Abstract
The broad diversity of cell types within vertebrates arises from a unique genetic blueprint by combining intrinsic cellular information with developmental and other extrinsic signals. Lying at the interface between cellular signals and the DNA is chromatin, a dynamic nucleoprotein complex that helps mediate gene regulation. The most basic subunit of chromatin, the nucleosome, consists of DNA wrapped around histones, a set of proteins that play critical roles as scaffolding molecules and regulators of gene expression. Growing evidence indicates that canonical histones are commonly replaced by protein variants prior to and during cellular transitions. Here we highlight exciting new results suggesting that histone variants are essential players in the control of cellular plasticity during development and in the adult nervous system.
Keywords: Histone variants, cellular plasticity, development, neuron, brain, epigenetics
Since the groundbreaking discovery of a distinct isoform of histone H3 in calf thymus [1], numerous histone variants have been discovered in eukaryotes [2, 3]. Histone variants are non-allelic isoforms of the canonical histone proteins H1, H2A, H2B, H3, and H4 [3] (see Glossary). Whereas canonical histones are generally produced in coordination with DNA replication from mRNAs containing short 3’- stem loop tails [4], many variants are translated from mRNAs with conventional poly-A tails outside of S-phase and incorporated into the chromatin, often with help of special chaperones [5]. The synthesis of histone variants outside of S-phase and in specific tissues enables them to perform specialized functions, such as DNA repair (H2A.X) [6], conversion of chromatin to nucleoprotamine during spermatogenesis (TSH2B) [7], and kinetochore assembly (CENP-A) [8]. Moreover, exciting new studies have revealed prominent roles for a subset of histone variants in regulating cellular plasticity, broadly defined as a cell’s capacity to undergo changes in its structural or functional properties.
Biochemical and structural alterations to chromatin are associated with an everexpanding array of histone post-translational modifications (PTMs; Box 1) [9, 10]. One might therefore wonder what additional features can be provided by the replacement of canonical histones with histone variants. Clearly, canonical histones are insufficient to sustain life in metazoans, perhaps due to the evolution of specific developmental and physiological processes in these organisms, which may require unique chromatin landscapes for specialized gene expression programs. At a biochemical level, some variants, such as macroH2A and H2A.Z, have highly divergent polypeptide sequences, which enable major changes in chromatin structure and function [11]. Other variants, such as H3.3, provide more subtle differences that can nevertheless cause critical changes in post-translational modifiability [12] and in interactions with chaperones [13] and chromatin ‘readers’ [14]. In addition to increasing functional diversity, histone variants also offer a means to reduce it. For example, the transient removal of H3.3 from the maternal genome following fertilization may enable the global resetting of pre-existing PTMs to create a totipotent embryo [15]. Whether histone variants provide additional yet uncovered functions is an exciting question.
Box 1. Histone post-translational modifications (PTMs).
Histones are subject to a wide array of PTMs (Table I) that are generated and removed by specific modifiers, which are precisely regulated in their genomic localization and activity (reviewed in [10, 109]. To date, only a fraction of known PTMs have been analyzed with respect to their genomic localization and fewer still are understood on a functional level. Of the PTMs that have been analyzed extensively, most appear to be enriched within specific gene regions. Moreover, PTM enrichment is strongly correlated with the transcriptional status of a gene. In some cases, PTMs are known to influence chromatin structure directly and/or through recruitment of effector molecules, and knockout of PTM modifiers or effectors have revealed severe developmental defects. Despite our incomplete understanding of PTMs, it is clear that they play pivotal roles in organizing chromatin and regulating gene expression during development and throughout life.
Despite decades of study, the functions and expression patterns of many vertebrate histone variants remain poorly defined due to high levels of sequence identity between histone species. Major advances in high-resolution and quantitative chromatin analysis [16, 17], combined with loss of function analysis [18, 19], have expanded inroads for the study of chromatin. In particular, advances have revealed that a set of vertebrate histone variants play critical roles in developmental transitions [20], when cells lose plasticity and acquire defined and stable identities [21]. In addition, critical roles have emerged for histone variants during cellular transitions that occur throughout life, particularly in the nervous system, where the modulation of gene expression drives experience-dependent cellular changes [22, 23]. Here we review recent studies of the roles played by histone variants in regulating cellular plasticity during early development and within the nervous system.
Histone variants are key regulators of developmental plasticity
Programmed differentiation, which begins at the earliest stages of development in the totipotent zygote and pluripotent embryonic stem cells (ESCs) [24] and continues throughout development and into adulthood [25], involves the coordinated activation of lineage-specific genes and the stable repression of genes underlying developmental multipotency. The balance between stem cell self-renewal and differentiation, as well as the process of differentiation itself, are tightly controlled at the level of chromatin [26]. Investigations of these processes have historically focused on the functions of transcription factors and PTMs. However, intriguing recent evidence indicates that critical aspects of developmental plasticity in the early embryo may be regulated by histone variants.
TH2A and TH2B help establish developmental plasticity in the early embryo
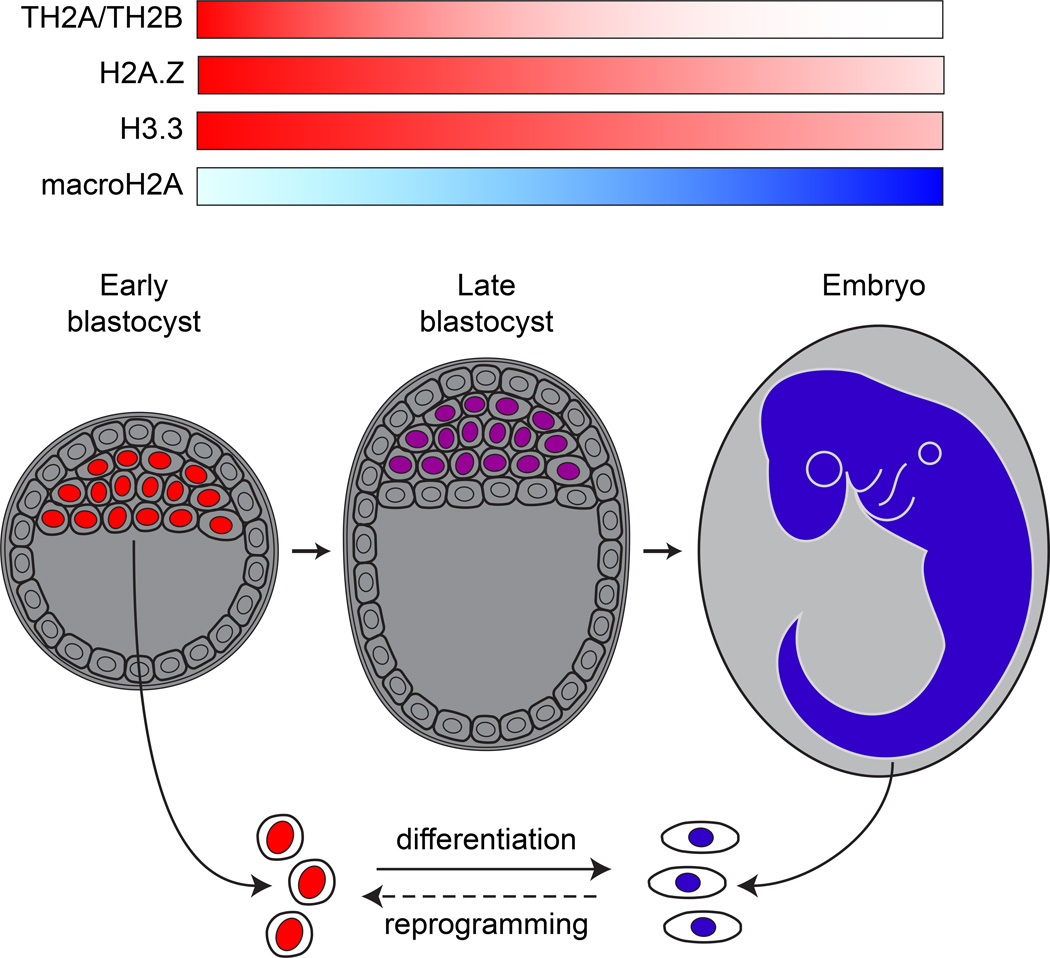
Histone variants TH2A and TH2B, which differ from canonical H2A and H2B by 15 and 16 amino acids, respectively [2], were first identified in testis tissue [27, 28]. Unexpectedly, high levels of these variants were also recently found in oocytes and zygotes, with decreasing levels observed as differentiation proceeds to the blastocyst stage [29] (Fig. 1). Interestingly, oocytes derived from TH2A/TH2B knockout mice showed a significantly reduced ability to reach the blastocyst stage following fertilization compared to wild type counterparts as well as defects in paternal genome activation [29]. These observations, together with findings that endogenous levels of TH2A and TH2B increase dramatically during ‘reprogramming’ of somatic cells to a pluripotent state, suggest that TH2A and TH2B might play a role in the induction of pluripotency [29]. Indeed, coexpression of TH2A and TH2B, together with the oocyte-specific histone chaperone NPM, enhanced the efficiency of reprogramming mouse embryonic fibroblasts (MEFs) 18-fold [29]. Moreover, reprogramming efficiency was reduced six-fold in MEFs obtained from TH2A/TH2B knockout mice, further evidence that endogenous TH2A and TH2B facilitate the induction of pluripotency [29].
Figure 1.
Expression of histone variants in embryonic cells during early development. ESCs within the inner cell mass of the early blastocyst have high levels of TH2A, TH2B, H2A.Z, and H3.3, and low levels of macroH2A. By contrast, differentiated cells have relatively low levels of TH2A, TH2B, H2A.Z, and H3.3, and high levels of macroH2A. Intermediate expression of both variants is extrapolated for epiblast stem cells of the late blastocyst, as depicted. Schematic depiction of histone variant expression levels are based on [33, 54, 55, 72].
How do TH2A and TH2B facilitate reprogramming? DNaseI sensitivity assays revealed that expression of the variants during reprogramming promotes an open chromatin structure [29], a hallmark of pluripotency [30]. Further, chromatin localization experiments showed that the variants are distributed widely throughout the genome of MEFs undergoing reprogramming, suggesting that their effects are genome-wide [29]. Future experiments will be needed to understand the mechanism by which amino acid differences within TH2A and TH2B promote an open chromatin structure and the role of this structure in establishing pluripotency.
H2A.Z is an essential facilitator of developmental plasticity
First identified in the ciliate Tetrahymena thermophila [31], H2A.Z is highly conserved among eukaryotes [32]. Early studies of H2A.Z function in mice showed that it is strictly required during embryogenesis, as H2A.Z-knockout embryos fail to proceed past ~E4.5. These observations, together with recent findings that H2A.Z expression peaks in ESCs and decreases upon differentiation [33](Fig. 1), suggest that the developmental failure of H2A.Z knockouts may be due in part to an inability of ESCs to self-renew. Indeed, depletion of H2A.Z in cultured ESCs has been shown to cause aberrant down-regulation of genes involved in pluripotency [34] and upregulation of genes involved in differentiation [34–36], and impaired colony morphology [34]. Consistent with these findings, H2A.Z-depleted ESCs showed an impaired ability to differentiate in vitro [34–36], to form normal embryos in a tetraploid complementation assay, and to contribute to chimeras following blastocyst injection [35]. Thus, H2A.Z appears to play a critical role both in establishing pluripotency and in facilitating differentiation.
How does H2A.Z participate in establishing ESC pluripotency? Genomic localization studies in ESCs have shown that H2A.Z is enriched within the promoter regions of genes displaying the ‘active’ H3K4me3 mark [34, 37], including a subset that also contain the ‘silent’ H3K27me3 mark [34, 35, 37] and are thus ‘bivalent,’ meaning silent but poised for activation [38] (Box 1; Fig. 2A). In support of the hypothesis that H2A.Z is required to create or maintain these domains, H2A.Z-depleted ESCs exhibited reduced levels of the H3K27me3 and H3K4me3 modifications and their respective deposition machinery [34, 35]. Interestingly, H2A.Z-depleted ESCs also exhibited a reduced genome-wide occupancy of Oct4 [34], a transcription factor that plays a crucial role in ESC pluripotency [39], an effect that may contribute to the ensuing impairment of ESC self-renewal [34, 35]. Thus, H2A.Z appears to play a critical role in organizing the chromatin in a manner compatible with pluripotency.
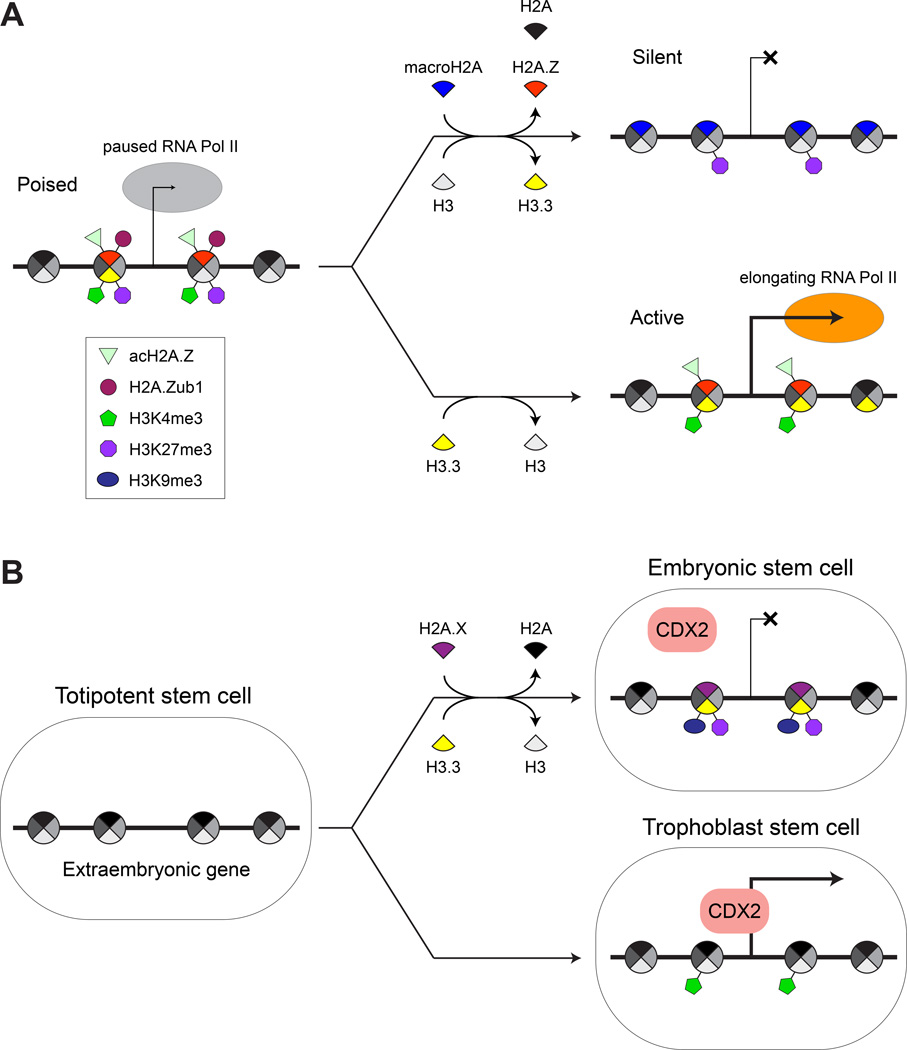
Figure 2.
Models for histone variant and PTM dynamics in ESC differentiation and repression of extraembryonic genes. A. In ESCs, lineage specific differentiation genes are frequently marked by H2A.Z and bivalent chromatin PTMs, and are therefore silent but poised for future activation or stable repression, depending on the differentiation pathway. Evidence indicates that stable gene repression entails the replacement of H2A and H2A.Z with macroH2A and H3.3 with H3, and the removal of active PTMs (e.g., H3K4me3 and acH2A.Z). Gene activation entails the replacement of canonical H3 with H3.3 in gene bodies and the removal of repressive PTMs (e.g., H3K27me3 and H2A.Zub1). B. The inhibition of extraembryonic gene expression in ESCs appears to require the deposition of histone variants H2A.X and H3.3, which in turn enable the repressive PTMs H3K9me3 and H3K27me3, respectively. Recent evidence suggests that these pathways are altered in trophoblast stem cells, thus enabling the activation of extraembryonic genes.
In turn, how does H2A.Z facilitate ESC differentiation? Studies of the retinoic acid (RA)-induced differentiation of ESCs into neural progenitors showed that H2A.Z occupies the regulatory regions of RA-responsive genes prior to differentiation and is removed upon RA activation [40]. Furthermore, depletion of H2A.Z was found to impair RA-induced binding of RA-receptor-alpha within RA-responsive genes [34]. Similar analyses of growth factor-induced differentiation of ESCs to the endoderm/hepatic lineage demonstrated that, prior to differentiation, H2A.Z is enriched within gene regulatory regions that, upon differentiation, become bound by the transcription factor Foxa2 and depleted of nucleosomes [36]. The reductions in H2A.Z levels observed upon ESC differentiation [33] correlate with dramatic reductions in chromatin openness [30], suggesting a possible causal relationship. In support of this hypothesis, chromatin nuclease accessibility analyses have revealed a strong association between H2A.Z occupancy and chromatin openness in ESCs [34, 36]. Moreover, the depletion of H2A.Z in ESCs results in chromatin changes similar to those observed upon ESC differentiation [34, 36], including increased global nucleosome occupancy and reduced chromatin accessibility [34].
Much attention has been devoted to the mechanism by which H2A.Z affects chromatin structure [41]. Early structural studies indicated that H2A.Z’s more extensive ‘acidic patch,’ a domain on the surface of the histone octamer, may strengthen intra-fiber interactions, thereby enhancing local compaction, but weaken inter-fiber interactions, thereby reducing global compaction [42, 43]. Indeed, substitution of H2A.Z’s acidic patch with an H2A-like counterpart in ESCs was found to cause increased nucleosome turnover and defects in the silencing of bivalent genes, indicating that this domain is important for maintaining a specialized chromatin structure [44]. H2A.Z also appears to affect nucleosome stability, but in a highly context-dependent manner. In ‘homotypic’ nucleosomes, which contain two copies of H2A.Z, the variant’s distinct L1 loop was anticipated to enhance nucleosome stability [42, 45], a prediction supported by some in vitro chromatin reconstitution studies [46]. However, recent studies in Drosophila cells indicate that H2A.Z homotypic nucleosomes may actually enhance transcriptional elongation via weakened docking interactions between H2A.Z and (H3/H4)2 tetramers [47, 48]. Interestingly, ‘heterotypic’ nucleosomes, in which L1 loop clashes between H2A.Z and H2A were predicted to destabilize nucleosomes, are nevertheless enriched in gene regulatory regions of mouse trophoblast stem cells [49], where they are thought to enhance chromatin accessibility [45]. Moreover, unstable ‘hybrid’ nucleosomes, containing both H2A.Z and variant H3.3, have been found in similar regions in human cells [50], leading to the suggestion that heterotypic and hybrid nucleosomes may be one and the same [49]. Aside from nucleosome context, H2A.Z appears to affects chromatin structure via its PTMs. Consistent with their opposing effects on chromatin accessibility [43, 51], acetylation on the N-terminal tail [34, 37] and ubiquitination on the C-terminal tail [37] have been found within active and silent genes, respectively, both in ESCs and differentiated cells (Box 1). Intriguingly, bivalent chromatin is reportedly enriched with H2A.Z containing both modifications [37]. Considering the importance of chromatin context, it is perhaps unsurprising that studies have reached differing conclusions regarding H2A.Z’s affects on chromatin structure [43, 45, 48]. Nevertheless, a consensus appears to be emerging that H2A.Z facilitates specialized chromatin environments that are critical for cellular plasticity.
Histone variant H3.3 is a key facilitator of developmental plasticity
H3.3, first characterized in mammalian tissues nearly four decades ago [52], remains intensively studied today. H3.3 differs from the canonical H3 histones by four or five amino acids and is expressed throughout the cell cycle [53]. Like TH2A and TH2B, H3.3 is expressed in oocytes from maternal transcripts and from zygotic transcripts after the two-cell stage [54] (Fig. 1), when it appears to play a critical role in development. In Xenopus, depletion of H3.3 or its chaperone HIRA caused developmental arrest and the misexpression of mesodermal differentiation genes at gastrulation, which follows a peak expression of H3.3 (Fig. 1) [55]. In mice, inactivation of both H3.3-encoding genes, H3f3a and H3f3b, uncovered a strict requirement for the variant in gametogenesis [19]. Less severe phenotypes were observed upon inactivation of a subset of the four alleles encoding H3.3, including reduced viability, delayed growth, infertility, and impaired chromosome segregation [19, 56, 57]. Providing further insights into the role of the variant in development, H3.3 depletion in fertilized mouse zygotes resulted in developmental arrest at the morula stage, along with chromosome over-condensation and dramatically reduced levels of the H4K16Ac and H3K36me2 PTMs [58], which together appear to inhibit chromatin compaction [59, 60].
Genomic localization studies in ESCs have found that H3.3 is enriched at active genes, such as those encoding the pluripotency factors Nanog and Oct4, and the promoters of bivalent genes [61] (Fig. 2A). Upon cellular differentiation, H3.3 appears within the bodies of bivalent genes that become activated, but disappears from pluripotency genes and regulatory regions of bivalent genes that become silenced [61]. This raises the question of whether H3.3 is an important component of active and/or bivalent domains. Indeed, H3.3-depleted ESCs were found to have ~50% fewer bivalent promoters due to lower levels of H3K27me3, as well as reduced nucleosome turnover at active and bivalent promoters [62]. Interestingly, H3.3-depleted ESCs were recently found to exhibit reduced silencing of endogenous retroviral elements, underscoring the complexity of H3.3 function in pluripotent cells [63].
Does H3.3 help to establish pluripotency? A study involving reprogramming of differentiated cells to pluripotency by somatic cell nuclear transfer (SCNT) to Xenopus oocytes found that the H3.3 chaperone HIRA is necessary for changes in transcription that are associated with reprogramming [64]. Moreover, SCNT into enucleated, H3.3-depleted mouse oocytes resulted in developmental arrest at the late morula or early blastocyst stage following parthenogenetic activation, and impaired activation of pluripotency-associated genes [54]. These findings indicate that H3.3 is an important facilitator of cellular plasticity.
The similarities of H3.3 and H2A.Z function in establishing and/or maintaining chromatin domains that facilitate developmental plasticity are striking and, as mentioned above, may reflect the formation of hybrid H3.3/H2A.Z nucleosomes [49, 50]. How these histone variants may otherwise influence or be influenced by each other or other variants is an exciting area of ongoing research.
H2A.X and H3.3 inhibit extraembryonic plasticity in ESCs
To maintain proper developmental potential, ESCs must be strictly prevented from giving rise to extraembryonic tissues, through molecular mechanisms that are not well understood. Recent studies have found that, surprisingly, a key component of this process is H2A.X, a histone variant best known for its role in the response to DNA damage [6, 65]. Within ESCs, H2A.X has been found specifically targeted to genes expressed in extraembryonic tissues [66] (Fig. 2B). Similar H2A.X localization patterns were found in reprogrammed pluripotent stem cells that were capable of fully supporting embryonic development in vivo [66, 67], but not in trophoblast stem cells, which give rise to extraembryonic tissues, or MEFs [66]. Moreover, ESCspecific H2A.X localization sites were found to overlap strongly with binding sites for CDX2 [66], a key transcription factor in extraembryonic lineage specification. Deletion of the gene encoding H2A.X in ESCs was found to cause the aberrant upregulation of genes controlled by extraembryonic lineage specification factors, concomitant with reduced levels of H3K9me3, a PTM associated with constitutive heterochromatin, at the binding sites of these factors. Remarkably, tetraploid complementation experiments found that H2A.X-knockout ESCs contribute aberrantly to the trophectoderm layer of blastocysts at a significantly greater frequency than control ESCs [66]. These results suggest that H2A.X plays a key role in restricting access to extraembryonic lineages by ESCs.
Strikingly similar results have been observed in H3.3-depleted ESCs, which likewise have been found to exhibit an aberrant upregulation of transcripts specific to trophectoderm and an enhanced propensity to form trophectoderm tissue in teratoma xenograft and embryoid body formation experiments [62]. Thus, like H2A.X, H3.3 appears necessary for establishing and/or maintaining a chromatin organization that restricts the developmental potential of ESCs to embryonic lineages. The molecular mechanisms by which these variants function in this capacity represent a fascinating area of future research.
MacroH2A is a key inhibitor of developmental plasticity
The loss of plasticity that accompanies embryogenesis must be stable in order to maintain proper tissue differentiation and function. Mechanisms that lock cells into a differentiated state have long been known to be controlled at the level of chromatin [26] and recent studies have implicated the histone variant macroH2A as an important component of this process. First identified in rat liver [68], vertebrate macroH2A consists of two ~80% identical isoforms, macroH2A.1 and macroH2A.2 [68, 69]. MacroH2A contains an N-terminal domain that is similar to canonical H2A and a large C-terminal “macrodomain” that protrudes from the nucleosome [11] and is thought to bind ligands, compact chromatin, and inhibit transcription [43]. Studies of macroH2A in zebrafish versus mice have come to disparate conclusions regarding its importance during development. While profound morphological defects were found in macroH2A.2-depleted zebrafish embryos, particularly within the brain [70], mice lacking both macroH2A isoforms appear to have only slight reductions in body size, perinatal survival, and fertility [18]. These differences may reflect distinct roles for macroH2A in fish and mice or may be due to the fact that defects in mice are masked by compensatory mechanisms [71].
Recent immunofluorescence analysis of macroH2A’s expression in mouse embryos revealed that the variant is expressed with a temporal pattern nearly opposite to that of H2A.Z [72] (Fig. 1). While low and undetectable levels of macroH2A.1 were observed in pluripotent stem cells at E4.5 and E6.5, respectively, high levels were observed throughout all differentiated cells by E9.5 [72]. Similarly, levels of macroH2A have been observed to rise markedly upon differentiation of cultured pluripotent cells [33, 71, 73–75], as well as adult neural stem cells [73]. Moreover, high levels of macroH2A appear to be a general feature of differentiated cells [33, 71, 73, 74].
Does the accumulation of macroH2A during differentiation contribute to the various chromatin modifications that prevent the reversion of somatic cells back to pluripotency [26, 76]? Supporting this hypothesis, strong down-regulation of the variant has been observed upon nuclear reprogramming [33, 72, 73](Fig. 1). Moreover, a screen for factors predictive of incomplete X-chromosome reactivation following SCNT into Xenopus oocytes identified macroH2A as the most critical [77]. Consistent with these results, depletion of macroH2A in neural stem cells [72], dermal fibroblasts [33], and keratinocytes [73] significantly enhanced reprogramming efficiency via the overexpression of pluripotency factors. Moreover, embryoid bodies derived from reprogrammed macroH2A-deficient cells showed an aberrant propensity to revert back to pluripotency [33]. Complementing these results, overexpression of macroH2A was found to strongly inhibit reprogramming [33, 72, 73].
MacroH2A’s identified function as a barrier to cellular plasticity might predict a possible role in preventing cancer, which can be characterized as a state of dedifferentiation [78]. Indeed, the levels of both macroH2A.1 and macroH2A.2 have been found to be strongly predictive of survival from numerous cancer types [79–82]. Moreover, macroH2A-depleted low-malignancy melanoma cells exhibited enhanced tumor growth, cell motility, and metastasis, whereas overexpression of macroH2A in highly malignant cells had the opposite effects [80].
How does macroH2A block cellular plasticity? Building on earlier findings that showed macroH2A’s selective localization to heterochromatic regions [83], recent high-resolution analyses have found that the variant is enriched at silent loci containing the H3K27me3 PTM [33, 73] and depleted at actively-transcribed loci containing H3K27Ac [33], H3K36me3, H3K4me3, and RNAPII [73]. Consistent with these findings are observations that macroH2A occupancy correlates inversely with gene expression [70, 73, 84, 85]. In differentiated cells, macroH2A has been localized to bivalent [73] and pluripotency promoting genes (e.g. Oct4 and Nanog) [33, 73]. Thus, macroH2A appears to function via the occupancy of silent chromatin domains (Fig. 2A).
Histone variants are regulators of neuronal plasticity
Cellular plasticity is most conspicuous during early development, but it is not restricted to this period. Indeed, life-long cellular plasticity enables multipotent stem cells to differentiate and integrate into mature tissues [25] and permits post-mitotic cells to functionally adapt based on changing environments. Such cellular plasticity is particularly salient to neuronal function. Early insights into the importance of neuronal plasticity for proper brain function came from seminal experiments demonstrating the critical role of visual stimulation of the two eyes during perinatal development for the proper organization of ocular dominance columns in the visual cortex [86]. Subsequently, a number of experience-dependent changes in cellular composition and connectivity have been identified throughout the vertebrate nervous systems, both in early life [87] and adulthood [88]. These changes entail stimulus-dependent transcriptional modulation [22, 23], mediated largely at the level of chromatin [89, 90]. Intriguingly, several histone variants, including H3.3, H2A.Z, macroH2A, and H2A.X, have been shown to accumulate in post-mitotic neurons [91]. Here we review recent evidence suggesting that histone variants, including several yet poorly characterized [2] may play important roles in experience-dependent neuronal plasticity.
H3.3 facilitates activity-dependent neuronal gene expression and plasticity
The mechanisms by which stimuli regulate neuronal gene expression, although known to involve changes in chromatin, are incompletely understood [89, 90]. A potential role for H3.3 in this process was recently investigated indirectly via the activity-dependent modulation of its incorporation by DAXX [92]. DAXX is an H3.3 chaperone [93, 94] found in abundance within neuronal nuclei and enriched near regulatory regions of activity-responsive genes [92]. Comparison of the chromatin occupancy of H3.3 in neurons either lacking or expressing DAXX suggested that the chaperone is required for the activity-dependent loading of H3.3 within regulatory regions of the immediate early genes Bdnf and Fos, which play diverse and critical roles in activity-dependent neuronal plasticity [23], as well as for the proper expression of these genes [92].
A direct investigation of H3.3’s role in neuronal plasticity was recently made through analysis and manipulation of the variant’s dynamics in cultured neurons and in living mice (Maze et al., 2015). In the neurons of adult mice, H3.3 was found to remain highly dynamic, with >30% of the total H3.3 pool replaced over a period of four weeks, despite its accumulation to near saturating levels (Maze et al., 2015). Interestingly, the turnover of H3.3 was found to depend upon neuronal activity, transcription of the H3f3b (but not H3f3a) gene, and incorporation by the euchromatic chaperone HIRA (Maze et al., 2015). Remarkably, H3.3 dynamics were found to be highly correlated with late-responsive activity-dependent gene expression in embryonic neurons (Maze et al., 2015). Moreover, nascent-H3.3 knockdown experiments revealed that turnover of the variant plays a crucial role in regulating activity-dependent gene expression, synaptic connectivity, and experience-dependent changes in behavior (Maze et al., 2015). The precise mechanism by which H3.3 turnover facilitates neuronal gene expression and plasticity, as well as the multitude of other factors that are likely involved in these processes remain to be determined. Nevertheless, H3.3 incorporation and turnover appear to represent key components of activity-dependent neuronal plasticity.
Underscoring the prominent role of H3.3 in brain development and function, mutations of Lys27 and Gly34 within the N-terminal tail of H3.3 (and, less frequently, H3.1) have recently been implicated in a large fraction of human pediatric high-grade astrocytomas, which are particularly aggressive and lethal brain tumors [95, 96]. Mechanistic studies of one identified mutant of H3.3, Lys27 to methionine, have uncovered a remarkable mechanism in which the mutation dramatically inhibits Polycomb Repressive Complex 2 (PRC2) activity and, consequently, the methylation of Lys27 residues within non-mutated H3 histones [97]. These and related recent findings will undoubtedly lead to new approaches for treating this deadly cancer.
H2A.Z regulates fear memory
Neuronal plasticity associated with the formation and storage of memories is a hallmark of vertebrate nervous systems [88, 98]. A potential role for H2A.Z in this process was recently investigated following initial observations that levels of the variant are strongly reduced in the pyramidal cell layer of the CA1 region of the hippocampus 30 minutes after mice received a foot-shock while exploring a novel test chamber [99]. Remarkably, short-hairpin RNA mediated H2A.Z depletion within the hippocampus was shown to cause significantly enhanced fear memory, both 24 hours and 30 days following fear conditioning [99]. Moreover, similar long-term memory enhancements were observed when H2A.Z was depleted in the prefrontal cortex, an area involved in long-term memory storage [99]. Although the precise mechanism by which H2A.Z enhances the consolidation of fear memory (or inhibits fear extinction) remains to be determined, this study opens a fascinating window onto a role for this variant in learning and memory.
H2B.E shapes the olfactory sensory neuron population based on activity
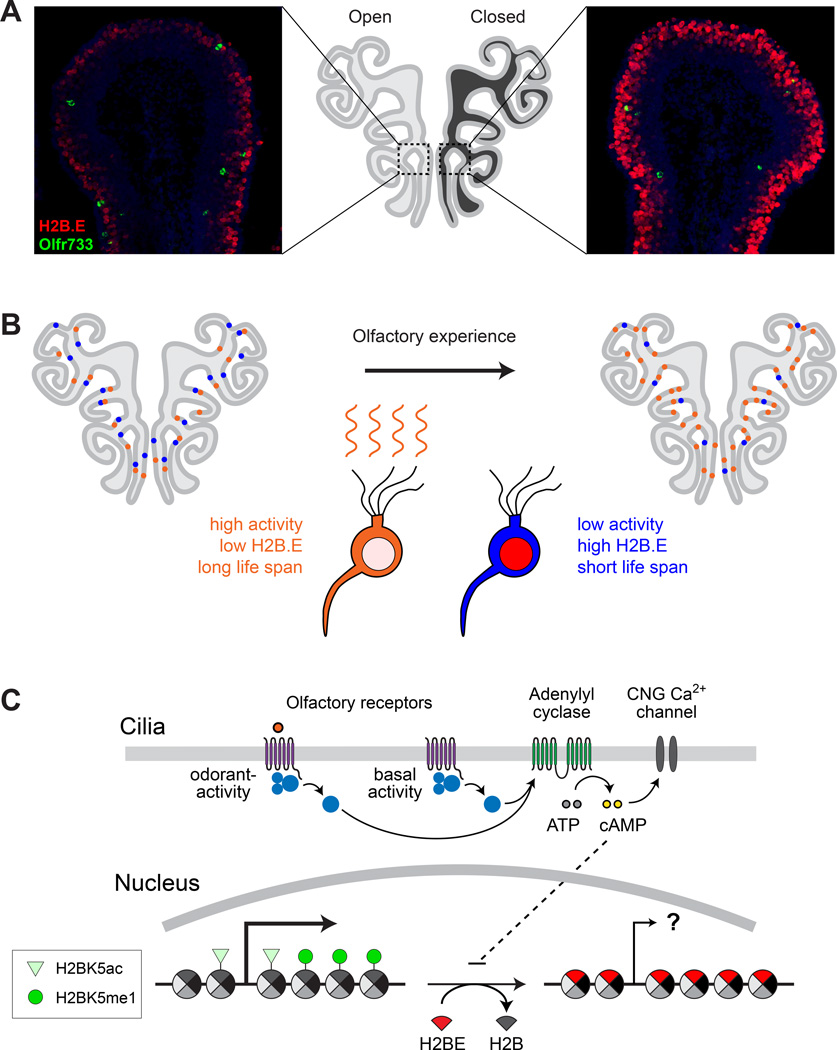
In addition to connectivity and function, plasticity of the nervous system can occur through changes in the composition of neuronal populations that undergo turnover in adults [100]. In mammals, these populations include olfactory sensory neurons (OSNs) [101], whose longevities are regulated in part by olfactory activity [102] (Box 2). Initial indications that a histone variant might participate in this regulation came from the discovery in the mouse of an olfactory-specific H2B variant called H2B.E that is expressed in an activity-dependent manner [103]. Levels of H2B.E appear heterogeneous among OSNs of the olfactory epithelium, but stereotyped according to the identity of the olfactory receptor (OR) gene expressed by a given OSN. Remarkably, olfactory deprivation by unilateral naris occlusion caused the widespread elevation of H2B.E in OSNs deprived of activity (Fig. 3A), while odor stimulation reduced H2B.E levels in OSNs expressing the corresponding OR genes [103]. These results indicate that the level of H2B.E within a given OSN is inversely tied to its level of neuronal activity (Fig. 3B). Analyses of H2B.E-knockout and - overexpressing mice found the average OSN life span to be significantly longer in H2B.E-knockouts and shorter in H2B.E-overexpressing mice, providing direct evidence that the H2B.E variant regulates OSN longevity [103]. Gene expression analysis further revealed significant and opposing changes in OR expression frequencies in each mutant, indicating that changes in neuronal longevity alter the representation of OSN subtypes [103]. Thus, a lack of OR stimulation appears to drive the incorporation of H2B.E, shortening the life span and reducing the abundance of inactive OSNs (Fig. 3B).
Box 2. Mammalian olfactory sensory neurons (OSNs).
OSNs, which reside in the main olfactory epithelium (MOE), are the primary chemosensory detectors of volatile odorants in mammals. Detection of olfactory information by OSNs mediates behavioral processes that are critical for life, including the avoidance of predators, identification of food, and mating. The following are properties of mammalian OSNs that are relevant to this review (for further review of the mammalian main olfactory system, see [112]):
Olfactory receptor (OR) gene expression. In mammals, each OSN expresses a single OR protein from ~1000 different OR genes (in mice; ~400 in humans) that are located in clusters within the genome. The identity of the OR that an OSN expresses is controlled through a remarkable process known as “OR choice,” which results in the selection and stable expression of a single OR allele for the lifetime of the neuron [113]. The identity of the chosen OR protein determines the odorants to which the OSN responds as well as the precise position within the olfactory bulb to which its axon projects (see below).
OSN turnover and organization in the MOE. The MOE is organized in a laminar structure, with neural stem cells located on the basal aspect of the epithelium and mature sensory neurons located near the apical aspect. Immature neurons, which are derived from stem cells, undergo OR choice and then migrate apically as they mature. Mature OSNs live for finite and non-uniform periods of time that have been reported to depend in part on levels of olfactory stimulation [102, 103]. Neurogenesis within the MOE occurs throughout adulthood to maintain an approximately constant number of OSNs [101].
Olfactory activity-dependent gene expression. Binding of odorants to OR proteins results in a signal transduction cascade that entails the sequential production of cyclic adenosine-monophosphate (cAMP), influx of calcium, and neuronal depolarization. In addition to their roles in mediating neuronal depolarization, OR-stimulated cAMP and calcium signals control gene expression programs, which are critical for axon guidance, refinement, and cell survival. Importantly, OR-derived cAMP signaling appears to occur in the absence of odorant stimulation due to spontaneous OR activation, and vary according to OR identity.
Figure 3.
The role of histone variant H2B.E in the activity-dependent plasticity of the olfactory sensory neuron population. A. H2B.E levels are regulated by olfactory activity, as demonstrated by the effects of unilateral naris occlusion for 10 days on H2B.E levels in MOE sections. Colocalization of Olfr733+ neurons (green cells) allows comparison of H2B.E levels in specific neuron subtypes (adapted from [103]). B. Model for the effects of neuronal activity on H2B.E level, lifespan, and neuronal representation in the MOE. Neurons that are active (orange) incorporate low levels of H2B.E and have longer lifespans, on average, compared to neuron types that are inactive (blue). Over time, active neurons increase in abundance within the MOE, while inactive decrease. C. H2B.E replacement of canonical H2B over time. The PTMs shown were found to be absent or substantially reduced on H2B.E [103]. The consequences of H2B.E replacement on cellular transcription are unknown, but the strong association of H2BK5me1 and H2BK5ac with active transcription suggests that H2B.E may negatively affect transcription through replacement of canonical H2B and altered post-translational modifiability.
Although H2B.E differs from canonical H2B at only five amino acid positions, studies of H3.3 demonstrate that subtle differences can profoundly affect a histone’s functional properties. A possible clue to the functional significance of H2B.E’s differences came from the discovery of dramatically reduced modifiability at H2B.E lysine 5 [103], a position in canonical H2B whose monomethylation and acetylation are strongly associated with transcriptional activity [104] (Box 1). Thus, H2B.E accumulation may function to dampen transcription (Fig. 3C), a hypothesis supported by gene expression analyses of H2B.E-knockout and -overexpressing mice [103].
What is the physiological function of H2B.E? Experience-dependent and H2B.E-mediated changes in the OSN population may facilitate adaptation of an individual’s OR repertoire to its specific olfactory environment and thus enhance the performance of tasks that are crucial for survival, such as finding food and avoiding predators. This type of adaptation may be uniquely important in the olfactory system, which, unlike other sensory systems, must process information from a near limitless array of complex odors [105] that can vary dramatically from one environment to the next.
Concluding remarks
Cellular changes require enhanced plasticity during periods of transition, followed by reduced plasticity to stabilize cellular changes. The involvement of histone variants in regulating cellular plasticity suggests that their functional properties may be particularly well suited to this purpose. The high evolutionary conservation of variants such as H2A.Z may reflect that their functional properties cannot be readily achieved by other means, while rapidly evolving variants such as H2A.B and H2A.L, which are involved in spermatogenesis [3], may reflect a need for rapid changes in chromatin functionality in systems that are under intense evolutionary pressure. The newly-discovered primate-specific variants H3.X and H3.Y may represent additional examples of the importance of rapidly generating new functionality [106]. The prospect of discovering additional variants with specialized functions seems likely considering the large number that remain to be characterized [2]. Understanding the function of these variants will require detailed analyses of the cell types and conditions under which they are expressed. Such efforts will benefit from advances in technologies that enable the detection of subtle sequence differences at the single cell level, including high-throughput sequencing [107], quantitative proteomics [16], and in situ hybridization [108]. In addition, the development and application of methods that enable analyses of histone localization and dynamics as well as the isolation of genetically-defined populations of nuclei [17] will greatly facilitate future discoveries of the functions of histone variants in cellular plasticity and beyond. Many open questions still remain (Box 3).
Box 3. Outstanding Questions.
What are the expression patterns and functions of the many histone variants encoded within mammalian genomes that have yet to be characterized?
Will uncharacterized histone variants be associated with new functions, particularly during development and in the nervous system?
Will unique chromatin structural features provided by histone variants remain to be discovered?
To what extent do phenotypes resulting from the loss of histone variants reflect a requirement for functionality provided by the variants versus a generic need for histone incorporation outside of S-phase, when canonical histones are not available?
What are the mechanisms by which chaperones establish distinct genomic distribution patterns of histone variants in different cell types and at different developmental stages?
To what extent does the facilitation of cellular plasticity by H2A.Z and H3.3 depend on their co-participation in hybrid nucleosomes? Is the formation of hybrid nucleosomes important for the functions of other histone variants?
Does H2A.Z affect fear memory by enhancing its formation or, rather, by inhibiting its extinction? Do histone variants play other roles in neural plasticity associated with memory and cognition?
How are neuronal H2B.E levels controlled by activity? What are the molecular mechanisms and physiological functions of H2B.E-mediated modulation of olfactory sensory neuron life span? Do histone variants play similar roles in other neurogenic regions of the adult nervous system?
How are histone variant incorporation patterns maintained, in some cases for months or a lifetime? What are the turnover rates of these variants and do they self-perpetuate?
Table I.
PTMs discussed in this review
| H3K4me3 | Promoter regions of active and poised genes | Transcriptional activation or poising in active or bivalent domains, respectively; catalyzed by enzymes of the Trithorax group: MLL1–4, SETD1A, and SETD1B |
| H3K27me3 | Promoter regions of silent and poised genes; large heterochromatin blocks | Transcriptional repression or poising in silent or bivalent domains, respectively; catalyzed by the Polycomb Repressive Complex 2 (PRC2), which contains EZH2 (or EZH1), SUZ12, and EED |
| H3K9me3 | Constitutive heterochromatin | Transcriptional repression |
| acH2A.Z | Promoter regions of active and poised genes a | Transcriptional activation or poising in active or bivalent domains, respectively |
| H2A.Zub1 | Promoter regions of poised genes a | Transcriptional repression or poising in silent or bivalent domains, respectively; modified by the Ring1B component of Polycomb Repressive Complex 1 (PRC1) [37, 110] |
| H2BK5me1 | Bodies of active genes, especially those with metabolic roles; enhancers; telomeres b | Transcriptional elongation |
| H2BK5ac | Promoter regions of active genes b | Transcriptional activation |
Highlights.
Canonical histone proteins are replaced with specific variants prior to and during periods of cellular transition
Histone variants regulate cellular plasticity during development and in the adult nervous system
Variants TH2A, TH2B, H2A.Z, and H3.3 facilitate cellular plasticity in early development
Variants H2A.X, H3.3, and macroH2A inhibit developmental plasticity following cellular differentiation
Variants H3.3, H2A.Z, and H2B.E regulate cellular plasticity in the adult nervous system
Abbreviations
- Bivalent chromatin
region of chromatin that contains both activating and repressive modifications and is thus silent but poised for potential future activation
- Embryonic stem cell (ESC)
cell derived from the inner cell mass of a blastocyst (an early-stage embryo) that is pluripotent and can be grown indefinitely in culture
- Heterotypic nucleosome
nucleosome that contains one copy of a particular histone variant and one copy of a canonical histone from the same family
- Histone post-translational modification (PTM)
covalent chemical modification of a histone protein that affects its structure and function
- Histones
five families of positively charged proteins (H1, H2A, H2B, H3, and H4) that package DNA into nucleosomes within eukaryotic cell nuclei and play central roles in regulating gene expression
- Histone variant
non-allelic isoform of a canonical histone protein
- Homotypic nucleosome
nucleosome that contains two copies of a particular histone variant
- Hybrid nucleosome
nucleosome that contains multiple distinct histone variants
- Multipotency
the ability of a cell to divide and produce more than one differentiated cell type
- Neuronal activity
electrical and chemical signaling that occurs upon activation of neuronal receptors
- Nuclear reprogramming
process in which a differentiated cell is converted to pluripotency
- Nucleosome
the basic unit of chromatin in eukaryotes, consisting of DNA wrapped around an octamer of histone proteins (two subunits from each of the H2A, H2B, H3, and H4 families)
- Pluripotency
the ability of a cell to divide and produce all of the differentiated cell types in an organism
- S-phase
phase of the cell cycle in which DNA is replicated and packaged largely with canonical histones
- Somatic cell nuclear transfer (SCNT)
process in which the nucleus from a differentiated cell is reprogrammed following transfer to an oocyte
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marzluff WF, Jr, et al. Two chemically and metabolically distinct forms of calf thymus histone F3. The Journal of biological chemistry. 1972;247(7):2026–2033. [PubMed] [Google Scholar]
- 2.Marzluff WF, et al. The human and mouse replication-dependent histone genes. Genomics. 2002;80(5):487–498. [PubMed] [Google Scholar]
- 3.Talbert PB, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics & chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature reviews. Genetics. 2008;9(11):843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20(1):14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS letters. 2010;584(17):3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montellier E, et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes & development. 2013;27(15):1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rop V, Padeganeh A, Maddox PS. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma. 2012;121(6):527–538. doi: 10.1007/s00412-012-0386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, et al. Quantitative proteomic analysis of histone modifications. Chemical reviews. 2015;115(6):2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos EI, Reinberg D. Histones: annotating chromatin. Annual review of genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 11.Ausio J. Histone variants--the structure behind the function. Briefings in functional genomics & proteomics. 2006;5(3):228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- 12.Hake SB, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(18):6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsasser SJ, et al. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491(7425):560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama T, et al. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS genetics. 2011;7(10):e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britton LM, et al. Breaking the histone code with quantitative mass spectrometry. Expert review of proteomics. 2011;8(5):631–643. doi: 10.1586/epr.11.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nature reviews. Genetics. 2014;15(12):814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 18.Pehrson JR, et al. Mice without macroH2A histone variants. Molecular and cellular biology. 2014;34(24):4523–4533. doi: 10.1128/MCB.00794-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang MC, et al. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS genetics. 2015;11(2):e1004964. doi: 10.1371/journal.pgen.1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Developmental cell. 2010;19(5):662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez Alvarado A, Yamanaka S. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell. 2014;157(1):110–119. doi: 10.1016/j.cell.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465(7299):728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor perspectives in biology. 2011;3(6) doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrode N, et al. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis. 2013;51(4):219–233. doi: 10.1002/dvg.22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas-Rios P, Gonzalez-Reyes A. Concise review: The plasticity of stem cell niches: a general property behind tissue homeostasis and repair. Stem cells. 2014;32(4):852–859. doi: 10.1002/stem.1621. [DOI] [PubMed] [Google Scholar]
- 26.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nature reviews. Molecular cell biology. 2009;10(8):526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 27.Trostle-Weige PK, et al. Isolation and characterization of TH2A, a germ cellspecific variant of histone 2A in rat testis. The Journal of biological chemistry. 1982;257(10):5560–5567. [PubMed] [Google Scholar]
- 28.Shires A, Carpenter MP, Chalkley R. A cysteine-containing H2B-like histone found in mature mammalian testis. The Journal of biological chemistry. 1976;251(13):4155–4158. [PubMed] [Google Scholar]
- 29.Shinagawa T, et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell stem cell. 2014;14(2):217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Gaspar-Maia A, et al. Open chromatin in pluripotency and reprogramming. Nature reviews. Molecular cell biology. 2011;12(1):36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allis CD, et al. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- 32.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nature reviews. Molecular cell biology. 2010;11(4):264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 33.Gaspar-Maia A, et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nature communications. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, et al. H2A.Z Facilitates Access of Active and Repressive Complexes to Chromatin in Embryonic Stem Cell Self-Renewal and Differentiation. Cell stem cell. 2013;12(2):180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creyghton MP, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135(4):649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151(7):1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ku M, et al. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome biology. 2012;13(10):R85. doi: 10.1186/gb-2012-13-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes & development. 2013;27(12):1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 40.Amat R, Gudas LJ. RARgamma is required for correct deposition and removal of Suz12 and H2A.Z in embryonic stem cells. Journal of cellular physiology. 2011;226(2):293–298. doi: 10.1002/jcp.22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Current opinion in genetics & development. 2011;21(2):175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suto RK, et al. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature structural biology. 2000;7(12):1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 43.Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic acids research. 2012;40(21):10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian V, et al. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS genetics. 2013;9(8):e1003725. doi: 10.1371/journal.pgen.1003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soboleva TA, et al. Histone variants at the transcription start-site. Trends in genetics : TIG. 2014;30(5):199–209. doi: 10.1016/j.tig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16(2):166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nature structural & molecular biology. 2010;17(12):1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes & development. 2014;28(7):672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nekrasov M, et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nature structural & molecular biology. 2012;19(11):1076–1083. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- 50.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nature genetics. 2009;41(8):941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thambirajah AA, et al. New developments in post-translational modifications and functions of histone H2A variants. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87(1):7–17. doi: 10.1139/O08-103. [DOI] [PubMed] [Google Scholar]
- 52.Franklin SG, Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 53.Filipescu D, Muller S, Almouzni G. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annual review of cell and developmental biology. 2014;30:615–646. doi: 10.1146/annurev-cellbio-100913-013311. [DOI] [PubMed] [Google Scholar]
- 54.Wen D, et al. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(20):7325–7330. doi: 10.1073/pnas.1406389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell reports. 2012;1(6):730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 56.Couldrey C, et al. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Human molecular genetics. 1999;8(13):2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- 57.Bush KM, et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics & chromatin. 2013;6(1):7. doi: 10.1186/1756-8935-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140(17):3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell O, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. The EMBO journal. 2007;26(24):4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell stem cell. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155(1):107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elsasser SJ, et al. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015 doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jullien J, et al. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics & chromatin. 2012;5(1):17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu T, et al. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell stem cell. 2014;15(3):281–294. doi: 10.1016/j.stem.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Buganim Y, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell stem cell. 2014;15(3):295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257(5075):1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 69.Costanzi C, Pehrson JR. MACROH2A2, a new member of the MARCOH2A core histone family. The Journal of biological chemistry. 2001;276(24):21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- 70.Buschbeck M, et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nature structural & molecular biology. 2009;16(10):1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- 71.Buschbeck M, Di Croce L. Approaching the molecular and physiological function of macroH2A variants. Epigenetics : official journal of the DNA Methylation Society. 2010;5(2):118–123. doi: 10.4161/epi.5.2.11076. [DOI] [PubMed] [Google Scholar]
- 72.Pasque V, et al. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. Journal of cell science. 2012;125(Pt 24):6094–6104. doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrero MJ, et al. Macrohistone Variants Preserve Cell Identity by Preventing the Gain of H3K4me2 during Reprogramming to Pluripotency. Cell reports. 2013 doi: 10.1016/j.celrep.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 74.Tanasijevic B, Rasmussen TP. X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS One. 2011;6(6):e21512. doi: 10.1371/journal.pone.0021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Creppe C, et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Molecular and cellular biology. 2012;32(8):1442–1452. doi: 10.1128/MCB.06323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322(5909):1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 77.Pasque V, et al. Histone variant macroH2A confers resistance to nuclear reprogramming. The EMBO journal. 2011;30(12):2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO reports. 2014;15(3):244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sporn JC, et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28(38):3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- 80.Kapoor A, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Novikov L, et al. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Molecular and cellular biology. 2011;31(20):4244–4255. doi: 10.1128/MCB.05244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sporn JC, Jung B. Differential regulation and predictive potential of MacroH2A1 isoforms in colon cancer. The American journal of pathology. 2012;180(6):2516–2526. doi: 10.1016/j.ajpath.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Millar CB. Organizing the genome with H2A histone variants. The Biochemical journal. 2013;449(3):567–579. doi: 10.1042/BJ20121646. [DOI] [PubMed] [Google Scholar]
- 84.Changolkar LN, et al. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Molecular and cellular biology. 2010;30(23):5473–5483. doi: 10.1128/MCB.00518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gamble MJ, et al. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes & development. 2010;24(1):21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hubel DH, Wiesel TN. Ferrier lecture. Functional architecture of macaque monkey visual cortex. Proceedings of the Royal Society of London. Series B, Containing papers of a Biological character. Royal Society. 1977;198(1130):1–59. doi: 10.1098/rspb.1977.0085. [DOI] [PubMed] [Google Scholar]
- 87.Hensch TK. Critical period regulation. Annual review of neuroscience. 2004;27:549–579. doi: 10.1146/annurev.neuro.27.070203.144327. [DOI] [PubMed] [Google Scholar]
- 88.May A. Experience-dependent structural plasticity in the adult human brain. Trends in cognitive sciences. 2011;15(10):475–482. doi: 10.1016/j.tics.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Cortes-Mendoza J, et al. Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2013;31(6):359–369. doi: 10.1016/j.ijdevneu.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 90.Guan JS, Xie H, Ding X. The role of epigenetic regulation in learning and memory. Experimental neurology. 2014 doi: 10.1016/j.expneurol.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 91.Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Developmental biology. 1987;123(1):51–58. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- 92.Michod D, et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74(1):122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drane P, et al. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes & development. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lewis PW, et al. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 96.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nature reviews. Neuroscience. 2009;10(9):647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 99.Zovkic IB, et al. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature. 2014;515(7528):582–586. doi: 10.1038/nature13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70(4):687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brann JH, Firestein SJ. A lifetime of neurogenesis in the olfactory system. Frontiers in neuroscience. 2014;8:182. doi: 10.3389/fnins.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watt WC, et al. Odorant stimulation enhances survival of olfactory sensory neurons via MAPK and CREB. Neuron. 2004;41(6):955–967. doi: 10.1016/s0896-6273(04)00075-3. [DOI] [PubMed] [Google Scholar]
- 103.Santoro SW, Dulac C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife. 2012;1:e00070. doi: 10.7554/eLife.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomas-Danguin T, et al. The perception of odor objects in everyday life: a review on the processing of odor mixtures. Frontiers in psychology. 2014;5:504. doi: 10.3389/fpsyg.2014.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wiedemann SM, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. The Journal of cell biology. 2010;190(5):777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Telese F, et al. "Seq-ing" insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77(4):606–623. doi: 10.1016/j.neuron.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Itzkovitz S, van Oudenaarden A. Validating transcripts with probes and imaging technology. Nature methods. 2011;8(4 Suppl):S12–S19. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews. Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 110.Sarcinella E, et al. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Molecular and cellular biology. 2007;27(18):6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS computational biology. 2009;5(11):e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci. 2011;34:467–499. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- 113.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155(2):321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]