Significance
Methane, the major natural-gas component, can be converted industrially to syngas (a mixture of CO2, CO, and H2), which is used to produce methanol, an important industrial precursor for producing commodity and specialty chemicals. Unlike methanol’s chemical conversion, which requires high temperature and pressure, its biological conversion, proceeding through its oxidation to formaldehyde, takes place at ambient conditions but suffers from relative low rates. As metabolite biosynthesis by industrial organisms utilizes NADH as an electron donor, methanol oxidation must be carried out by NAD-dependent methanol dehydrogenases, which have poor thermodynamic characteristics at mesophilic conditions. To solve this problem, we developed an engineering strategy for supramolecular enzyme assembly to dramatically enhance the carbon flux from methanol to the key intermediate fructose-6-phosphate.
Keywords: methane, methylotophs, supramolcular, scaffold, substrate channeling
Abstract
Methanol is an important feedstock derived from natural gas and can be chemically converted into commodity and specialty chemicals at high pressure and temperature. Although biological conversion of methanol can proceed at ambient conditions, there is a dearth of engineered microorganisms that use methanol to produce metabolites. In nature, methanol dehydrogenase (Mdh), which converts methanol to formaldehyde, highly favors the reverse reaction. Thus, efficient coupling with the irreversible sequestration of formaldehyde by 3-hexulose-6-phosphate synthase (Hps) and 6-phospho-3-hexuloseisomerase (Phi) serves as the key driving force to pull the pathway equilibrium toward central metabolism. An emerging strategy to promote efficient substrate channeling is to spatially organize pathway enzymes in an engineered assembly to provide kinetic driving forces that promote carbon flux in a desirable direction. Here, we report a scaffoldless, self-assembly strategy to organize Mdh, Hps, and Phi into an engineered supramolecular enzyme complex using an SH3–ligand interaction pair, which enhances methanol conversion to fructose-6-phosphate (F6P). To increase methanol consumption, an “NADH Sink” was created using Escherichia coli lactate dehydrogenase as an NADH scavenger, thereby preventing reversible formaldehyde reduction. Combination of the two strategies improved in vitro F6P production by 97-fold compared with unassembled enzymes. The beneficial effect of supramolecular enzyme assembly was also realized in vivo as the engineered enzyme assembly improved whole-cell methanol consumption rate by ninefold. This approach will ultimately allow direct coupling of enhanced F6P synthesis with other metabolic engineering strategies for the production of many desired metabolites from methanol.
Fossil fuels are recognized as a limited energy source given the world’s growing population and energy demands (1–3). Biofuels and bio-based chemicals have gained tremendous traction over the past decade as a means to produce both an alternative energy source as well as to replace bulk chemical production that is heavily reliant upon petrochemicals. Methanol, which can be produced inexpensively from natural gas or renewably through reduction of carbon dioxide by hydrogen (2, 4), can serve as a feedstock to produce biofuels, amino acids, and polymers (5, 6). Biological conversion of methanol to value-added chemicals may provide more favorable, economic operating conditions (i.e., lower temperature and pressure) than more traditional catalytic conversions.
Methylotrophic bacteria consist of a wide array of organisms that can use one-carbon (C1) compounds, and notably methane and methanol, as their sole source of carbon and energy (7). The ability to use methanol is initiated by its oxidation to formaldehyde by methanol dehydrogenase (Mdh) encoded by an mdh gene (8). There are two classes of Mdhs among methylotrophic bacteria: the well-studied pyrroloquinoline quinone (PQQ)-containing Mdhs, which are found in the most ubiquitous, Gram-negative, strictly aerobic methylotophs, and the NAD(P)+-dependent, cytoplasmic Mdhs commonly found in thermophilic Gram-positive bacteria (9). Generation of most useful metabolites produced by industrial organisms requires electrons in the form of NADH and culture conditions largely microaerobic or anaerobic (7, 10). Thus, organisms that can grow anaerobically or microaerobically and NAD-dependent Mdhs are essential for effective conversion of methanol to desirable metabolites (7). Although NAD(P)+-dependent Mdhs are the best candidates for engineering synthetic methylotrophs like Escherichia coli, these enzymes have poor predicted thermodynamic properties and are thus dependent on maintaining low intracellular formaldehyde and NAD(P)H concentrations to ensure the reaction continues to favor methanol oxidation (7). Formaldehyde, produced from the oxidation of methanol, is subsequently assimilated via distinct methylotrophic pathways, among which the ribulose monophosphate (RuMP) pathway is bioenergetically the most efficient (7). The best characterized NAD(P)+-dependent Mdhs were isolated from the thermotolerant Bacillus methanolicus MGA3 strain, which harbors two chromosomal mdh genes and one plasmid-borne mdh gene (6, 9). In particular, MGA3 Mdh3, a chromosomal mdh, is a decameric enzyme and has been shown to possess broad substrate specificity when tested with a range of primary alcohols (9, 11, 12).
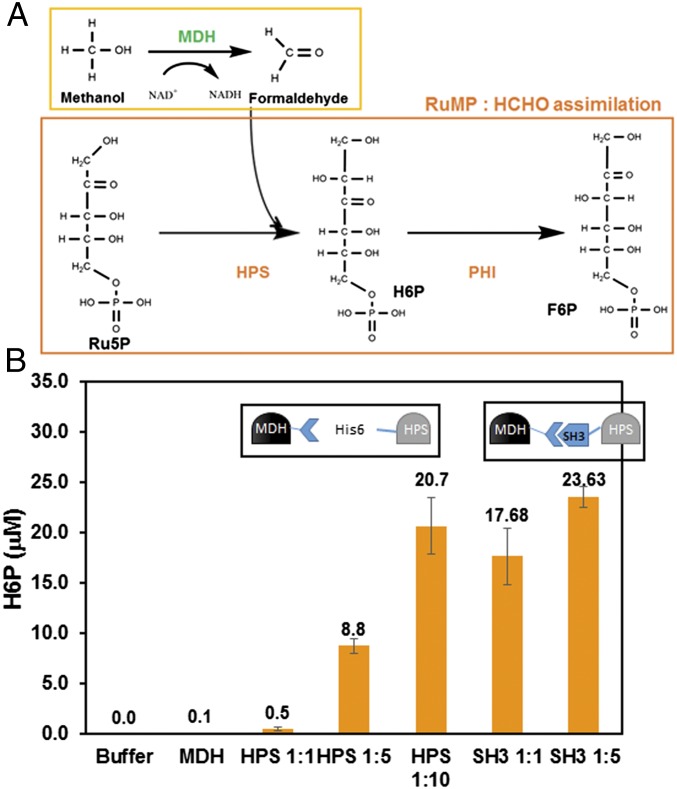
In the widespread methylotrophic RuMP pathway for formaldehyde assimilation (Fig. 1A) (7, 13), d-arabino-3-hexulose 6-phosphate (6HP) is first produced by a condensation reaction between formaldehyde and ribulose 5-phosphate (Ru5P) by 3-hexulose-6-phosphate synthase (Hps) (6, 13, 14). This is followed by the isomerization of heluxose 6-phosphate (H6P) by 6-phospho-3-hexuloseisomerase (Phi) (6, 13, 14) into fructose 6-phosphate (F6P), which can either enter central metabolism for biosynthesis (14) or be converted back to Ru5P to allow the pathway to cycle through (6, 14).
Fig. 1.
(A) A schematic of the Mdh–RuMP pathway for methanol assimilation. (B) H6P formation with increasing ratios of Mdh3–sSH3lig:Hps or Mdh3–sSH3lig–SH3–Hps enzyme complexes. Purified Mdh–sSH3lig was mixed with equal molar ratios of purified Hps ranging from 1:1–1:10. H6P was assayed according to the previously established method by Arfman (39). Error bars represent the SD of at least three replicate experiments.
Recent work has demonstrated the ability to transfer methylotrophic genes to other organisms to enable nonnative methanol oxidation (15–17). Methanol utilization and 13C-methanol–derived labeling of intracellular metabolites were first demonstrated in Corynebacterium glutamicum by expressing the mdh gene from B. methanolicus and the hps and phi genes from Bacillus subtilis (16, 17). A similar approach was also demonstrated in E. coli, where Mdh2 from B. methanolicus PB1 was coexpressed with Hps and Phi from B. methanolicus MGA3 to increase the production of 13C-labeled intracellular metabolites compared with the control strain expressing Mdh3 alone (15). Although these reports provide evidence that the RuMP pathway can be transferred to nonnative methylotrophs, the rate of methanol consumption is significantly lower than those reported for native methyltrophs. Detailed kinetics analyses revealed that the reverse formaldehyde reductase activity for Mdh is at least 1,000-fold higher than methanol oxidation (9), and the irreversible sequestration of formaldehyde by Hps and Phi serves as a driving force to pull the pathway equilibrium toward the desired direction. In B. methanolicus MGA3, expression of both hps and phi is more highly up-regulated than mdh upon methanol addition, and carefully coordinated expression of mdh and the RuMP pathway genes is required for efficient methanol utilization (18). In the same organism, overexpression of hps and phi results in a higher growth rate on methanol, again indicating the importance of direct coupling between the reversible methanol oxidation and the irreversible formaldehyde assimilation (18).
Multienzyme supramolecular complexes, which self-assemble into spatially defined architectures, have been shown to be a promising approach to improve the efficiency of cascade reactions (19–22). This spatial organization provides more stable enzyme structures, facilitates substrate channeling between active sites, and prevents the buildup of toxic intermediates (23–25). The use of driving forces such as ATP generation or NADH consumption as a kinetic control element to enable directional operation of highly reversible enzyme reactions has also been reported (26, 27). We hypothesize that engineered supramolecular enzyme assemblies can be similarly created as a kinetic trap to enable fast and efficient methanol utilization. Unlike other reports based on the use of a scaffold (24, 28), which tend to produce large and disordered multienzyme complexes, we take advantage of the decameric nature of Mdh3 from B. methanolicus to construct a scaffoldless supramolecular enzyme complex composed of Mdh3 and a previously validated Hps–Phi fusion from Mycobacterium gastri to enhance methanol conversion to F6P. Our ability to enhance F6P synthesis is ideally suited for the recently proposed methanol condensation cycle (29), which combines the enzymes from the RuMP pathway and the nonoxidative glycolysis pathway for the in vitro synthesis of platform chemicals from methanol.
Results
High Ratio of Hps to Mdh Enhances in Vitro Methanol Utilization.
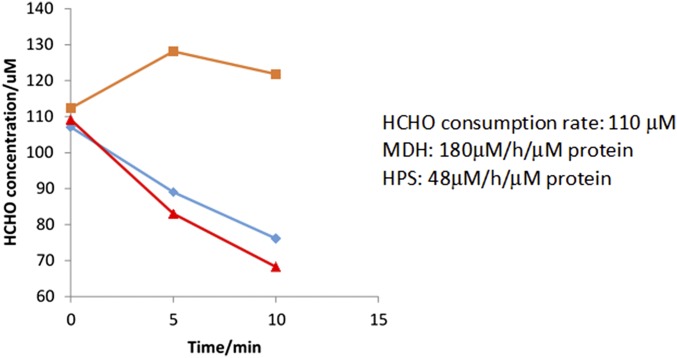
B. methanolicus MGA3 Mdh3 has a reported kcat/Km value roughly 1,000-fold lower for methanol oxidation than for formaldehyde reduction (9), indicating that a significant kinetic barrier must be overcome to achieve efficient methanol utilization. In methylotrophs, the irreversible conversion of formaldehyde by Hps and Phi is the driving force behind shifting the pathway equilibrium toward F6P formation. In B. methanolicus, it has been suggested that the level of Hps/Phi can impact methanol consumption and cell growth (18), and the stoichiometric effect of Mdh to Hps:Phi on the methanol consumption pathway remains unexplored either in native or synthetic methylotrophs. To investigate this issue, purified Mdh3 was mixed with different ratios of a highly active Hps from M. gastri (30) to create a kinetic trap to consume formaldehyde and increase H6P production. As expected, no H6P was detected in the absence of Hps. Even when Hps and the Mdh3 were mixed at a 1:1 molar ratio, only a small amount of H6P was detected over the background (Fig. 1B). This result is consistent with the roughly fourfold faster formaldehyde consumption rate by Mdh3 compared with Hps (Fig. S1). Increasing the molar ratio of Mdh3:Hps from 1:1 to 1:5 resulted in a significant increase in H6P production (Fig. 1B), supporting the notion that formaldehyde must be quickly and irreversibly removed to shift the pathway equilibrium. Although a higher level of H6P was achieved by further increasing the Mdh3:Hps ratio to 10, such a high ratio is difficult to maintain in vivo without resulting in a heavy cellular burden, especially if the same high ratio is needed for all subsequent enzymes in the pathway. One way to bypass this limitation is to directly couple these enzymes into a supramolecular enzyme complex to facilitate the rapid transfer of intermediates toward F6P formation.
Fig. S1.
Comparison of formaldehyde consumption rate by MDH (blue) and HPS (red). Virtually no consumption was detected without enzymes (orange).
Engineered Supramolecular Enzyme Complexes Enhance H6P and F6P Production.
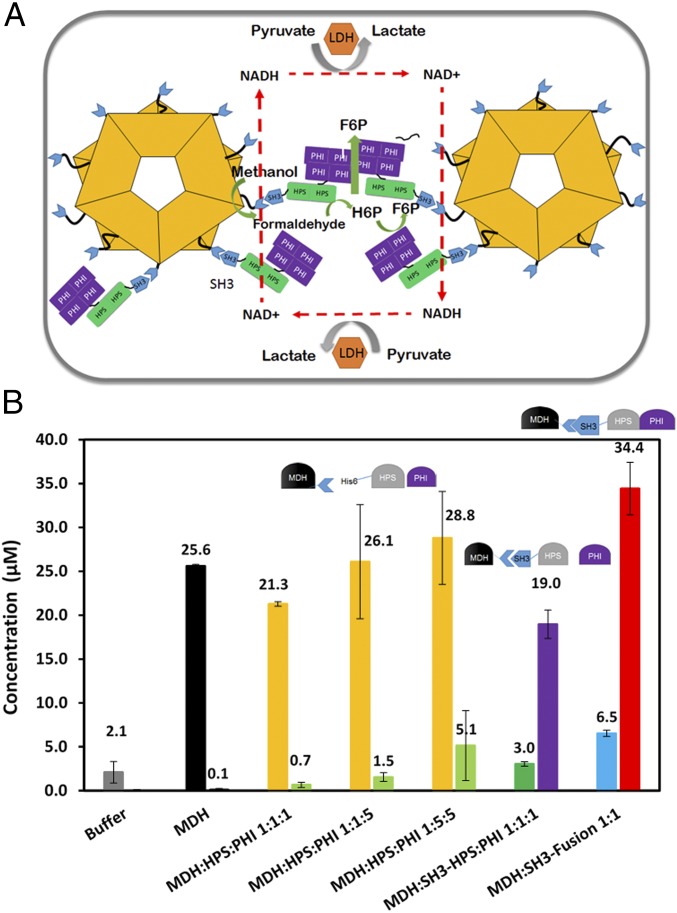
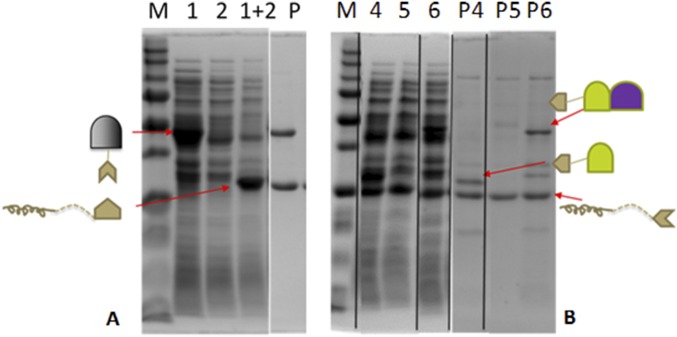
Protein scaffolding is one of the most successful strategies used to create supramolecular enzyme complexes (24, 31). However, enzyme cascades involving multimeric enzymes are often difficult to assemble, as they tend to result in large and disordered structures because of the random cross-linking between enzymes and scaffolds (31). An alternative approach is to exploit the ability of multimeric proteins to self-assemble into defined nanostructures by engineering appropriate interaction domains (32). Here, we took advantage of the decameric nature of Mdh3 as a natural base for assembling Hps and Phi into a functional multienzyme cascade (Fig. 2A). A well-known interaction pair, SH3 (Src homology 3 domain from the adaptor protein CRK), and its ligand (sSH3lig, PPPALPPKRRR), which have a reported Kd value of 0.1 µM (24), were used for protein assembly. The Mdh3–sSH3lig, SH3–Hps, and SH3–Hps–Phi fusions were solubly expressed in E. coli (Fig. S2). The functionality of both interaction domains was first confirmed by the coprecipitation of the fusion proteins with their respective elastin-like polypeptide (ELP) binding partners using the thermally induced phase transition property of ELP (33) (Fig. S2).
Fig. 2.
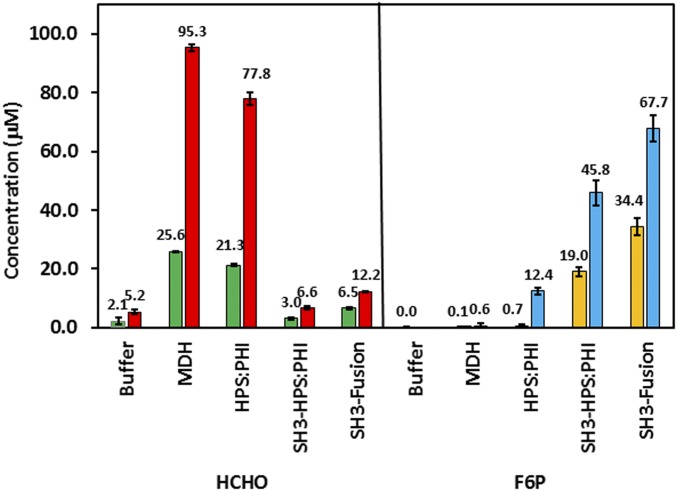
(A) A schematic of the Mdh3–Hps–Phi supramolecular enzyme complex and the corresponding cascade reactions. Ldh was used as an NADH Sink to further minimize formaldehyde reduction. (B) Formaldehyde (first bar) and F6P (second bar) formation with and without SH3-tethered enzymes. Purified Mdh–sSH3lig was mixed with either Hps and Phi, SH3–Hps and Phi, or the SH3–Hps–Phi fusion. Formaldehyde (HCHO) was assayed using the Nash reagent. Error bars represent the SD of at least three replicate experiments.
Fig. S2.
SDS/PAGE pull-down assay. Pull-down assays were performed to test the functionality of the individual SH3 interaction components when fused to enzymes. Briefly, (A) MDH–sSH3lig (1) and ELP–SH3 (2) were independently expressed in BL21(DE3), and soluble lysates were collected and mixed together at a 1:1 mass ratio (1+2). One round of ELP purification was performed and the purification product (P) was assessed by SDS/PAGE and commassie blue staining. (B) The same experiment was performed with SH3–HPS and ELP–sSH3lig (4), with SH3–HPS_PHI and ELP–sSH3lig (6), and with an empty vector lysate with ELP–sSH3lig (5). One round of ELP purification was performed, and purification products (P4, P5, and P6) for each condition assayed were examined by SDS/PAGE and commassie blue staining.
We next investigated the ability of the assembled Mdh3–Hps complex to drive H6P production. Even a 1:1 ratio of the two proteins resulted in higher levels of H6P production (Fig. 1B), which was roughly 35-fold higher than using nonassembled Mdh3 and Hps. More importantly, the level of H6P achieved was already twice the level achieved using a 1:5 ratio of Mdh3 and Hps. The use of a 1:5 ratio of Mdh3–sSH3lig to SH3–Hps improved H6P production further, although the impact was less significant than the 1:1 ratio (Fig. 1B). These results confirm the importance of enzyme assembly and substrate channeling in driving the conversion of methanol to H6P.
To investigate whether similar success could be achieved toward F6P production, Phi was added to the enzyme cascade. Consistent with the results with H6P, a barely detectable amount of F6P was produced using only a mixture of unassembled Mdh3, Hps, and Phi (Fig. 2B). Increasing the amount of Phi had virtually no impact on F6P production, whereas a higher amount of Hps to the other enzymes elevated the F6P level by over sevenfold. This result again highlights the importance of Hps toward irreversible removal of formaldehyde to shift the pathway equilibrium. Direct coupling of Mdh3 with Hps again resulted in a more than 27-fold enhancement in F6P production even at a 1:1 ratio (Fig. 2B). More importantly, only the direct coupling of Hps resulted in a significant reduction in the formaldehyde level, indicating the efficient channeling of formaldehyde to H6P rather than back to methanol. Clustering Mdh3 with a bifunctional Hps–Phi fusion, which has been previously shown to be more active than an equimolar mixture of the individual enzymes (30), further improved F6P production by another twofold, resulting in an overall 50-fold improvement over the uncomplexed enzyme mixture (Fig. 2B). Compared with the unassembled enzyme system, a much lower level of formaldehyde was detected (Fig. 2B), and only a small amount of H6P was accumulated (Fig. S3), indicating the effective channeling of formaldehyde toward F6P by the supramolecular enzyme complex due to improved molecular proximity.
Fig. S3.
H6P and F6P production by purified Mdh–sSH3lig and the SH3–Hps–Phi fusion in the presence or absence of LDH. F6P production was assayed as described earlier. The total concentration of H6P and F6P was determined by first deactivating the enzymes by acid treatment. The difference between the two is the net amount of H6P accumulated. In all cases, only a very small quantity of H6P accumulation was detected.
Dynamic Light Scattering Identification and TEM Visualization of the Supramolecular Enzyme Assemblies.
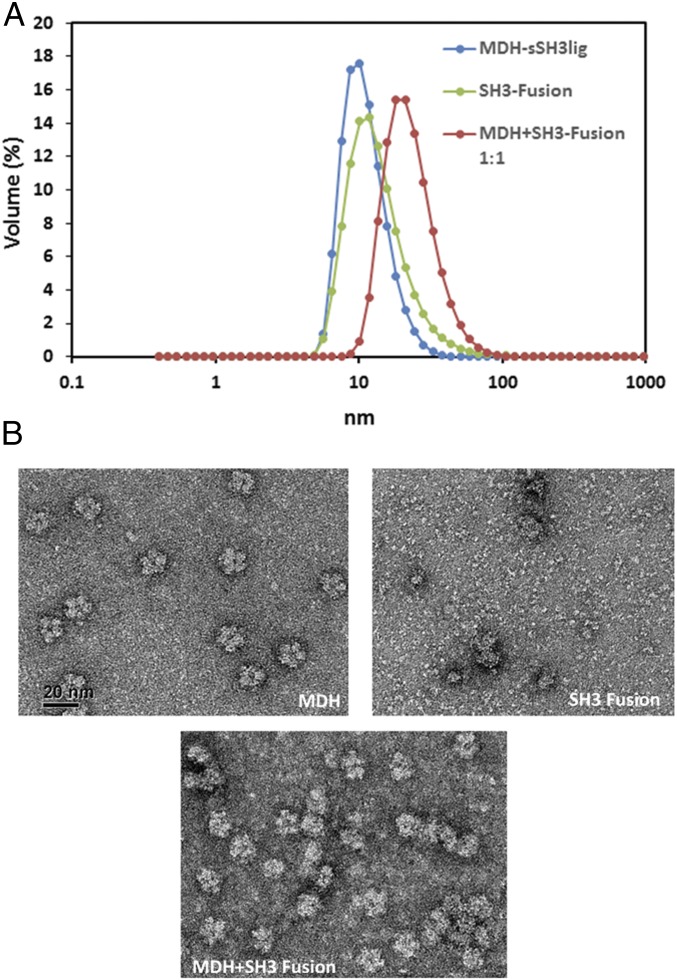
To confirm that the increased F6P production observed was the result of the formation of supramolecular enzyme complexes, we carried out biophysical characterization experiments to evaluate the enzyme assembly. Dynamic light scattering (DLS) was first used to identify the sizes of the individual enzymes and that of the enzyme complex. Mdh3–sSH3lig had a narrow size distribution with a peak at 11.6 ± 0.3 nm, consistent with the decameric nature of Mdh3 (Fig. 3A). The fusion enzyme, SH3–Hps–Phi, showed a much broader peak with an average diameter of 14.9 ± 1.2 nm (Fig. 3A) because of the formation of larger enzyme complexes between the dimeric Hps (30, 34) and the tetrameric Phi (30). Formation of a larger enzyme assembly with an average peak of 24.0 ± 1.9 nm was detected by mixing Mhd3–sSH3lig and SH3–Hps–Phi (Fig. 3A). This increase in size suggests that large, multienzyme complexes are not being formed, as the average size is roughly the same as the two individual components added together. To investigate this further, transmission electron microscopy (TEM) was used to visualize the complex formation. Individual Mdh3–sSH3lig enzymes have a decameric structure, composed of two planes with mirror symmetry, each composed of five monomers in a ring-like shape (12) (Fig. 3B). From the TEM images, it appears that the SH3–Hps–Phi enzyme fusion is globular and heterogeneous in size with little distinction between each of the components (Fig. 3B). When Mdh3–sSH3lig and SH3–Hps–Phi were mixed before TEM analysis, association between the two protein fusions is evident (Fig. 3B). In some instances, multiple Mdh3–sSH3lig proteins were found to decorate a single SH3–Hps–Phi, and large multimeric complexes were also visualized. Taken together, these data confirm that the beneficial effects on F6P production are due to successful enzyme assembly.
Fig. 3.
Characterization of the Mdh3–Hps–Phi supramolecular enzyme complex. (A) DLS was used to characterize the size of the individual enzyme components and the 1:1 Mdh3–sSH3lig to SH3–Hps–Phi enzyme conjugates using the Zetasizer Nano ZS instrument. Three measurements were made at 25 °C, and the volume average is reported. (B) TEM of enzyme complexes. The TEM micrograph for Mdh3–sSH3lig showed the previously reported pentagonal shape of B. methanolicus MGA3 Mdh3, whereas the SH3–Hps–Phi fusion proteins were mostly larger and nonuniform enzyme clusters. Complex formation was clearly demonstrated by mixing the two proteins at a 1:1 molar ratio. Error bars represent the SD of at least three replicate experiments.
Lactate Dehydrogenase as An “NADH Sink” Further Enhances Methanol Consumption and F6P Production.
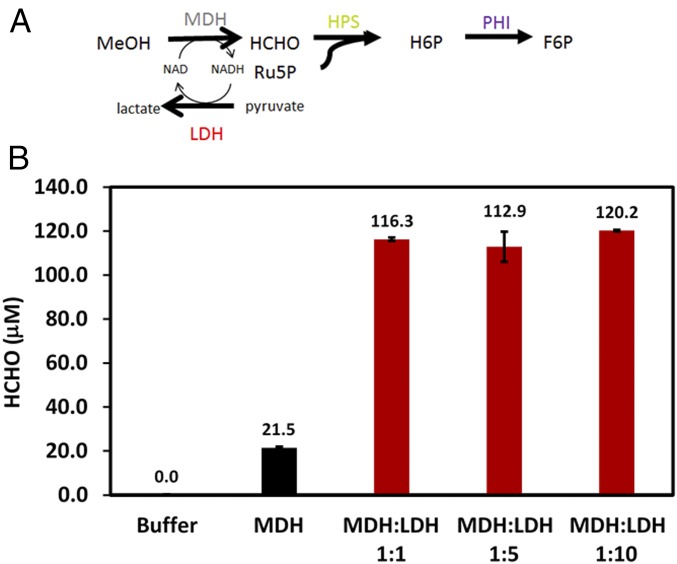
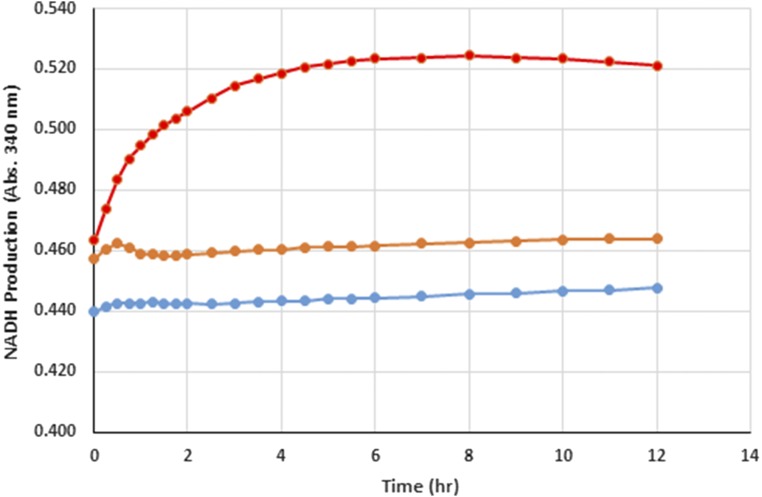
Although F6P production was significantly increased through complexation of Mdh3 with the bifunctional Hps–Phi fusion, a parallel approach to further improve MeOH utilization and F6P production is to prevent the reduction of formaldehyde back to methanol by consuming all of the NADH using an NADH Sink (Fig. 4A). Lactate dehydrogenase (Ldh) consumes NADH during the conversion pyruvate to lactate and can be used as an NADH Sink in synthetic pathways. E. coli Ldh was cloned from genomic DNA, expressed, and purified. Addition of Ldh to Mdh3 even at a 1:1 molar ratio improved methanol consumption and formaldehyde formation by 3.6-fold (Fig. 4B). This enhancement is the direct result of the efficient NADH recycling by Ldh as no net accumulation of NADH was detected (Fig. S4). Increasing the Ldh-to-Mdh3 ratio further resulted in only a marginal improvement in methanol consumption (Fig. 4B), likely due to the very high specific activity of Ldh (Table S1).
Fig. 4.
(A) A schematic illustrating the concept of NADH Sink using E. coli Ldh. (B) Effects of NADH Sink on methanol (CH3OH) conversion. Purified Mdh3–sSH3lig was mixed with increasing ratios of purified E. coli Ldh and assayed for formaldehyde (HCHO) according to the Nash method. Error bars represent the SD of at least three replicate experiments.
Fig. S4.
NADH production during methanol consumption by MDH in the presence (orange) or absence of LDH (red). NADH production was monitored by the increase in absorbance at 340 nm. A control without enzyme (blue) produced no NADH.
Table S1.
Specific activities of purified Mdh3
| Enzyme | Specific activity, nmol product/min/nmol enzyme | Reaction | Substrate concentration, mM |
| B. methanolicus MGA3 Mdh3 | 0.06 | CH3OH → HCHO | 500 |
| B. methanolicus MGA3 Mdh3 | 3.3 | HCHO → CH3OH | 0.1 |
| E. coli LDH | 221.2 | Pyruvate → Lactate | 5 |
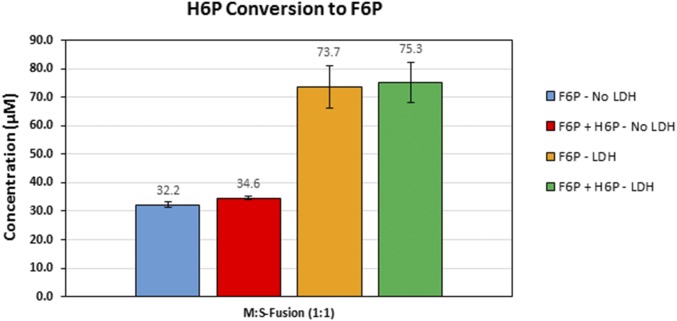
We next investigated whether this substantial improvement in methanol consumption using the NADH Sink concept could be coupled with the enzyme clustering approach. Different enzyme combinations were evaluated as described earlier, except Ldh was also added at a ratio of 1:1 to Mdh3 (Fig. 5). In all cases, F6P production was increased in the presence of Ldh. Although the F6P level was elevated by more than 20-fold for the uncomplexed enzyme mixture, a significant amount of formaldehyde remained unprocessed. This result again suggests the inability of the uncomplexed Hps to effectively compete for formaldehyde to form H6P against the formaldehyde conversion back to methanol by Mdh3. A completely different phenomenon was observed when the engineered enzyme assembly was used. In both cases, virtually all formaldehyde formed was diverted to F6P, albeit at a lower efficiency without using the Hps–Phi fusion. This result highlights that the synergistic action between the two parallel pathways, one in preventing reverse methanol formation and one in irreversibly removing formaldehyde toward F6P, is necessary not only to increase methanol consumption but also for the subsequent conversion to F6P. Overall, 67.7 µM F6P was produced using a combination of NADH Sink and enzyme assembly, which represents a 97-fold increase over the uncomplexed enzyme mixture.
Fig. 5.
Combinatorial effect of enzyme assembly and NADH Sink on methanol consumption and F6P production. Purified Mdh3–sSH3lig was mixed at a 1:1 molar ratio with either Hps and Phi, SH3–Hps and Phi, or SH3–Hps–Phi with or without an equal molar of Ldh. The HCHO and F6P concentration in the absence (first bar) and presence (second bar) of Ldh are shown. Error bars represent the SD of at least three replicate experiments.
In Vivo Engineered Enzyme Assembly Enhances Methanol Consumption by E. coli.
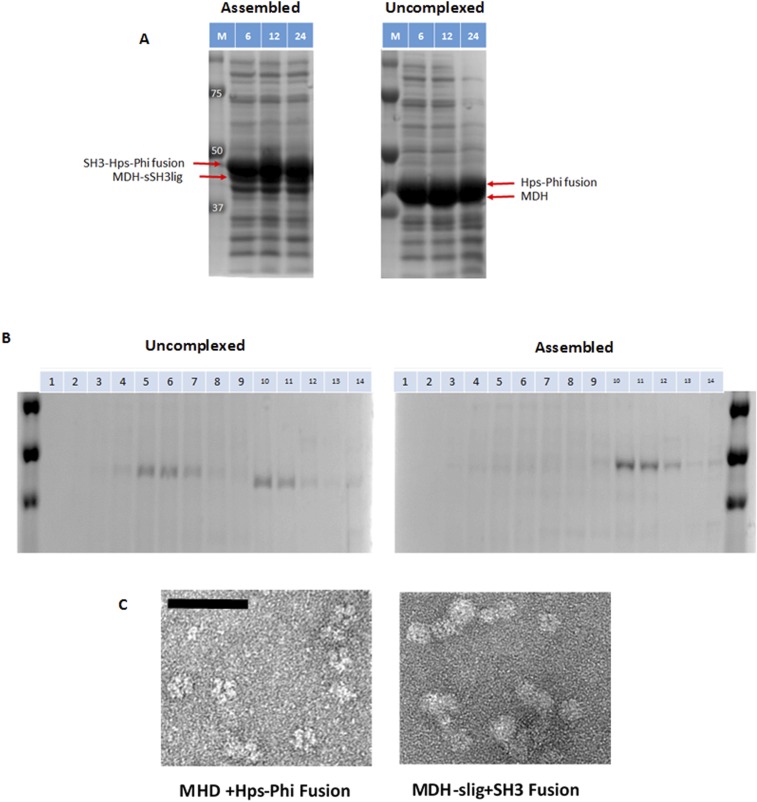
For the in vitro experiments, all substrates were provided in excess in order to investigate only the influence of enzyme kinetics on F6P production, a condition very different from the in vivo setting. To investigate whether the supramolecular enzyme assembly can impact in vivo methanol consumption in a similar fashion, two recombinant strains were created where Mdh3 was either assembled with Hps–Phi or expressed as uncomplexed enzymes. A ∆frmA strain was used to eliminate the native formaldehyde oxidation system from competing with the RuMP pathway (35, 36). A high level of enzyme expression was first demonstrated by SDS/PAGE (Fig. S5A). To confirm the intracellular formation of enzyme complexes, cell lysates were fractionated by sucrose gradient centrifugation. Mdh3 was detected only in the later fractions (fractions 10–12), indicating the presence of intact larger protein particles (Fig. S5B). More importantly, TEM analyses confirmed the formation of protein complexes for cells expressing the assembled enzymes, whereas mainly individual Md3 proteins were detected for the uncomplexed enzymes (Fig. S5C).
Fig. S5.
Analysis of in vivo enzyme assembly. (A) SDS/PAGE analysis of enzyme expression at different time points. The respective protein bands are highlighted. (B) Separation of enzyme complexes by sucrose gradient. (C) Formation of enzyme complexes by TEM analysis. (Scale bar: 50 nm.)
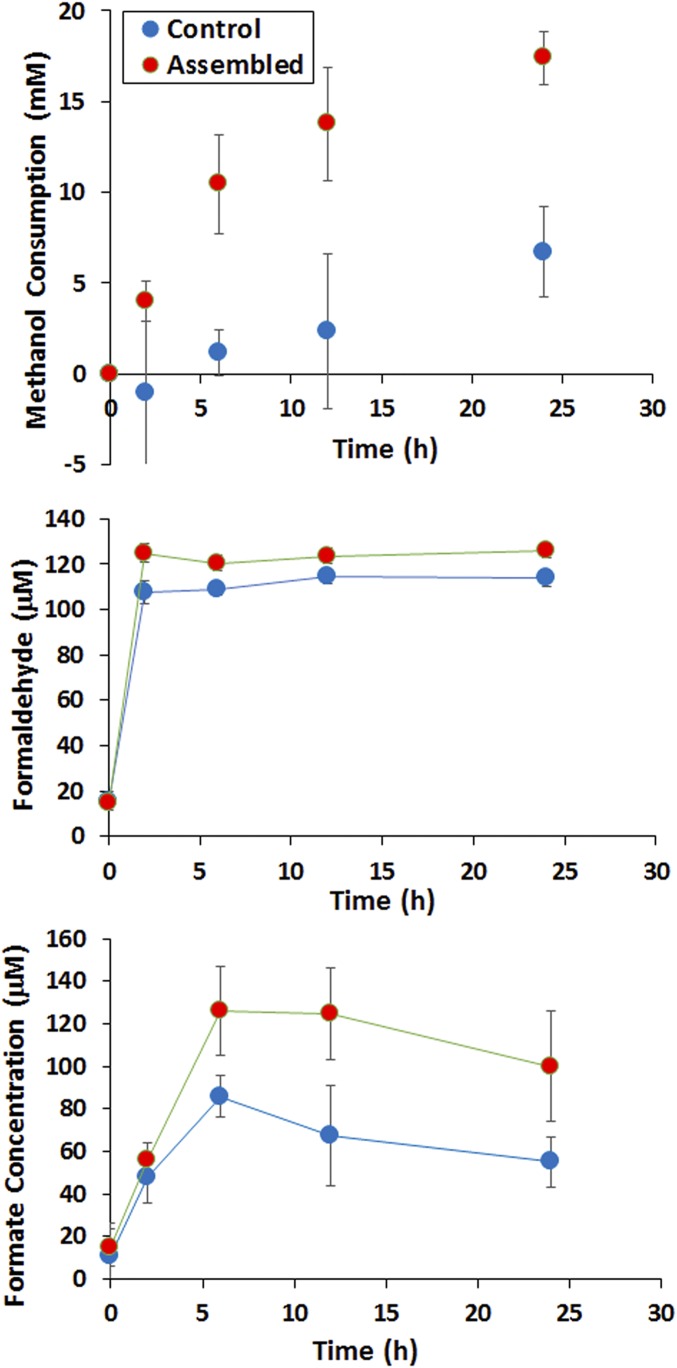
After confirming complex formation, in vivo methanol consumption was compared using resting cell cultures in the presence of 500 mM methanol. Similar to the in vitro results, methanol consumption was detected only for cells expressing Mdh3, even after 24 h of cultivation (<0.5 mM evaporation within 24 h). Although the extent of improvement is lower for the in vivo system, the initial methanol consumption rate was roughly 9 times faster for the recombinant strain expressing both Mdh3–sSH3lig and SH3–Hps–Phi (1.7 mM/h) than the control strain without the SH3 interaction components (0.19 mM/h) (Fig. 6). This whole-cell methanol consumption rate of 1.7 mM/h is also 14 times higher than the 0.12 mM/h reported by Müller et al. in a similar resting cell assay (15). It should be noted that the overall enhancement in methanol consumption was only 2.3-fold after 24 h incubation, likely due to limitations in R5P in resting cell cultures. The consumed methanol was clearly diverted further down the RuMP pathway, as only micromolar levels of formaldehyde were detected in both cases. The majority of the carbon flux is likely channeled toward CO2 formation via native glycolysis, although a small amount was diverted toward formate (Fig. 6). Collectively, these results indicate that the beneficial effects of enzyme assembly can be maintained in vivo and help improve methanol utilization by E. coli.
Fig. 6.
In vivo methanol consumption and utilization. Two recombinant ∆frmA strains were tested for methanol consumption for 24 h. One strain harboring Mdh3 and Hps–Phi without the docking components (Control) and a second strain harboring Mdh3–sSH3lig and SH3–Hps–Phi fusion (Assembled). Cells were grown to an OD600 of 1, washed, and resuspended in M9 medium and 500 mM methanol and incubated at 37 °C for 24 h. Methanol consumption was calculated as the difference between the starting and final methanol concentration. Formaldehyde accumulation was measured with the Nash reagent by harvesting cells and testing the clarified supernatant. Formate formation was determined by HPLC analysis of culture supernatants. Error bars represent SD; n = 4.
Discussion
Engineering E. coli as a synthetic methylotroph has been a challenging problem to date, as very little methanol utilization and no growth on methanol have been demonstrated (15). One of the key obstacles is the unfavorable kinetic properties of NAD-dependent Mdhs, such as Mdh3 (9), making the reversible reduction of formaldehyde far more favorable than methanol oxidation. Although the irreversible sequestration of formaldehyde by the RuMP pathway enzymes serves as a driving force to pull carbon flux to the synthesis of F6P, a 1:10 ratio of Mdh3 to Hps is necessary to drive a significant level of H6P production (Fig. 1B). This very high enzyme ratio is difficult to maintain, particularly if all of the downstream enzymes must be expressed at this high level.
The use of engineered enzyme assembly has been shown to substantially improve the efficiency of cascade reactions (19–22). We argue that this method is ideally suited for pulling formaldehyde toward F6P production because of the improved substrate channeling. Unlike other reports using a protein scaffold for enzyme assembly, we chose a scaffoldless self-assembly approach to cluster three multimeric enzymes. Our engineered supramolecular enzyme assembly yielded a 50-fold improvement in methanol conversion to F6P, a key metabolite that is capable of entering E. coli central metabolism. Although the improvement in F6P production was extremely promising, the overall improvement in methanol conversion remained modest. This observation suggested that an additional mechanism to slow down formaldehyde reduction was needed. This was accomplished by installing an NADH Sink to irreversibly consume NADH required for formaldehyde reduction using E. coli Ldh, which catalyzes the essentially unidirectional reduction of pyruvate (37). Addition of Ldh successfully eliminated any NADH accumulation and led to over twofold more methanol oxidation and F6P production.
We next investigated whether the beneficial effects of supramolecular enzyme assembly can be similarly realized in vivo, as the intracellular substrate availability is significantly different. Although the overall enhancement is lower, we demonstrated that enzyme assembly improved methanol consumption and increased formate production by 4.5- and eightfold, respectively. Optimization of protein expression and fermentation conditions will likely lead to further enhancement in productivity, as reported by others (33).
In summary, we have demonstrated an engineered system that couples a supramolecular enzyme assembly with an NADH Sink to provide a highly effective strategy for overcoming essential obstacles associated with methanol utilization. The approach provides a platform for the direct coupling of enhanced F6P synthesis with many other metabolic engineering strategies such as the methanol condensation cycle (29) for biological conversion of methanol to higher value chemicals by E. coli.
Materials and Methods
Chemicals and Reagents.
All reagents were purchased from Sigma-Aldrich, unless otherwise indicated. The following enzymes were also purchased from Sigma-Aldrich: glucose-6-phosphate dehydrogenase (G6dh) (Saccharomyces cerevisiae), phosphoglucose isomerase (Pgi) (S. cerevisiae), and phosphoribose isomerase (Rpi) (Spinacia oleracea). Phusion DNA polymerase, T4 DNA ligase ,and all restriction endonucleases were purchased from NEB.
Cloning of Methanol Utilization Genes.
All primer sequences are listed in Table S2. Detailed procedures for constructing the different expression vectors are described in SI Materials and Methods. Briefly, the gene encoding B. methanolicus MGA3 Mdh3 (9) was synthesized by Genscript. For the pull-down experiments, ELP-sSH3lig was created using SLIC (38) with 18 bp of homology on the ends of the linearized plasmid.
Table S2.
Primers used in this study
| Primer | Sequence |
| BmMDH3-sSH3lig_F | gatataCATATGAAAAATACGCAGTCCGCATTCTAC |
| BmMDH3-sSH3lig_R1 | TCCTCCAGACCCTCCGCCGCCCATCGCGTTTTTGATGATCTGGAT |
| BmMDH3-sSH3lig_R2 | CGGAAGAGCTGGGGGAGGGGAGCCCCCACCACCTGAACCGCCTCCTCCAGACCCTCCGCC |
| BmMDH3-sSH3lig_R3 | accagaCTCGAGTTACAAGTCCTCTTCAGAAATAAGCTTTTGTTCGGATCCCCTTCGTCTTTTTGGCGGAAGAGCTGGGGGAGG |
| Ptac_his6-HPS_F | gatataCATATGGGCAGCCATCATCATCATCATCACAGCGGCAAGCTCCAAGTCGCCATC |
| Ptac_his6-HPS_R | gctgcgACTAGTTTATTACAAGTCCTCTTCAGAAATAAGCTTTTG |
| Ptac_his6-PHI_F | gatataCATATGGGCAGCCATCATCATCATCATCACGGCAGCACGCAAGCCGCAGAAGCC |
| Ptac_his6-PHI_F | gctgcgACTAGTTTATTACTCGAGGTTGGCGTGGCG |
| XbaI_SH3-HPS_F1 | GATATAATGGGCAGCCATCATCATCATCATCACAGCGGCATGGCAGAGTATGTGCGGGCC |
| XbaI_SH3-HPS_F2 | ttccccTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATAATGGGCAGCCATCATCATCATCATCAC |
| NdeI_SH3-HPS_R | gagcttCATATGACCAGAACCACCACCAGAACCACCATACTTCTCCACGTAAGGGACAGGAATCAT |
| SLIC-ELPs1-F1 | cccgccgggcggcagcCCgcGGACTAGTCCTCCCCCAGCTCTTCCGCCAAAAAGACGAAGG |
| SLIC-ELPs1-R1 | cagtggtggtggtggtggtGCTCGAGCCTTCGTCTTTTTGGCGGAAGAGCTGGGGGAGGACTAGTCC |
| SacII-SH3 FP | atatCCGCGGATGGCAGAGTATGTGCGG |
| XhoI-SH3 RP | atatCTCGAGATACTTCTCCACGTAAGGGAC |
Shown are the primers used to clone all of the enzymes in this study as well as the SH3 domain and the sSH3ligand (as a fusion to MDH). Underlined sequences represent restriction enzyme cut sites.
Methanol Conversion to F6P.
A 1-mL reaction contained 50 mM Hepes (pH 7.4), 5 mM MgCl2, 1 mM NAD+, 5 mM Ru5P, 500 mM MeOH, and purified enzymes at concentrations between 1 and 10 μM. When Ldh was used, 2 mM pyruvate was added to the reaction mixture. The reaction was initiated with the addition of MeOH and incubated at 37 °C for 14 h. The reaction was stopped by the addition of 1 M HClO4 and neutralized with 1 M KOH. Following neutralization, reactions were analyzed for formaldehyde and F6P (39). Formaldehyde concentration was determined according to Nash (40).
In Vivo Testing of Enzyme Clustering.
Characterization of the genes required for methanol assimilation in E. coli was performed in a BW25113 ∆frmA host strain. Methanol assimilation genes were expressed heterologously using the pETM6 vector that has been modified to contain the tac promoter using PCR and standard cloning procedures. Detailed culture conditions are described in SI Materials and Methods. Resting cell cultures were incubated at 37 °C with aeration for 24 h, and methanol, formaldehyde, and secreted metabolites were quantified. Methanol and formate concentrations were measured via high performance liquid chromatography (Agilent Technologies) using an Aminex HPX-87H column (Bio Rad), 0.005 M H2SO4 as the mobile phase, and a flow rate of 0.5 mL/min. Formaldehyde in the culture supernatant was analyzed as previously described (40).
SI Materials and Methods
Cloning of Methanol Utilization Genes.
The amplified Mdh3 gene was ligated into the pETM6 vector using NdeI and XhoI. The Mdh3–sSH3lig–myc gene fusion was amplified by overlap extension PCR using pETM6–MGA3_Mdh3 as template, and the PCR product was ligated into pETM6 using NdeI and XhoI. Mdh3–sSH3lig–myc was amplified using the primers BmMDH3–sSH3lig_F, BmMDH3–sSH3lig_R1, BmMDH3–sSH3lig_R2, and BmMDH3–sSH3lig_R3. Genes encoding a fusion between Hps and Phi from M. gastri MB19 were cloned into pETM6 with an N-terminal His6-tag. The Hps gene was amplified using primers Ptac_his6-Hps_F and Ptac_his6-Hps_R; the Phi gene was amplified using Ptac_his6-Phi_F and Ptac_his6-Phi_R. Both genes were ligated into pETM6 using NdeI and SpeI restriction sites. An SH3 domain was added in-frame within the MCS of pET21a by PCR using primers XbaI_SH3–Hps_F1, XbaI_SH3–Hps_F2, and NdeI_SH3–Hps_R that were encoded for the SH3 domain. The amplified gene product was then ligated into pET21a using XbaI and NdeI restriction sties. SH3–enzyme fusions were created by amplifying via PCR the genes encoding either Hps, Phi, or the Hps–Phi gene fusion and ligating the PCR product in-frame using NdeI and SpeI. ELP–SH3 was constructed by amplifying the SH3 domain via PCR using SacII–SH3 FP and XhoI–SH3 RP and ligated into linearized pET24 [KV8F40]-IgA-his6 using the same restriction sites. ELP–sSH3lig was created using SLIC (38) with 18 bp of homology on the ends of the linearized plasmid. pET24 [KV8F40]-IgA-his6 was again linearized using SacII and XhoI.
Enzyme Expression and Purification.
All plasmids were transformed into E. coli strain BL21(DE3) for protein expression. Overnight cultures grown in lysogeny broth (LB) (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) supplemented with either 50 μg/mL kanamycin or 50 μg/mL ampicillin were used to inoculate expression cultures at a 1:50 dilution. Expression cultures were grown to an OD600 of 0.5–1.0 at 37 °C, induced with 0.2 mM isopropyl-thiogalactopyranoside (IPTG), and grown for an additional 3 h at 37 °C. Cells were harvested by centrifugation at 5,000 × g for 10 min, resuspended in PBS (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, KH2PO4, pH 7.4) to an OD600 pf 20, and lysed by sonication. The soluble fraction was isolated by centrifugation at 15,000 × g for 15 min. The soluble fraction was then applied to a column packed with HisPur Cobalt Resin (Thermo Scientific) for purification via the N-terminal His6 tag. Purified enzymes were eluted by three washes with increasing imidazole concentrations of 300 mM, 400 mM, and 500 mM imidazole. The soluble and purified fractions were loaded onto a 10% (wt/vol) SDS/PAGE gel, stained with Coomassie blue, and analyzed for expression and purification. Purified fractions of His6–Hps, His6–Phi, His6–SH3–Hps, and His6–SH3–Hps_Phi were dialyzed overnight into Assay Buffer (50 mM Hepes, 150 mM NaCl, 5 mM MgCl2). All protein concentrations were determined spectroscopically by absorbance at 280 nm.
ELP Pull-Down.
Soluble lysates containing either ELP–sSH3lig, ELP–SH3, His6–SH3–HPS, His6–SH3–HPS_PHI, MDH–sSH3lig, or pET24 (Empty) were mixed together at a 1:1 ratio by mass. The mixtures were ELP–SH3 and MDH–sSH3lig, ELP–SH3 and pET24 (Empty), ELP–sSH3lig and His6–SH3–HPS, ELP–sSH3lig and His6–SH3–HPS_PHI, and ELP–sSH3lig and pET24 (Empty). Na2SO4 was added to the lysate mixtures to a final concentration of 750 mM, and the samples were incubated at 37 °C for 15 min. The mixtures were then centrifuged at 16,000 × g for 15 min at room temperature. Supernatants were removed, and the remaining pellets were resuspended in 1 mL ice-cold PBS. The resuspensions were centrifuged at 16,000 × g for 10 min at 4 °C. The supernatants were collected and loaded onto a 10% (wt/vol) SDS/PAGE gel, stained by Coomassie blue, and analyzed.
H6P and F6P Assay.
Both H6P and F6P were assayed according to the protocol established by Arfman et al. (39). Briefly, a 1-mL reaction contained 50 mM Hepes (pH 7.4), 2 mM NADP+, 3 mM MgCl2, 5 U G6Pdh, 0.4 U Pgi, and 400 μL of the sample. The reaction was initiated by the addition of Pgi after absorbance at 340 nm had stabilized. The formation of NADPH was monitored at 340 nm for 5 min using a UV-1800 spectrophotometer (Shimadzu).
H6P Conversion Assay.
Similar to the MeOH to F6P assay, a 1-mL reaction contained 50 mM Hepes (pH 7.4), 5 mM MgCl2, 1 mM NAD+, 5 mM Ru5P, 500 mM MeOH, and purified enzymes at a concentration of 1 μM. When Ldh was used, 5 mM pyruvate was added to the reaction mixture. The reaction was initiated with the addition of MeOH and incubated at 37 °C for 14 h. The enzymes were heat inactivated at 65 °C for 20 min, and the reaction was aliquoted into two equal volumes. One sample was then stopped by the addition of 1 M HClO4 and neutralized with 1 M KOH, whereas 1 µM of PHI was added to the other aliquot and incubated at 37 °C for 2 h for continued conversion of H6P to F6P. After the 2-h incubation, the second aliquot was stopped and neutralized. The two samples were then assayed for F6P as described above.
Sucrose Gradient for TEM.
We layered 10%, 20%, 30%, and 40% (wt/vol) sucrose solutions in TN150 [10 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl] in 11 × 60 mm centrifuge tubes (Beckman Coulter). Each sucrose layer was 860 µL, and 560 µL of sample lysate was layered on top of the step gradient, for a total volume of 4 mL. The tubes were centrifuged for 12 h at 50,000 rpm at 4 °C in a Beckman SW 60 Ti rotor in a Optima l-100 XP ultracentrifuge (Beckman Coulter). Following centrifugation, 14 × 286 µL fractions were collected and analyzed by SDS/PAGE and TEM.
DLS.
A Zetasizer Nano ZS instrument (Malvern Instruments) was used to measure complex formation between MDH–sSH3lig and SH3–Hps_Phi. Purified proteins in assay buffer were filtered using 0.1-μm syringe filters, then mixed at a 1:1 molar ratio, and incubated on ice for 15 min before analysis. Each measurement was made three times at 25 °C.
TEM.
Four hundred mesh carbon-coated copper grids were glow-discharged in a Pelco EasiGlow glow discharge unit to render the surfaces of the grids hydrophilic. The freshly glow-discharged grids were floated on drops of sample for several seconds, washed on four drops of Nanopure water, and then negative-stained with 2% (wt/vol) uranyl acetate (aq). After drying, the grids were examined with a Zeiss Libra 120 transmission electron microscope operating at 120 kV. Images were acquired with a Gatan Ultrascan 1000 CCD camera.
In Vivo Testing of Enzyme Clustering.
E. coli strains were routinely cultured in LB media or M9 minimal media for methanol consumption and/or metabolite analysis. Preculture conditions for the methanol assimilating strain were as follows: A single colony was picked from a plate and grown in LB broth overnight at 37 °C with aeration. Overnight cultures were pelleted, washed, and resuspended at an OD600 of ∼1.0 in M9 minimal media supplemented with 500 mM methanol and 0.1 mM IPTG.
Acknowledgments
We thank Shannon Modla of the Bioimaging Center at the Delaware Biotechnology Institute for her assistance in acquiring TEM images and R. Kyle Bennett for help with in vivo experiments. Funding was provided by ARPA-E REMOTE Contract DE-AR0000432.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601797113/-/DCSupplemental.
References
- 1.Xu C, Bell L. Global reserves, oil production show increases for 2014. Oil and Gas Journal. 2014;112(12):30–31. [Google Scholar]
- 2.Olah GA. Beyond oil and gas: The methanol economy. Angew Chem Int Ed Engl. 2005;44(18):2636–2639. doi: 10.1002/anie.200462121. [DOI] [PubMed] [Google Scholar]
- 3.Usman M, Daud WMAW. Recent advances in the methanol synthesis via methane reforming processes. RSC Advances. 2015;5(28):21945–21972. doi: 10.1039/d0ra90096f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olah GA. Towards oil independence through renewable methanol chemistry. Angew Chem Int Ed Engl. 2013;52(1):104–107. doi: 10.1002/anie.201204995. [DOI] [PubMed] [Google Scholar]
- 5.Schrader J, et al. Methanol-based industrial biotechnology: Current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009;27(2):107–115. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Brautaset T, Jakobsen M ØM, Flickinger MC, Valla S, Ellingsen TE. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J Bacteriol. 2004;186(5):1229–1238. doi: 10.1128/JB.186.5.1229-1238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitaker WB, Sandoval NR, Bennett RK, Fast AG, Papoutsakis ET. Synthetic methylotrophy: Engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr Opin Biotechnol. 2015;33:165–175. doi: 10.1016/j.copbio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Arfman N, et al. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol. 1989;152(3):280–288. doi: 10.1007/BF00409664. [DOI] [PubMed] [Google Scholar]
- 9.Krog A, et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS One. 2013;8(3):e59188. doi: 10.1371/journal.pone.0059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papoutsakis ET. Reassessing the progress in the production of advanced biofuels in the current competitive environment and beyond: What are the successes and where progress eludes us and why. Ind Eng Chem Res. 2015;54(42):10170–10182. [Google Scholar]
- 11.Hektor HJ, Kloosterman H, Dijkhuizen L. Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus. J Biol Chem. 2002;277(49):46966–46973. doi: 10.1074/jbc.M207547200. [DOI] [PubMed] [Google Scholar]
- 12.Vonck J, et al. Electron microscopic analysis and biochemical characterization of a novel methanol dehydrogenase from the thermotolerant Bacillus sp. C1. J Biol Chem. 1991;266(6):3949–3954. [PubMed] [Google Scholar]
- 13.Kato N, Yurimoto H, Thauer RK. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci Biotechnol Biochem. 2006;70(1):10–21. doi: 10.1271/bbb.70.10. [DOI] [PubMed] [Google Scholar]
- 14.Mitsui R, Sakai Y, Yasueda H, Kato N. A novel operon encoding formaldehyde fixation: The ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19. J Bacteriol. 2000;182(4):944–948. doi: 10.1128/jb.182.4.944-948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller JEN, et al. Engineering Escherichia coli for methanol conversion. Metab Eng. 2015;28:190–201. doi: 10.1016/j.ymben.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Witthoff S, et al. Metabolic engineering of Corynebacterium glutamicum for methanol metabolism. Appl Environ Microbiol. 2015;81(6):2215–2225. doi: 10.1128/AEM.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leßmeier L, et al. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate. Appl Microbiol Biotechnol. 2015;99(23):10163–10176. doi: 10.1007/s00253-015-6906-5. [DOI] [PubMed] [Google Scholar]
- 18.Jakobsen OM, et al. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J Bacteriol. 2006;188(8):3063–3072. doi: 10.1128/JB.188.8.3063-3072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: Mimicking nature’s synergy. Curr Opin Biotechnol. 2008;19(5):492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J Biol Chem. 1999;274(18):12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 21.Chen R, et al. Biomolecular scaffolds for enhanced signaling and catalytic efficiency. Curr Opin Biotechnol. 2014;28:59–68. doi: 10.1016/j.copbio.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Siu K-H, et al. Synthetic scaffolds for pathway enhancement. Curr Opin Biotechnol. 2015;36:98–106. doi: 10.1016/j.copbio.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Banta S, Chen W. Functional assembly of a multi-enzyme methanol oxidation cascade on a surface-displayed trifunctional scaffold for enhanced NADH production. Chem Commun (Camb) 2013;49(36):3766–3768. doi: 10.1039/c3cc40454d. [DOI] [PubMed] [Google Scholar]
- 24.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, Yang S, Zhao C, Ren Y, Wei D. Artificial multienzyme supramolecular device: Highly ordered self-assembly of oligomeric enzymes in vitro and in vivo. Angew Chem Int Ed Engl. 2014;53(51):14027–14030. doi: 10.1002/anie.201405016. [DOI] [PubMed] [Google Scholar]
- 26.Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7(4):222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 27.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol. 2011;77(9):2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrado RJ, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40(4):1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogorad IW, et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle. Proc Natl Acad Sci USA. 2014;111(45):15928–15933. doi: 10.1073/pnas.1413470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orita I, Sakamoto N, Kato N, Yurimoto H, Sakai Y. Bifunctional enzyme fusion of 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase. Appl Microbiol Biotechnol. 2007;76(2):439–445. doi: 10.1007/s00253-007-1023-8. [DOI] [PubMed] [Google Scholar]
- 31.Moon TS, Dueber JE, Shiue E, Prather KLJ. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng. 2010;12(3):298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Bai Y, et al. Highly ordered protein nanorings designed by accurate control of glutathione S-transferase self-assembly. J Am Chem Soc. 2013;135(30):10966–10969. doi: 10.1021/ja405519s. [DOI] [PubMed] [Google Scholar]
- 33.Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat Biotechnol. 1999;17(11):1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 34.Kato N, Miyamoto N, Shimao M, Sakazawa C. 3-hexulose phosphate synthase from a new facultative methylotroph, Mycobacterium gastri MB 19. Agric Biol Chem. 1988;52(10):2659–2661. [Google Scholar]
- 35.Herring CD, Blattner FR. Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR. J Bacteriol. 2004;186(20):6714–6720. doi: 10.1128/JB.186.20.6714-6720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez CF, et al. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J Biol Chem. 2006;281(20):14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 37.Tarmy EM, Kaplan NO. Kinetics of Escherichia coli B D-lactate dehydrogenase and evidence for pyruvate-controlled change in conformation. J Biol Chem. 1968;243(10):2587–2596. [PubMed] [Google Scholar]
- 38.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4(3):251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 39.Arfman N, Bystrykh L, Govorukhina NI, Dijkhuizen L. 3-Hexulose-6-phosphate synthase from thermotolerant methylotroph Bacillus C1. Methods Enzymol. 1990;188:391–397. doi: 10.1016/0076-6879(90)88062-f. [DOI] [PubMed] [Google Scholar]
- 40.Nash T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J. 1953;55(3):416–421. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]