Significance
To date, simple membrane pore formation resulting in cytoplasmic leakage is the prevailing model for how membrane-active antibiotics kill bacteria and also is one of the main explanations for the activity of the membrane-binding antibiotic daptomycin. However, such models, typically derived from model membrane studies, often depict membranes as homogenous lipid bilayers. They do not take into account the complex architecture of biological membranes, with their many different membrane proteins, or the presence of microdomains with different fluidity properties. Here we report that daptomycin perturbs fluid microdomains in bacterial cell membranes, thereby interfering with membrane-bound cell wall and lipid synthesis processes. Our results add a different perspective as to how membrane-active antibiotics can kill bacteria.
Keywords: antibiotics, daptomycin, membrane potential, cell wall biosynthesis, Bacillus subtilis
Abstract
Daptomycin is a highly efficient last-resort antibiotic that targets the bacterial cell membrane. Despite its clinical importance, the exact mechanism by which daptomycin kills bacteria is not fully understood. Different experiments have led to different models, including (i) blockage of cell wall synthesis, (ii) membrane pore formation, and (iii) the generation of altered membrane curvature leading to aberrant recruitment of proteins. To determine which model is correct, we carried out a comprehensive mode-of-action study using the model organism Bacillus subtilis and different assays, including proteomics, ionomics, and fluorescence light microscopy. We found that daptomycin causes a gradual decrease in membrane potential but does not form discrete membrane pores. Although we found no evidence for altered membrane curvature, we confirmed that daptomycin inhibits cell wall synthesis. Interestingly, using different fluorescent lipid probes, we showed that binding of daptomycin led to a drastic rearrangement of fluid lipid domains, affecting overall membrane fluidity. Importantly, these changes resulted in the rapid detachment of the membrane-associated lipid II synthase MurG and the phospholipid synthase PlsX. Both proteins preferentially colocalize with fluid membrane microdomains. Delocalization of these proteins presumably is a key reason why daptomycin blocks cell wall synthesis. Finally, clustering of fluid lipids by daptomycin likely causes hydrophobic mismatches between fluid and more rigid membrane areas. This mismatch can facilitate proton leakage and may explain the gradual membrane depolarization observed with daptomycin. Targeting of fluid lipid domains has not been described before for antibiotics and adds another dimension to our understanding of membrane-active antibiotics.
Daptomycin is a lipopeptide antibiotic with excellent activity against Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA). It is used to treat skin infections caused by Gram-positive bacteria, S. aureus bacteremia, and right-sided endocarditis. It is a last-resort antibiotic for the treatment of infections caused by multidrug-resistant Gram-positive pathogens and is one of the few peptide antibiotics that can be administered systemically (1). Despite its important clinical role, the mechanism of action of daptomycin is still not well understood.
Daptomycin is produced by the Gram-positive soil bacterium Streptomyces roseosporus (2, 3) and consists of a macrolactone core containing 10 amino acid residues and three exocyclic amino acids linked to a decanoyl fatty acid tail (Fig. 1A) (4, 5). The antibiotic interacts with the cell membrane via its lipid tail, which is inserted between the fatty acyl chains of the membrane bilayer (6, 7). Daptomycin is negatively charged, and its activity depends on the presence of Ca2+ ions that stimulate daptomycin oligomerization and reduce the overall negative charge of the peptide (7–10). The Ca2+–daptomycin complex has an increased affinity for negatively charged phospholipids, including phosphatidylglycerol (PG) (7, 11), which are highly prevalent in bacterial membranes.
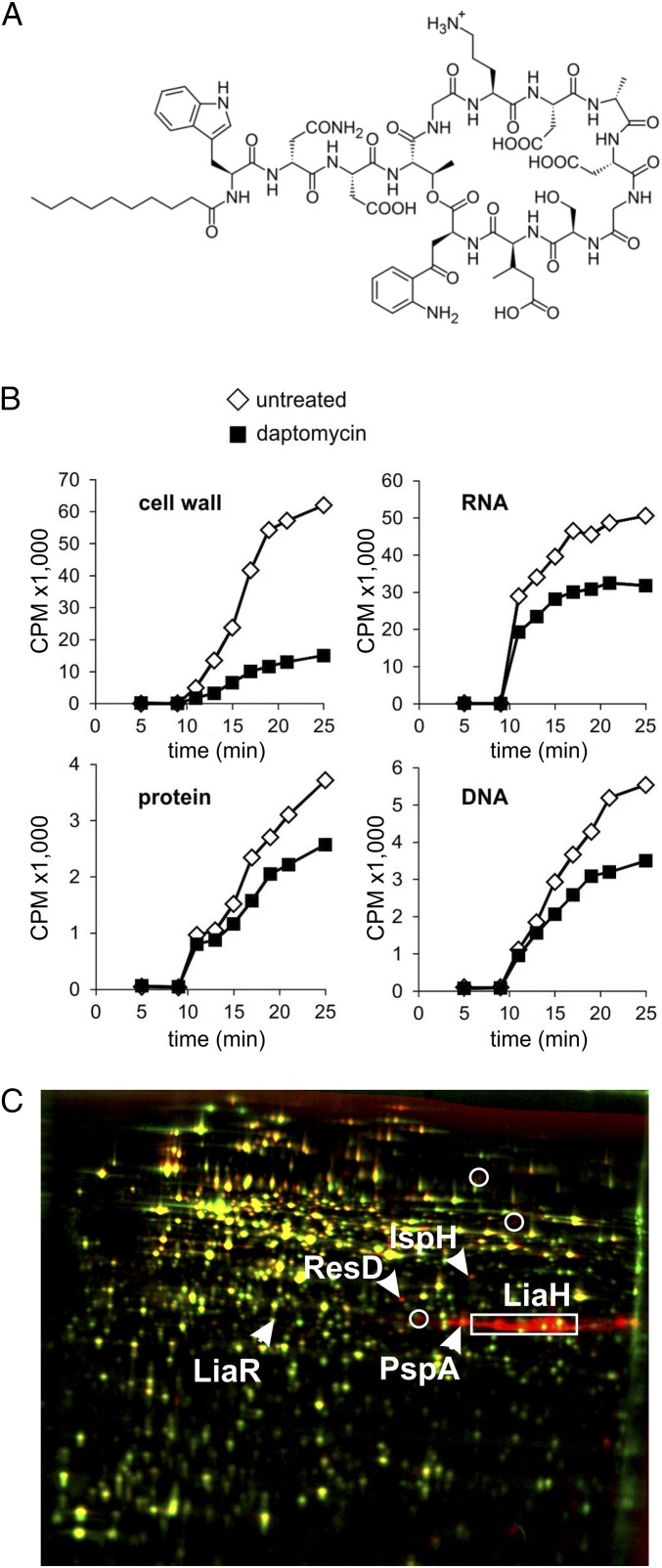
Fig. 1.
Influence of daptomycin on B. subtilis physiology. (A) Structure of daptomycin. Note its large peptide part and short acyl tail. (B) Incorporation of radioactively labeled precursors into the main cellular macromolecules. (C) Autoradiographs of cytosolic proteome extracts of B. subtilis 168 treated with 3.5 µg/mL daptomycin (red) were overlaid with those of untreated controls (green). Up-regulated proteins appear red; down-regulated proteins appear green. Proteins synthesized at equal rates appear yellow.
Early studies suggested that the peptidoglycan biosynthesis pathway is the main target of daptomycin (4, 12), although a direct interaction with the cell wall synthesis machinery has not been confirmed so far (13). Interference with lipoteichoic acid biosynthesis also has been proposed (14) but was subsequently disproven (15). Alternatively, different membrane-perturbing mechanisms have been suggested for daptomycin, including pore formation, potassium efflux, and membrane depolarization (16, 17). However, transcriptome and proteome analyses of daptomycin-treated cells pointed to cell wall stress rather than membrane stress (18, 19). More recently, studies using the Gram-positive model organism Bacillus subtilis showed that daptomycin causes membrane deformations that attract the conserved cell-division protein DivIVA, which binds specifically to negatively curved (concave) membrane areas (20, 21). From these results it was concluded that daptomycin insertion generates membrane areas with altered curvature, resulting in an aberrant recruitment of essential cell-envelope proteins. This recruitment would lead to dramatic cell wall and membrane defects and eventually to cell death (22). However, this mode of action does not explain why daptomycin appears to block the synthesis of cell envelope components preferentially or why it seems to have membrane pore-forming capacities.
The variety of explanations for the mechanism of action of daptomycin might be related to the variety of experiments and model organisms used in the different studies. Therefore we initiated a comprehensive mode-of-action study using the model organism B. subtilis. One of the advantages of this organism is the availability of a wide variety of GFP–protein fusions, enabling investigation of antibiotic-induced effects at the single-cell level. Using fluorescence light microscopy combined with a diverse set of biochemical and proteomic approaches, we found that daptomycin neither forms discrete membrane pores nor induces membrane areas with altered curvature. Instead, we found that the antibiotic binds to and clusters fluid lipids, i.e., lipids with short, branched, and/or unsaturated fatty acyl chains. This change in the cell membrane structure affects the binding of several important peripheral membrane proteins, most prominently binding of the essential phospholipid synthase PlsX and lipid II synthase MurG, explaining the effect of daptomycin on cell envelope synthesis. The targeting of fluid lipid microdomains, thereby delocalizing essential peripheral membrane proteins, is different from current working models of membrane-targeting antibiotics and explains why over the years such different killing mechanisms have been ascribed to daptomycin.
Results
Daptomycin Triggers Cell Envelope Stress.
The inhibiting effect of daptomycin on cell wall synthesis has been measured in S. aureus and Enterococcus faecium (4, 14). Therefore, we first checked whether this inhibition is also seen in B. subtilis. Indeed, macromolecular incorporation studies showed a strong impairment of cell wall synthesis by daptomycin, whereas DNA, RNA, and protein synthesis were much less affected (Fig. 1B and SI Appendix, Fig. S1). To arrive at a better understanding of the mechanism by which daptomycin hampers cell wall synthesis, we compared the proteome stress response of daptomycin-treated cells with a proteome reference library containing the unique stress profiles of B. subtilis treated with more than 60 different antibiotics (23, 24). Log-phase cultures were treated with sublethal daptomycin concentrations, and newly synthesized proteins were pulse-labeled with [35S]-l-methionine followed by 2D SDS/PAGE. Fig. 1C shows an overlay of autoradiographs from untreated and daptomycin-treated cultures. Proteins that were up-regulated more than twofold in three independent biological replicate experiments were defined as marker proteins (Table 1). We observed a very strong up-regulation of the phage shock protein PspA, which is thought to counteract proton leakage upon membrane damage (25), and an even stronger up-regulation of the PspA homolog LiaH. In addition, we observed up-regulation of the lia response regulator LiaR, ResD, which is involved in regulation of anaerobic respiration, and IspH, which is involved in the synthesis of the lipid carrier isopentenyl phosphate and the menaquinone precursor isoprenoid (26–29).
Table 1.
Marker proteins up-regulated after treatment with daptomycin
| Protein | IF | Protein function | Mass, Da | PI | Peptide count | Mascot score |
| LiaH | 4.1 | Similar to phage shock protein | 25,682 | 6.2 | 12 | 153 |
| LiaR | 13.9 | Two-component response regulator, regulation of the liaIHGFSR operon | 29,020 | 6.32 | 10 | 261 |
| PspA | 3.0 | Phage shock protein A | 25,125 | 5.87 | 14 | 160 |
| ResD | 10.6 | Two-component response regulator, regulation of aerobic/anaerobic respiration | 27,468 | 5.75 | 11 | 201 |
| IspH | 13.0 | Isopentenyl diphosphate biosynthesis | 34,902 | 5.68 | 14 | 323 |
Peptide count represents the number of tryptic peptides. IF, induction factor; PI, isoelectric point.
Up-regulation of LiaH is a specific marker for compounds that inhibit the membrane-bound steps of lipid II synthesis, and PspA induction is part of a specific response seen with membrane stress (23). Up-regulation of these marker proteins is also observed with the cell envelope-targeting antibiotics nisin and gallidermin (Table 2), both of which bind the peptidoglycan precursor lipid II. Binding of lipid II by nisin results in target-mediated pore formation in B. subtilis (23). However, other marker proteins that are characteristic for the treatment with cell wall synthesis-inhibiting antibiotics were not found (Table 2). Also, several marker proteins for antibiotics that cause membrane damage [e.g., gramicidin S, MP196, and aurein 2.3 (23)] were not observed in the daptomycin stress profile (Table 2). In fact, the proteome profile of daptomycin appears to be unique and does not match any compound in the proteome reference compendium. The up-regulation of ResD and IspH is found only after treatment with daptomycin and is indicative of a disturbance of respiration. Indeed, we measured a decrease in resazurin reduction, indicating lower respiratory chain activity, and a moderate reduction of ATP levels (SI Appendix, Fig. S2 A and B).
Table 2.
Overlap of marker proteins up-regulated by daptomycin with the proteome response library
| Marker protein | Daptomycin | Nisin | Gallidermin | Mersacidin | Valinomycin | Gramicidin A | Gramicidin S | MP196 | Aurein 2.3 |
| LiaH (CWB marker) | x | x | x | x | x | x | x | ||
| LiaR | x | ||||||||
| PspA (CM marker) | x | x | x | x | x | x | x | x | |
| ResD | x | ||||||||
| IspH | x | ||||||||
| Total markers | 8 | 8 | 21 | 13 | 20 | 5 | 17 | 27 | 33 |
CM, cell membrane; CWB, cell wall biosynthesis. LiaH is a specific marker for inhibition of membrane-bound steps of lipid II synthesis, and PspA is a specific marker for membrane stress. Note that in contrast to daptomycin, other membrane-active antibiotics typically have a higher number of total marker proteins.
Daptomycin Does Not Form Discrete Membrane Pores.
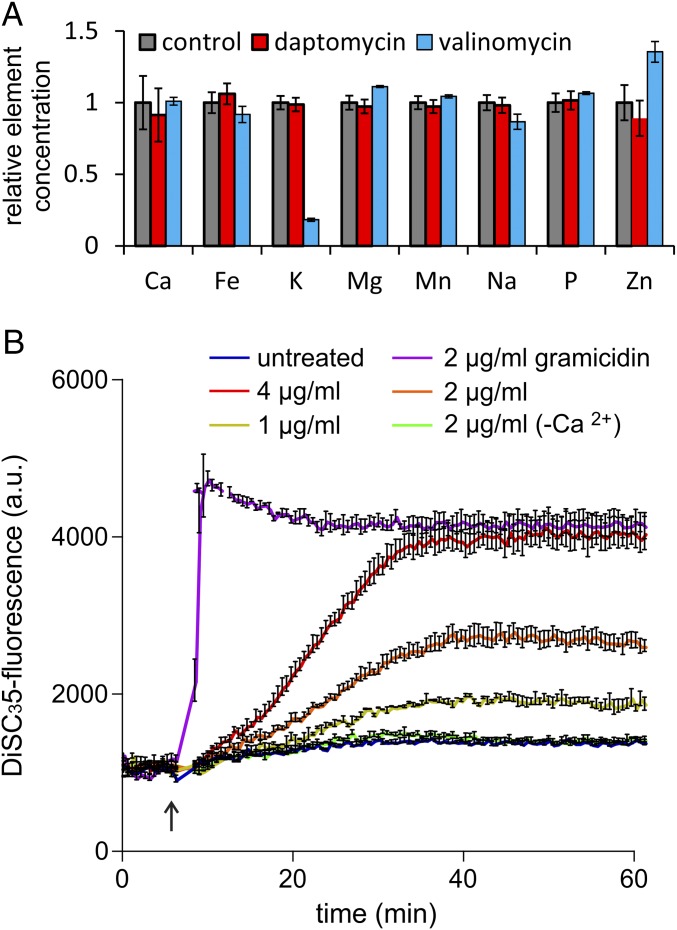
Based on the observed potassium leakage with daptomycin-treated S. aureus cells, it was concluded that daptomycin forms discrete ion-conducting pores in bacterial membranes (16, 30). This notion was later supported by studies using artificial lipid membranes (17). However, our proteome analysis showed no particular match with pore-forming peptides such as nisin or aurein 2.3 or specific cation ionophores such as gramicidin A or valinomycin (Table 2). Moreover, treatment with growth-inhibiting concentrations of daptomycin did not result in the uptake of the large fluorescent dye propidium iodide, an indicator of pore formation, or in severe membrane disruption (SI Appendix, Fig. S3) (31). To test whether daptomycin causes leakage of specific cations in B. subtilis, we measured the cellular element concentration using inductively coupled plasma optical emission spectroscopy (ICP-OES). Cells were incubated with a growth-inhibiting but sublytic daptomycin concentration. As shown in Fig. 2A, treatment with daptomycin did not reduce the levels of any element measured. This finding is in sharp contrast to the leakage of potassium caused by the specific potassium ionophore valinomycin (Fig. 2A) and the leakage of potassium, magnesium, iron, and manganese observed with the membrane-disrupting peptide gramicidin S (32).
Fig. 2.
Assessing the membrane pore-forming activity of daptomycin. (A) Cellular element concentrations after antibiotic treatment were measured by ICP-OES. Cells were washed with EDTA before resuspension in Tris buffer and were incubated with 3.5 µg/mL daptomycin or 10 µg/mL valinomycin. (B) Depolarization of the B. subtilis 168 cell membrane was measured with the membrane potential-sensitive fluorescent probe DiSC3(5). Gramicidin ABCD was used as positive control. The arrow indicates the time point of antibiotic addition. As a negative control, 2 µg/mL daptomycin without Ca2+ was used. a.u., arbitrary units.
Although daptomycin does not seem to form discrete ion pores in B. subtilis, several studies with S. aureus have shown that daptomycin affects the membrane potential (16, 30). Therefore we measured the dissipation of the membrane potential using the fluorescence potentiometric probe DiSC3(5). Daptomycin concentrations were used that allowed cells to continue to grow (1 µg/mL), that inhibited growth but did not cause cell lysis (2 µg/mL), or that induced lysis (4 µg/mL) (SI Appendix, Fig. S2C). As shown in Fig. 2B, concentrations that block cell growth (2 µg/mL) led to incomplete depolarization, and only the highest (lytic) daptomycin concentration was sufficient to dissipate the membrane potential completely. Even then, depolarization of the membrane was gradual and complete only after ∼25 min. This gradual depolarization is in sharp contrast to the almost immediate depolarization caused by the K+/Na+ channel-forming peptide mix gramicidin ABCD, a standard control for depolarization (Fig. 2B) (33). Together, these results show that daptomycin does not form distinct membrane pores.
Protein Delocalization Assay.
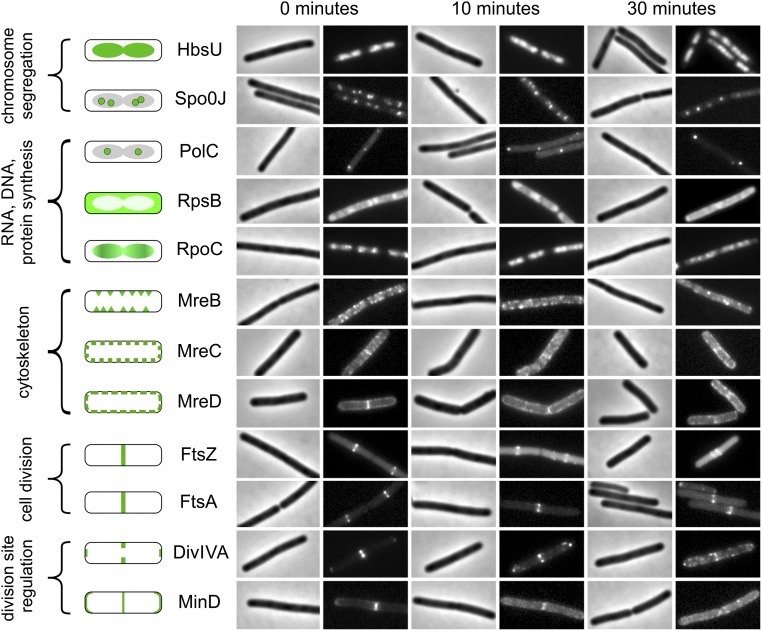
The results so far indicate that cell envelope synthesis is the primary target of daptomycin and that disruption of the membrane barrier function is a secondary effect. To obtain more information on the mechanism by which daptomycin affects cell envelope synthesis, we monitored the cellular localization of a set of marker proteins. Many proteins show a specific cellular localization pattern that is related to their activities, and inactivation by specific antibiotics results in protein delocalization that can be monitored by fluorescence light microscopy (34, 35). We compiled a broad set of B. subtilis reporter strains expressing GFP-tagged proteins involved in chromosome compaction (HbsU), DNA replication (Spo0J and PolC), transcription (RpoC), translation (RpsB), cell wall synthesis and cell shape (MreB, MreC, and MreD), cell division (FtsZ and FtsA), and cell division regulation (MinD and DivIVA). As shown in Fig. 3, growth-inhibitory but sublytic daptomycin concentrations (2 µg/mL) did not affect the localization of proteins involved in chromosome compaction, DNA, RNA, and protein synthesis. This finding is in line with precursor incorporation studies that revealed only marginal effects on the synthesis of DNA, RNA, and proteins (Fig. 1B). However, a gradual delocalization of the cell division proteins FtsZ, FtsA, DivIVA, and MinD and the cell shape-determining proteins MreB, MreC, and MreD was apparent. The peripheral membrane proteins MinD, FtsA, and MreB require the presence of the membrane potential for correct cellular localization (34). The gradual delocalization of these proteins probably is caused by the slow depolarization of the membrane by daptomycin. This depolarization will also affect the localization of FtsZ, MreC, and MreD indirectly, because these proteins require either FtsA or MreB for their localization (34).
Fig. 3.
The influence of daptomycin on marker protein localization. B. subtilis strains (listed in SI Appendix, Table S1) were grown in LB medium supplemented with 1.25 mM CaCl2 and treated with 2 µg/mL daptomycin. The left panel schematically shows the normal localization patterns of the different GFP fusions. Samples for fluorescence light microscopy were taken after 10 and 30 min of incubation. Field width, 6 μm.
Daptomycin Does Not Cause Negative Membrane Curvature.
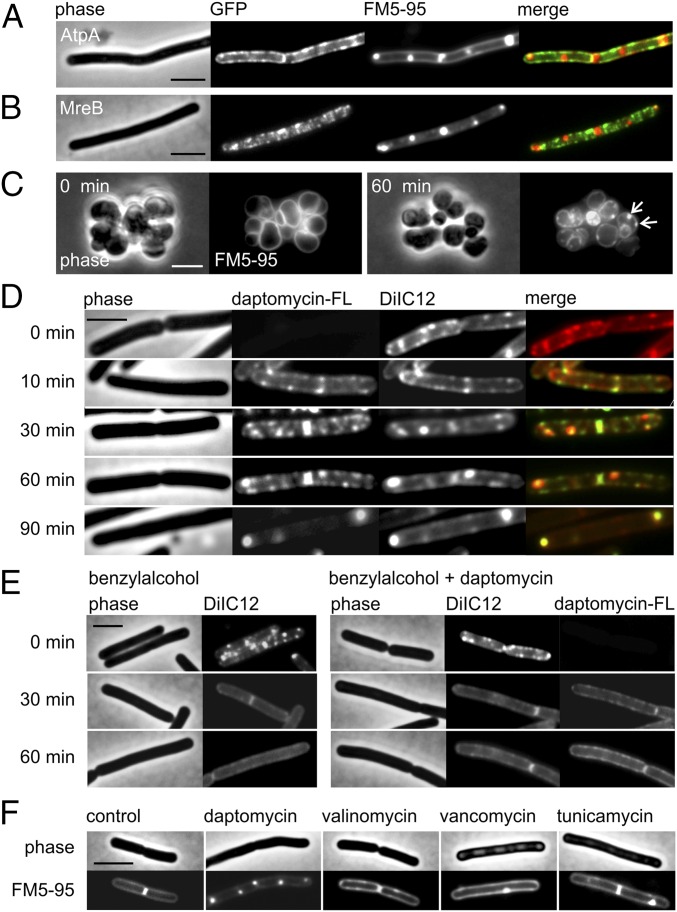
Fig. 3 also shows delocalization of the cell division regulator DivIVA. Previously, it has been shown that daptomycin-treated B. subtilis cells stained with the fluorescent membrane dye FM4-64 develop strongly fluorescent membrane patches (22). These fluorescent membrane areas, which occur after approximately 1 h (SI Appendix, Fig. S4A), attract DivIVA-GFP (22). Because DivIVA binds specifically to negatively curved (concave) membranes (20, 21), and because insertion of a conical molecule such as daptomycin into the membrane can induce membrane curvature (36, 37), it was concluded that these fluorescent patches are areas where the cell membrane is strongly negatively curved (22). This curvature can be caused by invagination of excess membrane material, resulting in an increased fluorescence membrane dye signal. To test whether daptomycin generates membrane invaginations, we looked at the localization of the transmembrane ATPase complex that can be used as a reporter for abnormal membrane shapes because of its uniform membrane localization (38). As shown in Fig. 4A, the localization of the ATP synthase subunit AtpA is unaffected by daptomycin treatment, and the reporter protein does not accumulate at areas that show a strong fluorescence signal of the general membrane dye FM5-95, indicating that these regions are not membrane invaginations. In fact, it turned out that the binding of DivIVA-GFP to these fluorescent membrane areas is unrelated to curvature but instead is an artifact caused by the propensity of GFP to form dimers (Discussion).
Fig. 4.
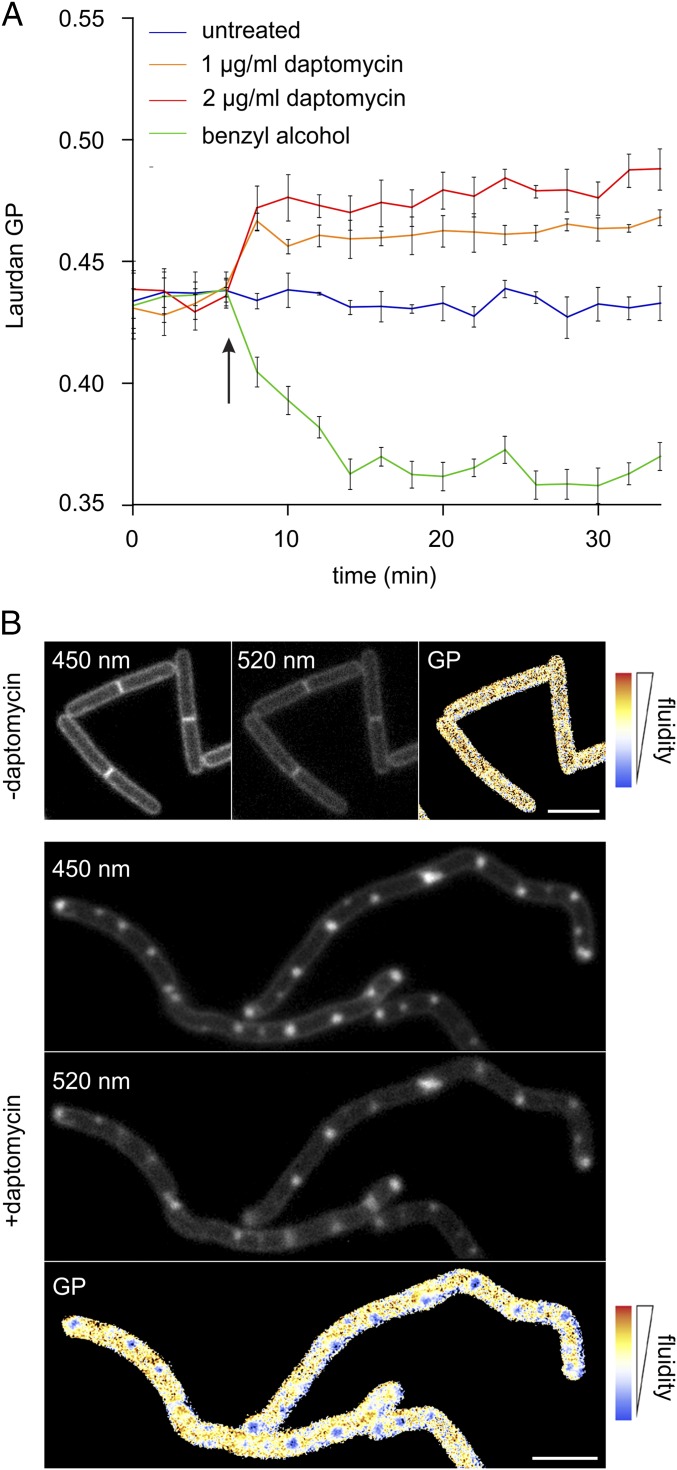
Daptomycin does not cause membrane invaginations but clusters fluid lipid domains. (A) Colocalization of daptomycin-induced membrane patches and AtpA–GFP. The membrane was stained with FM5-95. AtpA is distributed uniformly in the cell membrane and is not affected by daptomycin, demonstrating that the strongly fluorescent FM dye foci are not caused by membrane invaginations. (B) Colocalization of daptomycin-induced lipid patches and GFP–MreB. Membrane patches stained with FM5-95 do not overlap with MreB, suggesting that the protein is not involved in the generation of lipid patches. (C) The MreB triple-knockout mutant (ΔmreB Δmbl ΔmreBH) treated with daptomycin and stained with FM5-95. Without the rod shape-determining MreB proteins, the organization of lateral cell wall synthesis is disturbed, resulting in round cells. The presence of lipid patches (arrows) demonstrates independence from the RIF-organizing activity of MreB. (D) B. subtilis 168 costained with the fluorescent RIF dye DiIC12 and fluorescently labeled daptomycin-BODIPY (3 µg/mL). Daptomycin (1 µg/mL) causes native RIFs to deteriorate and fuse into large domains that overlap with daptomycin-BODIPY. (E) The importance of RIFs for daptomycin activity. (Left) The membrane fluidizer benzyl alcohol diminishes RIFs in the B. subtilis membrane. (Right) Pretreatment with benzyl alcohol (10 min) reduces daptomycin clustering in the membrane and prevents the accumulation of fluid lipids into large daptomycin-fluid lipid domains. (F) Lipid patches stained with FM5-95 are characteristic for daptomycin and do not occur after treatment with antibiotics that dissipate the membrane potential (valinomycin) or inhibit lipid II synthesis (vancomycin and tunicamycin). Unless otherwise stated, B. subtilis strains were treated with 2 µg/mL daptomycin for 60 min. (Scale bars: 2 µm.)
Clustering of Fluid Lipids.
The fluorescence of membrane dyes is influenced by the physical properties of the lipid environment (39–42). Because the strongly fluorescent membrane patches cannot be explained by excess cell membrane, it is likely that they constitute a different physical membrane environment. In a previous study we have shown that polymers formed by the actin homolog MreB associate with lipids that are in a fluid, liquid-disordered state. These are lipids that contain short, branched, and/or unsaturated fatty acyl chains. MreB polymers organize these fluid lipids in microscopically visible regions of increased fluidity (RIFs) (38). It has been speculated that the liquid-disordered state of these RIFs attracts more membrane dye molecules and/or increases their fluorescence quantum yield, resulting in enhanced fluorescence (39). In B. subtilis, depolarization of the membrane leads to clustering of MreB polymers together with RIFs, resulting in a strong focal fluorescent signal of the membrane dye (38). However, as shown in Fig. 4B, the strongly fluorescent membrane patches formed by daptomycin do not colocalize with MreB. In fact, when a B. subtilis mutant devoid of MreB and its homologs was incubated with daptomycin, fluorescent membrane patches were still observed (Fig. 4C).
Although MreB polymers do not seem to be involved in the daptomycin-induced formation of fluorescent membrane patches, it is still possible that these patches are formed by high concentrations of fluid lipids. To examine this possibility, we used the lipid-mimicking dye DiIC12, which localizes at fluid lipid regions because of its short fatty acyl chains (38, 43). Indeed, DilC12 showed the same staining pattern as FM5-95 (SI Appendix, Fig. S4B), indicating that the large fluorescent membrane patches are enriched in fluid lipids. Previously, fluorescently labeled daptomycin was reported to colocalize with the strongly fluorescent membrane patches stained with FM dye (22). Thus, daptomycin might be attracted to fluid lipids and cluster with them. To study this possibility, we costained cells with DiIC12 and fluorescently labeled daptomycin (daptomycin-BODIPY). As shown in Fig. 4D, the normally distinct RIFs began to deteriorate after ∼10 min of incubation and clustered into larger foci after ∼30 min, culminating in large fluorescent membrane patches after ∼60 min. During this process, the fluorescent daptomycin signal exhibits a clear overlap with the DilC12 signal, indicating that the antibiotic prefers to insert into fluid membrane domains and causes them to cluster together. Although these clusters often appear at the cell poles, they also are formed along the long axis of the cell (SI Appendix, Fig. S6). As a control, cells were pretreated with the membrane fluidizer benzyl alcohol (44) that disperses RIFs. Indeed, the presence of benzyl alcohol hampered the formation of daptomycin clusters and the formation of large fluid lipid domains (Fig. 4E and SI Appendix, Fig. S6).
To examine whether this clustering of fluid lipids is unique to daptomycin or also occurs with other cell envelope-targeting antibiotics, we treated B. subtilis cells with the potassium ionophore valinomycin and antibiotics inhibiting cell wall biosynthesis, vancomycin and tunicamycin. Interestingly, only slight alterations of the membrane stain were detected with these antibiotics, and we never observed the large fluorescent patches that occur when cells are incubated with daptomycin (Fig. 4F and SI Appendix, Fig. S7).
Daptomycin Affects Membrane Fluidity.
To analyze how the clustering of fluid lipids by daptomycin affects the overall fluidity of the cell membrane, we used the membrane fluidity-sensitive dye laurdan. This probe changes its fluorescence emission wavelength depending on the amount of water molecules between lipid head groups, thus providing a measure for lipid head group density and fatty acid chain flexibility (40). As shown in Fig. 5A, daptomycin induces a rapid increase in laurdan general polarization (GP) values, reflecting rapid membrane rigidification. The membrane fluidizer benzyl alcohol shows a similarly rapid but opposite effect on laurdan GP (Fig. 5A). The decrease in membrane fluidity occurs in less than 2 min, suggesting that it is a direct effect of daptomycin insertion into the bilayer rather than a cellular adaptation.
Fig. 5.
Daptomycin rigidifies the cell membrane. (A) Spectroscopic measurement of membrane fluidity with laurdan. Laurdan was excited at 330 ± 5 nm, and fluorescence emission was recorded at 460 ± 5 nm and 500 ± 5 nm. The membrane fluidizer benzyl alcohol was used as positive control. (B) B. subtilis 168 was treated with 1 µg/mL daptomycin for 60 min. Laurdan was excited at 360 nm, and fluorescence emission was captured at 522 nm and 450 nm. Color-coded GP maps were generated with Wolfram Mathematica 7. (Scale bars: 2 µm.)
Laurdan GP also can be used to visualize differences in membrane fluidity microscopically in bacterial cells (40). Indeed, laurdan GP microscopy of daptomycin-treated cells showed a clear decrease in overall membrane fluidity (Fig. 5B). Interestingly, the large fluorescent membrane patches, indicative of fluid lipids clustered together by daptomycin, showed a clearly reduced laurdan GP. This observation suggests that the interaction of the lipids with daptomycin reduces their acyl chain mobility, resulting in relatively rigid membrane domains.
Daptomycin Delocalizes MurG and PlsX.
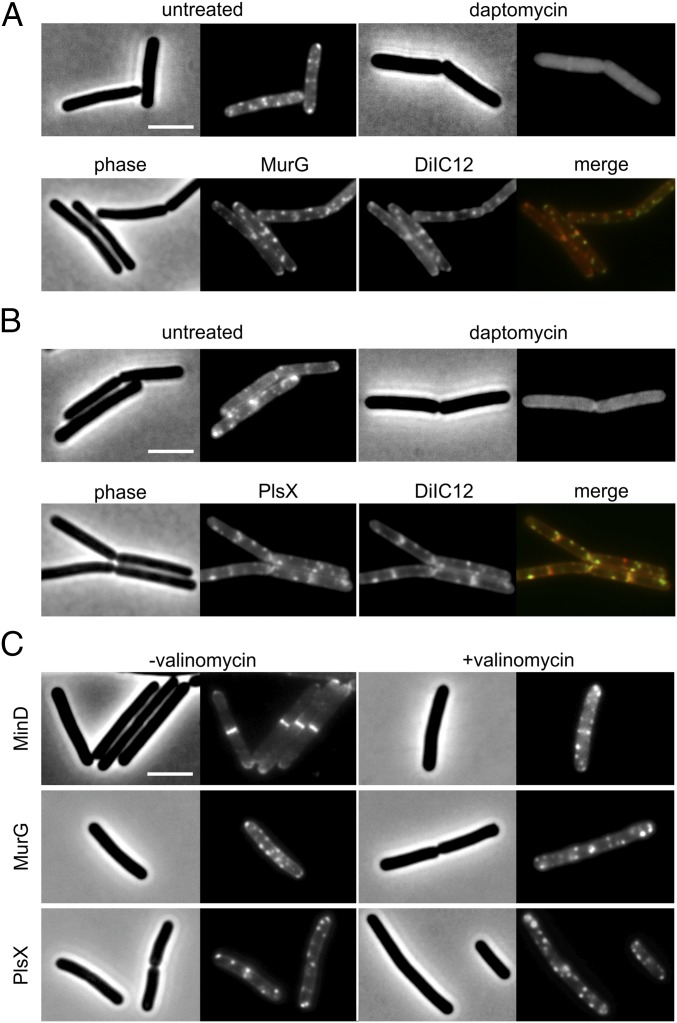
Although the effect of daptomycin on fluid lipid distribution is apparent, the question of how daptomycin inhibits cell envelope synthesis remains. Binding of many peripheral membrane proteins to the cell membrane is stimulated by fluid lipids, because their unsaturated short and/or branched-chain fatty acids provide the space and flexibility necessary for the insertion of the relatively bulky membrane-binding domains such as amphipathic helices (45). The binding of fluid lipids by daptomycin and the possible reduction of their fluid (liquid disorder) properties therefore might interfere with the attachment of peripheral membrane proteins to the membrane. In fact, several peripheral membrane proteins (MreB, MinD, and DivIVA) showed an aberrant localization in cells treated with daptomycin (Fig. 3). Interestingly, the synthesis of peptidoglycan, which appeared to be strongly affected in our precursor incorporation studies, requires one essential peripheral membrane protein, MurG. MurG is an N-acetylglucosamine transferase responsible for the last synthesis step of the peptidoglycan precursor lipid II (46). Remarkably, the addition of daptomycin resulted in a rapid (<2 min) and almost complete dissociation of MurG from the membrane into the cytosol (Fig. 6A and see SI Appendix, Fig. S8 for line scans). This dramatic effect might suggest that MurG associates strongly with RIFs that are targeted by daptomycin. Indeed, as shown in Fig. 6A, the MurG–GFP fusion showed a clear colocalization with DilC12-stained RIFs, demonstrating a preference of MurG for fluid lipid domains. The detachment of MurG from the cell membrane explains why daptomycin blocks cell wall synthesis.
Fig. 6.
Daptomycin delocalizes MurG and PlsX. (A, Upper) Localization of MurG after treatment with 2 µg/mL daptomycin for 10 min. (Lower) MurG localizes to natural RIFs in log-phase B. subtilis. (B, Upper) Localization of PlsX after treatment with 2 µg/mL daptomycin for 10 min. (Lower) PlsX localizes to natural RIFs in log-phase B. subtilis. (C) Localization of MurG (TNVS177, murG-msfgfp) and PlsX (TNVS29D, plsX-sfgfp) after treatment with the potassium ionophore valinomycin (10 µg/mL). MinD (LH131, gfp-minD), which is sensitive for the membrane potential (34), served as positive control. (Scale bars: 2 µm.)
To test whether daptomycin is specific for MurG, we examined the localization of another peripheral membrane protein (47), the essential acyltransferase PlsX, which is involved in phospholipid synthesis (48). Interestingly, this protein also dissociates rapidly from the cell membrane upon daptomycin treatment (Fig. 6B), and PlsX–GFP showed a clear colocalization with RIFs (Fig. 6B). The almost immediate delocalization of two different enzymes indicates that daptomycin does not target a specific protein but rather targets the specific fluid lipid-enriched membrane areas to which these enzymes bind.
Finally, it has been reported that the peripheral membrane proteins MinD, FtsA, and MreB delocalize quickly when the membrane potential is dissipated (34). However, we found that daptomycin does not cause an immediate depolarization of the membrane. To confirm that the observed detachment of MurG and PlsX from the cell membrane is not a consequence of membrane depolarization, we incubated cells with the membrane potential-dissipating antibiotic valinomycin. After 5 min of incubation the normal localization of MinD was abolished completely, whereas both MurG and PlsX still showed a peripheral localization (Fig. 6C).
Discussion
Revised Daptomycin Working Model.
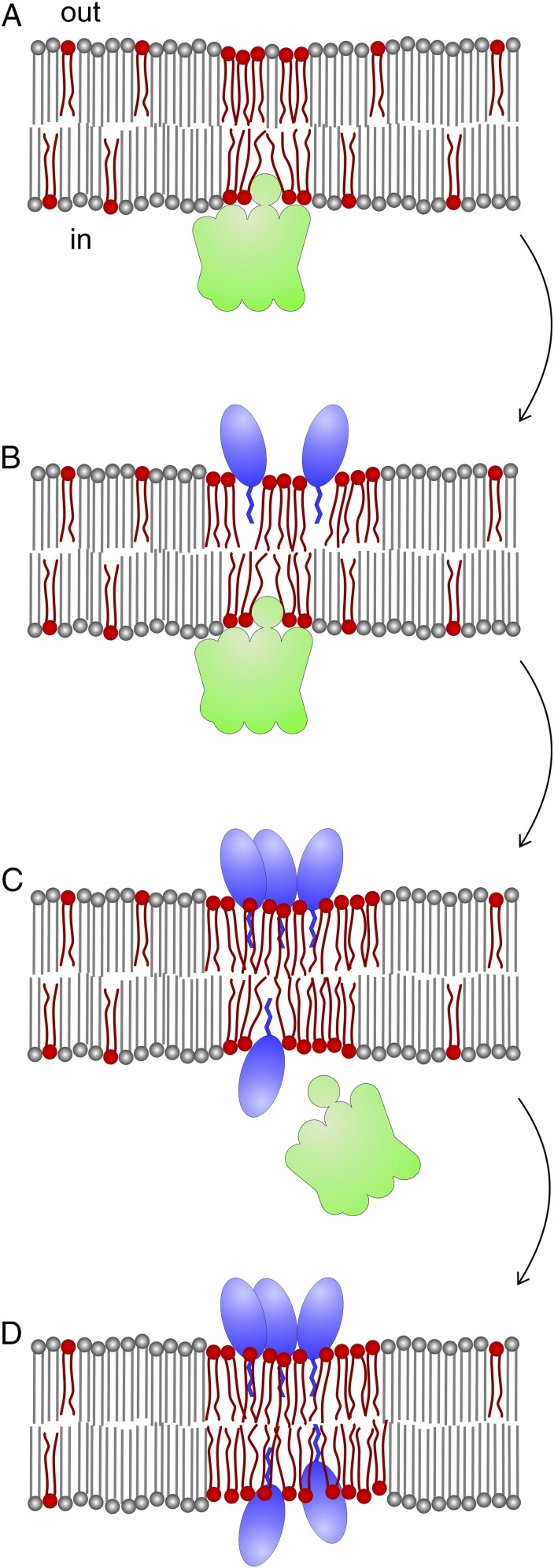
Based on our experiments and those of others (6–8, 22), we arrived at the following model for the mechanism of daptomycin (schematically explained in Fig. 7). When daptomycin reaches the cell membrane, it inserts its short lipid tail (C10) between the fatty acyl chains of phospholipid molecules (6), thereby strongly disturbing the regular fatty acid packing because of its large peptide ring structure (10 amino acids in the macrolactone core and three exocyclic amino acids) (7, 8, 49, 50). Ca2+-stimulated oligomerization of daptomycin leads to further bilayer distortion. Oligomerization of daptomycin has been described as occurring in two steps: first a loose aggregation involving a first Ca2+ ion and insertion of ornithine-6 into the membrane, followed by a second step involving the binding of a second Ca2+ ion, leading to the formation of a tightly associated oligomer and deeper insertion of tryptophane-1 and kynurenine-13 into the bilayer (51). The submersion of the peptide head group into the bilayer greatly enhances membrane distortion. Because lipids with short, branched and/or unsaturated fatty acyl chains are more flexible and therefore are better able to surround the bulky daptomycin complex, the antibiotic will associate preferably with membrane areas with a high concentration of these fluid lipids, i.e., RIFs. In fact, we observed a strong preference of daptomycin-BODIPY for RIFs when Ca2+ was added to the medium, in contrast to the poor membrane binding and more uniform distribution pattern in medium in the absence of Ca2+ (SI Appendix, Fig. S9). Oligomerization, together with the preference for fluid lipids, will further stimulate clustering of large daptomycin-fluid lipid complexes. Laurdan GP microscopy showed that these complexes have reduced fluidity, suggesting that daptomycin restricts the chain flexibility of lipids within these complexes. In addition, the attraction of fluid lipids by daptomycin complexes will reduce fluid lipids in the rest of the membrane, thereby contributing to the decrease in overall membrane fluidity. Thus, daptomycin has a dramatic effect on the liquid-order/disorder balance of the cell membrane. Several studies have shown that there is a coupling between the bilayer leaflets and that local disorder in one leaflet stimulates local disorder of the opposite leaflet (52–54). Thus, the distortion of lipids in the outer leaflet by daptomycin likely attracts fluid lipids in the inner leaflet as well. It also has been shown that daptomycin flips between the two membrane leaflets (55), and it is likely that high concentrations of fluid lipids facilitate this flipping. The change in the biophysical properties of the cell membrane, especially of RIFs, caused by daptomycin might impair the attachment of peripheral proteins such as MurG and PlsX to the membrane. However, an alternative explanation is that the bulky daptomycin displaces RIF-associated proteins and blocks access to fluid lipids that are required for binding of MurG and PlsX (Fig. 7).
Fig. 7.
Model of daptomycin interaction with fluid lipid domains. (A) Peripheral membrane proteins involved in cell wall (e.g., MurG) and lipid (e.g., PlsX) synthesis localize to RIFs indicated by a high concentration of fluid lipids (red). “Out” and “in” represent the cell wall side and the cytoplasmic side, respectively. (B) Daptomycin inserts into the bacterial membrane. Because of daptomycin’s bulky nature, insertion into regions with a high concentration of fluid lipids, such as RIFs, is favored. The combination of daptomycin’s bulky head group and its short hydrocarbon tail forces lipids apart and creates bilayer distortions, which are counterbalanced by the attraction of fluid lipids to the sites of daptomycin insertion. These fluid lipids are required to compensate for gaps in the lipid bilayer. (C) Calcium ions stimulate daptomycin oligomerization, potentiating this effect; as a result more fluid lipids are attracted. This attraction sets in first in the outer membrane leaflet and then causes a similar redistribution of lipids in the inner leaflet, facilitating the flipping of daptomycin molecules through the bilayer to the inner leaflet. As a consequence, peripheral membrane proteins are displaced from RIFs. (D) Daptomycin blocks access to fluid lipids in the inner leaflet, which are required for the association of these proteins, and clusters them into inflexible domains, resulting in the withdrawal of fluid lipids from the bulk and overall increased membrane rigidity.
Effect on Cell Wall Biosynthesis.
Daptomycin completely delocalizes the peripheral membrane protein MurG. MurG is essential and catalyzes the last synthesis step of the peptidoglycan precursor lipid II. Interestingly, it has been shown that fluorescently labeled vancomycin, which binds lipid II, localizes to the strongly fluorescent membrane patches induced by daptomycin (22), indicating the presence of lipid II in these arrested fluid lipid–daptomycin clusters. Lipid II is likely to accumulate at RIFs, because its long bactoprenol moiety prefers a fluid lipid environment (56). These observations are in good agreement with our finding that MurG preferentially binds to RIFs and further supports the notion that RIFs are membrane microdomains associated with lateral cell wall biosynthesis (38, 57, 58). The detachment of MurG from the cell membrane prevents efficient peptidoglycan synthesis and explains (i) why incorporation of radioactive precursors in the cell wall is quickly impaired after daptomycin addition, (ii) why daptomycin-treated cells primarily suffer cell envelope stress (18, 19, 59–61), and (iii) why the proteome response of B. subtilis to daptomycin treatment is dominated by the up-regulation of LiaH, which is a specific proteomic marker for the inhibition of membrane-bound lipid II synthesis (18, 23). Finally, a number of integral membrane proteins are involved in peptidoglycan synthesis, but the localization of these proteins (MraY, PBP1, and PBP2) was unaffected by daptomycin (SI Appendix, Fig. S10).
In B. subtilis, MurG is the only peripheral membrane protein involved in lipid II synthesis. However, the situation can be different in other bacteria. For example, S. aureus possesses additional peripheral membrane proteins involved in cell wall synthesis, such as FemXAB, that attach the peptide side chain to lipid II (62). Possibly, these proteins are affected by daptomycin as well. In addition to peptidoglycan, another important cell wall component in Gram-positive bacteria is teichoic acid. The synthesis of teichoic acids also requires the bactoprenol carrier molecule (63) and relies on the activity of peripheral membrane proteins, e.g., TagB in B. subtilis (64). Studies with S. aureus and E. faecium showed that daptomycin impairs the incorporation of teichoic acid polymers into the cell wall (14). This impairment would fit with our finding that daptomycin does not target one specific protein but instead delocalizes different peripheral membrane proteins, possibly including those involved in teichoic acid polymer synthesis.
Effect on Lipid Synthesis.
The phospholipid synthase PlsX also binds to RIFs and delocalizes upon treatment with daptomycin. PlsX couples the cytosolic fatty acid synthesis with the membrane-bound phospholipid synthesis steps (65). Therefore, its delocalization from the membrane will strongly impair its function. Many Gram-positive bacteria adapt their membrane fluidity by two different strategies: (i) slow adaptation via de novo synthesis of fluidizing lipids containing anteiso rather than iso branched-chain fatty acids, and (ii) fast adaptation by desaturating fatty acid chains via Des, a lipid desaturase induced upon cold adaptation (66–68). Slow adaptation requires PlsX, and it is tempting to speculate that the delocalization of this protein hampers the adaptation of the cell to the effects of daptomycin. Indeed, we did not observe any proteomic marker that suggests an adaptation of the phospholipid composition, such as proteins belonging to the standard fatty acid biosynthesis (Fab) pathway (69), alternative fatty acid synthases (70), or induction of the yuaF-floT-yuaI operon, which is controlled by the membrane stress-responsive alternative sigma factor W (71). In line with these observations, we did not observe a recovery to normal fluidity levels with laurdan GP for at least 90 min after addition of daptomycin (SI Appendix, Fig. S11), also suggesting that a quick adaptation of the membrane fluidity through the activity of the lipid desaturase Des is either impaired by daptomycin or is insufficient to counteract daptomycin-induced changes in the membrane. This apparent inability to adapt membrane fluidity could be one of the key factors contributing to the low development of resistance against daptomycin.
Effect on the Membrane Barrier Function.
Daptomycin further weakens the cell by slow depolarization of the membrane (Fig. 2B) (16). Phase segregation of lipids has been proposed to cause phase-boundary defects leading to membrane permeabilization (72–75), and it has been speculated that hydrophobic mismatches at the interface between fluid (thinner) and rigid (thicker) lipid domains compromise the barrier capacity of membranes (76). Transitions between the fluid lipid–daptomycin clusters (rigidified through tight clustering of fluid, short-chain lipids) and the more rigid (thicker) bulk of the cell membrane likely constitute such weak spots through which proton leakage can occur and might explain the gradual depolarization of the membrane. It should be mentioned that phase-boundary defects as a cause for membrane permeabilization have been studied only in vitro, and it remains to be seen how phase separation affects the barrier function of bacterial membranes that naturally contain lipid domains of different fluidity (38, 77, 78). Nonetheless, daptomycin-induced changes in bilayer organization lead to a slow increase in the passive permeability of the membrane, which may be why membrane pore formation has been considered the main function of daptomycin in former studies (16, 17, 31).
Delocalization of DivIVA.
An unanswered question is why the DivIVA-GFP reporter fusion binds to fluid lipid–daptomycin clusters although these areas do not show an apparent membrane curvature. This binding characteristic was found to be an artifact caused by GFP dimerization. GFP is known to have a tendency to form dimers that can lead to localization artifacts because of the stabilization of the protein complex (79). When we fused DivIVA to a monomeric version of GFP (80), a normal localization pattern was observed; however, the fusion protein no longer accumulated at the fluid lipid clusters when cells were incubated with daptomycin (SI Appendix, Fig. S12A). Interestingly, the cellular localization of this fusion protein also became much more sensitive to dissipation of the membrane potential (SI Appendix, Fig. S12B), which was not observed with the old nonmonomeric GFP fusion (34). Why GFP dimerization stimulates the binding of the DivIVA-GFP fusion to daptomycin-generated fluid membrane areas remains a matter of speculation but is unrelated to membrane curvature.
Daptomycin Resistance.
Although daptomycin is highly effective in the clinic, several nonsusceptible S. aureus and Enterococcus strains have been isolated. The most important mutations are related to either cell wall or membrane adaptation and occur in the teichoic acid alanylation enzymes DltABCD, the PG synthase PgsA, the lysyl-PG synthase MprF, the cardiolipin synthase Cls, the cell envelope stress-responsive two-component system LiaRS, and the cell wall-related two-component system WalKR (YycFG). Mutations in dltABCD increase d-alanylation of teichoic acids in the cell wall and are thought to limit the access of daptomycin to the membrane by charge repulsion (82, 83). Loss-of-function mutations in the PG synthase PgsA and gain-of-function mutations in the lysyl-PG synthase MprF lead to a reduction in negatively charged PG in the membrane. PG is needed for daptomycin–membrane interaction and antibacterial activity (81, 84–86). Mutations in the cardiolipin synthase Cls2 are associated with daptomycin resistance in S. aureus, Enterococcus faecalis, and E. faecium (81). Synthesis of cardiolipin consumes two molecules of PG, leading to the assumption that such mutations contribute to resistance by decreasing the PG content (81). However, it should be mentioned that replacing PG with cardiolipin does not alter the overall membrane charge, as happens when PG is replaced by lysyl-PG. Interestingly, cardiolipin also has been suggested to increase bilayer rigidity (87) and was shown to prevent flipping of daptomycin from the outer to the inner membrane leaflet in model membranes (55). Increased rigidity could be another means of preventing membrane insertion and the flipping of daptomycin and fits well with our observation that daptomycin complexes preferably integrate into fluid lipid domains. Furthermore, we observed strong up-regulation of LiaR and LiaH (Fig. 1C), indicating cell envelope stress (26, 88). Alterations in liaFSR are associated with daptomycin resistance (89), and deletion of liaR restores the susceptibility of daptomycin-resistant E. faecalis (59). Although the molecular function of the lia system is unclear, it appears to be related to cardiolipin distribution in the membrane of E. faecalis (81). The two-component system walKR is involved in cell wall synthesis and homeostasis (90). Although its role in daptomycin resistance is not yet characterized, it can be speculated that a mutated WalKR system influences cell wall synthesis, thereby reducing the effects of impaired lipid II synthesis caused by the delocalization of MurG.
Conclusion
By interfering with the lipid organization of the cell membrane, daptomycin impairs multiple cellular processes, of which cell wall synthesis is the most strongly affected. Because peptidoglycan-remodeling autolysins are still active, impaired cell wall synthesis leads to breaches in the cell wall and eventually to cell lysis. In addition to cell wall synthesis, daptomycin affects phospholipid synthesis and after prolonged incubation also affects respiration, membrane potential, and cell division. These multiple effects on cell physiology, together with such a common target as fluid lipids, might explain why daptomycin is so effective and why resistance to daptomycin does not occur easily.
Thus far it has been assumed that daptomycin functions as an ion pore. However, this mode of action does not explain its high selectivity for bacterial cells, because all other antibiotics that form ion pores exhibit some degree of cytotoxicity and can be applied only topically (91). It is tempting to speculate that daptomycin’s mechanism of inhibiting cell envelope synthesis by modifying fluid lipid domains is one of the main reasons underlying its remarkable selectivity.
Materials and Methods
Details on strains, plasmids, and primers used in this study are provided in SI Appendix, Tables S1–S3. Experimental details on minimal inhibitory concentration, growth experiments, proteome and element analysis, radioactive precursor incorporation, membrane potential, resazurin, ATP, fluidity measurements, and microscopy can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ute Krämer, Plant Physiology, Ruhr University Bochum, for providing access to OES; Petra Düchting and Michaele Josten for excellent technical support; Maki Kawai for construction of BMK21; and Edward de Koning for construction of EKB44. This work was funded by the Netherlands Organization for Scientific Research Grant STW-Vici 12128 (to L.W.H.), German Research Foundation Grant SCHN1284/1-2 (to T. Schneider), the German Center for Infection Research (T. Schneider, A.M., and F.G.), and a grant from the German federal state of North Rhine-Westphalia and the European Union (European Regional Development Fund “Investing in your future”) (to J.E.B. and H.-G.S.). H.S. was supported by Wellcome Trust Institutional Strategic Support Funds Grant 105617/Z/14/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611173113/-/DCSupplemental.
References
- 1.Baltz RH, Miao V, Wrigley SK. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat Prod Rep. 2005;22(6):717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 2.Debono M, et al. A21978C, a complex of new acidic peptide antibiotics: Isolation, chemistry, and mass spectral structure elucidation. J Antibiot (Tokyo) 1987;40(6):761–777. doi: 10.7164/antibiotics.40.761. [DOI] [PubMed] [Google Scholar]
- 3.Baltz RH, McHenney MA, Hosted TJ. Genetics of lipopeptide antibiotic biosynthesis in Streptomyces fradiae A54145 and Streptomyces roseosporus A21978C. In: Baltz RH, Hegeman GD, Skatrud PL, editors. Developments in Industrial Microbiology. Society for Industrial Microbiology; Fairfax, VA: 1997. p. 93. [Google Scholar]
- 4.Allen NE, Hobbs JN, Alborn WEJ., Jr Inhibition of peptidoglycan biosynthesis in gram-positive bacteria by LY146032. Antimicrob Agents Chemother. 1987;31(7):1093–1099. doi: 10.1128/aac.31.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber FM, Pieper RL, Tietz AJ. The formation of daptomycin by supplying decanoic acid to Streptomyces roseosporus cultures producing the antibiotic complex A21978C. J Biotechnol. 1988;7(4):283–292. [Google Scholar]
- 6.Straus SK, Hancock REW. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758(9):1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Jung D, Rozek A, Okon M, Hancock REW. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol. 2004;11(7):949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Scott WRP, Baek S-B, Jung D, Hancock REW, Straus SK. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim Biophys Acta. 2007;1768(12):3116–3126. doi: 10.1016/j.bbamem.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 9.Rotondi KS, Gierasch LM. A well-defined amphipathic conformation for the calcium-free cyclic lipopeptide antibiotic, daptomycin, in aqueous solution. Biopolymers. 2005;80(2-3):374–385. doi: 10.1002/bip.20238. [DOI] [PubMed] [Google Scholar]
- 10.Ball L-J, Goult CM, Donarski JA, Micklefield J, Ramesh V. NMR structure determination and calcium binding effects of lipopeptide antibiotic daptomycin. Org Biomol Chem. 2004;2(13):1872–1878. doi: 10.1039/b402722a. [DOI] [PubMed] [Google Scholar]
- 11.Hachmann AB, et al. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother. 2011;55(9):4326–4337. doi: 10.1128/AAC.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mengin-Lecreulx D, Allen NE, Hobbs JN, van Heijenoort J. Inhibition of peptidoglycan biosynthesis in Bacillus megaterium by daptomycin. FEMS Microbiol Lett. 1990;57(3):245–248. doi: 10.1016/0378-1097(90)90074-z. [DOI] [PubMed] [Google Scholar]
- 13.Schneider T, et al. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother. 2009;53(4):1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canepari P, Boaretti M, Lleó MM, Satta G. Lipoteichoic acid as a new target for activity of antibiotics: Mode of action of daptomycin (LY146032) Antimicrob Agents Chemother. 1990;34(6):1220–1226. doi: 10.1128/aac.34.6.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laganas V, Alder J, Silverman JA. In vitro bactericidal activities of daptomycin against Staphylococcus aureus and Enterococcus faecalis are not mediated by inhibition of lipoteichoic acid biosynthesis. Antimicrob Agents Chemother. 2003;47(8):2682–2684. doi: 10.1128/AAC.47.8.2682-2684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman JA, Perlmutter NG, Shapiro HM. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47(8):2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T, Muraih JK, MacCormick B, Silverman J, Palmer M. Daptomycin forms cation- and size-selective pores in model membranes. Biochim Biophys Acta. 2014;1838(10):2425–2430. doi: 10.1016/j.bbamem.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Wecke T, et al. Daptomycin versus Friulimicin B: In-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob Agents Chemother. 2009;53(4):1619–1623. doi: 10.1128/AAC.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hachmann A-B, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53(4):1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28(15):2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106(32):13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194(17):4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wenzel M, et al. Proteomic response of Bacillus subtilis to lantibiotics reflects differences in interaction with the cytoplasmic membrane. Antimicrob Agents Chemother. 2012;56(11):5749–5757. doi: 10.1128/AAC.01380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandow JE, Brötz H, Leichert LI, Labischinski H, Hecker M. Proteomic approach to understanding antibiotic action. Antimicrob Agents Chemother. 2003;47(3):948–955. doi: 10.1128/AAC.47.3.948-955.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66(1):100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- 26.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50(5):1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 27.Sun G, et al. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178(5):1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julsing MK, Rijpkema M, Woerdenbag HJ, Quax WJ, Kayser O. Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl Microbiol Biotechnol. 2007;75(6):1377–1384. doi: 10.1007/s00253-007-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leatherbarrow AJ, Yazdi MA, Curson JP, Moir A. The gerC locus of Bacillus subtilis, required for menaquinone biosynthesis, is concerned only indirectly with spore germination. Microbiology. 1998;144(Pt 8):2125–2130. doi: 10.1099/00221287-144-8-2125. [DOI] [PubMed] [Google Scholar]
- 30.Alborn WEJ, Jr, Allen NE, Preston DA. Daptomycin disrupts membrane potential in growing Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35(11):2282–2287. doi: 10.1128/aac.35.11.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenzel M, et al. Analysis of the mechanism of action of potent antibacterial hetero-tri-organometallic compounds: A structurally new class of antibiotics. ACS Chem Biol. 2013;8(7):1442–1450. doi: 10.1021/cb4000844. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel M, et al. Antimicrobial Peptides from the Aurein Family Form Ion-Selective Pores in Bacillus subtilis. ChemBioChem. 2015;16(7):1101–1108. doi: 10.1002/cbic.201500020. [DOI] [PubMed] [Google Scholar]
- 33.Te Winkel JD, Gray DA, Seistrup KH, Hamoen LW, Strahl H. Analysis of antimicrobial-triggered membrane depolarisation using voltage sensitive dyes. Front Cell Dev Biol. 2016;4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107(27):12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19(4):710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koller D, Lohner K. The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Biochim Biophys Acta. 2014;1838(9):2250–2259. doi: 10.1016/j.bbamem.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Zweytick D, Tumer S, Blondelle SE, Lohner K. Membrane curvature stress and antibacterial activity of lactoferricin derivatives. Biochem Biophys Res Commun. 2008;369(2):395–400. doi: 10.1016/j.bbrc.2008.01.176. [DOI] [PubMed] [Google Scholar]
- 38.Strahl H, Bürmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kucherak OA, et al. Switchable nile red-based probe for cholesterol and lipid order at the outer leaflet of biomembranes. J Am Chem Soc. 2010;132(13):4907–4916. doi: 10.1021/ja100351w. [DOI] [PubMed] [Google Scholar]
- 40.Parasassi T, Gratton E. Membrane lipid domains and dynamics as detected by Laurdan fluorescence. J Fluoresc. 1995;5(1):59–69. doi: 10.1007/BF00718783. [DOI] [PubMed] [Google Scholar]
- 41.Andrich MP, Vanderkooi JM. Temperature dependence of 1,6-diphenyl-1,3,5-hexatriene fluorescence in phophoslipid artificial membranes. Biochemistry. 1976;15(6):1257–1261. doi: 10.1021/bi00651a013. [DOI] [PubMed] [Google Scholar]
- 42.Vanderkooi JM, Landesberg R, Selick H, 2nd, McDonald GG. Interaction of general anesthetics with phospholipid vesicles and biological membranes. Biochim Biophys Acta. 1977;464(1):1–18. doi: 10.1016/0005-2736(77)90366-2. [DOI] [PubMed] [Google Scholar]
- 43.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sci. 2006;7(6):539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 44.Friedlander G, Le Grimellec C, Giocondi MC, Amiel C. Benzyl alcohol increases membrane fluidity and modulates cyclic AMP synthesis in intact renal epithelial cells. Biochim Biophys Acta. 1987;903(2):341–348. doi: 10.1016/0005-2736(87)90224-0. [DOI] [PubMed] [Google Scholar]
- 45.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi K, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100(8):4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sastre DE, Bisson-Filho A, de Mendoza D, Gueiros-Filho FJ. Revisiting the cell biology of the acyl-ACP:phosphate transacylase PlsX suggests that the phospholipid synthesis and cell division machineries are not coupled in Bacillus subtilis. Mol Microbiol. 2016;100(4):621–634. doi: 10.1111/mmi.13337. [DOI] [PubMed] [Google Scholar]
- 48.Takada H, et al. An essential enzyme for phospholipid synthesis associates with the Bacillus subtilis divisome. Mol Microbiol. 2014;91(2):242–255. doi: 10.1111/mmi.12457. [DOI] [PubMed] [Google Scholar]
- 49.Grünewald J, Sieber SA, Mahlert C, Linne U, Marahiel MA. Synthesis and derivatization of daptomycin: A chemoenzymatic route to acidic lipopeptide antibiotics. J Am Chem Soc. 2004;126(51):17025–17031. doi: 10.1021/ja045455t. [DOI] [PubMed] [Google Scholar]
- 50.Lohani CR, Taylor R, Palmer M, Taylor SD. Solid-phase total synthesis of daptomycin and analogs. Org Lett. 2015;17(3):748–751. doi: 10.1021/acs.orglett.5b00043. [DOI] [PubMed] [Google Scholar]
- 51.Taylor R, et al. Two successive calcium-dependent transitions mediate membrane binding and oligomerization of daptomycin and the related antibiotic A54145. Biochim Biophys Acta. 2016;1858(9):1999–2005. doi: 10.1016/j.bbamem.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 52.Blosser MC, Honerkamp-Smith AR, Han T, Haataja M, Keller SL. Transbilayer Colocalization of Lipid Domains Explained via Measurement of Strong Coupling Parameters. Biophys J. 2015;109(11):2317–2327. doi: 10.1016/j.bpj.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Putzel GG, Uline MJ, Szleifer I, Schick M. Interleaflet coupling and domain registry in phase-separated lipid bilayers. Biophys J. 2011;100(4):996–1004. doi: 10.1016/j.bpj.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiessling V, Wan C, Tamm LK. Domain coupling in asymmetric lipid bilayers. Biochim Biophys Acta. 2009;1788(1):64–71. doi: 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T, et al. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem. 2014;289(17):11584–11591. doi: 10.1074/jbc.M114.554444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganchev DN, Hasper HE, Breukink E, de Kruijff B. Size and orientation of the lipid II headgroup as revealed by AFM imaging. Biochemistry. 2006;45(19):6195–6202. doi: 10.1021/bi051913e. [DOI] [PubMed] [Google Scholar]
- 57.Muchová K, Wilkinson AJ, Barák I. Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan. FEMS Microbiol Lett. 2011;325(1):92–98. doi: 10.1111/j.1574-6968.2011.02417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schirner K, et al. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol. 2015;11(1):38–45. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyes J, et al. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis. 2015;211(8):1317–1325. doi: 10.1093/infdis/jiu602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camargo ILB da C, Neoh H-M, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother. 2008;52(12):4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother. 2008;52(3):980–990. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider T, Sahl HG. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol. 2010;300(2-3):161–169. doi: 10.1016/j.ijmm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Weidenmaier C, Peschel A. Teichoic acids and related cell wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6(4):276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 64.Bhavsar AP, D’Elia MA, Sahakian TD, Brown ED. The amino terminus of Bacillus subtilis TagB possesses separable localization and functional properties. J Bacteriol. 2007;189(19):6816–6823. doi: 10.1128/JB.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paoletti L, Lu Y-J, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189(16):5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chazarreta-Cifre L, Martiarena L, de Mendoza D, Altabe SG. Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J Bacteriol. 2011;193(16):4043–4048. doi: 10.1128/JB.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Mendoza D, Cronan JE. Thermal regulation of membrane lipid fluidity in bacteria. Trends Biochem Sci. 1983;8(2):49–52. [Google Scholar]
- 68.Beranová J, Jemioła-Rzemińska M, Elhottová D, Strzałka K, Konopásek I. Metabolic control of the membrane fluidity in Bacillus subtilis during cold adaptation. Biochim Biophys Acta - Biomembr. 2008;1778(2):445–453. doi: 10.1016/j.bbamem.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 69.Wenzel M, et al. Proteomic signature of fatty acid biosynthesis inhibition available for in vivo mechanism-of-action studies. Antimicrob Agents Chemother. 2011;55(6):2590–2596. doi: 10.1128/AAC.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wenzel M, et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci USA. 2014;111(14):E1409–E1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kingston AW, Subramanian C, Rock CO, Helmann JD. A σW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol Microbiol. 2011;81(1):69–79. doi: 10.1111/j.1365-2958.2011.07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Epand RF, Schmitt MA, Gellman SH, Epand RM. Role of membrane lipids in the mechanism of bacterial species selective toxicity by two α/β-antimicrobial peptides. Biochim Biophys Acta - Biomembr. 2006;1758(9):1343–1350. doi: 10.1016/j.bbamem.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 73.Epand RF, et al. Dual mechanism of bacterial lethality for a cationic sequence-random copolymer that mimics host-defense antimicrobial peptides. J Mol Biol. 2008;379(1):38–50. doi: 10.1016/j.jmb.2008.03.047. [DOI] [PubMed] [Google Scholar]
- 74.Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta. 2009;1788(1):289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 75.Jean-François F, et al. Aggregation of cateslytin β-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry. 2008;47(24):6394–6402. doi: 10.1021/bi800448h. [DOI] [PubMed] [Google Scholar]
- 76.Cruzeiro-Hansson L, Mouritsen OG. Passive ion permeability of lipid membranes modelled via lipid-domain interfacial area. Biochim Biophys Acta. 1988;944(1):63–72. doi: 10.1016/0005-2736(88)90316-1. [DOI] [PubMed] [Google Scholar]
- 77.Bramkamp M, Lopez D. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev. 2015;79(1):81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bach JN, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88(6):1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- 79.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat Methods. 2012;9(5):480–482. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296(5569):913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 81.Tran TT, Munita JM, Arias CA. Mechanisms of drug resistance: Daptomycin resistance. Ann N Y Acad Sci. 2015;1354:32–53. doi: 10.1111/nyas.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bertsche U, et al. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation. Antimicrob Agents Chemother. 2011;55(8):3922–3928. doi: 10.1128/AAC.01226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bertsche U, et al. Increased cell wall teichoic acid production and D-alanylation are common phenotypes among daptomycin-resistant methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates. PLoS One. 2013;8(6):e67398. doi: 10.1371/journal.pone.0067398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peleg AY, et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7(1):e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S-J, Mishra NN, Rubio A, Bayer AS. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother. 2013;57(11):5658–5664. doi: 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khatib TO, Stevenson H, Yeaman MR, Bayer AS, Pokorny A. Binding of Daptomycin to Anionic Lipid Vesicles Is Reduced in the Presence of Lysyl-Phosphatidylglycerol. Antimicrob Agents Chemother. 2016;60(8):5051–5053. doi: 10.1128/AAC.00744-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewis RNAH, McElhaney RN. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim Biophys Acta. 2009;1788(10):2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 88.Wolf D, et al. In-depth profiling of the LiaR response of Bacillus subtilis. J Bacteriol. 2010;192(18):4680–4693. doi: 10.1128/JB.00543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller C, et al. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother. 2013;57(11):5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70(6):1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 91.Hurdle JG, O’Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011;9(1):62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.