malekzadeh R, Alavian SM, Saberfar E, et al. The prevalence of hepatitis B virus surface antigen (HBsAg) variations and correlation with the clinical and serologic pictures in chronic carriers from Khorasan Province, North-East of Iran. Acta Med Iran. 2012;50(4):265–72. [PubMed] [Google Scholar]
- 41.Panigrahi R, Biswas A, De BK, Chakrabarti S, Chakravarty R. Characterization of antiviral resistance mutations among the Eastern Indian Hepatitis B virus infected population. Virol J. 2013;10:56. doi: 10.1186/1743-422X-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ismail AM, Samuel P, Eapen CE, Kannangai R, Abraham P. Antiviral resistance mutations and genotype-associated amino acid substitutions in treatment-naive hepatitis B virus-infected individuals from the Indian subcontinent. Intervirology. 2012;55(1):36–44. doi: 10.1159/000323521. [DOI] [PubMed] [Google Scholar]
- 43.Delaney WE, Yang H, Westland CE, Das K, Arnold E, Gibbs CS, et al. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77(21):11833–41. doi: 10.1128/JVI.77.21.11833-11841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan J, Degertekin B, Wong SN, Husain M, Oberhelman K, Lok AS. Tenofovir monotherapy is effective in hepatitis B patients with antiviral treatment failure to adefovir in the absence of adefovir-resistant mutations. J Hepatol. 2008;48(3):391–8. doi: 10.1016/j.jhep.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 45.van Hemert FJ, Berkhout B, Zaaijer HL. Differential binding of tenofovir and adefovir to reverse transcriptase of hepatitis B virus. PLoS One. 2014;9(9):106324. doi: 10.1371/journal.pone.0106324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahabadi M, Norouzi M, Alavian SM, Samimirad K, Azad TM, Saberfar E, et al. Drug-related mutational patterns in hepatitis B virus (HBV) reverse transcriptase proteins from Iranian treatment-naive chronic HBV patients. Hepat Mon. 2013;13(1):6712. doi: 10.5812/hepatmon.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karatayli E, Karatayli SC, Cinar K, Gokahmetoglu S, Guven K, Idilman R, et al. Molecular characterization of a novel entecavir mutation pattern isolated from a multi-drug refractory patient with chronic hepatitis B infection. J Clin Virol. 2012;53(2):130–4. doi: 10.1016/j.jcv.2011.10.011. [DOI] [PubMed] [Google Scholar]