Significance
The positive transcription elongation factor (P-TEFb) is required for transcription of most genes by RNA polymerase II. Hexim proteins associated to 7SK, a noncoding RNA, bind to P-TEFb and reversibly inhibit its activity. This transcription factor comprises the kinase Cdk9 and a cyclin T. Using a photoreactive amino acid incorporated into proteins, we provide the first evidence of a direct interaction between Cdk9 and Hexim1, in live cells, in cell extracts and in in vitro assembled complexes. An evolutionary conserved Hexim1 peptide, the “PYNT” sequence, cross-links to the Cdk9 activation segment that controls access to the catalytic cleft. Interference with binding of substrates accounts for kinase inhibition.
Keywords: cyclin-dependent kinase inhibition, transcription factor regulation, regulatory noncoding RNA, benzoyl phenylalanine, protein–protein cross-linking
Abstract
The positive transcription elongation factor (P-TEFb) is required for the transcription of most genes by RNA polymerase II. Hexim proteins associated with 7SK RNA bind to P-TEFb and reversibly inhibit its activity. P-TEFb comprises the Cdk9 cyclin-dependent kinase and a cyclin T. Hexim proteins have been shown to bind the cyclin T subunit of P-TEFb. How this binding leads to inhibition of the kinase activity of Cdk9 has remained elusive, however. Using a photoreactive amino acid incorporated into proteins, we show that in live cells, cell extracts, and in vitro reconstituted complexes, Hexim1 cross-links and thus contacts Cdk9. Notably, replacement of a phenylalanine, F208, belonging to an evolutionary conserved Hexim1 peptide (202PYNTTQFLM210) known as the “PYNT” sequence, cross-links a peptide within the activation segment that controls access to the Cdk9 catalytic cleft. Reciprocally, Hexim1 is cross-linked by a photoreactive amino acid replacing Cdk9 W193, a tryptophan within this activation segment. These findings provide evidence of a direct interaction between Cdk9 and its inhibitor, Hexim1. Based on similarities with Cdk2 3D structure, the Cdk9 peptide cross-linked by Hexim1 corresponds to the substrate binding-site. Accordingly, the Hexim1 PYNT sequence is proposed to interfere with substrate binding to Cdk9 and thereby to inhibit its kinase activity.
The positive transcription elongation factor (P-TEFb) is required for the transcription of most genes by RNA polymerase II (1, 2). In particular, it antagonizes the negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF), which promote transcriptional pausing shortly after the initiation of transcription. P-TEFb comprises a kinase subunit, Cdk9, and a cyclin T. The activity of P-TEFb is regulated. In metazoan cells, 7SK RNA and Hexim proteins are dynamically associated with an inactive form of P-TEFb (3–6).
7SK RNA is a noncoding RNA that stably associates Larp7 and Mepce proteins to form a core 7SK snRNP (7–9). Although the total amount of 7SK RNA does not vary in live cells, the level of 7SK RNA available for Hexim binding is modulated (10–12). As a consequence, the overall P-TEFb activity increases on inhibition of transcription (3, 4). Thus, in addition to other more specific functions (13–15), 7SK RNA serves as a sensor of transcriptional activity through a feedback loop, fine-tuning P-TEFb activity (10). Understanding how this noncoding RNA mechanistically regulates the activity of P-TEFb, a central transcription factor, remains challenging.
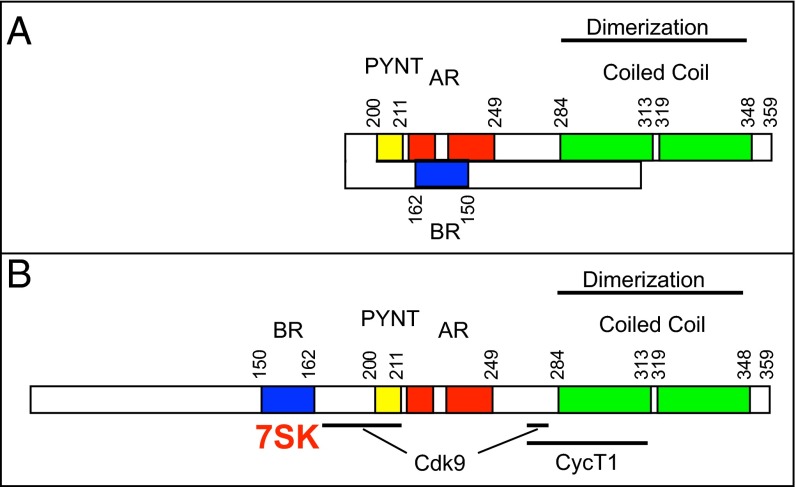
Hexim proteins have been conserved throughout evolution from mammals to nematodes (16–19). Most mammals have two distinct genes coding for the cognate Hexim1 and Hexim2 proteins. Both proteins share similar properties and functions, but most previous studies have been performed with human Hexim1. The binding of 7SK snRNP to a Hexim1 dimer creates an inactive P-TEFb complex comprising two P-TEFb modules (19–21). Studies using yeast two-hybrid analysis, GST pull-downs, analytical gel filtration, and isothermal calorimetry indicate that Hexim1 interacts directly with cyclin T (5, 22, 23). The cyclin T-binding domain is located in the C-terminal part of Hexim1, overlapping a coiled-coil dimerization domain (24, 25) (Fig. 1A, green). Furthermore, an evolutionary conserved motif in the center of the protein, the “PYNT” sequence, is critical for P-TEFb binding and inhibition (20, 21, 26). This motif, distinct from the cyclin T-binding domain, is located between a basic region (BR) involved in 7SK RNA binding and an acidic region (AR) (Fig. 1A).
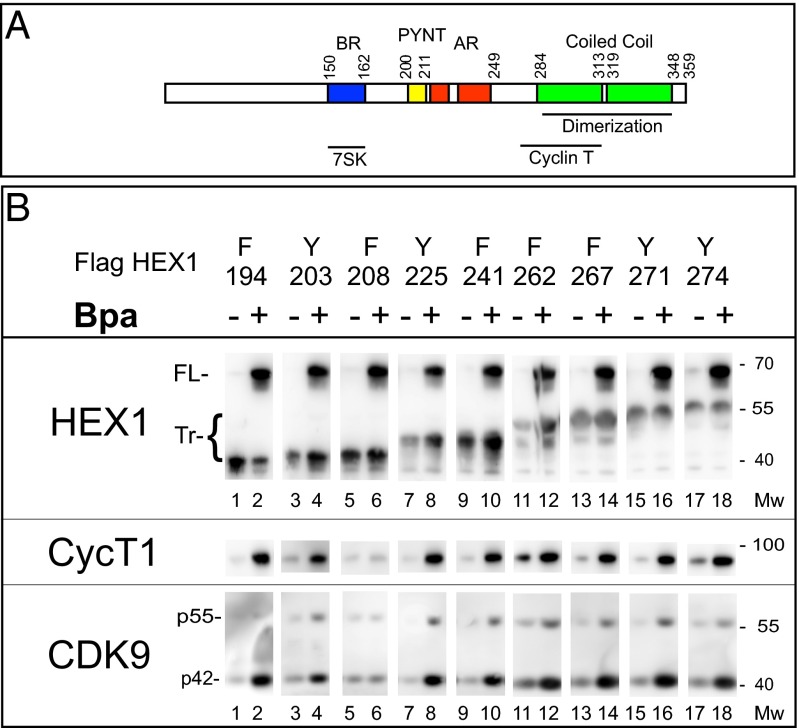
Fig. 1.
Coimmunoprecipitation of P-TEFb with Hexim1.Bpa. (A) Conserved functional domains in human Hexim1. The BR (blue) interacts with 7SK RNA, the AR (red) can interact with the BR, and the evolutionary conserved PYNT sequence (yellow) is located between the AR and BR regions. The coiled-coil dimerization domain (green) overlaps the cyclin T-binding domain. (B) HEK 293 cells were cotransfected with suppressor tRNA, Bpa synthetase, and a Hexim1 cDNA with a TAG stop codon replacing different amino acid codons (the corresponding amino acid position numbers are noted at the top). N-terminal Flag-tagged Hexim1 was immunoprecipitated from extracts of cells treated with (+) or without (−) 2 mM Bpa. Western blots were probed with antibodies directed against cyclin T1 (CycT1), Cdk9, or the N terminus of Hexim1 (HEX1) that detected both truncated (Tr) and FL Hexim1 proteins. Both Cdk9 (p42) and Cdk9 (p55) isoforms (39) are detected (even lanes).
Despite these insights, how Hexim1 binding inhibits Cdk9 kinase activity remains unclear. Hypotheses might be formulated based on our knowledge of other Cdk inhibitors. Hexim1 might compete with ATP to prevent it from interacting with the kinase active site like the p27Kip1 protein inhibitor of Cdk2/cyclin A (27). Hexim1 might induce conformational changes weakening cyclin binding to the kinase subunit, distorting the ATP binding site and/or misaligning catalytic residues like the Ink4 inhibitors of Cdk4/6/cyclin D (28, 29).
To characterize the mechanism underlying the inhibition of P-TEFb activity by Hexim1, we investigated possible direct interactions with Cdk9. Protein–protein contacts were probed by covalent protein–protein coupling using a photoreactive amino acid incorporated at defined positions into protein partners (30–32). When photoreactive amino acids are used, proteins in close contact can be covalently photocross-linked (33). In the present study, we demonstrate that the same Hexim1 residues cross-link Cdk9 in live cells, in cell extracts, and in in vitro reconstituted complexes. Cross-linked peptides generated by proteolytic digestion can be characterized by mass spectrometry (34–36). Hexim1 cross-links a Cdk9 peptide overlapping the activation segment that controls access to the catalytic cleft (37). These findings support the model that Hexim1 inhibits P-TEFb activity through interference with the substrate-binding surface in the catalytic site of Cdk9.
Results
Coimmunoprecipitation of P-TEFb with Hexim1.Bpa Proteins from Transfected Mammalian Cell Lysates.
Incorporation of an unnatural amino acid is driven by coexpression of a suppressor tRNA (tRNASup) and an aminoacyl tRNA synthetase specific for both the tRNASup and the unnatural amino acid (30–32). Aminoacyl tRNA synthetases adapted to the photoreactive amino acids, p-benzoyl-phenylalanine (Bpa) and p-azido-phenylalanine (Azp), have been developed (38). To synthesize proteins incorporating these residues, amber stop codons (TAG) are introduced at desired positions in the coding sequence. HEK 293 cells are cotransfected with a tRNASup for amber codons and the appropriate amino acyl tRNA synthetase, and also with a Hexim1 cDNA fused to an N-terminal Flag epitope. We focused on the C-terminal half of Hexim1, which contains sequences required for P-TEFb interactions (Fig. 1A) (5). Aromatic residues were selected to minimize potential structure alterations. Proteins coimmunoprecipitated from cell extracts with a Flag-antibody were analyzed by Western blot. In the absence of the photoreactive amino acid in the culture medium, mostly truncated Hexim1 proteins (Tr) were detected (Fig. 1B, odd lanes); however, when the photoreactive amino acid was added to the culture medium, the full-length (FL) protein increased sharply (even lanes). Thus, Hexim1.Bpa proteins are generated with good efficiency in transfected mammalian cells.
The presence of FL Hexim1.Bpa proteins coincides with a marked increase in coimmunoprecipitation of P-TEFb subunits (Cdk9 and cyclin T1) (Fig. 1B). Replacement of F208 is an exception in that the increase in immunoprecipitated P-TEFb was weaker (lane 6). Notably, a small amount of P-TEFb coprecipitates with the truncated proteins in the absence of Bpa (odd lanes). Expression of the suppressor tRNA and synthetase promote a weak suppression resulting in synthesis of some full-length Flag-HEXIM1 protein.
Position-dependent cross-links in live cells between Cdk9 and Hexim1.
To generate protein–protein cross-links, transfected cell lysates were irradiated and immunoprecipitated with anti-Flag beads. Immunoprecipitates were probed by Western blot analysis. After irradiation, high molecular weight bands (>130 kDa) were detected with a Hexim1 antibody (Fig. 2A, Upper). The most significant cross-links are observed upon Bpa replacement of Hexim1 Y167 or W172 or F262 or F267 or Y274. Two types of profiles are distinguished. Replacement of Y167 and W172 leads to a smear between 130 and 170 kDa apparent molecular weight, whereas F262, F267, and Y274 replacement results mainly in bands >170 kDa.
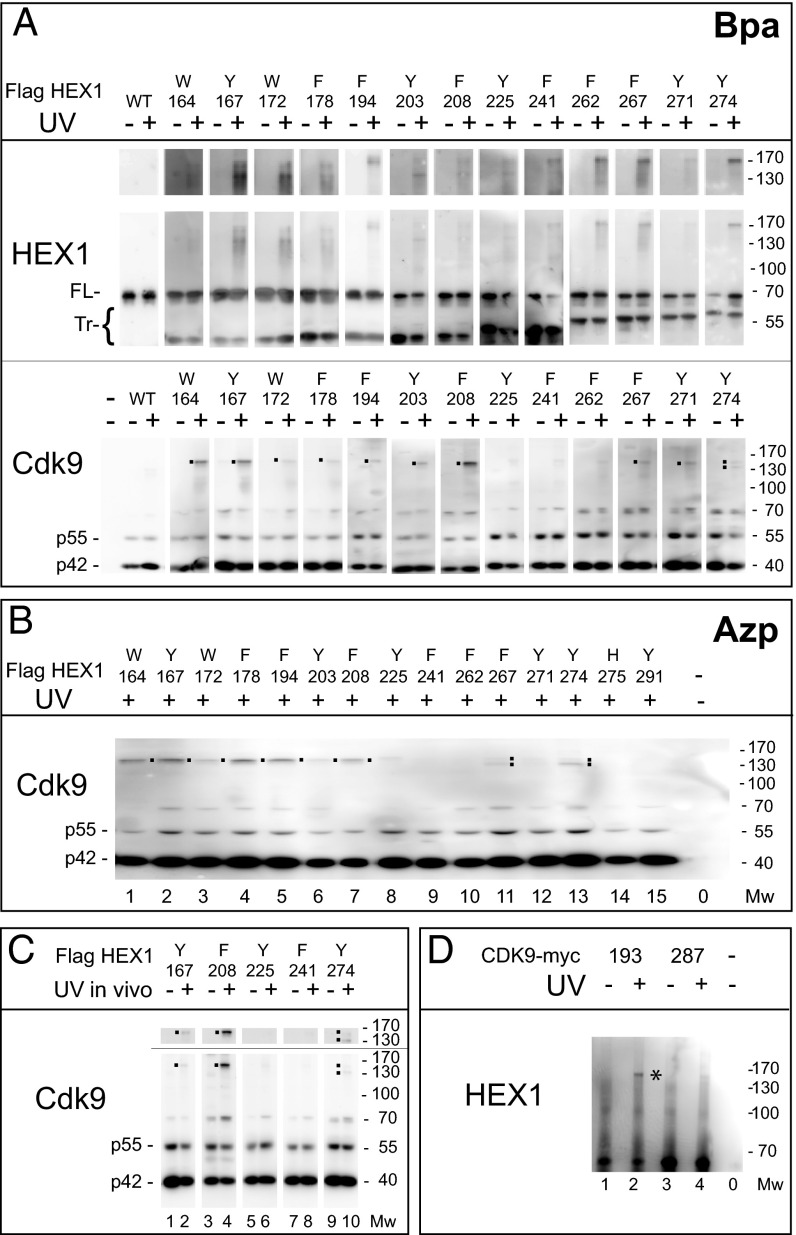
Fig. 2.
Immunoprecipitation of Cdk9 × Hexim1.Bpa cross-linked species. (A and B) Flag immunoprecipitation of N-terminal Flag-tagged Hexim1 from UV irradiated (+) or not (−) cell extracts. (C) Cells expressing various Hexim.Bpa proteins were UV-irradiated alive (+) under ice-cold PBS. Flag immunoprecipitation was performed from cell extracts. (D) Cells were transfected with myc-tagged Cdk9, followed by myc immunoprecipitation of C-terminal myc-tagged Cdk9 from UV irradiated (+) or not (−) cell extracts. In A, C, and D, HEK 293-transfected cells were exposed to Bpa. In B, transfected cells were exposed to Azp. Stronger exposures of the cross-linked species are shown in the upper parts of A and C. Western blots were probed with antibodies directed against Cdk9 or the N terminus of Hexim1 (HEX1).
Anti-Cdk9 antibodies detect UV generated cross-linked species between 130 and 170 kDa when Hexim1 residues between W164, Y167, and F208 were replaced with Bpa (Fig. 2A, Lower). Weaker signals were seen with other positions. Notably, two distinct bands were detected with Y274Bpa suggesting two different Hexim1 × Cdk9 cross-linked species. F208Bpa substitution showed the strongest cross-link efficiency; the cross-linked band intensity was close to that of uncross-linked Cdk9 p55 isoform. The same experiment was repeated with another photoreactive amino acid, Azp (Fig. 2B). Azp is smaller than Bpa, but the azido group is charged. Again, strong cross-links were detected with Azp substituting residues between positions 164 and 208 and with Y274Azp. The latter cross-link migrates faster than with other positions. The major bands detected by Cdk9 antibodies differ from the major bands detected with Hexim1 antibodies; however, it should be kept in mind that Hexim1 might cross-link with itself in dimers or with any of its numerous identified (or not) protein partners.
To demonstrate that the foregoing cross-links are not lysis artifacts, live cells were UV-irradiated while still attached to their Petri dishes and covered with chilled PBS. Flag Hexim1.Bpa is immunoprecipitated from extracts of cells irradiated alive. As in cell extracts, Hexim1 F208Bpa provided the strongest cross-link with Cdk9 (Fig. 2C, lane 4). Cross-links are undetectable with Y225Bpa and F241Bpa. Weak cross-links were detected with Hexim1 Y167Bpa and Y274Bpa (Fig. 2C, lanes 2 and 10). Like in cell extracts, two Y274Bpa cross-linked species (lane 10) were seen. The stronger species migrates faster than other positions. Cross-link profiles obtained with cell extracts match those of live cells.
Cross-linking in vitro reconstituted P-TEFb.Hexim1.7SK RNA complexes.
The amount of material recovered by immunoprecipitation from transfected mammalian cells is insufficient for a detailed analysis of the cross-linked peptides by mass spectrometry; however, larger amounts of inactive P-TEFb complexes can be reconstituted by the addition of recombinant Hexim1 and P-TEFb and in vitro-transcribed 7SK RNA (20). Microgram amounts of Hexim1.Bpa are readily purified from bacteria and P-TEFb from baculovirus-infected cells. The addition of recombinant Larp7 stabilizes 7SK RNA against degradation, which improves the efficiency and reproducibility of the reconstitution experiments.
We focused on Hexim1 F208Bpa, because this position has the highest cross-linking efficiency in human cell extracts and it belongs to an evolutionary-conserved peptide sequence (202PYNTTQFLM210), the PYNT motif, which was previously shown to be involved in P-TEFb inhibition (20, 21, 26). P-TEFb was added to purified F208Bpa Hexim1.Larp7.7SK RNA complex to reconstitute an inactive P-TEFb.Hexim1.Larp7.7SK RNA complex. UV irradiation generates a protein band (Fig. 3A, lane 2, band X) with a molecular weight of 130–170 kDa, like that of the Flag-Hexim1 × Cdk9 cross-linked species described above obtained from cell lysates. In contrast, UV irradiation has no effect on purified F208Bpa Hexim1 or on F208Bpa Hexim1 regardless of the presence of the Larp7.7SK RNA complex (Fig. S1). This experiment was repeated with Hexim1 with Bpa incorporated at other positions. Cross-linked species were also observed but formed with weaker efficiencies. The major bands obtained after mixing Hexim1 F208Bpa and P-TEFb in the presence of Larp7.7SK (Fig. 3A, lanes 1 and 2) were excised from the gel, submitted to double digestion by trypsin and GluC endopeptidases and analyzed by nano-liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and quantified by MS/MS extracted ion chromatography (XIC). The highest scores for bands C corresponded to Cdk9, for bands H to Hexim1, for bands L to Larp7 and for bands T to cyclin T1 (Fig. 3B). Scores for both Cdk9 and Hexim1 increased markedly in band 2X after irradiation. Actually, both proteins had by far the highest scores in band 2X. Scores for cyclin T1, Larp7, keratins, and trypsin in this band were much lower. Thus, we may conclude that Hexim1 F208Bpa very efficiently cross-links to Cdk9 in vitro.
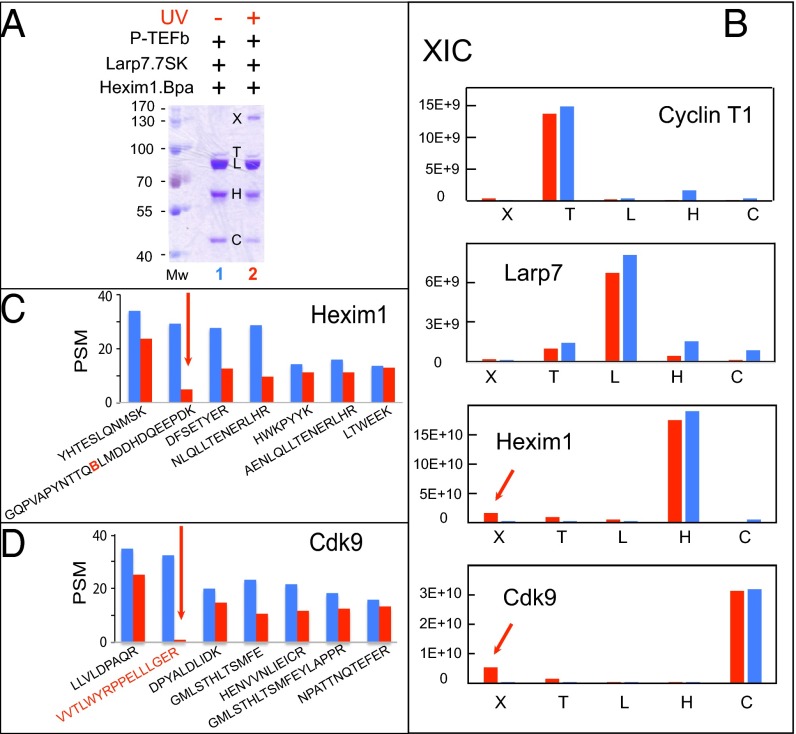
Fig. 3.
Mass spectrometry analysis of cross-linked in vitro reconstituted P-TEFb.Hexim1 F208Bpa.7SK complexes. (A) In vitro-reconstituted P-TEFb.Hexim1 208Bpa.Larp7.7SK complexes, both UV-irradiated (lane 2) and not (lane 1). Polyacrylamide gels were stained by Coomassie blue. (B) Histograms of MS/MS-peak XIC scores obtained from double-digested (trypsin and GluC V8 endopeptidase) gel bands. (C) Histograms of the top-seven validated PSMs for Hexim1 from band 1H (blue) or band 2X (red). (D) Histograms of the top-seven validated PSMs for Cdk9 peptides from band 1H (blue) or band 2X (red). Sequences of different charges or modification state but the same mass were averaged together over three LC-MS/MS runs. Lists of validated peptides are provided in Datasets S1–S3.
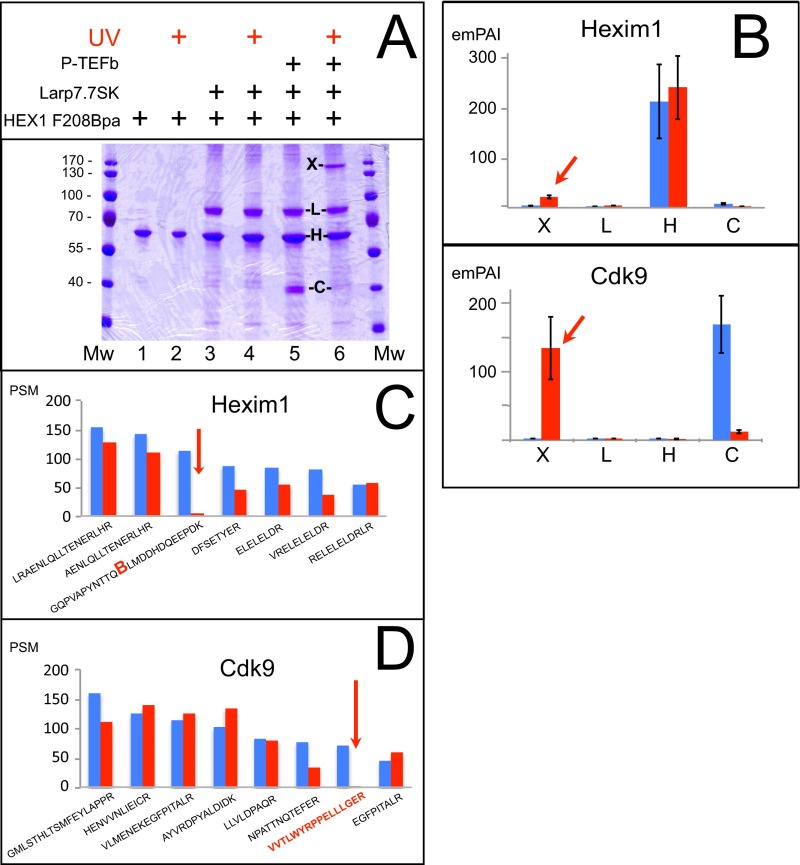
Fig. S1.
(A) In vitro-reconstituted P-TEFb.Hexim1 F208Bpa.Larp7.7SK complexes were UV-irradiated or not. Polyacrylamide gels stained by Coomassie blue. (B) Histograms of the emPAI for Hexim1and Cdk9 in bands X, L, H, and C digested by trypsin. Note the decrease in intensity of a 40-kDa band corresponding to Cdk9 (band C). (C) Histograms of the top-seven validated tryptic PSMs for Hexim1 from band 5C (blue) or band 6X (red). (D) Histograms of the top-eight validated tryptic PSMs for Cdk9 from band 5C (blue) or band 6X (red).
Identification of the Cdk9 target peptide of Hexim1 F208Bpa.
To identify the target peptide of Hexim1 F208Bpa, we ranked more than 100 double-digested peptides attributed to Cdk9 or to Hexim1 according to the number of validated peptide sequence matches (PSMs). Analysis of un–cross-linked Hexim1 from band 1H (Dataset S1) showed that the Hexim1 peptide containing Bpa (B, underscored) replacing F208, GQPVAPYNTTQBLMDDHDQEEPDK, had the second-highest number (Fig. 3C, blue histogram). In contrast, this peptide was hardly detected in the cross-linked band 2X (Dataset S2) (Fig. 3C, red histogram). Such a decrease was expected because Bpa forms a covalent bond with another peptide, thereby generating a new peptide shared by the cross-linked proteins. We also found that the VVTLWYRPPELLLGER Cdk9 peptide was ranked seventh in the number of validated matches in band C corresponding to un–cross-linked Cdk9 (Dataset S3) (Fig. 3D, blue histogram). In contrast, this peptide was hardly detected in band 2X (Dataset S2) (Fig. 3D, red histogram).
We repeated this experiment several times independently with samples from other reconstitution experiments, and found similar results (Fig. S1). Both the Hexim1 Bpa-containing peptide GQPVAPYNTTQBLMDDHDQEEPDK and the VVTLWYRPPELLLGER Cdk9 peptide disappear from gel bands digested by trypsin alone; thus, we conclude that the VVTLWYRPPELLLGER Cdk9 peptide cross-links to Hexim1 F208Bpa.
Hexim1 cross-links with a tryptophan residue located in the catalytic cleft of Cdk9.
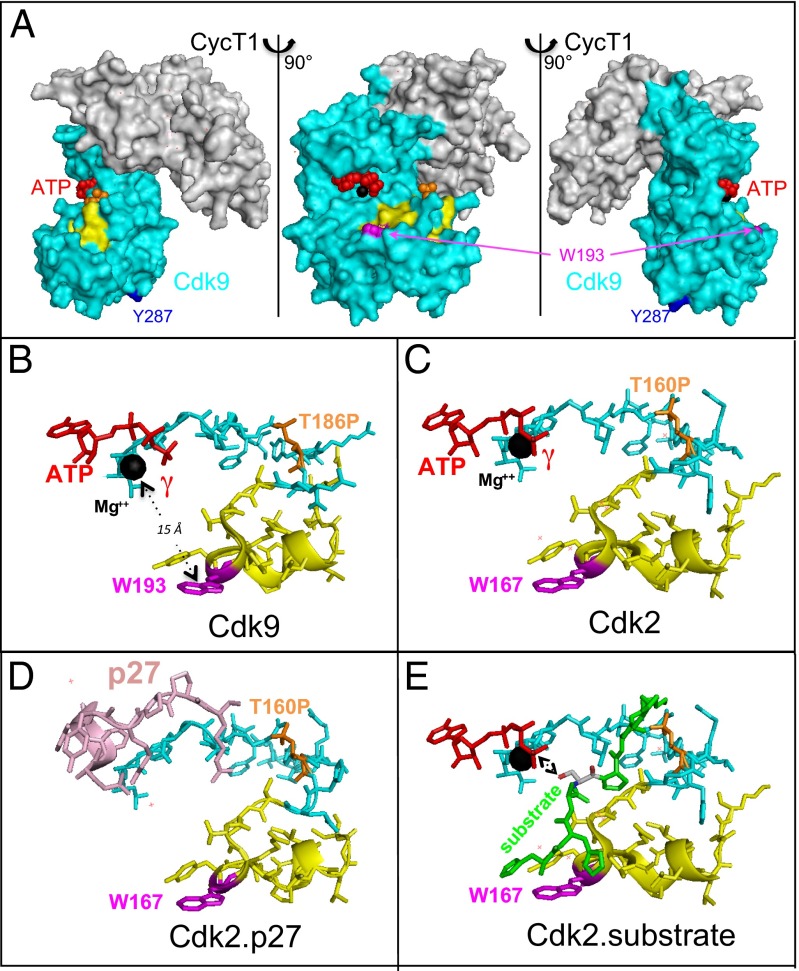
Three-dimensional structures of Cdk9 bound to cyclin T1 have been determined by X-ray crystallography (40, 41). The 189VVTLWYRPPELLLGER204 peptide borders the catalytic cleft (Fig. 4A, yellow) that binds ATP (red) and the catalytic magnesium atom (black sphere). It overlaps with the activation segment (residues 167–198) that controls the accessibility of substrates to the catalytic cleft (40, 42).
Fig. 4.
Localization of a Cdk9 peptide that cross-links Hexim1 F208Bpa on the P-TEFb 3D structure. (A) Surface representation of the Cdk9.cyclin T1 complex [Protein Data Bank (PDB) IDB code 3BLQ], where CDK9 is shown in cyan and cyclin T1 is in gray. Three views corresponding to 90° rotations are shown. (B) Cdk9 catalytic site peptide backbone corresponding to the activation segment (PDB ID code 3BLQ). (C) Cdk2 catalytic site peptide backbone corresponding to the activation segment (PDB ID code 1GMZ). (D) Cdk2 bound to p27Kip (in pink) (PDB ID code 1JSU). (E) Cdk2 bound to the model peptide substrate HHASPRK (in green, except for the serine phosphate acceptor, which is in gray with its phosphorylable oxygen atom in red) (PDB ID code 1QMZ). The Cdk9 189VVTLWYRPPELLLGER204 peptide that cross-links Hexim1 F208Bpa is stained in yellow, except for W193, which is magenta. Color codes for Cdk2 sequences correspond to those in the homologous Cdk9 sequences. ATP (red spheres) and its associated Mg2+ ion (black sphere) mark the catalytic site. Phosphorylated T186, in the T-loop, is in orange, and Y287 is in deep blue.
Our attention was attracted to tryptophan W193 in Cdk9 (Fig. 4, magenta), which is exposed to the solvent and evolutionary conserved in cyclin-dependent kinases. To test a possible reciprocal cross-link of Cdk9 to Hexim1, we replaced W193 by Bpa in a c-myc–tagged Cdk9 construct tagged. When lysates with Cdk9 W193Bpa are irradiated, a high molecular weight species (between 130 and 170 kDa) is immunoprecipitated with a c-myc antibody and detected by Hexim1 antibodies (Fig. 2D). As a negative control, we also replaced Y287, a residue largely exposed to the solvent but positioned on a Cdk9 surface opposite to the cyclin T1 contact interface (Fig. 4, deep blue). No high molecular weight species was detected after UV irradiation of Cdk9 Y287Bpa-containing lysates. This finding demonstrates that Hexim1 contacts the activation segment that controls access to the catalytic cleft of Cdk9.
Discussion
In this work, we show that Hexim1 molecules containing a photoreactive amino acid photocross-link to the Cdk9 subunit of P-TEFb. Importantly, cross-links are observed in live cells, cell extracts, and in vitro-reconstituted complexes. Our results provide proof of a direct interaction between Cdk9 and Hexim1. A cross-linked peptide that overlaps the activation segment that controls access to the catalytic cleft of Cdk9 was identified. This finding provides insight into the mechanism of Cdk9 inhibition by Hexim1.
The Cdk9-Binding Domain Partially Overlaps the Cyclin T1-Binding Domain.
Interaction between cyclin T1 and Hexim1 is well established (5, 22, 23). A cyclin T1-binding domain has been identified overlapping the coiled-coil dimerization domain of Hexim1 (24) (Fig. 1A). In previous work, we mapped cyclin T1 mutations in 11 residues that abolish interactions with Hexim1 in a yeast two-hybrid assay (23); however, only mutations in either one of three cyclin T1 residues (L133 or Y175 or K168) have a detectable effect on the formation of P-TEFb.Hexim1.7SK complexes in human cells. In this paper, we show that several Hexim1 residues cross-link to Cdk9. If Hexim1 contacts both Cdk9 and cyclin T1, then both contacts would be expected to contribute to the stability of the interaction between Hexim1 and P-TEFb. The two-hybrid assay lacks Cdk9. In human cells, cyclin T1 mutations that affect Hexim1 binding in the two-hybrid system might be compensated for by interactions between Cdk9 and Hexim1.
Irradiation of Bpa cleaves one bond in the carbon-oxygen double bond of the benzophenone moiety and generates a diradical, on the carbon atom and on its neighboring oxygen (43). The oxygen radical captures a hydrogen atom on a neighboring C-H bond usually on the peptide backbone, thereby generating another carbene. When they are within a few Ångstroms from one another, the two carbenes react together to form a carbon–carbon bond. Thus, the efficient formation of cross-links demonstrates that Hexim1 and Cdk9 are in very close contact.
Several Hexim1 residues cross-link efficiently with Cdk9. Many of these are positioned between the BR involved in 7SK RNA binding and an acidic region AR (Fig. 5). The Cdk9-binding domain is distinct from the cyclin T1-binding domain that spans between Hexim1 residues F262 and L310 (5, 23, 24). Hexim1 residues F267 and Y274 belong to this sequence. The F267L point mutation supresses cyclin T1 interaction in a two-hydrid assay and P-TEFb interaction in human cells (23). Phosphorylation of Y274 destabilizes Hexim1 binding to P-TEFb (44). Because these residues cross-link Cdk9, they might contact both Cdk9 and cyclin T1. To summarize, the Cdk9-binding domain is bipartite and partially overlaps the cyclin T1-binding domain.
Fig. 5.
Schemes for two Hexim1 functional conformations. (A) The BR (blue) interacts with the AR (red). (B) Binding of 7SK RNA (red) to the BR releases its association with the AR, thereby unmasking Cdk9- and cyclin T1-binding sites. The cyclin T-binding domain overlaps a Cdk9-binding site, as well as the coiled-coil dimerization domain (green).
The Hexim1 PYNT Sequence Interferes with Substrate Binding to Cdk9.
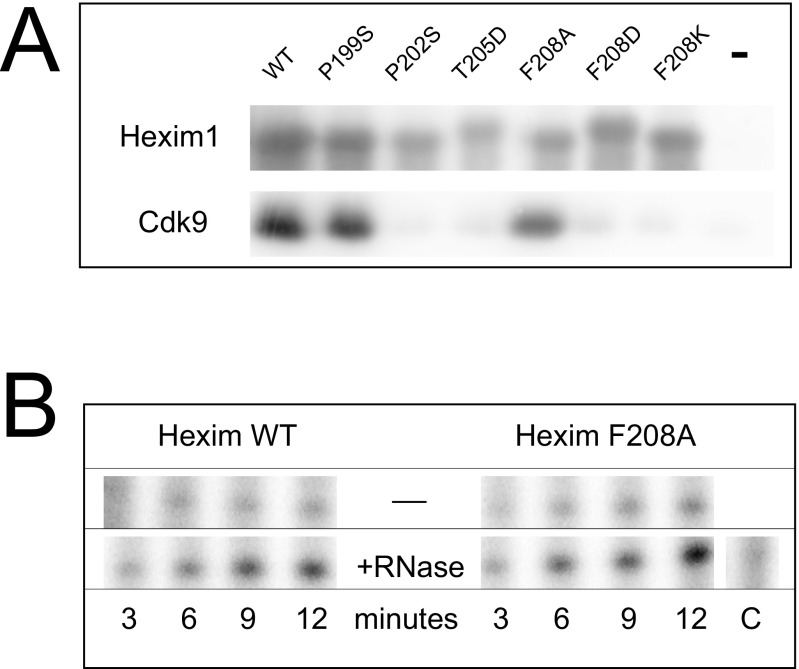
A very efficient cross-link was seen with Hexim1 F208Bpa in live cells, cell lysates and in vitro-reconstituted P-TEFb.Hexim1.7SK complexes. Reciprocally, replacing a tryptophan in this peptide with a photoreactive amino acid also allowed photocross-linking of Cdk9 to Hexim1. Phenylalanine F208 belongs to the PYNT sequence (201APYNTTQFL209), the most evolutionary conserved motif among Hexim proteins (16). This hydrophobic sequence has long been known to play an important role in the inhibition of P-TEFb (19–21). Hexim1 mutations in the PYNT sequence, P202S, T205D, F208D, and F208K prevented the formation of P-TEFb.Hexim1.7SK complexes in human cells (Fig. S2). The F208A mutation was of little consequence, but replacement of F208 by Bpa decreased the efficiency in P-TEFb coprecipitation (Fig. 1B, lane 6). Because Bpa is bulkier than phenylalanine, it has been suggested that F208 is positioned within a small hydrophobic pocket between Cdk9 and Hexim1. Importantly, truncated Hexim1 molecules lacking their N-terminal sequence up to V200 were found to efficiently inhibit P-TEFb in an in vitro kinase assay (26), whereas truncated Hexim1 molecules lacking their N-terminal sequence up to Q207 do not inhibit P-TEFb. Seven residues, including the PYNT motif, make all of the difference.
Fig. S2.
Flag-Hexim1 mutants are expressed in HEK293 cells. (A) Flag immunoprecipitates were probed by Western blot with anti-Hexim1 or anti-Cdk9 antibodies. (B) Flag immunoprecipitates were incubated with a CTD4 peptide substrate in the presence of radioactive ATP as described previously (3). Incorporation of radioactive phosphate was followed as a function of reaction time. The addition of RNase to the beads degraded 7SK and released active P-TEFb in the reaction mixture.
Proteins containing the PYNT sequence, the cyclin T-binding domain, and the AR but not the BR do not require 7SK RNA for complete inhibition (26). In contrast, P-TEFb inhibition by the FL Hexim1 protein, with both the AR and the BR, has an absolute requirement for 7SK RNA. Both regions might interact electrostatically with each other to prevent Hexim1 binding to P-TEFb (45) (Fig. 5A). Spatial proximity between the AR and BR is supported by the likely interactions between the 7SK G42AUC45//G64AUC67 repeated motif and the BR (46) and between 7SK uridine U30 and a Hexim1 sequence from amino acids 210–220 in the AR (47). 7SK RNA binding to the BR might neutralize its positive charges and consequently release its interaction with the AR, thereby unmasking a Cdk9-binding domain (Fig. 5B).
Efficient formation of cross-links with Hexim1 F208Bpa or Cdk9 W193Bpa indicates that Hexim1 F208 in the PYNT sequence is in very close contact with Cdk9.Hexim1 F208Bpa cross-links to the 189VVTLWYRPPELLLGER204 CDK9 peptide (Fig. 4B, yellow chain) that borders the catalytic site. Most importantly, replacing Cdk9 W193 by Bpa in this sequence leads to efficient cross-linking of Cdk9 to Hexim1; thus, Cdk9 W193 is in very close contact with Hexim1. The 189VVTLWYRPPELLLGER204 Cdk9 peptide overlaps the activation segment (from D167 to E198) that controls access to the catalytic site; thus, Hexim1 is in close contact with Cdk9 peptide segment controlling its kinase activity. Such contact between Hexim1 and the Cdk9 activation segment accounts for its inhibition. Importantly, phosphorylation of S175 and T186 in the “T-loop” is required for P-TEFb assembly with Hexim1 and 7SK RNA (21, 48, 49). Like Hexim1, the HIV viral protein Tat interacts mainly with cyclin T1 and is known to compete with Hexim1 for association with P-TEFb (5, 22). Tat also contacts the T-loop within the activation segment of Cdk9, and this contact induces a significant conformational change in Cdk9, modifying its substrate-binding surface (41); however, in contrast to Hexim1, Tat enhances the catalytic potential of Cdk9.
The 3D structure of Cdk2 bound to its inhibitors or substrates underlines the importance of the substrate-binding surface for Cdk9 inhibition by Hexim1. Indeed, the 3D structure of the Cdk2 catalytic site is highly similar to that of Cdk9 (Fig. 4C). The 189VVTLWYRPPELLLGER204 Cdk9 peptide (Fig. 4B, yellow chain) is a homolog of the 163VVTLWYRAPILLGCK178 Cdk2 peptide (Fig. 4C, yellow chain). Thus, Cdk2 W167 is the homolog of Cdk9 W193 (Fig. 4C, in magenta). These large aromatic hydrophobic residues are located at an extremity of the activation segments and exposed to the solvent. Importantly, the 163VVTLW167 Cdk2 peptide is in close contact with the HHASPRK model peptide substrate of Cdk2 (42) (Fig. 4E, in green). Of note, the peptidic hydrogen atom on the alpha carbon of the first histidine is engaged in a Pi-hydrogen bond with Cdk2 W167. By analogy, Cdk9 W193 would be expected to be in close contact with peptide substrates. Inhibition of Cdk2 by p27Kip involves an overlap and block of the ATP-binding site by a p27Kip peptide chain (Fig. 4D). Given that Cdk9 W193 is 15 Å away from the catalytic Mg2+ ion, inhibition of Cdk9 by Hexim1 may follow a different pathway. We propose that the evolutionary conserved Hexim1 PYNT sequence interferes with substrate binding to Cdk9 to inhibit its kinase activity.
Methods
Cross-Linking in Live Mammalian Cells or Lysates, and Immunoprecipitation.
HEK 293 cells were cotransfected with polyethyleneimine (Polysciences) with Bst-Yam suppressor tRNA plasmid, a plasmid coding for either Bpa-tRNA or Azp-tRNA synthetase (38), and mutated derivatives of pAdRSV-Flag-Hexim1 (5) or pCDNA-3myc-Cdk9 (a gift from Shona Murphy, Sir William Dunn School of Pathology, Oxford) providing N-terminal tagged proteins. At 4 h after transfection, Bpa (IRIS Biotech) or Azp (IRIS Biotech) (0.8 M stock solutions in 1 M NaOH) was added to culture media at a final concentration of 2 mM. Cells were lysed at 48 h after transfection in chilled HKM200 buffer (10 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2, and 200 mM NaCl ) with protease inhibitor mixture (Sigma-Aldrich) and 0.5% Igepal (Sigma-Aldrich). Clarified cell lysates (centrifuged at 20,000 × g) were irradiated on ice in a Petri dish at 365 nm for 45 min (4 J/cm2). Alternatively, live cells were irradiated while still attached to their 15-cm Petri dish under a 1 mM solution of glucose in chilled PBS. Lysis then proceeded as above.
Proteins were immunoprecipitated on anti-Flag M2 agarose beads (Sigma-Aldrich) or on protein G- coated Dynabeads after the addition of anti-myc 9E10 monoclonal antibody (Santa Cruz Biotechnology). Rabbit polyclonal antibodies N2 and C4 were made against Hexim1 N-terminal and C-terminal peptides, respectively. Anti-Cdk9 (C-20) and cyclin T1 (H-245) were obtained from Santa Cruz Biotechnology.
Reagents for Reconstitution Experiments.
Recombinant Hexim1.Bpa was made in BL21 cotransfected with pET-Hexim1-C-StrepT and p3tRNA.BpARS (50). pET-Hexim1-C-StrepT was derived from pET21-Hexim1 (20) by replacing the C-terminal His tag with a C-terminal streptavidin tag, and amber stop codons were introduced by targeted mutagenesis. Bacteria induced overnight by isopropyl β-d-1-thiogalactopyranoside (IPTG) at 23 °C in the presence of 1 mM Bpa were pelleted and then sonicated on ice in TNE100 buffer (100 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1.4 mM mercaptoethanol). Strep-tagged Hexim1.Bpa proteins were retained on Strep Tactin beads (IBA). Desthiobiotin (Sigma-Aldrich) eluates were fractionated with an AKTA purifier 10 fitted with a Superdex 200 10/300 GL column (GE Healthcare) and equilibrated in HKM500 buffer (500 mM NaCl). Fractions corresponding to Hexim dimers were used. Recombinant Larp7 was made from pnEA-HV-Cter-larp7-FL.
Bacteria induced overnight by IPTG at 23 °C were pelleted and sonicated on ice in TNE100 buffer. His-tagged Larp7 was retained on nickel agarose beads and eluted with 300 mM imidazole. The histidine tag was removed by overnight tobacco etch virus proteolysis. The resulting protein was purified further on a heparin column eluted with a 0–350 mM NaCl gradient in 20 mM Hepes pH 7.2 and 2 mM DTT. To obtain P-TEFb, Cdk9 cDNA fused to an N-terminal strep tag and full-length cyclin T1 cDNA were cloned into a PKL MultiBac vector, under the control of polyhedrin and p10 promoters (51). Sf21-infected cells were pelleted at 48 h postinfection and then lysed by sonication on ice in 20 mM Hepes pH 7.5, 250 mM NaCl, and 1 mM DTT with protease inhibitors (Sigma-Aldrich P8340). The lysate was clarified by centrifugation at 14,000 × g for 30 min. P-TEFb retained on Strep-Tactin Sepharose beads (IBA) was eluted with 2.5 mM desthiobiotin. 7SK RNA was prepared by T7 polymerase in vitro transcription (52).
In vitro P-TEFb.Hexim1.7SK RNA complex reconstitution and cross-linking.
P-TEFb.Hexim1.7SK RNA reconstitution was adapted from a previously published procedure (20). Larp7 was added first to 7SK RNA that had been denatured at 95 °C and renatured at 4 °C in 200 mM cacodylate pH 6.5, 40 mM MCl2, and 10 mM EDTA. All reagents were subsequently equilibrated in HKM150 buffer (150 mM NaCl) using Microspin G-50 columns (GE Healthcare). The Larp7.7SK RNA was then added to an excess of Hexim1.Bpa. P-TEFb was added last. The mixture was left for 30 min at room temperature. Irradiation was performed on ice for 45 min at 365 nm (4 J/cm2).
LC-MS/MS.
Reconstituted P-TEFb.HEXIM1.7SK RNA complexes, cross-linked or not, were adsorbed on Strep-Tactin beads. SDS/PAGE was stained with Coomassie brilliant blue. Bands of interest were cut off, alkylated with 55 mM iodoacetamide after reduction with 10 mM DTT, and then digested sequentially in PBS using 10 ng/μL endoproteinase GluC (Roche), followed by 40 ng/μL sequencing-grade trypsin (Promega Gold). For Fig. S1 B–D, the endoproteinase GluC digestion step was omitted. Proteolytic peptides were separated on a Thermo Fisher Scientific U3000 RSLC system fitted with a C18 column (75 μm i.d. × 50 cm long) and coupled to a Q Exactive Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific).
Data acquisition involved a top-10 experimental design. Each full-scan MS (range, 400–2,000 m/z; resolution, 70,000) is followed by 10 higher-energy collisional dissociation MS/MS on the 10 most intense species (resolution, 17,500) with dynamic exclusion. Mascot Server 2.5.1 (Matrix Science) and SwissProt (2016-06 551385 sequences) were used for database searches and quantitation by the Exponentially Modified Protein Abundance Index (emPAI) or SEQUEST HT (Thermo Fisher Scientific) Proteome Discoverer 2.1 for precursor area calculation (XIC label-free quantitation), with up to six miscleavages, carbamidomethylation on Cys, oxidation on Met, deamidation on Asn and Gln, and replacement of Y or F by Bpa included as variable modifications. Mass tolerances were 10 ppm for proteolytic peptides and 20 mDa for peptide fragments. Results were filtered on human taxonomy.
Supplementary Material
Acknowledgments
We thank Anne Catherine Dock-Bregeon, Isabelle Barbosa, Dorian Miremont, Shona Murphy, Van Trung Nguyen, Shixin Ye-Lehman, and Shigeyuki Yokoyama for reagents, help and discussion and the Institut de Génétique et de Biologie Moléculaire et Cellulaire’s structural biology platform and baculovirus service for help with insect cell production. This work was supported by the Association pour la Recherche sur le Cancer (Fellowship ARC 1010, to L.K.); the Agence Nationale pour la Recherche (Grant ANR-12-BSV5-0018 DynamIC); the “Investissements d’Avenir” program launched by the French Government, implemented by Grants ANR‐10-LABX-54 MEMOLIFE and ANR-11-IDEX-0001‐02 PSL* Research University; and the French Infrastructure for Integrated Structural Biology (Grant ANR-10-INSB-05-01) and INSTRUCT, as part of the European Strategy Forum on Research Infrastructures. Mass spectrometry equipment was supported by the SESAME program of the Conseil Régional d'Ile de France.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612331113/-/DCSupplemental.
References
- 1.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 5.Michels AA, et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23(14):4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen D, et al. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012;40(12):5283–5297. doi: 10.1093/nar/gks191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He N, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb–dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29(5):588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markert A, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9(6):569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27(20):6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Herreweghe E, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26(15):3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13(6):868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castelo-Branco G, et al. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013;14(9):R98. doi: 10.1186/gb-2013-14-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn RA, et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol. 2016;23(3):231–238. doi: 10.1038/nsmb.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNamara RP, et al. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol Cell. 2016;61(1):39–53. doi: 10.1016/j.molcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marz M, et al. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26(12):2821–2830. doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 17.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem. 2005;280(16):16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 18.is Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005 doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 19.Dulac C, et al. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J Biol Chem. 2005;280(34):30619–30629. doi: 10.1074/jbc.M502471200. [DOI] [PubMed] [Google Scholar]
- 20.Michels AA, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23(13):2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, et al. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280(31):28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 22.Schulte A, et al. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J Biol Chem. 2005;280(26):24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 23.Verstraete N, et al. A cyclin T1 point mutation that abolishes positive transcription elongation factor (P-TEFb) binding to Hexim1 and HIV tat. Retrovirology. 2014;11(50):50. doi: 10.1186/1742-4690-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dames SA, et al. Structure of the cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci USA. 2007;104(36):14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schönichen A, et al. A flexible bipartite coiled coil structure is required for the interaction of Hexim1 with the P-TEFB subunit cyclin T1. Biochemistry. 2010;49(14):3083–3091. doi: 10.1021/bi902072f. [DOI] [PubMed] [Google Scholar]
- 26.Czudnochowski N, Bösken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 27.Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382(6589):325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 28.Brotherton DH, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395(6699):244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 29.Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev. 2000;14(24):3115–3125. doi: 10.1101/gad.851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 31.Hino N, et al. Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. J Mol Biol. 2011;406(2):343–353. doi: 10.1016/j.jmb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 33.Hino N, et al. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2(3):201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 34.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25(4):663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 35.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14(8):696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012;11(4):M111.012088. doi: 10.1074/mcp.M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376(6538):313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 38.Ye S, et al. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem. 2008;283(3):1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 39.Shore SM, Byers SA, Maury W, Price DH. Identification of a novel isoform of Cdk9. Gene. 2003;307(5):175–182. doi: 10.1016/s0378-1119(03)00466-9. [DOI] [PubMed] [Google Scholar]
- 40.Baumli S, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27(13):1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tahirov TH, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465(7299):747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1(7):438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 43.Deseke E, Nakatani Y, Ourisson G. Intrinsic reactivities of amino acids towards photoalkylation with benzophenone: A study preliminary to photolabelling of the transmembrane protein glycophorin A. Eur J Org Chem. 1998;(2):243–251. [Google Scholar]
- 44.Mbonye UR, Wang B, Gokulrangan G, Chance MR, Karn J. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 promotes release of P-TEFb from the 7SK snRNP complex and enhances proviral HIV gene expression. Proteomics. 2015;15(12):2078–2086. doi: 10.1002/pmic.201500038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barboric M, et al. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24(24):4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebars I, et al. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010;38(21):7749–7763. doi: 10.1093/nar/gkq660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bélanger F, Baigude H, Rana TM. U30 of 7SK RNA forms a specific photo-cross-link with Hexim1 in the context of both a minimal RNA-binding site and a fully reconstituted 7SK/Hexim1/P-TEFb ribonucleoprotein complex. J Mol Biol. 2009;386(4):1094–1107. doi: 10.1016/j.jmb.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen R, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22(10):1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ammosova T, et al. Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS One. 2011;6(4):e18985. doi: 10.1371/journal.pone.0018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato S, et al. Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry. 2011;50(2):250–257. doi: 10.1021/bi1016183. [DOI] [PubMed] [Google Scholar]
- 51.Abdulrahman W, et al. The production of multiprotein complexes in insect cells using the baculovirus expression system. Methods Mol Biol. 2015;1261:91–114. doi: 10.1007/978-1-4939-2230-7_5. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Zapien D, et al. Intermolecular recognition of the non-coding RNA 7SK and HEXIM protein in perspective. Biochimie. 2015;117:63–71. doi: 10.1016/j.biochi.2015.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.