Abstract
Obesity is associated with increased classically activated M1 adipose tissue macrophages (ATMs) and decreased alternatively activated M2 ATMs, both of which contribute to obesity-induced inflammation and insulin resistance. However, the underlying mechanism remains unclear. We find that inhibiting DNA methylation pharmacologically using 5-aza-2′-deoxycytidine or genetically by DNA methyltransferase 1 (DNMT1) deletion promotes alternative activation and suppresses inflammation in macrophages. Consistently, mice with myeloid DNMT1 deficiency exhibit enhanced macrophage alternative activation, suppressed macrophage inflammation, and are protected from obesity-induced inflammation and insulin resistance. The promoter and 5′-untranslated region of peroxisome proliferator-activated receptor γ1 (PPARγ1) are enriched with CpGs and are epigenetically regulated. The saturated fatty acids stearate and palmitate and the inflammatory cytokine TNF-α significantly increase, whereas the TH2 cytokine IL-4 significantly decreases PPARγ1 promoter DNA methylation. Accordingly, inhibiting PPARγ1 promoter DNA methylation pharmacologically using 5-aza-2′-deoxycytidine or genetically by DNMT1 deletion promotes macrophage alternative activation. Our data therefore establish DNA hypermethylation at the PPARγ1 promoter induced by obesity-related factors as a critical determinant of ATM proinflammatory activation and inflammation, which contributes to insulin resistance in obesity.
Obesity-induced DNA hypermethylation at the PPARγ1 promoter is a critical determinant of adipose tissue macrophage proinflammatory activation and inflammation.
Introduction
Chronic inflammation is a key link between obesity and insulin resistance/type 2 diabetes (1, 2). An important feature of obesity-induced inflammation is the infiltration of macrophages into adipose tissue (3, 4). Adipose tissue macrophages (ATMs) can be separated into M1 or M2 macrophages by their differential expression of surface markers F4/80, CD11c, CD206/mannose receptor 1 (MRC1), or CD301 (5–8). ATMs from lean animals show an alternatively activated, M2 phenotype (CD206+ or CD301+) (5, 6, 9). These M2 macrophages are normally induced by the Th2 cytokines IL-4 and IL-13, and typically have downregulated inducible nitric oxide synthase (iNOS), and upregulated expression of arginase 1 (ARG1) and other antiinflammatory factors (IL-10 and IL-1 receptor antagonist [IL1RA]), thereby attenuating inflammation (10, 11). The induction of M2 macrophage polarization is mediated by signal transducer and activator of transcription 6 (STAT6) (2), while the nuclear receptors peroxisome proliferator-activated receptor γ (PPARγ) and PPARδ also play important roles in this process (12–17). Obesity is associated with deregulated ATM polarization, characterized by increased classically activated M1 ATMs (F4/80+CD11c+) and reduced alternatively activated M2 ATMs (5–9), which eventually tips the balance of ATMs toward a more proinflammatory phenotype and contributes to obesity-induced inflammation and insulin resistance.
While numerous studies have been devoted to the evaluation of genetic factors related to obesity and its associated complications, much less is known about epigenetic regulation, which serves as a molecular link between environmental factors (e.g., diet) and complex diseases, including obesity and diabetes. DNA methylation of cytosines at primarily CpG dinucleotides is the most common epigenetic modification. CpGs are often enriched in the promoter and the first exon/5′-untranslated region of genes. Promoters of transcriptionally active genes are typically hypomethylated, whereas DNA hypermethylation results in gene silencing by affecting the binding of methylation-sensitive DNA binding proteins and/or by further interacting with various histone modifications and corepressors that alter DNA accessibility to transcriptional factors (18–21).
DNA methylation is catalyzed by DNA methyltransferases (DNMTs), including DNMT1, DNMT3a, and DNMT3b. DNMT1, which is responsible for DNA methylation maintenance, binds to methyl groups in hemimethylated DNA strands during DNA replication, whereas de novo DNMT3a and DNMT3b add methyl groups to CpG dinucleotides of unmethylated DNA (19). However, it was recently suggested that DNMT1 may also have a role in de novo DNA methylation (22).
It is well documented that maternal or early-life nutrition has profound impact on the metabolic phenotypes in offspring and in later life (23, 24). Alterations of DNA methylation globally or in specific genes have also been observed during the development of obesity (25, 26). These data imply a significant role for DNA methylation in obesity. DNA methylation may also play a role in chronic inflammation–related diseases (27, 28). However, it is not known whether DNA methylation regulates obesity-induced ATM phenotypic switch and inflammation.
In the present study, we tested the hypothesis that epigenetic regulation of macrophage polarization by DNA methylation regulates obesity-induced inflammation and determines whole-body insulin sensitivity. We found that inhibiting DNA methylation by myeloid deletion of DNMT1 profoundly promoted macrophage alternative activation, which resulted in suppressed macrophage inflammation. Consistently, mice with myeloid deletion of DNMT1 exhibited enhanced macrophage alternative activation, suppressed inflammation, and were protected from obesity-induced insulin resistance. Mechanistically, this involves epigenetic regulation of the PPARγ1 promoter by DNA methylation.
Results
DNA methylation regulates alternative M2 macrophage polarization.
Although ATMs can be generally separated into M1 (F4/80+CD11c+) and M2 (CD206+ or CD301+) populations (5–9), recent data indicate that ATMs are more heterogeneous. For example, F4/80+CD11c+CD206+ or F4/80+CD11c+CD301+ ATMs (denoted hereafter as double+ ATMs) are found to express both M1 and M2 markers (29, 30), whereas F4/80+CD11c–CD206– ATMs (denoted hereafter as double– ATMs) have also been identified (31). We therefore characterized M1 and M2 marker expression profiles in different ATM populations based on the surface expression of F4/80, CD11c, and CD206 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.87748DS1).
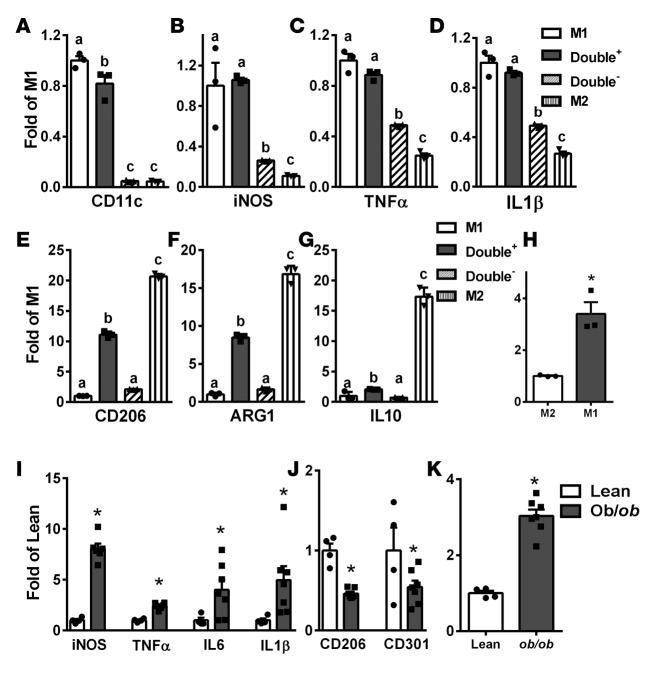
The expression profile of M1 and M2 markers was distinct between F4/80+CD11c+CD206– (M1) and F4/80+CD11c–CD206+ (M2) ATMs, with higher expression of the proinflammatory M1 markers (CD11c, iNOS, TNF-α, and IL-1β) in M1 (Figure 1, A–D) and higher expression of M2 markers (ARG1, CD206, and IL-10) in M2 ATMs (Figure 1, E–G). Levels of M1 markers in double+ ATMs were similar to those in M1 (Figure 1, A–D), whereas levels of M2 markers in double+ ATMs were intermediate between M1 and M2 ATMs (Figure 1, E–G). The double– ATMs expressed low levels of both M1 and M2 markers (Figure 1, A–G). Interestingly, DNMT1 expression was much lower in M2 ATMs than that in M1 ATMs (Figure 1H).
Figure 1. DNA methyltransferase 1 (DNMT1) expression is lower in M2 than in M1 adipose tissue macrophages (ATMs) and is upregulated in isolated ATMs from ob/ob mice.
(A–G) Expression of M1 and M2 markers, including CD11c (A), iNOS (B), TNF-α (C), IL-1β (D), CD206 (E), ARG1 (F), and IL-10 (G), in M1, M2, double+ and double– ATMs isolated from adipose tissue of 8- to 10-week-old male C57BL/6J mice. Cells from 3 to 4 mice were pooled for RNA isolation and gene expression measurements. M1, F4/80+CD11c+CD206–; M2, F4/80+CD11c–CD206+; double+, F4/80+CD11c+CD206+; double–, F4/80+CD11c–CD206–. (H) DNMT1 expression in M1 and M2 ATMs. (I–K) M1 (I) and M2 (J) markers and DNMT1 (K) expression in ATMs isolated from 14-week-old lean and ob/ob mice. Cells from 3 to 4 lean mice were pooled, whereas cells from individual ob/ob mice were used for RNA isolation and gene expression measurements. Data are expressed as the mean ± SEM. n = 3–7. Groups labeled with different letters are statistically different from each other. *P < 0.05. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
We have also studied gene expression profiles in isolated ATMs from ob/ob or diet-induced obese (DIO) mice. As shown in Supplemental Figure 2, A and B, F4/80 expression was significantly enriched in ATMs from ob/ob and DIO mice, indicating successful isolation of F4/80+ ATMs. ATMs from ob/ob (Figure 1, I and J) or DIO mice (Supplemental Figure 2, C and D) displayed higher proinflammatory M1 marker (e.g., iNOS, TNF-α, IL-6, and IL-1β) and lower M2 marker (e.g., CD206, CD301, and ARG1) expression compared with that of control mice. Interestingly, DNMT1 expression was significantly elevated in ATMs isolated from ob/ob mice compared with that of lean controls (Figure 1K). Similar results, albeit to a lesser extent, were observed in ATMs isolated from DIO mice (Supplemental Figure 2E).
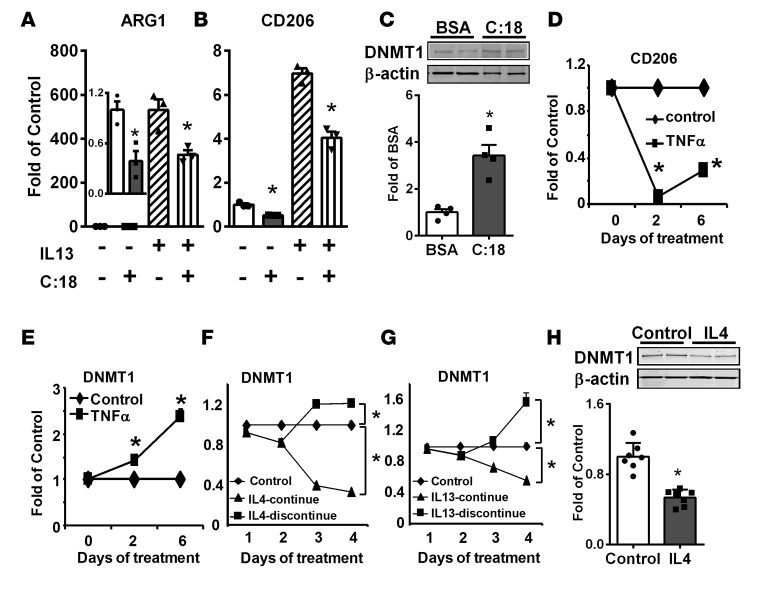
Dietary saturated fatty acids (SFAs) promote metabolic disorders partly by increased recruitment of proinflammatory M1 macrophages into inflamed adipose tissue (1, 2, 32, 33). Interestingly, the SFA stearate (C:18) significantly suppressed basal and IL-13–induced M2 marker expression in bone marrow–derived macrophages (BMDMs) (Figure 2, A and B) and the Raw264.7 murine macrophage cell line (Supplemental Figure 2, F and G), including ARG1 and CD206. This was associated with increased DNMT1 protein levels in BMDMs (Figure 2C) and Raw264.7 macrophages (Supplemental Figure 2H). Similarly, the inflammatory cytokine TNF-α significantly suppressed CD206 expression in BMDMs (Figure 2D), which was associated with significantly upregulated DNMT1 expression (Figure 2E).
Figure 2. DNA methyltransferase 1 (DNMT1) expression is increased by stearate and TNF-α and decreased by IL-4 and IL-13 in macrophages.
(A and B) ARG1 (A) and CD206 (B) expression in bone marrow–derived macrophages (BMDMs) stimulated with IL-13 with or without stearate (C:18). BMDMs were pretreated with 200 μM stearate for 4 days and then treated with 10 ng/ml IL-13 for an additional day. (C) DNMT1 protein levels in BMDMs treated with 200 μM stearate (C:18) for 4 days. (D and E) CD206 (D) and DNMT1 (E) expression in BMDMs treated with 10 ng/ml TNF-α for indicated times. (F and G) DNMT1 expression in RAW264.7 macrophages treated with 10 ng/ml IL-4 (F) or IL-13 (G) continuously or withdrawal after 2 days of treatment. (H) DNMT1 protein levels in RAW264.7 macrophages treated with 10 ng/ml IL-4 for 4 days. Data are expressed as the mean ± SEM. n = 3–8. Protein levels in C and H were normalized to β-actin. *P < 0.05. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
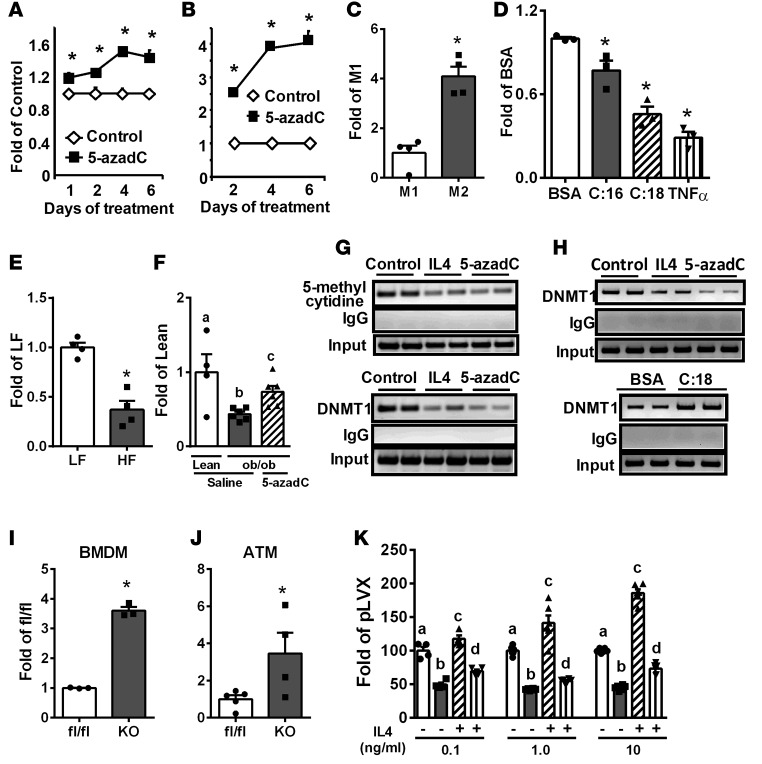
In contrast, IL-4 and IL-13 significantly inhibited DNMT1 expression in Raw264.7 macrophages (Figure 2, F and G). Importantly, this inhibition was specific to IL-4 and IL-13, as continuous IL-4 and IL-13 treatment in macrophages resulted in progressive inhibition of DNMT1 expression, whereas withdrawal of IL-4 and IL-13 after 2 days of treatment resulted in gradual reversal of DNMT1 expression to or above that of the control level (Figure 2, F and G). IL-4 also inhibited DNMT1 protein levels in RAW264.7 macrophages (Figure 2H). These data suggest that DNA methylation may be important in the regulation of macrophage alternative activation.
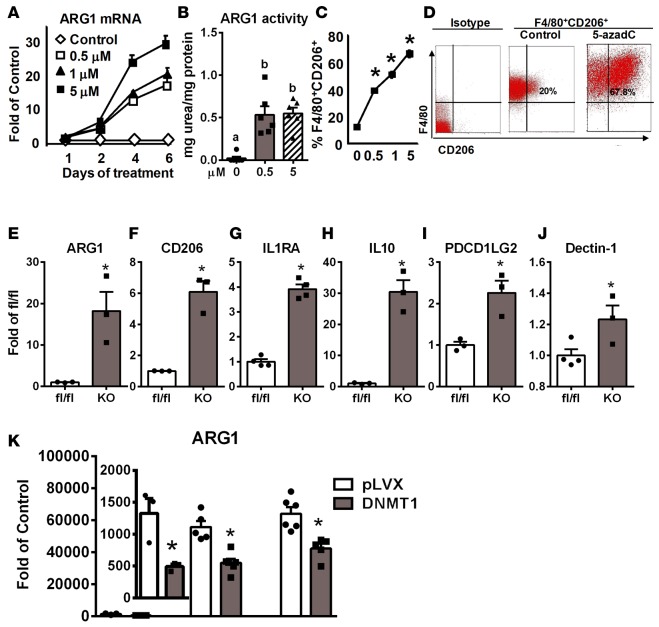
5-Aza-2′-deoxycytidine (5-azadC) is a nucleoside-based DNMT inhibitor widely used to study the role of DNA methylation in cancer development (34, 35). We found that 5-azadC treatment in macrophages resulted in rapid depletion of not only DNMT1, but also DNMT3a and 3b protein levels (Supplemental Figure 3), similar to previous reports (34, 36, 37). Interestingly, 5-azadC dose-dependently increased ARG1 expression and activity in BMDMs (Figure 3, A and B) or RAW264.7 macrophages (Supplemental Figure 4, A–C) when treated for up to 6 days. Other M2 markers induced by 5-azadC include CD206, IL-10, IL-4 receptor α (IL4Rα), and IL1RA, whose expression was upregulated in a dose- (Supplemental Figure 4, D–F) and time- (Supplemental Figure 4, G–J) dependent manner. 5-azadC also dose-dependently increased the percentage of F4/80+CD206+ M2 macrophages analyzed by FACS (Figure 3, C and D).
Figure 3. Inhibiting DNA methylation pharmacologically by 5-aza-2′-deoxycytidine or genetically by DNA methyltransferase 1 (DNMT1) deletion promotes macrophage alternative activation.
(A) Arginase 1 (ARG1) expression in bone marrow–derived macrophages (BMDMs) treated with 5-aza-2′-deoxycytidine (5-azadC) at the indicated doses and times. n = 4. (B) ARG1 activity in BMDMs treated with 5-azadC at the indicated doses for 4 days. (C) FACS analysis of percentage of F4/80+CD206+ M2 macrophages in RAW264.7 macrophages treated with 5-azadC at the indicated doses for 4 days. (D) Representative FACS analysis of F4/80+CD206+ M2 macrophages in RAW264.7 macrophages treated with 5 μM 5-azadC for 4 days. (E–J) M2 marker expression including ARG1 (E), CD206 (F), IL1RA (G), IL-10 (H), PDCD1LG2 (I), and dectin-1 (J) in BMDMs isolated from myeloid-specific DNMT1 knockout (MD1KO) and fl/fl control mice. (K) ARG1 expression in RAW264.7 macrophages transfected with control vector (pLVX) and DNMT1-overexpressing plasmids and treated with IL-4 at the indicated doses for 4 days. Data are expressed as the mean ± SEM. n = 3–6. *P < 0.05. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
We generated myeloid-specific DNMT1 knockout mice (MD1KO) by crossing DNMT1-floxed mice (38) with lysozyme Cre (Lys-Cre) mice (39). DNMT1 protein level was decreased by 80% in BMDMs isolated from MD1KO mice (Supplemental Figure 5A), whereas there was no change in DNMT3a and DNMT3b protein levels (Supplemental Figure 5B). Interestingly, macrophage DNMT1 deficiency led to significant upregulation of M2 markers, such as ARG1, CD206, IL1RA, IL-10, programmed cell death 1 ligand 2 (PDCD1LG2), and dectin-1 (Figure 3, E–J). In contrast, overexpressing DNMT1 in macrophages significantly inhibited IL-4–induced ARG1 expression (Figure 3K). These data collectively suggest that DNA methylation catalyzed by DNMT1 may be critical in macrophage alternative activation.
Inhibiting DNA methylation in macrophages suppresses inflammation and improves insulin sensitivity in cocultured 3T3-L1 adipocytes.
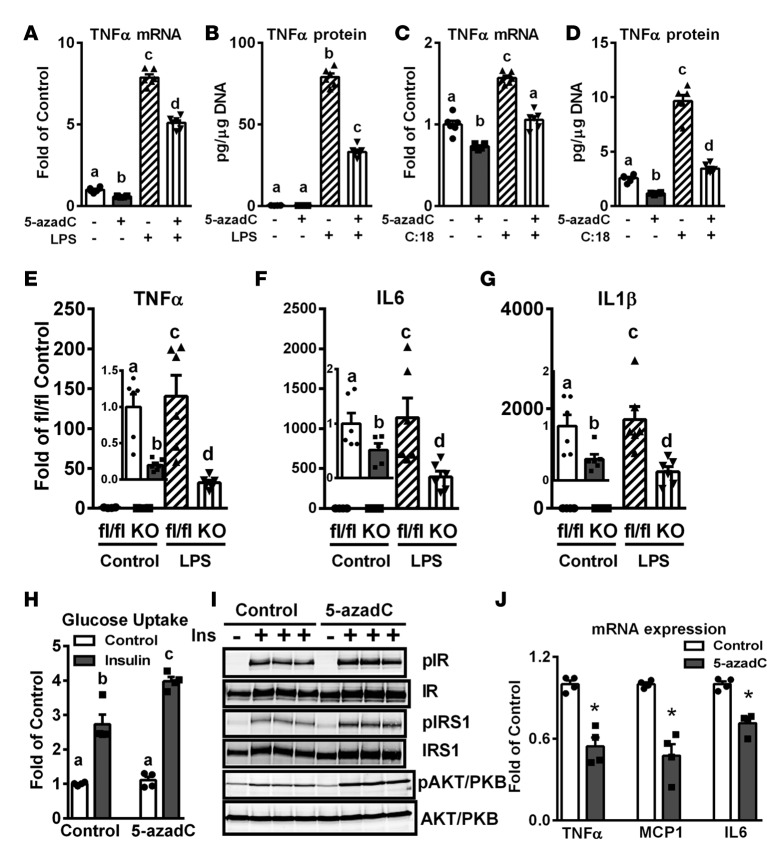
Pretreating Raw264.7 macrophages with 0.5 μM 5-azadC for 4 days, which did not affect cell viability (data not shown), significantly inhibited LPS- and stearate-induced inflammation, including the expression and secretion of TNF-α (Figure 4, A–D). Importantly, BMDMs isolated from MD1KO mice also exhibited lower basal and LPS-stimulated proinflammatory cytokine expression, including TNF-α, IL-6, and IL-1β (Figure 4, E–G). In addition, insulin-stimulated glucose uptake (Figure 4H) and signaling events, including tyrosyl phosphorylation of the insulin receptor (IR) and IR substrate 1 (IRS1) and serine phosphorylation of Akt/PKB (Figure 4I), were significantly improved, whereas the expression of proinflammatory cytokines and chemokines, including TNF-α, monocyte chemoattractant protein 1 (MCP1), and IL-6 (Figure 4J) was significantly decreased in differentiated 3T3-L1 adipocytes cocultured with 5-azadC–pretreated macrophages.
Figure 4. Inhibiting DNA methylation in macrophages suppresses inflammation in macrophages and improves insulin sensitivity in cocultured 3T3-L1 adipocytes.
(A and B) TNF-α expression (A) and secretion (B) in RAW264.7 macrophages pretreated with 0.5 μM 5-aza-2′-deoxycytidine (5-azadC) for 4 days and then challenged with 10 ng/ml LPS for 4 hours. (C and D) TNF-α expression (C) and secretion (D) in RAW264.7 macrophages pretreated with 0.5 μM 5-azadC for 4 days and then challenged with 200 μM stearate (C:18) for 4 hours. (E–G) TNF-α (E), IL-6, (F) and IL-1β (G) expression in bone marrow–derived macrophages isolated from myeloid-specific DNA methyltransferase 1 knockout (MD1KO) and fl/fl control mice treated with 10 ng/ml LPS for 4 hours. (H–J) Glucose uptake (H), insulin signaling events (I), and inflammatory cytokine and chemokine expression (J) in 3T3-L1 adipocytes cocultured with RAW264.7 macrophages pretreated with or without 0.5 μM 5-azadC for 4 days. Data are expressed as the mean ± SEM. n = 4–6. *P < 0.05. Groups labeled with different letters are statistically different from each other. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
Inhibiting DNA methylation by 5-azadC ameliorates obesity-induced inflammation and insulin resistance in ob/ob mice by promoting ATM alternative activation and suppressing ATM inflammation.
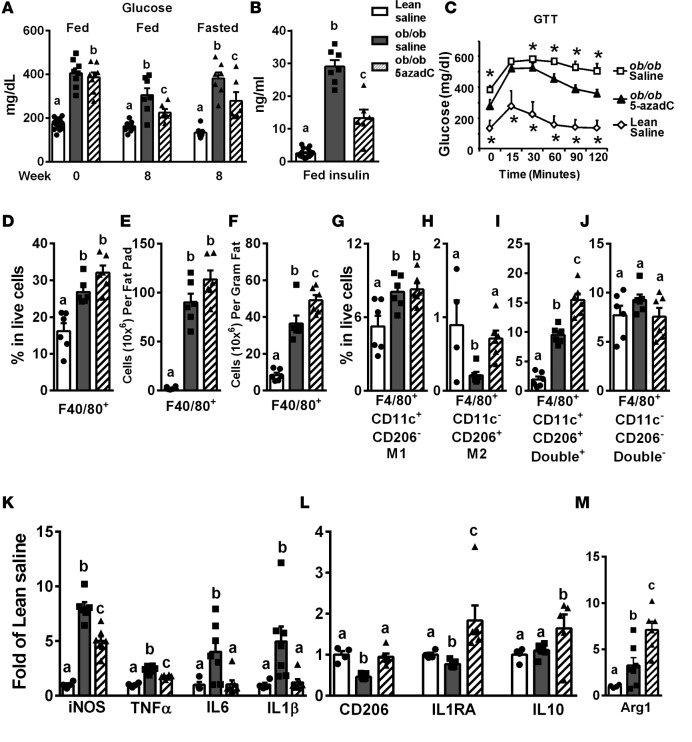
Treating ob/ob mice with a low dose of 5-azadC (0.25 mg/kg) i.p. 3 times per week for 8 weeks did not change body weight, fat mass, and food intake (Supplemental Figure 6, A–C), indicating this low dose of 5-azadC exerted no toxicity in animals, similar to a previous report (40). Importantly, 5-azadC treatment in ob/ob mice significantly reduced blood glucose (Figure 5A) and insulin (Figure 5B) levels, and improved insulin sensitivity as measured by glucose tolerance test (GTT) compared with saline-treated ob/ob mice (Figure 5C).
Figure 5. Inhibiting DNA methylation by 5-aza-2′-deoxycytidine improves insulin sensitivity, promotes adipose tissue macrophage (ATM) alternative polarization and suppresses ATM inflammation in ob/ob mice.
(A) Blood glucose levels in ob/ob and lean mice before and 8 weeks after 5-aza-2′-deoxycytidine (5-azadC) treatment (0.25 mg/kg 3 times per week). (B) Serum insulin levels in ob/ob and lean mice after 8 weeks of 5-azadC treatment. (C) Glucose tolerance test (GTT) in ob/ob and lean mice after 8 weeks of 5-azadC treatment. (D and F) Percentage of F4/80+ ATMs in live stromal vascular fraction (SVF) cells (D), F4/80+ ATM content per epididymal fat pad (E), and F4/80+ ATM content per gram of fat (F) in ob/ob and lean mice treated with saline or 5-azadC for 8 weeks. (G–J) Percentage of F4/80+CD1c+CD206– M1 ATMs (G), F4/80+CD1c–CD206+ M2 ATMs (H), F4/80+CD1c+CD206+ double+ ATMs (I), and F4/80+CD1c–CD206– double– ATMs in live SVF cells isolated from ob/ob and lean mice treated with saline or 5-azadC for 8 weeks. (K–M) M1 and M2 marker expression in ATMs isolated from adipose tissue of ob/ob and lean mice treated with saline or 5-azadC for 8 weeks. Data are expressed as the mean ± SEM. n = 6–16. *P < 0.05 vs. other groups. Groups labeled with different letters are statistically different from each other. Differences between groups were analyzed for statistical significance by Student’s t test, ANOVA with Fischer’s probable least-squares difference post hoc test, or ANOVA with repeated measures.
To explore mechanisms mediating the insulin-sensitizing effects of 5-azadC, we characterized ATM composition and gene expression profiles in ob/ob mice treated with saline or 5-azadC according to ATM surface expression of F4/80, CD11c, and CD206 (Figure 5 and Supplemental Figure 7).
As expected, ob/ob mice exhibited increased F4/80+ ATMs compared with lean controls, as demonstrated by an increased percentage of F4/80+ ATMs in live stromal vascular fraction (SVF) cells (Figure 5D and Supplemental Figure 7B), and increased F4/80+ ATMs in whole epididymal fat pad (Figure 5E) and per gram of fat (Figure 5F). However, 5-azadC did not significantly affect ATM content, as there was no significant change in F4/80+ ATM percentage in live SVF cells (Figure 5D and Supplemental Figure 7B) and in whole epididymal fat pad (Figure 5E), and even a slight increase in F4/80+ ATMs per gram of fat (Figure 5F) in 5-azadC–treated versus saline-treated ob/ob mice. In addition, the percentage of F4/80+CD206+ or F4/80+CD11c+ cells in live SVF cells was significantly increased in saline-treated ob/ob mice compared with lean controls (Supplemental Figure 7, C and D, and Supplemental Figure 8, A and B). 5-azadC treatment further increased F4/80+CD206+ cells in SVF cell populations. Unexpectedly, however, it also further increased F4/80+CD11c+ cells (Supplemental Figure 7, C and D, and Supplemental Figure 8, A and B).
To gain further insight into how inhibiting DNA methylation might change ATM phenotypes, we characterized M1 and M2 ATM composition in saline- and 5-azadC–treated lean and ob/ob mice. Although the percentage of CD206– M1 ATMs in the F4/80+CD11c+ gate was increased in saline-treated ob/ob mice compared with lean controls (Supplemental Figure 7E), because of significantly increased F4/80+ and F4/80+CD11c+ ATMs in saline-treated ob/ob mice (Figure 5D and Supplemental Figure 7, B and D), the percentage of F4/80+CD11c+CD206– M1 ATMs in live SVF cells was significantly increased (Figure 5G). On the other hand, both the percentage of CD206+ M2 ATMs in the F4/80+CD11c– gate (Supplemental Figure 7F) and percentage of F4/80+CD11c–CD206+ M2 ATMs in live SVF cells (Figure 5H) were significantly decreased in saline-treated ob/ob mice compared with lean controls. 5-azadC did not change the M1 ATM percentage in live SVF cells between saline- and 5-azadC–treated ob/ob mice (Figure 5G). On the other hand, 5-azadC completely restored the percentage of M2 ATMs in live SVF cells in ob/ob mice to that of the lean control (Figure 5H).
Obesity increases double+ ATMs, which are proinflammatory and may contribute to adipose tissue inflammation (30). The increased F4/80+CD206+ ATMs in saline-treated ob/ob mice (Supplemental Figure 7, C and D, and Supplemental Figure 8, A and B) may reflect the increase of these double+ ATMs. Indeed, our data show that the double+ ATM content was increased in saline-treated ob/ob mice compared with lean controls (Figure 5I). However, 5-azadC treatment in ob/ob mice resulted in further increase of the double+ ATMs compared with that of saline-treated ob/ob mice (Figure 5I), whereas there was no difference in the double– ATM content among the 3 treatment groups (Figure 5J). Accordingly, 5-azadC treatment in ob/ob mice increased CD206+ ATMs independently of CD11c expression, whereas it did not affect CD206– ATM populations (Supplemental Figure 8B). This resulted in increased F4/80+CD206+ ATMs in whole epididymal fat pad (Supplemental Figure 8C) and per gram of fat (Supplemental Figure 8D), whereas there was no difference in F4/80+CD206– ATMs in whole epididymal fat pad (Supplemental Figure 8E) and per gram of fat (Supplemental Figure 8F).
The double+ ATMs expressed high levels of M1 markers, similar to those of M1 ATMs (Figure 1, A–D), whereas those of M2 markers were intermediate between M1 and M2 ATMs (Figure 1, E–G). To better understand mechanistically whether and how the 5-azadC–induced ATM phenotypic switch contributes to improved insulin sensitivity observed in ob/ob mice, we measured inflammatory gene expression in M1, M2, and double+ ATMs isolated from saline- and 5-azadC–treated lean and ob/ob mice. The M1, M2, and double+ ATMs were isolated using a method similar to that described in Supplemental Figure 1. The expression of TNF-α and IL-6 was significantly increased in all 3 ATM populations in saline-treated ob/ob mice compared with that of lean controls (Supplemental Figure 8, G and H). 5-azadC specifically reduced TNF-α and IL-6 expression in CD206+ ATM populations, namely M2 and double+ ATMs, but not in CD206– M1 ATMs (Supplemental Figure 8, G and H).
Consistent with these results, ATMs isolated from saline-treated ob/ob mice exhibited increased expression of inflammatory M1 markers, including iNOS, TNF-α, IL-6, and IL-1β (Figure 5K), as well as deregulated expression of M2 markers, including reduced expression of CD206 and IL1RA, unchanged expression of IL-10, and even increased expression of ARG1 (Figure 5, L and M). The latter may be due to increased double+ ATMs in saline-treated ob/ob mice (Figure 5I). However, ATMs from 5-azadC–treated ob/ob mice exhibited significantly reduced M1 marker expression, and restored or further enhanced M2 marker expression (Figure 5, K–M). Our data suggest that inhibiting DNA methylation by 5-azadC in ob/ob mice significantly increased CD206+ ATM content and suppressed inflammatory gene expression in CD206+ ATMs without affecting those of CD206–ATMs. These 2 effects may collectively contribute to suppressed ATM inflammation and increased insulin sensitivity in 5-azadC–treated ob/ob mice.
Myeloid DNMT1 deletion promotes ATM alternative activation, suppresses adipose tissue inflammation, and improves whole-body insulin sensitivity.
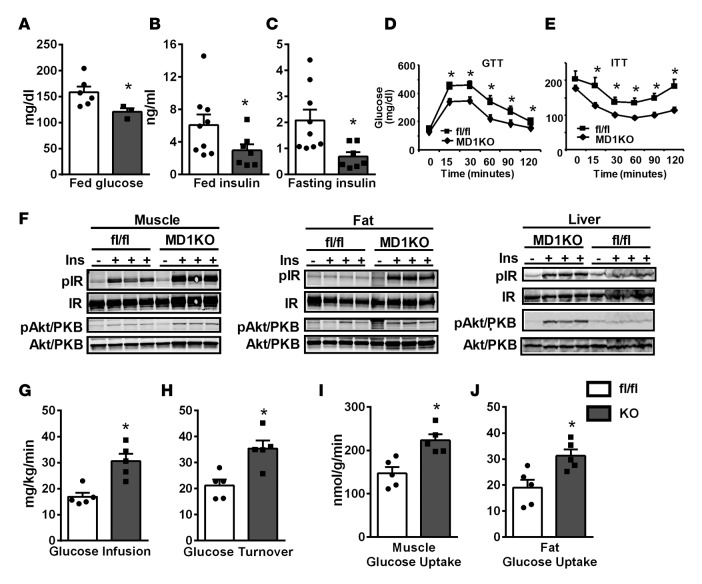
We further characterized metabolic phenotypes in control fl/fl and MD1KO mice fed a high-fat (HF) diet. There was no difference in body weight, fat pad mass, and liver and spleen weight between MD1KO and fl/fl mice fed an HF diet (Supplemental Figure 9, A and B). Interestingly, MD1KO mice displayed significantly decreased fed glucose levels (Figure 6A) and fed and fasting insulin levels (Figure 6, B and C). In addition, MD1KO mice exhibited improved glucose tolerance and insulin sensitivity as assessed by GTT and insulin tolerance test (ITT) (Figure 6, D and E), respectively. Consistently, insulin-stimulated tyrosyl phosphorylation of the IR and serine phosphorylation of Akt/PKB were significantly increased in skeletal muscle, fat, and liver of MD1KO mice compared with fl/fl control mice (Figure 6F). During a hyperinsulinemic-euglycemic clamp study, glucose infusion and turnover rates were significantly increased in MD1KO mice compared with fl/fl mice (Figure 6, G and H), and insulin-stimulated glucose uptake into skeletal muscle and fat was also increased in MD1KO mice (Figure 6, I and J). These data demonstrate that myeloid DNMT1 determines whole-body insulin sensitivity without affecting body weight and adiposity, and mice with myeloid deletion of DNMT1 are protected from obesity-induced insulin resistance and diabetes.
Figure 6. Mice with myeloid-lineage deletion of DNMT1 (MD1KO) exhibit improved insulin sensitivity.
(A–C) Fed glucose (A), fed insulin (B), and fasted insulin (C) in MD1KO and fl/fl control mice fed a high-fat (HF) diet for 12 to 14 weeks. (D and E) Glucose tolerance test (GTT, D) and insulin tolerance test (ITT, E) in MD1KO and fl/fl mice fed an HF diet for 18 to 22 weeks. (F) Insulin signaling events in muscle, fat, and liver in MD1KO and fl/fl mice fed an HF diet for 24 weeks. (G–J) Glucose infusion rate (G), glucose turnover rate (H), and muscle (I) and fat (J) glucose uptake in MD1KO and fl/fl mice fed an HF diet for 24 weeks as assessed by hyperinsulinemic-euglycemic clamp. Data are expressed as the mean ± SEM. n = 3–9. *P < 0.05. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with repeated measures.
We further studied whether myeloid DNMT deletion altered circulating immune cell composition. We analyzed peripheral blood immune cell composition using FACS analysis according to their surface expression of CD4, CD8, and B220 for CD4+ and CD8+ T lymphocytes, and B lymphocytes, respectively (Supplemental Figure 10A). We also analyzed the circulating monocyte percentage according to their surface expression of CD11b and the monocyte/macrophage-specific surface marker F4/80 (41) (Supplemental Figure 10B). FACS analysis showed that there was no difference in circulating CD4+ and CD8+ T lymphocytes and B220+ B lymphocytes between MD1KO and fl/fl mice (Supplemental Figure 10, C–E). In addition, there was no difference in circulating CD11b+F4/80+ monocytes between MD1KO and fl/fl mice (Supplemental Figure 10F), nor was there any difference in 7/4lo resident and 7/4hi inflammatory monocyte subset composition (Supplemental Figure 10, G and H), the latter being the monocyte subset that infiltrates inflamed tissues (42).
We also studied ATM subset composition in fl/fl and MD1KO mice fed an HF diet using FACS analysis according to the surface expression of F4/80, CD11c, and CD206 (Supplemental Figure 11). We found that myeloid DNMT1 deficiency did not affect ATM content in adipose tissue, as the percentage of F4/80+ ATMs in live SVF cells was not different between MD1KO and fl/fl mice (Supplemental Figure 11F), nor was there any difference in F4/80+CD11c+ or F4/80+CD11c– ATM percentage in live SVF cells between MD1KO and fl/fl mice (Supplemental Figure 11, G and H).
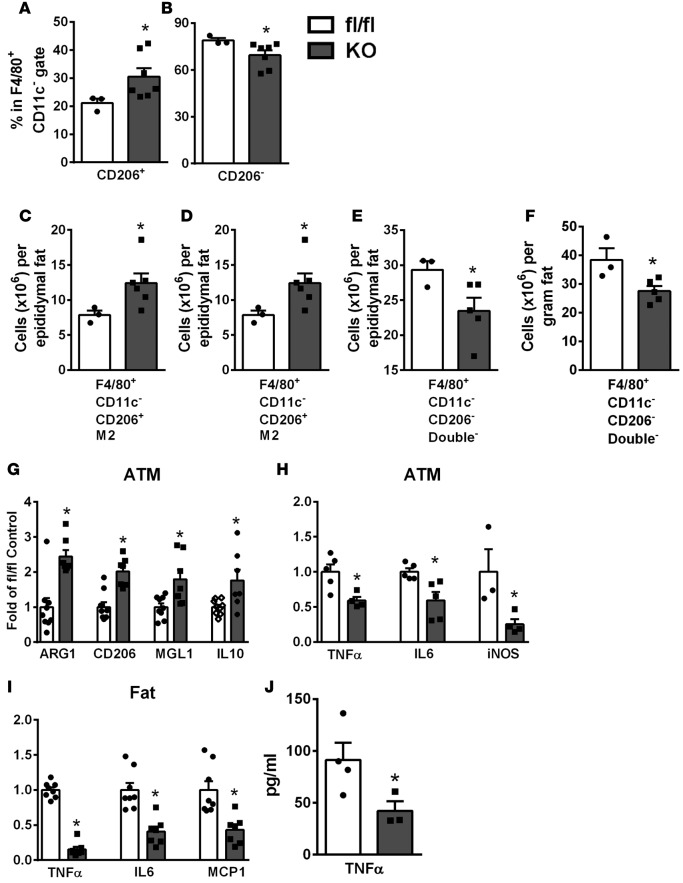
Unlike 5-azadC–treated ob/ob mice, where there was a significant increase in CD206+ ATMs in both F4/80+CD11c+ and F4/80+CD11c– ATM populations (Figure 5 and Supplemental Figure 8), there was no difference in CD206+ or CD206– ATM content in F4/80+CD11c+ ATMs between MD1KO and fl/fl mice (double+ and M1 ATMs, respectively, Supplemental Figure 11, I and J). However, the percentage of CD206+ ATMs was significantly increased in F4/80+CD11c– ATM populations (i.e., M2 ATMs), which was associated with a reciprocal reduction of CD206– ATM content in F4/80+CD11c– ATM populations (i.e., double– ATMs) (Figure 7, A and B). This corresponded to increased M2 ATMs in whole epididymal fat pad (Figure 7C) or per gram of fat (Figure 7D), and reciprocally decreased double– ATMs in whole epididymal fat pad (Figure 7E) or per gram of fat (Figure 7F). Consistently, the expression of M2 markers, including ARG1, CD206, MGL1, and IL-10 was significantly increased, whereas the expression of inflammatory cytokines and chemokines, including TNF-α, IL-6, and iNOS was significantly decreased, in ATMs isolated from MD1KO mice compared with fl/fl mice (Figure 7, G and H). This corresponded to reduced TNF-α, IL-6, and MCP1 expression in adipose tissue (Figure 7I), and reduced serum TNF-α levels of MD1KO mice compared with fl/fl mice (Figure 7J). These data suggest that myeloid deletion of DNMT1 specifically increases F4/80+CD11c–CD206+ M2 ATMs, which contribute to suppressed adipose tissue and systemic inflammation, thereby protecting MD1KO mice from developing obesity-induced insulin resistance.
Figure 7. Mice with myeloid-lineage deletion of DNMT1 (MD1KO) exhibit increased M2 adipose tissue macrophage (ATM) polarization and suppressed adipose tissue and systemic inflammation.
(A and B) Percentage of CD206+ (A) and CD206– (B) ATMs in the F4/80+CD11c– FACS gate in stromal vascular fraction cells isolated from the epididymal fat pad of MD1KO and fl/fl mice fed a high-fat (HF) diet for 24 weeks. (C and D) F4/80+CD11c–CD206+ M2 ATMs in whole epididymal fat pad (C) and per gram of fat (D) of MD1KO and fl/fl control mice fed an HF diet for 24 weeks. (E and F) F4/80+CD11c–CD206– double– ATMs in whole epididymal fat pad (E) and per gram of fat (F) of MD1KO and fl/fl mice fed an HF diet for 24 weeks. (G) M2 marker expression in ATMs of MD1KO and fl/fl mice fed an HF diet for 24 weeks. (H and I) Proinflammatory gene expression in ATMs (H) and epididymal fat tissue (I) of MD1KO and fl/fl mice fed an HF diet for 24 weeks. (J) Serum TNF-α levels in MD1KO and fl/fl mice fed an HF diet for 24 weeks. Data are expressed as the mean ± SEM. n = 3–8. *P < 0.05. Differences between groups were analyzed for statistical significance by Student’s t test.
DNA methylation at the PPARγ1 promoter regulates M2 macrophage activation.
The induction of M2 macrophage polarization is mediated by STAT6 (2), while the nuclear receptors PPARγ and PPARδ also play important roles in this process (12–17). We found that 5-azadC treatment did not change STAT6 phosphorylation or PPARδ expression in BMDMs and RAW264.7 macrophages (data not shown). However, 5-azadC significantly increased PPARγ1 expression in BMDMs (Figure 8A) and RAW264.7 macrophages (Figure 8B). There are 2 isoforms of PPARγ, namely PPARγ1 and PPARγ2 (43–45). Whereas PPARγ1 is more broadly expressed in various cell types including adipocytes and macrophages, PPARγ2 is only expressed in adipocytes (46). Consistently, we could not detect PPARγ2 expression in macrophages (data not shown), similar to a previous report (46). These data suggest that PPARγ1 may be a potential target for epigenetic regulation by DNA methylation in macrophages.
Figure 8. PPARγ expression is regulated by DNA methylation.
(A–F) PPARγ expression in 0.5 μM 5-aza-2′-deoxycytidine (5-azadC)–treated bone marrow–derived macrophages (BMDMs) (A) and RAW264.7 macrophages (B), in M1 (F4/80+CD11c+CD206–) and M2 (F4/80+CD11c–CD206+) adipose tissue macrophages (ATMs) (C), in BMDMs treated with 200 μM palmitate, 200 μM stearate, and 10 ng/ml TNF-α for 6 days (D), in ATMs isolated from low-fat-fed (LF-fed) and high-fat-fed (HF-fed) mice (E) and in ATMs isolated from saline- and 5-azadC–treated ob/ob and lean mice (F). (G) 5-Methyl cytidine levels and DNA methyltransferase 1 (DNMT1) binding at the PPARγ1 promoter in RAW264.7 macrophages treated with 10 ng/ml IL-4 and 0.5 μM 5-azadC for 4 days. (H) DNMT1 binding at the PPARγ1 promoter in BMDMs treated with 10 ng/ml IL-4, 0.5 μM 5-azadC, or 200 μM stearate (C:18) for 4 days. (I and J) PPARγ expression in BMDMs (I) and ATMs (J) isolated from myeloid-specific DNMT1 knockout (MD1KO) and fl/fl control mice. (K) PPARγ expression in RAW264.7 macrophages transfected with control (pLVX) or DNMT1-overexpressing plasmid and treated with IL-4 at the indicated doses for 4 days. Data are expressed as the mean ± SEM. n = 3–6. *P < 0.05. Groups labeled with different letters are statistically different from each other. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
Interestingly, PPARγ1 expression was significantly higher in M2 versus M1 macrophages (Figure 8C), suppressed in BMDMs exposed to stearate (C:18), palmitate (C:16), and TNF-α (Figure 8D), reduced in ATMs isolated from HF-fed mice compared with that of low-fat-fed (LF-fed) mice (Figure 8E), reduced in ATMs isolated from saline-treated ob/ob mice compared with that of lean controls, but was largely restored to that of the lean mice level by 5-azadC treatment (Figure 8F). Considering the importance of PPARγ in the regulation of macrophage alternative activation and whole-body insulin sensitivity, this obesity-induced downregulation of PPARγ expression could explain the disturbed balance between M2 and M1 ATMs and abnormal inflammation in obesity.
The proximal promoter and 5′-untranslated region of PPARγ1 is enriched with CpG sites (Supplemental Figure 12A). IL-4 and 5-azadC significantly reduced 5-methyl cytidine levels and DNMT1 binding at the PPARγ1 promoter in RAW264.7 macrophages in methylated DNA immunoprecipitation (MeDIP) assays and chromatin immunoprecipitation (ChIP) assays, respectively (Figure 8G). In addition, IL-4 and 5-azadC significantly decreased, whereas stearate significantly increased, DNMT1 binding at the PPARγ1 promoter in BMDMs (Figure 8H). Moreover, PPARγ1 expression was significantly upregulated in BMDMs (Figure 8I) and ATMs (Figure 8J) isolated from MD1KO compared with fl/fl mice, whereas DNMT1 overexpression in macrophages profoundly suppressed IL-4–induced PPARγ1 expression (Figure 8K). These data suggest that DNA methylation at the PPARγ1 promoter is dynamically regulated, presumably by DNMT1. Consistent with this, PPARγ1 promoter–driven luciferase activity of the unmethylated promoter constructs were 200-fold higher than that of fully methylated promoter constructs (Figure 9A), further confirming that PPARγ1 promoter activity is highly regulated by DNA methylation.
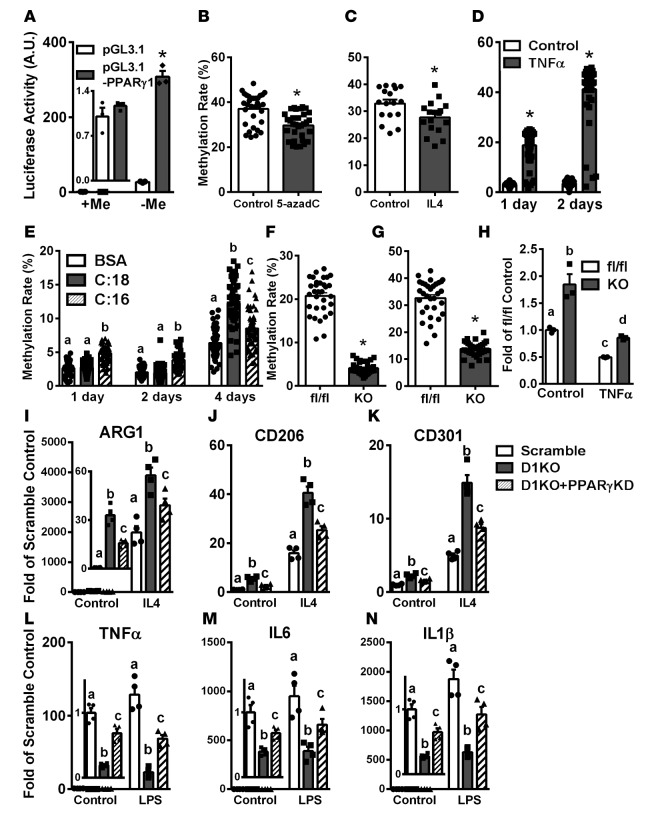
Figure 9. PPARγ is necessary for M2 macrophage polarization induced by DNA methyltransferase 1 (DNMT1) deletion.
(A) PPARγ1 promoter luciferase activity in fully methylated (+Me) and unmethylated (–Me) constructs. n = 3. (B–G) PPARγ1 promoter DNA methylation rate in RAW264.7 macrophages treated with 0.5 μM 5-aza-2′-deoxycytidine (5-azadC) (B) or 10 ng/ml IL-4 for 4 days (C), in bone marrow–derived macrophages (BMDMs) treated with 10 ng/ml TNF-α for 1 and 2 days (D), in BMDMs treated with 200 μM stearate (C:18) and 200 μM palmitate (C:16) for the indicated times (E), in thioglycolate-elicited peritoneal macrophages isolated from myeloid-specific DNMT1 knockout (MD1KO) and fl/fl mice on a low-fat (LF) diet (F), or adipose tissue macrophages isolated from MD1KO and fl/fl mice fed a high-fat (HF) diet for 24 weeks (G). n per group ranges from 17 in C, 32 in B, F, and G, and 48–50 in D and E. (H) PPARγ expression in BMDMs isolated from MD1KO and fl/fl mice stimulated with TNF-α for 1 day. n = 3. (I–K) M2 marker expression including ARG1 (I), MRC1 (J), and MGL1 (K) in BMDMs isolated from MD1KO and fl/fl mice with PPARγ siRNA knockdown and stimulated with 10 ng/ml IL-4 for 2 days. n = 4. (L–N) M1 marker expression including TNF-α (L), IL-6 (M), and IL-1β (N) in BMDMs isolated from MD1KO and fl/fl mice with PPARγ siRNA knockdown and stimulated with 10 ng/ml LPS for 4 hours. n = 4. Data are expressed as the mean ± SEM. *P < 0.05. Groups labeled with different letters are statistically different from each other. Differences between groups were analyzed for statistical significance by Student’s t test or ANOVA with Fischer’s probable least-squares difference post hoc test.
We measured the DNA methylation level at the PPARγ1 promoter/5′-untranslated region using bisulfite conversion and pyrosequencing. The average methylation rate at the PPARγ1 promoter was approximately 33%–36% in Raw264.7 macrophages (Figure 9, B and C), which was moderately but significantly reduced by 5-azadC (Figure 9B and Supplemental Figure 12, B and C) and IL-4 (Figure 9C and Supplemental Figure 12D). In contrast, the basal DNA methylation rate at the PPARγ1 promoter was approximately 3% in BMDMs (Figure 9, D and E). TNF-α profoundly increased the DNA methylation rate at the PPARγ1 promoter in BMDMs to 18% after 1 day of treatment and 40% after 2 days of treatment (Figure 9D and Supplemental Figure 13, A and B). In addition, stearate and palmitate significantly increased DNA methylation at the PPARγ1 promoter in BMDMs when treated for up to 4 days (Figure 9E and Supplemental Figure 13, C and D). These data strongly suggest that PPARγ1 promoter/5′-untranslated region DNA methylation is dynamically regulated by SFAs and inflammatory cytokines. In addition, the substantially higher basal DNA methylation rate at the PPARγ1 promoter in RAW264.7 macrophages may also partly explain the more robust stimulation of M2 marker expression by 5-azadC observed in RAW264.7 macrophages than that of BMDMs (Figure 3 and Supplemental Figure 4).
More importantly, DNA methylation at the PPARγ1 promoter was significantly reduced in peritoneal macrophages isolated from MD1KO compared with that of fl/fl mice (Figure 9F and Supplemental Figure 14A). Similarly, ATMs isolated from MD1KO mice fed an HF diet also exhibited significantly reduced DNA methylation at the PPARγ1 promoter (Figure 9G and Supplemental Figure 14B). Consistent with reduced DNA methylation at the PPARγ1 promoter in MD1KO macrophages, BMDMs from MD1KO mice exhibited significantly higher basal PPARγ1 expression (Figure 9H). In addition, PPARγ1 expression was profoundly suppressed by TNF-α in BMDMs from fl/fl mice, but was largely restored in TNF-α–treated BMDMs from MD1KO mice (Figure 9H).
Finally, BMDMs isolated from MD1KO mice exhibited significant increases in basal and IL-4–induced M2 marker expression compared with that of fl/fl mice, including ARG1, CD206, and CD301 (Figure 9, I–K). This effect was largely blocked by PPARγ knockdown (Figure 9, I–K). Similarly, BMDMs isolated from MD1KO mice exhibited significant inhibition of basal and LPS-induced inflammatory gene expression compared with that of 5-azadC–treated fl/fl macrophages, including TNF-α, IL-6, and IL-1β (Figure 9, L–N). This effect was also largely blocked by PPARγ knockdown (Figure 9, L–N). These data demonstrate that the promotion of alternative activation and the suppression of inflammation in macrophages exerted by DNMT1 deletion require PPARγ1.
Discussion
This study was designed to address the hypothesis that epigenetic regulation of macrophage activation by DNA methylation is an underlying mechanism mediating obesity-induced ATM phenotypic switch and inflammation. The plausibility of this hypothesis derives from several prior observations. First, obesity results in increased classically activated M1 ATMs and reduced alternatively activated M2 ATMs, which eventually tips the balance toward a proinflammatory environment in adipose tissue and is a central feature of obesity contributing to insulin resistance/type 2 diabetes and other metabolic diseases (47). However, a critical question is how obesity leads to defective ATM alternative activation. Second, PPARγ is a key transcriptional factor that controls macrophage alternative activation and whole-body insulin sensitivity. PPARγ activation promotes macrophage alternative activation (12–14), whereas myeloid PPARγ deletion results in impaired macrophage alternative activation and insulin resistance (16). However, whether and how deregulation of PPARγ leads to altered macrophage polarization in obesity is not known.
Here, we have investigated these important questions with an epigenetic perspective. We demonstrate that DNA methylation plays a key role in the regulation of macrophage alternative activation and inflammation, which involves DNA methylation at the PPARγ1 promoter by DNMT1. This is not only mechanistically important but also physiologically relevant.
Most complex diseases, including obesity and diabetes, are results of gene and environment interactions. One of the mechanisms underlying the ability of environmental factors such as diet to affect gene expression involves their capacity to reprogram the epigenome (48, 49). We found that dietary SFAs and proinflammatory cytokines (e.g., TNF-α), levels of which are elevated in obesity, significantly upregulated DNMT1 expression in macrophages. Most importantly, SFAs and TNF-α also significantly increased DNA methylation levels at the PPARγ1 promoter, leading to suppressed PPARγ1 expression in macrophages. Dietary SFAs have been shown to differentially regulate DNA methylation patterns in human blood leukocytes compared with those of mono- or polyunsaturated fatty acids (50), whereas dietary palmitate also dynamically alters DNA methylation patterns in human islets (51), further supporting an epigenetic regulation of gene expression by dietary SFAs. Therefore, our findings provide a mechanistic basis for how obesity results in a macrophage phenotypic switch to a more proinflammatory M1 phenotype, leading to increased inflammation and eventually the development of obesity-associated metabolic diseases, including insulin resistance.
DNMT1 has been traditionally viewed as a maintenance enzyme, as it binds to methyl groups in hemimethylated DNA strands during DNA replication, thereby mainly functioning in proliferating cells (19). However, myeloid deletion of DNMT1 did not affect hematopoiesis, as there was no difference in circulating lymphocyte and monocyte populations in MD1KO and fl/fl mice (Supplemental Figure 10). Circulating monocytes are heterogeneous, as defined by their differential expression of surface markers. Ly-6Chi (or 7/4hi) inflammatory monocytes give rise to M1 macrophages when entering an inflamed site, whereas Ly-6Clo (or 7/4lo) resident monocytes typically differentiate into M2 macrophages when migrating into tissues (42, 52). However, there was no difference in 7/4hi inflammatory and 7/4lo resident monocyte subset composition in MD1KO and fl/fl mice (Supplemental Figure 10), thus excluding the reprogramming of monocyte subsets by myeloid DNMT1 deficiency during hematopoiesis in bone marrow or when circulating in the bloodstream in MD1KO mice.
On the other hand, myeloid DNMT1 deletion specifically reprogrammed ATMs by increasing the CD206+ subset in F4/80+CD11c– ATMs (M2 ATMs) when fed an HF diet, with a reciprocal reduction of the CD206– subset in F4/80+CD11c– ATMs (double– ATMs), whereas that of F4/80+CD11c+ ATMs (including M1 and double+) was not affected (Figure 7). Interestingly, a recent study demonstrated that whereas tissue M1 macrophages were typically recruited from blood monocytes, M2 macrophages in tissues mainly came from local macrophage proliferation rather than recruitment from blood (53). Therefore, our data suggest that increased F4/80+CD11c–CD206+ M2 ATM subsets in inflamed adipose tissue in HF-fed MD1KO mice may be due to local proliferation of M2 ATMs. Alternatively, since we observed a reciprocal reduction of double– ATMs in adipose tissue, it is possible that the increased M2 ATMs in MD1KO mice may be derived from reprogramming and conversion from double–ATMs. This is supported by recent evidence demonstrating that DNMT1 may also participate in de novo DNA methylation, thereby catalyzing DNA methylation not only in proliferating cells, but also in nonproliferating postmitotic cells (22).
Similarly, inhibiting DNA methylation with the demethylating agent 5-azadC in ob/ob mice significantly increased CD206+ subsets both in F4/80+CD11c– (M2) and F4/80+CD11c+ (double+) ATM populations without significantly affecting CD206– ATM subsets (Figure 5). In addition, 5-azadC also reprogrammed CD206+ ATM subsets to a more antiinflammatory phenotype, as shown by reduced inflammatory cytokine expression in CD206+ but not CD206– ATM subsets (Supplemental Figure 8). We observed more robust effects on reprogramming ATMs by 5-azadC than myeloid DNMT1 deletion, as 5-azadC–treated ob/ob mice exhibited increases in CD206+ subsets independent of CD11c expression, whereas MD1KO mice exhibited increases in the CD206+ subset only in F4/80+CD11c– ATM populations. Inhibiting DNA methylation by 5-azadC depleted all 3 DNMTs examined: DNMT1, DNMT3a, and DNMT3b (Supplemental Figure 3) (34, 36, 37). Interestingly, our recent data suggest that DNMT3b, but not DNMT3a, may exert similar effects on macrophage polarization and inflammation (54). Thus, it is possible that other DNMTs, such as DNMT3b, may also contribute to the regulation of macrophage alternative activation.
Theoretically, reducing DNA methylation at gene promoters is associated with gene activation (19). However, global gene activation was not observed in the current study when ob/ob mice were treated with 5-azadC or myeloid DNMT1–deficient mice were fed an HF diet. For example, the effect of inhibiting DNA methylation by DNMT1 deletion or 5-azadC treatment on inflammatory gene expression, including TNF-α, IL-6, and IL-1β, is inhibition, rather than activation. It is possible that these genes may not be regulated at the level of DNA methylation, but rather by other mechanisms. On the other hand, we observed selective demethylation and activation of PPARγ1, a key regulator of macrophage alternative activation. The increased PPARγ expression may in turn downregulate gene expression involved in inflammation. Therefore, our data suggest that inhibiting DNA methylation in macrophages specifically regulates DNA methylation patterns of genes involved in certain pathways, for example, macrophage alternative activation and antiinflammation, but may not change global DNA methylation.
The role of DNA methylation in tumorigenesis has been relatively well studied, whereas that in metabolic disease is less understood. It is well accepted that whereas there is an overall decrease in the level of cytosine methylation in the genome of tumor cells, the CpG sites of certain groups of genes, including those of tumor suppressors, become partially or fully methylated, which in turn leads to gene silencing and can be reactivated by demethylation agents, such as 5-azadC (34). It is possible that similar dynamic changes in DNA methylation may also occur during the development of obesity and insulin resistance. For example, during the development of metabolic disease, certain pathway-specific genes may become partially or fully methylated and silenced, similar to that observed in tumorigenesis. These genes could remain hypomethylated and activated in mice with myeloid deletion of DNMT, or can be reactivated by treating obese mice with demethylating agents, such as 5-azadC. Further studies are needed to thoroughly elucidate the role of DNA methylation and other epigenetic mechanisms in the development of metabolic diseases.
In summary, we demonstrate that DNA methylation by DNMT1 plays an important role in the regulation of macrophage polarization and inflammation. Myeloid DNMT1 deficiency leads to decreased macrophage PPARγ1 promoter DNA methylation, increased ATM alternative activation, and decreased ATM inflammation, which results in improved insulin sensitivity in obese animals. In contrast, DNMT1-mediated PPARγ1 promoter DNA methylation in macrophages is significantly enhanced by obesity-associated factors, such as SFAs and proinflammatory cytokines. Therefore, deregulated DNA methylation at the PPARγ1 promoter in obesity, presumably in response to nutritional cues (e.g., SFAs) and inflammatory stimuli (e.g., TNF-α), may result in deregulated macrophage polarization, thereby contributing to obesity-induced inflammation and insulin resistance. These studies could also help guide the development of epigenetic regulation as new therapeutic targets in the prevention and treatment of obesity-induced insulin resistance/type 2 diabetes.
Methods
Animals.
MD1KO mice were created by crossing DNMT1-floxed mice (Mutant Mouse Regional Resource Centers) (38) with lysozyme Cre (Lys-Cre) mice (Jackson Laboratory), where Cre is specifically expressed in myeloid lineage cells including macrophages and monocytes (39). The DNMT1-floxed mouse was created by inserting 2 loxP sites flanking exons 4 and 5, which causes a frame shift and lack of the motifs for the catalytic domain (38). MD1KO mice and their fl/fl littermates were put on an HF diet (D12492, 60% calories from fat, Research Diets Inc.) for 24 weeks starting at 6 weeks of age. The ob/ob mice and their lean controls (+/?) were fed a regular chow diet (LabDiet 5P00, fat content 14% by calorie) and were treated with 5-azadC (0.25 mg/kg body weight) via i.p. injection 3 times per week for 8 weeks starting at 6 weeks of age. This dose of 5-azadC has been shown to effectively reduced tumorigenesis without any visible adverse effects by chronic i.p. injection for 18 weeks (40). In some experiments, 6- to 8-week-old male C57BL/6J mice were fed an LF diet (D12450B, 10% calories from fat, Research Diets Inc.) or HF diet (D12492, 60% calories from fat, Research Diets Inc.) for 16 weeks.
Metabolic measurements.
Body weight was monitored weekly. Blood glucose was measured using a OneTouch Ultra glucose meter (Lifescan). Serum insulin levels were measured using rat insulin enzyme-linked immunosorbent assay (ELISA) kits (Crystal Chem). Plasma and medium TNF-α levels were measured by ELISA kits (R&D Systems). GTTs and ITTs were performed as previously described (55–57). For GTTs, mice were fasted overnight, and blood glucose was measured immediately before and 15, 30, 60, 90, and 120 minutes after an i.p. injection of glucose (1.2–1.8 g/kg of body weight). For ITTs, mice were injected i.p. with 1–1.8 U/kg of human insulin (Humulin R, Eli Lilly) after a 6-hour food removal, and glucose levels were measured at different time points (0, 15, 30, 60, and 120 minutes). For insulin signaling studies, mice were fasted overnight. Human insulin (10 U/kg of body weight; Humulin R) was injected i.p.; 10 minutes later, mice were euthanized by CO2, and tissues were quickly collected and snap-frozen in liquid nitrogen. Tissues were stored at –80°C until processing.
Hyperinsulinemic-euglycemic clamp studies.
Hyperinsulinemic-euglycemic clamp studies were performed essentially as previously described (55–57). Mice were implanted with the indwelling catheters and allowed to recover for 5 days. After an initial 5-μCi bolus, [3-3H]-glucose was infused at 0.05 μCi/min for 2 hours to measure basal glucose turnover. A 2-hour hyperinsulinemic-euglycemic clamp was conducted with a prime and continuous infusion of insulin at a rate of 2.5 mU/kg/min, coupled with a variable infusion of 40% glucose to maintain blood glucose at 6 mM. Blood glucose was measured via tail bleed every 5 minutes in the first hour to achieve stable blood glucose levels and every 10 minutes until the end of the 2-hour clamp to maintain constant blood glucose levels. The rate of whole-body glucose turnover was estimated using a continuous infusion of [3-3H]-glucose at 0.1 μCi/min. Tissue-specific glucose uptake was estimated by a bolus administration of 10 μCi 2-deoxy-D-[1-14C]-glucose 45 minutes before the end of clamp experiments.
Cell culture.
Raw264.7 macrophages were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen) containing 10% heat-inactivated FBS. BMDMs and peritoneal macrophages were cultured as previously described (55–57). Briefly, BM was flushed from the femur and tibia, dispersed, and cultured in DMEM containing 10% FBS and 30% L929 conditional medium for 8 days. For peritoneal macrophage isolation, 8- to 10-week-old fl/fl and MD1KO mice on regular chow diet (LabDiet 5P00, fat content 14% by calorie) were i.p. injected with 2 ml 3% thioglycollate to elicit peritoneal macrophages. Mice were euthanized by CO2 inhalation 48–72 hours later and cells were collected by lavaging the peritoneal cavity and used for pyrosequencing analysis.
For fatty acid treatment, stearate (C:18) and palmitate (C:16) (Sigma-Aldrich) were conjugated with BSA at a 4:1 molar ratio. Stearate and palmitate were first dissolved in 95% ethanol at 60°C and then were mixed with prewarmed BSA (10%) to yield a stock concentration of 3.75 mM.
For 5-azadC treatment, it has been reported that 5-azadC led to rapid loss of DNMT activity in cell culture at a dose low enough to avoid triggering cell death (58). We also found that similarly low doses of 5-azadC (up to 2 μM) did not affect cell viability (data not shown). Therefore, we used a low dose of 5-azadC (0.5 μM) in most of our cell culture experiments except for those used in dose-dependence studies.
Macrophage and 3T3-L1 adipocyte coculture.
Coculture studies were performed as previously described (59). Briefly, murine 3T3-L1 preadipocytes were propagated in DMEM containing 10% FBS (growth medium). Confluent preadipocytes were induced to differentiate with DMEM containing 10% FBS supplemented with 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 200 nM insulin (differentiation medium). Cells were incubated in the differentiation medium for 2 days and then cultured in growth medium containing 200 nM insulin for another 2 to 3 days. Subsequently, cells were maintained in growth medium until 8 days after differentiation.
For the macrophage-adipocyte coculture experiment, RAW264.7 macrophages were pretreated with 5-azadC (0.5 μM) for 4 days. The cells were then washed and plated in a transwell insert (0.4-μm porous membrane) and placed into wells containing 3T3-L1 adipocytes at day 8 after the induction of differentiation. Four days after coculture, adipocytes were used for insulin signaling and glucose transport assays as described previously (60, 61).
For insulin signaling studies, cocultured adipocytes were stimulated with 100 nM insulin for 5 minutes. Cells were harvested and homogenized in modified radioimmunoprecipitation assay (RIPA) buffer. IR (Tyr1162/Tyr1163), IRS-1 (Tyr612), Akt/PKB (Ser473) phosphorylation and total IR, IRS1, and Akt/PKB were measured by immunoblotting.
For glucose uptake assays, cocultured adipocytes were incubated in KRH buffer (129 mM NaCl, 4.8 mM KCl, 1.2 mM MgSO4, 1.8 mM CaCl2, and 20 mM HEPES, pH 7.4) with or without 100 nM insulin for 20 minutes. To initiate glucose transport, 1 μCi of 2-deoxy-[3H]glucose was added to each well and incubated for 10 minutes. To stop the transport, cells were rinsed with ice-cold KRH buffer 3 times and lysed with 0.1% SDS. Cell lysates were added to scintillation vials with 5 ml of scintillation liquid and counted. One well of cells from each treatment group included 50 μM cytochalasin B to determine nonspecific transport, which was subtracted from each sample.
Isolation of adipose tissue SVF cells.
Isolation of adipose tissue SVF cells was performed as previously described (55, 57). Briefly, 1–2 g of epididymal fat was placed in KRH buffer containing 10 mg/ml fatty acid–poor BSA (Sigma-Aldrich), minced, and digested in 1 mg/ml collagenase type I (Worthington Biochemical) by shaking (70 Hz) at 37°C for up to 60 minutes. Samples were filtered through a 300-μm nylon mesh (Spectrum Laboratories), and the resulting suspension was centrifuged at 400 g for 10 minutes to separate SVF cells from adipocytes.
FACS analysis.
Adipose tissue SVF cells were incubated in erythrocyte lysis buffer (eBioScience) for 5 minutes and then washed with KRH-BSA buffer. Cells were then resuspended in FACS buffer (eBioscience). A portion of the cells was counted with a hemocytometer. Based on trypan blue exclusion, the percentage of live cells per sample was usually greater than 95% (55, 57). Cells were incubated in the dark on a shaker with FcBlock (eBioscience) for 30 minutes at 4°C and further incubated for 1 hour with allophycocyanin (APC)-F4/80 (clone A3-1), phycoerythrin (PE)-CD206 (clone MR5D3), FITC-CD301 (clone ER-MP23) (all AbD Serotec), and PE-Cy7-CD11c (BD Pharmingen, clone HL3) antibodies. After incubation, cells were washed with FACS buffer, fixed in 4% paraformaldehyde and analyzed with a BD FACSCalibur or BD FACSCantoII Flow Cytometer. FACS data were analyzed using CellQuest software or FACSDiva software (both BD).
Isolation of ATMs and M1 and M2 macrophages.
SVF cells from adipose tissue were incubated with rat anti–mouse F4/80 polyclonal antibodies (AbD Serotec), followed by pulling down F4/80-positive cells with sheep anti-rat microbeads using a magnetic-activated cell sorting (MACS) system according to the manufacturer’s instructions (Miltenyi Biotec). Isolated F4/80+ ATMs were used for gene expression and pyrosequencing analysis. In another experiment, SVF cells were labeled with APC-F4/80, PE-Cy7-CD11c, and PE-CD206 as described above, and M1/M2 macrophage subsets were isolated using a BD FACSAria.
Analysis of blood leukocyte composition.
Peripheral blood was collected for leukocyte composition analysis (62). Red blood cells were lysed using ACK lysing buffer (Lonza). The remaining white blood cells were incubated with the following antibodies: Pacific blue-B220 (clone RA3-6B2), PerCP-Cy5-CD4 (clone RM4-5), APC-Cy7-CD8 (clone 53-6.7), PE-Cy7-CD11c (clone HL3) (all BD Pharmingen), PE-CD11b (eBioscience, clone M1/70), APC-F4/80, and FITC-7/4 (AbD Serotec, clone 7/4). Data were acquired by FACS using a BD FACSCanto II and were analyzed using CellQuest or FACSDiva software. Blood monocytes were gated based on their surface expression of CD11b and the monocyte/macrophage-specific marker F4/80 (41), and resident and inflammatory monocyte subsets were distinguished by the expression of surface marker 7/4 (42).
ChIP and MeDIP assays.
ChIP was conducted using a ChIP assay kit (Upstate) as previously described (59). Briefly, cells were fixed with 1% formaldehyde and then harvested in cell lysis buffer (5 mM PIPES, 85 mM KCl, and 0.5% NP-40, supplemented with protease inhibitors, pH 8.0). The lysates were sonicated to shear genomic DNA to an average fragment length of 200–1,000 bp. Lysates were centrifuged, and the supernatants were collected. The supernatants underwent overnight immunoprecipitation with anti-DNMT1 antibodies (Santa Cruz Biotechnology, clone H-300), elution, reverse cross-linking, and protease K digestion. The DNA recovered from phenol/chloroform extraction was used for PCR amplification. Two pairs of PCR primers for the PPARγ1 promoter were used, which amplify PCR fragments of ~289 and ~210 bp, respectively: primer pair 1, forward 5′-GGCTGTGAGGAGCAAGGCGG-3′ and reverse 5′-CCGGGGCGACTCTGACCTGA-3′; primer pair 2, forward 5′-TCAGGTCAGAGTCGCCCCGG-3′ and reverse 5′-CCCCTCAACCGAGACCCCGT-3′.
MeDIP was conducted as previously described (63). Briefly, genomic DNA was isolated by phenol/chloroform extraction and was immunoprecipitated with anti–5-methyl-cytidine antibodies (AnaSpec, clone 33D3). The immunocomplex was washed and digested with protease K. The recovered DNA was used for PCR amplification as described for the ChIP assay.
Plasmid overexpression and siRNA knockdown.
The full-length DNMT1 cDNA clone was obtained from Open Biosystems and further subcloned into the pLVX lentiviral expression vector (Clontech). The vector and DNMT1-overexpressing plasmids were transfected into Raw264.7 macrophages using electroporation (Nucleofector, Lonza). Transfected cells were selected with puromycin (10 μg/ml) for 10 days and surviving cells were pooled and treated with IL-4 at the indicated concentrations and times.
For siRNA-mediated knockdown, BMDMs were isolated from fl/fl and MD1KO mice as described above and were transfected with PPARγ siRNA (SMART pool, Dharmacon, Thermo Scientific) using Nucleofector. After 2 days of transfection, cells were treated with IL-4 or LPS as indicated.
PPARγ1 promoter cloning and luciferase reporter assays.
A 1.5-kb fragment covering the PPARγ1 proximal promoter and part of the 5′-untranslated region was PCR amplified from a PPARγ1 bacterial artificial chromosome with primers as follows: PPARγ1 forward-1 (F1), 5′-GTCTGGTACCTCTGGTGAGGATGGTTTGTA-3′; PPARγ1 reverse-1 (R1), 5′-CCGGGGCGACTCTGACCTGA-3′; PPARγ1 forward-2 (F2), 5′-GGCTGTGAGGAGCAAGGCGG-3′; PPARγ1 reverse-2 (R2), 5′-CCCTAGATCTTTGTCTGTCACACAGTCCTG-3′. The primer sets F1/R1 and F2/R2 amplify 2 overlapping fragments on the PPARγ1 promoter (see Figure 5A). The 2 PCR fragments were digested with KpnI/ApalI and ApalI/BglII, respectively. The digested fragments were ligated to pGL3-Basic at the KpnI/BglII sites to generate pGL3-PPARγ1. The constructs were confirmed by sequencing. To obtain unmethylated promoter, the reporter constructs were transformed into the dam–/dcm– E. coli strain (New England Biolabs). To obtain fully methylated reporter, constructs were incubated with 3 U/μg SssI methylase (New England Biolabs) in the presence of 160 μM S-adenosylmethionine at 37°C for 3 hours (64, 65). Methylation were confirmed by checking the resistance of reporter constructs to HpyCH4IV digestion (64). The unmethylated or methylated PPARγ1 reporter constructs were transfected into RAW264.7 cells using a SuperFect Transfection Reagent kit (Qiagen) and luciferase activity was measured using a Dual-Luciferase Reporter Assay kit (Promega).
Immunoblotting.
Tissues or cells were homogenized in a modified RIPA buffer (55–57, 59). For immunoblotting, protein lysate (50–100 μg) was resolved by SDS-PAGE, and the phosphorylation and total levels of specific proteins were measured by immunoblotting. After incubating with primary antibodies, blots were incubated with Alexa Fluor 680–conjugated anti-rabbit (catalog A-21109), anti-mouse (catalog A-21058), and anti-goat (catalog A-21084) secondary antibodies (all Invitrogen) and developed with an Odyssey Infrared Imager system (Li-COR Biosciences). Rabbit anti-IR (catalog sc-711) and goat anti–β-actin (catalog sc-1616) polyclonal antibodies were from Santa Cruz Biotechnology. Rabbit anti–IRS-1 polyclonal antibody was from Upstate (catalog 06-248). Rabbit anti–phospho-IR Tyr1162/Tyr1163 (catalog 44-804G) and anti–phospho-IRS1 Tyr612 (catalog 44-816G) polyclonal antibodies were from Invitrogen. Rabbit anti–phospho-Akt/protein kinase B (PKB) Ser473 (catalog 9271) and anti–total Akt/PKB (catalog 9272) polyclonal antibodies were from Cell Signaling Technology.
RNA extraction and reverse transcription (RT)-PCR.
RNA was extracted using TriReagent (Molecular Research Center, Inc.). The mRNA levels of target genes were quantified by quantitative real-time RT-PCR as described previously using a Stratagene Mx3000 (Agilent Technologies) (55–57, 59). Primer and probe sequences for cyclophilin were: 5′-GGTGGAGAGCACCAAGACAGA-3′ (forward), 5′-GCCGGAGTCGACAATGATG-3′ (reverse), and 5′-TCCTTCAGTGGCTTGTCCCGGCT-3′ (probe). TaqMan primer/probes for all other genes were purchased from Applied Biosystems.
Arginase activity.
Arginase activity was measured as previously described (6, 66, 67). Briefly, cells were lysed in 12.5 mM Tris-HCl (pH 7.5) containing 0.1% Triton X-100, 10 mM MnCl2, 1 μM pepstatin A, and 1 μM leupeptin. Arginase activity was measured by the addition of L-arginine substrate to 250 mM final concentration. The reaction was incubated at 37°C for 30 minutes and stopped by the addition of H2SO4 /H3PO4/H2O (1:3:7, v/v/v). After addition of 3% α-isonitrosopropiophenone (dissolved in absolute ethanol), samples were maintained in the dark for 10 minutes. Urea production was then measured by absorbance at 540 nm using a spectrophotometer and normalized to total protein content. The standard curve was prepared with increasing amounts of urea between 1.5 and 120 μg.
Bisulfite conversion and pyrosequencing.
Genomic DNA was prepared by phenol/chloroform extraction. Bisulfite conversion was performed using an EpiTech Bisulfite Kit (Qiagen). The primers that were used to amplify PPARγ1 promoter/5′-untranslated region covering CpG sites (Supplemental Figure 5A) were designed by EpiGexDX. Bisulfite-converted DNA (~1 μg) was amplified by PCR and pyrosequencing was performed by EpiGenDX.
Statistics.
Differences between groups were analyzed for statistical significance by Student’s t test, ANOVA with Fischer’s probable least-squares difference post hoc test, or ANOVA with repeated measures as appropriate.
Study approval.
The animal studies were approved by the institutional animal care and use committees of the Wake Forest University School of Medicine and Georgia State University. All animals were housed with a 12-hour light/dark cycle in a temperature-controlled facility and had free access to water and food.
Author contributions
Hang Shi and BX designed the experiments and supervised experiments and manuscript writing. XW and QC performed the experiments, analyzed/interpreted the data, and contributed to manuscript writing. LY and Huidong Shi contributed helpful suggestions and comments to the experiments and manuscript.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R01DK084172 (to Hang Shi), R01HL107500 and R01DK107544 (to B.X.), R01DK085176 (to L.Y.), American Heart Association grants 10SDG3900046 (to B.X.) and 11GRNT7370080 (to Hang Shi), and American Diabetes Association grant 7–13-BS-159 (to Hang Shi).
Footnotes
Conflict of interest: The authors have declared no conflict of interest.
Reference information:JCI Insight. 2016;1(19):e87748. doi:10.1172/jci.insight.87748.
Contributor Information
Xianfeng Wang, Email: xwang@wakehealth.edu.
Qiang Cao, Email: QCAO@GSU.EDU.
Liqing Yu, Email: lyu123@umd.edu.
Huidong Shi, Email: hshi@gru.edu.
Bingzhong Xue, Email: bxue@gsu.edu.
Hang Shi, Email: hshi3@gsu.edu.
References
- 1.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 2.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 3.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujisaka S, et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes. 2009;58(11):2574–2582. doi: 10.2337/db08-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen MT, et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem. 2007;282(48):35279–35292. doi: 10.1074/jbc.M706762200. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, et al. CD11c expression in adipose tissue and blood and its role in diet-induced obesity. Arterioscler Thromb Vasc Biol. 2010;30(2):186–192. doi: 10.1161/ATVBAHA.109.198044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57(12):3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 11.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Bouhlel MA, et al. Unlike PPARgamma, PPARalpha or PPARbeta/delta activation does not promote human monocyte differentiation toward alternative macrophages. Biochem Biophys Res Commun. 2009;386(3):459–462. doi: 10.1016/j.bbrc.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 13.Bouhlel MA, et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Charo IF. Macrophage polarization and insulin resistance: PPARgamma in control. Cell Metab. 2007;6(2):96–98. doi: 10.1016/j.cmet.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Kang K, et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7(6):485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odegaard JI, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7(6):496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäckdahl L, Bushell A, Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int J Biochem Cell Biol. 2009;41(1):176–184. doi: 10.1016/j.biocel.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 19.Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44(3):143–154. [PubMed] [Google Scholar]
- 20.Maunakea AK, Chepelev I, Zhao K. Epigenome mapping in normal and disease states. Circ Res. 2010;107(3):327–339. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 22.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Vickers MH. Developmental programming and transgenerational transmission of obesity. Ann Nutr Metab. 2014;64 Suppl 1:26–34. doi: 10.1159/000360506. [DOI] [PubMed] [Google Scholar]
- 24.Vickers MH. Early life nutrition, epigenetics and programming of later life disease. Nutrients. 2014;6(6):2165–2178. doi: 10.3390/nu6062165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan C, et al. Genome-wide screen of promoter methylation identifies novel markers in diet-induced obese mice. Nutr Hosp. 2014;30(1):42–52. doi: 10.3305/nh.2014.30.1.7521. [DOI] [PubMed] [Google Scholar]
- 26.Marco A, Kisliouk T, Weller A, Meiri N. High fat diet induces hypermethylation of the hypothalamic Pomc promoter and obesity in post-weaning rats. Psychoneuroendocrinology. 2013;38(12):2844–2853. doi: 10.1016/j.psyneuen.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Kröger H, Dietrich A, Grätz R, Wild A, Ehrlich W. The effect of tryptophan plus methionine, 5-azacytidine, and methotrexate on adjuvant arthritis of rat. Gen Pharmacol. 1999;33(2):195–201. doi: 10.1016/S0306-3623(98)00273-0. [DOI] [PubMed] [Google Scholar]
- 28.Stenvinkel P, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261(5):488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 29.Shaul ME, Bennett G, Strissel KJ, Greenberg AS, Obin MS. Dynamic, M2-like remodeling phenotypes of CD11c+ adipose tissue macrophages during high-fat diet--induced obesity in mice. Diabetes. 2010;59(5):1171–1181. doi: 10.2337/db09-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wentworth JM, et al. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes. 2010;59(7):1648–1656. doi: 10.2337/db09-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeyda M, Gollinger K, Kriehuber E, Kiefer FW, Neuhofer A, Stulnig TM. Newly identified adipose tissue macrophage populations in obesity with distinct chemokine and chemokine receptor expression. Int J Obes (Lond) 2010;34(12):1684–1694. doi: 10.1038/ijo.2010.103. [DOI] [PubMed] [Google Scholar]
- 32.Hotamisligil GS. Inflammation and endoplasmic reticulum stress in obesity and diabetes. Int J Obes (Lond) 2008;32 Suppl 7:S52–S54. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborn O, Sears DD, Olefsky JM. Fat-induced inflammation unchecked. Cell Metab. 2010;12(6):553–554. doi: 10.1016/j.cmet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 35.Patra SK, Bettuzzi S. Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2′-deoxycytidine and DNA-demethylation. Biochemistry Mosc. 2009;74(6):613–619. doi: 10.1134/S0006297909060042. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Bartlett DL, Gorry MC, O’Malley ME, Guo ZS. Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol Pharmacol. 2009;76(5):1072–1081. doi: 10.1124/mol.109.056291. [DOI] [PubMed] [Google Scholar]
- 37.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2(1):62–72. [PubMed] [Google Scholar]
- 38.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27(1):31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 39.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 40.McCabe MT, Low JA, Daignault S, Imperiale MJ, Wojno KJ, Day ML. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006;66(1):385–392. doi: 10.1158/0008-5472.CAN-05-2020. [DOI] [PubMed] [Google Scholar]
- 41.Gordon S, Hamann J, Lin HH, Stacey M. F4/80 and the related adhesion-GPCRs. Eur J Immunol. 2011;41(9):2472–2476. doi: 10.1002/eji.201141715. [DOI] [PubMed] [Google Scholar]
- 42.Rosas M, Thomas B, Stacey M, Gordon S, Taylor PR. The myeloid 7/4-antigen defines recently generated inflammatory macrophages and is synonymous with Ly-6B. J Leukoc Biol. 2010;88(1):169–180. doi: 10.1189/jlb.0809548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Alvares K, Huang Q, Rao MS, Reddy JK. Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. J Biol Chem. 1993;268(36):26817–26820. [PubMed] [Google Scholar]
- 45.Zhu Y, et al. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc Natl Acad Sci USA. 1995;92(17):7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefterova MI, et al. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30(9):2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris DL, Singer K, Lumeng CN. Adipose tissue macrophages: phenotypic plasticity and diversity in lean and obese states. Curr Opin Clin Nutr Metab Care. 2011;14(4):341–346. doi: 10.1097/MCO.0b013e328347970b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115(9):1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voisin S, Almén MS, Moschonis G, Chrousos GP, Manios Y, Schiöth HB. Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of Greek preadolescents. Eur J Hum Genet. 2015;23(5):654–662. doi: 10.1038/ejhg.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall E, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332(6035):1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Wang X, Liu D, Yu L, Xue B, Shi H. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol Endocrinol. 2014;28(4):565–574. doi: 10.1210/me.2013-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Yang Z, Xue B, Shi H. Activation of the cholinergic antiinflammatory pathway ameliorates obesity-induced inflammation and insulin resistance. Endocrinology. 2011;152(3):836–846. doi: 10.1210/en.2010-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z, et al. Regulation of insulin and leptin signaling by muscle suppressor of cytokine signaling 3 (SOCS3) PLoS ONE. 2012;7(10):e47493. doi: 10.1371/journal.pone.0047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, et al. The full capacity of AICAR to reduce obesity-induced inflammation and insulin resistance requires myeloid SIRT1. PLoS ONE. 2012;7(11):e49935. doi: 10.1371/journal.pone.0049935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;257(4):2041–2048. [PubMed] [Google Scholar]
- 59.Yang Z, Kahn BB, Shi H, Xue BZ. Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem. 2010;285(25):19051–19059. doi: 10.1074/jbc.M110.123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116(11):3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H, Tzameli I, Bjørbaek C, Flier JS. Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J Biol Chem. 2004;279(33):34733–34740. doi: 10.1074/jbc.M403886200. [DOI] [PubMed] [Google Scholar]
- 62.Brown AL, et al. Omega-3 fatty acids ameliorate atherosclerosis by favorably altering monocyte subsets and limiting monocyte recruitment to aortic lesions. Arterioscler Thromb Vasc Biol. 2012;32(9):2122–2130. doi: 10.1161/ATVBAHA.112.253435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thu KL, Vucic EA, Kennett JY, et al. Methylated DNA immunoprecipitation. J Vis Exp. 2009;(23):935. doi: 10.3791/935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shore A, Karamitri A, Kemp P, Speakman JR, Lomax MA. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia. 2010;53(6):1164–1173. doi: 10.1007/s00125-010-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Ara RM, et al. Diagnostic performance of arginase activity in colorectal cancer. Clin Exp Med. 2002;2(1):53–57. doi: 10.1007/s102380200007. [DOI] [PubMed] [Google Scholar]
- 67.Munder M, Eichmann K, Morán JM, Centeno F, Soler G, Modolell M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol. 1999;163(7):3771–3777. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.