Abstract
Background
Changes in susceptibility patterns of bacterial pathogens isolated from urinary tract infections emphasize the need for regional surveillance to generate information that can be used in management of patients. Knowledge on the current status of antimicrobial resistance in uropathogens, and the prevalence of expanding spectrum beta-lactamases (ESBLs) in the isolates will guide policy formulations and encourage prudent use of antimicrobials.
Objective
Identify bacterial pathogens causing UTI and determine the association between the pathogens isolated from patients attending KNH. Determine antimicrobial susceptibility patterns of the UTI pathogens and the prevalence of ESBL in the isolated pathogens.
Design
Laboratory-based study.
Setting
Department of Medical Microbiology University of Nairobi and Kenyatta National Hospital microbiology laboratory, Nairobi, Kenya.
Subjects
Nine hundred and forty eight patients presenting directly to the Kenyatta National Hospital’s diagnostic lab. Patients were only classified as in-patients if at the time of specimen collection they were being admitted to one of KNH wards.
Results
Out of the 948 urine samples processed, 189 in-patients and 37 out-patients samples had significant bacterial growth. The uropathogens identified from in-patient specimens were Escherichia coli (56), Klebsiellapneumoniae (33), Enterococcus spp. (34) and Entrobacter (16) making up 30%, 18%, 18% and 9% respectively. ESBL isolates were found to be resistant to the locally administered antibiotics; Augmentin (37%), Levofloxacin (37%), Cefoperazone (37%), Ampicillin (39%), Doxycyline (41%), Gentamicin (30%) and Nalidixic Acid (38%).
Conclusion
The increased prevalence of multidrug resistant ESBL pathogens poses challenges for health care providers at KNH and signifies the need for new approach to treat UTI. It would be prudent for laboratories to include specialized tests for detection of ESBL producing pathogens from isolates obtained from in-patients. Further studies on the mechanisms and pathways utilized by these bacteria to cause UTI will highlight other avenues in patient management.
INTRODUCTION
Urinary Tract Infection (UTI) is a general term that refers to infection/inflammation of any part of the urinary tract caused by bacteria. UTI is one of the most common bacterial infections encountered by clinicians worldwide (1). Moreover, since reporting of antibiotic susceptibility results in suspected cases of UTI takes at least 48 hours following sampling, the condition is treated empirically, based on available data reflecting antibiotic resistance (2).
UTI is classified into two major categories, namely complicated infection or uncomplicated infection. The term complicated refers to the fact that an individual’s urinary tract has been in some way obstructed or is abnormal. Uncomplicated infections are far more common and apply to infections within a normal, unobstructed tract (3). If UTI is not detected in early stages and treated adequately, it can result in chronic illness and long term renal damage (4). Uncomplicated UTI in patients can be treated since the causative microbes are highly predictable: 80%–90% are caused by Escherichia coli (5).
Beta-lactam antibiotics are among the safest and most frequently prescribed antimicrobial agents in the United States and worldwide (6). However, the evolution of beta-lactam resistance via the extended spectrum beta-lactamase (ESBL), an enzyme produced by beta-lactam resistant bacteria in clinically encountered pathogens has greatly limited their utilization. ESBLs are capable of bacterial resistance against common UTI antibiotics such as penicillin, cephalosporin, cephamycins and monobactams by hydrolyzing the antibiotics (7,8).
Primary ESBL-producing bacteria are from Enterobacteriaceae family of Gram-negative organisms, especially Klebsiellapneumoniae and Escherichia coli (9,10). Proteus mirabilis, Pseudomonas aeruginosa, Acinetobacter spp., Serratiaspp, and gram-positive bacteria such as Enterococcusspp, and Staphylococcus spp. are also known to cause UTI (11). Understanding these common uropathogens and their susceptibility patterns will help to improve prescription decisions for both complicated and uncomplicated UTI (6).
ESBL infections also are more aggressive than non-ESBL infections. Patients suffering from ESBL induced UTI often encounter: delay in the initiation of appropriate therapy, significantly longer hospital stays, higher medical costs, recurrent UTI and increased risk of clinical failure and death in comparison to non-ESBL induced UTI (12). One class of drugs successfully used to treat ESBL bacteria are carbapenems (13). However, due to the widespread use of these antibiotics in the treatment of UTI, researchers and clinicians fear that ESBLs will soon become resistant to the drugs.
In Africa, there is no comprehensive data regarding ESBL producing Enterobacteriaceae (10). However, there is sufficient evidence to suggest that there is ESBL-producing uropathogens in Africa (10). Also, there had been reports of UTIs owing to ESBL-producing bacteria being on the rise. One longitudinal study in Turkey showed an increase from 13 UTI cases due to ESBLs to 111 ESBL cases at the end of five years (13). These numbers suggest that ESBLs are outcompeting non- ESBLs and are replacing the latter as the dominant uropathogens. Thus, studies to increase knowledge on pathogens causing UTI and their antibiotic resistance patterns regionally are very important to guide clinicians in empiric treatment (2).
METHODS AND MATERIALS
The study protocol was approved with a waiver of informed consent by the Institutional Review Board of the University of Alabama at Birmingham and the Kenyatta National Hospital Ethical Review Committee before its implantation. Study investigators had no direct contact with the patients and were provided with urine samples containing only identification numbers by the hospital laboratory staff. All urine samples had undergone routine processing by microscopy and culture on the urinalysis bench and patients were treated according to the results obtained.
This laboratory-based study was conducted in the Department of Medical Microbiology University of Nairobi from May to August 2013. Urine samples from both in and outpatients were cultured and subjected to antibiotics susceptibility testing. Antimicrobial agents used were: Augmentin, Levofloxacin, Meropenem, Ceftazidime, Nalidixic Acid, Gentamicin, Cefoperazone, Ceftriazone, Imipenem, Doxycycline and Ampicillin. Sensitivity and resistance were determined by measuring the diameter of zones around each drug according to Clinical Laboratory Standard Institute (CSLI) guidelines (14).
Isolates were streaked on CHROMagar ESBL media plates to determine whether they were ESBL positive or negative, according to the manufacturer’s instructions. CHROMagar ESBL was obtained directly from the manufacturer (CHROMagar, New Jersey, U.S.A). All components were provided as dehydrated powders and media were prepared in-house according to the manufacturer’s instruction; CHROMagar Orientation (15) was used as the base medium for preparation of CHROMagar ESBL CHROMagar. Quality control for the media was performed using stock organisms of Klebsiellapneumoniae, Escherichia coli, Proteus spp., Pseudomonas spp. and Staphylococcus aureus. Samples streaked on CHROMagar media were incubated overnight at 37 °C. ESBL producing pathogens were determined by full growth on plates i.e. growth past initial inoculation streak coupled with colonies. ESBL negative samples failed to form colonies and/or grow past the initial inoculation streak.
Statistical data analysis was performed to determine the significance in our findings. Data from study was inserted into two separate MS Excel spreadsheets that were later on merged using DigDB to ensure correct match of specimen information with unique patient ID. The present study focused on analyzing the 189 in-patient samples that had bacterial growth. Goodness-of-fit statistics (Pearson’s Chi-square test) was used to compare the fits of different variables using p-values of α-level of 0.05. Descriptive statistics was also employed to explain the outcome of variables.
RESULTS
In our study, we recorded significant bacterial growth among the most common uropathogens. Out of the 948 urine samples that the KNH lab processed, 189 in-patient samples had bacterial growth. Among the isolates, the most common organisms identified were Escherichia coli (56), Klebsiella pneumoniae (33), Enterococcus spp. (34) and Entrobacter (16) making up 30%, 18%, 18% and 9% respectively (see Figure 1).
Figure 1.
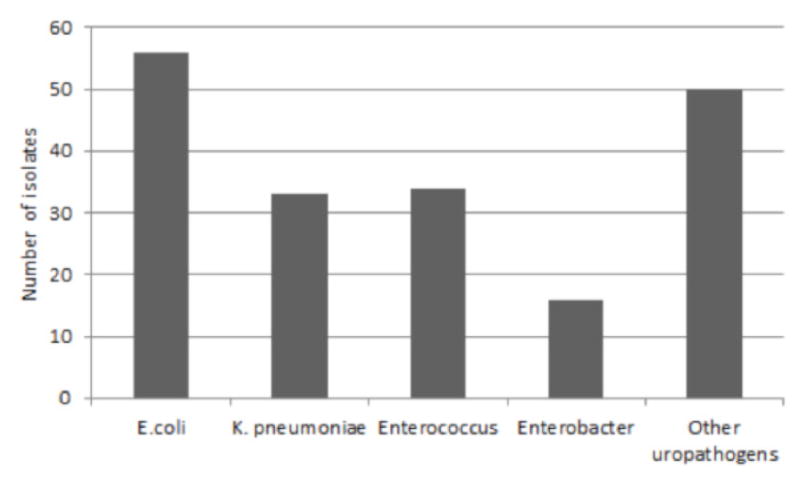
Frequency of uropathogens isolated in the study (n = 189)
The other uropathogens in the chart represents Acinebacter spp., Citrobacter spp., Protes spp., Pseudonomas spp. and Staphylococcus spp.
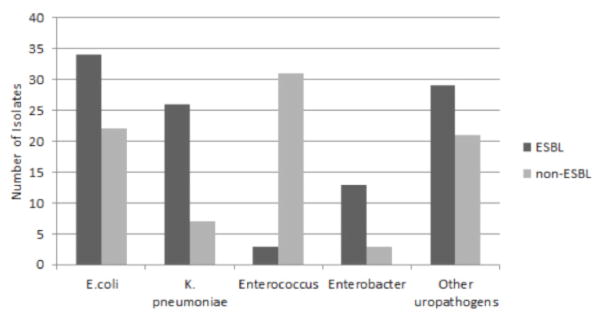
Among E.coli, 34 specimens be were found to ESBL positive while 22 were ESBL negative. Enterobacter spp. Showed 13 ESBL positive and 3 ESBL negative samples. K. pneumoniae contained 26 ESBL positive and 7 ESBL negative samples. Within Enterococcus spp., 3 samples were ESBL positive while 31 samples were ESBL negative. Of the less common organisms isolated including: Citrobacter spp., Proteus spp., Pseudomonas spp. and Staphylococcus spp. 29 were ESBL positive and 21 were ESBL negative isolates. These data are further illustrated in Figure 2.
Figure 2.
Prevalence of ESBLs and non-ESBL producing uropathogens isolated in the study
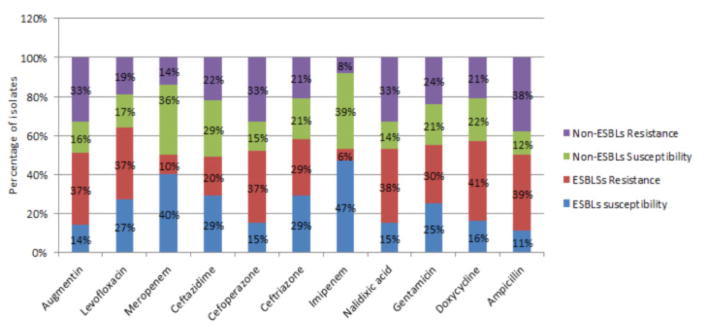
Figure 3 outlines patterns of susceptibility and resistance among ESBL and non-ESBL pathogens to 11 commonly used antibiotics. In 8 out of 11 antibiotics, ESBLs had a greater percentage of resistance when compared to non-ESBL resistance. Meropenem, Ceftazidime and Imipenem has about the non-ESBLs resistance representing 14%, 22% and 8% versus the10%, 20% and 6% of ESBLs respectively. The highest percentages of susceptibility among ESBLs were in response to Imipenem (47%), Meropenem (40%), Cefoperazone (29%), Levofloxacin (27%), Gentamicin (25%) and Ceftazidime (29%). Susceptibility patterns also show that non-ESBL pathogens are susceptible to Imipenem (39%), Meropenem (36%), Cefoperazone (21%), and Doxycycline (22%), Gentamicin (21%) and Ceftazidime (29%).
Figure 3.
Antibiotic susceptibility profile of ESBLs and non-ESBLs uropathogens to commonly used antibiotics at KNH, Nairobi
DISCUSSION
E.coli, K. pneumoniae and Enterococcus spp. were the primary isolates found to cause UTI from our study at KNH. These findings are in agreement with the results from previous studies from Cameroon, Pakistan, Israel and Turkey (2,7,16,13) that found E. coli and K. pneumoniae were the most significant causers of UTIs. However, the development of Enterococcus spp. as a cause of UTI shows that other pathogens may be starting to cause UTI.
Additionally, ESBL positive species within E.coli and Enterobacter spp. constituted 61% and 81% respectively while K. pneumoniae had a 79% to 21% split between ESBL and non-ESBL isolates respectively. Our results indicate that ESBL pathogens were very significant (P<0.0001) across these groups: E.coli, Enterobacterspp. and K.pneumoniae. Even though there is no national surveillance figures for ESBL-producing uropathogensin Kenya, there have been reports of ESBL caused UTI cases (17,18). The fact that some of the uropathogens investigated in this study; E.coli, Enterobacter and K. pneumoniae have a higher prevalence of ESBL pathogens in comparison to the non-ESBLs within the same bacterial species indicates that ESBL pathogens out-compete their non-ESBL counterparts in order to cause UTI. The high percent of those ESBL producing uropathogense specially E. coli and K. pneumoniae have also been reported in previous similar studies (7,16).
ESBL pathogens are often known to be multidrug resistant. Our results showed ESBLs were highly resistant (over 30%) to Augmentin, Levofloxacin, Cefoperazone Ampicillin, Doxycycline, Nalidixic Acid and Gentamicin. The resistant to these antibiotics: Augmentin, Levofloxacin and Ampicillin were found to be significant (P<0.005) among the ESBL-producing uropathogens. These data shows that ESBLs are multidrug resistant and pose challenges to clinicians in determining which drugs to use to effectively treat UTI. The emergence of ESBL-producing uropathogens demonstrates the remarkable ability of bacteria to evolve and survive in the world of antibiotics. Our Clinicians face clinical and economic challenges on managing UTI due to such resistant isolates, the need to perform specialised laboratory tests for detection, and treatment with more expensive antibiotic agents will be necessary in some cases.
Our study focused primarily on the in-patient samples mainly because of the smaller outpatient population size (15.7%). A comparatively higher outpatient population size is necessary to determine if UTI caused by ESBLs from our in-patient study resulted from nosocomial or was community acquired infection. A large scale study with balanced in-patients and out-patients is needed to resolve the cause of the infection.
Lack of access to previous patient history of UTI hindered our assessment of how current ESBL causing pathogens differ from previously acquired ESBL strains.
Acknowledgments
We thank all of the members of the Medical Microbiology Lab at KNH that helped us execute this study. MeghaJha helped in the statistical analysis of the data.
FUNDING
This study was supported by the Minority Health International Research Training(MHIRT) grant no. T37-MD001448 from the National Institute on Minority Health and Health Disparities, National Institutes of Health, Bethesda, MD, USA, the University of Nairobi and the Kenyatta National Hospital, Nairobi, Kenya.
Footnotes
COMPETING INTERESTS
‘The authors declare that they have no competing interests’.
AUTHORS’ CONTRIBUTIONS
IK, HM and MO prepared ESBL media. MO provided instruction on experimental methods. IK and HM carried out ESBL experiments, collected data and drafted manuscript. HM carried out statistical analysis. WJ, PJ, and SO conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Contributor Information
H. I. Magale, The University of Arizona College of Medicine, 1501 N Campbell Ave, Tucson, Arizona, United States
I. A. Kassim, The Brody School of Medicine at East Carolina University, 600 Moye Boulevard, Greenville, North Carolina, United States
S. A. Odera, Department of Medical Microbiology, University of Nairobi, P. O. Box 19676-00202, Nairobi, Kenya
M. J. Omolo, Department of Medical Microbiology, University Of Nairobi, P. O. Box 19676-00202, Nairobi, Kenya
W. G. Jaoko, Department of Medical Microbiology, University of Nairobi, P. O. Box 19676-00202, Nairobi, Kenya, Kenya AIDS Vaccine Initiative (KAVI), School of Medicine, University of Nairobi, P.O. Box 19676-00202, Nairobi, Kenya
P. E. Jolly, Department of Epidemiology, University of Alabama in Birmingham School of Public Health, 1665 University Blvd, Birmingham, Alabama, United States
References
- 1.Biadglegn F, Abera B. Antimicrobial resistance patterns of bacterial isolates from urinary tract infections at Felege Hiwot Referral Hospital, Ethiopia. Ethiop J Health Dev. 2009;23:236–238. [Google Scholar]
- 2.Akoachere JF, Yvonne S, Akum NH, Seraphine EN. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes. 2012;5:219. doi: 10.1186/1756-0500-5-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielubowicz GR, Mobley HL. Host–pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 4.Adjei O, Opoku C. Urinary tract infections in infants. Int J Antimicr Agents. 2004;24:32–34. doi: 10.1016/j.ijantimicag.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 5.DeBusscher J, Zhang L, Buxton M, Foxman B, Barbosa-Cesnik C. Persistent extended-spectrum β-lactamase urinary tract infection. Emerging infectious diseases. 2009;15:1862. doi: 10.3201/eid1511.081501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong-Beringer A. Therapeutic Challenges Associated with Extended-Spectrum, β-Lactamase-Producing Escherichia coli and Klebsiellapneumoniae. Pharmacother: J Hum Pharmacol Drug Therap. 2001;21:583–592. doi: 10.1592/phco.21.6.583.34537. [DOI] [PubMed] [Google Scholar]
- 7.Ejaz H, Zafa A, Mahmood S, Javed MM. Urinary tract infections caused by extended spectrum β-lactamase (ESBL) producing Escherichia coli and Klebsiellapneumoniae. African Journal of Biotechnology. 2013;10:16661–16666. [Google Scholar]
- 8.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended spectrumbeta-lactamase-producing Escherichia coli and Klebsiellapneumoniae: risk factors for infection and impact of resistance on outcomes. Clinical Infectious Diseases. 2001;32:1162–71. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- 9.Paterson DL, Bonomo RA. Extended-spectrumβ-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhillon RHP, Clark J. ESBLs: A clear and present danger? Crit Care Res Pract. 2012;2012:1–11. doi: 10.1155/2012/625170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby GA, Munoz-Price LS. The new beta-lactamases. New England Journal of Medicine. 2005;352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 12.Meyer KS, Urban C, Eagan JA, Berger BJ, Rahal JJ. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Annals of Internal Medicine. 1993:119. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 13.Özçakar ZB, Yalçınkaya F, Kavaz A, Kadıoğlu G, Elhan AH, Aysev D, … Ekim M. Urinary tract infections owing to ESBLi producing bacteria: microorganisms change–clinical pattern does not. Acta Paediatrica. 2011;100:e61–e64. doi: 10.1111/j.1651-2227.2011.02262.x. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. Seventeenth informational supplement. 2007:M100–S16. [Google Scholar]
- 15.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J Clin Microbiol. 1996;34:1788–1793. doi: 10.1128/jcm.34.7.1788-1793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qureshi AM. Organisms causing urinary tract infection in pediatric patients at Ayub Teaching Hospital Abbottabad. J Ayub Med Coll Abbottabad. 2005;17:72–74. [PubMed] [Google Scholar]
- 17.Kiiru J, Kariuki S, Goddeeris BM, Butaye P. Analysis of β-lactamase phenotypes and carriage of selected β-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC microbiology. 2012;1:155. doi: 10.1186/1471-2180-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maina D, Makau P, Nyerere A, Revathi G. Antimicrobial resistance patterns in extended-spectrum β-lactamase producing Escherichia coli and Klebsiellapneumoniae isolates in a private tertiary hospital, Kenya. Microbiology Discovery. 2013;1:5. [Google Scholar]