Significance
The molecular basis of radial migration of cortical neurons is a well-studied process showing prominent roles for axon guidance, cell adhesion, cell polarity, and cytoskeleton remodeling. Remarkably, knowledge about transcriptional control of such processes is scarce. In this study, we show that the forkhead transcription factor FoxO6 influences Plexin A4 (Plxna4) expression, a key component of the Semaphorin signaling pathway, known for its role in axonal guidance and cortical migration. FoxO6 knockdown animals show a hampered migration of embryonic day 14.5-born neurons, which can be rescued by recombinant Plxna4 expression constructs. Altogether, our data provide insights into the molecular mechanisms whereby transcriptional programs influence cortical development.
Keywords: FoxO6, cortex, development, radial migration, Plxna4
Abstract
The forkhead transcription factor FoxO6 is prominently expressed during development of the murine neocortex. However, its function in cortical development is as yet unknown. We now demonstrate that cortical development is altered in FoxO6+/− and FoxO6−/− mice, showing migrating neurons halted in the intermediate zone. Using a FoxO6-directed siRNA approach, we substantiate the requirement of FoxO6 for a correct radial migration in the developing neocortex. Subsequent genome-wide transcriptome analysis reveals altered expression of genes involved in cell adhesion, axon guidance, and gliogenesis upon silencing of FoxO6. We then show that FoxO6 binds to DAF-16–binding elements in the Plexin A4 (Plxna4) promoter region and affects Plxna4 expression. Finally, ectopic Plxna4 expression restores radial migration in FoxO6+/− and siRNA-mediated knockdown models. In conclusion, the presented data provide insights into the molecular mechanisms whereby transcriptional programs drive cortical development.
The FoxO (forkhead box O) family of transcription factors is known to be involved in a wide array of biological processes, including apoptosis, cellular survival, differentiation, and stress resistance. FoxOs are part of the conserved PI3K-AKT-FoxO signaling pathway, which has been shown to convey extracellular signals to intracellular transcriptional programs. Insulin and insulin-like growth factors (IGFs; e.g., Igf1) regulate FoxO transcriptional activity mainly via posttranslational modifications, resulting in the nuclear exclusion of the protein, thereby rendering it inactive (1–5). In humans, de novo mutations in genes involved in the PI3K/AKT pathway result in a wide array of cortical malformations, including cortical dysplasia, megalencephaly-polymicrogyria-polydactyly-hydrocephalus (MPPH), and megalencephaly-capillary malformation-polymicrogyria syndrome (MCAP) (6, 7). Studying the downstream effectors of this pathway (e.g., FoxO proteins) provides in-depth insights into the molecular mechanisms underlying these cortical malformations.
Recent studies have shown that FoxOs are required for the coordinated regulation of postnatal neuronal stem cell (NSC) homeostasis, demonstrating a prominent role for FoxO3 in NSC proliferation and renewal (8–10). In a combined FoxO1,3,4-deficient mouse model, an initial increase in brain size was observed followed by a decline in the adult NSC pool. Furthermore, it was shown, both in vivo and in vitro, that the single-FoxO3–deficient animal results in a similar decline in NSC number (10). FoxO6 was shown to be required for the regulation, in the hippocampus, of a set of genes involved in synaptic function (11) and is essential in Pak1-mediated cellular polarity in the cerebellum (12), demonstrating the importance of this forkhead member in the mammalian brain.
Apart from the functions of FoxO6 in the postnatal hippocampus and cerebellum, FoxO6 is likely to play a role during the embryonic development of the cortex, judging from its specific temporal and spatial expression pattern (13). From embryonic day (E)12.5 during murine embryonic development, FoxO6 expression is found mainly in the hippocampus, cortex, and striatum (14). This pattern is conserved at later stages until birth, when expression of FoxO6 is mainly found in the hippocampus (14). Importantly, in early developmental stages, expression is observed in proliferating areas in the cortex whereas at later stages FoxO6 is also prominently expressed in the postmitotic cortical plate, suggesting different functions for FoxO6 during development.
During cortical development, neuroepithelial cells, located in the cortical ventricular zone of the neural tube, will elongate at mouse E11 and assume a radial glial morphology. These radial glial cells divide asymmetrically to generate new radial glial cells and intermediate progenitor cells, destined to become postmitotic neurons (15, 16). Postmitotic cells undergo outward migration to form the upper layers of the cortical plate and eventually generate the six-layered adult neocortex. Although several factors, such as guidance molecules and cell-adhesion molecules (10, 17, 18), have been implicated in coordinating migration, molecular programming underlying these processes remains poorly understood.
In this study, we show that FoxO6 affects radial migration events during cortical development. Transcriptome analysis revealed that Plexin A4 (Plxna4) is regulated by FoxO6 in different mouse models. Moreover, we show that FoxO6 occupies DAF-16–binding sites present in the Plxna4 promoter and is able to regulate its transcription. Finally, in FoxO6 knockdown models, radial migration is restored by ectopic overexpression of Plxna4. These data suggest that FoxO6 acts in a transcriptional program directed toward correct migration events during cortical development.
Results
Global Cortical Markers Are Unchanged in FoxO6−/− Cortices.
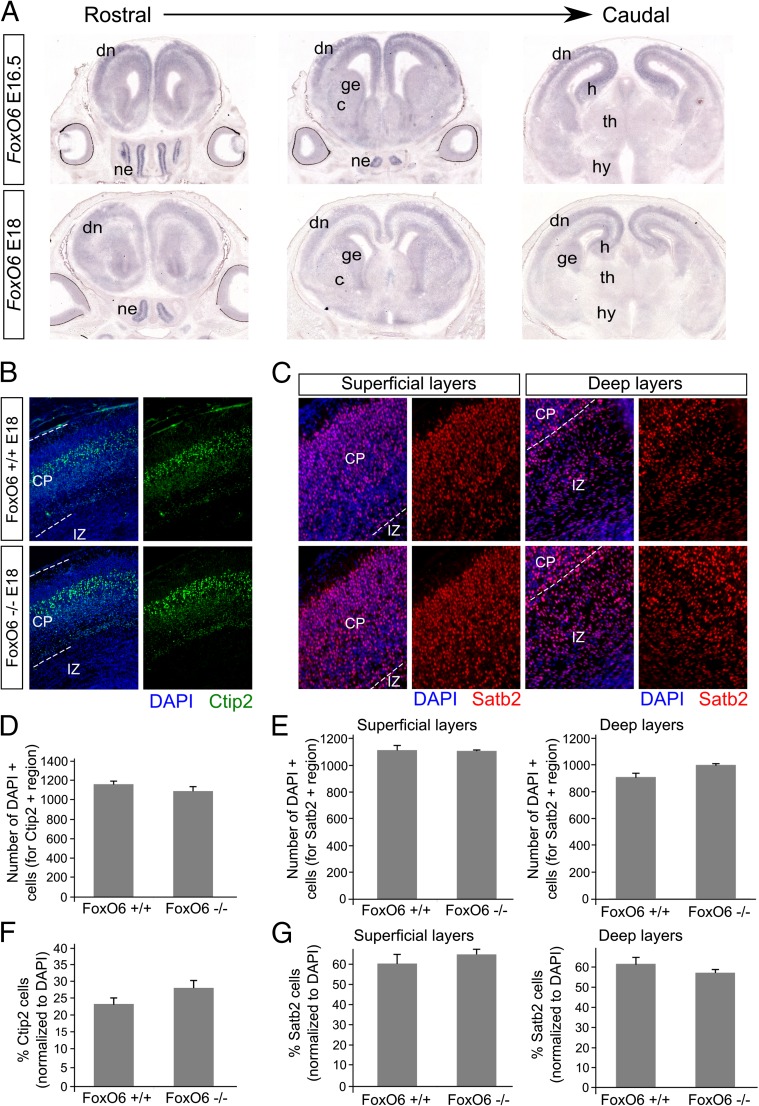
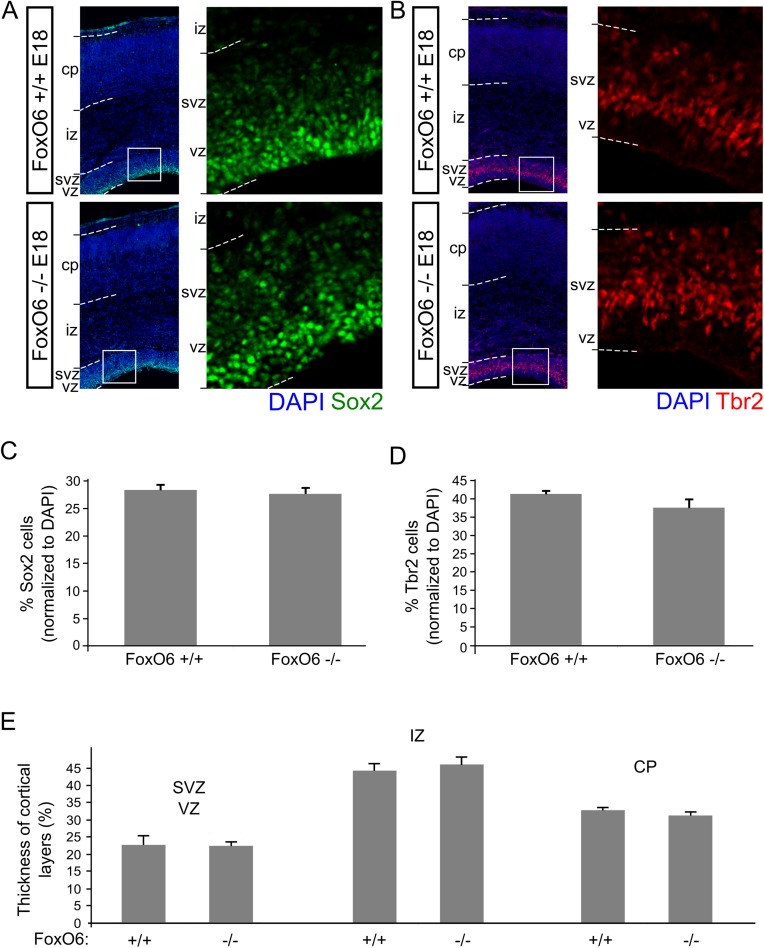
During cortical development, FoxO6 has a regionalized expression pattern, mapping to different cortical subregions at different developmental stages, suggesting FoxO6 functions in multiple neuronal developmental processes (14). At early developmental stages (E12.5), FoxO6 is expressed in the proliferating ventricular zone (VZ) and subventricular zone (SVZ), suggesting FoxO6 may be involved in neuronal stem cell proliferation and/or stem cell maintenance, as has been suggested previously for other mammalian FoxOs (8–10). At E14.5, FoxO6 remains expressed in the VZ and SVZ and expression is now also observed in the outer layer of the cortical plate (CP) (14). To substantiate these data and investigate expression at later time points of cortical development, we performed ISH experiments on WT brain material of E16.5 and E18 animals (Fig. 1A). We show that at E16.5, throughout the rostral-to-caudal axis, FoxO6 was predominantly expressed in the developing ganglionic eminences, caudate putamen, and dorsal neocortex. Notably, FoxO6 was mostly detected in the developing hippocampus, as described previously (11, 14). At E18, FoxO6 expression was more strictly confined to the developing telencephalon, showing no expression in the thalamic and hypothalamic regions surrounding the third ventricle. In addition, FoxO6 is expressed throughout the VZ, SVZ, and CP. To investigate cortical development in the absence of FoxO6, we analyzed the distribution of global cortical markers in the cortices of FoxO6−/− and wild-type mice at E18 by immunohistochemistry. To do so, we visualized the apical and basal progenitor cells [respectively marked by Sox2 and eomesodermin (Tbr2)] and quantified the positive cells for a fixed region, covering the VZ and SVZ, representing the amount as the percentage of total cells in that region. Our analysis revealed that the amount of multipotent progenitor cells, marked by Sox2, was not significantly different between the two genotypes (Fig. S1 A and C; n = 4 per condition). The amount of intermediate progenitor cells in the SVZ, marked by Tbr2, was also unaffected in FoxO6−/− mice (Fig. S1 B and D; n = 4 per condition). To investigate whether the postmitotic regions of the developing cortex were affected by the loss of FoxO6 at E18, we performed immunohistochemistry for B cell leukemia/lymphoma 11B (Ctip2) and special AT-rich sequence binding protein 2 (satb2) to identify the subcortical and callosal projecting pyramidal neurons (Fig. 1 B and C). The amount of subcortical projection neurons, located in the deep layers V and VI (marked by Ctip2), was not significantly altered in FoxO6−/− cortices at E18 (Fig. 1 D and F; n = 4 per condition). We next divided the Satb2-positive cortical region into two bins covering the upper-layer Satb2-positive neurons and the deep-layer, still migrating, Satb2-positive neurons, revealing both populations to be not significantly changed (Fig. 1 E and G; n = 4 per condition). Supporting the results described above, the gross morphology of the developing layers was unaffected in FoxO6−/− cortices as shown by an unchanged thickness of the VZ/SVZ, intermediate zone (IZ), and CP at E18 (Fig. S1E; n = 3 per condition).
Fig. 1.
FoxO6 is present in the developing cortex, and ablation does not lead to abnormalities in layering as shown by Ctip2 and Satb2 distribution. (A) During cortical development, FoxO6 is broadly expressed, showing expression in the proliferating (sub)ventricular zone and the postmitotic cortical plate at E16.5 and E18. Next to the cortical expression of FoxO6, expression is primarily found in the nasal epithelium and the ganglionic eminences at E16.5 and E18. (B) Immunohistochemistry for Ctip2 reveals no clear differences in the amount of subcortical projection neurons in FoxO6−/− cortices. Cortices were counterstained with DAPI. (C) Immunohistochemistry for Satb2 reveals no clear differences in the amount of callosal-projecting pyramidal neurons in FoxO6−/− cortices. Satb2-expressing area was divided into two bins covering the upper layers and deep Satb2-expressing layers. Cortices were counterstained with DAPI. (D) Quantification of the number of DAPI-positive cells for the Ctip2 region in FoxO6+/+ and FoxO6−/− cortices. (E) Quantification of the number of DAPI-positive cells for a fixed region covering the upper and deep Satb2-expressing layers in FoxO6+/+ and FoxO6−/− cortices. (F) Quantification of the number of Ctip2-positive cells for a fixed region covering layers V and VI in FoxO6+/+ and FoxO6−/− cortices. Data were normalized against the number of DAPI cells in that region. (G) Quantification of the number of Satb2-positive cells for a fixed region covering the upper and deep Satb2-expressing layers in FoxO6+/+ and FoxO6−/− cortices. Data were normalized against the number of DAPI cells in that region. All quantifications were performed on n = 4 per condition. The error bars show the SEM. Two-tailed Student’s t test. c, caudate putamen; dn, dorsal neocortex; ge, ganglionic eminences; h, hippocampus; hy, hypothalamus; ne, nose epithelium; th, thalamus.
Fig. S1.
Numbers of apical and basal progenitor cells are unchanged in FoxO6−/− cortices. (A) Immunostaining for Sox2 reveals no clear difference in the amount of multipotent apical progenitor cells in FoxO6−/− cortices. Cortices were counterstained with DAPI. (B) Immunostaining for Tbr2 reveals that the pool of basal progenitors is not clearly affected in FoxO6−/− cortices. Cortices were counterstained with DAPI. (C) Quantification of the number of Sox2-positive cells for a fixed region covering the VZ and SVZ in FoxO6+/+ and FoxO6−/− cortices. Data were normalized against the number of DAPI cells in that region. (D) Quantification of the number of Tbr2-positive cells for a fixed region covering the VZ and SVZ in FoxO6+/+ and FoxO6−/− cortices. Data were normalized against the number of DAPI cells in that region. (E) Quantification of the average width of the subventricular zone/ventricular zone, intermediate zone, and cortical plate. Previously performed DAPI counterstaining was used to identify and measure the width of the indicated layers in E18 FoxO6+/+ and FoxO6−/− brains. The error bars show the SD. Two-tailed Student’s t test; no significant differences were observed.
FoxO6−/− Mice Display Hampered Radial Migration During Cortical Development, Resulting in a Decreased Amount of Cut-Like Homeobox 1-Positive Cells.
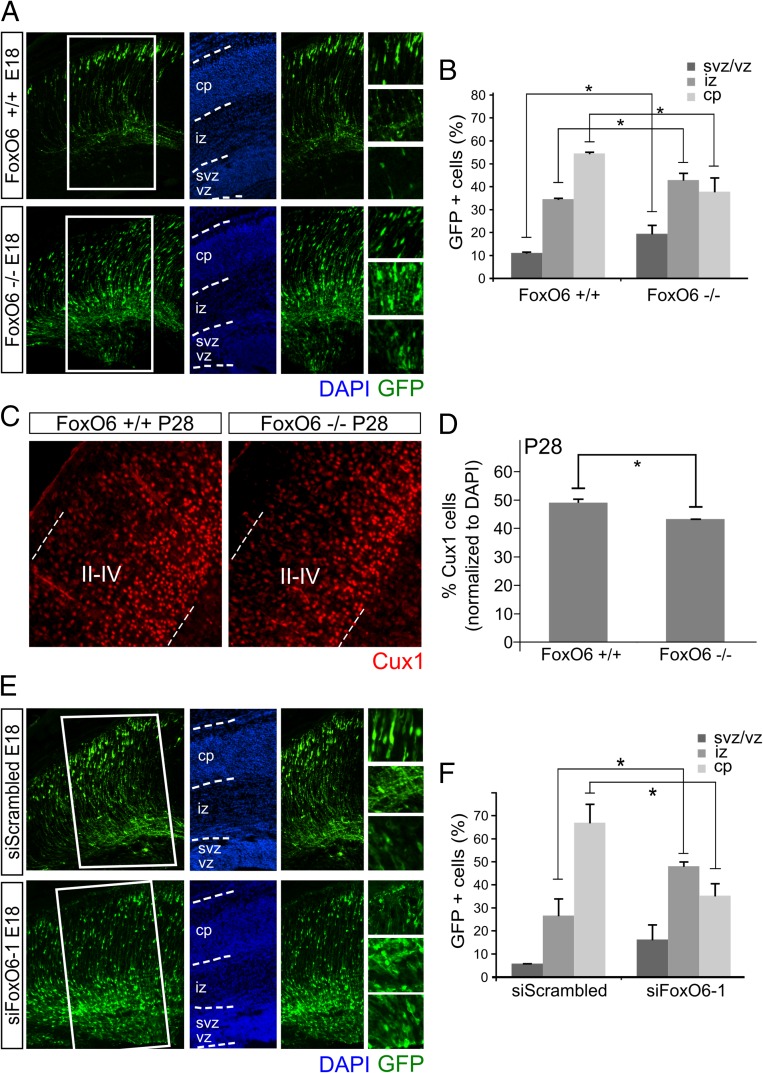
Because global cortical markers did not reveal significant changes in developing FoxO6−/− cortices, we set out to detect more subtle deviations in the developing cortex caused by FoxO6 ablation. Therefore, we introduced a GFP expression plasmid in E14.5 FoxO6−/− and FoxO6+/+ cortices by in utero electroporation, thereby tracing neurons born at the peak of neurogenesis. At E18, GFP-positive transfected cells in control cortices (Fig. 2A) migrated outward, reaching the cortical plate. Interestingly, in FoxO6−/− cortices (Fig. 2A), the proportion of GFP-positive cells reaching the cortical plate was decreased significantly. Quantification revealed that a majority of the GFP-positive cells (54.40%; SD ±0.53) in wild-type control cortices reached the cortical plate, whereas only 37.2% (SD ±6.1; P < 0.05) of these cells in FoxO6−/− cortices reached the cortical plate (Fig. 2B; n = 4 per condition). In line with these findings, 42.83% (SD ±3.10) of GFP-positive cells in FoxO6−/− cortices are located in the IZ compared with 34.50% (SD ±0.35; P < 0.05) in wild-type littermate controls (Fig. 2B). Furthermore, we observed a significantly increased ratio of GFP-labeled cells in the VZ of FoxO6−/− cortices (19.4%; SD ±3.8) compared with FoxO6+/+ controls (11%; SD ±0.35; P = 0.01) (Fig. 2B), indicating the necessity of proper FoxO6 function for the migration of these developing neurons. Although FoxOs have been implicated to regulate cell polarity in the cerebellum (12), magnifications of the different cortical bins revealed that GFP-labeled cells, ablated for FoxO6, display a normal bipolar morphology (Fig. 2A, Right), which makes polarity changes unlikely to be related to the observed phenotype. These data, together with the observation that global cortical markers are unaffected in FoxO6−/− cortices, suggest that only a relatively small proportion of cells born at E14.5 is affected by the loss of FoxO6. In line with this, we analyzed the amount of cut-like homeobox 1 (Cux1)-positive cells in postnatal day (P)28 FoxO6−/− and FoxO6+/+ cortices (Fig. 2 C and D). As shown, the amount of Cux1-positive cells in the upper layers of the FoxO6−/− cortex is decreased significantly (12%; P < 0.05; n = 3 per condition), indicating that ablation of FoxO6 influences a proportion of cortical neurons born at E14.5.
Fig. 2.
FoxO6−/− mice display hampered radial migration during cortical development, which is exemplified by the altered presence of Cux1-positive callosal projection neurons. (A) Tracing newborn neurons by in utero electroporation (IUE) at E14.5 with a GFP expression vector revealed an altered distribution of cortical cells in FoxO6−/− cortices compared with FoxO6+/+ cortices at E18. Magnifications are shown for the cortical plate, intermediate zone, and ventricular/subventricular zone. (B) Quantification of triplicate experiments as shown in A. Ratios between the ventricular zone, intermediate zone, and cortical plate are shown. (C) Immunostaining for Cux1 in FoxO6−/− cortices compared with FoxO6+/+ cortices. (D) Quantification of the number of Cux1-positive cells for a fixed region covering layers II–IV in FoxO6+/+ and FoxO6−/− cortices. Data were normalized against the number of DAPI cells in that region. (E) Cortices in utero electroporated with siFoxO6-1 at E14.5 and cotransfected with a GFP expression vector display altered neuronal distribution at E18 compared with siScrambled (control) electroporated cortices. Magnifications are shown for the cortical plate, intermediate zone, and ventricular/subventricular zone. (F) Quantification of triplicate experiments as shown in E. Ratios between the ventricular zone, intermediate zone, and cortical plate are shown. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05 (Bonferroni-corrected significance level in B and F).
siRNA-Mediated Down-Regulation of FoxO6 Expression in the Embryonic Cortex Phenocopies the Radial Migration Defect Observed in FoxO6−/− Mice.
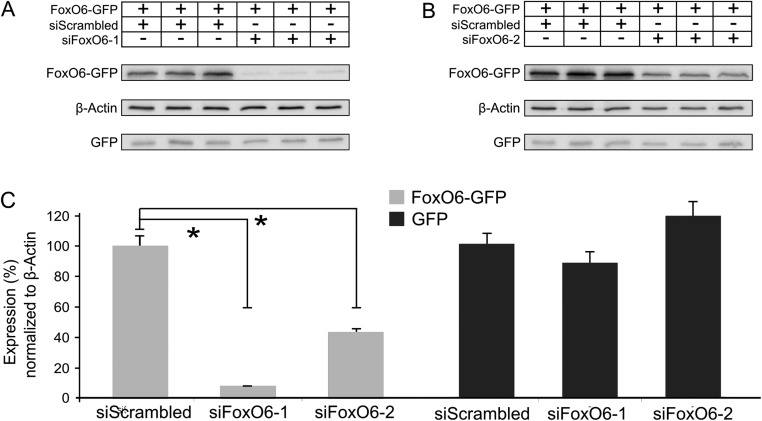
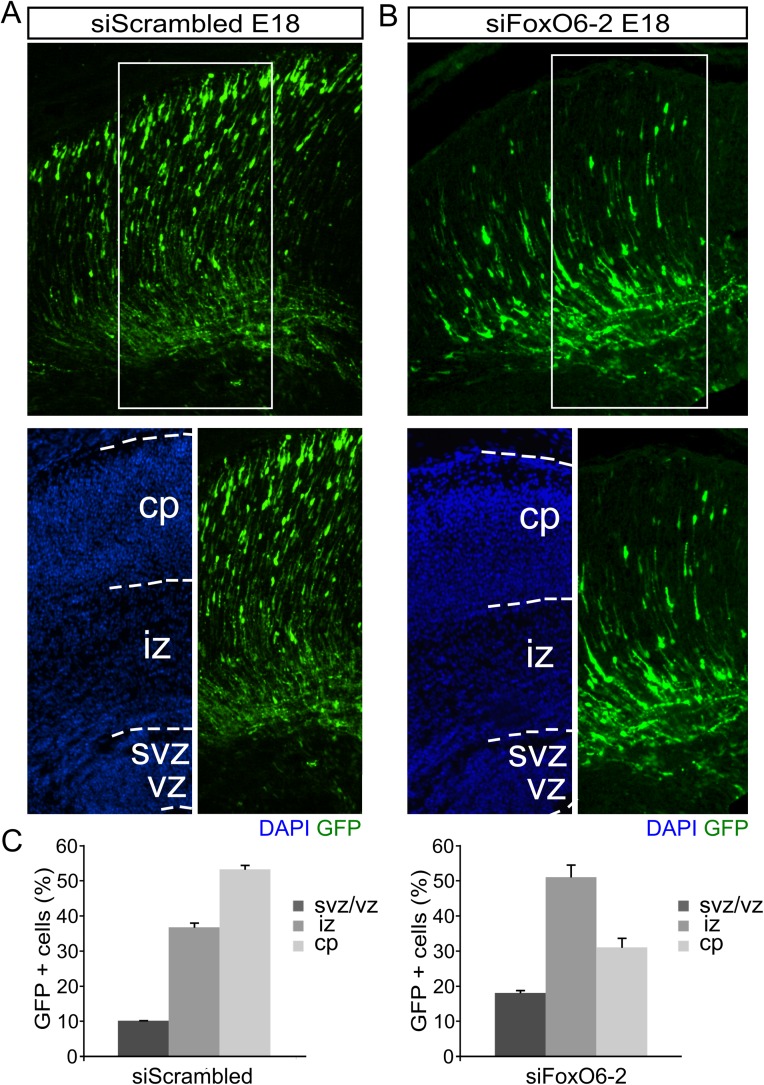
To substantiate the cell-autonomous role of FoxO6 in cortical migration, we followed an acute knockdown strategy in which FoxO6 expression in the embryonic cortex was down-regulated via in utero electroporation of siRNAs specifically directed against FoxO6. Out of two different siRNAs directed against FoxO6, we used the most efficient one in these experiments (siFoxO6-1; Fig. S2). To monitor radial migration, we coelectroporated a GFP expression vector and analyzed these cortices at E18. Mimicking FoxO6−/− animals, a hampered radial migration was observed at E18 when cortices were electroporated with siFoxO6-1 at E14.5 (Fig. 2 E and F). At E18, 3.5 d postelectroporation, cortices electroporated with scrambled control siRNA showed 27% (SD ±2.5) of transfected cells located in the intermediate zone compared with 48% (SD ±1.1; P < 0.001) in cortices down-regulated for FoxO6. Furthermore, only 35.2% (SD ±1.4) of the transfected cells in cortices electroporated with siFoxO6-1 reached the cortical plate, compared with 66.9% (SD ±1.2; P < 0.001) in controls (Fig. 2 E and F). Furthermore, we observed an increased ratio of GFP-labeled cells in the ventricular and subventricular zones in cortices down-regulated for FoxO6 (16.2% SD ±6.6) compared with control cortices (5.7%; SD ±2.2l; P < 0.05). Taken together, the phenotype as observed in FoxO6−/− electroporated cortices resembles the FoxO6 siRNA-mediated knockdown phenotype. To elaborate upon the specificity of the siRNA, we introduced the siRNA targeting FoxO6 in FoxO6−/− cortices. As expected, in these cortices, the distribution of the GFP-labeled cells was unaltered (Fig. S3). In addition, to exclude the possibility that part of the observed siRNA-mediated phenotype is due to off-target effects, we performed similar in utero electroporation experiments but now using a different siRNA directed against FoxO6 (siFoxO6-2) targeting a different part of the FoxO6 mRNA. Importantly, cortices electroporated with siFoxO6-2 showed an identical phenotype as cortices electroporated with siFoxO6-1 (Fig. S4), indicating that the phenotype observed is due to the down-regulation of FoxO6. Taken together, siRNA-mediated knockdown of FoxO6 in the developing cortex starting at E14.5 results in hampered radial migration similar to the effects observed in the full FoxO6 knockout.
Fig. S2.
siFoxO6-1 and siFoxO6-2 down-regulate FoxO6-GFP protein expression. (A and B) HEK293 cells were transfected with FoxO6-GFP and either siScrambled, siFoxO6-1, or siFoxO6-2. Western blot revealing down-regulation of FoxO6-GFP upon treatment with siFoxO6-1 and siFoxO6-2. β-Actin was used as a loading control. GFP was used to show that the siRNAs target FoxO6 and not GFP. (C) Densitometric analysis of the Western blot analysis; n = 3 vs. n = 3. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05.
Fig. S3.
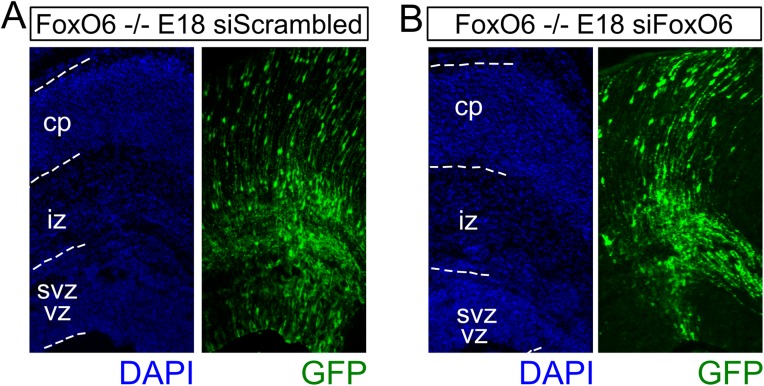
siRNA-mediated down-regulation of FoxO6 does not induce additional migration defects in FoxO6−/− cortices. (A) E18 FoxO6−/− cortices in utero electroporated with siScrambled at E14.5 and cotransfected with a GFP expression vector display altered neuronal distribution at E18. (B) E18 cortices in utero electroporated with siFoxO6 at E14.5 and cotransfected with a GFP expression vector did not display additional migration deficits.
Fig. S4.
Knockdown of FoxO6 via siFoxO6-2 results in defective radial migration, confirming data of siFoxO6-1. (A and B) GFP protein expression at E18 in cortices in utero electroporated with siScrambled or siFoxO6-2, cotransfected with a GFP expression vector. siFoxO6-2 displays an altered distribution of GFP-positive cells at E18 compared with siScrambled (control) electroporated cortices. (C) Quantification of duplicate experiments as shown in A and B. Ratios of GFP+ cells between the ventricular zone, intermediate zone, and cortical plate are shown. Note the reduction in cortical plate-positioned neurons upon treatment with siFoxO6-2, resembling the phenotype observed with siFoxO6-1. The error bars show the SD.
FoxO6 Knockdown Results in Altered Expression of Genes Involved in Cell Adhesion, Axon Guidance, and Gliogenesis.
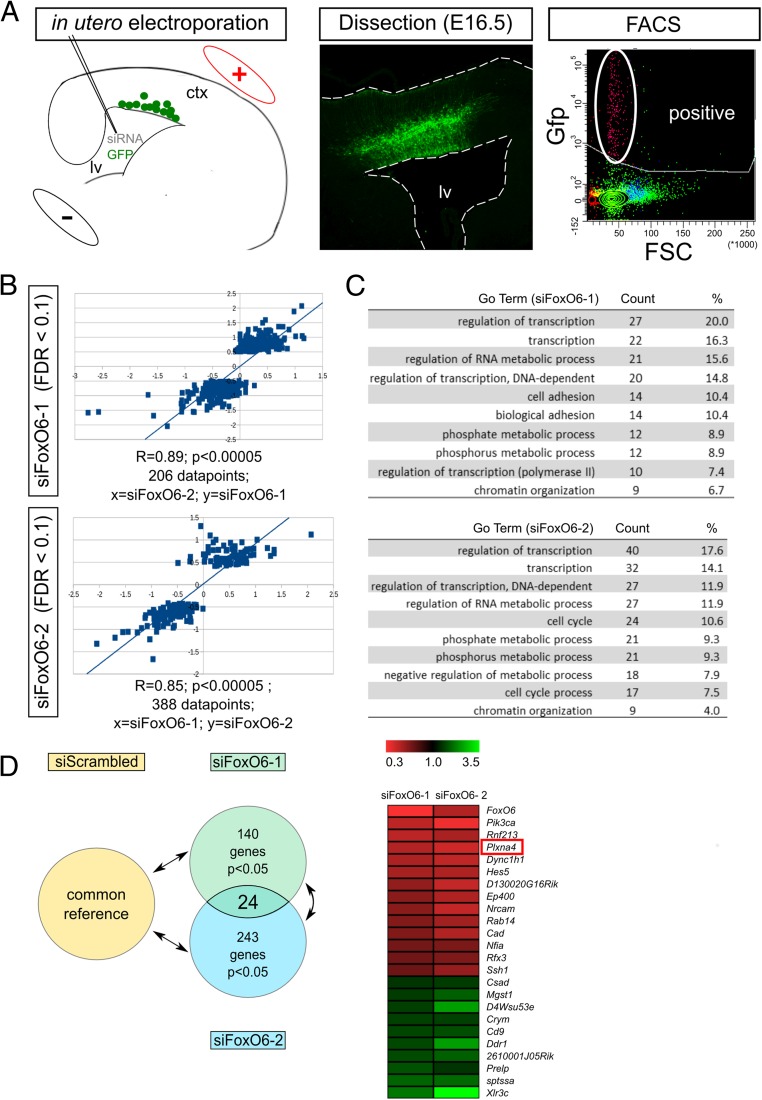
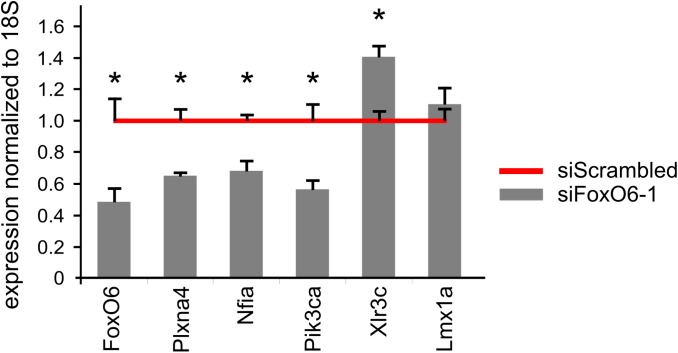
To gain insight into the acute molecular mechanisms governing FoxO6-mediated cortical radial migration, genome-wide transcriptome analysis was performed. To this end, we specifically silenced FoxO6 expression in the cortex at E14.5 by in utero electroporation of siFoxO6-1 together with a GFP expression plasmid. GFP-positive cells were FACsorted at E16.5, which is at the peak of radial migration and neurogenesis, followed by RNA isolation (Fig. 3A). We used two different siRNAs targeting FoxO6 (siFoxO6-1 and siFoxO6-2) and independently compared the results with a control siRNA (siScrambled, common reference) to minimize off-target effects and to select the most significant genes regulated by FoxO6 activity (Table S1). For each condition at least three cortices were pooled (randomized over litters) to minimize the possibility of finding initial differences due to the electroporation procedure itself or due to between-nest variation. In total, 12 cortices were used, whereby for each biological replicate 3 cortices were pooled, generating a total 4 replicates. Electroporated cortices from different litters were pooled to minimize effects due to electroporation and litter differences. To investigate whether the different siRNAs and the separate in utero electroporations provided similar data, we performed a correlation analysis on the false discovery rate (FDR)-corrected values (FDR cutoff 0.1). The correlation coefficient (R) was at least 0.85 (siFoxO6-2, R = 0.85, P < 0.001; siFoxO6-1, R = 0.89, P < 0.001), indicating that the gene expression profile as a result of the FoxO6 knockdown by the two different siRNAs is highly comparable (Fig. 3B). Subsequently, we performed a Gene Ontology analysis (19) on the separate datasets, showing that similar processes are highly enriched (Fig. 3C). The results from siFoxO6-2 (n = 3) and siFoxO6-1 (n = 3) and control siRNA (common reference) (n = 9) were used in a three-way comparison, thereby generating two independent datasets that contained ±200 and ±300 FoxO6-regulated genes (P < 0.05 cutoff) (FDR) (Fig. 3D). Overlap of these two restricted datasets (P < 0.05, for separate list selection) yielded a list of 24 regulated genes (Fig. 3D). We confirmed the transcription modulation of several genes in independently electroporated and FACsorted cortices using quantitative (q)PCR [Fig. S5; LIM homeobox transcription factor 1 alpha (Lmx1a) was used as an unchanged reference], thereby confirming the validity of the transcriptome analysis.
Fig. 3.
Genome-wide transcriptional profiling of FoxO6 knockdown neurons. (A) A graphical representation of the methodology used. FoxO6 is down-regulated by in utero electroporation at E14.5 using two different siRNAs targeting FoxO6 expression. Brains are dissected and dissociated at E16.5. Cortices are coelectroporated with GFP to FACsort the transfected cells followed by RNA isolation and microarray analysis. Three electroporated cortices were pooled for each N per condition to minimize the effects due to electroporation differences and between-litter differences. (B) Regulated genes by FoxO6 (FDR cutoff 0.1), identified as a result of the two different siRNAs, display a strong correlation indicating that in both datasets similar genes are represented, which are changed in a similar direction; siFoxO6-2, R = 0.85, P < 0.01; siFoxO6-1, R = 0.89, P < 0.01. (C) GO term analysis reveals that similar processes are regulated by FoxO6 knockdown through the use of siFoxO6-2 and siFoxO6-1. (D, Left) Graphical representation of the microarray analysis setup and resulting amount of regulated genes (243, siFoxO6-2; 140, siFoxO6-1) and the exact overlap (24 genes) in the P < 0.05 FDR-restricted datasets. (D, Right) Heatmap of the 24 genes that are present in such an overlap, including FoxO6. ctx, cortex; FACS, fluorescence-activated cell sorting; FSC, forward scatter; GFP, green fluorescent protein; lv, lateral ventricle.
Fig. S5.
Validation of a selection of genes as identified in the FoxO6 knockdown genome-wide transcriptome analysis. In an independent set of experiments, cortices were in utero electroporated with siFoxO6-1 and cotransfected with a GFP expression vector, followed by FACsorting and RNA isolation. Genes significantly regulated according to the transcriptome data were investigated by qPCR. Significant down-regulation of FoxO6, Plxna4, Nfia, and Pik3ca was confirmed. Significant up-regulation of Xlr3c was also confirmed. Lmx1a was used as an unchanged control. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05.
FoxO6 Affects Radial Cortical Migration in a Plxna4-Dependent Manner.
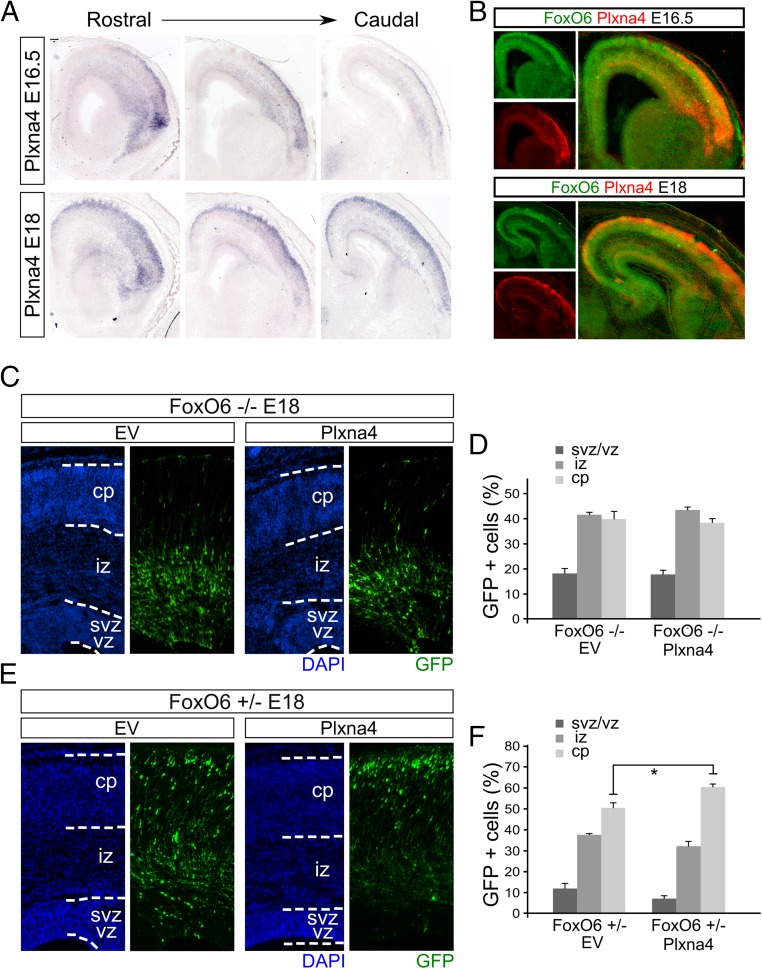
One of the identified FoxO6-regulated genes, Plxna4 (Fig. 3D), is known to be required for proper radial migration of newborn neurons in the developing cortex. Plxna4 plays a part in the Semaphorin signaling cascade, which is disrupted upon Plxna4 ablation, resulting in an impaired radial migration of cortical neurons (20). Importantly, the observed defect found in FoxO6-deficient cortices, with neurons halted in the deeper layers of the developing cortex, phenocopies the defect observed when rat embryonic cortices are electroporated with siRNA specifically directed against Plxna4 (20). To further explore the possibility that the correct radial migration of neurons in the developing cortex is influenced by a FoxO6-mediated regulation of Plxna4, we first analyzed the spatial and temporal expression profile of Plxna4 by performing in situ hybridization of E16.5 and E18 cortices (Fig. 4A). At E16.5, from rostral to caudal, Plxna4 is most prominently expressed in the superficial layers of the cortical plate and intermediate zone with an apparent gradient from lateral to medial regions. At E18, expression is not restricted to the lateral regions of the developing cortex but is now prominently expressed in the entire outer layer of the cortical plate (Fig. 4A), resembling, in part, the expression domain of FoxO6 (Fig. 4B), as shown in Fig. 1. To substantiate the possible functional interaction of Plxna4, FoxO6, and cortical migration, we designed an in utero electroporation experiment in which we overexpress Plxna4 in FoxO6-deficient animals.
Fig. 4.
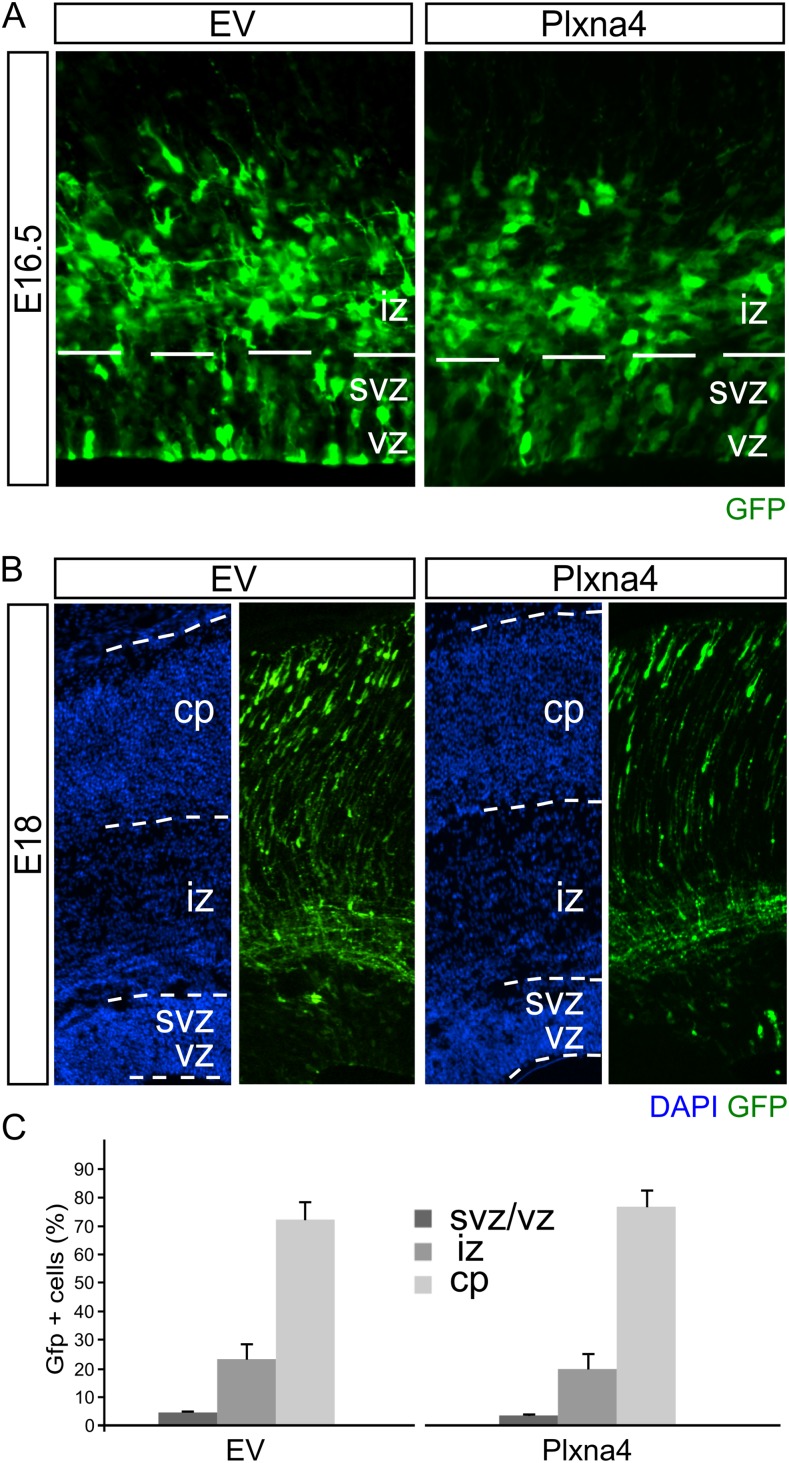
Plxna4 is expressed in the developing cortex and restores radial migration defects in FoxO6+/− animals. (A) In situ hybridization for Plxna4 at E16.5 and E18. At E16.5, from rostral to caudal, Plxna4 is most prominently expressed in the superficial layers of the cortical plate and intermediate zone with an apparent gradient from lateral to medial regions. At E18, expression is not restricted to the lateral regions of the developing cortex but is now prominently expressed in the entire outer layer of the cortical plate (20). (B) Pseudooverlay of adjacent sections analyzed for FoxO6 and Plxna4 expression indicate regional overlap. (C) GFP-traced neurons in FoxO6−/− cortices of E18 embryos in utero electroporated with a GFP expression vector and either empty vector control or Plxna4 expression vector. Cortices were counterstained with DAPI. (D) The distribution of the transfected cells was quantified for the indicated layers for at least three brains per condition using multiple sections per brain. (E) GFP-traced neurons in FoxO6+/− cortices of E18 embryos in utero electroporated with a GFP expression vector and either empty vector control or Plxna4 expression vector. Cortices were counterstained with DAPI. (F) The distribution of the transfected cells was quantified for the indicated layers for at least three brains per condition using multiple sections per brain. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05.
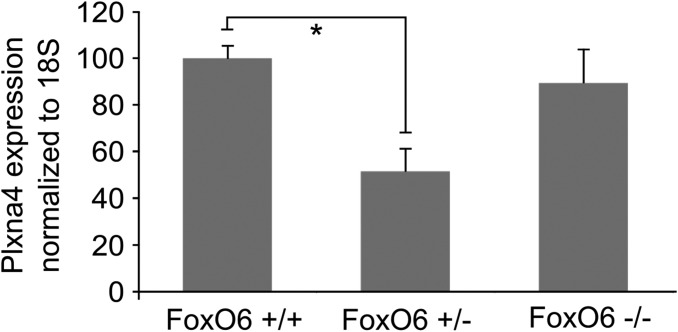
As demonstrated earlier, in FoxO6−/− cortices, radial migration is altered (Fig. 2 A and B), showing GFP-labeled cells primarily in the IZ of the developing cortex. To investigate whether Plxna4 is related to the disrupted radial migration in FoxO6−/− animals, we expressed Plxna4 in these cortices at E14.5 and analyzed the distribution of GFP-labeled cells at E18. Our results show that in FoxO6−/− cortices, ectopic expression of Plxna4 was insufficient to restore normal radial migration (Fig. 4 C and D). To investigate whether expression of Plxna4 is altered in full FoxO6 mutant cortices similar to neurons silenced for FoxO6 (Fig. 3D), we introduced a GFP expression plasmid at E14.5 in cortices and FACsorted the resulting GFP-labeled cells from FoxO6−/−, FoxO6+/−, and FoxO6+/+ cortices at E16.5, followed by qPCR for Plxna4. We did not observe a significant difference in the expression of Plxna4 in FoxO6+/+ and FoxO6−/− cortices. However, we did detect a significant reduction of Plxna4 in FoxO6+/− cortices (Fig. S6; P < 0.05). This indicates that full FoxO6 knockout animals, which lack FoxO6 from the beginning, have compensated for this loss of functionality, suggesting that the migration defect is not caused by limiting levels of Plxna4 in FoxO6−/− animals. However, in developing cortices that do contain FoxO6, but in limiting levels, a reduction of Plxna4 is observed, which would suggest that in such animals Plxna4 overexpression might influence the radial migration events. To investigate this further, we performed a rescue experiment in FoxO6+/− cortices. To this end, we coelectroporated E14.5 cortices with expression plasmids for GFP and Plxna4 or an empty vector (EV) control (Fig. 4 E and F). Importantly, overexpression of Plxna4 in FoxO6+/− cortices significantly increased the amount of GFP-positive cells in the CP (P < 0.05).
Fig. S6.
Plxna4 expression is significantly reduced specifically in FoxO6+/− FACsorted cortical cells. FoxO6+/+, FoxO6+/−, and FoxO6−/− cortices were in utero electroporated with GFP at E14.5, and labeled cells were FACsorted at E16.5 and followed by RNA isolation. Plxna4 expression was measured by qPCR. Only in FoxO6+/− animals was a significant down-regulation of Plxna4 observed. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05.
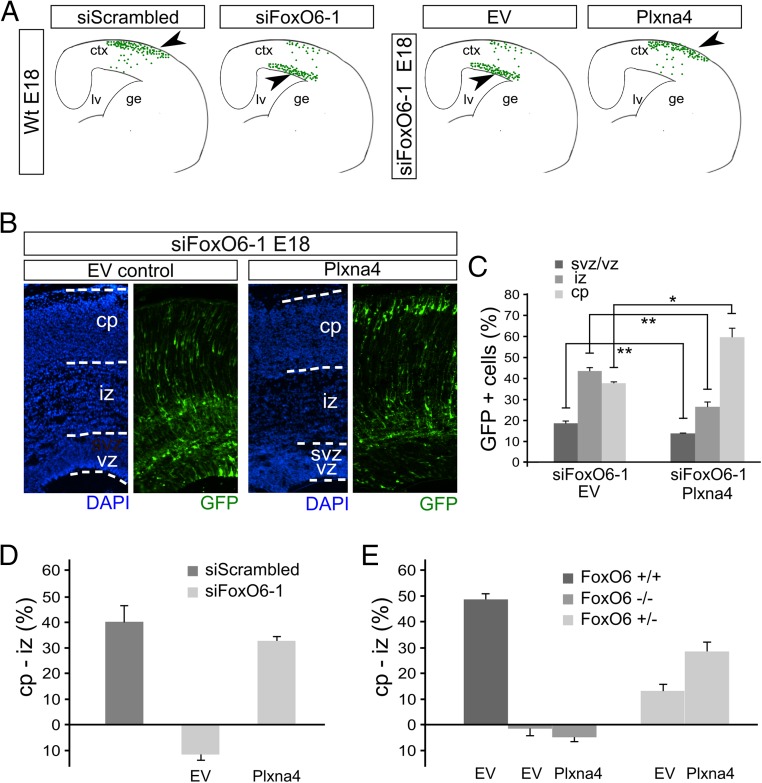
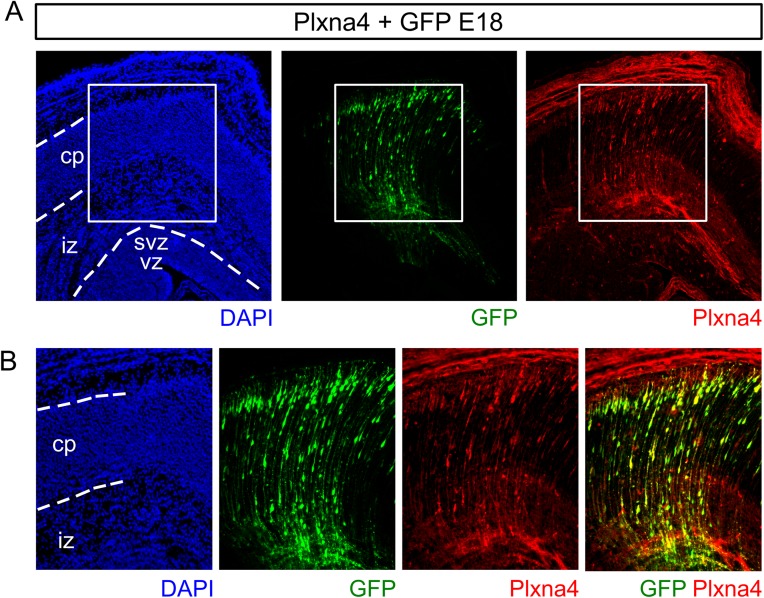
To further substantiate the role of FoxO6 in modulating Plxna4 expression, we performed similar rescue experiments in cortices silenced for FoxO6 expression via siRNA-mediated knockdown. To this end, we coelectroporated E14.5 cortices with siFoxO6-1 and an expression vector for Plxna4 or an EV control. We hypothesized that Plxna4 is able to, at least partly, rescue radial migration in cells deficient for FoxO6, resulting in restored radial migration of transfected cells (Fig. 5A). The overexpression of Plxna4 was confirmed by immunohistochemistry (Fig. S7). Confirming earlier results, cortices electroporated with siFoxO6-1 and EV control (n = 4) showed 18.8% (SD ±1.2) of transfected cells located in the ventricular zone compared with 14.0% (SD ±0.36; P < 0.001) in cortices coelectroporated with siFoxO6-1 and the Plxna4 expression vector (Fig. 5 B and C; n = 4). Furthermore, only 26.6% (SD ±2.4) of the transfected cells in cortices electroporated with Plxna4 were located in the IZ compared with 43.5% (SD ±1.7; P < 0.001) in cortices coelectroporated with siFoxO6-1 and the EV control plasmid. Finally, 59.4% (SD ±4.4) of the GFP-labeled cells reached the cortical plate when coelectroporated with Plxna4 compared with 37.7% (SD ±0.8; P < 0.01) in EV controls (Fig. 5 B and C). In conclusion, our data suggest that cortices electroporated with siFoxO6-1 and a Plxna4 expression vector display restored migration of developing neurons toward the cortical plate compared with FoxO6 knockdown controls. To test the possibility that overexpression of Plxna4 results in improved migration of wild-type neurons by itself, wild-type E14.5 cortices were transfected with Plxna4 or EV control plasmids. Analysis of these cortices at E16.5 and E18 did not show an enhanced migration phenotype (Fig. S8), indicating that the induction of Plxna4 expression above normal levels does not alter radial migration in wild-type neurons. To compare all used genetic models, we chose to calculate the difference in percentage of the GFP cells in the CP and IZ because these two parameters are mainly changed upon FoxO6 modulation (Fig. 5 D and E). This representation clearly shows that the siRNA-mediated knockdown of FoxO6 results in a negative value, indicating that more cells reside in the IZ compared with the CP. A similar pattern is observed in FoxO6−/− animals. However, in FoxO6+/− cortices, the amount of cells is higher in the CP compared with the IZ. This indicates that a mild phenotype is present in FoxO6+/− animals. Introduction of Plxna4 increases the migration to the CP in the knockdown and FoxO6+/− cortices in concordance with the lower Plxna4 levels in both the knockdown and FoxO6+/− cortices. Taken together, our data indicate that radial migration events, in the developing murine cortex, are influenced by FoxO6.
Fig. 5.
Plxna4 restores radial migration in FoxO6 knockdown-induced radial migration defects. (A) Schematic overview of cortical migration phenotypes in FoxO6+/+ cortices transfected with the indicated siRNAs showing hampered radial migration in cortices transfected with siFoxO6-1. Schematic representation of the hypothesis that Plxna4 rescues altered radial migration in cells down-regulated for FoxO6 (Right). Arrowheads point to the hypothesized positions of the neurons in the different treatment situations. (B) GFP-traced neurons in cortices of E18 embryos in utero electroporated with siFoxO6-1, a GFP expression vector, and either empty vector control (20) or Plxna4 expression vector. Cortices were counterstained with DAPI. (C) The distribution of the transfected cells was quantified for the indicated layers for at least three brains per condition using multiple sections per brain. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05 and **P < 0.001. (D) Meta-analysis of the distribution of GFP-labeled cells in cortices treated with siScrambled, siFoxO6-1, and either empty vector control or Plxna4 expression vector. The analysis represents the CP minus IZ value (%). (E) Similar representation as in D for FoxO6+/+, FoxO6+/−, and FoxO6−/−, electroporated with either EV or Plxna4. Introducing Plxna4 restores in part the percentage of GFP-labeled cells in the cortical plate in FoxO6+/− cortices. The error bars show the SD. Wt, wild type.
Fig. S7.
Validation of Plxna4 protein overexpression after IUE with a Plxna4 expression plasmid. (A) Cortices were electroporated with both GFP and Plxna4 expression plasmids. Immunostaining for GFP and Plxna4 reveals that both genes are expressed in transduced cortical cells. (B) Magnifications showing GFP and Plxna4 colocalization.
Fig. S8.
Wild-type cortices overexpressing Plxna4 are unaffected in cortical migration. (A) Cortical migration is unaffected at E16.5 in embryos in utero electroporated at E14.5 with a vector expressing Plxna4 compared with empty vector control. (B) Cortical migration is unaffected at E18 in embryos in utero electroporated at E14.5 with a vector expressing Plxna4 compared with empty vector control. (C) Quantification of duplicate experiments as shown in B. Ratios of GFP+ cells between the (sub)ventricular zone, intermediate zone, and cortical plate are shown. The error bars show the SD.
FoxO6 Interacts with Specific Daf-16–Binding Elements Within the Plxna4 Promoter, Regulating Plxna4 Gene Expression.
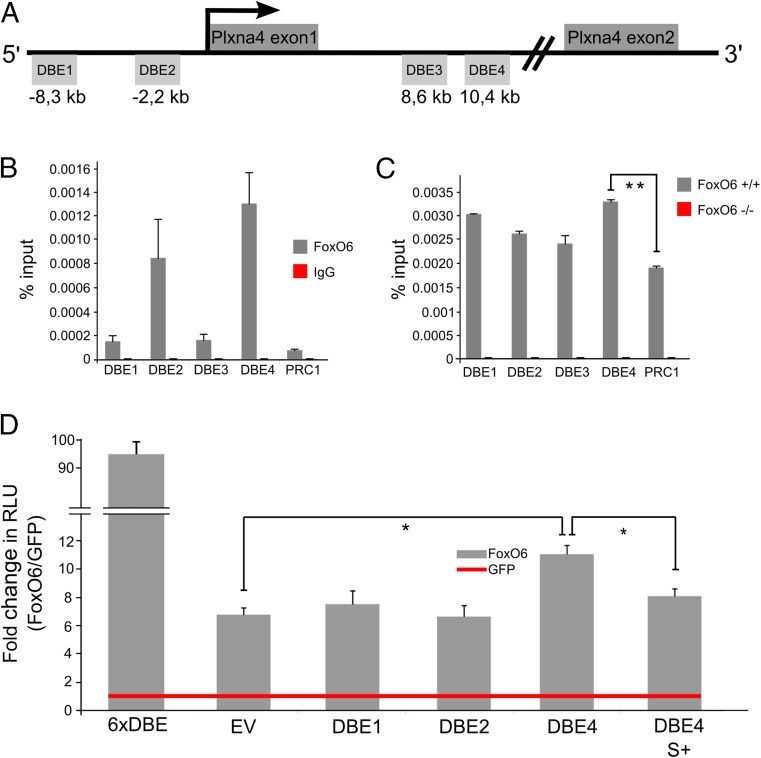
The results as described above indicate that the cortical migration defect, as observed in FoxO6 knockdown models, is mediated by a decrease of Plxna4 expression. To investigate whether Plxna4 transcription is directly regulated by FoxO6 via interaction with four in silico identified Daf-16–binding elements (DBEs) (21) located within 10 kb up- and downstream of the mouse Plxna4 promoter (Fig. 6A), we performed chromatin immunoprecipitation (ChIP) analysis.
Fig. 6.
FoxO6 binds and is able to regulate the Plxna4 promoter via an identified FoxO6 DNA-binding element. (A) Schematic representation of four putative FoxO DBEs in a 10-kb region surrounding the mouse Plxna4 transcription start site. (B) In vivo FoxO6 ChIP-qPCR in E16.5 cortices showing enrichment of DBE2 and DBE4 (n = 2). (C) FoxO6 ChIP-qPCR showing highest enrichment (n = 3) at DBE4 of the Plxna4 promoter. (D) Dual-luciferase assays in 3T3 cells with the different DBEs showing increased activity for DBE4 (n = 3, P < 0.05), which is diminished by the addition of serum (n = 3). Values are normalized to Renilla. DBE, DAF-16–binding element; S+, serum added. The error bars show the SD. Two-tailed Student’s t test; *P < 0.05 and **P < 0.001.
To this end, we performed two independent in vivo FoxO6 ChIP-qPCR experiments on E16.5 cortices. First, we compared the FoxO6 IP and the control IgG IP in FoxO6+/+ cortices. Using this approach, we measured the highest enrichment of FoxO6 at DBE4 (Fig. 6B; n = 2). We then performed a FoxO6 IP on E16.5 FoxO6+/+ and FoxO6−/− cortices. Within this experimental design, FoxO6−/− cortices served as the negative control. Here we observed an enrichment of FoxO6 on each DBE, showing the highest enrichment on the locus covering DBE4 (Fig. 6C; n = 3). We then asked whether binding of FoxO6 to DBEs located in the Plxna4 promoter is able to regulate Plxna4 transcription. To this end, we used part of the Plxna4 promoter with and without confirmed FoxO6-binding elements and cloned this 5′ of a luciferase reporter (pGL3). In this approach, luminescence (measured in relative light units; RLUs) is a readout for the activity of the cloned promoter regions. In these experiments, a positive control (pGL3-6xDBE) (5) showed a large increase in RLUs (FoxO6-GFP/GFP), confirming our approach (Fig. 6D). The pGL3-EV showed a 6.5-fold increase in luminescence (FoxO6-GFP/GFP) (Fig. 6D), suggesting that FoxO6 may regulate transcription in general, as has previously been suggested (22). Importantly, promoter elements containing the confirmed in vivo FoxO6-binding element (pGL3-DBE4) show a significant increase in RLUs (P < 0.01) (FoxO6-GFP/GFP) compared with the control (pGL3-EV) (Fig. 6D). Together, these data indicate that FoxO6 affects Plxna4 transcription through binding to the identified DBE located within the Plxna4 promoter.
It has been described that transcriptional activity of FoxO transcription factors is negatively regulated via insulin and insulin-like growth factors (3–5, 13). This would imply that treatment with insulin, or IGFs in general, would negatively regulate the RLUs activated via the Plxna4 DBE4 enhancer element, thereby resembling the FoxO6 DNA-binding mutant used in the ChIP experiment described above. To investigate this possibility, we repeated the reporter experiments as described above under conditions with and without serum-mediated repression of FoxO6 transcriptional activity. To this end, cells were grown in serum-free media and treated with 10% heat-inactivated FCS (repression of FoxO activity) or kept under serum-free conditions (3, 4, 13). Interestingly, luciferase levels significantly dropped to ∼27% (Fig. 6D) in 3T3 cells grown under serum-containing conditions, compared with luciferase levels in cells grown under serum-free conditions. These results indicate that serum-mediated signaling can modulate DBE4-driven Plxna4 transcription.
Discussion
In this study, we established evidence that FoxO6, a forkhead transcription factor predominantly expressed during cortical development, affects radial migration. We show that in siRNA-mediated knockdown and in FoxO6+/− animals the level of FoxO6 expression is linked to the expression level of Plxna4, a critical component in cortical migration (20). However, in full FoxO6 knockouts, we were unable to rescue the observed migration phenotype, which was confirmed by the absence of a down-regulation of Plxna4. Our data suggest that, although the migration phenotype in FoxO6−/− animals resembles the migration phenotype in neurons conditionally silenced for FoxO6, the underlying molecular mechanisms differ. The cause could lie in the fact that FoxO6 expression in the KO is ablated in neural progenitors before E14.5, the time point of the in utero electroporation. This could affect the molecular signature of early progenitors expressing FoxO6 and as a consequence lead to cortical abnormalities as observed.
Our data demonstrate that in FoxO6−/− cortices a subpopulation of E14.5 neurons fail to localize to the correct position in the developing cortex. Although we did not observe a defect in global cortical markers, we did detect a significant difference in the number of Cux1-positive cells in the upper layers, which supports the notion that a small subgroup is affected by ablation of FoxO6.
The genome-wide transcriptome analysis performed in this study has identified 24 genes that were deregulated upon silencing of FoxO6. Next to Plxna4, the down-regulation of Pik3 catalytic subunit alpha (Pik3ca) was of particular interest. In humans, de novo mutations in core components of the PI3K/AKT pathway, such as Pik3ca, result in a wide array of cortical malformations, including cortical dysplasia, MPPH, and MCAP (6, 7). These mutations generally lead to a gain of function and activation of the PI3K/AKT pathway, which, in turn, results in the phosphorylation of FoxOs, thereby rendering them inactive (23). Moreover, a subset of patients with mutations in PIK3CA suffers from autism or display autistic features, which may imply a role for FoxO6 in the development of autism (6).
Plxna4, the coreceptor for Plxna/NP1 receptors, has been known to be part of the Sema3/6 signaling pathway. The hampered radial migration as observed upon FoxO6 silencing resembles to a great extent the disrupted migration upon the in utero silencing of Plxna2 and Plxna4 (20, 24, 25). This is in line with our hypothesis that FoxO6 may influence radial migration through Plxna4 regulation in the developing cortex.
Studies investigating the role of FoxO transcription factors in mitotic regions of the adult mammalian cortex have illustrated the general importance of FoxO factors in maintaining the population of NSCs (8, 10). Studies in FoxO3-deficient mice have shown that adult NSC homeostasis is regulated by FoxO3, mainly via the regulation of genes involved in quiescence (10). In a combined FoxO1,3,4-deficient mouse model, a progressive decline in adult NSC numbers was observed, also showing deregulation of genes linked to human brain size and the control of cellular proliferation and differentiation (8). Therefore, it is plausible that in the absence of FoxO6 proliferation and NSCs, homeostasis is altered. The numbers of NSCs and intermediate progenitor cells (IPCs) were unchanged in FoxO6+/+ and FoxO6−/− cortices (Fig. S1), suggesting that this effect is subtle and not likely to contribute to the hampered radial migration.
It is known that FoxO proteins are regulated by the PI3K/AKT signaling pathway. In the presence of insulin or IGFs, FoxO proteins are phosphorylated at specific amino acids (3), resulting in decreased DNA binding and shuttling to the cytoplasm (3–5, 13). It is noteworthy that these upstream repressors of FoxO activity are normally present in embryonic cerebrospinal fluid (CSF) (reviewed in ref. 26). Hypothetically, this hints toward a mechanism in which extrinsic factors, present in the embryonic CSF, regulate the transcriptional activity of FoxO proteins, thereby regulating corticogenesis via downstream target genes such as Plxna4.
In addition to the down-regulation of Plxna4, our results revealed an altered expression of a diverse set of genes via which FoxO6 potentially executes its functions during corticogenesis. Remarkably, several of these additional identified genes are known to be involved in the process of neuronal migration, including Dync1h1, a dynein motor protein, and Nrcam, a neuronal cell-adhesion protein associated with psychiatric disorders such as autism (27–29). The expression of Nfia, a family member of Nfix, which has been shown to be involved in regulating the rostral migratory stream of SVZ-derived neuroblasts (30), was, like Plxna4, significantly decreased upon siRNA-mediated knockdown of FoxO6.
The molecular basis of radial migration of cortical neurons is a well-studied process showing prominent roles for axon guidance (20, 31, 32), cell adhesion (33, 34), cell polarity (35, 36), and cytoskeleton remodeling (28, 37, 38). Remarkably, knowledge about transcriptional control of genes involved in radial migration is scarce. Transcription factors known to be important in radial migration include Brn1 and Brn2, regulating Cdk5-mediated cortical lamination (39); Aff3, controlling Mdga2-mediated cortical migration (40); RP58, a zinc finger transcription factor, which negatively regulates Rho family GTPase 2 (Rnd2) (41); COUP-TFI, which also regulates Rnd2 during mid to late corticogenesis (42); and FoxP1, found to regulate morphogenesis of cortical cells, thereby also critical for neuronal migration (43). In this perspective, the study as presented here is important, as it identifies a player in the transcriptional control of neuronal migration during cortical development.
Materials and Methods
Animals.
The C57BL/6J mouse line was used for in utero electroporations silencing FoxO6 expression and rescuing Plxna4 expression. Adult FoxO6 mutant animals (−/−) were backcrossed to the C57BL/6J line and used in heterozygous breeding generating FoxO6 wild-type (+/+) and mutant progeny (−/−). Embryos were collected at E14.5, E16.5, and E18.5 (E0.5 is defined as day of copulatory plug). Genotyping was performed by PCR analysis using the following primers: forward FoxO6-1, 5′-ccagtttgtccagcctcct-3′; reverse FoxO6-1, 5′-cagaggccaggtacacgag-3′; and reverse FoxO6-2, 5′-ctaaagcgcatgctccagac-3′. The FoxO6 wild-type allele produced a band of 257 bp, and the FoxO6−/− allele produced a band of 174 bp in a PCR. All procedures were according to and fully approved by the Dutch Ethical Committees for Animal Experimentation (University Medical Center Utrecht and University of Amsterdam).
In Situ Hybridization.
In short, coronal brain sections (16-μm) were collected using a Leica CM3050 S cryostat. Sections were fixed in 4% (wt/wt) paraformaldehyde (PFA) dissolved in PBS and blocked by acetylation. Next, sections were prehybridized for 2 h in prehybridization solution [50% (vol/vol) deionized formamide, 5× SSC, 5× Denhardt’s solution, 250 μg/mL baker’s yeast tRNA, 500 μg/mL sonicated salmon sperm DNA]. Hybridization was performed overnight at 68 °C with 400 ng probe per mL hybridization mix. The probes used were as follows: FoxO6, base pairs 1514–2430 of mouse coding sequence (CDS) and 3′ UTR; and Plxna4, base pairs 1976–3232 of mouse CDS. The slides were washed in 2× SSC at 68 °C followed by 0.2× SSC at room temperature (RT). Slides were blocked in 10% (wt/wt) heat-inactivated (HI) FCS and incubated overnight at 4 °C with anti-DIG alkaline phosphatase, diluted 1:5,000 in TBS containing 1% HI FCS. Next, the slides were treated with nitro-blue tetrazolium/5-bromo-4-chloro-3’-indolyphosphate solution. Incubation took 4 to 16 h, depending on the probe. The color reaction was stopped in 10 mM Tris⋅HCl, 5 mM EDTA, and the slides were dehydrated with ethanol and embedded using Entellan.
Immunohistochemistry.
E16.5 and E18.0 heads were fixed in 4% (wt/wt) PFA in PBS for 4 and 8 h, respectively, followed by incubation in 30% (wt/wt) sucrose in PBS for at least 48 h. Coronal brain sections (16-μm) were collected using a Leica CM3050 S cryostat. Antigen retrieval was performed for several antibodies. In short, sections were incubated in 10 mM sodium citrate (pH 6.0) for 15 min at 90 °C. Sections were blocked with 4% (wt/wt) HI FCS in THZT (50 mM Tris, 500 mM NaCl, 0.5% Triton X-100, pH 7.6). Primary antibody was incubated overnight at 4 °C. Primary antibodies and dilutions were as follows: rabbit anti-GFP (1:1,000; ab290; Abcam), chicken anti-GFP (1:1,000; ab13970; Abcam), rabbit anti-Sox2 (1:1,000; ab97959; Abcam), rabbit anti-Tbr2 (1:1,000; ab23345; Abcam), rabbit anti-Ctip2 (1:1,000; ab28488; Abcam), mouse anti-Satb2 (1:1,000; ab51502; Abcam), rabbit anti-Cux1 (1:500; sc-13024; Santa Cruz Biotechnology), and Armenian hamster anti-Plxna4 (kindly provided by F. Suto, National Center of Neurology and Psychiatry, Tokyo). Secondary antibodies were incubated at room temperature for 2 h (Molecular Probes; goat anti-rabbit IgG Alexa Fluor 488; goat anti-rabbit IgG Alexa Fluor 594; goat anti-mouse IgG Alexa Fluor 594; goat anti Armenian hamster IgG Alexa Fluor 594). Sections were counterstained with DAPI and embedded in FluorSave. Fluorescent images were taken with a Leica microscope (DFC310 FX).
Transfection and Western Blot.
To test the efficiency of two siRNAs targeting FoxO6, we transfected HEK293 cells, using Lipofectamine 2000, with a plasmid expressing a translational fusion of FoxO6 and GFP, a GFP expression plasmid, and either siScrambled, siFoxO6-1, or siFoxO6-2. At one day posttransfection, the cells were lysed directly in sample buffer [62.5 mM Tris⋅HCl, 2% (wt/wt) SDS, 10% (vol/vol) glycerol, 50 mM DTT, 0.01% bromophenol blue] and sonicated for 5 min for optimal lysis. The lysates were centrifuged at 13,300 × g for 20 min. The supernatant was collected and denatured at 95 °C. Proteins were separated by 10% (wt/wt) SDS/PAGE and blotted onto a 0.2-μm nitrocellulose membrane at 100 V for 2 h at 4 °C. The blot was blocked for 1 h at room temperature in Tris-buffered saline (TBS)-Tween (0.1%) containing 5% (wt/wt) milk, followed by overnight incubation at 4 °C with rabbit anti-GFP (1:50,000; ab290; Abcam). Goat anti-rabbit HRP-conjugated secondary antibody was incubated for 1 h at RT before visualization with ECL detection substrate. After detection of GFP, the blots were washed extensively with TBS-Tween (0.1%). Blots were blocked as described above before incubation with mouse anti–β-actin diluted 1:5,000 in TBS-Tween (0.1%). Goat anti-mouse HRP-conjugated secondary antibody was incubated for 1 h at RT before visualization with ECL substrate. The densitometric analysis was performed using LI-COR Image Studio Lite software.
Quantification and Statistical Analysis.
All cell quantifications represent the average values of experiments performed on multiple animals originating from multiple litters, using at least three sections per brain. Data indicate the means with SD unless stated otherwise. GFP-labeled cortices were subdivided into three bins covering the cortical plate, intermediate zone, and ventricular zone. All GFP-positive cells were counted per bin manually. Statistical analysis was performed by Student’s t test (two-way, unpaired). Bonferroni correction was applied to correct for multiple testing. Sox2, Tbr2, Ctip2, Satb2, and Cux1 countings were performed digitally with ImageJ software (NIH) on 400× magnified thresholded images using optimized parameters. Sox2 quantifications are represented as the percentage of Sox2-positive cells per DAPI-positive cells for a set region covering the VZ and SVZ; n = 4 was used for each condition. Tbr2 quantifications are represented as the percentage of Tbr2-positive cells per DAPI-positive cells for a set region covering the SVZ; n = 4 was used for each condition. Ctip2 quantifications are represented as the percentage of Ctip2-positive cells per DAPI-positive cells for a set region covering layers V and VI; n = 4 was used for each condition. Satb2 quantifications are represented as the percentage of Satb2-positive cells per DAPI-positive cells for a set region covering upper- and deep-layer Satb2-positive pyramidal neurons; n = 4 was used for each condition. Cux1 quantifications are represented as the percentage of Cux1-positive cells per DAPI-positive cells for a fixed region covering upper layers II, III, and IV; n = 3 was used for each condition. qPCR results represent the average values of experiments performed on four biological samples for each condition, originating from multiple litters. Data indicate means with SD unless stated otherwise. Statistical analysis was performed by Student’s t test (two-way, unpaired). The average width of the cortex was measured using ImageJ software. Previously performed DAPI counterstaining was used to identify the width of the indicated layers in E18 FoxO6+/+ and FoxO6−/− brains. The width of the cortex was measured at three fixed locations per section and then averaged per section.
In Utero Electroporation.
C57BL/6J mice at 14 d of gestation were anesthetized by injecting a mixture of ketamine/medetomidin/atropine intraperitoneally. The uteruses of these mice were exposed and a mixture containing 1 μg/μL siRNA (siFoxO6-1 or siScrambled), 1 μg/μL expression vector containing GFP and/or Plxna4, and 0.01% Fast Green dissolved in 20 mM Tris⋅HCl (pH 8) was injected through pulled glass capillaries, using a microinjector, into one lateral ventricle of each embryo. We used custom-made siRNAs (Thermo Scientific) to silence FoxO6 (siFoxO6-2, 5′-NNACAUGCCAGUGACGACUAC-3′; siFoxO6-1, 5′-NNCCAUCAGCCUUAUGACGCU-3′) and a nontargeting siRNA (Thermo Scientific; D-001210-03-20). The expression vectors used were pCAG-AFP 2424 (GFP) and pcDna3.1 (-) Plexin-A4 (kindly provided by R. J. Pasterkamp, Brain Center Rudolf Magnus, University Medical Center Utrecht). Next, electric pulses were generated by an ECM 830 Square Wave Electroporation System and applied to the cerebral wall at five repeats of 33 V for 50 ms with an interval of 950 ms. After the electroporation the uterus was placed back and the mouse was taken care of. Depending on the analysis, the embryos developed to stage E16.5 or stage E18 before they were killed.
Microarray and qPCR Validation.
Embryos were electroporated at E14.5 with either siFoxO6-2 or siFoxO6-1, both targeting FoxO6 specifically or the control siScrambled. All cortices were cotransfected with GFP. At E16.5, embryos were harvested, and the cortex was isolated and dissociated using a papain kit according to the manufacturer’s protocol (Worthington Biochemical) followed by FACsorting. Sorted cells were subjected to RNA isolation using the phenol-chloroform extraction method. Electroporated cortices from different nests were pooled to minimize effects due to electroporation differences and nest differences. All RNA samples were analyzed using a 2100 Bioanalyzer (Agilent Technologies) to ensure the quality of the RNA. Microarray analysis was performed using four experimental samples (three cortices pooled) in utero electroporated with siFoxO6-1 and four experimental samples in utero electroporated with siFoxO6-2. These samples were compared with a reference pool of RNA derived from eight siScrambled electroporated cortices. Per siRNA, two samples were labeled normal and two in a dye swap against the common reference, and hybridized on a mouse expression microarray. Agilent Mouse Whole Genome Gene Expression Microarray V1 (Agilent Technologies) sets were used for all hybridizations, in a 4 × 44 K layout. Hybridized slides were scanned on an Agilent scanner (G2565BA) at 100% laser power, 30% photomultiplier. Data were analyzed using ANOVA (R version 2.2.1/MAANOVA version 0.98-7; www.r-project.org/). P values were determined by a permutation F2 test. Genes with P < 0.05 after family-wise error correction (or Benjamini–Hochberg correction/FDR control) were considered significantly changed. The data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE71954 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71954). Top hits from the array were validated with qPCR. Analysis was performed on a LightCycler 480 II (Roche) using a One Step SYBR Green Kit (Qiagen) according to the manufacturer’s protocol. Independent in utero electroporated samples were FACsorted, followed by RNA isolation. Water was used as a nontemplate control. All samples were normalized to an 18S reference. Primers used for qPCR amplification were as follows: 18S (60 °C): forward 5′-aaacggctaccacatccaag-3′, reverse 5′-cctccaatggatcctcgtta-3′; FoxO6 (60 °C): forward 5′-agagcgccccggacaagaga-3′, reverse 5′-gccgaatggagttcttccagcc-3′; Plxna4 (60 °C): forward 5′-gcggattgaaccagaatggagc-3′, reverse 5′-ttcctccatgcttggcacggat-3′; Nfia (60 °C): forward 5′-agttgagccgagttggaaatg-3′, reverse 5′-tggggcagaagtgcttcaatg-3′; Pik3ca (60 °C): forward 5′-ttatgaaccagtaggcaaccg-3′, reverse 5′-gctatgaggcgagttgagatcc-3′; Xlr3c (60 °C): forward 5′-gaaacatgccgaaacactctct-3′, reverse 5′-agcgatggaatttccccga-3′; and Lmx1a (60 °C): forward 5′-aaccagcgagccaagatgaa-3′, reverse 5′-cccgcattcccactaccatt-3′.
ChIP-qPCR.
In vivo ChIP was performed on E16.5 FoxO6+/+ and FoxO6−/− cortices. Cells were cross-linked using 1% PFA followed by sonication (optimized for neuronal tissue to generate 400- to 1,000-bp DNA fragments). Cross-linked samples were incubated overnight with either rabbit anti-FoxO6 (kindly provided by A. Brunet) or rabbit IgG control followed by incubation with preblocked protein A agarose beads. Beads were extensively washed (nine times) in different salt washing buffers, and DNA–histone complexes were eluted using elution buffer (1% SDS, 0.1 M NaHCO3). Samples were reverse–cross-linked and treated with proteinase K. DNA was purified and used for qPCR analysis. qPCR primers are specified below. In vitro ChIP was performed similarly. In short, different mouse cell lines (3T3) were transfected with either pcDNA3.1 FoxO6-Myc or N1-FoxO6-GFP using Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Life Technologies). Cells were cross-linked using 1% PFA followed by sonication. The latter sonication procedure was optimized for each cell line to generate DNA fragments of 200 to 1,000 bp. Cross-linked samples were incubated overnight with either rabbit anti-Myc or rabbit anti-GFP and the rabbit IgG control followed by incubation with preblocked protein A agarose beads. Beads were extensively washed in different salt washing buffers, and DNA–histone complexes were eluted using elution buffer (1% SDS, 0.1 M NaHCO3). Samples were reverse–cross-linked and treated with proteinase K. DNA was purified and used for qPCR analysis. In silico search for FoxO DBEs revealed four FoxO DBEs in the 10 kb surrounding the transcription start site (TSS) of Plxna4. Primers used for qPCR amplification were as follows: DBE_plexina4_2.2kbupstream_TSS (60 °C): forward 5′-tgcacattctttccctgtca-3′, reverse 5′-gtgtgtgtcctgatggatgg-3′; DBE_plexina4_8.6kbdownstream_TSS (60 °C): forward 5′-tcaatacctttccctgatgttg-3′, reverse 5′-accacacgtcaatactcaccac-3′; DBE_Plexina4_8.3kbupstream_TSS (60 °C): forward 5′-ccctttctcctctctccaca-3′, reverse 5′-gccactaggtaaggctggaa-3′; DBE_Plexina4_10.4kbdownstream_TSS (60 °C): forward 5′-cccagctgcacttaaggaaa-3′, reverse 5′-ccttcattccccttgaggat-3′; 6xDBE (60 °C): forward 5′-tacgcggtgctagcccg-3′, reverse 5′-gcaggcatgcaagcttactt-3′.
Luciferase Assay.
A total of ∼400 bp surrounding the DBEs was independently cloned into the pGL3 promoter vector. 3T3 cells (neuronal cell lines died when overexpressing FoxO6) were grown in 24-well plates and transfected with 2 μg plasmid DNA per well (N1-FoxO6-GFP or N1-GFP) and cotransfected with 0.2 μg pGL3 promoter vector and 0.02 μg CMV-Renilla using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were serum-starved to activate FoxO6. After 8 h, cells were lysed and assayed for luciferase activities using a Dual-Luciferase Reporter Assay (Promega; E1910) according to the manufacturer’s protocol. All experiments were performed in triplicate. Data represent average values.
FACsorting.
Freshly dissected cortices were dissociated using a papain dissociation system (Worthington Biochemical), and cells were sorted on a Cytopeia Influx or BD FACSAria III cell sorter. Sort gates were set on forward scatter versus side scatter (life cell gate), forward scatter versus pulse width (elimination of clumps), and forward scatter versus fluorescence channel 1 (528/38 filter; GFP fluorescence). Cells were sorted using a 100-μm nozzle at a pressure of 15 or 20 psi (BD FACSAria III) with an average speed of 5,000 cells per s and collected in TRIzol-LS reagent (Invitrogen).
Supplementary Material
Acknowledgments
We are grateful to Prof. Dr. O. Reiner for the aGFP construct used in the in utero electroporations; Prof. Dr. B. M. Burgering for the pGL3-6xDBE vector; Prof. Dr. R. J. Pasterkamp for the Plxna4 vector; Prof. Dr. A. Brunet for the FoxO6−/− mouse model; and Dr. F. Suto for providing the Armenian hamster anti-Plxna4 antibody. We thank Dr. F. M. J. Jacobs for critical reading of the manuscript, and Dr. F. van der Kloet for help with statistical analysis. This work was supported by a Dutch Brain Foundation (HSN) grant (to M.P.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE71954).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609111113/-/DCSupplemental.
References
- 1.Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol Cell Biol. 2001;21(10):3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Heide LP, Smidt MP. Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem Sci. 2005;30(2):81–86. doi: 10.1016/j.tibs.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.van der Heide LP, Jacobs FM, Burbach JP, Hoekman MF, Smidt MP. FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo-cytoplasmic shuttling. Biochem J. 2005;391(Pt 3):623–629. doi: 10.1042/BJ20050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: Mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380(Pt 2):297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kops GJ, Burgering BM. Forkhead transcription factors: New insights into protein kinase B (c-Akt) signaling. J Mol Med (Berl) 1999;77(9):656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 6.Mirzaa GM, Rivière JB, Dobyns WB. Megalencephaly syndromes and activating mutations in the PI3K-AKT pathway: MPPH and MCAP. Am J Med Genet C Semin Med Genet. 2013;163C(2):122–130. doi: 10.1002/ajmg.c.31361. [DOI] [PubMed] [Google Scholar]
- 7.Rivière JB, et al. Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ro SH, Liu D, Yeo H, Paik JH. FoxOs in neural stem cell fate decision. Arch Biochem Biophys. 2013;534(1–2):55–63. doi: 10.1016/j.abb.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salih DA, et al. FoxO6 regulates memory consolidation and synaptic function. Genes Dev. 2012;26(24):2780–2801. doi: 10.1101/gad.208926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Torre-Ubieta L, et al. A FOXO-Pak1 transcriptional pathway controls neuronal polarity. Genes Dev. 2010;24(8):799–813. doi: 10.1101/gad.1880510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs FM, et al. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278(38):35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 14.Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6(2):134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31(5):727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 16.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 17.Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007;448(7156):901–907. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- 18.Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999;22(2):277–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, et al. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11(1):36–44. doi: 10.1038/nn2018. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349(Pt 2):629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaves I, et al. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol. 2014;24(11):1248–1255. doi: 10.1016/j.cub.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Hay N. Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta. 2011;1813(11):1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Gunput RA, Pasterkamp RJ. Semaphorin signaling: Progress made and promises ahead. Trends Biochem Sci. 2008;33(4):161–170. doi: 10.1016/j.tibs.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Homman-Ludiye J, Bourne JA. The guidance molecule Semaphorin3A is differentially involved in the arealization of the mouse and primate neocortex. Cereb Cortex. 2014;24(11):2884–2898. doi: 10.1093/cercor/bht141. [DOI] [PubMed] [Google Scholar]
- 26.Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiorillo C, et al. Novel dynein DYNC1H1 neck and motor domain mutations link distal spinal muscular atrophy and abnormal cortical development. Hum Mutat. 2014;35(3):298–302. doi: 10.1002/humu.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirier K, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45(6):639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marui T, et al. Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int J Neuropsychopharmacol. 2009;12(1):1–10. doi: 10.1017/S1461145708009127. [DOI] [PubMed] [Google Scholar]
- 30.Heng YH, et al. NFIX regulates proliferation and migration within the murine SVZ neurogenic niche. Cereb Cortex. 2015;25(10):3758–3778. doi: 10.1093/cercor/bhu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polleux F, Morrow T, Ghosh A. Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature. 2000;404(6778):567–573. doi: 10.1038/35007001. [DOI] [PubMed] [Google Scholar]
- 32.Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383(6600):525–528. doi: 10.1038/383525a0. [DOI] [PubMed] [Google Scholar]
- 33.Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol. 2008;18(3):245–250. doi: 10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonosaki M, et al. L1cam is crucial for cell locomotion and terminal translocation of the soma in radial migration during murine corticogenesis. PLoS One. 2014;9(1):e86186. doi: 10.1371/journal.pone.0086186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saillour Y, et al. Beta tubulin isoforms are not interchangeable for rescuing impaired radial migration due to Tubb3 knockdown. Hum Mol Genet. 2014;23(6):1516–1526. doi: 10.1093/hmg/ddt538. [DOI] [PubMed] [Google Scholar]
- 36.Sapir T, et al. Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons. J Neurosci. 2013;33(29):11932–11948. doi: 10.1523/JNEUROSCI.5425-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 38.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19(7):347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295(5559):1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 40.Moore JM, et al. Laf4/Aff3, a gene involved in intellectual disability, is required for cellular migration in the mouse cerebral cortex. PLoS One. 2014;9(8):e105933. doi: 10.1371/journal.pone.0105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng JI, et al. The zinc finger transcription factor RP58 negatively regulates Rnd2 for the control of neuronal migration during cerebral cortical development. Cereb Cortex. 2015;25(3):806–816. doi: 10.1093/cercor/bht277. [DOI] [PubMed] [Google Scholar]
- 42.Alfano C, et al. COUP-TFI promotes radial migration and proper morphology of callosal projection neurons by repressing Rnd2 expression. Development. 2011;138(21):4685–4697. doi: 10.1242/dev.068031. [DOI] [PubMed] [Google Scholar]
- 43.Li X, et al. Foxp1 regulates cortical radial migration and neuronal morphogenesis in developing cerebral cortex. PLoS One. 2015;10(5):e0127671. doi: 10.1371/journal.pone.0127671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.