ABSTRACT
The cerebral cortex of primates has evolved massively and intricately in comparison to that of other species. Accumulating evidence indicates that this is caused by changes in cell biological features of neural stem cells (NSCs), which differentiate into neurons and glial cells during development. The fate of NSCs during rodent cortical development is stringently regulated by epigenetic factors, such as histone modification enzymes, but the role of these factors in human corticogenesis is largely unknown. We have recently discovered that a lysine-specific demethylase 1 (LSD1), which catalyzes the demethylation of methyl groups in the histone tail, plays a unique role in human fetal NSCs (hfNSCs). We show that, unlike the role previously reported in mice, LSD1 in hfNSCs is necessary for neuronal differentiation and controls the expression of HEYL, one of the NOTCH target genes, by modulating the methylation level of histones on its promoter region. Interestingly, LSD1-regulation of Heyl expression is not observed in mouse NSCs. Furthermore, we first demonstrated that HEYL is able to maintain the undifferentiated state of hfNSCs. Our findings provide a new insight indicating that LSD1 may be a key player in the development and characterization of the evolved cerebral cortex.
KEYWORDS: cerebral cortex, epigenetics, HEYL, histone methylation, LSD1, neural stem cell, neurogenesis, notch
In the mammalian fetal cerebral cortex, neural stem cells (NSCs) proliferate symmetrically to increase in number1-3 and then differentiate into neurons through asymmetric division.4,5 The differentiation program of NSCs is tightly regulated by epigenetic modifications such as DNA methylation and histone modification.6-11 For instance, several histone methyltransferases participate in the neuronal differentiation of NSCs by controlling chromatin reconstitution in the regulatory region of neurogenic genes in the mouse fetal cortex.9,10 Although numerous studies using rodent models have indicated that many epigenetic factors contribute to cortical development, the detailed functions of these factors in primate cortical development has been largely unknown.
A recent report has shown that cortical NSCs in the fetal brain have distinct features across species.12 In rodents, NSCs in the fetal cortex can differentiate into excitatory neurons, but not inhibitory neurons, while human NSCs can differentiate into both of these neuronal types (Fig. 1A).13 In addition, primate basal radial glia (bRG) can proliferate at the outer subventricular zone (oSVZ), resulting in an increase in the number of neurons, and ultimately leading to the establishment of the large primate neocortex (Fig. 1A).14-21 Understanding the molecular mechanism underlying these functional differences in NSCs between the species may not only shed light on the molecular machinery involved in human corticogenesis, but also provide important insights into the pathogenic mechanism underlying neurological and psychiatric disorders, such as autism spectrum disorders (ASD) and schizophrenia.
Figure 1.
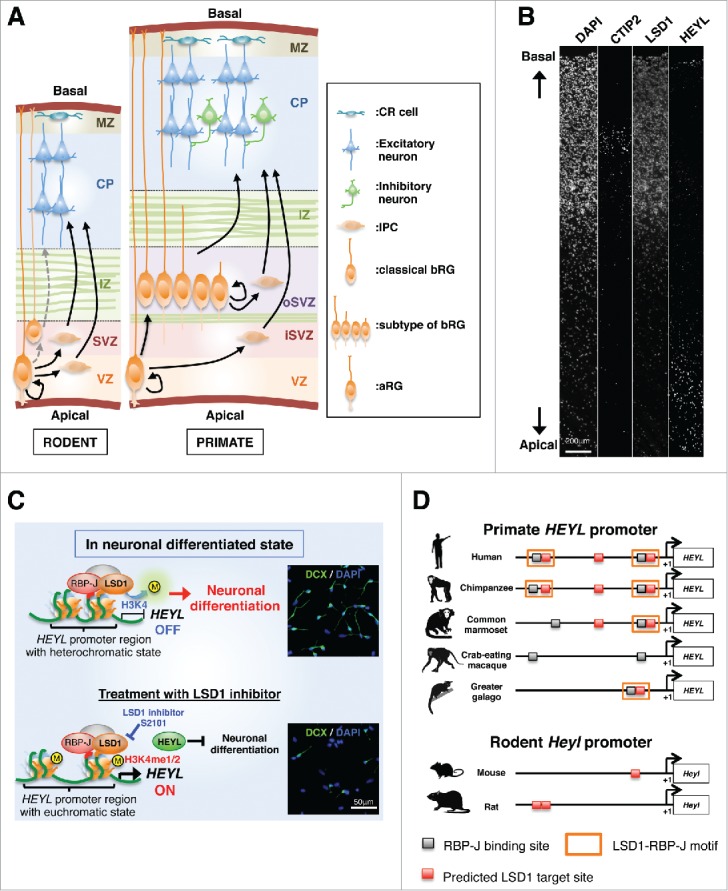
Role of LSD1 in the developing cerebral cortex. (A) Comparisons of rodent and primate cortical development. Radial glia (RG) cells comprise the major population of neural stem cells (NSCs), and are found in both rodent (left) and primate (right) developing cortex. The RG proliferates in the ventricular zone (VZ; referred to as apical RGs [aRGs]), and generates an intermediate progenitor cell (IPC) and basal RGs (bRGs) to yield neurons. bRGs, with their soma located in the subventricular zone (SVZ), or in the outer SVZ (oSVZ) in primates, lack contact with the apical surface, but maintain their basal processes on the pial surface.21,32-34 In addition to this classical bRG type, recent studies have found other bRG subtypes that have features that depend on the presence of basal or apical processes, or both.18,19 In the primate developing cortex, all types of bRGs self-renew and provide neurons directly or through the IPC. More bRGs than aRGs are present in primates, and bRGs are rare do not proliferate in rodents.14-17,21 (B) Immunohistochemistry of human fetal cerebral cortex at 26 gestational weeks for CTIP2, LSD1, and HEYL. (C) During neuronal differentiation, LSD1 is associated with the HEYL proximal promoter, along with RBP-J, and represses the expression of HEYL by inducing a switch to the heterochromatic state, which then promotes neurogenesis. On the other hand, LSD1 inhibitor treatment blocks the effect of LSD1 on the HEYL promoter, resulting in high levels of HEYL, which may repress the expression of genes such as CTIP2 and DLX5, directly or indirectly, and prevent neuronal differentiation. hfNSCs were differentiated with or without an LSD1 inhibitor for 7days. After that, immature neurons were stained with antibody against doublecortin (DCX, green). DAPI (blue) was used to stain the nucleus. (D) Cross-species comparison of HEYL proximal promoter regions (−1000 to +1). Gray and red boxes indicate the consensus RBP-J-binding site and the predicted LSD1-target site,29 respectively. Abbreviations: IZ, intermediate zone; CP, cortical plate; MZ, marginal zone; CR cell, Cajal-Retzius cell; M, methyl group.
Lysine-specific demethylase 1 (LSD1, also known as KDM1A) is the first enzyme identified as a histone demethylase, and catalyzes the demethylation of histone H3 that is mono- and di-methylated at lysine 4 (H3K4me1/2) and lysine 9 (H3K9me1/2).22-24 LSD1 is also a member of the flavin-containing amine oxidase family; i.e., its activity depends on flavin adenine dinucleotide (FAD).22 Recent evidence has linked the functional disruption of LSD1 to various human diseases, such as cancer, metabolic syndrome, and genetic disorders, including neurodevelopmental disorders.
To date, numerous studies have reported the role and function of LSD1 in brain development. For example, in the mouse fetal cortex, LSD1 is required for maintenance of neural progenitor/precursor cells through the regulation of the Atrophin1 gene (Atn1).25 Other studies have also suggested the importance of the LSD1/CoREST complex for neuronal differentiation, migration, and morphology during mouse corticogenesis.26,27 Interestingly, it has been reported that there are 4 functional splice variants of LSD1 in mammals,28 and 2 variants that are specifically expressed in neurons form a complex with supervillin (SVIL); this complex plays a crucial role in the expression of neuronal genes in human neuroblastoma cells.29 Moreover, this report also indicates that enrichment of LSD1 in neuronal cells is observed at the binding site of several transcriptional factors, suggesting that the target gene of LSD1 might be determined by the binding partner of LSD1 in each type of cell. However, the function of LSD1 in cortical development in the human fetus has not yet been reported in detail. Thus, our current study focused on its role in the neuronal differentiation of NSCs isolated from the human fetal cortex.30
Neurogenesis of human fetal NSCs requires LSD1 catalytic activity
The mammalian developing cerebral cortex consists of several germinal compartments, such as the ventricular zone (VZ), subventricular zone (SVZ), intermediate zone, subplate, cortical plate, and marginal zone (MZ), in order from apical to basal layers. In the primate fetal cortex, the SVZ is separated into 2 distinct germinal compartments, known as the inner SVZ (iSVZ) and the oSVZ (Fig. 1A).31-34 In our current study, we first investigated the expression pattern of LSD1 in the human fetal cortex.30 Immunohistochemistry using an LSD1 antibody showed that LSD1-expressing cells were distributed in the whole region of the neocortex from VZ through MZ in sections of human fetal brain (Fig. 1B). To examine LSD1 expression in neural stem cells and their differentiated neurons in further detail, we used an in vitro culture system of human fetal NSCs (hfNSCs) derived from the fetal cortex. In this culture system, the undifferentiated and differentiated cells exhibited LSD1 expression, supporting the observations in human fetal brain sections. These results indicated that LSD1 is expressed in all neural cells in the developing fetal cortex, and implied that it contributes to corticogenesis.
A recent screening study of chemical inhibitors targeting epigenetic factors for use as anti-cancer drugs led to the discovery of S2101, which exhibited a strong inhibitory effect against LSD1 catalytic activity, mediated by blocking the association between LSD1 and FAD.35 In our study, treatment of cells with S2101 at the induction of differentiation of hfNSCs, resulted in a marked reduction of the number of neurons as compared with untreated cells.30 Notably, this reduction was observed in both CTIP2-positive excitatory neurons and GAD65-positive inhibitory neurons. Considering that this treatment upregulated the number of SOX2-positive undifferentiated cells and did not induce glial differentiation and apoptosis, our findings indicated that LSD1 activity is necessary for neuronal differentiation of hfNSCs (Fig. 1C).
Interestingly, the neuronal differentiation of mouse NSCs derived from the embryonic cortex were not repressed by LSD1 catalytic inhibition, demonstrating a functional difference in LSD1 between human and mouse NSCs during corticogenesis.
HEYL is a novel target gene of LSD1 during human neurogenesis
We next attempted to identify the target gene of LSD1 in hfNSCs. In the mammalian developing cortex, NOTCH signaling plays an essential role in the fate specification of NSC, and inactivation of its signaling induces differentiation of NSCs.34 Upon ligand binding to NOTCH in NSCs, the intracellular domain of NOTCH (NICD) is released from the plasma membrane and translocates into the nucleus, where it converts the CBF1 (RBP-J) repressor complex into an activator complex. This NICD/RBP-J activator complex targets genes such as hairy/enhancer of split-1/5 (HES1/5) and Hes-related family of bHLH-type transcription factors with YRPW motif-1/2 (HEY1/2), which inhibit the expression of proneural genes, and suppress the neuronal differentiation of NSCs.37 It has been reported that LSD1 is associated with a repressor complex, including RBP-J, and participates in the repression of NOTCH target genes in Drosophila.38 Moreover, the direct interaction of LSD1 with RBP-J in the embryonic cortex of mice has also recently been reported by another group.27 Similar to their study, our preliminary experiment revealed the interaction between LSD1 and RBP-J in hfNSCs [unpublished data]. Therefore, we examined the expression of NOTCH target genes, such as the genes in the HES and HEY families, in differentiated hfNSCs under S2101 treatment. We found that HEY-like (HEYL) gene expression was significantly increased in response to treatment. Recently, another group has identified the consensus sequence enriched in LSD1 as the LSD1 target site by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in human neuroblastoma.29 This sequence is located close to the RBP-J binding consensus sequence39 (named RBP-J binding site) on the HEYL proximal promoter, and we showed association of LSD1 to the LSD1 target site using a ChIP assay. Moreover, administration of S2101 to hfNSCs during the induction of neuronal differentiation prevented demethylation of H3K4me1/2 at the HEYL proximal promoter region. Demethylation of H3K4me1/2 at the regulatory region causes chromatin condensation and transcriptional repression of the target gene.24 Therefore, our results suggest that LSD1, together with RBP-J, acts as a transcriptional repressor of HEYL via chromatin reconstitution in NSCs in the human fetus cortex (Fig. 1C).
Although not much has been reported on the function of HEYL in neuronal development in the fetal brain, our findings implied that HEYL is probably involved in the repression of neurogenesis in hfNSCs. Indeed, immunohistochemistry of human fetal brain in our current study revealed that HEYL-expressing cells were distributed in the apical side of the developing cortex, where undifferentiated NSCs are located (Fig. 1B). Thus, to examine whether HEYL expression is able to suppress neurogenesis, we performed overexpression of HEYL in hfNSCs and induced differentiation. In this experiment, hfNSCs overexpressing HEYL displayed an undifferentiated state, even under neuronal differentiation-inducing culture condition, suggesting that HEYL expression is sufficient to repress neurogenesis in hfNSCs.30
Furthermore, we showed that excessive expression of HEYL significantly reduced the expression of CTIP2 and DLX5, which are excitatory and inhibitory neuron markers, respectively [unpublished data]. Additionally, the inhibition of neurogenesis of hfNSCs by S2101 treatment was abrogated by HEYL knockdown, indicating that HEYL is responsible for the repression of neuronal differentiation upon LSD1 inactivation. Taken together, our findings demonstrated that the control of HEYL expression by demethylation of H3K4me1/2, which is dependent on LSD1 activity, is essential for adequate neurogenesis of NSCs during the development of the human fetal cortex.
We also found that Heyl expression was not increased by LSD1 inactivation in mouse NSCs, unlike in human NSCs. To investigate the cause of this difference, we compared the genome sequences of the HEYL proximal promoter in rodents and primates (Fig. 1D). The sequence revealed the presence of a set of LSD1- and RBP-J-associating sites, termed an LSD1−RBP-J motif, in most primates, but not in rodents. Thus, it is likely that the difference in the HEYL proximal promoter determines the difference in responsiveness to LSD1 inactivation in NSCs of individual species.
Conclusion and future direction
Our study has elucidated a novel molecular mechanism underlying LSD1 activity in the neurogenesis of cortical NSCs in the human fetus.30 Furthermore, we have demonstrated that this machinery is clearly different between humans and mice. This difference may be caused by changes in the LSD1 target genes, accompanied by the emergence of an LSD1−RBP-J motif in the HEYL promoter during mammalian evolution. This hypothesis is supported by a recent report that genetic variation in the promoter region, in accordance with mammalian evolution, attains a primate-specific chromatin status through histone modification, including acetylation and methylation.40,41 Moreover, Reilly et al. have recently identified promoters and enhancers that have epigenetically gained activity in fetal cortex of humans. This was demonstrated by comparative epigenetic profiling, including genome-wide mapping of H3K4me2 in the embryonic cortex of humans and other species, and suggested that these gains may be linked to the evolutionary expansion and elaboration of the human cerebral cortex. Furthermore, it has also been reported that chromatin-remodeling factors, including histone-modification enzymes, control cerebral cortical size and thickness.42 Considering these reports and our findings, a primate-specific chromatin status that is regulated by such factors, including LSD1, may have allowed the primate brain to become large and intricate, and enabling primates to exert higher-order brain function.
One of the unique features of human NSCs in the developing cortex is that they possess a potential for differentiating into not only excitatory neurons, but also inhibitory neurons.13 Understanding the mechanism that characterizes this feature may help to the elucidate the pathogenic mechanism underlying neurodevelopmental disorders, such as ASD, in which the balance of excitatory and inhibitory neurons is disrupted.43-45 Our study showed that LSD1 inactivation in hfNSCs caused repression of differentiation into inhibitory neurons as well as excitatory neurons, which implies that LSD1 contributes to the characteristics of human NSCs and generation of the appropriate numbers of both types of neurons during cortical development. Therefore, LSD1 may be an attractive potential drug target for neurodevelopmental diseases.
In conclusion, our current study in hfNSCs has suggested a new role for LSD1 in neurogenesis. A further study using induced pluripotent stem cells established from patients with neurodevelopmental disorder may provide significant insights into the function of histone-modification enzymes, and may contribute to a further understanding of the pathogenic mechanisms of such disorders.
Abbreviations
- aRG
apical redial glia
- bRG
basal radial glia
- CP
cortical plate
- CR cell
Cajal-Retzius cell
- hfNSC
human fetal neural stem cell
- IPC
intermediate progenitor cell
- iSVZ
inner SVZ
- IZ
intermediate zone
- LSD1
lysine-specific demethylase 1
- MZ
marginal zone
- oSVZ
outer SVZ
- SVZ
subventricular zone
- VZ
ventricular zone
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Editage for their editorial support of manuscript.
Funding
This work was supported by a Grant-in-Aid for Young Scientists (B) (grant no. JP15K21661) and a Grant-in-Aid for Scientific Research (C) (grant no. JP26430083) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- [1].Fujita S. The matrix cell and cytogenesis in the developing central nervous system. J Comp Neurol 1963; 120:37-42; PMID:13960118; http://dx.doi.org/ 10.1002/cne.901200104 [DOI] [PubMed] [Google Scholar]
- [2].Fujita S. Transitory differentiation of matrix cells and its functional role in the morphogenesis of the developing vertebrate CNS. Curr Top Dev Biol 1986; 20:223-42; PMID:3514136; http://dx.doi.org/ 10.1016/S0070-2153(08)60666-3 [DOI] [PubMed] [Google Scholar]
- [3].Fujita S. The discovery of the matrix cell, the identification of the multipotent neural stem cell and the development of the central nervous system. Cell Struct Funct 2003; 28:205-28; PMID:14586132; http://dx.doi.org/ 10.1247/csf.28.205 [DOI] [PubMed] [Google Scholar]
- [4].Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001; 409:714-20; PMID:11217860; http://dx.doi.org/ 10.1038/35055553 [DOI] [PubMed] [Google Scholar]
- [5].Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 2004; 7:136-44; PMID:14703572; http://dx.doi.org/ 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- [6].Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res 2007; 61:58R-63R; PMID:17413844; http://dx.doi.org/ 10.1203/pdr.0b013e3180457635 [DOI] [PubMed] [Google Scholar]
- [7].Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci 2008; 363:2099-109; PMID:18375376; http://dx.doi.org/ 10.1098/rstb.2008.2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Juliandi B, Abematsu M, Nakashima K. Epigenetic regulation in neural stem cell differentiation. Dev Growth Differ 2010; 52:493-504; PMID:20608952; http://dx.doi.org/ 10.1111/j.1440-169X.2010.01175.x [DOI] [PubMed] [Google Scholar]
- [9].Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 2010; 11:377-88; PMID:20485363; http://dx.doi.org/ 10.1038/nrn2810 [DOI] [PubMed] [Google Scholar]
- [10].Imamura T, Uesaka M, Nakashima K. Epigenetic setting and reprogramming for neural cell fate determination and differentiation. Philos Trans R Soc Lond B Biol Sci 2014; 369:1652; http://dx.doi.org/ 10.1098/rstb.2013.0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adefuin AM, Kimura A, Noguchi H, Nakashima K, Namihira M. Epigenetic mechanisms regulating differentiation of neural stem/precursor cells. Epigenomics 2014; 6:637-49; PMID:25531257; http://dx.doi.org/ 10.2217/epi.14.53 [DOI] [PubMed] [Google Scholar]
- [12].Borrell V, Reillo I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev Neurobiol 2012; 72:955-71; PMID:22684946; http://dx.doi.org/ 10.1002/dneu.22013 [DOI] [PubMed] [Google Scholar]
- [13].Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature 2002; 417:645-49; PMID:12050665; http://dx.doi.org/ 10.1038/nature00779 [DOI] [PubMed] [Google Scholar]
- [14].Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci 2011; 14:555-61; PMID:21478886; http://dx.doi.org/ 10.1038/nn.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci 2011; 31:3683-95; PMID:21389223; http://dx.doi.org/ 10.1523/JNEUROSCI.4773-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr Opin Neurobiol 2012; 22:747-53; PMID:22487088; http://dx.doi.org/ 10.1016/j.conb.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lewitus E, Kelava I, Huttner WB. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front Hum Neurosci 2013; 7:424; PMID:23914167; http://dx.doi.org/ 10.3389/fnhum.2013.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 2013; 80:442-57; PMID:24139044; http://dx.doi.org/ 10.1016/j.neuron.2013.09.032 [DOI] [PubMed] [Google Scholar]
- [19].Pilz GA, Shitamukai A, Reillo I, Pacary E, Schwausch J, Stahl R, Ninkovic J, Snippert HJ, Clevers H, Godinho L, et al.. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun 2013; 4:2125; PMID:23839311; http://dx.doi.org/ 10.1038/ncomms3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 2014; 141:2182-94; PMID:24866113; http://dx.doi.org/ 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- [21].De Juan Romero C, Borrell V. Coevolution of radial glial cells and the cerebral cortex. GLIA 2015; 63:1303-19; PMID:25808466; http://dx.doi.org/ 10.1002/glia.22827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941-53; PMID:15620353; http://dx.doi.org/ 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- [23].Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437:436-9; PMID:16079795 [DOI] [PubMed] [Google Scholar]
- [24].Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437:432-5; PMID:16079794 [DOI] [PubMed] [Google Scholar]
- [25].Zhang F, Xu D, Yuan L, Sun Y, Xu Z. Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat Commun 2014; 5:5815; PMID:25519973; http://dx.doi.org/ 10.1038/ncomms6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fuentes P, Cánovas J, Berndt FA, Noctor SC, Kukuljan M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb Cortex 2012; 22:1431-41; PMID: 21878487; http://dx.doi.org/ 10.1093/cercor/bhr218 [DOI] [PubMed] [Google Scholar]
- [27].Lopez CI, Saud KE, Aguilar R, Berndt FA, Cánovas J, Montecino M, Kukuljan M. The chromatin modifying complex CoREST/LSD1 negatively regulates Notch pathway during cerebral cortex development. Dev Neurobiol 2016; In press; PMID:27112428 [DOI] [PubMed] [Google Scholar]
- [28].Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci 2010; 30:2521-32; PMID:20164337; http://dx.doi.org/ 10.1523/JNEUROSCI.5500-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, et al.. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell 2015; 57:957-70; PMID:25684206; http://dx.doi.org/ 10.1016/j.molcel.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hirano K, Namihira M. LSD1 Mediates Neuronal Differentiation of Human Fetal Neural Stem Cells by Controlling the Expression of a Novel Target Gene, HEYL. Stem Cells 2016; 34:1872-82; PMID:27018646; http://dx.doi.org/ 10.1002/stem.2362 [DOI] [PubMed] [Google Scholar]
- [31].Tyler WA, Haydar TF. A new contribution to brain convolution: progenitor cell logistics during cortex development. Nat Neurosci 2010; 13:656-7; PMID:20498684; http://dx.doi.org/ 10.1038/nn0610-656 [DOI] [PubMed] [Google Scholar]
- [32].Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al.. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci 2010; 13:690-9; PMID:20436478; http://dx.doi.org/ 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- [33].Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010; 464:554-61; PMID:20154730; http://dx.doi.org/ 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- [34].Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 2011; 21:1674-94; PMID:21127018; http://dx.doi.org/ 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- [35].Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry 2010; 49:6494-503; PMID:20568732; http://dx.doi.org/ 10.1021/bi100299r [DOI] [PubMed] [Google Scholar]
- [36].Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev 2002; 16:846-58; PMID:11937492; http://dx.doi.org/ 10.1101/gad.975202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 2011; 69:840-55; PMID:21382546; http://dx.doi.org/ 10.1016/j.neuron.2011.02.031 [DOI] [PubMed] [Google Scholar]
- [38].Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al.. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 2011; 42:689-99; PMID:21596603; http://dx.doi.org/ 10.1016/j.molcel.2011.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res 1994; 22:965-71; PMID:8152928; http://dx.doi.org/ 10.1093/nar/22.6.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al.. Enhancer evolution across 20 mammalian species. Cell 2015; 160:554-66; PMID:25635462; http://dx.doi.org/ 10.1016/j.cell.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 2015; 347:1155-9; PMID:25745175; http://dx.doi.org/ 10.1126/science.1260943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell 2013; 25:256-69; PMID:23643363; http://dx.doi.org/ 10.1016/j.devcel.2013.04.005 [DOI] [PubMed] [Google Scholar]
- [43].Pizzarelli R, Cherubini E. Alterations of GABAergic signaling in autism spectrum disorders. Neural Plast 2011; 2011:297153; PMID:21766041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vallianatos CN, Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 2015; 7:503-19; PMID:26077434; http://dx.doi.org/ 10.2217/epi.15.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al.. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015; 162:375-90; PMID:26186191; http://dx.doi.org/ 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]


