Significance
Terminal commitment of differentiating cells is fundamental to multicellular life but remains the least characterized phase of development. Using Anabaena, a multicellular cyanobacterium that irreversibly commits 10% of cells to specialized nitrogen-fixing heterocysts, we report the identification of four genes that regulate commitment timing and efficacy in a cyanobacterium, including two that delay commitment: a unique finding across developmental model systems. Through protein–protein interactions, cell type-specific and -nonspecific expression patterns, and epistatic relationships, we present evidence that these four genes function together in a hierarchy to control correct timing of the commitment decision. This work illustrates the importance of Anabaena as a model system for studying the genetic underpinnings controlling the process of cellular differentiation.
Keywords: commitment, heterocyst, Anabaena, hetP, cellular differentiation
Abstract
The commitment of differentiating cells to a specialized fate is fundamental to the correct assembly of tissues within a multicellular organism. Because commitment is often irreversible, entry into and progression through this phase of development must be tightly regulated. Under nitrogen-limiting conditions, the multicellular cyanobacterium Anabaena sp. strain PCC 7120 terminally commits ∼10% of its cells to become specialized nitrogen-fixing heterocysts. Although commitment is known to occur 9–14 h after the induction of differentiation, the factors that regulate the initiation and duration of this phase have yet to be elucidated. Here, we report the identification of four genes that share a functional domain and modulate heterocyst commitment: hetP (alr2818), asl1930, alr2902, and alr3234. Epistatic relationships between all four genes relating to commitment were revealed by deleting them individually and in combination; asl1930 and alr3234 acted most upstream to delay commitment, alr2902 acted next in the pathway to inhibit development, and hetP acted most downstream to drive commitment forward. Possible protein–protein interactions between HetP, its homologs, and the heterocyst master regulator, HetR, were assessed, and interaction partners were defined. Finally, patterns of gene expression for each homolog, as determined by promoter fusions to gfp and reverse transcription–quantitative PCR, were distinct from that of hetP in both spatiotemporal organization and regulation. We posit that a dynamic succession of protein–protein interactions modulates the timing and efficiency of the commitment phase of development and note that this work highlights the utility of a multicellular cyanobacterium as a model for the study of developmental processes.
The developmental programs of multicellular organisms share a common structure that ensures proper assembly of the adult organism. First, inducing signals from the environment or physiological niche initiate the program, and then cells are patterned to place each cell type within the organism, sometimes with strikingly regular periodicity. Expanding on Turing’s chemical basis of morphogenesis, Meinhardt and Gierer proposed a mechanism for the emergence of biological periodic patterns out of homogeneity, which they termed lateral inhibition. Lateral inhibition is borne out in the activator–inhibitor model of pattern formation (1–4) in which an autocatalytic activator of differentiation is expressed in and remains near a source cell. The activator also initiates the production of a developmental inhibitor, which moves away from the source cell laterally to prevent differentiation of neighboring tissue. The spatial interplay between a gradient of inhibitor and an autoregulatory activator thereby generates a reproducible distance between differentiating cells. This model has been found to be a common mechanism of specifying the placement of differentiating cells for a wide variety of periodic biological patterns across entire domains of life (5–9), but does not determine whether differentiation will be completed. This decision is governed by the next step in development: commitment.
Commitment is a sustained pattern of altered gene expression resulting in a cell that will complete differentiation irrespective of changes to the inducing signals that initiated its developmental program. The time at which commitment initiates and the duration needed to complete commitment can be determined empirically by removing the inducing signal at progressively later time points in development and assessing the completion or cessation of morphogenesis. The new morphological state can be categorized as either terminal (permanent) or nonterminal (reversible). Broadly, eukaryotes exhibit both terminal and nonterminal differentiation with many proposed commitment mechanisms such as balancing positive and negative regulators (10), chromatin remodeling (11, 12), and cascades of transcription factors (13). Prokaryotes with morphologically and physiologically distinct cell types have also been examined as model developmental systems and several have been investigated in mechanistic detail. Bacterial sporulation, competence for DNA uptake, flagellar motility, and the switch to a dormant persister state are all developmental decisions that hinge on the accumulation of an autoregulatory activator, either a transcription factor or σ factor, relative to some minimum stoichiometric threshold (14–20). Importantly, however, these prokaryotic model systems all exhibit nonterminal differentiation and rely on cell-autonomous mechanisms of fate choice. In contrast, the multicellular cyanobacterium Anabaena sp. strain PCC 7120 (hereafter Anabaena) undergoes patterned commitment to a terminally differentiated cell fate. Anabaena thus provides a powerful, yet simple, genetic system for the study of location-dependent multicellular patterning, similar to that seen in eukaryotic cellular differentiation.
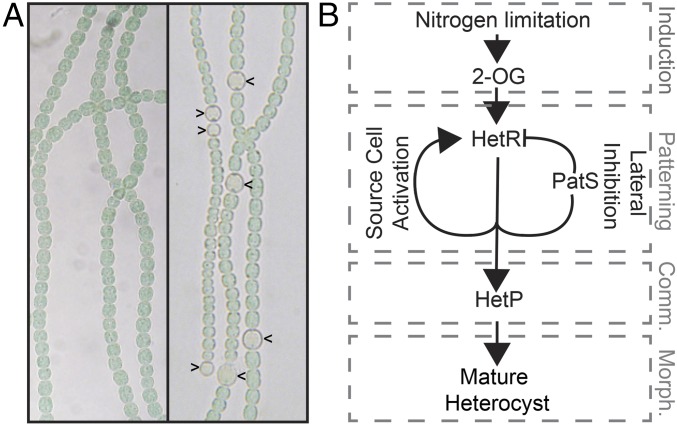
Anabaena grows as filaments of hundreds of undifferentiated photosynthetic vegetative cells (Fig. 1A, Left) (21–23). When combined nitrogen in the environment is low or absent, a developmental program is induced that results in the terminal differentiation of specialized nitrogen-fixing heterocyst cells over the course of ∼24 h (Fig. 1A, carets in Right). Heterocysts are nondividing, nonphotosynthetic, microoxic chambers for the oxygen-labile nitrogenase enzyme complex to convert atmospheric dinitrogen into a bioactive form. The cascade of developmental events governing this transition can be grouped into four stages: the induction of differentiation, biological pattern formation, commitment to a differentiated cell fate, and morphogenesis (Fig. 1B). The inducing signal for development, nitrogen starvation, is perceived by the cell as a transient increase in the metabolic intermediate, 2-oxoglutarate (24, 25), which activates a series of transcription factors that ultimately promote production of the master regulator of differentiation, HetR (22). HetR is an autocatalytic transcriptional regulator required for heterocyst differentiation in an otherwise wild-type background (26, 27) that promotes the expression of its own inhibitor, patS (28). The interaction of the PatS inhibitor, which diffuses laterally from source cells and signals degradation of HetR (29), and the HetR activator, which remains in the source cell and directly controls gene expression, places cells that can become heterocysts in a one-dimensional pattern along filaments (2, 3). These patterned cells now have the capacity to become mature heterocysts but are not yet irreversibly committed to their fate.
Fig. 1.
Heterocyst differentiation in Anabaena in various conditions. (A) In combined nitrogen-replete conditions, Anabaena grows in multicellular filaments of hundreds of undifferentiated cells (Left), but after 24 h of nitrogen starvation, a periodic pattern of terminally differentiated heterocysts (indicated by carets in Right) is elaborated. (B) The developmental program directing heterocyst differentiation can be simplified and summarized as a genetic hierarchy with an activator–inhibitor module at its center. Arrows indicate activation. T bars indicate repression. Gray dashed boxes correspond to the stages of development indicated by gray text: induction, patterning, commitment (comm.), and morphogenesis (morph.).
In wild type, commitment occurs 9–14 h postinduction by nitrogen starvation, after which the readdition of a source of fixed nitrogen, such as nitrate or ammonia, will not prevent the completion of heterocyst development (30). Although the timing of commitment has been observed, the regulation of this terminal transition is not understood. Several pieces of indirect evidence, however, implicate the hetP gene as a potential regulator of the commitment stage of development. First, a hetP mutant successfully executes the heterocyst developmental program through the patterning stage, but fails to complete morphogenesis for most patterned cells (31, 32). Second, overexpression of hetP is capable of bypassing the master regulator of patterning, hetR, to produce partially functional heterocysts in a ΔhetR mutant that is otherwise incapable of differentiation (32). Finally, the hetP promoter is HetR dependent and is activated in patterned cells. Although heterocyst regulatory protein HetP contains no domains of known function, it shares homology with asl1930, alr2902, and alr3234; it has therefore been suggested that these genes may serve similar functions.
In this work, we define hetP as a regulator of heterocyst commitment in Anabaena. Furthermore, we identify asl1930, alr2902, and alr3234 as sharing a conserved functional domain with HetP and characterize their genetic relationships and roles in commitment through epistasis analysis, gene expression studies, and protein–protein interactions. Taken together, we propose a gene network that functions to govern the timing, duration, and efficacy of the commitment phase of development in a multicellular bacterium.
Results
Overexpression of hetP Produces Heterocysts in Normally Repressive Conditions.
Heterocyst function is an energy-intensive process, and heterocysts do not divide. As such, repressing inappropriate development, either environmentally or genetically, is important for the survival of the population as a whole. The presence of a combined nitrogen source, such as ammonia, and the action of the PatS inhibitor, whether expressed during pattern formation or added exogenously to cultures (PatS5) (33), have strong repressive effects. Previous work indicates that hetP functions downstream of hetR and patterning during differentiation and suggests that overexpression of hetP could bypass both the presence of fixed nitrogen as well as PatS5 (32).
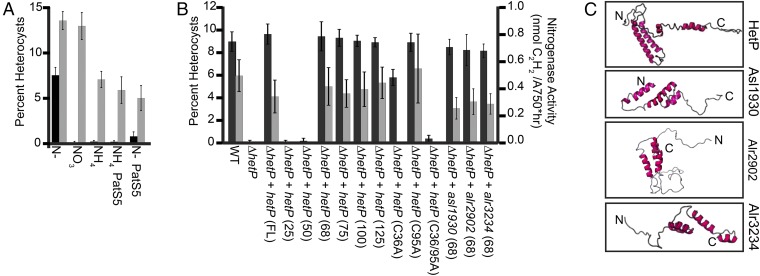
To test the ability of hetP to bypass known environmental and genetic cues, vectors carrying the copper-inducible petE promoter alone (PpetE; vector) or PpetE with hetP (PpetE-hetP) were introduced into wild type and assessed for their ability to drive heterocyst development in normally repressive conditions. Heterocyst formation with vector alone was ∼7% when starved for fixed nitrogen (N−) and was repressed to < 0.1% by the addition of nitrate (NO3), ammonia (NH4), or PatS5 peptide (Fig. 2A, black bars). The induction of PpetE-hetP resulted in the accumulation of supernumerary heterocysts during nitrogen starvation (N−) and in every repressive condition tested, including the simultaneous presence of ammonia and PatS5 peptide (Fig. 2A, gray bars). The overexpression of hetP did not, however, abolish pattern formation but resulted in decreased distances between heterocysts (SI Appendix, Fig. S1). This phenotype was similar to that seen resulting from mutation of patN, a gene involved in heterocyst differentiation in the related cyanobacterium Nostoc punctiforme strain ATCC 29133 (34). Taken together, we conclude that hetP is capable of bypassing both nitrogen status and PatS-mediated inhibition of differentiation, and we infer that hetP functions downstream of the induction and patterning stages of development.
Fig. 2.
Bypass and functional complementation by alleles of hetP and hetP homologs. (A) Overexpression of hetP in repressive conditions results in heterocyst formation. Average percent heterocysts out of 500 cells after 24 h of growth in the condition specified below the x axis: BG-11 media without fixed nitrogen (N−), BG-11 media with nitrate (NO3), and BG-11 media with ammonia (NH4). Media were supplemented with 10 μM PatS5 peptide as indicated. Black bars represent wild type carrying the empty PpetE vector (pPJAV213) and gray bars represent wild type carrying the copper-inducible PpetE-hetP (pSMC224). (B) Heterocyst percentage (left axis, black bars) and nitrogenase activity (right axis, gray bars). Numbers indicate length of HetP in amino acids from the N terminus or full length (FL). Strains used are wild type (WT), ΔhetP (UHM158), ΔhetP PhetP-hetPFL (pPJAV304), ΔhetP PhetP-hetP25 (pPJAV298), ΔhetP PhetP-hetP50 (pPJAV299), ΔhetP PhetP-hetP68 (pPJAV300), ΔhetP PhetP-hetP75 (pPJAV301), ΔhetP PhetP-hetP100 (pPJAV302), ΔhetP PhetP-hetP125 (pPJAV303), ΔhetP PhetP-hetP(C36A) (pPJAV305), ΔhetP PhetP-hetP(C95A) (pPJAV306), ΔhetP PhetP-hetP(C36A, C95A) (pPJAV307), ΔhetP PhetP-asl193068 (pPJAV353), ΔhetP PhetP-alr290268 (pPJAV354), and ΔhetP PhetP-alr323468 (pPJAV358). All measurements were conducted in triplicate and expressed as the average ± SD. (C) Predicted protein structures for HetP (Alr2818), Alr2902, Alr3234, and Asl1930. All models were generated by the RaptorX tertiary structure prediction program. N indicates the N terminus, whereas C indicates the C terminus of the protein.
A Shared Region Constitutes a Functional Domain of HetP and Its Homologs.
Proteins with a high degree of homology to HetP are broadly distributed across filamentous heterocystous, filamentous nonheterocystous, and unicellular cyanobacteria (SI Appendix, Fig. S2A). Notably, a 60- to 70-aa region at the N terminus of these homologs is particularly well conserved, whereas the C terminus varies greatly in both length and sequence (SI Appendix, Fig. S2B). To define the minimum functional size of the HetP protein in Anabaena, a series of truncated alleles were created, introduced into a hetP deletion mutant (UHM158), and assayed for their ability to complement heterocyst development and function. Whereas alleles of hetP encoding only the N-terminal 25 or 50 aa failed to complement either characteristic, the first 68, 75, 100, and 125 aa all fully complemented the hetP mutant equivalent to using the full-length (FL) hetP gene (Fig. 2B). Overexpression of the full-length hetP gene produced functional heterocysts in the wild type but not in a hetR mutant (32), suggesting that a HetR-dependent pathway is required for hetP to facilitate proper differentiation. Additionally, mutation of the C-terminal C95 HetP residue to alanine had no effect on function, whereas mutation of the N-terminal C36 residue, or mutation of both C36 and C95, impaired heterocyst formation and function (SI Appendix, Fig. S2B, and Fig. 2B).
Comparison of the amino acid sequence between HetP and its homologs, Asl1930, Alr2902, and Alr3234 (32), shows a conserved amino acid region corresponding to the minimum length of HetP needed for complementation (SI Appendix, Fig. S2C). This conserved region is predicted to consist of helices with the remainder of each protein being disordered or poorly predicted by tertiary structure modeling (Fig. 2C). To determine whether the conserved 68-aa regions of asl1930, alr2902, and alr3234 had shared function, their homologous domains were expressed from the hetP promoter, individually introduced into a hetP mutant, and assayed for complementation as above. This conserved region from each homolog restored both heterocyst percentage and function to near wild-type levels (Fig. 2B). To test for differences in function of the full-length proteins, each homolog was expressed from the petE promoter and introduced into both hetR and hetP mutant backgrounds. None of these genes were able to bypass the need for hetR or fully complement a hetP mutant, with only alr2902 achieving partial recovery of heterocyst development (SI Appendix, Fig. S3A) (32). We conclude that asl1930, alr2902, alr3234, and hetP contain a functionally redundant domain and suggest that the remainder of these proteins serve different functions.
Epistasis Analysis of hetP, asl1930, alr2902, and alr3234.
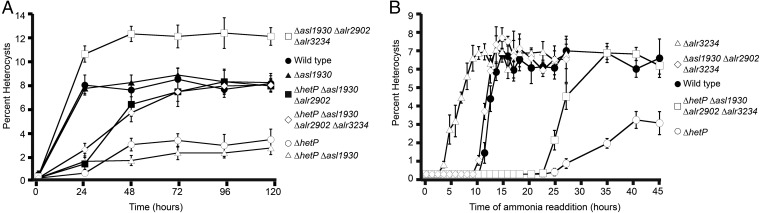
The functional redundancy of the homologous domains of asl1930, alr2902, alr3234, and hetP led us to test the requirement of each homolog for heterocyst development and their epistatic relationship to hetP. To accomplish this, asl1930, alr2902, and alr3234 were mutated by cleanly deleting each ORF from the genome singly, pairwise, together as a triple mutant, and with hetP. Although wild-type Anabaena accumulated ∼8% heterocysts by 24 h and maintained that percentage over 120 h of nitrogen starvation (Fig. 3A, filled circles), a hetP mutant was delayed in differentiating heterocysts until 48 h and only accumulated ∼3% heterocysts over the 120-h time course (Fig. 3A, open circles). All single and double mutants lacking a hetP mutation displayed wild-type heterocyst formation and function (Fig. 3A, filled triangles; SI Appendix, Table S1). Similar to the hetP mutant, each double mutant that included a hetP mutation (ΔhetP Δasl1930, ΔhetP Δalr2902, ΔhetP Δalr3234) was delayed in heterocyst development and failed to accumulate a wild-type percentage of heterocysts. We conclude that neither asl1930, nor alr2902, nor alr3234 is independently required for heterocyst development and that hetP is epistatic to each homolog individually.
Fig. 3.
Epistasis analysis and the commitment timing of hetP, asl1930, alr2902, and alr3234 mutant strains. (A) Plots of the percentage of heterocysts formed as a function of time after nitrogen stepdown. For simplicity, one representative curve is shown from each epistasis group. The full dataset can be seen in SI Appendix, Table S1. Wild type, ΔhetP (UHM158), single-mutant representative (Δasl1930 UHM295), double-mutant representative (ΔhetP Δasl1930 UHM288), triple-mutant representative (ΔhetP Δasl1930 Δalr2902 UHM282), triple homolog mutant Δasl1930 Δalr2902 Δalr3234 (UHM282), and quadruple mutant ΔhetP Δasl1930 Δalr2902 Δalr3234 (UHM333). Error bars represent the SD of three replicates. (B) Dynamics of commitment to a heterocyst fate. Cultures were stepped down to media lacking combined nitrogen to induce heterocyst development and then harvested at the time points indicated and supplemented with ammonia. Each data point represents the percent heterocyst accumulation 24–48 h after supplementation with ammonia. Error bars represent the SD for three replicates. For simplicity, one representative of each commitment phenotype is graphed. The full dataset can be seen in SI Appendix, Table S1. Strains depicted are as follows: wild type, ΔhetP (UHM158), Δalr3234 (UHM336), and ΔhetP Δasl1930 Δalr2902 Δalr3234 (UHM333).
Given these results, we predicted that a strain triply mutant for hetP and two of the homologs (ΔhetP Δasl1930 Δalr2902, UHM282; ΔhetP Δasl1930 Δalr3234, UHM283; ΔhetP Δalr2902 Δalr3234, UHM284), or quadruply mutant for all four genes (ΔhetP Δasl1930 Δalr2902 Δalr3234, UHM333) would display the same phenotype as the hetP single mutant. Contrary to our predictions, the quadruple mutant and two of the three triple mutant backgrounds recovered heterocyst formation and function to wild-type levels after 48–72 h and displayed normal pattern formation (Fig. 3A, filled squares, open diamonds; SI Appendix, Table S1 and Fig. S4). Furthermore, a strain with only alr2902 remaining intact (ΔhetP Δasl1930 Δalr3234) completely failed to form heterocysts or exhibit patterned gene expression (SI Appendix, Table S1 and Fig. S4). Importantly, these mutant backgrounds revealed that the timing of development is genetically separable from the magnitude of development with hetP acting as a positive regulator and the three homologs collectively acting in an opposing, inhibitory manner. If true, a strain with hetP but lacking all three homologs (Δasl1930 Δalr2902 Δalr3234) should produce the highest number of heterocysts of the genotypes examined. Indeed, this strain yielded ∼50% more heterocysts than wild type by 24 h of development (Fig. 3A, open squares).
In light of the complexity of the genetic relationships described above, we next used a quantitative genetic approach to aid in epistasis interpretation. The two-allele two-phenotype algorithm uses the fitness of each strain, calculated from the highest percentage of heterocysts produced relative to wild type, to quantify the effect of genes on a given phenotype (35, 36). The four genes mutated singly or triply displayed antagonistic interactions, whereas synergistic interactions were observed from the mutation of hetP in combination with each homolog (SI Appendix, Table S2). Using a multiple-regression analysis, mutation of all four genes and all possible interactions were entered as predictors and accounted for 96% of the variance in heterocyst production (R2 = 0.96, R2Adjusted = 0.94). Of the four mutations examined, only hetP produced a significant main effect (β = −4.40, P < 0.001), indicating that hetP alone has a significant impact on phenotype irrespective of mutation of the three other genes. Significant interaction effects were seen for hetP, asl1930, and alr2902 (β = 4.20, P < 0.01) and hetP, alr2902, and alr3234 (β = 5.00, P < 0.001), which demonstrates that mutating these genes in combination produces effects beyond those that would be expected based on the effects of the single mutations. Taken together, the qualitative and quantitative assessment of epistasis between these four genes suggests that, although they each play distinct roles in regulating the timing and efficacy of heterocyst development, there is likely a hierarchy of complex interactions among loci rather than a simple linear pathway.
Assessment of the Individual Contributions of HetP, Asl1930, Alr2902, and Alr3234 to Heterocyst Development.
The epistasis results above indicate that HetP and its homologs may have related but opposing functions. To gain further evidence for the individual functions of hetP, asl1930, alr2902, and alr3234 irrespective of native transcriptional regulation or the potentially antagonistic functions of the other homologs, each gene was expressed from the heterologous petE promoter in wild type, a hetP mutant, and the quadruple mutant described above (UHM333). Consistent with the placement of hetP downstream of the three homologs, overexpression of hetP resulted in the formation of supernumerary heterocysts in all strains tested (SI Appendix, Fig. S3 B–D). A notable exception, however, was the introduction of PpetE-asl1930 into wild type and UHM333, which produced opposing phenotypes. Overexpression in the wild type suppressed heterocyst differentiation for 24 h after induction (SI Appendix, Fig. S3B and Table S3). Conversely, PpetE-asl1930 nearly complemented UHM333 by producing 5.7% heterocysts 24 h after nitrogen stepdown (SI Appendix, Fig. S2C). These results suggest that, although Asl1930 might have a positive function in isolation, it normally operates in opposition to the differentiation machinery in the wild-type context.
HetP, Asl1930, Alr2902, and Alr3234 Modulate Commitment to a Heterocyst Fate.
Although we had described a set of genetic interactions related to heterocyst development, the phase of differentiation during which these genes functioned remained unclear. A simple explanation for the dynamic changes in the timing and magnitude of heterocyst development seen in the genetic backgrounds described above could be the result of changes in the execution of initial pattern formation; however, expression of the patterning reporter PpatS-gfp was wild type for timing and localization in all backgrounds tested that produced heterocysts (SI Appendix, Fig. S4) (30, 32). Alternatively, regulation at the level of the commitment stage of development could account for both a delay in morphogenesis as well as changes to the percent of mature heterocysts produced. A delay in commitment could manifest as a delay in heterocyst formation, and a decrease in the efficiency of advancing patterned cells past commitment could appear as a decrease in the magnitude of heterocysts produced.
To test whether the phenotypes observed were due to changes in either the timing or efficiency of commitment, both characteristics were tested. Commitment can be operationally defined as the time in development at which the removal of the inducing signal no longer stops the progression of differentiation. In the case of Anabaena heterocyst development, the inducing signal is an absence of combined nitrogen, and this signal can be removed by the addition of a combined nitrogen source, such as ammonia (30). To assess the contribution of genes to the commitment phase of heterocyst development, strains were induced for differentiation and samples of each culture were harvested at 1-h intervals, supplemented with ammonia, and the total number of morphologically distinct heterocysts that developed in 24–48 h was determined. If commitment had not yet occurred, the readdition of ammonia to induced cells would suppress development and no heterocysts would result. If commitment had occurred, however, the readdition of ammonia would fail to suppress development and heterocysts would form.
Wild-type Anabaena displayed a commitment phase spanning 9–13 h after induction, which is consistent with previous work (Fig. 3B, closed circles; SI Appendix, Table S1) (30). By 13 h after induction, the addition of ammonia failed to repress differentiation and these populations developed to wild-type levels. In contrast to wild type, repression of differentiation by ammonia in the ΔhetP mutant was lost beginning at 22 h after induction and the transition between this precommitment phase and postcommitment spanned around 15 h rather than the roughly 4-h span observed in the wild type (Fig. 3B, open circles; SI Appendix, Table S1). Differentiation of the quadruple mutant (UHM333) was repressed by ammonia readdition for 20 h after induction, equivalent to the ΔhetP mutant (Fig. 3B, open squares; SI Appendix, Table S1). However, repression was sharply attenuated 22–24 h after induction. By 30 h, the addition of ammonia failed to repress differentiation and these populations developed numbers of heterocysts equivalent to wild type. Given these phenotypes, we conclude that hetP is required for both advancing the initiation of and accelerating progress through the commitment phase of heterocyst differentiation. Furthermore, we infer that one or more of the three homologs of hetP function to modulate the dynamics of the commitment phase.
To clarify the roles of the three homologs of hetP in commitment, we tested strains mutated for asl1930, alr2902, and alr3234 individually and in combination triply by the readdition of ammonia to developing cultures as above. Similar to wild type, Δalr2902 underwent commitment over a 4-h time span between 8 and 12 h after induction (SI Appendix, Table S1). Δalr3234 and Δasl1930 also underwent commitment during an approximately 4-h time span; however, both Δalr3234 and Δasl1930 started commitment notably earlier, initiating this phase at 2 and 4 h after induction, respectively (Fig. 3B, open triangles; SI Appendix, Table S1). We conclude that asl1930 and alr3234 both function to delay the initiation of commitment but are not required for normal progression through this phase.
Finally, to determine the relationship between asl1930, alr2902, and alr3234, the commitment phenotype of a strain mutant for all three genes (Δasl1930 Δalr2902 Δalr3234) was assessed. The triple mutant displayed a timing and duration of commitment that was equivalent to the alr2902 single mutant and wild type (SI Appendix, Table S1). Among the three homologs, therefore, we find that asl1930 and alr3234 normally function to delay the initiation of commitment, with alr2902 being epistatic to both genes. Taken together, these data suggest that all four genes containing the common HetP functional domain modulate the commitment phase of heterocyst development. Asl1930 and Alr3234 appear to be most upstream in the pathway, and both function to delay commitment initiation. Alr2902 acts downstream of Asl1930 and Alr3234 and is a potent inhibitor of development. Finally, HetP acts downstream of all three homologs, driving the initiation of commitment and promoting the efficient conversion of patterned cells into an irreversibly committed heterocyst.
PatS Is Not Responsible for Commitment Timing.
There are several possible mechanisms that could explain the observed differences in the timing of commitment. One possibility invokes the fact that the cell that differentiates is also the cell that produces the inhibitory signal that represses differentiation in neighboring cells. Therefore, to complete the developmental program, a differentiating cell must resolve the paradox of being both the source of the inhibitor and the cell that will differentiate. Becoming immune to the inhibitor allows the cell to complete differentiation while maintaining the pattern of nondifferentiated cells around it. In the case of Anabaena heterocyst development, we hypothesized that if immunity to the patS inhibitor is the sole requirement for commitment to occur, a ΔpatS mutant would commit as early as or earlier than Δasl1930 and Δalr3234 mutants (4 and 2 h after induction, respectively). Contrary to our prediction, a ΔpatS mutant initiated commitment 8–9 h after induction in a manner similar to wild type, and a ΔpatS ΔhetP double mutant was delayed in commitment and produced an increased percentage of heterocysts similar to a ΔpatS mutant (SI Appendix, Table S1). In addition, heterocyst development was inhibited in early-committing strains (Δasl1930 and Δalr3234) by either overexpression of patS from an inducible promoter or exogenous addition of the PatS5 peptide. We conclude that neither the absence of nor immunity to the PatS inhibitory signal is sufficient to initiate early commitment in developing heterocysts.
The Genes asl1930, alr2902, and alr3234 Are Regulated Differently than hetP.
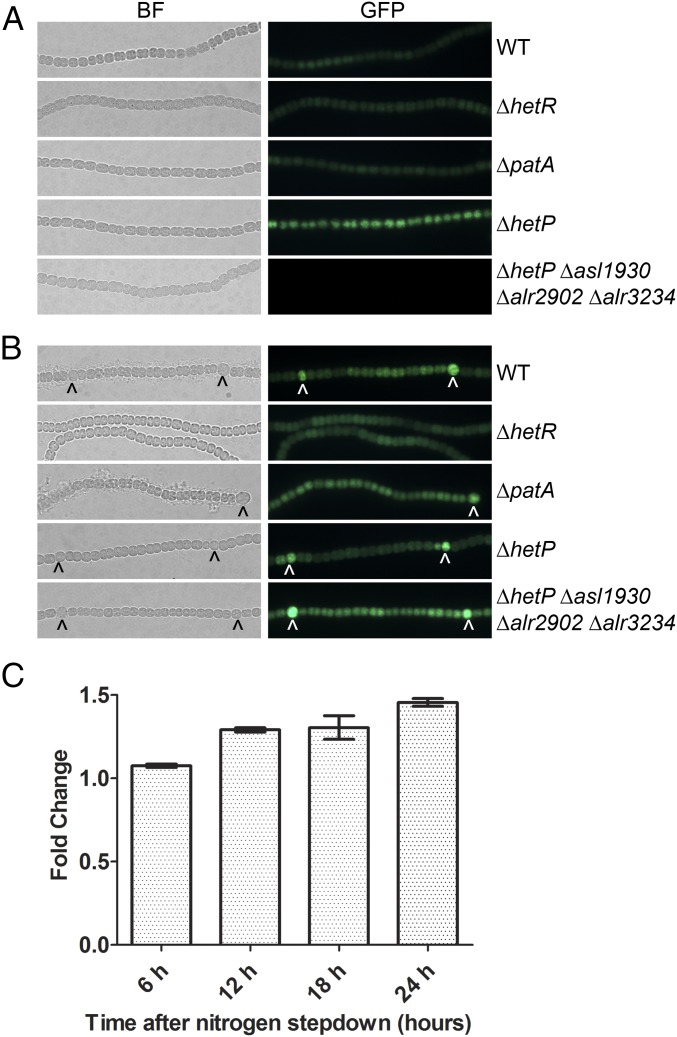
The hetP gene is initially up-regulated 6 h after nitrogen stepdown and expression is increased in heterocysts by 24 h after induction (SI Appendix, Fig. S5C) (31, 32, 37). Expression of hetP is directly regulated by HetR, which binds to a defined inverted repeat in the hetP promoter (SI Appendix, Fig. S6A) (32, 38). To investigate the transcriptional profiles of asl1930, alr2902, and alr3234, reverse transcription–quantitative PCR (RT-qPCR) was conducted and fusions were created between the promoter region of each homolog and gfp. Fluorescence was then observed in the wild type, a patA mutant known to have high levels of HetR protein, a hetR mutant with no HetR protein, a hetP mutant, and the quadruple hetP homolog mutant (UHM333). In all growth conditions, fluorescence from Palr2902-gfp failed to accumulate above background (SI Appendix, Fig. S5A). Its expression, however, was detected by RT-qPCR at low levels in the wild type (SI Appendix, Fig. S5C). Fluorescence from Palr3234-gfp was low, but detectable, in the wild type, in both the presence and absence of a combined nitrogen source. Expression did not change significantly from the uninduced level of expression over a 24-h time period as measured by RT-qPCR (SI Appendix, Fig. S5 B–D). Expression of alr2902 and alr3234 displayed decreased expression in a hetR mutant at time 0 but not 24 h after nitrogen removal, which indicates that HetR or a member of its regulon may regulate these genes in nitrogen-replete conditions (SI Appendix, Fig. S6 C and D). Fluorescence was uniformly increased in ΔhetP and UHM333; however, we have noted that fluorescence in backgrounds lacking hetP is higher than other strains, irrespective of the construct or fluorophore being used, and likely represents a general response rather than specific activation of these promoters. In contrast, expression from Pasl1930-gfp was only observed in ΔhetP and not UHM333 during growth in a combined nitrogen source (Fig. 4A). Following nitrogen stepdown, fluorescence from Pasl1930-gfp was notably increased in a pattern corresponding to differentiated heterocysts in the wild type, ΔpatA, and UHM333, as well as in cells likely capable of differentiation in ΔhetP (Fig. 4B). Consistent with this observation, RT-qPCR revealed that asl1930 expression increased significantly (P < 0.05) in the wild type by 6 h of development and continued to increase throughout the differentiation process (Fig. 4C). A gradient of fluorescence emanating from heterocysts in a ΔpatA strain, like that of the HetR-dependent hetP promoter, was not observed (32). Consistent with this, fluorescence from Pasl1930-gfp in ΔhetR was observed at levels comparable to that of vegetative cells of the other strains used, and expression of asl1930 did not vary between low and high HetR backgrounds as was seen for hetP when measured by RT-qPCR (SI Appendix, Fig. S6 A and B). We infer that, although each homolog contains a functionally redundant protein domain, transcriptional regulation is distinct from that of hetP and the expression profiles of these genes may control their roles in differentiation.
Fig. 4.
Cell type-specific expression of asl1930. A plasmid containing a Pasl1930-gfp transcriptional fusion was introduced into wild type (WT), ΔhetR (UHM103), ΔpatA (UHM101), ΔhetP (UHM158), and ΔhetP Δasl1930 Δalr2902 Δalr3234 (UHM333). (A) The resulting strains were grown in nitrogen-replete conditions and imaged (bright-field, left columns, and GFP, right columns). (B) The same strains were then stepped down to media lacking combined nitrogen and imaged after 24 h (WT, ΔhetR, ΔpatA) or 48 h (ΔhetP, ΔhetP Δasl1930 Δalr2902 Δalr3234). (C) Expression of asl1930 in wild type at 6, 12, 18, and 24 h after nitrogen stepdown relative to expression at time 0 as measured by RT-qPCR. Error bars represent the SD for three replicates. All bars are significantly different (t test, P < 0.05) from wild-type expression at time 0.
HetP Homolog Interactions.
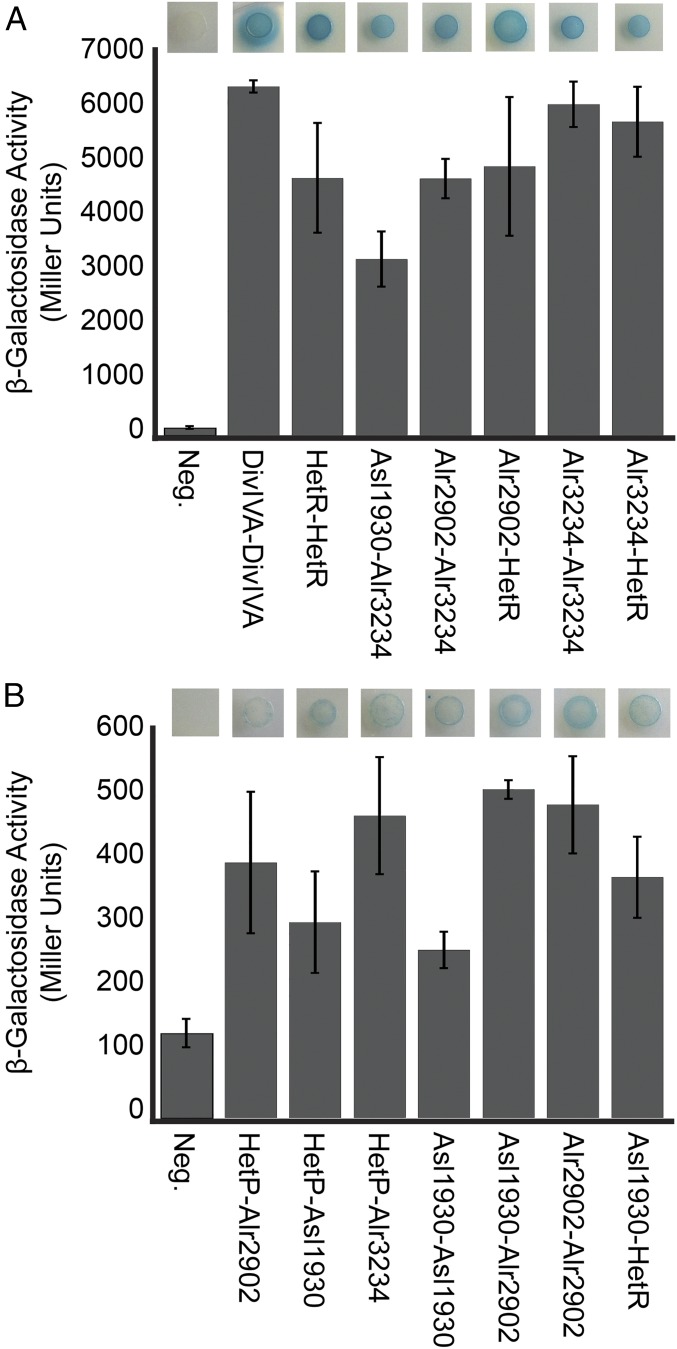
To gain insight into possible mechanisms of commitment regulation and provide evidence for the epistatic and expression analysis presented above, we tested the network of potential interactions between HetR, HetP, Alr1930, Alr2902, and Alr3234, using a bacterial two-hybrid assay (BACTH). Each protein was translationally fused to separate catalytic domains of the adenylate cyclase protein that, when brought together by protein–protein interaction, result in expression of the β-galactosidase enzyme (39). All possible combinations of N- or C-terminal orientation and 18- or 25-kDa adenylate cyclase domain were tested. Any combination that produced blue color on solid media containing X-gal was then measured by β-galactosidase assay (SI Appendix, Table S4). Several protein combinations resulted in robust β-galactosidase activity including HetR/HetR, HetR/Alr2902, HetR/Alr3234, Asl1930/Alr3234, Alr2902/Alr3234, and Alr3234/Alr3234 (Fig. 5A). A second group of protein combinations resulted in lower yet significant (P < 0.05) levels of β-galactosidase activity including HetP/Asl1930, HetP/Alr2902, HetP/Alr3234, Asl1930/Asl1930, Asl1930/Alr2902, Asl1930/HetR, and Alr2902/Alr2902 (Fig. 5B). These results suggest that Asl1930, Alr2902, and Alr3234 are capable of direct protein interactions with each other as well as with HetR and HetP. Although the exact function of the common domain of HetP and its homologs remains unknown, these results suggest that the remaining nonhomologous portion of each protein may provide interaction specificity for a subset of protein partners.
Fig. 5.
Bacterial two-hybrid assay for protein–protein interactions. Positively interacting combinations of HetR, HetP, Asl1930, Alr2902, and Alr3234 are represented qualitatively as a colony growing on media supplemented with X-gal (top image) and quantitatively as a Miller unit value (bar graph). (A) Strong interactions (>1,000 Miller units) were observed using the following plasmid combinations in the order presented: pST579-pST572, pRO180-pRO191, pRO189-pRO186, pST565-pRO187, pRO189-pRO191, and pRO189-pST558. (B) Weak interactions (200–600 Miller units) were observed using the following plasmid combinations in the order presented: pRO185-178, pRO181-pRO170, pRO189-178, pRO180-pRO183, pRO180-pRO187, pRO184-pRO187, and pST565-pRO183. All interactions are in comparison with the activity resulting from two empty BACTH vectors (negative control) using pT18-N-link-pT25-N-link. Error bars represent the SD for three replicates. Interactions that were significantly higher than the negative control (Student’s t test, P < 0.05) are shown. The full dataset can be seen in SI Appendix, Table S4.
In sum, we propose that cell fate commitment in Anabaena is modulated by a hierarchy of four homologous proteins: one that promotes differentiation and three that oppose it. The wild-type timing of the commitment phase of heterocyst development (beginning 9–11 h after induction) and the duration of this phase (1–4 h) are therefore a balance of the activity of each factor with there likely being a dynamic set of protein–protein interactions that ultimately lead to irreversible heterocyst commitment.
Discussion
Anabaena is a multicellular cyanobacterium that undergoes patterned terminal differentiation of specialized nitrogen-fixing heterocyst cells. Although the hetP gene has been shown to be necessary for developing a normal percentage of heterocysts following the induction of differentiation, here we demonstrate that it is required specifically for the timing and efficiency of commitment to the heterocyst fate. We show that, despite sharing a conserved functional domain that is widely distributed across cyanobacteria, HetP, Asl1930, Alr2902, and Alr3234 play distinct and even opposing regulatory roles with respect to commitment. The heterocyst developmental program was found to be driven forward by hetP and epistasis analysis revealed that the three homologous genes act upstream of hetP to either delay (asl1930, alr3234) commitment or inhibit development (alr2902). Protein interactions of differing strengths were detected between subsets of these regulators and differing patterns of gene expression were observed. These results are consistent with a model in which a dynamic succession of protein interactions governs the entry into and completion of commitment.
The mechanism by which individual, reversible molecular interactions in developing heterocysts create irreversible commitment remains unknown. Among bacteria, the best understood commitment model is that of sporulation in Bacillus subtilis. Nutrient limitation induces the sporulation pathway, causing B. subtilis cells to undergo asymmetric division, followed by the engulfment of the smaller cell (forespore) by the larger cell (40). Within 2–4 h of initial nutrient limitation, the forespore irreversibly commits to completing sporulation irrespective of changes in the nutrient status of its environment (41). Progression toward commitment is gradual, resulting from an accumulation of the phosphorylated form of the master regulator of sporulation: the Spo0A transcription factor (42). The commitment decision occurs downstream of Spo0A, is promoted by the functionally redundant spoIIP and spoIIQ genes under control of the forespore-specific σ factor σF, and is switch-like rather than gradual (15, 43). Similarly, Anabaena commitment also occurs downstream of the master regulator of development (hetR) (Fig. 1B), requires functional redundancy among regulators (hetP, asl1930, alr2902, and alr3234) (Fig. 2B), and results in a switch-like output (Fig. 3B). This comparison highlights the similarities between the underlying methods by which cellular differentiation proceeds in prokaryotes. However, the work presented here describes both positive and negative regulators of commitment.
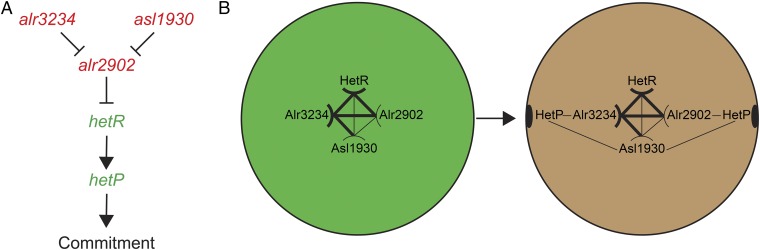
Epistasis analysis showed that hetP and its homologs exert genetically separable but synergistic forces on the timing and efficacy of commitment and indicated that heterocyst commitment is likely a hierarchical process consisting of at least three tiers (Fig. 6A). The genes asl1930 and alr3234 are probably the earliest elements in the hierarchy to provide lag time before commitment, as strains mutant for these genes committed far earlier than the wild type. Morphogenesis is an energetically taxing process that abolishes a cell’s lineage, so delaying terminal differentiation may reflect a strategy against short-term variations in combined nitrogen abundance. The epistasis analysis showed that strains mutant for alr2902 did not display differences from the parent strains in the timing of commitment, but a triple mutant that only retained an intact copy of alr2902 failed to pattern properly and did not produce heterocysts; the only genetic background in this study to do so. In addition to delaying commitment, therefore, Asl1930 and Alr3234 appear to normally attenuate the activity of this potent developmental inhibitor, placing it downstream of asl1930 and alr3234. Finally, based on the observations that hetP was the only gene studied here to create supernumerary heterocysts when overexpressed, bypass the need for hetR, and whose mutation resulted in a protracted and inefficient commitment phenotype, we conclude that, of the four genes examined, hetP is most proximal to the mechanism of commitment.
Fig. 6.
Genetic and spatial constraints required for commitment. (A) Model of genetic relationships among commitment regulators. Arrows represent a positive effect, and T bars represent a repressive effect. Red text indicates a negative influence on commitment progression. Green text indicates a positive influence on commitment progression. (B) Demonstrated interactions and accumulation patterns of proteins involved in commitment. Thick lines indicate a strong interaction, whereas thin lines indicate a weak interaction as measured by BACTH analysis. Curved lines indicate a self-interaction, whereas straight lines indicate non–self-interactions. Postinduction but precommitment (green circle), HetR, Asl1930, Alr2902, and Alr3234 are present in the cell. For commitment to proceed, increased cell type-specific expression of HetR, Asl1930, and HetP could alter the interaction partner network, ultimately resulting in the relocation of HetP to the poles. Heterocyst poles indicated by black ovals representative of cyanophycin granules.
The subcellular localization of HetP appears to change from the cytoplasm to the cell poles over the course of development (44). In light of our findings that define HetP as a regulator of commitment, this observation is intriguing. What drives such a change in subcellular localization and is it related to the global transcriptomic switch following the transition from patterning to morphogenesis? Bacterial two-hybrid analysis indicated that HetR and each of the HetP homologs (Asl1930, Alr2902, and Alr3234) were capable of interacting with themselves and with each other. Alr2902 and Alr3234 interacted with each other and with HetR with strengths similar to that of HetR self-interaction. In contrast, the interactions of Asl1930, Alr2902, and Alr3234 with HetP were far weaker and we saw no evidence of HetP self-interaction. Although these four homologs share a functional domain, this alone is insufficient to mediate the specificity and strength of protein–protein interactions. Indeed, RaptorX structure modeling identified both helical and disordered regions within each of the four homologs, which could allow for distinct topology depending on partner-specific interactions (Fig. 2C). It is possible that the changes in topology resulting from different binding partners may control the observed transition of HetP localization from the cytoplasm to cell poles. Because all of the HetP homologs interact with one another and HetR, it is tempting to hypothesize that the homologs act as a scaffold to fine-tune the timing of commitment via protein–protein interactions. The strong interaction of Alr2902 and Alr3234 may constitute the core upon which HetR, HetP, and Asl1930 can bind. HetR binds to DNA (28), interacts with RNA polymerase (45), and could act in a manner functionally equivalent to that of a σ factor. In this scenario, we speculate that polar HetP localization results from changes in interaction partners, driven by differences in the spatiotemporal gene expression patterns of hetP, its homologs, and hetR to indirectly tie HetP to the modulation of HetR availability.
Previous transcriptomic work and transcriptional fusions with luciferase have shown that hetP is up-regulated as early as 6 h after the removal of combined nitrogen (31, 37). Here, we show that both asl1930 and alr3234 act to delay commitment and are regulated differently from hetP. Given the potency of hetP, as demonstrated by the production of supernumerary heterocysts in even the most repressing conditions, it is not surprising to have multiple levels of control on it and therefore on commitment. The timing of gene expression and the role each gene plays in commitment support the scenario described above in which the shift in HetP localization contributes to the timing and efficacy of commitment. In this case, each of the homologs is expressed in vegetative cells and is available in the early stages of differentiation for protein–protein interactions, possibly with HetR (Fig. 6B). After patterning commences, patterned up-regulation of hetP and asl1930 results in the eventual shift from cytoplasmic to polar localization of HetP in developing heterocysts (34). It is entirely possible that the observed differences in spatiotemporal expression of hetP and its homologs contribute to the timing of commitment and the global transcriptional shift that takes place during morphogenesis via interactions with several protein partners.
How cells commit to differentiation is a question that is important across all of biology. Its study, however, is often complicated by the three-dimensional nature of patterning interactions, numerous tissue types, and long developmental timescales inherent in the growth of higher eukaryotes. With its combination of periodic patterning, differentiated cell types, and relative genetic and structural simplicity, Anabaena occupies an important niche within developmental biology research. The work presented here identifies regulators of commitment in a cyanobacterium and represents an important step in defining the components of patterned cellular differentiation, a process found to obey many of the same general mechanistic requirements shown for higher-order developmental systems. Because the most extreme defect in commitment still resulted in ∼3% of cells differentiating into heterocysts, it is apparent that additional positive factors remain to be elucidated. Our future work to define the precise mechanism governing heterocyst commitment to a terminally differentiated cell fate will likely have thematic, if not mechanistic, implications for a wide variety of developmental systems.
Materials and Methods
Bacterial Strains and Growth Conditions.
The growth of Escherichia coli and Anabaena sp. strain PCC 7120 (wild type) and its derivatives (listed in SI Appendix, Table S5), concentrations of antibiotics, the induction of heterocysts in media lacking a source of combined nitrogen, and conditions for photomicroscopy were as previously described (32, 46). Media for Anabaena growth on ammonia as the nitrogen source was prepared as previously described (47). Heterocyst percentages were determined as previously described, and all results are expressed as the average of three replicates ± SD (48). The vegetative cell interval between heterocysts was determined by counting 300 intervals as previously described (47). To inhibit heterocyst differentiation, the PatS-5 pentapeptide (RGSGR) was added to cultures to a final concentration of 10 μM. Transcription from the copper-inducible petE promoter was induced with the addition of copper to a final concentration of 2 μM (49). Plasmids were introduced into Anabaena strains by conjugation from E. coli as previously described (50). Fluorescence from green fluorescent protein (GFP) was imaged by an Olympus BX-51 epifluorescence microscopy with an excitation wavelength of 488 nm and an emission wavelength of 510 nm. Heterocyst commitment assays were performed as previously described (30), with specifics detailed in SI Appendix. The phylogenetic comparisons, assessment of protein structure, image analysis, mathematical epistasis analysis, and RNA isolation and RT-qPCR analysis are outlined in SI Appendix.
Plasmid and Strain Construction.
The plasmids and oligonucleotide primers used in this study are listed in SI Appendix, Tables S5 and S6, respectively, and their construction is described in SI Appendix.
Alcian Blue Staining, Acetylene Reduction Assays, Bacterial Two-Hybrid Assays, and β-Galactosidase Assays.
Heterocyst-specific exopolysaccharides were stained with Alcian blue as previously described (32). Aerobic and anaerobic acetylene reduction assays were performed as previously described (38, 46, 51). Bacterial two-hybrid assays and β-galactosidase assays were conducted as previously described with specifics detailed in SI Appendix (39, 52, 53).
Supplementary Material
Acknowledgments
We are very grateful to Sean Callahan (University of Hawaii at Manoa) for guidance, resources, and feedback; Kelly Higa and Sasa Tom (University of Hawaii at Manoa) for plasmid creation and technical assistance; Dan Kearns (Indiana University) for the plasmids pT18-N-link, pT18-C-link, pT25-N-link, and pT25-C-link; and Randall Wong (Molecular Biology Unit, University of Colorado Barbara Davis Center BioResources Core Facility) for performing the RT-qPCR. This work was supported by NSF-PRFB Award 1103610 (to L.M.C.) and an Illinois Wesleyan Artistic and Scholarly Development Grant (to L.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610533113/-/DCSupplemental.
References
- 1.Turing A. The chemical basis of morphogenesis. Philos Trans R Soc London Ser B. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12(1):30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 3.Meinhardt H. Models of biological pattern formation: From elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 4.Ball P. Forging patterns and making waves from biology to geology: A commentary on Turing (1952) “The chemical basis of morphogenesis.”. Philos Trans R Soc Lond B Biol Sci. 2015;370(1666):1666. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuong CM, Yeh CY, Jiang TX, Widelitz R. Module-based complexity formation: Periodic patterning in feathers and hairs. Wiley Interdiscip Rev Dev Biol. 2013;2(1):97–112. doi: 10.1002/wdev.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saga Y. The mechanism of somite formation in mice. Curr Opin Genet Dev. 2012;22(4):331–338. doi: 10.1016/j.gde.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Marcon L, Sharpe J. Turing patterns in development: What about the horse part? Curr Opin Genet Dev. 2012;22(6):578–584. doi: 10.1016/j.gde.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Robinson DO, Roeder AH. Themes and variations in cell type patterning in the plant epidermis. Curr Opin Genet Dev. 2015;32:55–65. doi: 10.1016/j.gde.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Joshi M, Buchanan KT, Shroff S, Orenic TV. Delta and Hairy establish a periodic prepattern that positions sensory bristles in Drosophila legs. Dev Biol. 2006;293(1):64–76. doi: 10.1016/j.ydbio.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35(9):565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Simmons AR, Bergmann DC. Transcriptional control of cell fate in the stomatal lineage. Curr Opin Plant Biol. 2016;29:1–8. doi: 10.1016/j.pbi.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25(8):789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moody SA, Klein SL, Karpinski BA, Maynard TM, Lamantia AS. On becoming neural: What the embryo can tell us about differentiating neural stem cells. Am J Stem Cells. 2013;2(2):74–94. [PMC free article] [PubMed] [Google Scholar]
- 14.Parker GF, Daniel RA, Errington J. Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology. 1996;142(Pt 12):3445–3452. doi: 10.1099/13500872-142-12-3445. [DOI] [PubMed] [Google Scholar]
- 15.Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121(3):401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Leisner M, Stingl K, Frey E, Maier B. Stochastic switching to competence. Curr Opin Microbiol. 2008;11(6):553–559. doi: 10.1016/j.mib.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Gamba P, Jonker MJ, Hamoen LW. A novel feedback loop that controls bimodal expression of genetic competence. PLoS Genet. 2015;11(6):e1005047. doi: 10.1371/journal.pgen.1005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76(2):273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Zhang Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob Agents Chemother. 2007;51(6):2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2(4):a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muro-Pastor AM, Hess WR. Heterocyst differentiation: From single mutants to global approaches. Trends Microbiol. 2012;20(11):548–557. doi: 10.1016/j.tim.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Wolk CP. Heterocyst formation. Annu Rev Genet. 1996;30:59–78. doi: 10.1146/annurev.genet.30.1.59. [DOI] [PubMed] [Google Scholar]
- 24.Laurent S, et al. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc Natl Acad Sci USA. 2005;102(28):9907–9912. doi: 10.1073/pnas.0502337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J-H, Laurent S, Konde V, Bédu S, Zhang C-C. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology. 2003;149(Pt 11):3257–3263. doi: 10.1099/mic.0.26462-0. [DOI] [PubMed] [Google Scholar]
- 26.Buikema WJ, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5(2):321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 27.Black TA, Cai Y, Wolk CP. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9(1):77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Dong Y, Zhao J. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc Natl Acad Sci USA. 2004;101(14):4848–4853. doi: 10.1073/pnas.0400429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risser DD, Callahan SM. Genetic and cytological evidence that heterocyst patterning is regulated by inhibitor gradients that promote activator decay. Proc Natl Acad Sci USA. 2009;106(47):19884–19888. doi: 10.1073/pnas.0909152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon H-S, Golden JW. PatS and products of nitrogen fixation control heterocyst pattern. J Bacteriol. 2001;183(8):2605–2613. doi: 10.1128/JB.183.8.2605-2613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández-Piñas F, Leganés F, Wolk CP. A third genetic locus required for the formation of heterocysts in Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176(17):5277–5283. doi: 10.1128/jb.176.17.5277-5283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higa KC, Callahan SM. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol. 2010;77(3):562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 33.Yoon H-S, Golden JW. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282(5390):935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 34.Risser DD, Wong FCY, Meeks JC. Biased inheritance of the protein PatN frees vegetative cells to initiate patterned heterocyst differentiation. Proc Natl Acad Sci USA. 2012;109(38):15342–15347. doi: 10.1073/pnas.1207530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouyos RD, Silander OK, Bonhoeffer S. Epistasis between deleterious mutations and the evolution of recombination. Trends Ecol Evol. 2007;22(6):308–315. doi: 10.1016/j.tree.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 36.MacLean RC. Predicting epistasis: An experimental test of metabolic control theory with bacterial transcription and translation. J Evol Biol. 2010;23(3):488–493. doi: 10.1111/j.1420-9101.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 37.Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Videau P, et al. Expanding the direct HetR regulon in Anabaena sp. strain PCC 7120. J Bacteriol. 2014;196(5):1113–1121. doi: 10.1128/JB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95(10):5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113(1):29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19(18):2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuchina A, et al. Temporal competition between differentiation programs determines cell fate choice. Mol Syst Biol. 2011;7:557. doi: 10.1038/msb.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrales-Guerrero L, Flores E, Herrero A. Relationships between the ABC-exporter HetC and peptides that regulate the spatiotemporal pattern of heterocyst distribution in Anabaena. PLoS One. 2014;9(8):e104571. doi: 10.1371/journal.pone.0104571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valladares A, Flores E, Herrero A. The heterocyst differentiation transcriptional regulator HetR of the filamentous cyanobacterium Anabaena forms tetramers and can be regulated by phosphorylation. Mol Microbiol. 2016;99(4):808–819. doi: 10.1111/mmi.13268. [DOI] [PubMed] [Google Scholar]
- 46.Borthakur PB, Orozco CC, Young-Robbins SS, Haselkorn R, Callahan SM. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 2005;57(1):111–123. doi: 10.1111/j.1365-2958.2005.04678.x. [DOI] [PubMed] [Google Scholar]
- 47.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA. 2011;108(50):20130–20135. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Videau P, Rivers OS, Higa KC, Callahan SM. ABC transporter required for intercellular transfer of developmental signals in a heterocystous cyanobacterium. J Bacteriol. 2015;197(16):2685–2693. doi: 10.1128/JB.00304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buikema WJ, Haselkorn R. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc Natl Acad Sci USA. 2001;98(5):2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elhai J, Wolk CP. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 51.Ernst A, et al. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J Bacteriol. 1992;174(19):6025–6032. doi: 10.1128/jb.174.19.6025-6032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 53.van den Ent F, et al. Dimeric structure of the cell shape protein MreC and its functional implications. Mol Microbiol. 2006;62(6):1631–1642. doi: 10.1111/j.1365-2958.2006.05485.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.