Summary
ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) is necessary and sufficient for root hair elongation in Arabidopsis thaliana. Root hair length is determined by the duration for which RSL4 protein is present in the developing root hair. The aim of this research was to identify genes regulated by RSL4 that affect root hair growth.
To identify genes regulated by RSL4, we identified genes whose expression was elevated by induction of RSL4 activity in the presence of an inhibitor of translation.
Thirty‐four genes were identified as putative targets of RSL transcriptional regulation, and the results suggest that the activities of SUPPRESSOR OF ACTIN (SAC1), EXOCSYT SUBUNIT 70A1 (EXO70A1), PEROXIDASE7 (PRX7) and CALCIUM‐DEPENDENT PROTEIN KINASE11 (CPK11) are required for root hair elongation.
These data indicate that RSL4 controls cell growth by controlling the expression of genes encoding proteins involved in cell signalling, cell wall modification and secretion.
Keywords: Arabidopsis cell growth, basic helix‐loop‐helix transcription factor, CALCIUM‐DEPENDENT PROTEIN KINASE11 (CPK11), EXOCSYT SUBUNIT 70A1 (EXO70A1), PEROXIDASE7 (PRX7), root hair, ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4), SUPPRESSOR OF ACTIN (SAC1)
Introduction
Root hairs are tubular projections that develop from the outermost layer of roots (epidermis or periderm). They can constitute up to c. 50% of the surface area of the root and extend the absorptive surface of the root into the surrounding soil, facilitating the uptake of nutrients, such as phosphate, with limited mobility in the soil. Growth is restricted to the tip of the root hair; no growth occurs along the shanks. New cell surface is generated though the localized secretion of new membrane and cell wall components from the endomembrane system to a spatially restricted area at the root hair tip (Carol & Dolan, 2002; Datta et al., 2011; Balcerowicz et al., 2015; Salazar‐Henao & Schmidt, 2016). Growth at the tip is coordinated by a number of activities including Ca2+, pH and reactive oxygen oscillations at the tip (Ishida et al., 2008; Datta et al., 2011; Salazar‐Henao & Schmidt, 2016).
The growth of root hairs is determined by the interaction of a plethora of external environmental signals with internal growth regulators. Phosphate availability is the best‐characterized external factor that influences root hair length (Bates & Lynch, 2000; Lopez‐Bucio et al., 2003; Zhang et al., 2003; Lee & Cho, 2013; Niu et al., 2013). In most species, root hairs are longer in roots grown in low‐phosphate conditions than when grown in replete phosphate conditions. This adaptive response to low phosphate availability increases the capacity for phosphate uptake because longer hairs can mine larger volumes of the soil for phosphate (Brown et al., 2013; Lopez‐Arredondo et al., 2014). Such phenotypic plasticity is characteristic of plant development and is often under strict transcriptional control (Rubio et al., 2001; Jain et al., 2012).
The ROOT HAIR DEFECTIVE SIX‐LIKE (RSL) class I basic helix‐loop‐helix (bHLH) transcription factors positively regulate the development of root hairs in angiosperms and rhizoids in liverworts and mosses (Menand et al., 2007; Yi et al., 2010; Proust et al., 2016). Arabidopsis thaliana plants deficient in RSL class I activity – root hair defective six rsl1 double mutants – do not develop root hairs (Menand et al., 2007). Similarly, no rhizoids develop in the rsl1 mutant of the liverwort Marchantia polymorpha or the Pprsl1 Pprsl2 double mutant of the moss Pyscomitrella patens (Menand et al., 2007; Proust et al., 2016). Arabidopsis thaliana RSL class I genes act, at least in part, by directly regulating the expression of RSL4, a closely related bHLH transcription factor (Yi et al., 2010). RSL4 is necessary for root hair elongation; few short root hairs develop on rsl4 mutants (Yi et al., 2010). Furthermore, RSL4 is also sufficient for root hair growth as constitutive expression of RSL4 results in the constitutive growth of root hairs (Yi et al., 2010). During wild‐type development, RSL4 is produced in a pulse at the beginning of root hair development and growth continues until RSL4 is removed from the cell through proteolysis (Datta et al., 2015). Mutated forms of RSL4 that are more stable than the wild‐type form programme growth for a longer time than the wild‐type form, leading to the development of longer root hairs than in the wild‐type (Datta et al., 2015). This demonstrates the role of RSL4 stability in determining root hair length. Taken together, these data indicate that RSL4 is a potentiator of root hair growth that is produced in a pulse just before growth initiates, and growth ceases once proteolysis removes the last of the RSL4 from the growing root hair.
Low phosphate‐induced root hair elongation is mediated, at least in part, by modulation of RSL4 expression (Svistoonoff et al., 2007; Yi et al., 2010; Koltai & Kapulnik, 2013; Lopez‐Arredondo et al., 2014). Low concentrations of phosphate in the growth media increase the steady‐state levels of RSL4 mRNA (Yi et al., 2010). This increase does not occur in mutants that are defective in phosphate sensing, indicating that the increase is a target of the phosphate‐sensing mechanism (Yi et al., 2010). The amount of RSL4 synthesized in a pulse just before root hair initiation is higher in low phosphate than in replete phosphate. Because the amount of RSL4 synthesized is greater, the duration of growth is increased, resulting in development of longer root hairs compared with plants grown in replete phosphate where less RSL4 is produced before root hair initiation (Datta et al., 2015). These data indicate that RSL4 is a key modulator of root hair growth and the target of pathways that modulate root hair elongation.
To determine how RSL4 controls root hair elongation, we set out to identify genes encoding proteins that are targets of RSL4 transcriptional regulation. Here, we identified 34 putative targets of RSL4 and demonstrated that four of these genes are required for root hair growth and overexpression of two of the genes increases root hair elongation. These data indicate that RSL4 regulates the expression of genes involved in cell signalling, vesicle transport and cell wall modification.
Materials and Methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana, (L.) Heynh.) ecotype Columbia (Col‐0) was used for all the experiments. The T‐DNA insertion mutant seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) and the Nottingham Arabidopsis Stock Centre (NASC). The Arabidopsis seeds were surface‐sterilized in 70% ethanol for 5 min followed by 5% (v/v) bleach for 10 min and rinsed with sterile distilled water three times. For the T‐DNA insertion lines and the Glucocorticoid Receptor (GR) inducible lines, the seeds were plated on Murashige and Skoog (MS) medium (pH 5.8) supplemented with 1% sucrose (w/v) and 0.5% Phytagel (w/v) (Sigma, UK). For the overexpression studies, at least four independent transgenic lines overexpressing the target genes were examined. In order to observe the root hair phenotype of the overexpression lines, the plants were grown vertically at a 45° angle in half‐strength Johnson medium with 0.5% Phytagel. After a period of stratification at 4°C for 3 d, the plants were grown vertically under continuous illumination at 25°C in the growth chamber. The roots were observed 6–7 d after sowing.
Generation of GR‐fusion constructs
To generate the N‐terminal GR fusion vector, the multisite gateway system was used. Gateway entry clones were generated using pDONR P4‐P1R containing the promoter plus the 5′ untranslated region (UTR) of the gene, pDONR P2R‐P3 cloned with the coding region of the gene plus 3′ UTR and terminator and the GR domain inserted into pDONR 207 as described (Yi et al., 2010). Gateway LR multisite reaction (Gateway LR clonase reaction mix; Invitrogen) was carried out with pDONR P4‐P1R, pDONR P2R‐P3, pDONR 207‐GR plus the binary destination vector pGWB multisite in order to generate the GR:RSL4 fusion vector. Primers used to check the GR:RSL4 constructs are listed in Supporting Information Table S1. The binary vector pGWB multisite was a gift from Matt Tomlinson (John Innes Centre, Norwich, UK) and was generated by replacing the R1‐CmR‐ccdB‐R2 cassette of pGWB1 with R4‐CmR‐ccdB‐R3 (pGWB1 was from Tsuyoshi Nakagawa, Shimane University, Japan). This construct was then transformed into the rsl2 rsl4 mutant background.
Plant transformation
The Agrobacterium tumefaciens‐mediated floral dip method was used to transform all the binary vectors into Arabidopsis (Clough & Bent, 1998). Plants with a few siliques and several immature buds were used for transformation. A volume of 250 ml of A. tumefaciens culture incubated at 28°C overnight was spun down and the pellet was re‐suspended in 200 ml of solution containing 5% (w/v) sucrose and 0.05% (v/v) Silwet L‐77. The floral tissues of the Arabidopsis plants were dipped into the bacterial mixture for 30 s, after which the plants were laid down on a tray with wet tissues and covered with cling film overnight. The dipped plants were then returned to the glasshouse.
Dexamethasone treatment
A 20 mM stock solution of dexamethasone (DEX) in ethanol was used for the treatment. T2 seeds of GR:RSL4 rsl2 rsl4 plants were first germinated in MS medium containing 25 μg ml−1 hygromycin and after 4 d the resistant seedlings were transferred to MS medium without antibiotics. Six‐day‐old seedlings were treated with 20 μM DEX by flooding with 20 ml of MS liquid medium containing 20 μl of DEX (DEX treated) or 20 μl of ethanol (mock treated). After treatment, the root tips were sampled at several time‐points for qRT‐PCR and microarray analysis.
Cycloheximide treatment
Cycloheximide (CHX) was dissolved in ethanol to make 20 mM stock solutions to be used in the experiments. Similar to the DEX treatment, 6‐d‐old seedlings of GR:RSL4 rsl2 rsl4 plants were used for the CHX treatment. For CHX treatment, seedlings were flooded with 20 ml of MS liquid containing 20 μl of CHX (CHX treated). The seedlings were further treated with 20 μM CHX and 20 μM DEX (DEX+CHX treated) by flooding with 20 ml of MS liquid medium containing 20 μl of DEX and 20 μl of CHX. After treatment, the root tips were sampled at several time‐points for qRT‐PCR and microarray analysis.
Microarray hybridization
Gene expression in root hairs of GR:RSL4 rsl2 rsl4 seedlings under different treatments (mock, DEX, and DEX+CHX treated) were compared using the Affymetrix (Santa Clara, CA, USA) Gene Chip Arabidopsis ATH1 Genome Array. Total RNA was isolated from three independent biological replicates. The RNA samples from the root tips of the RSL4 inducible lines under different treatments were quantified using a NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific, USA) and assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). cDNA was synthesized from 4 μg of total RNA using one‐cycle target labelling and control reagents (Affymetrix) to produce biotin‐labelled cDNA. Following fragmentation, the cDNA was hybridized to the Arabidopsis ATH1 Genome Array. Each microarray was washed and stained with streptavidin‐phycoerythrin in a Fluidics station 450 (Affymetrix) and scanned at 1.56‐μm resolution in a GeneChipScanner 3000 7G System (Affymetrix). All the analyses, including the hybridization, staining, washing and screening for quality of the arrays, were carried out at the Genomics Service of the Centro Nacional de Biotecnología, Madrid, Spain (Consejo Superior de Investigaciones Científicas).
Gene expression analysis
For background correction, normalization, and expression level summarization, the multi‐array analysis algorithm was used (Irizarry et al., 2003). Statistical analysis and graphical visualization of data were carried out using the interactive tool Fiesta http://bioinfogp.cnb.csic.es/tools/FIESTA/index.php). In order to determine changes in gene expression, an analysis of variance was used. P‐values were adjusted by false‐discovery rate to correct for multiple testing (Benjamini & Hochberg, 1995). Genes with adjusted P‐values of ≤ 0.001 were selected as statistically significant. They were considered to be differently expressed if the change in expression was > 2‐fold (up‐regulated). The values obtained for the DEX‐treated samples were compared with those obtained for mock‐treated samples to identify the genes induced by RSL4. Further, in order to determine if the gene expression of these RSL4‐induced genes are unaffected upon adding CHX, the values for the DEX+CHX‐treated samples were compared with those for the mock. The genes that exhibited similar values in both analyses were taken as putative direct target genes of RSL4.
Root hair length measurements
To measure the length of root hairs in plants homozygous for T‐DNA insertions, root hairs located 3 mm from the root tip of 6‐ to 7‐d‐old seedlings were imaged under the Leica DFC310 FX camera mounted on a Leica M165 FC stereo microscope. The digital images obtained were used directly for root hair length measurement using imagej software https://imagej.nih.gov/ij/. Twenty seedlings of each genotype were measured.
Results
The bHLH transcription factor RSL4 is necessary and sufficient for post‐mitotic cell growth in Arabidopsis root hairs (Yi et al., 2010). We hypothesized that the immediate targets of RSL4 would be required for root hair elongation. To identify downstream targets of RSL4, we developed an inducible system by expressing a glucocorticoid receptor:RSL4 fusion protein (GR:RSL4) in the root hairless rsl2 rsl4 double mutant background (Lloyd et al., 1994). The GR‐RSL4 protein is located in the cytoplasm and moves to the nucleus upon exposure to a synthetic steroid, DEX. Once in the nucleus, GR:RSL4 will be active and promote the expression of RSL4‐regulated genes and induce root hair elongation. MS+ethanol‐treated (mock) GR:RSL4 rsl2 rsl4 plants were root hairless (identical to rsl2 rsl4 double mutants), indicating that the GR:RSL4 protein is not transcriptionally active when mock treated (Fig. 1a–c). Upon DEX treatment, the GR:RSL4 rsl2 rsl4 seedlings developed root hairs (Fig. 1d), consistent with the induction of RSL4 activity by DEX. Root hairs were visible as bulges emerging from the epidermal surface of trichoblasts within 30 min after DEX treatment (Fig. 2). This indicates that DEX‐mediated induction of GR‐RSL4 activity is rapid. Taken together, these data indicate that root hair development can be induced by DEX treatment in the GR:RSL4 rsl2 rsl4 background.
Figure 1.
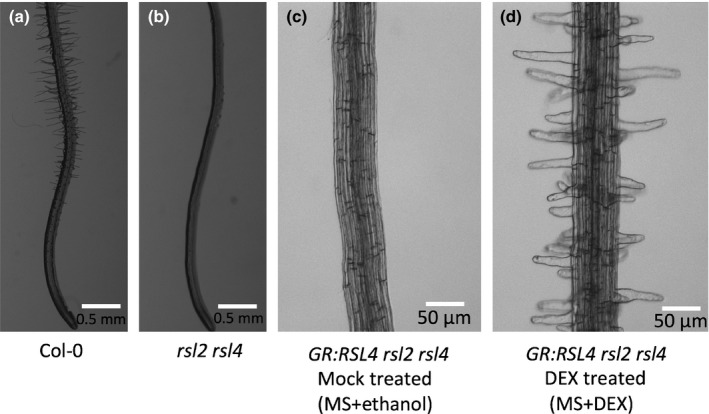
Dexamethasone (DEX) treatment induced root hair development in the Arabidopsis thaliana Glucocorticoid Receptor (GR):ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) rsl2 rsl4 background. (a, b) Root hair development in 6‐d‐old Arabidopsis thaliana Col‐0 and rsl2 rsl4 seedlings. (c, d) GR:RSL4 rsl2 rsl4 line 24 h after mock (MS + ethanol) treatment (c) and after DEX treatment (d).
Figure 2.
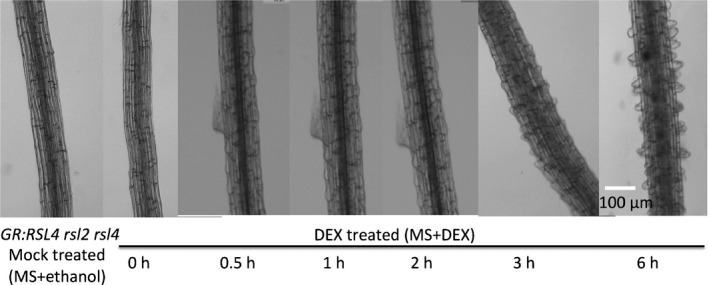
Dexamethasone (DEX) treatment induced root hair initiation in the Arabidopsis thaliana Glucocorticoid Receptor (GR):ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) rsl2 rsl4 background within 30 min. Images of root hair development in the A. thaliana GR:RSL4 rsl2 rsl4 root at different times after DEX treatment are shown compared with 6 h after mock (Murashige and Skoog (MS) + ethanol) treatment.
To identify the immediate RSL4 target genes that affect root hair growth, RSL4 activity was induced by DEX in the presence of the protein synthesis inhibitor CHX (Fig. 3). Only genes that do not depend on protein synthesis will be induced by the combined DEX and CHX treatment of GR:RSL4 rsl2 rsl4 plants; that is, those genes that are regulated by the GR:RSL4 fusion protein directly. The development of root hairs was inhibited upon addition of CHX, indicating that CHX prevents hair development by inhibiting the translation of proteins required for root hair growth (Fig. 3a). However, as gene transcription is not inhibited by CHX, comparing the transcriptional profiles of roots treated with DEX with roots treated with both DEX and CHX can identify putative direct targets of RSL4. Genes that are induced both by DEX treatment and by the combined DEX and CHX treatment but whose levels of induction remain unaltered in the two treatments are putative direct RSL4 targets. RNA was isolated 6 h after mock treatment, DEX treatment and the combined DEX and CHX treatment of GR:RSL4 rsl2 rsl4 roots. Steady‐state mRNA levels were measured using Affymetrix ATH1 arrays.
Figure 3.
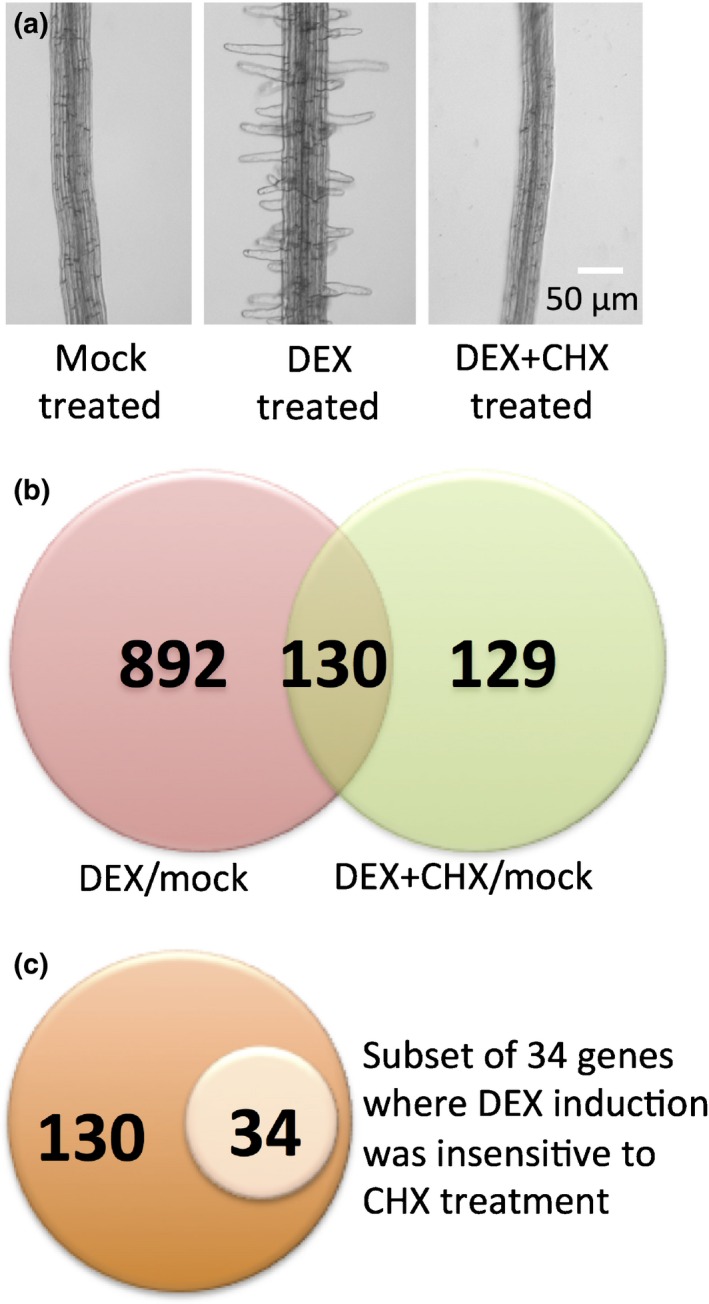
Identification of genes whose induction in the Glucocorticoid Receptor (GR):ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) rsl2 rsl4 background by dexamethasone (DEX) is insensitive to cycloheximide (CHX). (a) Development of root hairs 24 h after an Arabidopsis thaliana GR:RSL4 rsl2 rsl4 plant was treated with Murashige and Skoog (MS) medium containing 20 μM DEX. No root hairs developed after treatment of the seedling with DEX + CHX and MS + ethanol (mock). (b) Venn diagram of genes induced upon RSL4 activation under different treatments. Pink and green circles represent genes induced only in the DEX/mock and DEX + CHX/mock sample, respectively. The brown region represents genes (130) that were induced significantly (P ≤ 0.001) upon RSL4 activation both in DEX/mock and DEX + CHX/mock samples. (c) A subset of 34 genes of the 130 genes which exhibited a minimum two‐fold increase in expression upon DEX treatment and no change in expression levels upon addition of CHX were selected as putative direct targets of RSL4.
To identify genes whose expression was induced by DEX treatment, we selected genes whose expression was at least twice as high in the DEX‐ and DEX+CHX‐treated GR:RSL4 rsl2 rsl4 roots compared with mock‐treated GR:RSL4 rsl2 rsl4 roots. This identified 1022 genes whose expression was increased at least two‐fold by DEX treatment compared with mock treatment after applying a stringent filter, that is, P‐value < 0.0001 (Table S2). A total of 259 genes were more highly expressed in DEX+CHX compared with mock samples with P‐value < 0.001 (Table S3). The expression of 130 genes was significantly induced in both the DEX‐treated samples and the DEX+CHX‐treated samples; that is, 130 genes were common to both data sets (DEX/mock and DEX+CHX/mock) (Fig. 3b; Table S4). To identify the putative RSL4 direct target genes among this set of 130 genes, those DEX‐induced genes whose expression was insensitive to the addition of CHX were identified. The expression of 34 of the 130 genes (26% of the 130 genes selected) was identical in both DEX and DEX+CHX treatments (Fig. 3c). These 34 DEX‐inducible genes whose transcription was insensitive to CHX treatment were potential targets of RSL4 transcriptional regulation (Fig. 3c; Table 1).
Table 1.
Thirty‐four Arabidopsis thaliana genes whose expression was more than twice as high in dexamethasone (DEX)‐treated Glucocorticoid Receptor (GR):ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) rsl2 rsl4 plants as in mock‐treated plants and whose DEX induction was insensitive to cycloheximide (CHX) treatment
| Accession code | Gene symbol | Protein encoded/biological function |
|---|---|---|
| AT4G01120 | GBF2 | bZIP transcription factor that binds to the G‐box/blue light‐mediated development |
| AT5G02780 | GSTL1 | Glutathione transferase lambda 1/thiol transferase activity |
| AT1G21610 | UBN1 | Wound‐responsive family protein/response to wounding |
| AT2G27775 | Arabidopsis thaliana unknown protein | |
| AT5G58375 | Arabidopsis thaliana unknown protein | |
| AT5G43430 | ETFBETA | Electron transfer flavoprotein beta/electron carrier activity |
| AT1G27360 | SPL11 | Squamosa promoter‐like 11/regulate timing of transition from vegetative to reproductive phase, transcription factor |
| AT3G56210 | ARM repeat superfamily protein | |
| AT4G27620 | Arabidopsis thaliana unknown protein | |
| AT5G03540 | EXO70A1 | Exocyst subunit EXO70 family protein A1/exocytosis |
| AT1G01800 | NADBP | NAD(P)‐binding Rossmann‐fold superfamily protein/oxidoreductase activity |
| AT3G17090 | PP2C | Protein phosphatase 2C family protein/dephosphorylation |
| AT3G18670 | Ankyrin repeat family protein | |
| AT1G26940 | CLP | Cyclophilin‐like protein/protein folding, isomerization |
| AT1G35670 | CDPK2/CPK11 | Calcium‐dependent protein kinase/induced by drought and high salt, positive regulator of ABA signalling |
| AT3G27700 | Zinc finger (CCCH‐type) protein/DNA, RNA binding | |
| AT1G30870 | PER7 | Peroxidase superfamily protein/haem binding, metal ion binding, peroxidase activity |
| AT4G35890 | LARP 1C | LAM domain‐containing protein/leaf senescence |
| AT3G24880 | EAF 1A | Nuclear protein interacting with NuA4 histone acetyltransferase complex/DNA, protein binding |
| AT3G23540 | Alpha/beta‐hydrolase superfamily protein | |
| AT3G57570 | ARM repeat superfamily protein | |
| AT4G26400 | RING/U‐box protein/zinc ion binding, response to chitin | |
| AT2G22910 | NAGS1 | N‐acetyl‐l‐glutamate synthase 1/arginine biosynthesis |
| AT5G61570 | Protein kinase family protein/phosphorylation | |
| AT3G24350 | SYP32 | SYNTAXIN OF PLANTS 32/vesicle‐mediated transport |
| AT4G00830 | LIF2 | LHP1‐Interacting Factor 2, RNA‐binding partner of LHP1 (Polycomb complex component)/controls flowering, cell fate |
| AT1G75850 | VPS35B/LAZ4 | Vacuolar protein sorting‐associated protein 35/intracellular protein transport |
| AT5G65205 | NAD(P)‐binding Rossmann‐fold superfamily protein, oxidoreductase activity | |
| AT4G26240 | Arabidopsis thaliana unknown protein | |
| AT5G05987 | PRA1.A2 | PRENYLATED RAB ACCEPTOR 1.A2, vesicular transport |
| AT2G25570 | Arabidopsis thaliana unknown protein | |
| AT2G17670 | Tetratricopeptide repeat (TPR)‐like superfamily protein | |
| AT5G59870 | H2A.W.6/HTA6 | Histone H2A protein/chromatin binding, heterochromatin organization |
| AT1G22620 | SAC1 | SUPPRESSOR OF ACTIN (SAC) domain containing phosphatidylinositol‐4,5‐bisphosphate 5‐phosphatase/morphogenesis, cell wall synthesis, and actin organization |
EAF 1A, ESA1‐associated factor 1A; GBF2, G‐Box binding factor 2; LARP 1C, La‐related protein 1C; UBN1, Ubinuclein 1.
Genes whose expression is induced by DEX treatment in a CHX‐independent manner in rsl2 rsl4 GR:RSL4 plants encode proteins involved in a variety of cellular activities, including signalling, secretion and cell wall modification (Table 1). As RSL4 is a key regulator of root hair development, we predicted that at least some of these genes would be required for root hair elongation. We determined the root hair phenotype in plants homozygous for T‐DNA insertions in nine of the 34 putative RSL4 target genes. These nine genes were expressed at high levels in the root hair and/or other root tissues as reported in the Arabidopsis e‐FP Browser (Table S5) (Winter et al., 2007). They included G‐BOX BINDING FACTOR2 (GBF2) (AT4G01120), EXOCSYT SUBUNIT 70A1 (EXO70A1) (AT5G03540), NAD(P) BINDING PROTEIN (NADBP) (AT1G01800), PROTEIN PHOSPHATASE 2C FAMILY PROTEIN (PP2C) (AT3G17090), CYCLOPHILIN‐LIKE PROTEIN (CLP) (AT1G26940), CALCIUM‐DEPENDENT PROTEIN KINASE11 (CPK11/CDPK2) (AT1G35670), PEROXIDASE7 (PRX7) (AT1G30870), VACUOLAR PROTEIN SORTING‐ASSOCIATED PROTEIN 35B (VPS35B/LAZ4) (AT1G75850) and SUPPRESSOR OF ACTIN (SAC1) (AT1G22620). Plants with reduced function of four of these nine genes developed shorter root hairs than the wild‐type; root hair initiation was normal but the subsequent elongation by tip growth was defective (Fig. 4a,b). No difference was observed in root hair length between the wild‐type and plants homozygous for T‐DNA insertions in the remaining five genes (Figs 4a,b, S1).
Figure 4.
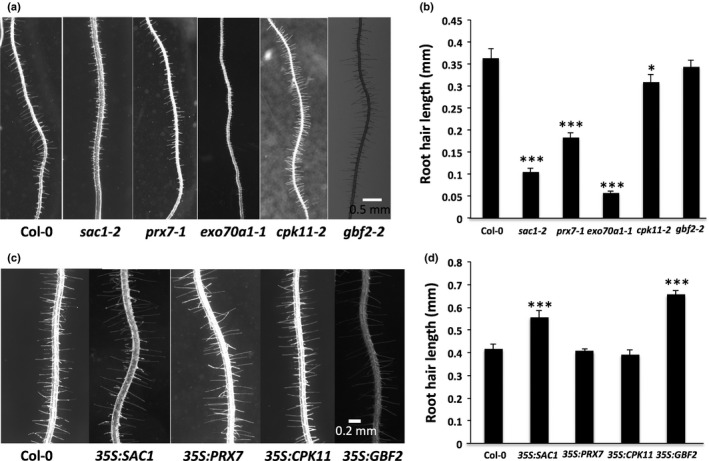
Targets of ROOT HAIR DEFECTIVE SIX‐LIKE4 (RSL4) regulation encode proteins involved in cell signalling, cell wall modification and secretion that are required root hair growth. (a) Seven‐day‐old Arabidopsis thaliana wild‐type (Col‐0 roots), suppressor of actin 1‐2 (sac1‐2), peroxidase7‐1 (prx7‐1), exocsyt subunit 70a1‐1 (exo70a1‐1),calcium‐dependent protein kinase11‐2 (cpk11‐2) and g‐box binding factor2‐2 (gbf2‐2) roots. (b) Root hair length measurements of 7‐d‐old wild‐type (Col‐0 roots), sac1‐2, prx7‐1, exo70a1‐1, cpk11‐2 and gbf2‐2 roots (n = 15). (c) Images of 7‐d‐old roots of wild‐type (Col‐0) and plants transformed with 35S:SAC1, 35S:PRX7, 35S:CPK11 and 35S:GBF2; (d) root hair length measurements of wild‐type (Col‐0) and plants transformed with 35S:SAC1, 35S:PRX7, 35S:CPK11 and 35S:GBF2 (n = 15). Error bars represent SEM. *, P < 0.05; ***, P < 0.001 by Student's t‐test.
SAC1 (AT1G22620) encodes a phosphatidylinositol (PI) phosphatase that dephosphorylates phosphotidylinositol 3,5 bisphosphate to form phosphotidylinositol 3‐phosphate (Zhong et al., 2005). While the root hairs initiated normally on plants that were homozygous for T‐DNA insertions in SAC1, root hair elongation was defective (Fig. 4a); wild‐type root hairs were 0.36 mm (± 0.02) (mean ± SEM) long while those root hairs that elongated in sac1 were 0.10 mm (± 0.01) (mean ± SEM) long (Fig. 4b). This suggests that SAC1 is required for the initiation of tip growth. As SAC1 is required for root hair elongation, we hypothesized that constitutive expression of SAC1 would lead to the formation of longer hairs compared with the wild‐type. To test this hypothesis, we constitutively expressed SAC1 by transforming wild‐type plants with a cauliflower mosaic virus (CaMV) 35S:SAC1 construct (Fig. 4c,d). Root hairs were 0.42 mm (± 0.02) (mean ± SEM) long in the wild‐type while root hairs were 0.56 mm (± 0.02) (mean ± SEM) long in 35S:SAC1 transformed lines (Fig. 4d). This indicates that not only is SAC1 required for root hair growth but that its activity is limiting for growth. As auxin induces RSL4 activity, we tested the effect of auxin on the expression of SAC1. We found that the mRNA levels of SAC1 were increased two‐fold upon 100 nM IAA treatment compared with controls (Fig. S2). These data are consistent with the hypothesis that auxin positively regulates SAC1 expression via RSL4.
PRX7 encodes a class III peroxidase that plays a role in the cleavage and crosslinking of the cell wall polysaccharides (Tognolli et al., 2002; Valerio et al., 2004). The root hairs of plants homozygous for T‐DNA insertions into PRX7 are almost half the length of wild‐type root hairs (Fig. 4a); prx7‐1 root hairs were 0.18 mm (± 0.01 mm) (mean ± SEM) and wild‐type root hairs were 0.36 mm (± 0.02) (mean ± SEM) (Fig. 4b). Root hair length was indistinguishable between the wild‐type and wild‐type plants transformed with the CaMV 35S:PRX7 construct (Fig. 4c,d). This indicates that PRX7 is required for root hair elongation but that increased levels of PRX7 expression are not sufficient for hair elongation.
EXO70A1 is a subunit of exocyst, a secretory vesicle tethering complex that spatially regulates exocytosis into the plasma membranes of growing cells (Li et al., 2010). Trichoblasts of plants homozygous for T‐DNA insertion in EXO70A1, exo70a1‐1, develop swellings that do not elongate, indicating a defect in the transition to tip growth (Fig. 4a); wild‐type root hairs were 0.36 ± 0.02 mm (± SEM) long whereas exo70a1‐1 mutant root hairs were 0.06 ± 0.01 mm in length (Fig. 4b). A similar root hair growth defect was seen in plants with another mutant allele, exo70a1‐2 (Fig. S3). Both these alleles have been previously reported to confer defects in root hair growth (Synek et al., 2006). The phenotypes of exo70a1 mutant root hairs suggest that EXO70A1 activity is required for the initiation of tip growth from the bulge that forms at the earliest stage of root hair development.
Calcium‐dependent protein kinases (CPKs) catalyse Ca2+‐dependent phosphorylation in cell signalling processes (Patil et al., 1995; Sanders et al., 1999). CPK11 was identified as a putative direct downstream target of RSL4. Loss‐of‐function cpk11‐2 mutants developed shorter root hairs than Col‐0 (Fig. 4a). cpk11‐2 root hairs were 0.31 mm ± 0.01 mm (mean ± SEM) long, compared with 0.36 mm ± 0.02 (mean ± SEM) in Col‐0 (Fig. 4b). To determine if overexpression of CPK11 increased root hair elongation, we transformed wild‐type plants with the 35S:CPK11 construct. Root hair length in plants transformed with 35S:CPK11 was identical to that of untransformed controls (Fig. 4c,d), indicating that the levels of CPK11 expression in wild‐type are not limiting for growth. Maintenance of a tip‐focused cytoplasmic calcium (Ca2+) gradient is indispensable for root hair growth and CPK11 may be a component of the calcium signalling pathway, regulating RSL4‐mediated root hair elongation.
One of the 34 putative targets of RSL4 encoded a protein with a putative role in transcription. GBF2 was induced by DEX in a CHX‐insensitive fashion by RSL4:GR. GBF2 has been reported to control blue light‐mediated growth and development in Arabidopsis. To determine if GBF2 is involved in root hair elongation, we generated loss‐of‐function and gain‐of‐function GBF2 lines. Root hair development in plants homozygous for a T‐DNA insertion in GBF2 (gbf2‐2) is identical to that of the wild‐type (Fig. 4a,b). However, root hairs of plants transformed with CaMV 35S:GBF2 were longer (0.66 ± 0.02 mm (SEM)) than those of the wild‐type (0.41 ± 0.02 mm (SEM)) (Fig. 4c,d). This indicates that RSL4‐regulated GBF2 expression promotes root hair elongation but it is likely that another gene can compensate for reduced GBF2 function.
Loss of function of four genes had no impact on root hair elongation; the length of nadbp‐1, pp2c‐1, clp‐1, vps35a‐1 mutant root hairs was identical to that of the wild‐type (Fig. S1). To identify a function in root hair elongation in the four genes for which loss‐of‐function mutants provided no functional information, we generated gain‐of‐function lines. Root hair length was indistinguishable from that of wild‐type controls (0.41 ± 0.02 mm (SEM)) in plants transformed with CaMV 35S:NADBP, 35S:PP2C, 35S:CLP and 35S:VPS35B constructs. This indicates that overexpression of none of these four genes is sufficient for root hair elongation. Therefore, we cannot define a role for these genes in root hair development. It is possible that these genes are not involved in root hair elongation, that genetic redundancy compensates for their loss of function or that these genes are not required for root hair elongation in the conditions used in this analysis.
Taken together, these results suggest that genes regulating secretion, cell wall modifications and signal transduction modulate RSL4‐mediated root hair growth.
Discussion
We have shown that RSL4 controls root hair elongation by regulating the transcription of genes that are required for cell growth. These include genes encoding proteins involved in vesicle trafficking, lipid signalling and those encoding RAB GTPase regulators among others. There are also genes that encode proteins involved in cellular activities associated with transcriptional regulation and chromatin components among the 34 putative targets. Four genes (12%) encode proteins of unknown function in plants and their function remains to be determined.
Among the genes required for root hair growth that are regulated by RSL4 is SAC1. While loss of SAC1 function results in short root hairs, gain of function results in longer root hairs than those of the wild‐type. Together, these gain‐of‐function and loss‐of‐function data are strong evidence that SAC1 is required for elongation during root hair cell morphogenesis. fragile fibre7 (fra7) mutants were identified because they have defects in the morphogenesis of fibre cells and the defect is caused by mutations in the SAC1 gene (Zhong et al., 2005). This indicates that FRA7/SAC1 is required for morphogenesis of diverse cell types in the plant. SAC1 encodes a PI phosphatase. PIs regulate several cellular processes such as signal transduction, vesicle trafficking and the organization of the actin cytoskeleton (Zhong et al., 2005). SAC domain‐containing phosphatases are a group of enzymes that were first identified in yeast and animals and have been shown to hydrolyse phosphates on several positions of the inositol head group of PI (Hughes et al., 2000). There are nine SAC domain‐containing PI phosphatase‐like proteins (SAC1−9) in Arabidopsis (Zhong & Ye, 2003). Mutations in the SAC7/ROOT HAIR DEFECTIVE H4 (RHD4) gene result in the formation of short, bulged and branched root hairs (Thole et al., 2008). Together, these data indicate that members of the SAC family of phosphatases are involved in root hair elongation.
The defective root hair elongation in the sac1‐2 mutant indicates that SAC1‐mediated phosphorylation of phosphotidylinositol phosphates (PIPs) is important for root hair growth. There is evidence that other proteins that catalyse the phosphorylation or dephosphorylation of PIs are also required for root hair elongation. It is hypothesized that SAC1 dephosphorylates phosphotidylinositol 3,5‐bisphosphate(PtdIns) (3,5)P2 to phosphatidylinositol 3‐phosphate PtdIns(3)P (Zhong et al., 2005). The reverse reaction is catalysed by a PtdIns3P 5‐kinase (FORMATION OF APLOID AND BINUCLEATE CELLS, FAB1), to form PtdIns(3,5)P2. Both FAB1 gain‐of‐function (overexpression) and loss‐of‐function mutants develop short root hairs indicative of the role of FAB1 in root hair development (Hirano et al., 2011). Other kinase enzymes that phosphorylate PI substrates have been implicated in root hair development. For example, VPS34 is a class III PI3‐kinase that phosphorylates PI to form PtdIns(3)P. Inhibition of VPS34 activity results in the development of shorter root hairs with slower growth rates than wild‐type (Lee et al., 2008). Taken together, these results demonstrate the importance of the PtdIns(3)P pathway in regulating root hair cell growth. RSL4‐regualted SAC1 transcription is a key element in the regulation of the pathway.
Secretion is necessary for cell growth; it delivers lipids, polysaccharides and proteins that constitute the new plasma membrane and cell wall of the growing cell. We report that the transcript levels of EXO70A1 are probably directly regulated by the bHLH transcription factor RSL4. EXO70A1 encodes a subunit of the exocyst protein complex (Synek et al., 2006). This complex regulates the first contact (tethering) between secretory vesicles and the target membrane. It plays a crucial role in the transport and secretion of new wall and membrane materials from the endo membrane system to the growing surface. Kulich et al. (2010) showed that EXO70A1 is essential for the polarized delivery of cell wall pectins during seed coat development (Kulich et al., 2010). EXO70A1 is also involved in the polar recycling of PIN1 and PIN2 and thus participates in polar auxin transport (Drdova et al., 2013). It has previously been reported that root hair elongation is defective in exo70a1 mutants with reduced function (Synek et al., 2006). exo70a1 mutants developed bulges from which root hairs did not elongate, suggesting that there is a defect in transition to tip growth in this background. The defective root hair elongation phenotype of exo70a1‐2 mutants is partially rescued by Napthalene Acetic Acid (NAA) treatments (Synek et al., 2006). The inability of NAA to completely rescue root hair elongation in this mutant indicates that auxin‐modulated root hair elongation acts least partially through EXO70A1. The partial rescue of the Exo70a1 mutant phenotype is probably attributable to auxin regulation of other activities that in part compensate for the loss of EXO70A1 function. In rice, OsEXO70A1 has been implicated in the regulation of nutrient assimilation which may be attributable to defects in root hair‐mediated nutrient uptake (Tu et al., 2015). Given that RSL4 activity is increased by low nutrient availability which induces root hair elongation, our data are consistent with a model in which RSL4‐mediated EXO70A1 transcription is required for tip growth.
Root hair growth is accompanied by the modification of the plant cell wall and our finding that PRX7, a class III peroxidase gene, is regulated by RSL4 suggests that RSL4 promotes root hair growth, in part by regulating PRX7‐modulated cell wall modification. Plant cell walls are dynamic structures that determine shape and mechanically support the cell. Several enzymes including class III peroxidases have been implicated in the cleavage and reassembly of the cell wall components during dynamic transformations. There are reports that peroxidases enable wall loosening by generating reactive oxygen species (ROS) (Passardi et al., 2004). PRX33 and PRX34 have been shown to be involved in root elongation by promoting cell wall loosening (Passardi et al., 2006). Mutations in two peroxidase genes, PRX44 and PRX57, resulted in root hair cell wall rupture indicative of defective cell walls at the growth tip where walls are thin and mechanically weakest (Kwon et al., 2015). Our finding that PRX7 acts downstream of RSL4 indicates that cell wall modifications play an important role in the RSL4‐mediated root hair elongation process.
We identified CPK11 as a potential direct target of RSL4 using an inducible RSL4 protein driven from its native promoter. CPK11 encodes a calcium‐dependent protein kinase that has been shown to regulate the abscisic acid signalling pathways in Arabidopsis (Zhu et al., 2007). CPKs are characterized by the presence of a C‐terminal calmodulin‐like regulatory domain with four calcium‐binding EF hands and their activity is regulated by Ca2+ signals (Patil et al., 1995; Sanders et al., 1999). Medicago truncatula CDPK1 has been shown to regulate root hair elongation. Silencing of CDPK1 by RNA interference resulted in the development of short root hairs (Ivashuta et al., 2005). Furthermore, the maintenance of tip‐focused cytoplasmic calcium (Ca2+) is indispensable for root hair growth. A local elevation in the concentration of cytoplasmic Ca2+ in the growing tip is required for root hair elongation (Bibikova et al., 1997). CPK11 may be a component of the calcium signalling pathway involved in RSL4‐mediated root hair elongation.
Five genes did not mutate to a root hair defective phenotype. This suggests that they are not involved in root hair elongation. However, it is possible that similar genes have overlapping functions and functionally compensate in the single mutants that we characterized here. Such genetic redundancy is thought to be common and the observation that there are many closely related genes in families supports this hypothesis. Genetic redundancy can be determined by generating lines in which there are mutations in more than one gene. It is also formally possible these genes are not involved in root hair growth. These genes may carry out other roles in the root hair that are not related to growth per se but some other function that is restricted to root hair cells, as RSL4 expression is restricted to this cell type.
Author contributions
P.V. carried out the transcriptional analysis under the supervision of S.D. P.V. and S.D. analysed the data of the transcriptional analysis. P.V., S.D. and L.D. analysed the complete data set of the study. L.D. conceived and managed the project. S.D. and L.D. wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Six‐day‐old Arabidopsis thaliana wild‐type Col‐0, nadbp‐1, pp2c‐1, clp‐1 and vps35a‐1 roots.
Fig. S2 SAC1 mRNA level in the absence and presence of exogenously applied 100 nM IAA.
Fig. S3 Six‐day‐old Arabidopsis thaliana wild‐type (Col‐0) and exo70a1‐2 mutants with short root hairs.
Table S1 List of primers used in this study
Table S5 List of Arabidopsis thaliana mutants used in this study with details of T‐DNA insertions
Table S2 1022 genes in Arabidopsis thaliana whose expression was more than twice as high in DEX‐treated GR:RSL4 rsl2 rsl4 plants as in mock‐treated plants with a P‐value < 0.0001
Table S3 259 genes in Arabidopsis thaliana whose expression was more than twice as high in DEX+CHX‐treated GR:RSL4 rsl2 rsl4 as in mock‐treated plants with a P‐value < 0.001
Table S4 130 genes in Arabidopsis thaliana whose expression was increased by more than two‐fold in both DEX‐ and DEX+CHX‐treated GR:RSL4 rsl2 rsl4 plants compared with mock treatment
Acknowledgements
This research was funded by a BBSRC grant (BBG0198941) and an ERC (EVO500; Project no: 25028) grant to L.D. S.D. received funding from EMBO LTF (ALTF 817‐2008) and EU Marie Curie FP7‐PEOPLE‐IEF‐2008 (Project no. 236802) and P.V. received funding through a Scholarship from Exeter College.
References
- Balcerowicz D, Schoenaers S, Vissenberg K. 2015. Cell fate determination and the switch from diffuse growth to planar polarity in Arabidopsis root epidermal cells. Frontiers in Plant Science 6: 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. 2000. Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). American Journal of Botany 87: 958–963. [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57: 289–300. [Google Scholar]
- Bibikova TN, Zhigilei A, Gilroy S. 1997. Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203: 495–505. [DOI] [PubMed] [Google Scholar]
- Brown LK, George TS, Dupuy LX, White PJ. 2013. A conceptual model of root hair ideotypes for future agricultural environments: what combination of traits should be targeted to cope with limited P availability? Annals of Botany 112: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Dolan L. 2002. Building a hair: tip growth in Arabidopsis thaliana root hairs. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences 357: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Datta S, Kim CM, Pernas M, Pires ND, Proust H, Tam T, Vijayakumar P, Dolan L. 2011. Root hairs: development, growth and evolution at the plant–soil interface. Plant and Soil 346: 1–14. [Google Scholar]
- Datta S, Prescott H, Dolan L. 2015. Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nature Plants 1: 15138. [DOI] [PubMed] [Google Scholar]
- Drdova EJ, Synek L, Pecenkova T, Hala M, Kulich I, Fowler JE, Murphy AS, Zarsky V. 2013. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant Journal 73: 709–719. [DOI] [PubMed] [Google Scholar]
- Hirano T, Matsuzawa T, Takegawa K, Sato MH. 2011. Loss‐of‐function and gain‐of‐function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiology 155: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ. 2000. Sac phosphatase domain proteins. Biochemical Journal 350: 337–352. [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer‐Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. 2008. A genetic regulatory network in the development of trichomes and root hairs. Annual Review of Plant Biology 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, VandenBosch KA, Vance CP, Harrison MJ, Gantt JS. 2005. RNA interference identifies a calcium‐dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17: 2911–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Nagarajan VK, Raghothama KG. 2012. Transcriptional regulation of phosphate acquisition by higher plants. Cellular and Molecular Life Sciences 69: 3207–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H, Kapulnik Y. 2013. Unveiling signaling events in root responses to strigolactone. Molecular Plant 6: 589–591. [DOI] [PubMed] [Google Scholar]
- Kulich I, Cole R, Drdová E, Cvrčková F, Soukup A, Fowler J, Žárský V. 2010. Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytologist 188: 615–625. [DOI] [PubMed] [Google Scholar]
- Kwon T, Sparks JA, Nakashima J, Allen SN, Tang Y, Blancaflor EB. 2015. Transcriptional response of Arabidopsis seedlings during spaceflight reveals peroxidase and cell wall remodeling genes associated with root hair development. American Journal of Botany 102: 21–35. [DOI] [PubMed] [Google Scholar]
- Lee Y, Bak G, Choi Y, Chuang W‐I, Cho H‐T, Lee Y. 2008. Roles of phosphatidylinositol 3‐kinase in root hair growth. Plant Physiology 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RD, Cho HT. 2013. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Frontiers in Plant Science 4: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, van Os GMA, Ren S, Yu D, Ketelaar T, Emons AMC, Liu C‐M. 2010. Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type‐specific exocytosis. Plant Physiology 154: 1819–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. 1994. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid‐inducible regulator. Science 266: 436–439. [DOI] [PubMed] [Google Scholar]
- Lopez‐Arredondo DL, Leyva‐Gonzalez MA, Gonzalez‐Morales SI, Lopez‐Bucio J, Herrera‐Estrella L. 2014. Phosphate nutrition: improving low‐phosphate tolerance in crops. Annual Review of Plant Biology 65: 95–123. [DOI] [PubMed] [Google Scholar]
- Lopez‐Bucio J, Cruz‐Ramirez A, Herrera‐Estrella L. 2003. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6: 280–287. [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. 2013. Responses of root architecture development to low phosphorus availability: a review. Annals of Botany 112: 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. 2004. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends in Plant Science 9: 534–540. [DOI] [PubMed] [Google Scholar]
- Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C. 2006. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223: 965–974. [DOI] [PubMed] [Google Scholar]
- Patil S, Takezawa D, Poovaiah BW. 1995. Chimeric plant calcium/calmodulin‐dependent protein kinase gene with a neural visinin‐like calcium‐binding domain. Proceedings of the National Academy of Sciences, USA 92: 4897–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H, Honkanen S, Jones VA, Morieri G, Prescott H, Kelly S, Ishizaki K, Kohchi T, Dolan L. 2016. RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants. Current Biology 26: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz‐Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar‐Henao JE, Schmidt W. 2016. An inventory of nutrient‐responsive genes in Arabidopsis root hairs. Frontiers in Plant Science 7: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF. 1999. Communicating with calcium. Plant Cell 11: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot‐Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. 2007. Root tip contact with low‐phosphate media reprograms plant root architecture. Nature Genetics 39: 792–796. [DOI] [PubMed] [Google Scholar]
- Synek L, Schlager N, Elias M, Quentin M, Hauser MT, Zarsky V. 2006. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant Journal 48: 54–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr, Nielsen E. 2008. root hair defective4 encodes a phosphatidylinositol‐4‐phosphate phosphatase required for proper root hair development in Arabidopsis thaliana . Plant Cell 20: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P. 2002. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana . Gene 288: 129–138. [DOI] [PubMed] [Google Scholar]
- Tu B, Hu L, Chen W, Li T, Hu B, Zheng L, Lv Z, You S, Wang Y, Ma B et al 2015. Disruption of OsEXO70A1 causes irregular vascular bundles and perturbs mineral nutrient assimilation in rice. Scientific Reports 5: 18609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio L, De Meyer M, Penel C, Dunand C. 2004. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 65: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix‐loop‐helix transcription factor controls cell growth and size in root hairs. Nature Genetics 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Lynch JP, Brown KM. 2003. Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. Journal of Experimental Botany 54: 2351–2361. [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Nairn CJ, Wood‐Jones A, Morrison WH 3rd, Ye ZH. 2005. Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17: 1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2003. The SAC domain‐containing protein gene family in Arabidopsis. Plant Physiology 132: 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S‐Y, Yu X‐C, Wang X‐J, Zhao R, Li Y, Fan R‐C, Shang Y, Du S‐Y, Wang X‐F, Wu F‐Q et al 2007. Two calcium‐dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Six‐day‐old Arabidopsis thaliana wild‐type Col‐0, nadbp‐1, pp2c‐1, clp‐1 and vps35a‐1 roots.
Fig. S2 SAC1 mRNA level in the absence and presence of exogenously applied 100 nM IAA.
Fig. S3 Six‐day‐old Arabidopsis thaliana wild‐type (Col‐0) and exo70a1‐2 mutants with short root hairs.
Table S1 List of primers used in this study
Table S5 List of Arabidopsis thaliana mutants used in this study with details of T‐DNA insertions
Table S2 1022 genes in Arabidopsis thaliana whose expression was more than twice as high in DEX‐treated GR:RSL4 rsl2 rsl4 plants as in mock‐treated plants with a P‐value < 0.0001
Table S3 259 genes in Arabidopsis thaliana whose expression was more than twice as high in DEX+CHX‐treated GR:RSL4 rsl2 rsl4 as in mock‐treated plants with a P‐value < 0.001
Table S4 130 genes in Arabidopsis thaliana whose expression was increased by more than two‐fold in both DEX‐ and DEX+CHX‐treated GR:RSL4 rsl2 rsl4 plants compared with mock treatment


