Significance
To form organs of correct size and proportion, growth must be tightly controlled. Previous studies have characterized how biochemical signals influence organ growth; this report describes an interrelationship between tissue mechanics and organ growth. We show that differential growth leads to accumulation of mechanical stress within tissues and describe both theoretically and experimentally how this mechanical stress can result in reduced tension within faster-growing cells. We show how this reduced tension can increase the activity of the Hippo signaling pathway, which decreases growth rates, and show that this mechanism influences patterns of cell proliferation in vivo. Our results support and extend a theoretical model, termed “mechanical feedback,” that described the relationship between growth rates and tissue mechanics.
Keywords: Ajuba, tissue mechanics, Drosophila development, cytoskeleton, Hippo
Abstract
Mechanical stress can influence cell proliferation in vitro, but whether it makes a significant contribution to growth control in vivo, and how it is modulated and experienced by cells within developing tissues, has remained unclear. Here we report that differential growth reduces cytoskeletal tension along cell junctions within faster-growing cells. We propose a theoretical model to explain the observed reduction of tension within faster-growing clones, supporting it by computer simulations based on a generalized vertex model. This reduced tension modulates a biomechanical Hippo pathway, decreasing recruitment of Ajuba LIM protein and the Hippo pathway kinase Warts, and decreasing the activity of the growth-promoting transcription factor Yorkie. These observations provide a specific mechanism for a mechanical feedback that contributes to evenly distributed growth, and we show that genetically suppressing mechanical feedback alters patterns of cell proliferation in the developing Drosophila wing. By providing experimental support for the induction of mechanical stress by differential growth, and a molecular mechanism linking this stress to the regulation of growth in developing organs, our results confirm and extend the mechanical feedback hypothesis.
Growth regulation is needed to form organs of correct size and proportion, but the mechanisms that define organ and organism size remain poorly understood (1). Cells in a developing organ are exposed to multiple growth factors, at concentrations that can vary depending upon cellular location, developmental stage, and nutrition. Signaling pathways that conduct these biochemical signals have been extensively studied, and in many cases their contributions to growth control are well characterized. However, in addition to the biochemical environment, cells in a developing organ also experience a mechanical environment in which they are subject to forces through their contact with neighboring cells and the extracellular matrix. The mechanical environment has also been proposed to modulate organ growth, yet how this occurs and what it contributes to in vivo growth regulation remains unclear.
The Hippo signaling pathway plays an essential role in regulating organ growth from arthropods through vertebrates (2, 3). One remarkable feature of Hippo signaling is its role as an integrator of growth control signals (Fig. 1A). Indeed Hippo signaling is influenced by or cross-talks with multiple biochemical pathways that can act as general growth regulators, or promote growth linked to positional information, nutritional status, or developmental stage. Hippo signaling is also affected by contacts with neighboring cells and the extracellular matrix, and by mechanical stress (4). However, most prior studies of Hippo pathway regulation by mechanical cues have examined it either in the context of in vitro models or used nonphysiological manipulations such as drugs or mutations that disrupt the cytoskeleton, leaving unanswered the question of how Hippo signaling might modulate growth in response to mechanical stresses that cells experience during developmental or physiological processes.
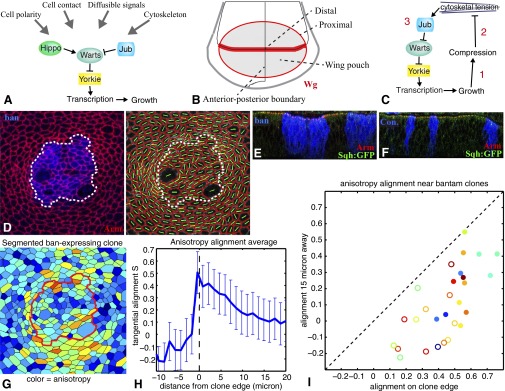
Fig. 1.
Overview, and tissue distortion induced by ban-expressing clones. (A) Simplified schematic of the Hippo pathway. (B) Schematic of the wing imaginal disc. (C) Schematic illustrating the key elements of the mechanical feedback loop: 1, growth can compress cells; 2, compression of cells can decrease cytoskeletal tension; and 3, resulting decrease in cytoskeletal tension is sufficient to trigger changes in the localization of Jub and Wts, and in Yki activity. (D) Cell anisotropy analysis of ban-expressing clones. Green lines indicate long axis, which just outside the clone tends to align parallel to the clone boundary. (E) Vertical section through a wing disc with ban-expressing clones, marked by BFP (blue). These clones typically induce distortions of the epithelium that include apical invaginations and lateral bulging. (F) Vertical section through a wing disc with control clones (BFP expressing). (G–I) Influence of ban-expressing clones on cell orientation. (G) Example of segmentation (Left, color scale shows anisotropy). (H) Average alignment of cell elongation axis relative to clone boundary as a function of the distance to clone boundary (Center, n = 29). (I) Summary analysis of clones, plotting average level of alignment at the clone edge versus 15 μm away for each clone. Color indicates relative clone area (as a heat map, red is bigger, blue is smaller), filled circles identify clones closer to the center of the wing pouch, and open circles identify clones farther from the center of the wing pouch. The analysis shows cells tend to align parallel to the clone boundary at the clone edge but not 15 μm away, and this effect is more pronounced for clones closer to the center of the wing.
Hippo signaling regulates growth by controlling a transcriptional coactivator protein, Yorkie (Yki) (5, 6) (Fig. 1A). Yki activity is down-regulated through phosphorylation by the Warts (Wts) kinase, which promotes cytoplasmic localization of Yki. Wts is regulated in several ways, including phosphorylation by Hippo (7), regulation of Wts abundance (8), regulation of Wts localization (9, 10), and regulation of Wts interaction with cofactors and inhibitors (11–15). We recently defined a mechanism for biomechanical regulation of Hippo signaling in which cytoskeletal tension induces recruitment of Ajuba LIM protein (Jub) to α-catenin, and thence recruitment of Wts to Jub (11). Jub is an inhibitor of Wts (12, 13), and this tension-dependent corecruitment of Jub and Wts to adherens junctions is associated with reduced Wts activity, and hence increased Yki activity (11), at least in part because it prevents Wts from moving to sites where Wts gets activated (9). Conversely, under conditions of lower cytoskeletal tension, recruitment of Wts and Jub to adherens junctions is decreased, Wts activity is increased, and consequently Yki activity is decreased.
A consideration of the mechanical stresses cells might experience in a growing organ and their influence on growth was provided by the mechanical feedback model (16). This model argued that differential growth could lead to local tissue compression as faster-growing cells push against surrounding slower-growing cells, and proposed that this compression might then decrease growth, thereby restoring even growth rates and reducing further compression. Mechanical feedback is thus a negative feedback that limits the extent to which a population of cells can overgrow, potentially providing a homeostatic mechanism that ensures cells proliferate at similar rates to minimize tissue distortion. In this context, it has been suggested that it might explain how cell proliferation in organs like the Drosophila wing can be homogeneous despite inhomogeneous distributions of growth factors. The hypothesis that growth-induced compression inhibits further growth, along with the observation that cells are more tightly packed at later stages of development, has also been suggested as an explanation for why organs stop growing when they reach their final size (16–19).
Although mechanical feedback provides an attractive hypothesis for contributions of mechanics to organ size control, it has lacked direct experimental support or a molecular mechanism. Here, we use the wing imaginal discs of Drosophila (Fig. 1B) to perform experimental tests of mechanical feedback. Wing disc cells form a pseudostratified epithelial monolayer, connected to each other at apical adherens junctions, which are attached to an actin–myosin network that is under tension (20). Our analysis takes advantage of recent progress in characterizing Jub-mediated biomechanical Hippo signaling to define three critical stages of mechanical feedback (Fig. 1C). We first test the prediction that differential growth can lead to mechanical stress, and observe that accumulation of stress is most pronounced under conditions where mechanical feedback has been bypassed. We then show that mechanical stress induced by differential growth is associated with reduced cytoskeletal tension within faster-growing cells, and explain this using a theoretical model of epithelial mechanics that incorporates adaptive tension. We next show that the Jub biomechanical Hippo pathway is influenced by differential growth through these associated reductions in cytoskeletal tension. Finally, we use this understanding of mechanical feedback to demonstrate its role in modulating cell proliferation during wing development. These studies confirm the mechanical feedback hypothesis, provide a mechanistic basis for mechanical feedback, and argue that mechanical feedback contributes to growth regulation in vivo.
Results
Reduced Cytoskeletal Tension Within Faster-Growing Clones.
One prediction of mechanical feedback is that a coherent population of cells growing at a faster rate than surrounding cells will become compressed (16). We tested this prediction by using UAS-Gal4–driven expression to create clones of faster-growing cells. The microRNA gene bantam (ban) promotes growth in Drosophila (21) (SI Appendix, Fig. S1A). Moreover, ban is a key downstream target of Yki, and ban can promote growth even in the absence of Yki (22, 23). Thus, if mechanical feedback acts through Yki, forced expression of ban under UAS-Gal4 control could bypass mechanical feedback, enabling growth to continue and tissue compression to accumulate. Indeed, ban-expressing clones in the wing disc deform surrounding cells, visible as anisotropic distortions with elongation aligned parallel to the edges of ban-expressing clones (Fig. 1 D and G–I). Radial compression and azimuthal elongation of cells is exactly the pattern of deformation expected outside of overgrowing regions, as follows from continuum elasticity considerations elaborated in SI Appendix, SI Text. Tissue distortion is also evident through a widening of clones between the apical and basal surfaces and invagination of the apical and basal surfaces (Fig. 1 E and F). All of these distortions are most evident near the center of the disc, where cells are already more compressed (24, 25). Similar distortions have been reported for clones of cells expressing activated Yki or mutant for wts (24–26), which should also bypass any mechanical feedback that depends upon regulation of Yki activity.
Overexpression of ban, or expression of an activated form of Ras (RasV12) that promotes growth (27), also result in a consistent reduction in apical accumulation of Non-Muscle Myosin II (myosin, visualized using GFP fusions to myosin regulatory light chain, Sqh, or to myosin heavy chain, Zip) (Fig. 2 A, F, and G and SI Appendix, Fig. S2B). Reduced myosin activity can also lead to reduced F-actin accumulation in wing discs (11), and consistent with this finding, we observed modest reductions in F-actin levels within faster-growing clones (SI Appendix, Fig. S2 B, E, and F). As myosin both creates and responds to tension (28), the reduction in its levels implies that cytoskeletal tension is lower within these fast-growing clones.
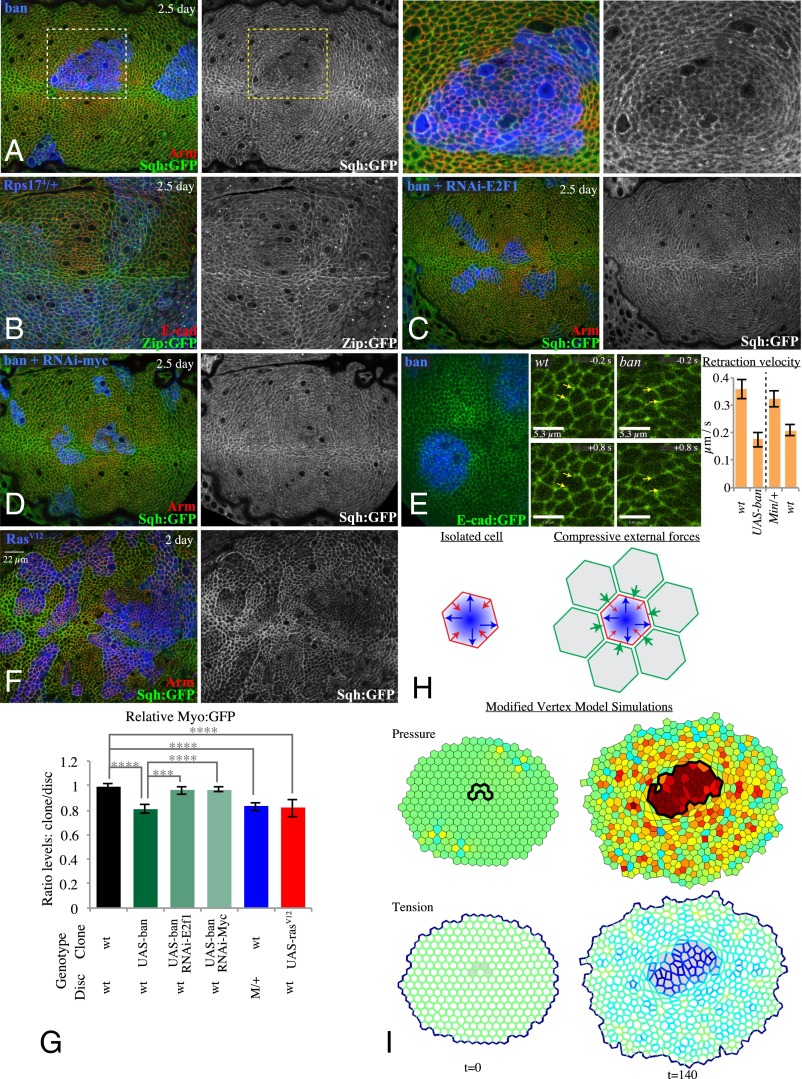
Fig. 2.
Reduced tension within faster-growing clones. (A) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and myosin labeled by Sqh:GFP (green/white), showing reduced junctional myosin within clones. (Right) Higher magnification of the boxed regions. (B) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by absence of β-gal marker (blue), with cell junctions labeled by E-cad (red) and myosin labeled by Zip:GFP (green/white). (C and D) Wing discs with clones coexpressing ban and RNAi-E2f1 (C) or RNAi-Myc (D) grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and myosin labeled by Sqh:GFP (green/white); reduction in junctional myosin is suppressed. (E) Assessment of junctional tension by laser cutting. Junctions were cut within live discs with clones labeled by mCD8:RFP and junctions labeled by E-cad:GFP, examples of regions of discs inside and outside of ban-expressing clones 0.2 s before and 0.8 s after cutting are shown (Movies S1 and S2); histogram shows mean retraction velocities measured from 46 (Left pair, UAS-ban clones in wild type) or 42 (Right pair, wild-type clones in Minute/+) pairs of cuts; error bars indicate 95% confidence interval. (F) Wing disc with clones of Rasv12-expressing cells grown for 2 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and myosin labeled by Sqh:GFP (green/white). (G) Histogram showing relative levels of Myo:GFP in cells within clones of the indicated genotypes, compared with cells outside of the clones at similar proximal–distal locations within the same wing disc. Values and numbers of clones analyzed are tabulated in SI Appendix, Fig. S2B. Comparisons of the significance (by one-way ANOVA) of differences between some of these mean ratios is indicated by the gray lines; ****P < 0.0001, ***P < 0.001. (H) Cartoon illustrating balancing forces that maintain cell shape. Internal cell pressure reflecting compression of nuclei and other cytoplasmic components generates an expanding force (blue arrows) that is balanced by myosin-generated cortical tension (red arrows). External forces provided by interaction with neighboring cells could compress (green arrows) cells, altering cell shapes, pressures, and tensions. (I) Snapshots of the modified vertex model-based simulation (Movies S3 and S4) showing altered cellular pressures and tensions that result from differences in growth rates. In this simulation (SI Appendix provides details), intrinsic cell area is larger for a clone of faster growing cells (identified by black line above and gray shading below). Pressure is increased and tension is reduced as cells are constrained within an area smaller than their intrinsic size. Relative pressures and tensions are indicated by color scale (red, high; blue, low).
Expression of ban or RasV12 in clones results in faster-growing cells surrounded by wild-type cells. Differential growth can also be introduced by creating a mosaic between wild-type cells and slow-growing cells. This can be done in Drosophila using Minute mutations, which are mutations in genes required for ribosome function that cause a dominant slow-growth phenotype (29). Moreover, because Minute mutations simply reduce the capacity for protein synthesis, they are not expected to directly increase myosin levels. Nonetheless, wild-type cells surrounded by cells heterozygous for Minute mutations exhibit lower apical and junctional myosin accumulation than their Minute/+ neighbors (Fig. 2 B and G and SI Appendix, Fig. S2B). Thus, cells growing at a faster rate than their neighbors consistently have reduced levels of myosin. Wild-type clones within Minute/+ discs are not, however, associated with anisotropy of neighboring cells or invaginations of the disc epithelium (SI Appendix, Fig. S1 B and C), which might reflect a sensitivity to mechanical feedback that reduces growth rates to prevent extreme tissue compression.
To confirm that the reduced myosin accumulation observed within faster-growing clones is associated with decreased junctional tension, we used a junction cutting assay in which a laser is used to sever cell–cell junctions (20). The retraction velocities of vertices neighboring the cut were lower within ban-expressing clones than in comparable regions of the wing disc without clones, and lower within wild-type clones than in comparable regions with Minute/+ cells (Fig. 2E and Movies S1 and S2). Thus, differences in growth rates between neighboring cell populations can lead to decreased tension along cell junctions within the faster-growing cells.
To further confirm that altered myosin levels occur as a consequence of differential growth, we suppressed the overgrowth of ban-expressing clones by using RNAi to decrease the expression of either of two different genes that are required for normal clone growth: E2f1 and Myc (30, 31). In both cases, the reduction of myosin in ban-expressing clones was suppressed (Fig. 2 C, D, and G and SI Appendix, Fig. S2B), as was distortion of the disc epithelium (SI Appendix, Fig. S1D). Conversely, RNAi of E2f1 or Myc in otherwise wild-type cells did not significantly affect myosin (SI Appendix, Fig. S2 B–D). Thus, reduced levels of myosin in ban-expressing clones can be attributed to their elevated growth rates.
A Model of Epithelial Mechanics That Produces Decreased Tension in Compressed Cells.
Our observations raised the question of whether compression of faster-growing cells could reduce their cytoskeletal tension. For an isolated cell, cell shape reflects a balance between (myosin-generated) cortical tension and intracellular pressure (reflecting compression of cellular components) (Fig. 2H). Within an epithelium, cells also experience external forces arising from interaction with neighboring cells in the tissue. If these external forces are compressive, then one would expect cell areas to become smaller than their intrinsic size (as defined by the balance between cortical tension and intracellular pressure in the absence of external forces). However, live tissue does not behave as a simple elastic medium as cells also adapt their mechanical properties to external forces. For example, when an external force pulls on cells, they increase actin–myosin contractility (28). Conversely, when subject to compressive forces, as is the case for faster-growing clones constrained by surrounding slower-growing cells, then we expect that the same cellular response would cause cells to reduce their cytoskeletal tension. In both cases, the compensating response reduces cell deformation; we demonstrate this explicitly by formulating and analyzing a physical model of cell area with adaptive response to external mechanical stress (SI Appendix, SI Text).
To illustrate the sufficiency of this model to explain the reduced tension within faster-growing clones, we developed a simulation of a 2D epithelial sheet containing a clone growing more rapidly than the surrounding cells. The simulation is based on a vertex model representing epithelial tissue as a polygonal array of cells, with cell geometry determined by minimization of mechanical energy (representing intracellular pressure, cortical tension, and mechanical interaction between adjacent cells). Cell growth and proliferation is represented by continuously increasing the intrinsic area of each cell followed, stochastically, by cell division. In contrast with earlier vertex models (20), we allow for an adaptive cytoskeletal response, decreasing cortical tension proportionally with increasing pressure acting on a cell. The model of adaptive cytoskeletal response used here is consistent with the recently proposed “active tension network” theory (32), which postulated that the rate of myosin recruitment (or release) into the actin–myosin cortex and concomitant changes in cortical tension is modulated by changes in mechanical stress. Model simulations considering cell growth and resulting heterogeneity of pressure confirm that differential clone growth leads to higher cellular pressure, and lower cortical tension, within faster-growing cells (Fig. 2I and Movies S3 and S4). This theoretical description of epithelial mechanics thus reproduces the lower tension observed within faster-growing clones. The model also illustrates the characteristic pattern of cellular deformation surrounding faster growing clones: cells close to the clone are strongly elongated parallel to the clone boundary. (In SI Appendix, SI Text we also provide an explicit mathematical description of elastic deformation surrounding an expanding disc, which helps to understand the observed pattern of cell anisotropy that surrounds clones.)
In the model simulations, tension along clone boundaries is intermediate between the lower tension in the interior of clones and higher tension outside of clones, and it is also influenced by clone and cell shapes (Fig. 2I). This relatively higher tension at clone edges compared with the clone interior is consistent with experimental observations that the area of reduced myosin sometimes (9/30 clones scored) appears smaller than the area of the clone (Fig. 2A).
Faster-Growing Cells Reduce Wts and Jub Localization to Adherens Junctions.
Changes in cytoskeletal tension induced by genetic manipulations can modulate Hippo signaling through the tension-dependent recruitment of Jub to adherens junctions (11). To determine whether the reduced tension observed within faster-growing clones is sufficient to alter Jub recruitment, we assayed Jub localization using a genomic Jub:GFP line (33). Indeed, Jub levels at adherens junctions were lower within clones of faster-growing cells compared with their neighbors. Reduction in Jub was most obvious within ban-expressing clones (Fig. 3 A and J and SI Appendix, Fig. S3J), but was also visible within Rasv12-expressing clones and wild-type clones surrounded by Minute/+ cells (Fig. 3 B, C, and J and SI Appendix, Fig. S3J). Thus, differences in growth rates between neighboring populations of disc cells are sufficient to reduce cytoskeletal tension within faster-growing clones in a manner and to a degree that decreases the recruitment of Jub to adherens junctions. As with the reduction in myosin, the area of lower Jub levels sometimes (7/28 clones scored) appeared slightly smaller than the clone (Fig. 3A). This finding is generally consistent with our modeling of tension in response to changes in pressure, which results in intermediate levels of tension along clone borders (Fig. 2I).
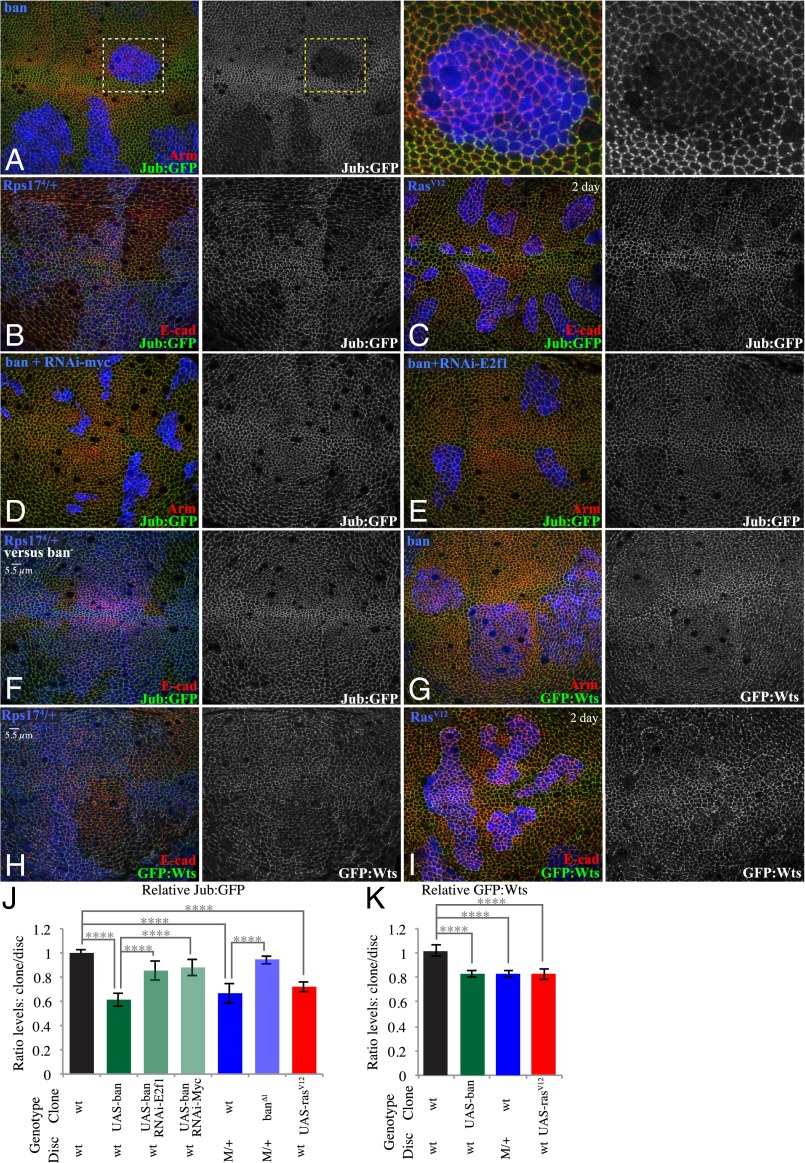
Fig. 3.
Reduced junctional Jub and Wts within faster-growing clones. (A) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and Jub labeled by Jub:GFP (green/white). (Right) Higher magnification of the boxed regions. (B) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by absence of β-gal marker (blue), with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (C) Wing disc with clones of RasV12-expressing cells grown for 2 d, labeled by coexpression of 2xBFP, with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (D and E) Wing disc with clones coexpressing ban and RNAi-Myc (D) or RNAi-E2f1 (E) grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and Jub labeled by Jub:GFP (green/white). (F) Minute heterozygous (Rps174) wing disc with clones of ban∆1 mutant cells grown for 2.5 d, labeled by absence of β-gal marker (blue), with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (G) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and Wts labeled by Wts:GFP (green/white). (H) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by absence of β-gal marker (blue), with cell junctions labeled by E-cad (red) and Wts labeled by Wts:GFP (green/white). (I) Wing disc with clones of RasV12-expressing cells grown for 2 d, labeled by coexpression of 2xBFP, with cell junctions labeled by E-cad (red) and Wts labeled by GFP:Wts (green/white). (J and K) Histograms showing relative levels of Jub:GFP (J) and GFP:Wts (K) in cells within clones of the indicated genotypes, compared with cells outside of the clones at similar proximal–distal locations within the same wing disc. Values and numbers of clones analyzed are tabulated in SI Appendix, Fig. S3J. Comparisons of the significance (by one-way ANOVA) of differences between some of these mean ratios is indicated by the gray lines; ****P < 0.0001.
We confirmed that this influence of ban-expressing clones on Jub is largely due to their elevated growth rates by suppressing their overgrowth through knockdown of Myc or E2f1. This manipulation largely reversed the lower levels of junctional Jub (Fig. 3 D, E, and J and SI Appendix, Fig. S3J), whereas knockdown of Myc or E2f1 on their own did not visibly influence Jub (SI Appendix, Fig. S3 E, F, and J). Similarly, suppressing the growth differential between wild-type clones and Minute/+ neighbors by replacing wild-type clones with ban∆1 mutant (slow growing) clones (21) suppressed the decrease in Jub levels associated with non-Minute clones in Minute/+ discs (Fig. 3 F and J and SI Appendix, Fig. S3J), whereas ban∆1 mutant clones in a wild-type background did not visibly affect Jub levels (SI Appendix, Fig. S3 D and J). These observations confirm that elevated growth rates in clones are sufficient to lower Jub recruitment to adherens junctions within the faster-growing cells.
To confirm that the reductions in junctional levels of Jub within faster-growing cells were also associated with a reduction in corecruitment of Wts, we monitored Wts localization using a genomic Wts:GFP line (11). Wts levels at adherens junctions were lower within ban-expressing clones, Rasv12-expressing clones, and wild-type clones surrounded by Minute/+ cells (Fig. 3 G–I and K and SI Appendix, Fig. S3J). Thus, a variety of manipulations that alter growth rates through distinct biochemical mechanisms all trigger a similar biomechanical response, which includes reduced junctional accumulation of both Jub and Wts.
To confirm that this decreased Jub and Wts accumulation within faster-growing clones is due to the decreased cytoskeletal tension that we detected, as opposed to other potential effects of these genotypes, we increased cytoskeletal tension within faster-growing clones by expressing an activated form of the myosin regulatory light chain (Sqh.EE) (34). Indeed, coexpression of Sqh.EE suppressed the reduction of junctional Jub within ban- or RasV12-expressing clones, as well as within wild-type clones in Minute/+ discs (Fig. 4 A–D and E). For wild-type clones in Minute/+ discs, we also confirmed that the reduction of junctional Wts is suppressed by expression of Sqh.EE (SI Appendix, Fig. S3 H–J).
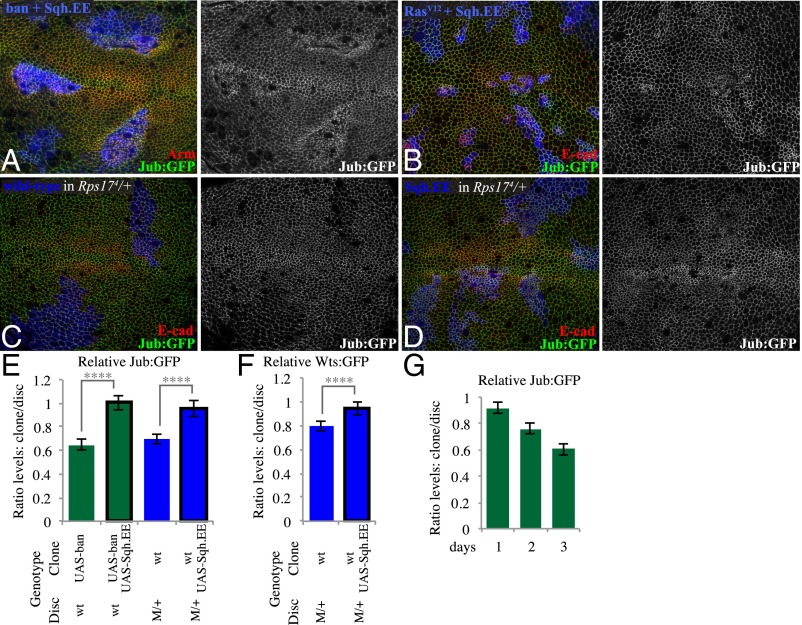
Fig. 4.
Increasing myosin activity suppresses reductions of Jub in faster-growing clones. (A) Wing disc with clones coexpressing ban and Sqh.EE grown for 2.5 d, labeled by coexpression of 2xBFP, with cell junctions labeled by Arm (red) and Jub labeled by Jub:GFP (green/white). (B) Wing disc with clones coexpressing RasV12 and Sqh.EE grown for 2 d, labeled by coexpression of 2xBFP, with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (C) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by presence of BFP marker (blue) using MARCM, with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (D) Minute heterozygous (Rps174) wing disc with clones of cells expressing Sqh.EE grown for 2.5 d, labeled by presence of BFP marker (blue) using MARCM, with cell junctions labeled by E-cad (red) and Jub labeled by Jub:GFP (green/white). (E and F) Histograms showing relative levels of Jub:GFP (E) and GFP:Wts (F) in cells within clones of the indicated genotypes, compared with cells outside of the clones at similar proximal–distal locations within the same wing disc. Values and numbers of clones analyzed are tabulated in SI Appendix, Fig. S3J. Comparisons of the significance (by one-way ANOVA) of differences between some of these mean ratios is indicated by the gray lines; ****P < 0.0001. (G) Quantitation of relative Jub levels over time (SI Appendix, Fig. S4), based on paired measurements inside and outside of ban-expressing clones, 1 (n = 14), 2 (n = 13), or 3 (n = 13) d after clone induction; error bars indicate confidence intervals.
Spatial and Temporal Pattern of Jub Regulation.
The tension-dependent recruitment of Jub and its inhibition of Wts position Jub as a key link connecting cytoskeletal tension to Hippo signaling (11). Thus, we focused on Jub localization as a marker for evaluation of how faster-growing clones, exemplified by ban-expressing clones, influence cytoskeletal tension relevant to Hippo signaling. ban-expressing clones tend to have a stronger reduction of Jub in more distal regions of the wing disc (i.e., nearer the center of the disc, Fig. 1B) than in more proximal regions (SI Appendix, Fig. S3B). Cells nearer the center of the wing disc are more compressed and have lower junctional tension than cells in more proximal regions (24, 25, 35). These observations suggest that mechanical feedback is more evident where surrounding cells are already more compressed. The influence of ban-expressing clones also increased with the duration of clone growth, being barely detectable after 1 d, clearly visible after 2 d, and pronounced after 3 d (Fig. 4G and SI Appendix, Fig. S4). For comparison, a GFP-ban sensor that is a direct target of ban (21) was affected similarly between distal and proximal regions of the wing, and at 2 or 3 d after clone induction (SI Appendix, Fig. S4). These observations are consistent with the inference that the reductions in Jub localization at junctions associated with faster-growing clones result from growth-induced compression, as compression is expected to continually increase within ban-expressing clones.
Faster-Growing Clones Reduce Yki Activity.
Decreased recruitment of Wts and Jub to adherens junctions induced by genetic inhibition of myosin activity is associated with increased Wts activity, and consequently decreased Yki activity (11). To confirm that Yki activity is lower within faster-growing clones, we examined both Yki localization and the expression of Yki target genes. We were unable to examine the influence of mechanical feedback on Yki within RasV12-expressing clones because RasV12 acts upstream of Yki and promotes growth at least in part through Yki activation (36–38).
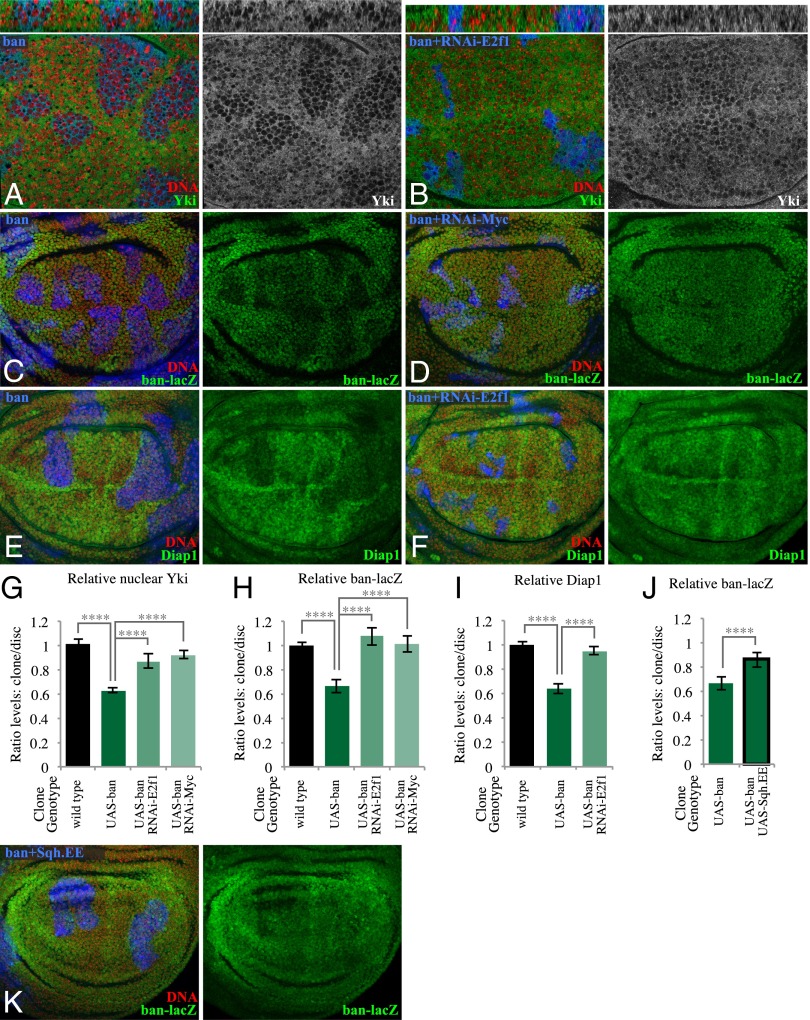
Nuclear localization of Yki was clearly lower within ban-expressing clones (Fig. 5 A and G and SI Appendix, Fig. S6E). Consistent with this finding, two different reporters of Yki’s transcriptional activity, ban-lacZ and Diap1, were also decreased within ban-expressing clones in the developing wing (Fig. 5 C, E, H, and I). We note that ban was previously reported not to influence Yki activity (22, 23), based mostly on analysis of a distinct reporter, ex-lacZ. In our hands the influence of ban-expressing clones on ex-lacZ was complex, as ex-lacZ was increased in proximal regions of the wing, but not in distal regions (SI Appendix, Fig. S5K). The observation that the influence of ban on ex-lacZ does not match its influence on Yki localization suggests that ex is also regulated by specific targets of the ban microRNA, and based on the consistent reductions in nuclear Yki, and in two out of three downstream target genes examined, we infer that Yki activity is lower within ban-expressing clones, consistent with the changes in Jub and Wts localization.
Fig. 5.
Influence of ban-expressing clones on Yki activity. (A) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of UAS-mRFP (blue), and stained for DNA (Hoechst, red) and Yki (green/white), showing reduced nuclear Yki in clones. Thin panels above show vertical sections. (B) Wing disc with clones of cells coexpressing ban and a UAS-RNAi construct targeting E2f1, grown for 2.5 d, labeled by coexpression of 2xBFP (blue), stained for DNA (red) and Yki (green/white). Thin panels above show vertical sections. (C) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of 2xBFP (blue), stained for DNA (red) and ban-lacZ (green). (D) Wing disc with clones of cells coexpressing ban and a UAS-RNAi construct targeting Myc, grown for 2.5 d, labeled by coexpression of 2xBFP (blue), stained for DNA (red) and ban-lacZ (green). (E) Wing disc with clones of ban-expressing cells grown for 2.5 d, labeled by coexpression of 2xBFP, and stained for expression of Diap1 (green). (F) Wing disc with clones of cells coexpressing ban and a UAS-RNAi construct targeting E2f1, grown for 2.5 d, labeled by coexpression of 2xBFP (blue), stained for DNA (red) and Diap1 (green). (G–J) Histograms showing relative levels of nuclear Yki (G), ban-lacZ (H and J), and Diap1 (I) in cells within clones of the indicated genotypes, compared with wild-type cells outside of the clones at similar proximal–distal locations within the same wing disc. Values and numbers of clones analyzed are tabulated in SI Appendix, Fig. S6E. Comparisons of the significance (by one-way ANOVA) of differences between some of these mean ratios is indicated by the gray lines; ****P < 0.0001. (K) Wing disc with clones coexpressing ban and Sqh.EE grown for 2.5 d, labeled by coexpression of 2xBFP (blue), and stained for DNA (red) and ban-lacZ (green); the reduction of ban-lacZ is suppressed.
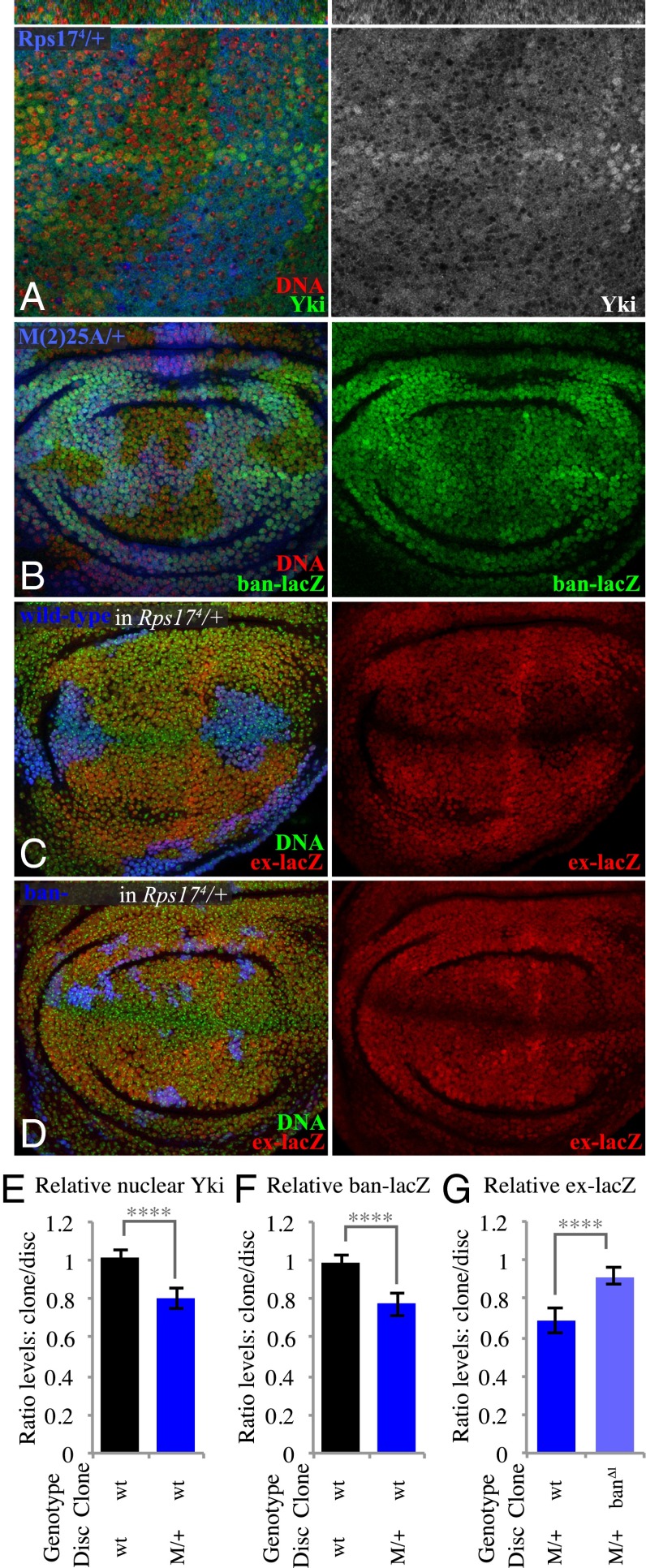
We also observed a reduction in Yki activity within wild-type clones in Minute/+ mosaic discs, visible both through decreased nuclear Yki and decreased expression of Yki target genes, including both ban-lacZ and ex-lacZ (Fig. 6 A–C and E–G and SI Appendix, Fig. S6E). Thus, two very different means of creating differential growth rates, ban-expressing clones in a wild-type disc and wild-type clones in a Minute/+ disc, both reduce Yki activity within the faster-growing cells. Together with the observations described above, this implies that faster-growing clones trigger a mechanical feedback that down-regulates Yki activity. Consistent with this conclusion, coexpression of Sqh.EE suppressed the decrease in Yki activity both within ban-expressing clones and within wild-type clones in Minute/+ discs (Fig. 5 J and K and SI Appendix, Fig. S6 A–C and E).
Fig. 6.
Influence of wild-type clones in Minute heterozygotes on Yki activity. (A) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by absence of β-gal marker (blue), and stained for DNA (Hoechst, red) and Yki (green/white), showing reduced nuclear Yki. Thin panels above show vertical sections. (B) Minute heterozygous (M(2)25A) wing disc with clones of wild-type cells grown for 2.5 d, labeled by absence of GFP marker (blue), and stained for DNA (red) and ban-lacZ (green). (C) Minute heterozygous (Rps174) wing disc with clones of wild-type cells grown for 2.5 d, labeled by presence of GFP marker (blue) using MARCM, and stained for DNA (green) and ex-lacZ (red). (D) Minute heterozygous (Rps174) wing disc with clones of ban∆1 mutant cells grown for 2.5 d, labeled by presence of GFP marker (blue) using MARCM, and stained for DNA (green) and ex-lacZ (red). (E–G) Histograms showing relative levels of nuclear Yki (E), ban-lacZ (F), and ex-lacZ (G) in cells within clones of the indicated genotypes, compared with nonclone cells outside of the clones at similar proximal–distal locations within the same wing disc. Values and numbers of clones analyzed are tabulated in SI Appendix, Fig. S6E. Comparisons of the significance (by one-way ANOVA) of differences between some of these mean ratios is indicated by the gray lines; ****P < 0.0001.
To further confirm this relationship between differential growth and regulation of Yki, we suppressed the overgrowth of ban-expressing clones compared with their wild-type neighbors, or the growth of wild-type clones compared with their Minute/+ neighbors. Within ban-expressing clones, RNAi of E2f1 or Myc suppressed the influence of ban-expressing clones on Yki activity (Fig. 5 B, D, F, and G–I and SI Appendix, Fig. S6E), whereas RNAi of these genes in otherwise wild-type cells does not significantly influence Yki activity (SI Appendix, Fig. S5). Similarly, reduction of the growth differential between Minute/+ cells and non-Minute cells using a ban mutation suppressed the reduced Yki activity within non-Minute cells (Fig. 6 D and G and SI Appendix, Fig. S6E), whereas ban mutant clones in a wild-type background do not increase Yki activity (SI Appendix, Fig. S6 D and E) (22, 23). These observations indicate that down-regulation of Yki activity is occurring within faster-growing clones as a consequence of their elevated growth rates.
Blocking Mechanical Feedback Alters Patterns of Cell Proliferation.
The results described above establish that mechanical feedback can be induced in vivo by creating clones of cells that grow at different rates. We next considered the question of how mechanical feedback might contribute to growth control during normal development, by genetically preventing mechanical feedback (Fig. 7A). As mechanical feedback acts on Yki, and ban promotes growth despite the absence of Yki (22, 23), forced expression of ban under UAS-Gal4 control effectively bypasses mechanical feedback. Because mechanical feedback acts through Jub-mediated regulation of Wts, knockdown of Wts should also block mechanical feedback. Forced activation of myosin could also suppress mechanical feedback, because it suppresses the down-regulation of myosin activity that would otherwise be introduced by tissue compression. Indeed, genetic activation of myosin suppressed the down-regulation of Jub and Yki that would otherwise occur within ban-expressing clones (Figs. 4 and 5).
Fig. 7.
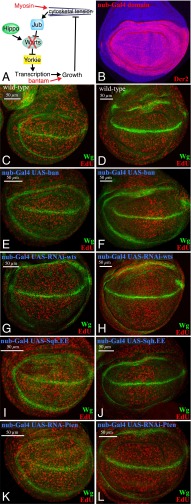
Influence of mechanical feedback on cell proliferation. (A) Schematic illustration of mechanical feedback loop, with points of experimental blockage or bypass in red. (B) Wing disc expressing UAS-Dcr2 under nub-Gal4 control, stained for Dcr2 (red), to illustrate the nub-Gal4 expression domain. (C–L) Wing discs subject to EdU labeling (red) for proliferating cells, and stained for Wingless (Wg, green), which is expressed along the dorsal/ventral boundary and also encircles the developing wing. Wild-type (C and D), nub-Gal4 UAS-ban (E and F), nub-Gal4 UAS-RNAi-wts (G and H), nub-Gal4 UAS-Sqh.EE (I and J), and nub-Gal4 UAS-Pten RNAi (K and L) are shown. (Left) (C, E, G, I, and K) Discs from midthird instar larvae; (Right) (D, F, H, J, and L) Discs from late third instar larvae. Cell proliferation is normally evenly distributed at these stages, but is relatively higher in the middle of the developing wing when mechanical feedback is bypassed (F, H, and J).
Thus, to assess the consequences of suppressing mechanical feedback, we expressed ban, a transgene directing RNAi against wts, or Sqh.EE. They were expressed under nub-Gal4 control, which drives expression throughout the developing wing (Fig. 7B). Normally, cell proliferation is relatively evenly distributed in wing discs, as can be visualized by EdU labeling (Fig. 7 C and D). In contrast, when mechanical feedback was blocked, cell proliferation at later larval stages was relatively higher in the medial region of the developing wing (Fig. 7 F, H, and J). Increasing growth induced through an alternate method not expected to block mechanical feedback, knockdown of the tumor-suppressor Pten did not result in the same spatial bias in disc cell proliferation (Fig. 7L). These observations imply that the mechanical feedback mechanism that we identified modulates patterns of cell proliferation during normal wing development. The pattern that emerged is relevant to a long-standing question in the field: How is it that cells near the center of the disc experience higher levels of the key growth factor Decapentaplegic (Dpp), yet proliferate at similar rates as cells far from the Dpp source (39)? A variety of models have been proposed to explain this observation, including one class of models that essentially invoke the mechanical feedback hypothesis (16–19). According to these models, uniform growth rates arise because higher mitogenic signaling in the center of the disc is counterbalanced by higher compression as cell numbers increase. Our observation that three distinct manipulations that suppress mechanical feedback lead to higher cell proliferation in the medial region of the wing disc, where Dpp signaling is higher, is consistent with this class of models.
Discussion
The hypothesis that organ growth is modulated by tissue mechanics in vivo has been popular, but untested. A key obstacle precluding attempts to investigate the role of mechanics in growth control has been the difficulty of distinguishing mechanical effects from influences on genetic or biochemical regulatory pathways. We have overcome this by identifying and characterizing biomechanical pathways that regulate growth, by analyzing combinations of genotypes whose only common feature is their influence on growth rates and by characterizing influences of differential growth rates on tissue mechanics.
To establish that mechanical stresses within the range of what cells normally experience in vivo can influence growth, we took advantage of the insight, predicted by the mechanical feedback hypothesis, that local differences in growth rates should lead to mechanical strain. Indeed we found that this mechanical strain is particularly evident under conditions where mechanical feedback is blocked, as, for example, when ban is overexpressed. Moreover, we observed reduced myosin accumulation and consequent effects on Hippo signaling components within wild-type cells, simply because surrounding cells have a reduced ribosome function, and hence reduced growth. This observation cannot be attributed to any direct, genetic regulation of myosin within clones. We assayed overgrowth clones induced using two independent genotypes (ban expressing and RasV12 expressing), each of which has a distinct relationship to Hippo signaling, yet found they share this same reduction in myosin, Jub, and Wts accumulation. It was recently reported that RasV12-expressing clones did not autonomously reduce myosin levels in the pupal notum under conditions where RasV12 expression was induced after clone growth was essentially completed (40); this observation further supports our conclusion that the reduction in myosin we observed is not due to RasV12 expression per se, but rather the promotion of clone growth. Moreover, we observed that the influence of ban-expressing clones (or wild-type clones in a slow-growth disc) could be suppressed by knocking down the expression of single genes that reduce growth, but do not themselves directly regulate myosin. Together, these observations (SI Appendix, Fig. S7A) indicate that Hippo signaling is activated within faster growing clones as a consequence of cellular compression, rather than through biochemical pathways dependent upon the various genotypes analyzed.
Quite generally, mechanical feedback regulation of growth is a homeostatic mechanism: by suppressing local overgrowth, it reduces tissue distortion. Indeed, genotypes that we now know could block mechanical feedback (expression of ban or activated Yki, or knockdown of Wts) produce an evident strain within the disc epithelium, presumably because they allow growth-induced compression to accumulate. Conversely, wild-type clones within Minute/+ discs, which are presumably susceptible to mechanical feedback, are not associated with equivalent distortions of the epithelium and also have weaker effects on myosin, Jub, Wts, and Yki than ban-expressing clones. Additional processes could also contribute to minimizing distortions of epithelial tissues in response to differential growth. For example, differential growth is sometimes associated with a process of cell competition, in which slower-growing “loser” cells become eliminated by apoptosis. This removal of neighboring cells effectively creates space for faster-growing cells to occupy and thus presumably reduces the compression, and ultimately mechanical feedback, that would otherwise be exerted on the faster-growing clones. The adaptive, mechanical stress-dependent recruitment of myosin into the cortical cytoskeleton that we invoke to explain reduced myosin within compressed cells serves as another, cell-level, homeostatic mechanism that reduces deformation in response to stress.
In addition to confirming the existence of mechanical feedback, our observations have identified a mechanism by which it occurs. This mechanism is initiated by a reduction in cytoskeletal tension within faster-growing clones, which we suggest stems from the responsiveness of myosin recruitment to stress exerted on actin–myosin networks (28). The decreased tension that results from cellular compression then triggers a biomechanical response that includes decreased recruitment of Jub and Wts to adherens junctions, and consequently decreased activity of Yki. Given the crucial role of Yki in promoting organ growth, this down-regulation of Yki should be sufficient to reduce growth rates, and thus altogether our observations establish a mechanism for compression-induced growth inhibition. The Jub-dependent biomechanical pathway that is modulated by mechanical feedback was discovered in the context of direct manipulations of myosin activity using transgenes or drugs (11). The present study extends our understanding of the regulation of this pathway by establishing that stress experienced by cells in vivo as a simple consequence of differential growth is both qualitatively and quantitatively sufficient to modulate the Jub biomechanical pathway (SI Appendix, Fig. S7B).
Mechanical feedback is a homeostatic mechanism, and to the extent that the growth of clones expressing an oncogene like RasV12 is suppressed, mechanical feedback could provide a tumor suppressor function. However, mechanical feedback is not limited to situations where cells have genetically determined differences in growth rates. It could also occur under any conditions of growth-induced compression and is an attractive mechanism to compensate for naturally occurring variation in growth rates. Indeed, by blocking mechanical feedback, we identified an influence on the normal patterning of cell proliferation within the developing wing. The elevated cell proliferation observed in the center of the wing disc, near the anterior–posterior compartment boundary, is consistent with the hypothesis that higher mitogenic signaling in the center of the disc is normally balanced by higher mechanical compression, which triggers mechanical feedback. Thus, when compression-induced growth suppression is bypassed by genetic manipulations that suppress mechanical feedback, higher cell proliferation is observed in the center of the wing disc. Thus, our observations also implicate mechanical feedback in the normal patterning of growth within developing organs.
Materials and Methods
Drosophila Culture.
Unless otherwise indicated, crosses were performed at 25 °C. For clone induction, larvae were grown on standard medium at 25 °C. In most cases, larvae at 108 ± 6 h after egg laying (AEL) were fixed and used in the study. Heat shocks were performed at 37 °C for 5–10 min, 2–3 d before dissection. For ban overexpressing clones grown for 1, 2, or 3 d, larvae at 108 ± 6 h AEL were dissected and heat shocked at 37 °C for 5–10 min. For wild-type, Sqh.EE-expressing, or ban∆1 clones in Minute/+ backgrounds, larvae of 120 ± 6 h AEL were used because of their developmental delay; and heat shocks were performed at 37 °C for 10 min, 2.5 d before dissection.
To induce ectopic expression clones, act > y+> Gal4 with UAS-2xBFP (described below) or UAS-mCD8:RFP (gift of G. Morata, Universidad Autónoma de Madrid, Madrid), or act > Cd2 > Gal4 UAS-nRFP (Bloomington 30558) were crossed to the following UAS transgenes with hs-Flp: UAS-bantam (21), Gs-Bantam (41), UAS-Myc (30), UAS-p35 [3-W] (gift of B. Hay, Caltech, Pasadena, CA), UAS-Yki:V5 (42), UAS- Rasv12 (27), UAS-sqh.E20E21 (34), UAS-Myc-RNAi [Vienna Drosophila RNAi Center (VDRC) 2948], UAS-E2F1-RNAi (VDRC 15886), and UAS-Dcr2. The UAS-2xBFP transgene was made by inserting two inframe copies of tagBFP into pUAST.
Protein or gene localization and expression was monitored using previously characterized transgenes: ex-lacZ, ban-lacZ, Jub:GFP (33), Wts:GFP (11), zip:GFP, sqh:GFP (43), and Ubi-Ecad:GFP (44).
For inducing wild-type clones in Minute/+ backgrounds, FLP–FRT-mediated recombination was performed using: y w hs-Flp; FRT80B, y w hs-Flp; FRT40A, y w hs-Flp; FRT80B Wts:GFP, y w hs-Flp; FRT80B Jub:GFP, y w hs-Flp; tub-Gal4 UAS-mCD8:RFP; Rps174 tub-Gal80 FRT80B/TM6B, w; Rps174 arm-lacZ FRT80B/TM6B (Bloomington 6358), hs-Flp tub-Gal4 UAS-GFP y w; Rps174 tub-Gal80 FRT80B/TM6B (Bloomington 42732), M(2)25A Ubi-GFP FRT40A/CyO, w hs-FLP; and arm-lacZ M(2)z FRT40A/CyO (FBst1001673). For making ban mutant clones, ban∆1 FRT80B/TM6B (45) was crossed to either hs-Flp; Rps174 arm-lacZ FRT80B/TM6B or y w hs-Flp; arm-lacZ FRT80B (Bloomington 6341). To increase myosin activity within wild-type clones surrounded by Minute/+ neighbors, mosaic analysis with a repressible cell marker (MARCM) clones were made using y w hs-Flp; tub-Gal4 UAS-mCD8:RFP; Rps174 tub-Gal80 FRT80B/TM6B, hs-Flp tub-Gal4 UAS-GFP y w; Rps174 tub-Gal80 FRT80B/TM6B and UAS-Sqh.EE; FRT80B.
For making bantam overexpressing and wild-type clones in the same disc, a TIE-DYE stock (46) was crossed to hs-Flp; UAS-bantam flies.
For the EdU labeling experiments, nub-Gal4 UAS-dcr2 was crossed to UAS-ban, UAS-wts-RNAi[vdrc101475], UAS-sqh.EE, or UAS-Pten-RNAi[vdrc9928].
Histology and Imaging.
For most experiments, discs were fixed in 4% (wt/vol) paraformaldehyde for 15 min at room temperature. Wts:GFP discs were fixed for 8 min, and Sqh:GFP or Zip:GFP discs were fixed for 12 min. Primary antibodies used were rabbit anti-Yki (1:400) (47), mouse anti–β-galacotsidase [1:200, Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Wg (1:400 DSHB), mouse anti-Diap1 (1:200, B. Hay), rat anti–E-cad (1:400 DCAD2; DSHB), mouse anti-Armadillo (1:200, DSHB), rabbit anti-cleaved Caspase (1:400 Dcp-1; Cell Signaling Technology). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and Invitrogen. F-actin was stained using Alexa Fluor 488-phalloidin (Life Technologies), and DNA was stained using Hoechst (Invitrogen) or TO-PRO-3 (Life Technologies). Confocal images were captured on a Leica SP5 or a PerkinElmer Ultraview.
For EdU labeling, larvae were dissected in Ringer’s solution and anterior halves were immediately placed in 250 μL WM1 (48). An equal volume of 20 μM EdU (Click-iT EdU Alexa Fluor 594 Imaging Kit; Life Technologies) in WM1 was added for a final concentration of 10 μM EdU and samples were incubated for 10 min. Tissue was then fixed for 20 min with 4% paraformaldehyde in PBS. Subsequent standard antibody staining protocol using mouse anti-WG (1:400 DSHB) was then followed by 30 min of EdU detection using 0.6 μL Alexa Fluor azide 594 per 500 μL Click-iT reaction mixture. Afterward, tissues were treated with Hoechst. Wing discs were removed and mounted on a slide in Vectashield.
Supplementary Material
Acknowledgments
We thank Cordelia Rauskolb for EdU labeling experiments, assistance with confocal microscopy, and comments on the manuscript and N. Baker, K. Basler, S. Cohen, A. Garcia-Bellido, B. Hay, L. Johnston, D. Kiehart, G. Morata, R. Padgett, J. Zallen, the Developmental Studies Hybridoma Bank, and the Bloomington Stock Center for antibodies and Drosophila stocks. This research was supported by NIH Grant R01 GM078620 and the Howard Hughes Medical Institute (K.D.I.), National Science Foundation (NSF) PHY-1220616, Gordan and Betty Moore Foundation 2919 (to B.I.S.), and NSF PHY11-25915.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615012113/-/DCSupplemental.
References
- 1.Hafen E, Stocker H. How are the sizes of cells, organs, and bodies controlled? PLoS Biol. 2003;1(3):E86. doi: 10.1371/journal.pbio.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu FX, Guan KL. The Hippo pathway: Regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng Z, Moroishi T, Guan K-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun S, Irvine KD. Cellular organization and cytoskeletal regulation of the Hippo signaling network. Trends Cell Biol. 2016;26(9):694–704. doi: 10.1016/j.tcb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh H, Irvine KD. Yorkie: The final destination of Hippo signaling. Trends Cell Biol. 2010;20(7):410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122(3):421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114(4):445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 8.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38(10):1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 9.Sun S, Reddy BVVG, Irvine KD. Localization of Hippo signalling complexes and Warts activation in vivo. Nat Commun. 2015;6:8402. doi: 10.1038/ncomms9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin F, et al. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154(6):1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158(1):143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 2011;9(6):e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20(7):657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai ZC, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120(5):675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Vrabioiu AM, Struhl G. Fat/Dachsous signaling promotes Drosophila Wing growth by regulating the conformational state of the NDR kinase Warts. Dev Cell. 2015;35(6):737–749. doi: 10.1016/j.devcel.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shraiman BI. Mechanical feedback as a possible regulator of tissue growth. Proc Natl Acad Sci USA. 2005;102(9):3318–3323. doi: 10.1073/pnas.0404782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aegerter-Wilmsen T, et al. Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development. 2012;139(17):3221–3231. doi: 10.1242/dev.082800. [DOI] [PubMed] [Google Scholar]
- 18.Hufnagel L, Teleman AA, Rouault H, Cohen SM, Shraiman BI. On the mechanism of wing size determination in fly development. Proc Natl Acad Sci USA. 2007;104(10):3835–3840. doi: 10.1073/pnas.0607134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124(4):318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Farhadifar R, Röper J-C, Aigouy B, Eaton S, Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr Biol. 2007;17(24):2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 21.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 22.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126(4):767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16(19):1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 24.Mao Y, et al. Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J. 2013;32(21):2790–2803. doi: 10.1038/emboj.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legoff L, Rouault H, Lecuit T. A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc. Development. 2013;140(19):4051–4059. doi: 10.1242/dev.090878. [DOI] [PubMed] [Google Scholar]
- 26.Heemskerk I, Lecuit T, LeGoff L. Dynamic clonal analysis based on chronic in vivo imaging allows multiscale quantification of growth in the Drosophila wing disc. Development. 2014;141(11):2339–2348. doi: 10.1242/dev.109264. [DOI] [PubMed] [Google Scholar]
- 27.Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125(1):1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Kasza KE, Zallen JA. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr Opin Cell Biol. 2011;23(1):30–38. doi: 10.1016/j.ceb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marygold SJ, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8(10):R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98(6):779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93(7):1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 32.Noll N, Mani M, Heemskerk I, Streichan S, Shraiman BI. 2015. Active Tension Network model of epithelial mechanics. arXiv:1508.00623v1. [DOI] [PMC free article] [PubMed]
- 33.Sabino D, Brown NH, Basto R. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J Cell Sci. 2011;124(Pt 7):1156–1166. doi: 10.1242/jcs.076711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105(1):81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 35.Nienhaus U, Aegerter-Wilmsen T, Aegerter CM. Determination of mechanical stress distribution in Drosophila wing discs using photoelasticity. Mech Dev. 2009;126(11–12):942–949. doi: 10.1016/j.mod.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Hong X, et al. Opposing activities of the Ras and Hippo pathways converge on regulation of YAP protein turnover. EMBO J. 2014;33(21):2447–2457. doi: 10.15252/embj.201489385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell. 2013;24(5):459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan R, Kim N-G, Gumbiner BM. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc Natl Acad Sci USA. 2013;110(7):2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Affolter M, Basler K. The Decapentaplegic morphogen gradient: From pattern formation to growth regulation. Nat Rev Genet. 2007;8(9):663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 40.Bosveld F, et al. Modulation of junction tension by tumor suppressors and proto-oncogenes regulates cell-cell contacts. Development. 2016;143(4):623–634. doi: 10.1242/dev.127993. [DOI] [PubMed] [Google Scholar]
- 41.Reddy BV, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 2011;138(23):5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28(17):1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol Biol Cell. 2004;15(2):838–850. doi: 10.1091/mbc.E03-06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001;114(Pt 3):493–501. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- 45.Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161(4):1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worley MI, Setiawan L, Hariharan IK. TIE-DYE: A combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development. 2013;140(15):3275–3284. doi: 10.1242/dev.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135(6):1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zartman J, Restrepo S, Basler K. A high-throughput template for optimizing Drosophila organ culture with response-surface methods. Development. 2013;140(3):667–674. doi: 10.1242/dev.088872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.