Significance
We describe a mechanism of regulating immune cell chemotaxis. We identified in serum a peptide derived from domain 5 of high-molecular-weight kininogen produced by coagulation factor XIIa cleavage that accelerates C-C chemokine receptor 7-mediated chemotaxis. Thus, we present a paradigm in which a humoral peptide functions as a chemotactic cofactor that links inflammation to immunity.
Keywords: lymphocyte, chemotaxis, CCR7, factor XII, kininogen
Abstract
Chemokines and their receptors play a critical role in immune function by directing cell-specific movement. C-C chemokine receptor 7 (CCR7) facilitates entry of T cells into lymph nodes. CCR7-dependent chemotaxis requires either of the cognate ligands C-C chemokine ligand 19 (CCL19) or CCL21. Although CCR7-dependent chemotaxis can be augmented through receptor up-regulation or by increased chemokine concentrations, we found that chemotaxis is also markedly enhanced by serum in vitro. Upon purification, the serum cofactor activity was ascribed to domain 5 of high-molecular-weight kininogen. This peptide was necessary and sufficient for accelerated chemotaxis. The cofactor activity in serum was dependent on coagulation factor XIIa, a serine protease known to induce cleavage of high-molecular-weight kininogen (HK) at sites of inflammation. Within domain 5, we synthesized a 24-amino acid peptide that could recapitulate the activity of intact serum through a mechanism distinct from up-regulating CCR7 expression or promoting chemokine binding to CCR7. This peptide interacts with the extracellular matrix protein thrombospondin 4 (TSP4), and antibodies to TSP4 neutralize its activity. In vivo, an HK domain 5 peptide stimulated homing of both T and B cells to lymph nodes. A circulating cofactor that is activated at inflammatory foci to enhance lymphocyte chemotaxis represents a powerful mechanism coupling inflammation to adaptive immunity.
Chemotaxis is directional movement in response to a specific chemical gradient and is necessary for immune homeostasis and inflammatory response, among other critical biologic processes (1). Numerous chemokines, along with their cognate receptors, provide a molecular mechanism to orchestrate chemotaxis of distinct cell types to diverse stimuli (2). C-C chemokine receptor 7 (CCR7) and its ligands, C-C chemokine ligand 19 (CCL19) and CCL21, comprise a signaling axis required for homing of T cells into and within lymphoid organs, as well as the movement of B cells and dendritic cells (3). CCR7-mediated chemotaxis is important in developing adaptive immunity, in addition to maintaining tolerance and memory (4). Alternatively, this physiologic mechanism of lymph node entry can be coopted by malignancies; tumor CCR7 expression is associated with greater metastatic potential and increased mortality (5, 6). In chronic disease, ectopic production of CCL19/21 from tertiary lymphoid structures within sites of inflammation retains immune cells in peripheral tissue, accelerating the progression of conditions such as multiple sclerosis, rheumatoid arthritis, and atherosclerosis (7–10). Thus, a deeper understanding of molecular regulators of CCR7–CCL19/21 chemotaxis would inform the therapeutic approach to a wide spectrum of diseases.
Chemokines are broadly grouped as homeostatic or inflammatory (11). For the latter, acutely increasing production is often sufficient to coordinate a chemotactic response. For homeostatic chemokines, such as CCL19/21, signal modulation occurs chiefly by altering receptor density or available ligand concentration. This effect can be achieved either by increasing receptor expression or by reducing available ligand concentration by atypical chemokine receptors (12). For instance, exposure to lipopolysaccharide (LPS) or prostaglandin E2 up-regulates CCR7 expression, and ACKR4, an atypical chemokine receptor, can scavenge CCL19/21 (13). Currently, there are no known humoral cofactors that regulate migration in response to CCL19/21 independent of receptor or ligand availability. We observed an enhanced chemotactic response of T cells to CCL19/21 in the presence of serum. The serum cofactor activity for CCR7 chemotaxis was purified and identified as a peptide derived from high-molecular-weight kininogen (HK). This abundant plasma protein has a well-known role in inflammation, particularly as the parent molecule of the nonapeptide bradykinin, and HK cleavage products have been shown to modulate angiogenesis and endothelial cell migration (14). We demonstrate that serum-accelerated chemotaxis is dependent on active coagulation factor XII (FXIIa), the canonical initiator of the contact activation system that is generated at inflammatory foci and promotes cleavage of HK (15). Our findings present a role for HK in lymphocyte chemotaxis and provide the basis for a paradigm of regulating immune cell chemotaxis through a humoral cofactor.
Results
Serum Fractionation Identifies an HK-Derived Peptide as a Chemotactic Cofactor.
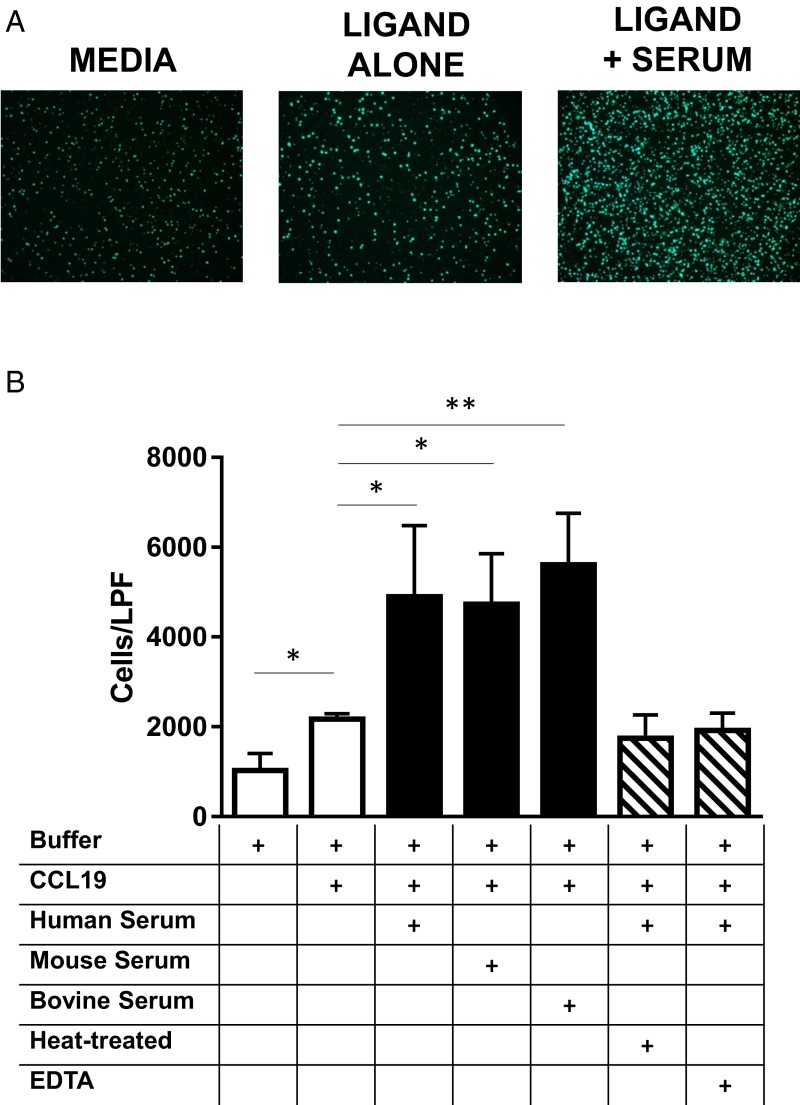
While interrogating CCR7/CCL19-dependent chemotaxis, we observed a more than fourfold increase in cell migration in the presence of serum (Fig. 1). Initial characterization of serum activity showed that it was thermolabile and could be inhibited by EDTA, implicating a protein and a metal cation (Fig. 1B). Additionally, this cofactor activity was evolutionarily conserved—sera from other mammalian species also elicited a robust increase in human T-cell migration in response to human CCL19 (Fig. 1B). We therefore sought to identify the cofactor responsible for promoting chemotaxis.
Fig. 1.
Serum enhances CCR7-dependent chemotaxis. (A) Direct visualization by fluorescence microscopy of the transwell insert as viewed from the lower chamber after 1 h, showing enhanced chemotaxis in the presence of serum (2%; bottom chamber). (B) Quantification of migrating cells. *P < 0.05; **P < 0.01. Pretreatment of serum with EDTA (2 mM) or heat (65 °C for 30 min) abolished chemotactic activity; sera (4%; bottom chamber) from various mammalian species accelerated chemotaxis.
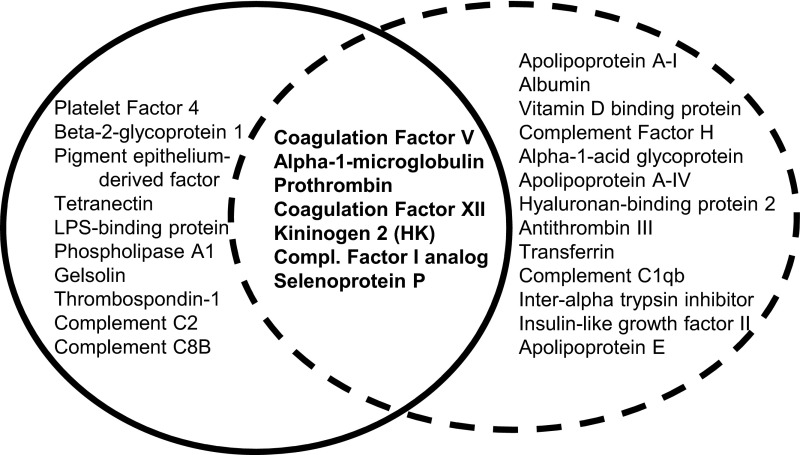
Fast protein liquid chromatography (FPLC) was used to isolate, identify, and characterize the putative migration cofactor. Preliminary studies demonstrated large increases in specific activity after cation exchange chromatography. A second observation was that serum treated with 8 M urea decreased the apparent molecular mass of the cofactor from ∼250 to ∼60 kDa. These findings led to two parallel purification strategies using multistep column chromatography (Tables 1 and 2). For the first algorithm (scheme 1), size exclusion was performed after urea treatment, whereas for the second algorithm (scheme 2), size-exclusion chromatography was initially used to isolate the larger form of the cofactor, followed by urea denaturation and repeat size-exclusion to capture the lower-molecular-mass form. Schemes 1 and 2 resulted in active fractions with specific activities >5,000-fold and >1,000-fold higher than the initial serum input, respectively. Mass-spectrometry analysis of these fractions identified an overlapping set of candidate proteins for the chemotactic cofactor (Fig. 2).
Table 1.
FPLC purification scheme 1
| Purification step | Protein output active fraction, mg | Specific activity, units/mg | Cumulative yield |
| Serum | 240,000 | 4.3 | 1 |
| Cation exchange | 220 | 196 | 0.0418 |
| Anion exchange | 30 | 525 | 0.0153 |
| Urea and size exclusion | 1.4 | 2,160 | 0.0029 |
| Cation exchange | 0.01 | 22,400 | 0.0002 |
Table 2.
FPLC purification scheme 2
| Purification step | Protein output active fraction, mg | Specific activity, units/mg | Cumulative yield |
| Serum | 80,000 | 4.3 | 1 |
| Cation exchange | 73 | 196 | 0.0418 |
| Ig depletion (Protein G) | 55 | 225 | 0.0360 |
| Size exclusion | 6 | 532 | 0.0093 |
| Urea and size exclusion | 0.06 | 4,410 | 0.0007 |
Fig. 2.
Venn diagram of protein candidates from mass-spectrometry analyses. The most abundant proteins in fraction with the highest specific activity from the primary purification (solid) and proteins present in active fraction from the secondary purification scheme (dashed) are shown.
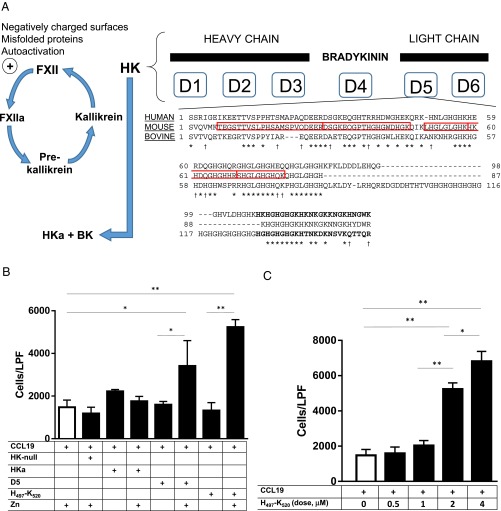
From these proteins, HK emerged as a likely candidate for several reasons. First, HK (120 kDa) circulates in plasma associated predominantly with prekallikrein (86 kDa). At sites of inflammation, a proteolytic cascade is initiated by (auto)activation of FXII to FXIIa which subsequently activates the zymogen prekallikrein to kallikrein, resulting in HK cleavage to yield bradykinin and other bioactive peptides (Fig. 3A) (16). Furthermore, five of six HK-derived peptides identified by mass spectrometry in the primary purification mapped to domain 5 (D5) within HK (Fig. 3A). D5 is known to bind cell surfaces and initiate signal transduction in a zinc-dependent manner, consistent with the inhibitory effect of EDTA (17). Finally, serum genetically deficient in HK was devoid of accelerated CCR7/CCL19 chemotactic activity (Fig. 3B). For other candidates, we tested the ability of purified proteins to accelerate chemotaxis in response to CCL19, but were unable to detect significant activity. Although it is possible that a subdomain of one or more of the other candidates may have chemotactic activity, we focused on HK and showed that a D5 peptide was capable of stimulating chemotaxis.
Fig. 3.
HK-derived peptides are sufficient for chemotaxis. (A) Schematic of contact system activation, driven by activation of FXII with subsequent reciprocal activation of FXII and prekallikrein, and ultimately kallikrein cleavage of HK to liberate bradykinin (BK). Domain 5 (D5) of HK is conserved at the protein level among human, bovine, and mouse species (* indicates amino acid identity, and † denotes biochemical similarity). Peptides within D5 identified by LC-MS/MS (boxed) and synthetic peptides used in subsequent studies (bold) are shown. (B) Chemotaxis after 1 h in response to HK-null sera (5%), HKa (2 μM), D5 (2 μM), or synthetic peptide H497–K520 (2 μM) in the presence or absence of zinc (75 μM). *P < 0.01; **P < 0.001. (C) Dose–response of accelerated chemotaxis after 1 h with H497–K520. *P < 0.01; **P < 0.001.
HK-Derived Peptides Are Necessary and Sufficient for Accelerated CCL19 Chemotaxis and Bind to Thrombospondin-4.
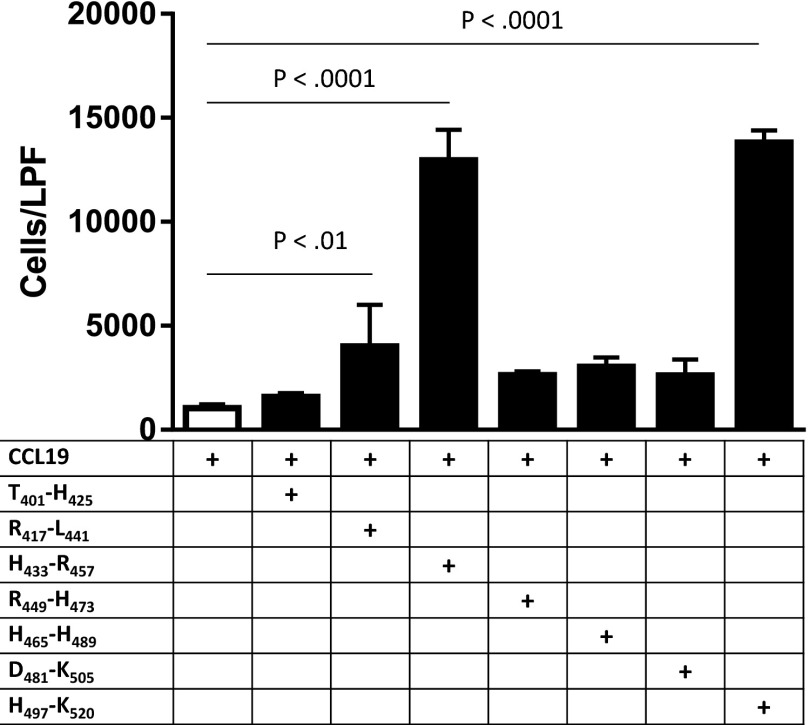
Intact HK did not have any effect on CCL19-dependent migration. However, bioactive HK cleavage products are generated by the contact system of inflammation in vivo, particularly at sites of inflammation (18). HK is processed by kallikrein to release the nonapeptide bradykinin, with resultant production of two-chain HK (HKa), although HKa, too, was ineffective at accelerating CCL19-dependent chemotaxis (Fig. 3B). We therefore decided to further investigate putative cleavage products of HK for chemotactic activity. As suggested by the D5-specific peptides detected by mass spectrometry, recombinant human D5 was sufficient to enhance chemotaxis (Fig. 3B). Consistent with prior studies of D5 surface-binding characteristics, zinc was necessary for accelerated chemotaxis. Using synthetic peptides to survey subdomains within human D5, we identified the 24-mer oligopeptide spanning HK residues histidine-497 through lysine-520 (H497–K520) as sufficient for enhanced chemotactic activity, as were other peptides within D5 (Fig. 3A and Fig. S1). Moreover, H497–K520 accelerated chemotaxis in a dose-dependent manner (Fig. 3C). H497–K520 is lysine-rich (9 of 24 residues), which may account for why it was not directly identified by mass spectrometry performed after trypsin digestion.
Fig. S1.
Chemotaxis assay using peptides within human D5.
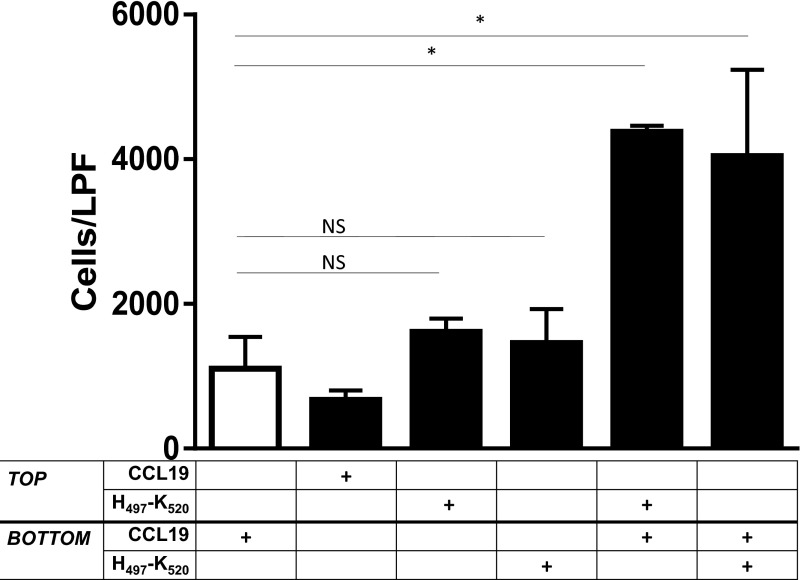
Using checkerboard-style analysis, we observed that, compared with CCL19 alone, there was no increase in migration with H497–K520 alone, irrespective of a H497–K520 gradient (Fig. 4). Furthermore, in the presence of CCL19, the cofactor activity requires a CCL19 gradient, but is independent of an H497–K520 gradient. Thus, this HK-derived peptide is not independently chemotactic or chemokinetic, but robustly increases chemotaxis toward a CCL19 gradient, regardless of its spatial distribution.
Fig. 4.
Checkerboard analysis. No enhanced migration after 1 h with H497–K520 in the absence of CCL19, with marked synergy in the presence of CCL19, irrespective of the H497–K520 gradient. *P < 0.001. NS, not significant.
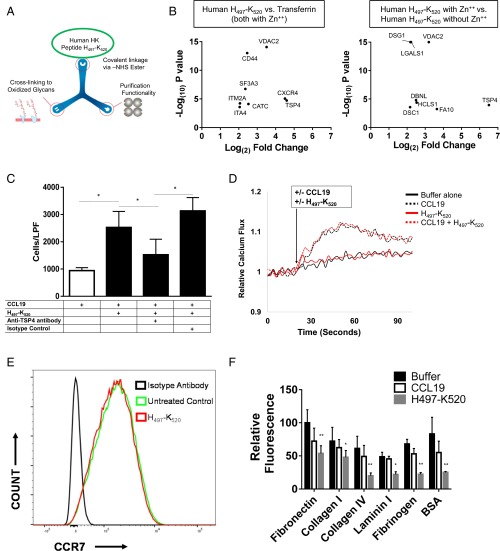
Prior studies have shown that D5 binds various cell surface receptors, including the urokinase receptor (UPAR), the globular head of the C1q receptor (gC1qR), and cytokeratin 1 (CK1) (19). We tested the ability of antibodies directed against these receptors to block the ability of H497–K520 to accelerate migration, but found no inhibitory effect. To identify potential receptors in an unbiased manner, we used ligand–receptor capture (LRC-TriCEPS) technology as a tool for detecting T-lymphocyte surface proteins that physically interact with H497–K520 (Fig. 5A) (20). Transferrin was used as a control ligand to eliminate nonspecific interactions with the TriCEPS reagent. Additionally, to control for nonspecific binding to H497–K520, TriCEPS coupled to H497–K520 was studied in the presence and absence of zinc. Analysis of captured peptides led to several candidate proteins, with thrombospondin-4 (TSP4) present in both analyses as the most enriched compared with both the transferrin and “no zinc” controls (Fig. 5B). The abundance of TSP4 isolated in the presence of H497–K520 and zinc was >80-fold higher than without H497–K520 or without zinc. Indeed, antibodies directed against TSP4 significantly blunted accelerated chemotaxis in response to H497–K520 (Fig. 5C). Voltage-dependent anion channel 2 (VDAC2) was also identified as significantly enriched compared with both controls, although antibodies directed against VDAC2 did not have an effect on accelerated chemotaxis.
Fig. 5.
H497–K520 binds to TSP4 and does not function through enhanced calcium flux or by altering CCR7 expression. (A) Schematic of TriCEPS ligand-capture reagent functional groups. (B) Enrichment plots of proteins captured with H497–K520 vs. transferrin (Left) or H497–K520 in the presence or absence of zinc (Right). (C) Chemotaxis assay with anti-TSP4 antibodies showing blunted acceleration compared with H497–K520 alone or isotype control. (D) T cells labeled with the calcium indicator fluo-4 were treated with CCL19 (600 ng/mL), H497–K520 (2 μM), the combination of the two, or buffer (HBSS/Hepes without calcium or magnesium). Results were indexed to the initial baseline measurements. (E) CCR7 surface expression determined by flow cytometry for untreated (green) or H497–K520 (2 μM)-treated (red) cells. (F) Adhesion assay with T cells incubated for 3 h with buffer alone, CCL19 (600 ng/mL), or H497–K520 (4 μM). Detection of adherent cells that remained after washing was determined by fluorescence. *P < 0.05; **P < 0.001.
To investigate whether H497–K520-accelerated chemotaxis was due to altered chemokine or receptor availability, we first evaluated its effect on intracellular calcium flux, a rapid indicator of chemokine binding. CCL19 is able to induce intracellular calcium flux independent of H497–K520 (Fig. 5D). Furthermore, H497–K520 has no effect on calcium flux alone, nor does it enhance calcium flux in combination with CCL19. Thus, although it can accelerate CCR7-mediated chemotaxis, H497–K520 does not appear to function by facilitating the presentation of ligand to CCR7. This finding is further supported by the absence of CCR7 in the candidate T-cell surface proteins that interact with H497–K520. Moreover, H497–K520 does not increase receptor expression of CCR7 (Fig. 5E). Because thrombospondins are adhesive glycoproteins, we tested the effect of H497–K520 in an adhesion assay (Fig. 5F). The HK-derived peptide significantly decreased T-cell adhesion to several extracellular matrix (ECM) proteins.
FXIIa Activity Is Required for Serum-Accelerated Chemotaxis.
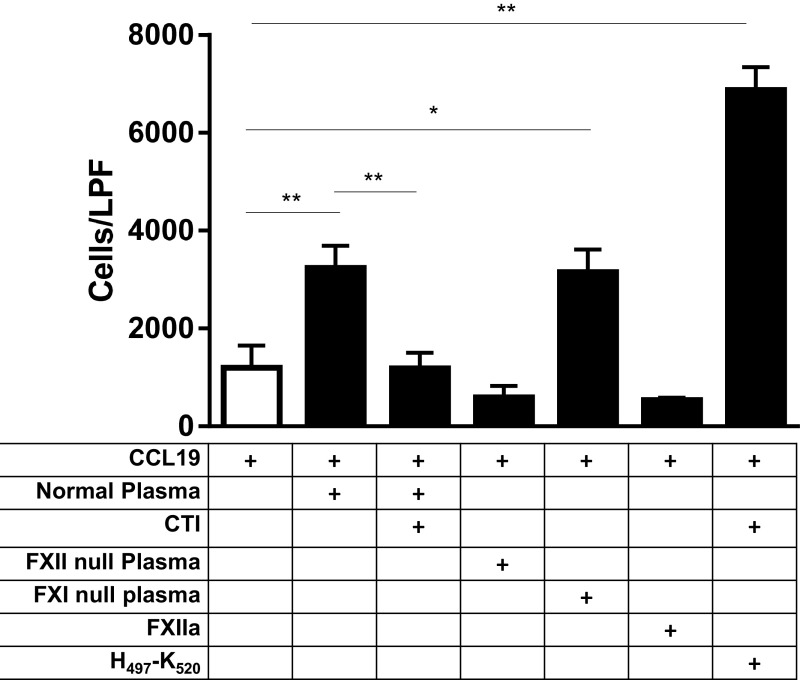
Because of its primacy in contact system activation, and hence HK cleavage, we tested the role of FXII in accelerated CCR7 chemotaxis. FXII-null plasma was unable to enhance chemotactic activity, as was normal plasma treated with the FXIIa-specific antagonist corn trypsin inhibitor (CTI; Fig. 6). H497–K520 rescued the inhibitory effect of CTI treatment (Fig. 6), consistent with H497–K520 alone being sufficient to accelerate chemotaxis toward CCL19. Although FXIIa initiates the intrinsic coagulation cascade via activation of factor XI, FXIIa-accelerated chemotaxis is independent of downstream mediators of clotting, because augmented chemotaxis was preserved in factor XI-null plasma. (Fig. 6). These studies indicate that HK-derived chemotactic peptides are not preformed to a significant degree in plasma. In addition, their generation is dependent on FXIIa.
Fig. 6.
The role of FXIIa in HK-accelerated chemotaxis. The FXIIa-specific antagonist CTI (3 μM) inhibits accelerated chemotaxis. Similarly, the absence of FXII, but not FXI (2.5% plasma), prevents accelerated chemotaxis. FXIIa alone has no effect on chemotaxis, whereas H497–K520 accelerates chemotaxis in the presence of CTI. *P < 0.01; **P < 0.001.
HK-Derived Peptides Promote Migration of Native Murine Lymphocytes both in Vitro and in Vivo.
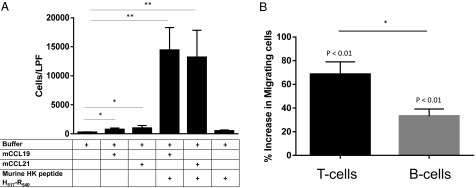
To test whether HK-derived peptides were relevant under more physiologic conditions, we first determined whether an analogous murine peptide accelerated chemotaxis of native murine lymphocytes. The 24-mer oligopeptide corresponding to a region within D5 of murine kininogen (murine H517–R540) potentiated the migration of murine lymphocytes in response to CCL19 and CCL21 (Figs. 3A and 7A).
Fig. 7.
Native murine lymphocytes respond to murine HK in vitro and in vivo. (A) Murine lymphocytes demonstrated accelerated chemotaxis toward murine CCL19 or CCL21 in the presence of H517–R540 (6 μM), a peptide corresponding to a similar region within domain 5 of murine HK (1.5 h), but not to H517–R540 alone. *P < 0.05; **P < 0.001. (B) Murine lymphocytes were fluorescently labeled and incubated with H517–R540 (8 μM) or vehicle (HBSS with 75 μM Zn) for 30 min, and injected into recipient mice (1 × 107 cells per animal). More donor cells were recovered from lymph nodes of mice that received H517–R540-treated lymphocytes vs. vehicle controls. The increased number of recovered donor cells shows a significantly greater proportion of donor T cells (CD3+CD19−) vs. B cells (CD19+CD3−) in recipient lymph nodes. *P < 0.05. Data were pooled from three independent experiments (n = 3 animals per group for each experiment).
A major physiologic role for CCL19/21 is to chemoattract T cells into lymphoid tissue. Correspondingly, the murine plt mutation is the deletion of the Ccl19 and Ccl21a loci, resulting in paucity of lymph-node T cells (21). Thus, murine H517–R540 would be predicted to increase the rate of entry of T cells from the periphery into lymph nodes by augmenting the in vivo response to CCL19/21. Indeed, treatment of lymphocytes with H517–R540 ex vivo, followed by adoptive transfer, resulted in significantly greater accumulation of donor T cells in recipient lymph nodes, compared with treatment with vehicle alone (69% increase vs. control; P < 0.01; Fig. 7B). There was also greater accumulation of donor B cells, although to a lesser degree than for T cells (33% increase vs. control; P < 0.01 vs. control and P < 0.05 in comparison with T cells), supporting a T-cell–dominant effect of the HK peptide.
Discussion
Peptides derived from the abundant plasma protein HK are novel cofactors for chemotaxis toward CCL19/21. HK-derived peptides are necessary and sufficient to mimic the effect of serum for accelerated chemotaxis and interact with cell surface TSP4. In plasma and sera, FXIIa activity is also required. An HK-derived peptide can also accelerate homing of native lymphocytes to lymph nodes, demonstrating the in vivo relevance of this novel mode of accelerating chemotaxis. As derivatives of contact system activation, these peptides localize chemotactic cofactor activity to foci of inflammation. Therefore, this cofactor mechanism delineates a distinct contribution of inflammation and coagulation to CCL19/21-dependent chemotaxis and provides a molecular basis for distinguishing and targeting accelerated vs. homeostatic chemotaxis.
CCL19 and CCL21 are produced by lymphoid organs to maintain homeostatic flux. As constitutively expressed chemokines, they permit a basal level of immune-cell movement through lymphoid tissue. The primary mechanism for increasing cellular responsiveness to this axis is through up-regulation of CCR7 expression, which is transcriptionally regulated (22). Our discovery of HK-derived peptides as chemotactic cofactors introduces a plausible mechanism of rapidly accelerating chemotaxis independent of altering CCR7 expression or levels of CCL19/21. Because the HK-derived peptides are not chemotactic themselves, but show strong chemokinetic properties in the presence of CCL19/21, exposure of immune cells to an HK peptide would be sufficient to accelerate migration of these cells along a basal CCL19 gradient. In this way, focal production of HK-derived peptides provides a mechanism for acutely accelerating the migration of immune cells from sites of inflammation to lymphoid organs. Furthermore, these cofactor peptides can be generated in minutes and do not rely on de novo transcription or protein synthesis. Local concentrations of HK-derived peptides could increase severalfold with inflammation and, by potentiating chemotaxis, provide another mechanism for coupling adaptive immunity to inflammation.
As the parent molecule of bradykinin, HK has a known role in inflammation. However, bradykinin represents a mere 9 of HK’s 626 amino acids, and other domains within HK participate in inflammatory effector pathways. Peptides from HK domains 3 and 5 stimulate monocyte cytokine production and inhibit neutrophil and monocyte adhesion (16, 23, 24). Consistent with these proinflammatory roles, our data demonstrate that D5 and peptides from within D5 promote T-cell chemotaxis, while decreasing T-cell adhesion. The requirement for zinc imparts selectivity for this pathway to operate at sites of inflammation. Plasma zinc concentrations are <10 μM, although free zinc levels can rise 10-fold when zinc is released by activated platelets and lymphocytes (25). In this way, zinc serves as a requisite second signal for HK-derived peptides to accelerate CCR7-mediated chemotaxis. Zinc also enhances autoactivation of FXII, which would increase the generation of HK peptides (26). However, neither zinc nor HK cleavage is necessary for basal chemotaxis, such as that required for immunosurveillance. This distinction between homeostatic chemotaxis and inflammation-driven chemotaxis offers insight into controlling overactive immunity while minimizing immunosuppression.
FXIIa initiates several proinflammatory cascades, including thrombosis and complement activation, in addition to kallikrein activation and bradykinin generation, although FXII activation does not necessarily lead to clotting (27). Although FXIIa and kallikrein reciprocally activate each other, FXII has been accorded the primary role in contact system activation because it can autoactivate and is also activated by several biologic stimuli, including misfolded proteins, LPS lipid A, and negatively charged surfaces, such as polyphosphates released by platelets at sites of vascular injury (28–30). Indeed, FXIIa can directly cleave HK and release bradykinin independent of kallikrein (31). Furthermore, studies of the FXII knockout mouse demonstrate that virtually all of the bradykinin produced as a result of contact activation is dependent on FXII (32). At foci of inflammation that ectopically produce CCL19/21, HK cleavage triggered by FXII activation may lead to enhanced recruitment of effector T cells and worsening disease progression. Enhanced T-cell chemotaxis can be a prominent feature of pathologic states, such chronic inflammation and autoimmune disease (33, 34). In fact, for several diseases, such as rheumatic disease, neuroinflammation, and atherosclerosis, CCL19/21 in afflicted sites promotes T-cell recruitment into tissue (7–9). In this context, FXIIa activity may be a modulator of immune-mediated disease activity, although convincing human data are lacking (35). Genetic deficiency of FXII is rare, and little is known about the resultant immune phenotype, although there are no reports of associated immunodeficiency.
TSP4 is a member of the thrombospondin family, comprising ECM proteins that participate in cell–cell and cell–ECM signaling (36). TSP4 itself is required for cell adhesion and migration of diverse cell types, including neurons and vascular smooth muscle cells (37, 38). Accordingly, TSP4 has been implicated in vascular inflammation and atherogenesis, acting at least in part by promoting macrophage recruitment into the neointima (39). Indeed, a missense, gain-of-function TSP4 variant (A387P) was associated with coronary artery disease in two cohorts (40, 41). Although a role for TSP4 in lymphocyte biology has not been reported, it does interact with integrins and CD44 (42, 43). These widely expressed proteins are common mediators of cell migration and adhesion with well-described roles in T-cell trafficking (44). We identified TSP4 using an unbiased screening approach targeting physical interactions. Known receptors for D5, including UPAR, CK1, and gC1qR, were absent from the list of candidate proteins using this method. This finding suggests that D5 may achieve diverse effector functions by signaling through an equally diverse repertoire of cellular receptors.
With this finding, several new questions have arisen that will require further study. The molecular mechanism by which extracellular HK peptides transduce their signal to accelerate cell migration requires further elucidation. In terms of cell specificity, we observed that T cells have a stronger response than B cells, and within these two lymphocyte classes, certain subsets may respond more vigorously than others. In a similar way, other CCR7-expressing cells, aside from lymphocytes, may exhibit accelerated chemotaxis in response to HK-derived peptides. Beyond CCR7, HK-derived peptides may accelerate chemotaxis for other chemokine systems as well. H497–K520 does not promote ligand binding to CCR7, as we showed with calcium flux studies. Therefore, the peptide’s action may be synergistic to, although distinct from, CCR7 signaling. In this way, H497–K520 signaling may be synergistic to migration along other chemokine gradients. Finally, to establish the in vivo relevance of this pathway, our ex vivo stimulation with H517–R540 enhanced lymph node homing, although the role of HK-derived peptides would be more definitively exemplified in an inflammatory model where HK peptides were generated in vivo.
Despite our evolving understanding, the need to mitigate morbidity and mortality from diseases associated with chronic inflammation remains significant (45). Regimens for treating immune-mediated diseases can have limited clinical utility—not from lack of efficacy, but because of immunosuppression. From this perspective, our finding of a humoral cofactor that regulates chemotactic activity distinct from chemokine binding to its receptor offers an additional target mechanism of action. For HK cleavage in particular, antagonizing FXIIa or neutralizing the interaction between D5 and TSP4 to modulate chemotaxis may confer specificity to pathologic sites of inflammation rather than interfere with homeostatic immune cell flux into lymphoid organs. These strategies would be more effective in preserving immunosurveillance than the current standard of care, such as glucocorticoids, which nonspecifically dampen immunity and have myriad off-target toxicities. Conversely, an agonist modeled after human H497–K520 to accelerate immune-cell chemotaxis may also hold promise for augmenting an immune response in immunocompromised individuals or to improve vaccine efficacy. Although much is known about the role of chemotactic axes in health and disease, there remains an unmet medical need to more precisely manipulate these systems, with potential for both antagonist and agonist approaches in the context of a chemotactic cofactor.
Materials and Methods
Cell Culture.
CCRF-CEM cells (ATCC) were maintained in RPMI 1640 supplemented with 25 mM Hepes and 10% (vol/vol) FBS (ATCC) at 37 °C with 5% (vol/vol) CO2. This cell line is an acute human T-cell lymphoblastic leukemia line with high levels of endogenous CCR7 expression (22).
Chemotaxis Assay.
Cells were labeled with 2.5 μM calcein-AM (Invitrogen) at 37 °C for 30 min, washed, and suspended to a concentration of 1 × 106 cells per mL in RPMI 1640 supplemented with 25 mM Hepes (Life Technologies). A total of 250 μL was seeded into FluoroBlok (Corning) 3-μm porous inserts, which served as the upper chamber of a Boyden assay within a 24-well plate. The lower chamber contained 750 μL of HBSS (Life Technologies) with Hepes (25 mM), CCL19 or CCL21 (600 ng/mL; PeproTech), 0.1% BSA (Sigma-Aldrich), and zinc chloride (75 μM; Sigma-Aldrich), unless otherwise indicated. For direct visualization of migrating cells, imaging of the underside of the insert was performed by using a Zeiss Axiovert 200 inverted fluorescent microscope with digital images captured with AxioVision software. We chose 1 h as the time point for evaluation, when >98% of migrating cells remained adherent to the membrane insert and had not passed into the lower-chamber solution.
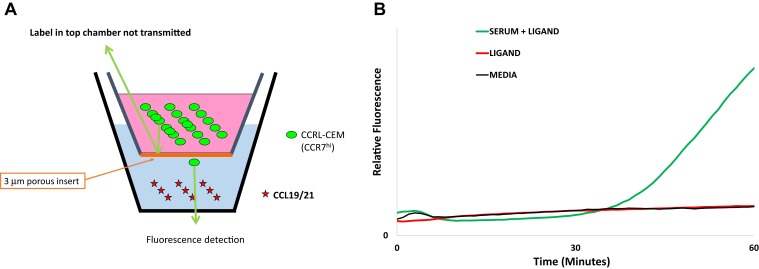
To facilitate evaluation of multiple conditions simultaneously during serum-fractionation studies, we adapted the transwell assay to permit real-time, continuous fluorimetric monitoring of migrating cells using a plate reader (Fig. S2A), where migrating cells could be quantified and fraction activity assessed by the rate of change in fluorescence over time. Fluorimetric measurements were made with an excitation wavelength of 485 nm and detection wavelength of 520 nm by using a SpectraMax M2e microplate reader (Molecular Devices), preheated to 37 °C. Kinetic curves were generated by using SoftMax Pro software. This method demonstrated an increase in signal with ligand in the presence of serum (Fig. S2B). We confirmed that the increase in fluorescence detected by the fluorimeter represented cells that were adherent to the underside of the transwell membrane.
Fig. S2.
(A) Schematic of the chemotaxis assay with fluorescent CCR7hi T lymphocytes in the upper chamber and CCL19 or CCL21 in the lower chamber, separated by a light-impermeant 3-μm porous insert. Chemotaxis is quantified in real time by measuring relative fluorescence in the bottom chamber. (B) Kinetic curves of fluorimeter-based measurements, consistent with data from direct visualization.
Chromatography.
Adult bovine serum (Sigma-Aldrich) was used for the initial input. Samples were injected into the following columns (all from GE Healthcare) as part of an AKTA Purifier apparatus (GE Healthcare) maintained at 4 °C: Superdex 200 (gel filtration); MonoQ (anion exchange); HiLoad Sepharose SP or MonoS (cation exchange); and HiTrap Protein G (Ig depletion). For gel filtration, columns were equilibrated with HBSS/Hepes (25 mM). For anion exchange, samples underwent buffer exchange into Tris (20 mM; pH 8.1) and NaCl (25 mM). Gradient or stepwise elution was performed with increasing NaCl concentrations (0–400 mM) of Tris-buffered (20 mM; pH 8.1) saline. For cation exchange, samples underwent buffer exchange into Hepes (25 mM; pH 7.4) and sodium chloride (25 mM). For input sera undergoing cation exchange, no buffer exchange was performed, and only Hepes (25 mM) was added before running the sera into the column. Gradient elution was performed with Hepes-buffered (25 mM; pH 7.4) saline (0–400 mM). Fractions were concentrated by using an Amicon Ultra centrifugal filter unit with a molecular mass cutoff of 10 kDa, and desalted into HBSS with Hepes (25 mM) using a HiTrap Desalting column (GE Healthcare) before testing activity in the chemotaxis assay.
Mass Spectrometry.
Select fractions, based on chemotactic activity, were submitted to The Rockefeller University Proteomics Resource Center. Proteins in solution were reduced by adding 10 mM DTT (Sigma-Aldrich) in 1 M triethylammonium bicarbonate (Sigma-Aldrich). After incubation for 30 min at 57 °C, proteins were alkylated by adding 20 mM iodoacetamide (Sigma-Aldrich). Proteins were trypsinized (Promega) overnight at 37 °C. Digestions were halted with formic acid, followed by desalting and concentration using Empore C18 (3 M) StAGE tips (46). Tryptic peptides were analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS): Q-Exactive or Orbitrap XL (Thermo), both connected to nano-LC (Dionex NCP3200RS LC setups; Thermo). A 75-μm × 12-cm (3-μm particles) C18 reversed-phase column (Nikkyo Technos) was used to resolve the peptides. The analytical gradient was generated at 200 nL/min, increasing from 5% solution B (0.1% formic acid in acetonitrile)/95% solution A (0.1% formic acid) to 45% solution B/55% solution A over 45–75 min, followed by column wash with 90% solution B/10% solution A. All LC-MS/MS experiments were performed in data-dependent (nontargeted) mode.
LC-MS/MS data were processed with ProteomeDiscoverer (Version 1.4; Thermo) and Mascot (Version 2.3) and queried against Uniprot’s Complete Bovine proteome protein database (January 2013) annotated for common contaminants (47). Precursor mass accuracy of 20 ppm was used, and fragment ion mass tolerance of 0.5 Da and 20 mDa were used for Orbitrap XL and Q-Exactive data, respectively. All cysteines were considered as alkylated with acetamide. Oxidation of methionine and protein N-terminal acetylation were chosen as variable modifications. Matched peptides were filtered by using a Percolator-based false discovery rate of 1% or better (48). The average area of the three most abundant peptides for a matched protein was used to gauge protein amounts within and in-between samples (49).
TriCEPS Ligand-Receptor Capture.
The TriCEPS reagent is a proprietary trifunctional molecule with three key moieties: (i) an amine-reactive group capable of binding a peptide ligand of interest, (ii) a cross-linking group capable of bonding to oxidized glycans, and (iii) an affinity tag for downstream extraction (20, 50). These studies were performed by DualSystems Biotech. Briefly, either human H497–K520 or transferrin was coupled to the TriCEPS reagent. The TriCEPS–ligand complex was added to CCRF-CEM cells in complete medium in the presence or absence of zinc (100 μM), after treatment with an oxidizing reagent to oxidize surface glycoproteins. Cells were then lysed and processed as described, including LC-MS/MS analysis of captured peptides (20). Comparisons between conditions were made in triplicate to determine the relative fold enrichment of a given protein. P values were determined for pairwise comparisons and adjusted for multiple comparisons.
Intracellular Calcium Flux.
CEM cells were loaded with Fluo-4 (2 μM; Life Technologies) for 1 h at 37 °C and then washed twice in Hepes-buffered (25 mM) HBSS without calcium/magnesium, supplemented with zinc (75 μM). Cells were suspended to a concentration of 1 × 107 cells per mL. Fluorescence was measured by using a Fluoroskan Ascent fluorimeter (Thermo Scientific).
Adhesion Assay.
CCRF-CEM cells were labeled with calcein-AM and resuspended in serum-free RPMI 1640 supplemented with zinc (75 μM) at a concentration of 1 × 106 per mL. A total of 150 μL was seeded into a plate precoated with various ECM proteins (Cell Biolabs). Human CCL19 or H497–K520 was added to the wells. After 3 h, the wells were washed five times with serum-free RPMI 1640, and fluorescence values of individual wells were determined. Each condition was performed in triplicate.
Animals.
Wild-type C57BL/6 mice were purchased from The Jackson Laboratories. Sera and tissue were obtained from animals between 8 and 10 wk of age. Animals were housed under specific pathogen-free conditions, and all study procedures were approved by Rockefeller University’s Institutional Animal Care and Use Committee.
Adoptive Transfer.
Spleens of wild-type C57BL/6 mice were surgically excised. Single-cell suspensions were prepared by grinding lymphoid tissue through a 40-μm mesh cell strainer (Fisher). Cells were treated with ACK red blood cell lysis buffer (BioWhittaker), washed with RPMI with 10% FBS, and labeled with CellTracker Green (Life Technologies). Cells were washed and suspended in Hepes-buffered HBSS with 75 μM zinc at a concentration of 1 × 108 cells per mL. Cells were incubated with a synthetic HK peptide, based on the mouse HK protein (H517–R540; HGHGHGHGKHTNKDKNSVKQTTQR; 8 μM) or vehicle at 37 °C for 30 min. A total of 100 μL of the cell suspension was injected i.v. into male wild-type C57BL/6 mice. After 60 min, inguinal lymph nodes from recipient mice were harvested and single-cell suspensions were prepared, as described above. Cells were washed and then stained with fluorochrome-conjugated anti-CD3 or -CD19 antibodies (BioLegend). Cells were washed and then treated with one-step Fix/Lyse Solution (eBioscience), and then analyzed by using a BD LSR II Flow Cytometer (BD Biosciences). Cells were gated for lymphocytes based on side scatter and forward scatter (FSC) measurements. Further gating on FSC-Area and FSC-Height was used to identify single cells. Cells were then classified as either CD3+ or CD19+, followed by gating for CellTracker-labeled cells to determine the proportion of donor cells that had accumulated in recipient lymph tissue as a percentage of lymphocytes.
Reagents.
Human HKa, human coagulation factor XIIa, and human plasma kallikrein were purchased from Enzyme Research Laboratories. Recombinant domain 5 of human HK (kininostatin) was purchased from R&D Systems. Human factor XI-null and factor XII-null plasma were purchased from George King Bio-Medical. Control plasma was purchased from Equitech-Bio. CTI was purchased from Haematologic Technologies. Kininogen-deficient serum was a generous gift from Alvin Schmaier, Case Western Reserve University, Cleveland. Antibodies against TSP4, VDAC2, and isotype control were purchased from Abcam, and those against CCR7 were from BioLegend. Human and mouse HK peptide syntheses were performed by the Proteomics Resource Center of The Rockefeller University, based on the Uniprot sequences for human kininogen (P01042) and mouse kininogen (O08677). The full sequence of the human H497–K520 peptide is HKHGHGHGKHKNKGKKNGKHNGWK.
Statistical Analysis.
For in vitro chemotaxis studies, each condition was performed in triplicate. Data shown from chemotaxis and calcium flux studies are representative of at least three independent experiments. For quantification based on direct visualization, digital images of low power fields (2.5×) were analyzed by using ImageJ software, using the “Analyze Particle” function, after uniform thresholding was applied to all images under comparison. Cell count metrics between conditions were analyzed by using GraphPad Prism software. Statistical tests were applied to replicates within an experiment, unless otherwise specified. Data are shown as mean values and SD of replicates within an experiment. Comparisons among three or more conditions were made by using one-way analysis of variance, and comparisons between two conditions were made by using the two-tailed Student’s t test.
Acknowledgments
We thank Drs. Henrik Molina and Henry Zebroski (Rockefeller Proteomics Resource Center) for assistance with mass spectrometry analyses and peptide synthesis; Dr. Juana Gonzalez (director of the Translational Technology Core Laboratory) for assistance with flow cytometry studies; Dr. Alvin Schmaier for providing kininogen-deficient serum; and Ms. Eliza Prangley for technical assistance and manuscript preparation. This work was supported by NIH Grants K08DK085154 (to M.P.P.) and 8 UL1 TR000043 (to The Rockefeller University); The Sackler Center for Biomedicine and Nutrition Research at The Rockefeller University; and the Robertson Therapeutic Development Fund at The Rockefeller University.
Footnotes
Conflict of interest statement: The authors note that The Rockefeller University has filed two provisional patent applications for small molecule inhibitors of Factor XIIa; however, none of these inhibitors were used in this paper.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615671113/-/DCSupplemental.
References
- 1.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: The multistep paradigm. Cell. 1994;76(2):301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 2.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 3.Comerford I, et al. A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Factor Rev. 2013;24(3):269–283. doi: 10.1016/j.cytogfr.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: Balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 5.Irino T, et al. CC-Chemokine receptor CCR7: A key molecule for lymph node metastasis in esophageal squamous cell carcinoma. BMC Cancer. 2014;14:291. doi: 10.1186/1471-2407-14-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Correlation effect of EGFR and CXCR4 and CCR7 chemokine receptors in predicting breast cancer metastasis and prognosis. J Exp Clin Cancer Res. 2010;29:16. doi: 10.1186/1756-9966-29-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moschovakis GL, Förster R. Multifaceted activities of CCR7 regulate T-cell homeostasis in health and disease. Eur J Immunol. 2012;42(8):1949–1955. doi: 10.1002/eji.201242614. [DOI] [PubMed] [Google Scholar]
- 8.Schieffer B, Luchtefeld M. Emerging role of chemokine receptor 7 in atherosclerosis. Trends Cardiovasc Med. 2011;21(8):211–216. doi: 10.1016/j.tcm.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Krumbholz M, et al. CCL19 is constitutively expressed in the CNS, up-regulated in neuroinflammation, active and also inactive multiple sclerosis lesions. J Neuroimmunol. 2007;190(1-2):72–79. doi: 10.1016/j.jneuroim.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Dieu-Nosjean MC, Goc J, Giraldo NA, Sautès-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 2014;35(11):571–580. doi: 10.1016/j.it.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11(9):597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 12.Nibbs RJ, Graham GJ. Immune regulation by atypical chemokine receptors. Nat Rev Immunol. 2013;13(11):815–829. doi: 10.1038/nri3544. [DOI] [PubMed] [Google Scholar]
- 13.Comerford I, et al. The atypical chemokine receptor CCX-CKR scavenges homeostatic chemokines in circulation and tissues and suppresses Th17 responses. Blood. 2010;116(20):4130–4140. doi: 10.1182/blood-2010-01-264390. [DOI] [PubMed] [Google Scholar]
- 14.Renné T. The procoagulant and proinflammatory plasma contact system. Semin Immunopathol. 2012;34(1):31–41. doi: 10.1007/s00281-011-0288-2. [DOI] [PubMed] [Google Scholar]
- 15.Long AT, Kenne E, Jung R, Fuchs TA, Renné T. Contact system revisited: An interface between inflammation, coagulation, and innate immunity. J Thromb Haemost. 2016;14(3):427–437. doi: 10.1111/jth.13235. [DOI] [PubMed] [Google Scholar]
- 16.Sainz IM, Pixley RA, Colman RW. Fifty years of research on the plasma kallikrein-kinin system: From protein structure and function to cell biology and in-vivo pathophysiology. Thromb Haemost. 2007;98(1):77–83. [PubMed] [Google Scholar]
- 17.LaRusch GA, et al. Factor XII stimulates ERK1/2 and Akt through uPAR, integrins, and the EGFR to initiate angiogenesis. Blood. 2010;115(24):5111–5120. doi: 10.1182/blood-2009-08-236430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan AP, Ghebrehiwet B. The plasma bradykinin-forming pathways and its interrelationships with complement. Mol Immunol. 2010;47(13):2161–2169. doi: 10.1016/j.molimm.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Pixley RA, et al. Interaction of high-molecular-weight kininogen with endothelial cell binding proteins suPAR, gC1qR and cytokeratin 1 determined by surface plasmon resonance (BiaCore) Thromb Haemost. 2011;105(6):1053–1059. doi: 10.1160/TH10-09-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frei AP, et al. Direct identification of ligand-receptor interactions on living cells and tissues. Nat Biotechnol. 2012;30(10):997–1001. doi: 10.1038/nbt.2354. [DOI] [PubMed] [Google Scholar]
- 21.Gunn MD, et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J Exp Med. 1999;189(3):451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buonamici S, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459(7249):1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan MM, et al. High-molecular-weight kininogen fragments stimulate the secretion of cytokines and chemokines through uPAR, Mac-1, and gC1qR in monocytes. Arterioscler Thromb Vasc Biol. 2006;26(10):2260–2266. doi: 10.1161/01.ATV.0000240290.70852.c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavakis T, et al. Regulation of leukocyte recruitment by polypeptides derived from high molecular weight kininogen. FASEB J. 2001;15(13):2365–2376. doi: 10.1096/fj.01-0201com. [DOI] [PubMed] [Google Scholar]
- 25.Vu TT, Fredenburgh JC, Weitz JI. Zinc: An important cofactor in haemostasis and thrombosis. Thromb Haemost. 2013;109(3):421–430. doi: 10.1160/TH12-07-0465. [DOI] [PubMed] [Google Scholar]
- 26.Schousboe I. Contact activation in human plasma is triggered by zinc ion modulation of factor XII (Hageman factor) Blood Coagulation Fibrinolysis. 1993;4(5):671–678. [PubMed] [Google Scholar]
- 27.Björkqvist J, Nickel KF, Stavrou E, Renné T. In vivo activation and functions of the protease factor XII. Thromb Haemost. 2014;112(5):868–875. doi: 10.1160/TH14-04-0311. [DOI] [PubMed] [Google Scholar]
- 28.Maas C, et al. Misfolded proteins activate factor XII in humans, leading to kallikrein formation without initiating coagulation. J Clin Invest. 2008;118(9):3208–3218. doi: 10.1172/JCI35424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmaier AH. The contact activation and kallikrein/kinin systems: Pathophysiologic and physiologic activities. J Thromb Haemost. 2016;14(1):28–39. doi: 10.1111/jth.13194. [DOI] [PubMed] [Google Scholar]
- 30.Morrison DC, Cochrane CG. Direct evidence for Hageman factor (factor XII) activation by bacterial lipopolysaccharides (endotoxins) J Exp Med. 1974;140(3):797–811. doi: 10.1084/jem.140.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiggins RC. Kinin release from high molecular weight kininogen by the action of Hageman factor in the absence of kallikrein. J Biol Chem. 1983;258(14):8963–8970. [PubMed] [Google Scholar]
- 32.Iwaki T, Castellino FJ. Plasma levels of bradykinin are suppressed in factor XII-deficient mice. Thromb Haemost. 2006;95(6):1003–1010. doi: 10.1160/TH06-03-0128. [DOI] [PubMed] [Google Scholar]
- 33.Kanda H, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116(6):1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damås JK, et al. Enhanced expression of the homeostatic chemokines CCL19 and CCL21 in clinical and experimental atherosclerosis: Possible pathogenic role in plaque destabilization. Arterioscler Thromb Vasc Biol. 2007;27(3):614–620. doi: 10.1161/01.ATV.0000255581.38523.7c. [DOI] [PubMed] [Google Scholar]
- 35.Kenne E, Renne T. Factor XII: A drug target for safe interference with thrombosis and inflammation. Drug Discov Today. 2014;19(9):1459–1464. doi: 10.1016/j.drudis.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Adams JC, Lawler J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011;3(10):a009712. doi: 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girard F, Eichenberger S, Celio MR. Thrombospondin 4 deficiency in mouse impairs neuronal migration in the early postnatal and adult brain. Mol Cell Neurosci. 2014;61:176–186. doi: 10.1016/j.mcn.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Lv L, et al. Thrombospondin-4 ablation reduces macrophage recruitment in adipose tissue and neointima and suppresses injury-induced restenosis in mice. Atherosclerosis. 2016;247:70–77. doi: 10.1016/j.atherosclerosis.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Frolova EG, et al. Thrombospondin-4 regulates vascular inflammation and atherogenesis. Circ Res. 2010;107(11):1313–1325. doi: 10.1161/CIRCRESAHA.110.232371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topol EJ, et al. Single nucleotide polymorphisms in multiple novel thrombospondin genes may be associated with familial premature myocardial infarction. Circulation. 2001;104(22):2641–2644. doi: 10.1161/hc4701.100910. [DOI] [PubMed] [Google Scholar]
- 41.Wessel J, Topol EJ, Ji M, Meyer J, McCarthy JJ. Replication of the association between the thrombospondin-4 A387P polymorphism and myocardial infarction. Am Heart J. 2004;147(5):905–909. doi: 10.1016/j.ahj.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Pluskota E, et al. Mechanism and effect of thrombospondin-4 polymorphisms on neutrophil function. Blood. 2005;106(12):3970–3978. doi: 10.1182/blood-2005-03-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Congote LF, Sadvakassova G, Dobocan MC, Difalco MR, Kriazhev L. Biological activities and molecular interactions of the C-terminal residue of thrombospondin-4, an epitome of acidic amphipathic peptides. Peptides. 2010;31(4):723–735. doi: 10.1016/j.peptides.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Jordan AR, Racine RR, Hennig MJ, Lokeshwar VB. The role of CD44 in disease pathophysiology and targeted treatment. Front Immunol. 2015;6:182. doi: 10.3389/fimmu.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabas I, Glass CK. Anti-inflammatory therapy in chronic disease: Challenges and opportunities. Science. 2013;339(6116):166–172. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 47.Bunkenborg J, García GE, Paz MI, Andersen JS, Molina H. The minotaur proteome: Avoiding cross-species identifications deriving from bovine serum in cell culture models. Proteomics. 2010;10(16):3040–3044. doi: 10.1002/pmic.201000103. [DOI] [PubMed] [Google Scholar]
- 48.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 49.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Frei AP, Moest H, Novy K, Wollscheid B. Ligand-based receptor identification on living cells and tissues using TRICEPS. Nat Protoc. 2013;8(7):1321–1336. doi: 10.1038/nprot.2013.072. [DOI] [PubMed] [Google Scholar]