Significance
Living cells need fuel in the form of adenosine triphosphate, or ATP, to stay alive. This fuel is generated by a molecular machine made of two motors joined by a rotor. One generates rotation by using energy provided by oxidative metabolism or photosynthesis; the other uses energy transmitted by the rotor to make ATP molecules from its building blocks, adenosine diphosphate, or ADP, and inorganic phosphate. The structure has been determined of a fungal machine, isolated from its cellular power stations, the mitochondria, where the machine operates. It provides unsuspected details of the blueprint of the machine and how it works. The working principles of the fungal machine apply to similar machines in all species.
Keywords: Pichia angusta, ATP synthase, structure, proton translocation
Abstract
The structure of the intact monomeric ATP synthase from the fungus, Pichia angusta, has been solved by electron cryo-microscopy. The structure provides insights into the mechanical coupling of the transmembrane proton motive force across mitochondrial membranes in the synthesis of ATP. This mechanism requires a strong and integral stator, consisting of the catalytic α3β3-domain, peripheral stalk, and, in the membrane domain, subunit a and associated supernumerary subunits, kept in contact with the rotor turning at speeds up to 350 Hz. The stator’s integrity is ensured by robust attachment of both the oligomycin sensitivity conferral protein (OSCP) to the catalytic domain and the membrane domain of subunit b to subunit a. The ATP8 subunit provides an additional brace between the peripheral stalk and subunit a. At the junction between the OSCP and the apparently stiff, elongated α-helical b-subunit and associated d- and h-subunits, an elbow or joint allows the stator to bend to accommodate lateral movements during the activity of the catalytic domain. The stator may also apply lateral force to help keep the static a-subunit and rotating c10-ring together. The interface between the c10-ring and the a-subunit contains the transmembrane pathway for protons, and their passage across the membrane generates the turning of the rotor. The pathway has two half-channels containing conserved polar residues provided by a bundle of four α-helices inclined at ∼30° to the plane of the membrane, similar to those described in other species. The structure provides more insights into the workings of this amazing machine.
The ATP synthases (F-ATPases) found in mitochondria, chloroplasts, and eubacteria are membrane-bound molecular machines with a rotary action. Our understanding of how they work has come mainly from single-molecule studies of rotation on bacterial F-ATPases (1) and from structures of their constituent domains determined predominantly with enzymes from mitochondria (2–5). The most complete atomic resolution structure contains about 85% of the bovine F-ATPase, including the membrane domain of the rotor. It is a mosaic built from substructures determined by X-ray crystallography (2) within the constraints of an overall structure determined by cryo-electron microscopy (cryo-EM) (6). In this mosaic structure, the enzyme’s rotor is defined clearly, but a detailed description of the remaining 15% in the membrane domain of the stator and details of its membrane extrinsic domain are still needed as the basis for a complete molecular understanding of the enzyme’s mechanism. A general outline of some of the features of the residual region has been provided by cryo-EM structures of the bovine (7) and an algal enzyme (8) and by the crystal structure of an intact bacterial enzyme (9), but none of them is at a sufficient resolution to provide a complete molecular description of the enzyme.
The mosaic structure (2) established that the rotor is an ensemble of a membrane intrinsic domain, made of a ring of c-subunits, attached to a globular, elongated, and asymmetric structure, known as the central stalk, made in mitochondria of single copies of subunits γ, δ, and ε. Its elongated region is a coiled-coil of α-helices in the N- and C-terminal regions of the γ-subunit, which penetrates into the globular α3β3-catalytic domain. The α3β3-domain is part of the stator, against which the rotor turns, and is linked by the peripheral stalk to the a-subunit and six associated supernumerary subunits in the membrane domain. The supernumerary subunits have no known direct role in ATP synthesis, but some of them mediate interactions between monomeric F-ATPase complexes in dimers (10–13) of the complex that are associated in rows along the edges of the mitochondrial cristae (14, 15). The peripheral stalk itself is made of single copies of the oligomycin sensitivity conferral protein (OSCP), subunit F6 (or the orthologous fungal h-subunit), and the b- and d-subunits (16–18). In the mosaic model (2), and reiterated in the bovine cryo-EM structure (7), the OSCP is bound to the N-terminal region of one of the α-subunits, and the peripheral stalk extends via a largely α-helical structure in the b-, d-, and F6-subunits along the external surface of the F1-domain into the membrane region of the enzyme. Here, the membrane sector of the b-subunit is thought to be bound to the a-subunit, keeping the static a-subunit and the rotating c-ring in contact and maintaining a specific pathway in their interface region for translocation of protons through the membrane. In the course of proton translocation, potential energy stored by the proton-motive force and the membrane capacitance is released, impelling the turning of the rotor at speeds of up to 350 cycles/s (19). During ATP synthesis, the turning of the rotor in an anticlockwise direction as viewed from above the complex modulates the conformation of the three catalytic sites in the α3β3-domain, taking each of them through a cycle of substrate binding, ATP formation, and product release. Thus, each 360° rotation produces three molecules of ATP.
As described here by cryo-EM, we have determined the structure of the entire monomeric F-ATPase complex from the mitochondria of the moderately thermophilic fungus, Pichia angusta, bound to the inhibitory region of the natural bovine inhibitor protein, IF1 (inhibitor of F1-ATPase). This structure fills significant gaps in our knowledge of the mechanism of the F-ATPase. First, it contributes to our understanding of the coupling mechanism by providing unsuspected details of the peripheral stalk and how it is attached to both the catalytic F1-domain and the membrane sector of the enzyme. Second, it provides an independent description of the transmembrane proton pathway for protons, helping both to define common features in the proton translocation mechanism in bacterial and mitochondrial F-ATPases and to explain human pathological mutations in the subunit.
Results
Structure Determination.
The structure was determined with the monomeric enzyme from P. angusta inhibited with residues 1–60 of bovine IF1 (20). Formation of the inhibited complex requires the hydrolysis of ATP, and both bovine and yeast IF1 bind to their orthologous F-ATPases at the same major site (3, 20). In structures of F1-IF1 complexes, the inhibitor protein is entrapped in the αDPβDP-catalytic interface, the most closed of the three catalytic interfaces of the enzyme (3–5, 21). In the intact F-ATPase, this site can be in any one of three positions relative to the peripheral stalk, separated from each other by 120° rotations about the central axis of the F1-domain. The purified inhibited complex contains all of the known subunits of the enzyme, excepting supernumerary subunits e, g, k, and l (20). Thus, the membrane domain of this preparation contains the c-ring and subunits a, f, j, ATP8, and the membrane domain of subunit b.
The inhibited F-ATPase complex was examined by single-particle cryo-EM (SI Appendix, Fig. S1). After refinement, motion correction, and B-factor weighting, the entire dataset of 100,724 particles resulted in a map with overall resolution of ∼7 Å. Following 3D classification of the weighted particles with the resolution of alignment limited to 12 Å, the particles were grouped into three states based on the position of the inhibitor protein (SI Appendix, Figs. S2 and S3). In state 1, the αDPβDP-catalytic interface with the bound inhibitor, is closest to the peripheral stalk (SI Appendix, Fig. S2). In states 2 and 3, the same interface and bound inhibitor are in the two other possible orientations relative to the peripheral stalk. As 44.5% of the particles are in state 1, the resolution of the corresponding map is superior to those of states 2 and 3, containing 22.8 and 17.4% of the particles, respectively (SI Appendix, Fig. S2). The local resolution of the F1 domain is better than that of the membrane domain (SI Appendix, Fig. S4), and the detergent-lipid annulus around the membrane domain has the lowest resolution. After refinement, as expected, the three states differed mainly in the central stalk and the F1-domain (SI Appendix, Fig. S2).
The map from state 1 was used to build a model of the F-ATPase, and states 2 and 3 were modeled from state 1. The state 1 map was interpreted with the structures of the F1-IF1 complex (3) and the c-ring from the F1-c10 subcomplex (22), both from Saccharomyces cerevisiae, and with the bovine peripheral stalk (18) and IF1 (21); as expected, α-helices are resolved more clearly than β-strands. The final model (Fig. 1; SI Appendix, Fig. S5) contains the following residues: chains αE, 5–509; αTP, 12–406 and 412–509; αDP, 7–509; βDP, 6–475; βE, 8–475; βTP, 7–475; γ, 1–59 and 71–276; δ, 11–23 and 27–137; ε, 1–49 and 53–61; bovine IF1 8–50; the c-subunits in the c10-ring, 3–75, 2–72, 1–72, 1–72; 1–73, 1–74, 1–74, 1–73, 1–73, 2–73; OSCP, 6–153, and 180–194; b-subunit 49–208; d-subunit 11–127, plus a segment of uncertain register modeled as poly-Ala (1,001–1,028); h-subunit, modeled as poly-Ala (1,001–1,021); a-subunit, 44 residues in α-helices aH2 and aH3 (all modeled as poly-Ala; 1,001–1,044); aH4 and aH5, 119–206; and aH6, 210–252. The model of the membrane domain also contains four unconnected segments of secondary structure that have been assigned, but with less certainty (see proposed identities in Fig. 1). They are the following: chain 1, 30 residues (1,001–1,030); chain 2, 25 residues (1,001–1,025); chain 3, 17 residues (1,001–1,017); and chain 4, 27 residues (1,001–1,027), all modeled as poly-Ala. All residues have been truncated to Cβ throughout. At the secondary structure level, there is excellent fit between the map and model (SI Appendix, Fig. S4), and the state 1 model was used in the interpretations below. The quality of the map is illustrated by bovine IF1 where residues 8–50 are well resolved (SI Appendix, Fig. S6). In the bovine cryo-EM structure (7), IF1 was not modeled.
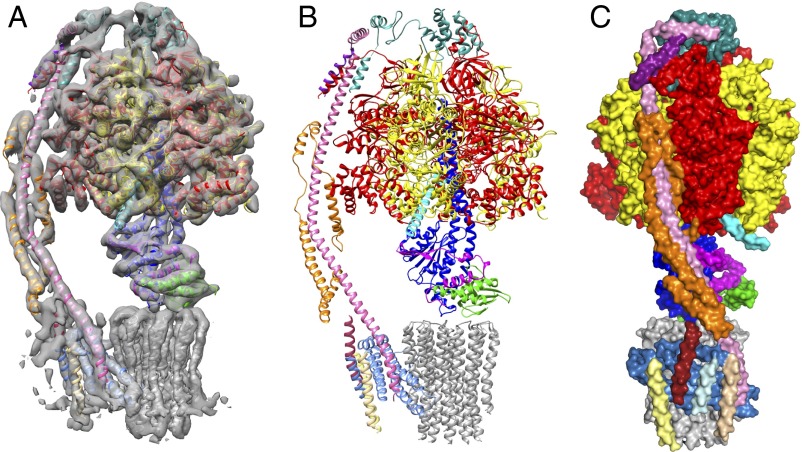
Fig. 1.
The structure of the F-ATPase from P. angusta. The enzyme was inhibited with residues 1–60 of the bovine inhibitor protein IF1. (A) The cryo-EM map of state 1 and structural model viewed from the side, with the peripheral stalk on the left, the catalytic domain at the top, attached by the central and peripheral stalks to the membrane domain below. (B and C) Side views of the enzyme-inhibitor complex in cartoon and surface representation, respectively. C is rotated to the right by 90° relative to A and B. The α-, β-, γ-, δ-, and ε-subunits forming the membrane extrinsic catalytic domain are red, yellow, royal blue, green, and magenta, respectively; the inhibitor protein is cyan; and the peripheral stalk subunits OSCP, b, d, and h are sea-green, pink, orange, and purple, respectively. In the membrane domain, the c10-rotor is gray, the resolved region of the associated subunit a is corn-flower blue. Chains Ch1–Ch4 are pale yellow, brick-red, pale cyan, and beige, respectively, and have been assigned as transmembrane α-helices in subunit f and, in ATP8, as aH1 and bH1, respectively. In SI Appendix, Fig. S5, the identities of subunits are placed directly on an enlarged version of C.
The Peripheral Stalk.
In the structure of bovine F1-ATPase with a truncated version of the peripheral stalk (referred to as bF1-tPS) (18), containing the OSCP and F6 subunits, and residues 99–214 and 1–118 of subunits b and d, respectively, the peripheral stalk is attached to the “top” of the F1-domain by the N-terminal region of the αE-subunit interacting with the N-terminal domain of the OSCP, and the equivalent region in the EM map of the bovine enzyme has been modeled similarly (7). Residues 5–17 and 7–20, respectively, of α-subunits in the bovine and P. angusta enzymes are predicted to be α-helical (SI Appendix, Fig. S7), but in models of bovine F1-ATPase alone, all three are largely unstructured (23–25). As in the bovine protein, the N-terminal domain of the OSCP in P. angusta consists of a bundle of six α-helices (OH1–OH6), and the N-terminal regions of two of the three α-subunits interact with it. Like bF1-tPS, an α-helix containing residues 12–21 of the αE-subunit occupies a groove between OH1 and OH5. In addition, a region of density attributed to the N-terminal segment of the αTP-subunit is associated with the region between OH3 and OH4 (Fig. 2). Similar interactions via the αE- and αTP-subunits and the δ-subunit, the bacterial ortholog of the OSCP, are present in the crystal structure of the F-ATPase from Paracoccus denitrificans (9).
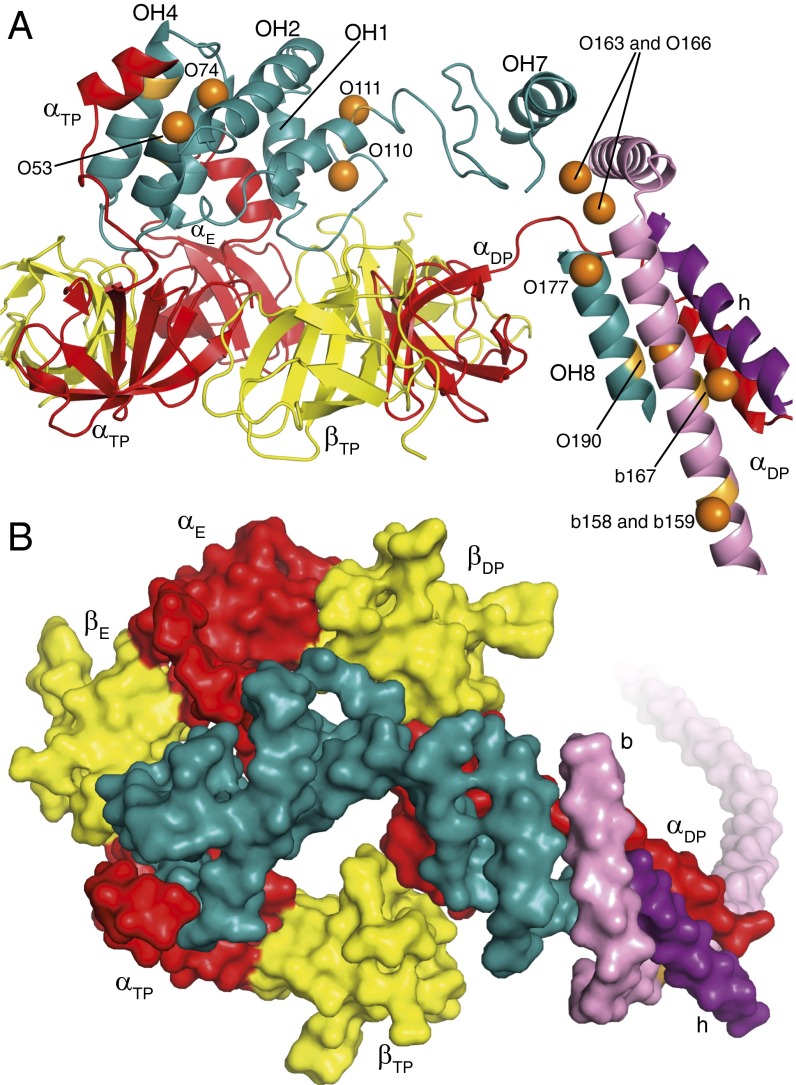
Fig. 2.
Attachment of the peripheral stalk to the crown of the F1-catalytic domain of of the F-ATPase from P. angusta. The diagrams are based on state 1. (A and B) Views from the side and from above the N-terminal crown of the F1-domain in cartoon and surface representation, respectively. The OSCP is sea-green, the three α-subunits are red, the three β-subunits are yellow, the b-subunit is pink, and subunit h is purple. In A, the N-terminal α-helical regions of the α-subunits are labeled αE, αDP, and αTP. In the OSCP, the positions of α-helices OH1, OH2, OH4, OH7, and OH8 are indicated. The orange spheres represent the positions of selenium atoms in the structure of bovine F1-ATPase with the truncated peripheral stalk (bF1-tPS), and orange patches indicate the positions of corresponding amino acids in the fungal OSCP. They are labeled according to the subunit where they reside, with the prefix O for OSCP and b for subunit b, followed by the residue number in the respective subunits. O53, O74, O110, O111, O190, b167, and b158–159 correspond to residues in resolved regions of the structure of the P. angusta F-ATPase. O163, O166, and O177 indicate the positions of the selenomethionine residues in bF1-tPS and indicate the positions of the equivalent P. angusta residues in unresolved regions of the current structure.
In P. angusta, and similarly in the bovine enzyme (SI Appendix, Fig. S8), the C-terminal domain of the OSCP is predicted to consist of a short β-strand, Oβ1 (residues 117–120), and two α-helices, OH7 and OH8 (residues 130–141 and 176–194, respectively) with intervening β-strands, Oβ2 and Oβ3 (residues 151–158 and 161–175). In the P. angusta structure, β1, OH7, and OH8 were resolved, but a region of density between OH7 and OH8 containing predicted Oβ2 and Oβ3 could not be interpreted (Fig. 2). However, the positions of selenomethionine residues, introduced biosynthetically into the truncated bovine peripheral stalk before its reassembly with bovine F1-ATPase to form bF1-tPS (18), coincide with the positions of equivalent residues in the structure of the N-terminal domain of the P. angusta OSCP; the residues are Val-53, Leu-74, Leu-110, and Asn-111 (labeled O53, O74, O110, and O111; Fig. 2; SI Appendix, Fig. S8). Others coincide with the structure of OH8 in the C-terminal domain (residue Leu-190; O190 in Fig. 2 and SI Appendix, Fig. S8) and in the b-subunit (Fig. 2; SI Appendix, Fig. S8) with b167 (Leu-167) and the adjacent pair b158/159 (Gln-158/Val-159). Therefore, O163, O166, and O177, which fall in regions of unmodeled density between OH7 and OH8, indicate where the corresponding residues (Leu-163 in the loop Oβ2-Oβ3, Leu-166 in Oβ3, and Leu-177 probably just preceding OH8) lie in the P. angusta OSCP (Fig. 2). Residues 25–30 of the αDP-subunit are immediately adjacent to where Oβ2 and Oβ3 are thought to be, and residues 11–22 of the αDP-subunit could interact with OH8, thus providing a third point of attachment of an α-subunit to the OSCP. The N-terminal region of the αDP-subunit probably also interacts with the b-subunit via bH2 and bH3 and with the h-subunit via hH1 (Fig. 2).
The C-terminal domain of the OSCP is also bound to the C-terminal region of the b-subunit. The membrane extrinsic part of the b-subunit from residues 70 to 204 is α-helical (SI Appendix, Fig. S9), and the α-helix in the bovine b-subunit is broken from residues 184 to 187 and is terminated by bH3 from residues 188 to 207 (18). A similar structure is found in the P. angusta enzyme, where bH3 (residues 184–202) is associated with density in the region between OH7 and OH8 (Fig. 2). Residues 165–179 at the C-terminal end of bH2 run alongside OH8, and the same region of subunit b is associated with the N-terminal α-helix of subunit h (hH1; residues 1–17). Subunit h is the fungal ortholog of bovine F6 (SI Appendix, Fig. S10), and the interpretation of hH1 depends upon the crystal structure of bF1-tPS (18). The remainder of subunit h is probably represented by unmodeled density that runs along the outside of bH2 (SI Appendix, Fig. S11). The rest of the membrane extrinsic region of subunit b represented by bH2 is about 160 Å long, and, in addition to subunit h, subunit d is also associated closely with it. Subunit d is predicted to be folded into seven α-helices, dH1–dH7 with extended segments (residues 49–57 and 77–86) between dH3 and dH4 and dH5 and dH6, respectively (SI Appendix, Fig. S12). α-helices dH2–dH6 have similar structures to those determined in the crystal structures of bF1-tPS and in the separate bovine peripheral stalk (known as bPS). α-Helices dH4 and dH5 form a hairpin with an intervening turn from residues 63–67. Both bPS and bF1-tPS lacked the 36 C-terminal residues of subunit d corresponding to dH7 (132–138). The d-subunit of P. angusta has a longer C-terminal region and is predicted to have an extended dH7 (residues 136–156) (SI Appendix, Fig. S12). Density for an α-helix adjacent to dH6 has been interpreted as dH7 running antiparallel to dH6, but the connection between them (residues 128–135) was not resolved. This region of the peripheral stalk, where α-helices in subunits b, d, and h lie parallel to each other, extends from near the “top” of the F1-domain, along the periphery of the F1-domain, to the lipid head group region of the membrane domain of the enzyme. However, the peripheral stalk is straighter in the P. angusta enzyme than in bF1-tPS, where it was bent toward the F1-domain by crystal lattice contacts (18).
The Membrane Domain.
Around residue 68, bH2 kinks and enters the inner membrane of the mitochondrion, forming a transmembrane α-helix from residues 51 to 68, in close agreement with the secondary structure and hydrophobicity (SI Appendix, Fig. S9). A second transmembrane α-helix, bH1 is also predicted, and unconnected density lying parallel to bH2, has been attributed to it, re-emerging from the matrix side of the membrane around residue 28. In the current model, α-helices bH1 and bH2 interact with the a-subunit (described below), providing the membrane region of the stator with integrity. Based on the conservation of hydrophobic segments in their sequences, it is likely that the a-subunit itself has six transmembrane α-helices (aH1–aH6) as described before (7–9) (SI Appendix, Fig. S13). In the current structure, a bundle of four α-helices, each inclined at 30° to the plane of the membrane, has been attributed to aH3–aH6; aH5 and aH6 consist of 36 and 37 residues, respectively, and they correspond to the exceptionally long hydrophobic segments 5 and 6 in their sequences (SI Appendix, Figs. S13 and S14). α-Helix aH5 contains the absolutely conserved residue, Arg-179. Its positively charged side-chain is an essential feature of the transmembrane proton translocation mechanism (26) and is known to be close to another essential feature, the carboxy side-chain of Glu-59 in the c-subunit (27). The shorter α-helices, aH3 and aH4, have correspondingly shorter hydrophobic regions in their sequences and are packed against aH5 and aH6. The biochemical data that support these and similar assignments in a bacterial F-ATPase have been discussed elsewhere (9). Similar conclusions have been reached in the bovine enzyme (7). Density attributed to aH2, joined by a loop to aH3, forms a relatively short α-helix that lies horizontally in the lipid head group region of membrane on the matrix side, and aH1 is probably represented by a vertical canonical α-helical segment (Ch3) packed against aH3 (Fig. 3).
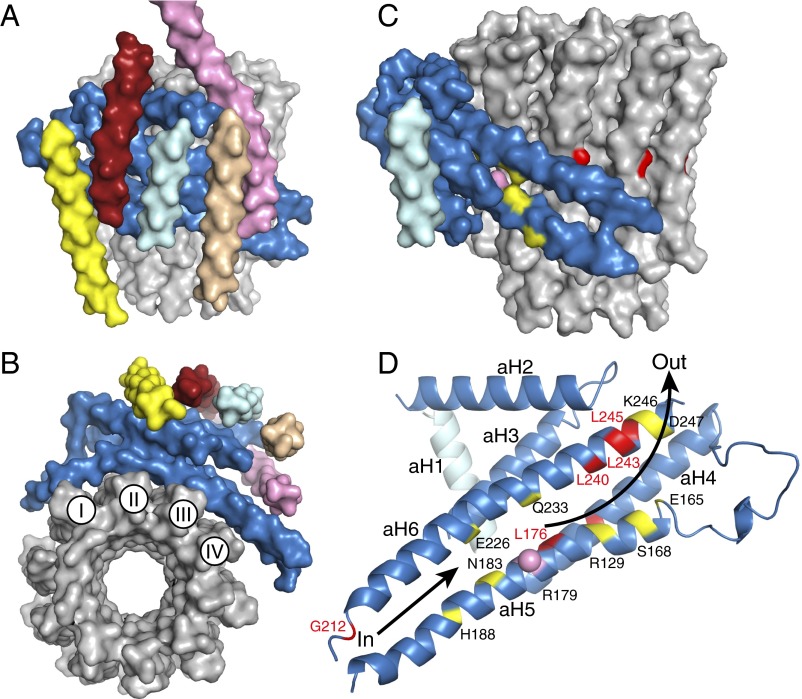
Fig. 3.
The membrane domain of the F-ATPase from P. angusta. In A–D, the a-subunit is corn-flower blue. (A and B) Views in solid representation from the side and below the membrane domain. The c10-ring is gray, the b-subunit (upper part not shown) is pink, and the pale yellow, brick-red, light cyan, and beige segments are transmembrane α-helices, Ch1–Ch4 assigned to subunit f, ATP8, aH1, and bH1, respectively. In the c-ring, I–IV indicate the four transmembrane C-terminal α-helices in contact with subunit a. (C and D) Views of the a-subunit in solid and cartoon representation viewed from outside and looking out from the interface with the c-ring, respectively, with aH1 in pale cyan. Conserved polar residues are yellow; the positions of human mutations associated with pathologies (SI Appendix, Table S1) are red. The pink sphere denotes the conserved Arg-179 in aH5 that is essential for proton translocation. The lower arrow indicates the inlet pathway for protons that transfer to Glu-59 in the C-terminal α–helix-II of the c-ring. They are carried around the ring by anticlockwise rotation, as viewed from above, until they arrive at Arg-179 where they enter the exit pathway, denoted by the upper arrow.
α-Helices aH5 and aH6 are intimately associated with the c-ring that forms the membrane region of the enzyme’s rotor. The c-ring and the tightly associated central stalk together constitute the rotor. As in S. cerevisiae (22), the c-ring has 10 identical c-subunits (Fig. 3), each with two transmembrane α-helices joined by a short loop on the matrix side of the inner mitochondrial membrane. The N- and C-terminal α-helices form inner and outer concentric rings, as viewed in cross-section (Fig. 3). Four c-subunits in the c-ring (I–IV in Fig. 3) are in contact with the subunit a, mainly via inclined aH5 and aH6.
The membrane domain of this particular preparation of the monomeric F-ATPase from P. angusta contains three additional subunits, ATP8, f, and j, each with a single predicted transmembrane α-helix (SI Appendix, Figs. S15 and S16) (20). The sequence of ATP8 from P. angusta is related weakly to bovine ATP8, and it is shorter (SI Appendix, Fig. S15). It is also related to the membrane domains of bacterial b-subunits (SI Appendix, Fig. S17). Its α-helical region has been ascribed to Ch2, brick red in Fig. 3A, with its C-terminal region protruding from the matrix of the inner mitochondrial membrane. The final feature resolved in the membrane domain of the P. angusta enzyme is another canonical transmembrane α-helix (Ch1, pale yellow in Fig. 3A). This α-helix has been ascribed to subunit f rather than subunit j because subunit f has an extensive membrane extrinsic domain (SI Appendix, Fig. S16), which would account for some of the unascribed density in region I in SI Appendix, Fig. S11. Subunit j has no such feature (SI Appendix, Fig. S16). Region I, the largest region of unascribed density, also contains the N-terminal region of the b-subunit and the membrane extrinsic region of ATP8. The membrane domain of the preparations of the P. angusta and bovine F-ATPases used in the cryo-EM studies differ in the compositions of their supernumerary subunits. The former lacks subunits e, g, k, and l (20). Bovine mitochondria have no subunits equivalent to k and l, but they contain orthologs of e and g, which were present in the preparation used in cryo-EM studies (7, 28). Therefore, a comparison of the P. angusta and bovine maps (SI Appendix, Fig. S18) indicates that the e- and g-subunits occupy an additional region of density in the bovine map that probably contributes to the formation of the dimeric enzyme in the mitochondrial cristae.
Discussion
The Mechanical Coupling Mechanism.
The peripheral stalk of the F-ATPase is a central component that ensures the mechanical coupling of the transmembrane proton-motive force to the synthesis of ATP, and in this capacity it fulfills two main roles. First, it provides the enzyme’s stator with integrity by connecting the α3β3-catalytic domain to the a-subunit in the membrane domain, and those interactions have to be sufficiently strong to survive the mechanical torque of the enzyme. Although the relevant interactions are still not fully resolved, the current structure demonstrates that the attachment to the catalytic domain is much more extensive and robust than had been thought, probably involving both the N- and C-terminal domains of the OSCP and the N-terminal regions of all three α-subunits. The structure has also provided evidence that, at the lower end of the peripheral stalk (distal from the F1-domain), the hydrophobic N-terminal region of the b-subunit is, as predicted, folded into two transmembrane α-helices, bH1 and part of bH2. It interacts with aH1 and aH2, and bH2 with aH5 and aH6, and most likely, in addition, with the loop between aH3 and aH4. It has also confirmed that the middle part of the peripheral stalk, consisting of the central α-helical pillar that is about 160 Å long and provided by bH2, is augmented by roughly parallel α-helices from subunit d and most likely from subunit h (as in the related bovine F6). Thus, it has the characteristics of a seemingly rigid and inflexible structure (29). The greatest uncertainty concerns part of the C-terminal domain of the OSCP and the region joining it to the central α-helical structure reaching down to the membrane, where the density is currently not well resolved. It has been proposed previously that this region could be flexible and provide an elbow or joint between the N-terminal domain of the OSCP plus its attachments to the subunits αE and αTP and the rigid α-helical region involving bH2 (18). Here bH3, which lies approximately orthogonal to bH2, could be part of a pivot together with OH7. The elbow, or joint, would allow the rigid α-helical pillar and attached membrane domain of the stator to adjust its position during a catalytic cycle, where the α3β3-domain is displaced from side-to-side by the turning of the asymmetrical upper region of the rotor. Therefore, it is of particular interest that the region has been proposed to be the site where benzodiazapine inhibitors bind (30, 31) and that Lys-139 of the OSCP becomes acetylated and deacetylated in response to nutrient- and exercise-induced stress (32, 33), suggesting a regulatory role for the region.
The second role of the peripheral stalk is to help keep the a-subunit in contact with the rotating c-ring, possibly by exerting lateral pressure toward the central axis, thereby ensuring the integrity of the transmembrane proton pathway. Subunit ATP8 may contribute here via its C-terminal region. It is known that the longer C-terminal region of bovine ATP8 extends from the membrane into the peripheral stalk, where it interacts with subunits b and d (34), thereby providing another brace in addition to subunit b to hold subunit a against the rotating c-ring.
Conservation of the Proton Pathway.
The structure of the P. angusta a-subunit and its interface with the c-ring are similar to those in the F-ATPases from P. denitrificans (9) and bovine mitochondria (7) (Fig. 3). The most striking feature in all three structures is an inclined bundle of four α-helices made from aH3–aH6 with aH5 and aH6 in contact with four adjacent canonical transmembrane α-helices representing the C-terminal regions of four c-subunits in the c-ring. α-Helices aH3 and aH4 are packed closely behind aH5 and aH6 (distal from the c-ring). As in the other structures (7, 9), aH5 contains the strictly conserved Arg-179 adjacent to Glu-59 (both P. angusta numbering) in C-terminal α-helix-III of the c-subunit. Both residues are known from studies in Escherichia coli to be essential for proton translocation (26, 27). Superimposition of the various structures shows their overall similarity (SI Appendix, Fig. S19). However, their various structures are required to interact with c-rings from c8 to c12, and the various ring sizes will require structural adjustments in the a-subunit that are not readily apparent at the current levels of resolution. Experimental evidence for the location of the proton path in the E. coli enzyme has been summarized previously (9). As noted before (9), the entry to the proton pathway is between aH5 and aH6, probably on the side distal from the c-ring, and the exit pathway also between aH5 and aH6 is probably more proximal to the rotor. Both pathways contain conserved polar residues (Fig. 3D). The proposed pathway is also consistent with human pathogenic mutations in subunit a (ATP6), which is encoded in the mitochondrial genome (35, 36) (SI Appendix, Table S1).
Perspectives.
A comparison of the current structures of the bacterial and mitochondrial enzymes (Fig. 4) illustrates their known similarities in overall architecture and in the detailed structures of the catalytic and proton translocating regions. Somewhat unexpectedly, the peripheral stalk regions of mitochondrial and bacterial enzymes are also similar, despite significant differences in subunit composition (17, 18, 37–41) and lack of similarity in sequence of their constituent subunits (excepting the orthologous mitochondrial OSCP and bacterial δ-subunits). The relationship between the ATP8 subunit and the membrane domains of the bacterial b-subunits (SI Appendix, Fig. S17) adds to this structural similarity. Thus, peripheral stalks from bacteria, chloroplasts, and eukaryotes have similar designs, and presumably similar physical properties, to allow them to perform their roles in ensuring the maintenance of an intact proton pathway in the interface between rotor and stator and in keeping the stator together. The current descriptions of the pathway itself are rudimentary, and a full understanding will require atomic resolution structures where positions of amino acid side-chains and participating water molecules are defined. Also to be taken into consideration is the role of lipids in the mechanism of the enzyme. Cardiolipin bearing two negative charges is an essential component of active F-ATPases in bacteria and mitochondria (42). It appears to be attracted to the c-ring selectively over phospholipids, where it binds transiently and repeatedly around basic residues in the head-group regions of both leaflets of the bilayer, suggesting a possible role in proton translocation (42).
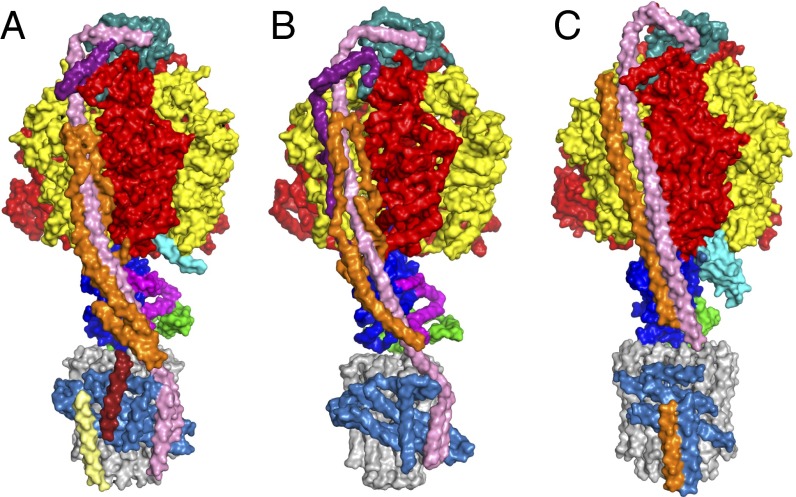
Fig. 4.
Comparison of the current structures of the F-ATPases. The structures of the three complexes are viewed from the side toward the peripheral stalk with the F1-catalytic domain above and the membrane domain beneath. Enzymes (A and B) from P. angusta and bovine mitochondria (7) and (C) from P. denitrificans (9). The α-, β-, and γ-subunits are red, yellow, and royal blue, respectively. The c-rings (made of 10, 8, and 12 subunits in A–C, respectively) are gray, and the adjacent a-subunits are corn-flower blue. In A and B, the OSCP subunits (Top), and in C, the orthologous δ-subunit, are sea-green; the δ-subunits (A and B) and orthologous ε-subunit in C are green. In A and B only, the ε-subunit (next to the green δ-subunit) is magenta. In A and B, in addition to the OSCP, the peripheral stalks contain the b-subunits (pink), the d-subunits (orange), and the orthologous h and F6-subunits (purple), respectively. In C, in addition to the δ-subunit, the peripheral stalk contains a b-subunit (pink) and a b′-subunit (orange). In A, the pale yellow and brick-red α-helices packed against the c-rings have been assigned tentatively to the f and ATP8 subunits, respectively.
As this paper neared completion, the structure of the dimeric F-ATPase from the fungus Yarrowia lipolytica determined by cryo-EM was published (43). There is excellent agreement between many of the features in the two enzymes. The current paper contains a more precise and accurate description of the attachment of the peripheral stalk to the catalytic domain, and the dimeric structure confirms the presence of the e- and g-subunits in the dimer interface and gives undiscovered details of how they are folded.
Materials and Methods
The F-ATPase from P. angusta was purified as described before (20). Images of single monomeric complexes were recorded with a Titan Krios electron microscope (FEI) with a Falcon II CMOS (complementary metal oxide semiconductor) direct electron detector using the EPU automated data acquisition software. For full details of these processes and the structure determination and analysis, see SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. A. Leslie and R. Henderson for their comments; S. Chen and C. Savva for help with electron microscopes; and J. Grimmett and T. Darling for help with computing. This work was supported by the Medical Research Council of the United Kingdom by Grant MC_U1065663150 and by Programme Grant MR/M009858/1, (both to J.E.W.) and by Grant MC_U105184322 (to K.R.V.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM maps have been deposited in the EMDataBank (accession nos. EMD-4102, EMD-4101, and EMD-4100). The atomic coordinates have been deposited in the Protein Data Bank (accession nos. 5LQZ, 5LQY, and 5LQX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615902113/-/DCSupplemental.
References
- 1.Watanabe R, Noji H. Chemomechanical coupling mechanism of F1-ATPase: Catalysis and torque generation. FEBS Lett. 2013;587(8):1030–1035. doi: 10.1016/j.febslet.2013.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41(1):1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 3.Robinson GC, et al. The structure of F₁-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF₁. Open Biol. 2013;3(2):120164. doi: 10.1098/rsob.120164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bason JV, Montgomery MG, Leslie AGW, Walker JE. Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc Natl Acad Sci USA. 2014;111(31):11305–11310. doi: 10.1073/pnas.1411560111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bason JV, Montgomery MG, Leslie AGW, Walker JE. How release of phosphate from mammalian F1-ATPase generates a rotary substep. Proc Natl Acad Sci USA. 2015;112(19):6009–6014. doi: 10.1073/pnas.1506465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proc Natl Acad Sci USA. 2012;109(29):11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 2015;4:e10180. doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521(7551):237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 9.Morales-Rios E, Montgomery MG, Leslie AGW, Walker JE. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc Natl Acad Sci USA. 2015;112(43):13231–13236. doi: 10.1073/pnas.1517542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: Identification of three dimer-specific subunits. EMBO J. 1998;17(24):7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paumard P, et al. Two ATP synthases can be linked through subunits in the inner mitochondrial membrane of Saccharomyces cerevisiae. Biochemistry. 2002;41(33):10390–10396. doi: 10.1021/bi025923g. [DOI] [PubMed] [Google Scholar]
- 12.Arselin G, et al. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur J Biochem. 2003;270(8):1875–1884. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 13.Fronzes R, Weimann T, Vaillier J, Velours J, Brèthes D. The peripheral stalk participates in the yeast ATP synthase dimerization independently of e and g subunits. Biochemistry. 2006;45(21):6715–6723. doi: 10.1021/bi0601407. [DOI] [PubMed] [Google Scholar]
- 14.Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP. Structure of dimeric ATP synthase from mitochondria: An angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 2005;579(25):5769–5772. doi: 10.1016/j.febslet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 15.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27(7):1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE. The F1Fo-ATPase complex from bovine heart mitochondria: The molar ratio of the subunits in the stalk region linking the F1 and Fo domains. Biochemistry. 1996;35(38):12640–12646. doi: 10.1021/bi960969t. [DOI] [PubMed] [Google Scholar]
- 17.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25(12):2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106(51):21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno H, Suzuki T, Kinosita K, Jr, Yoshida M. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc Natl Acad Sci USA. 2005;102(5):1333–1338. doi: 10.1073/pnas.0407857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, et al. The purification and characterization of ATP synthase complexes from the mitochondria of four fungal species. Biochem J. 2015;468(1):167–175. doi: 10.1042/BJ20150197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104(40):15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286(5445):1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 23.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 24.Kagawa R, Montgomery MG, Braig K, Leslie AGW, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23(14):2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J Biol Chem. 2007;282(19):14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 26.Lightowlers RN, Howitt SM, Hatch L, Gibson F, Cox GB. The proton pore in the Escherichia coli FoF1-ATPase: A requirement for arginine at position 210 of the a-subunit. Biochim Biophys Acta. 1987;894(3):399–406. doi: 10.1016/0005-2728(87)90118-6. [DOI] [PubMed] [Google Scholar]
- 27.Hoppe J, Sebald W. Amino acid sequence of the proteolipid subunit of the proton-translocating ATPase complex from the thermophilic bacterium PS-3. Eur J Biochem. 1980;107(1):57–65. doi: 10.1111/j.1432-1033.1980.tb04624.x. [DOI] [PubMed] [Google Scholar]
- 28.Runswick MJ, et al. The affinity purification and characterization of ATP synthase complexes from mitochondria. Open Biol. 2013;3(2):120160. doi: 10.1098/rsob.120160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sielaff H, et al. Domain compliance and elastic power transmission in rotary FoF1-ATPase. Proc Natl Acad Sci USA. 2008;105(46):17760–17765. doi: 10.1073/pnas.0807683105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson KM, et al. Identification and validation of the mitochondrial F1Fo-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem Biol. 2005;12(4):485–496. doi: 10.1016/j.chembiol.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Cleary J, Johnson KM, Opipari AW, Jr, Glick GD. Inhibition of the mitochondrial F1Fo-ATPase by ligands of the peripheral benzodiazepine receptor. Bioorg Med Chem Lett. 2007;17(6):1667–1670. doi: 10.1016/j.bmcl.2006.12.102. [DOI] [PubMed] [Google Scholar]
- 32.Wu YT, Lee HC, Liao CC, Wei YH. Regulation of mitochondrial FoF1 ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977bp deletion of mitochondrial DNA. Biochim Biophys Acta. 2013;1832(1):216–227. doi: 10.1016/j.bbadis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Vassilopoulos A, et al. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21(4):551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, et al. Organisation of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. J Biol Chem. 2015;290(21):13308–13320. doi: 10.1074/jbc.M115.645283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kucharczyk R, et al. Mitochondrial ATP synthase disorders: Molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793(1):186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Xu T, Pagadala V, Mueller DM. Understanding structure, function, and mutations in the mitochondrial ATP synthase. Microb Cell. 2015;2(4):105–125. doi: 10.15698/mic2015.04.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker JE, Saraste M, Gay NJ. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- 38.Dunn SD. The polar domain of the b subunit of Escherichia coli F1Fo-ATPase forms an elongated dimer that interacts with the F1 sector. J Biol Chem. 1992;267(11):7630–7636. [PubMed] [Google Scholar]
- 39.Collinson IR, et al. ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J Mol Biol. 1994;242(4):408–421. doi: 10.1006/jmbi.1994.1591. [DOI] [PubMed] [Google Scholar]
- 40.Karrasch S, Walker JE. Novel features in the structure of bovine ATP synthase. J Mol Biol. 1999;290(2):379–384. doi: 10.1006/jmbi.1999.2897. [DOI] [PubMed] [Google Scholar]
- 41.Dunn SD, McLachlin DT, Revington M. The second stalk of Escherichia coli ATP synthase. Biochim Biophys Acta. 2000;1458(2-3):356–363. doi: 10.1016/s0005-2728(00)00086-4. [DOI] [PubMed] [Google Scholar]
- 42.Duncan AL, Robinson AJ, Walker JE. Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc Natl Acad Sci USA. 2016;113(31):8687–8692. doi: 10.1073/pnas.1608396113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell. 2016;63(3):445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.