Significance
The findings of this study have revealed a potential molecular pathway for how fatty acid synthase (FASN) overexpression causes drug and radiation resistance and contributes to poor clinical prognosis of cancer diseases. FASN is the sole cytosolic enzyme responsible for de novo lipid synthesis, required for cancer cell survival but not for most normal nonadipose tissues. The finding that FASN regulates DNA repair by regulating specificity protein 1 and NF-κB in cancer cell responses to anticancer treatments will have a profound impact on designing future treatment strategies. It will also help establish FASN as a target for therapeutic discovery to sensitize drug and radiation resistance.
Keywords: fatty acid synthase, transcription regulation, DNA repair, drug resistance, radiation resistance
Abstract
Fatty acid synthase (FASN), the sole cytosolic mammalian enzyme for de novo lipid synthesis, is crucial for cancer cell survival and associates with poor prognosis. FASN overexpression has been found to cause resistance to genotoxic insults. Here we tested the hypothesis that FASN regulates DNA repair to facilitate survival against genotoxic insults and found that FASN suppresses NF-κB but increases specificity protein 1 (SP1) expression. NF-κB and SP1 bind to a composite element in the poly(ADP-ribose) polymerase 1 (PARP-1) promoter in a mutually exclusive manner and regulate PARP-1 expression. Up-regulation of PARP-1 by FASN in turn increases Ku protein recruitment and DNA repair. Furthermore, lipid deprivation suppresses SP1 expression, which is able to be rescued by palmitate supplementation. However, lipid deprivation or palmitate supplementation has no effect on NF-κB expression. Thus, FASN may regulate NF-κB and SP1 expression using different mechanisms. Altogether, we conclude that FASN regulates cellular response against genotoxic insults by up-regulating PARP-1 and DNA repair via NF-κB and SP1.
Fatty acid synthase (FASN) is the key mammalian enzyme required for de novo synthesis of palmitate. FASN expression and activity are largely suppressed by sufficient dietary fat in most normal nonadipose tissues but are abnormally elevated in many human cancers and associated with poor prognosis (1). FASN association with poor prognosis may derive in part from FASN function in drug resistance during chemotherapy. Indeed, it has been found that FASN expression and/or activity was increased in drug-selected and -resistant breast (2) and pancreatic (3) cancer cells. It was also found that FASN overexpression causes cellular resistance to DNA-damaging drugs such as doxorubicin and mitoxantrone but not to microtubule modulators such as vinblastine and paclitaxel (4). Decreased ceramide production following doxorubicin treatment via suppression of tumor necrosis factor (TNF)-α production is believed to be one of the mechanisms of FASN-induced resistance to doxorubicin (4).
The observation that FASN increases resistance to genotoxic drugs prompted us to hypothesize that FASN overexpression may up-regulate DNA damage response/repair pathways. In this study, we tested this hypothesis with a focus on the repair of DNA double-strand breaks (DSBs), which are commonly induced by the anticancer drugs doxorubicin and mitoxantrone and ionizing radiation. In mammalian cells, DSBs are repaired mainly via homologous recombination (HR) and nonhomologous end-joining (NHEJ) pathways. NHEJ is the predominant form of DSB repair because it occurs during all phases of the cell cycle whereas HR only initiates at late G1 and S phases (5). Hence, we examined NHEJ repair of DSBs and found that FASN up-regulates NHEJ activity and repair of DSBs by increasing poly(ADP-ribose) polymerase 1 (PARP-1) expression via increasing the expression of specificity protein 1 (SP1) and reducing the expression of NF-κB, which bind to the same composite element in the PARP-1 promoter. Furthermore, lipid deprivation suppresses SP1 expression, which is able to be rescued by palmitate supplementation. With these findings, we conclude that FASN may up-regulate DNA repair mechanisms by increasing PARP-1 expression via NF-κB and SP1, which in turn contributes to cellular resistance to genotoxic anticancer treatments. Thus, lipid metabolism likely plays an important role in cancer cell survival against genotoxic insults by regulating DNA repair pathways.
Results
FASN Overexpression Causes Resistance to Multiple Genotoxic Treatments.
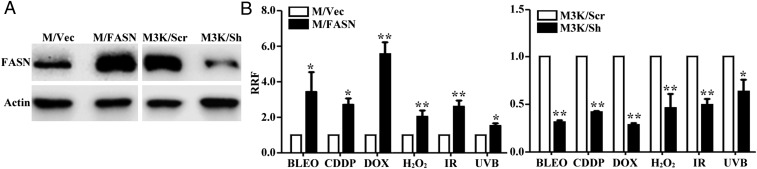
We previously reported that FASN overexpression increases cellular resistance to anticancer drugs that cause DSBs but not to non–DNA-damaging drugs (2, 4). To determine whether FASN overexpression causes resistance to genotoxic treatments that cause different types of DNA lesions, we tested the survival of a previously established stable MCF7 clone with FASN overexpression (M/FASN) in comparison with vector-transfected (M/Vec) control cells (4) (see also Fig. 1A) following treatments with bleomycin, cisplatin, doxorubicin, H2O2, ionizing radiation (IR), and UVB. As shown in Fig. 1B and Fig. S1A, M/FASN cells are significantly more resistant to all these treatments than M/Vec cells. Stable FASN knockdown (M3K/Sh) in a drug-resistant MCF7/AdVp3000 cell line (Fig. 1A), which has elevated endogenous FASN levels compared with the parental MCF7 cells (4), significantly reduced resistance levels to these treatments compared with the control cells (M3K/Scr) (Fig. 1B and Fig. S1B). Thus, FASN overexpression likely contributes to cellular resistance to multiple genotoxic treatments that cause different types of DNA lesions.
Fig. 1.
Effect of FASN on the cellular response to genotoxic treatments. (A) Western blot analysis of FASN expression in stable MCF7 cells with FASN overexpression (M/FASN), MCF7/AdVp3000 cells with stable FASN knockdown (M3K/Sh), and their respective control vector-transfected (M/Vec) and scrambled control shRNA-transfected (M3K/Scr) cells. (B) Survival assay. M/FASN, M3K/Sh, and their respective control cells were tested for resistance to bleomycin (BLEO), cisplatin (CDDP), doxorubicin (DOX), and H2O2 using an MTT assay, and to ionizing radiation and UVB using a colony-formation assay (n = 3; *P < 0.05, **P < 0.01; error bars, standard deviation).
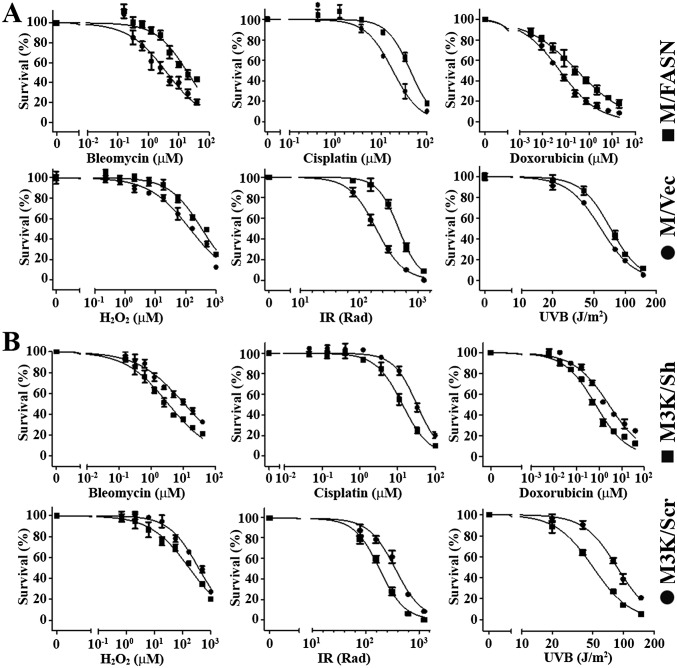
Fig. S1.
Representative dose–response curves for survival assays. M/FASN with FASN overexpression (A), M3K/Sh with FASN knockdown (B), and their respective control cells (M/Vec and M3K/Scr) were tested for resistance to bleomycin, cisplatin, doxorubicin, and H2O2 using an MTT assay and to ionizing radiation (IR) and UVB using a colony-formation assay. Representative survival curves are shown and expressed as % of survival to untreated controls. Each data point represents the mean ± SD for n = 6 replicates. Error bars, standard deviation.
FASN Overexpression Increases Repair of IR- and Doxorubicin-Induced DNA Damage.
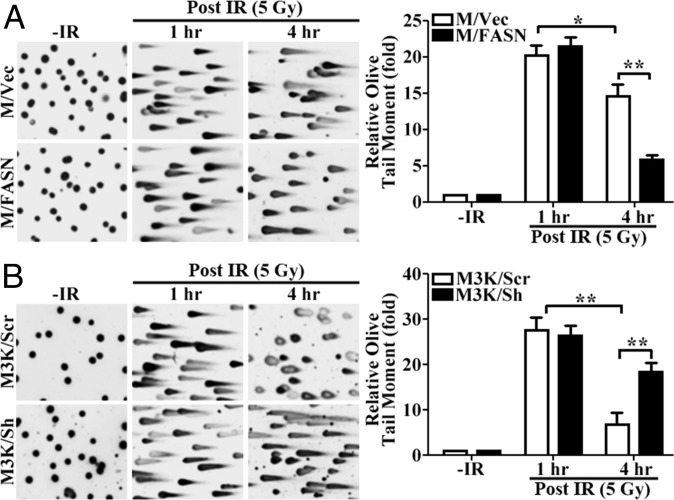
The above findings suggest that FASN overexpression may protect cells against DNA damage induced by these genotoxic treatments or by increasing the repair of this damage. To test this possibility, we focused on DSBs induced by IR for the convenience of monitoring repair and because DSBs appear to be mostly affected by FASN (Fig. 1B). We first performed a neutral comet assay with the two paired cell lines (M/FASN vs. M/Vec and M3K/Sh vs. M3K/Scr) following IR treatment. Fig. 2A shows that, at 1 h following IR treatment, the reported time of maximal DNA damage induction post IR exposure (6), both M/FASN and M/Vec cells formed similarly elongated tails with no difference in olive tail moment, indicating that the DNA in both cells is equally damaged by IR and that FASN overexpression may not protect cells from IR-induced DNA damage. Similar results were also observed for M3K/Sh and M3K/Scr cells (Fig. 2B). However, at 4 h after IR, a dramatic difference in olive tail moment was observed between M/FASN and M/Vec cells and between M3K/Sh and M3K/Scr cells. Whereas the olive tail moment was significantly lower in M/FASN cells, it was significantly higher in M3K/Sh cells than their respective control cells. These findings indicate that FASN may increase the cellular repair activity of DSBs induced by IR.
Fig. 2.
Effect of FASN on the accumulation of IR-induced DNA damage. Stable FASN-overexpressing (M/FASN) and control (M/Vec) cells (A) as well as stable FASN knockdown (M3K/Sh) and control (M3K/Scr) cells (B) were treated with or without 5 Gy IR and then subjected to a neutral comet assay at different times. The histograms show quantitative olive moment analysis from three independent experiments (*P < 0.05, **P < 0.01; error bars, standard deviation).
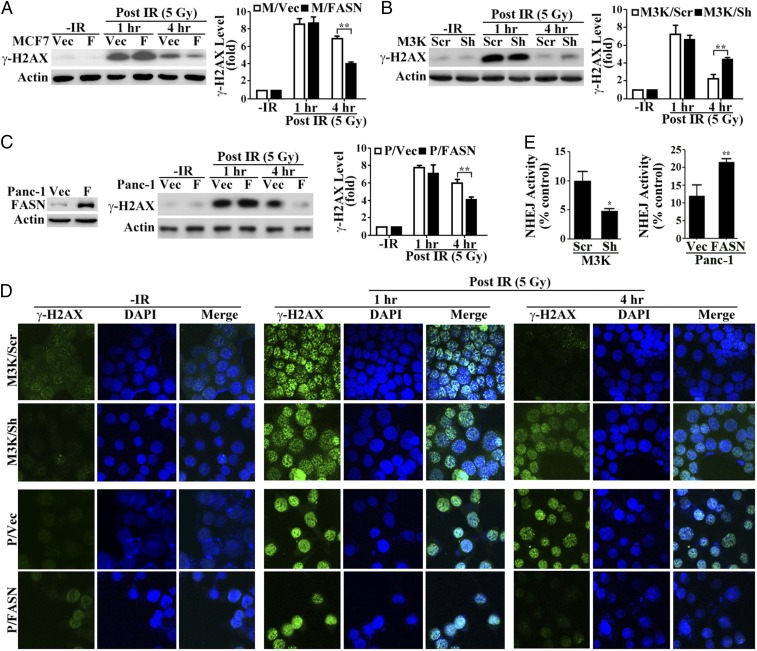
To verify the above findings, we next determined the level of γ-histone 2AX (γ-H2AX), an indicator of DSBs (7), in these paired cells at different recovery times following IR by Western blot analysis. As shown in Fig. 3 A and B, a similarly induced level of γ-H2AX was detected between M/FASN and M/Vec cells and between M3K/Sh and M3K/Scr cells at 1 h following IR compared with their respective untreated control cells. However, γ-H2AX levels decreased significantly more in M/FASN than M/Vec control cells and decreased significantly less in M3K/Sh than M3K/Scr control cells at 4 h following IR. These observations are consistent with the findings of the comet assay (Fig. 2). Immunofluorescence analysis of M3K/Sh and M3K/Scr cells showed equivalent increases in punctate staining of γ-H2AX in the nuclei of both cells at 1 h after IR compared with the control untreated cells (Fig. 3D). At 4 h after IR, the nuclear staining of γ-H2AX in M3K/Scr control cells reduced dramatically whereas it remained at high levels in M3K/Sh cells with FASN depleted. These observations are consistent with that shown by Western blot analysis of γ-H2AX expression.
Fig. 3.
Effect of FASN on DSB repair. (A–C) Western blot analysis of IR-induced γ-H2AX in FASN-overexpressing MCF7 (M/FASN) and its control (M/Vec) cells (A), stable FASN knockdown MCF7/AdVp3000 (M3K/Sh) and its control (M3K/Scr) cells (B), and FASN-overexpressing Panc-1 (P/FASN) and its control (P/Vec) cells (C). Histograms show relative γ-H2AX levels determined from three independent experiments. (D) Immunofluorescence staining of γ-H2AX in M3K/Sh, P/FASN, and their respective control M3K/Scr and P/Vec cells. DNA was stained by DAPI. (E) NHEJ activity assay. M3K/Sh and P/FASN cells along with their respective control M3K/Scr and P/Vec cells were subjected to an NHEJ host cell reactivation assay as described in Experimental Procedures (n = 4; *P < 0.05, **P < 0.01; error bars, standard deviation). F, FASN; Vec, vector.
Because M3K/Scr vs. M3K/Sh and M/Vec vs. M/FASN cells in the above studies are of the same MCF7 origin, the above observations may be specific to MCF7 cells. To eliminate this possibility, we tested another pair of cell lines of different genetic background, P/FASN (with ectopic FASN overexpression) and P/Vec (vector-transfected control), both derived from Panc-1 cells (8) (Fig. 3C). Whereas the γ-H2AX level was similarly high between P/FASN and P/Vec cells at 1 h following IR treatment, it was significantly reduced in P/FASN cells compared with P/Vec cells at 4 h following IR treatment (Fig. 3C). This finding was confirmed by the punctate staining of γ-H2AX in the nuclei of P/FASN and P/Vec cells (Fig. 3D). Thus, the function of FASN in promoting DSB repair may not be cell line-specific.
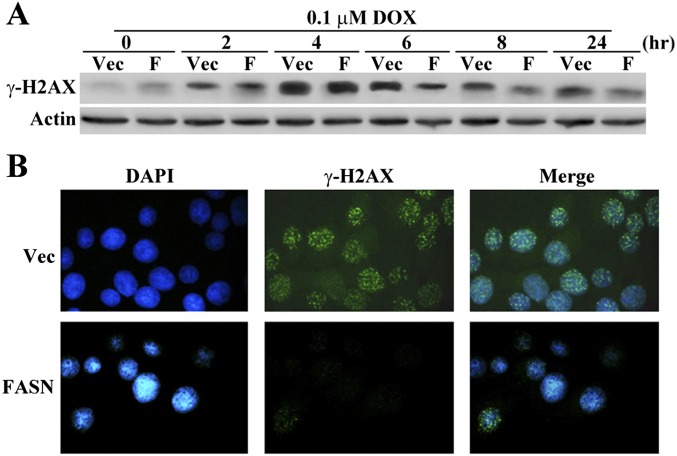
Because FASN overexpression contributes to cellular resistance to the anticancer drug doxorubicin (Fig. 1), which also induces DSBs, we next determined whether FASN overexpression has a similar effect on γ-H2AX due to DSBs induced by doxorubicin. As shown in Fig. S2A, at 4 h of treatment, γ-H2AX reached maximum levels in both M/FASN and control M/Vec cells. At 6 h and beyond, M/FASN cells have less γ-H2AX than the control M/Vec cells. This observation was also corroborated by immunostaining of γ-H2AX in the nuclei shown in Fig. S2B. Together, these findings suggest that higher FASN expression levels may increase the repair of IR- or doxorubicin-induced DSBs.
Fig. S2.
Effect of FASN on γ-H2AX level following doxorubicin treatment. Western blot analysis (A) and immunofluorescence staining (B) of doxorubicin (DOX)-induced γ-H2AX in MCF7-derived FASN-overexpressing (M/FASN) and control vector-transfected cells (M/Vec). Cells were treated with 0.1 μM doxorubicin and collected at the designated time points. Immunofluorescence staining was done at 24 h post doxorubicin treatment.
FASN Overexpression Increases NHEJ Repair Activity.
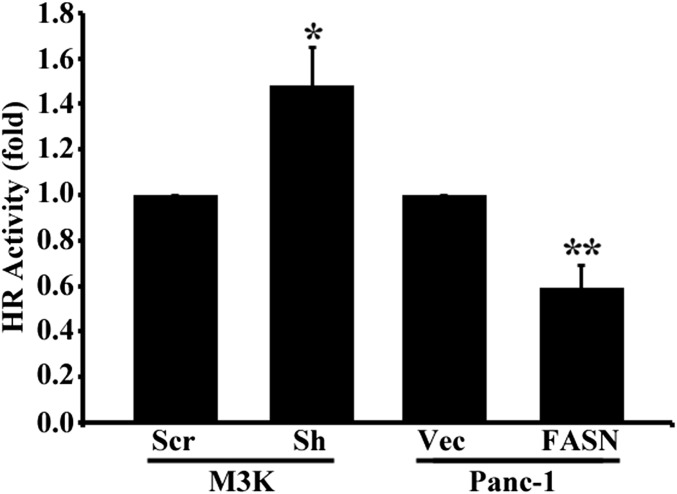
Next, we tested the possibility that FASN overexpression increases the repair of DSBs using a reporter-based host cell reactivation assay of both NHEJ and HR activities without drug or IR treatments. As shown in Fig. 3E, M3K/Sh cells with FASN knockdown had significantly reduced NHEJ activity than control M3K/Scr cells whereas P/FASN cells with FASN overexpression had significantly increased NHEJ activity over control P/Vec cells. However, HR activity was significantly increased and reduced, respectively, in M3K/Sh and P/FASN cells compared with their control cells (Fig. S3). These findings are very interesting and suggest that NHEJ but not HR may contribute to FASN-induced resistance to IR (Discussion).
Fig. S3.
Effect of FASN expression on HR activity. M3K cells with FASN knockdown (M3K/Sh) and Panc-1 cells with FASN overexpression (P/FASN) along with their respective control cells (M3K/Scr and P/Vec) were subjected to an HR assay as described in Experimental Procedures (n = 3; *P < 0.05, **P < 0.01; error bars, standard deviation).
FASN Overexpression Enhances Ku Protein Recruitment and DNA-PK Activity.
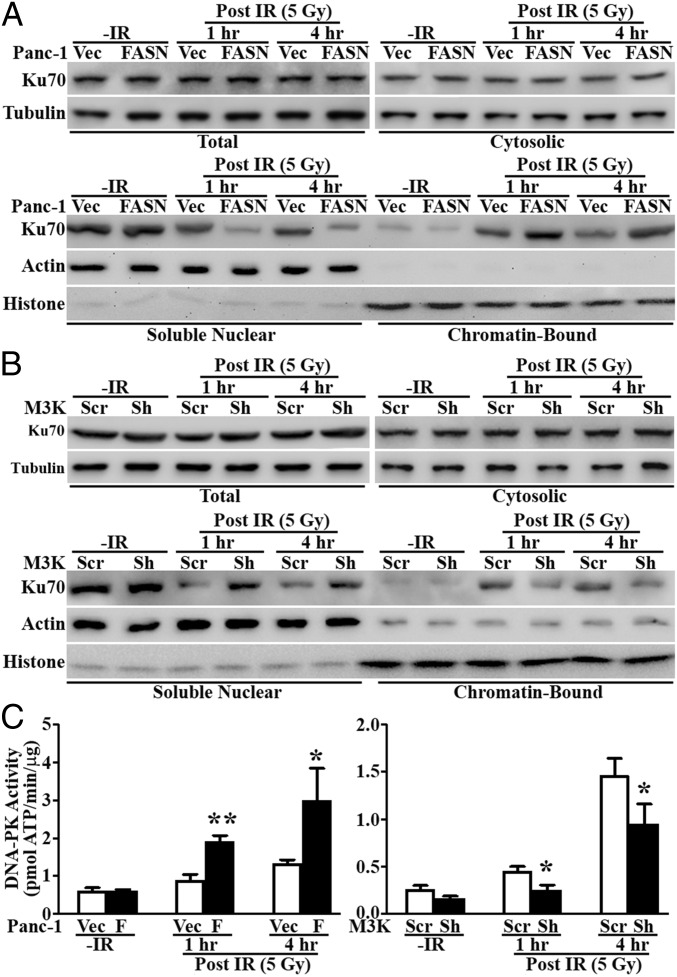
We next investigated the molecular basis of the up-regulated NHEJ activity by FASN overexpression. Because NHEJ repair is initiated by recruiting Ku70/80 dimers and DNA-PKcs to the damage sites and because increased Ku70 expression has been found to promote cellular resistance to doxorubicin and ionizing radiation (9–11), we tested the effect of FASN on Ku70 expression and recruitment to damaged chromatin. As shown in Fig. 4 A and B, M3K/Sh cells with FASN knockdown and P/FASN cells with FASN overexpression had similar total and cytoplasmic Ku70 levels as their respective control cells before and after IR. The chromatin-bound Ku70 was increased in all cells following IR treatment. However, the increase was greater in P/FASN cells and less in M3K/Sh cells compared with their respective control cells. These changes were accompanied by the corresponding lower and higher levels of Ku70 in the unbound (soluble) nuclear fractions than their respective controls. Further, more chromatin-bound Ku70 protein in P/FASN and less in M3K/Sh cells was accompanied by significantly augmented and reduced DNA-PK activity following IR compared with their respective controls (Fig. 4C). Thus, we conclude that FASN overexpression increases recruitment of Ku proteins to chromatin and DNA-PK activity following IR-induced DNA damage.
Fig. 4.
Effect of FASN on Ku recruitment and DNA-PK activity. (A and B) Western blot analysis of Ku70 distribution in different cellular fractions following IR in FASN-overexpressing Panc-1 cells (P/FASN) (A), FASN knockdown MCF7/AdVp3000 cells (M3K/Sh) (B), and their respective control cells (P/Vec and M3K/Scr). Tubulin, actin, and histone H3 were used as markers for cytosolic, soluble nuclear, and insoluble chromatin fractions, respectively. (C) DNA-PK activity assay. P/FASN and M3K/Sh cells together with their respective control P/Vec and M3K/Scr cells were treated without or with IR and harvested at the indicated times following IR for determination of DNA-PK activity (n = 3; *P < 0.05, **P < 0.01; error bars, standard deviation).
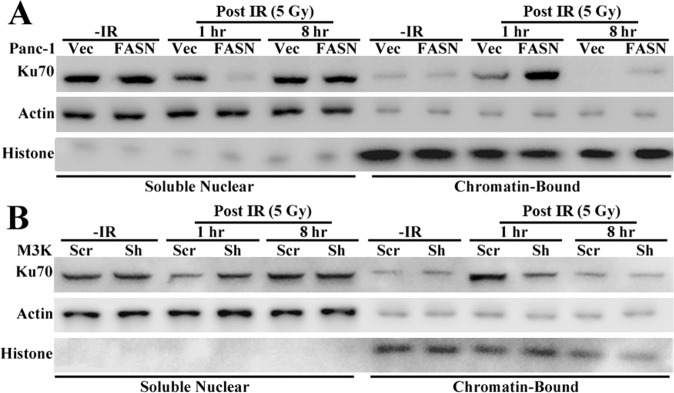
It is noteworthy that Ku70 binding to chromatin and DNA-PK activity were sustained at 4 h post IR, when most DNA damage was repaired, as evidenced by the significant decrease in γ-H2AX level/foci and reduction in olive tail moment (Figs. 2 and 3). It is possible that the sustained Ku70 retention on chromatin was due to the existence of residual DNA damage at 4 h post IR, as also indicated by γ-H2AX level/foci and olive tail moment (Figs. 2 A and B and 3 A–C). To test this possibility, we analyzed chromatin-bound Ku70 at 8 h post IR. As shown in Fig. S4, the chromatin-bound Ku70 was reduced to the basal level. Thus, the sustained chromatin retention of Ku70 and DNA-PK activity may be due to cellular activity to repair residual DNA damage.
Fig. S4.
Western blot analysis of Ku70 distribution in different nuclear fractions following IR in FASN-overexpressing Panc-1 (P/FASN) (A), FASN knockdown MCF7/AdVp3000 (M3K/Sh) (B), and their respective control cells (P/Vec and M3K/Scr). Actin and histone H3 were used as markers for soluble nuclear and insoluble chromatin fractions, respectively.
FASN Overexpression Promotes PARP-1 Expression, Which Mediates FASN Enhancement of NHEJ Repair and Cellular Resistance to IR.
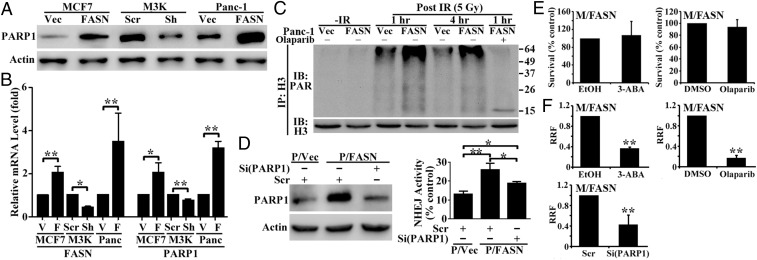
PARP-1 has emerged as a key player in the DSB repair pathway (12, 13) and was found to promote recruitment and retention of Ku at DSB sites via Ku70 binding to poly(ADP-ribosyl)ated (PARylated) nuclear proteins and facilitate NHEJ repair (14). Thus, we tested the possibility that FASN overexpression up-regulates PARP-1 expression, which in turn promotes Ku70 recruitment and increases NHEJ activity. Fig. 5A shows that PARP-1 protein is increased dramatically in M/FASN and P/FASN clones with FASN overexpression but is decreased in the stable M3K/Sh clone with FASN knockdown compared with their respective control cells. PARP-1 mRNA also changed similar to PARP-1 protein in these cells (Fig. 5B).
Fig. 5.
PARP-1 mediates FASN-associated DNA repair and genotoxic treatment resistance. (A and B) Effect of FASN on PARP-1 expression as determined by Western blot (A) and real-time RT-PCR (B) in stable FASN-overexpressing and knockdown cells. (C) Effect of FASN on IR-induced histone H3 PARylation. Cells were pretreated without or with 0.5 µM olaparib followed by treatment with or without IR and harvested at different times following IR for lysate preparation, immunoprecipitation (IP) of histone H3 in the presence of 100 µM poly(ADP-ribose) glycohydrolase inhibitor gallotannin, and Western blot analysis (IB) of PARylated and total histone H3. (D) Effect of PARP-1 knockdown on FASN-induced NHEJ up-regulation. FASN-overexpressing (P/FASN) and control (P/Vec) cells were transiently transfected with PARP-1 or scrambled control siRNA followed by Western blot analysis of PARP-1 (Left) and NHEJ activity assay (Right). (E) Effect of PARP-1 inhibition on survival. M/FASN cells were treated with 1 mM 3-ABA, 0.5 µM olaparib, or their vehicle control for 48 h followed by an MTT assay. (F) Effect of PARP-1 inhibition on doxorubicin resistance. M/FASN cells were pretreated for 6 h by 1 mM 3-ABA or 0.5 µM olaparib or transfected with control or PARP-1 siRNA followed by testing doxorubicin resistance using an MTT survival assay (n = 3–4; *P < 0.05, **P < 0.01; error bars, standard deviation).
We next examined whether increased PARP-1 expression due to FASN overexpression would lead to higher PARP activity following DNA damage by determining PARylated histone H3, a target of PARP-1 after DNA damage and relevant to DNA repair (15). For this purpose, the stable P/FASN clone and its P/Vec control cells were treated without or with 5 Gy IR, followed by immunoprecipitation of histone H3 and Western blot analysis of PARylated histone H3. As shown in Fig. 5C, PARylated histone H3 was increased in P/FASN cells compared with the control P/Vec cells following IR. Pretreatment with the PARP-1 inhibitor olaparib inhibited IR-induced PARylation with minimal remaining PARylation of histone H3 (Fig. 5C). Clearly, the increased PARP-1 expression is accompanied by increased PARP-1 activity.
To determine whether the increased PARP-1 mediates FASN enhancement of NHEJ activity, we transiently transfected P/FASN cells with PARP-1 siRNA to knock down PARP-1 expression followed by an NHEJ activity assay. Fig. 5D shows that the increased PARP-1 level in P/FASN cells due to FASN overexpression is successfully suppressed by PARP-1 siRNA. The increased NHEJ activity due to FASN overexpression in P/FASN cells is significantly reduced by PARP-1 knockdown. Together, these findings suggest that FASN up-regulates PARP-1 expression, which in turn increases NHEJ activity, possibly by promoting Ku recruitment and enhancing DNA-PK activity.
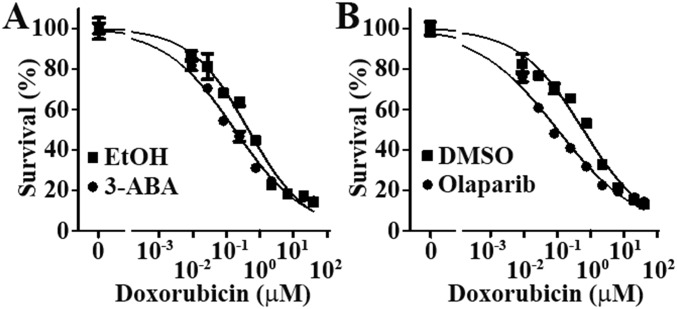
To determine whether FASN enhancement of PARP-1 likely mediates FASN function in cellular resistance to DNA damage, we performed a survival assay of P/FASN cells following doxorubicin treatment in the absence or presence of the PARP-1 inhibitors 3-ABA (3-aminobenzamide) and olaparib. As shown in Fig. 5E, either 1 mM 3-ABA or 0.5 µM olaparib alone had no effect on M/FASN cell survival. However, 1 mM 3-ABA and 0.5 µM olaparib significantly reduced the doxorubicin resistance level of M/FASN cells (Fig. 5F and Fig. S5). Similarly, knocking down PARP-1 using siRNA also significantly sensitized M/FASN cells to doxorubicin (Fig. 5F). These findings suggest that PARP-1 likely mediates FASN overexpression-induced resistance to DSBs, possibly by increasing NHEJ repair of DSBs.
Fig. S5.
Representative dose–response curves for survival assays. M/FASN cells were pretreated with 1 mM 3-ABA (A), 0.5 µM olaparib (B), or their respective vehicle controls before being subjected to an MTT assay in the presence of different concentrations of doxorubicin. Representative survival curves are shown and expressed as % of survival controls without doxorubicin. Error bars, standard deviation.
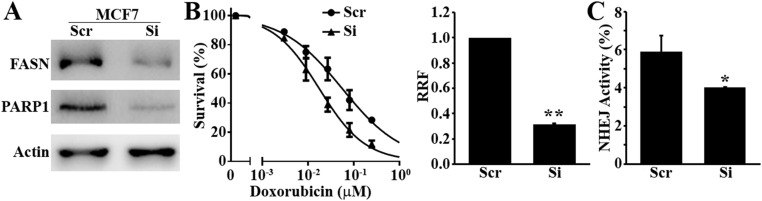
Both MCF7 and Panc-1 cells are known to express high levels of endogenous FASN, and M3K cells have further elevated endogenous FASN (2, 8, 16). To eliminate the possibility that the above observations may be due to forced increases in FASN expression either by ectopic transfection or drug selection, we transiently knocked down FASN in the parental MCF7 cells and tested its effect on doxorubicin sensitivity, NHEJ activity, and PARP-1 expression. As shown in Fig. S6, FASN knockdown in MCF7 cells significantly reduced doxorubicin resistance, NHEJ activity, and PARP-1 expression. These findings are consistent with our previous observation that FASN knockdown in MDA-MB-468 cells also significantly reduced doxorubicin and mitoxantrone resistance (2). Thus, endogenous levels of FASN in cancer cells likely regulate cellular response to DNA damage and NHEJ repair of DSBs.
Fig. S6.
Effect of FASN knockdown on doxorubicin response, NHEJ repair, and PARP-1 expression in MCF7 cells. MCF7 cells were transiently transfected with FASN siRNA followed by Western blot analysis of FASN, PARP-1, and loading control actin (A), MTT survival assay in the presence of doxorubicin (B), and NHEJ activity assay (C) (n = 3; *P < 0.05, **P < 0.01; error bars, standard deviation).
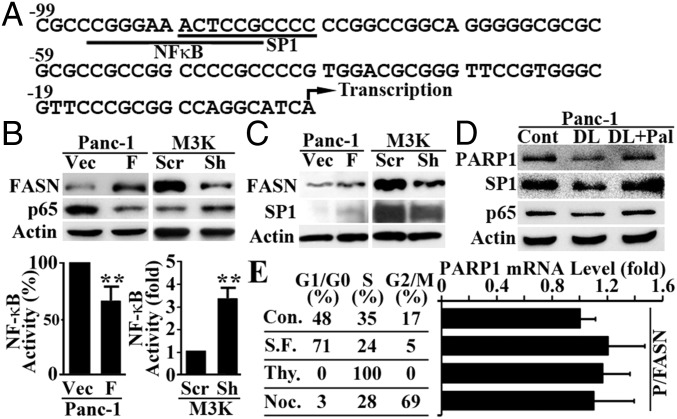
FASN Transcriptionally Regulates PARP-1 Expression Through Inhibition of NF-κB and Up-Regulation of SP1.
Fig. 5B shows that the mRNA level of PARP-1 was increased by FASN overexpression and decreased by FASN knockdown, suggesting that FASN may participate in regulating PARP-1 transcription. To understand how FASN promotes PARP-1 transcription, we examined the promoter sequence of human PARP-1 and found a composite element with overlapping binding sites for NF-κB and SP1 transcription factors (Fig. 6A). Previously, it has been shown that the rat PARP-1 gene is activated by SP1 (17) and inactivated by NF1, which competes with SP1 on a composite element in the promoter of rat PARP-1 (18). We have also shown that FASN down-regulates NF-κB expression (4). Fig. 6B shows that FASN overexpression in Panc-1 cells and knockdown in MCF7/AdVp3000 (M3K) cells also significantly decreased and increased, respectively, p65 expression as determined using Western blot and NF-κB activity using a reporter assay. On the other hand, FASN overexpression increased SP1 expression whereas FASN knockdown reduced SP1 expression (Fig. 6C). We also tested whether the effect of FASN on SP1 and NF-κB expression is possibly via its catalytic product palmitate by culturing cells in delipidated serum with or without palmitate supplementation. As shown in Fig. 6D, culturing Panc-1 cells in media with delipidated serum reduced the expression of SP1 and PARP-1 but not NF-κB. Supplementation of palmitate rescued both SP1 and PARP-1 expression from the suppression. However, altering the cell cycle in P/FASN cells had no effect on PARP-1 expression (Fig. 6E). Thus, palmitate may play an important role in mediating FASN regulation of SP1 expression but not NF-κB.
Fig. 6.
Effect of FASN on p65 and SP1 expression. (A) Putative composite element in the human PARP-1 proximal promoter. The overlapping underlined sequences represent NF-κB– and SP1-binding sites. The numbers indicate positions relative to the transcription start site (arrow). (B) Effect of FASN overexpression in Panc-1 cells or knockdown in MCF7/AdVp3000 cells on p65 expression and NF-κB activity. (C) Effect of FASN overexpression in Panc-1 cells or knockdown in MCF7/AdVp3000 cells on SP1 expression. (D) Effect of delipidation (DL) and palmitate supplementation (Pal) culture on SP1 and p65 expression in Panc-1 cells. (E) Effect of cell-cycle distribution on PARP-1 expression as determined using real-time RT-PCR. Different cell-cycle stages were achieved by culturing P/FASN cells in serum-free (S.F.) media for G0/G1, thymidine (Thy.) block for S, and nocodazole (Noc.) treatment for G2/M phases (n = 3; *P < 0.01; error bars, standard deviation).
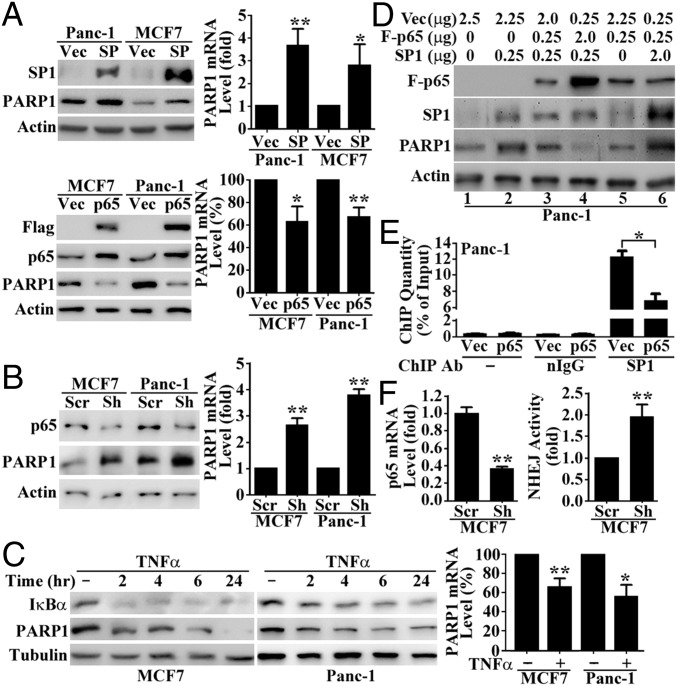
Based on the above studies, we hypothesized that FASN promotes PARP-1 expression by suppressing NF-κB and increasing SP1, which bind to the PARP-1 promoter in a mutually exclusive manner. To test this hypothesis, we first determined whether SP1 and NF-κB regulate PARP-1 expression by transiently transfecting MCF7 and Panc-1 cells with SP1 or p65 cDNA followed by determination of their effect on the expression of endogenous PARP-1. As shown in Fig. 7A, SP1 overexpression dramatically increased whereas Flag-p65 overexpression significantly reduced PARP-1 protein and mRNA levels. Knocking down p65 using shRNA significantly increased PARP-1 protein and mRNA levels (Fig. 7B), whereas activation of p65 with TNF-α had the contrary effect (Fig. 7C). Taken together, these results suggest that NF-κB and SP1 oppose each other in regulating PARP-1 transcription, with NF-κB functioning as a suppressor and SP1 as an activator.
Fig. 7.
NF-κB and SP1 regulate PARP-1 transcription by binding to the same site. (A) Effect of SP1 or Flag-p65 overexpression on PARP-1 expression in MCF7 and Panc-1 cells as determined by Western blot analysis (Left) and quantitative (q)RT-PCR (Right). (B) Effect of shRNA-induced p65 knockdown on PARP-1 expression in MCF7 and Panc-1 cells as determined using Western blot (Left) and qRT-PCR (Right). (C) Effect of NF-κB activation by TNF-α treatment on PARP-1 expression in MCF7 and Panc-1 cells as determined by Western blot and qRT-PCR analysis. (D) Competition between p65 and SP1 in regulating PARP-1 expression. Panc-1 cells were transiently transfected with various combinations of Flag-p65 and SP1 cDNA together with vector control to ensure a constant total level of DNA for each transfection. Cells were harvested at 48 h post transfection and subjected to Western blot analysis. (E) ChIP assay. Panc-1 cells were transiently transfected with Flag-p65 cDNA (p65) or a vector control (Vec) followed by ChIP without or with normal IgG control or SP1 antibody and real-time PCR analysis. Data are expressed as the percentage of signal detected with the nonimmunoprecipitated input (10% of total chromatin). (F) Effect of shRNA-mediated p65 knockdown on NHEJ activity in MCF7 cells (n = 3; *P < 0.05, **P < 0.01; error bars, standard deviation).
To determine whether NF-κB interferes with SP1 activation of PARP-1 expression and vice versa, we cotransfected varying amounts of Flag-p65 and SP1 cDNAs into Panc-1 cells followed by determination of PARP-1 expression. As shown in Fig. 7D, SP1 expression increased PARP-1 protein levels (compare lanes 1 and 2). However, an SP1-induced increase in PARP-1 protein level was diminished by p65 cDNA in a dose-dependent manner (compare lanes 2, 3, and 4, Fig. 7D). On the other hand, when p65 cDNA was kept constant, increasing SP1 cDNA from 0 to 2 μg resulted in a steady increase in PARP-1 level (compare lanes 5, 3, and 6).
To further determine whether NF-κB interferes with SP1 binding to and activation of the PARP-1 promoter, we performed a chromatin immunoprecipitation (ChIP) assay to quantify SP1-bound PARP-1 promoter following ectopic p65 overexpression in Panc-1 cells. As shown in Fig. 7E, the PARP-1 promoter immunoprecipitated by SP1 antibody was reduced significantly from ∼13% (relative to input) in vector-transfected control cells to ∼7% in p65-overexpressing cells. Collectively, these results suggest that NF-κB likely interferes with SP1 binding to and activation of the PARP-1 promoter for PARP-1 expression, possibly via competition for binding onto the composite element within the PARP-1 promoter.
Finally, to determine whether these transcription factors possibly play any role in up-regulating NHEJ activity, we analyzed the effect of p65 knockdown on NHEJ activity. As shown in Fig. 7F, p65 shRNA effectively knocked down p65 expression, which led to a significant increase in NHEJ activity in MCF7 cells. This finding suggests that downregulating NF-κB expression by FASN may contribute to the increased NHEJ repair of DSBs.
Discussion
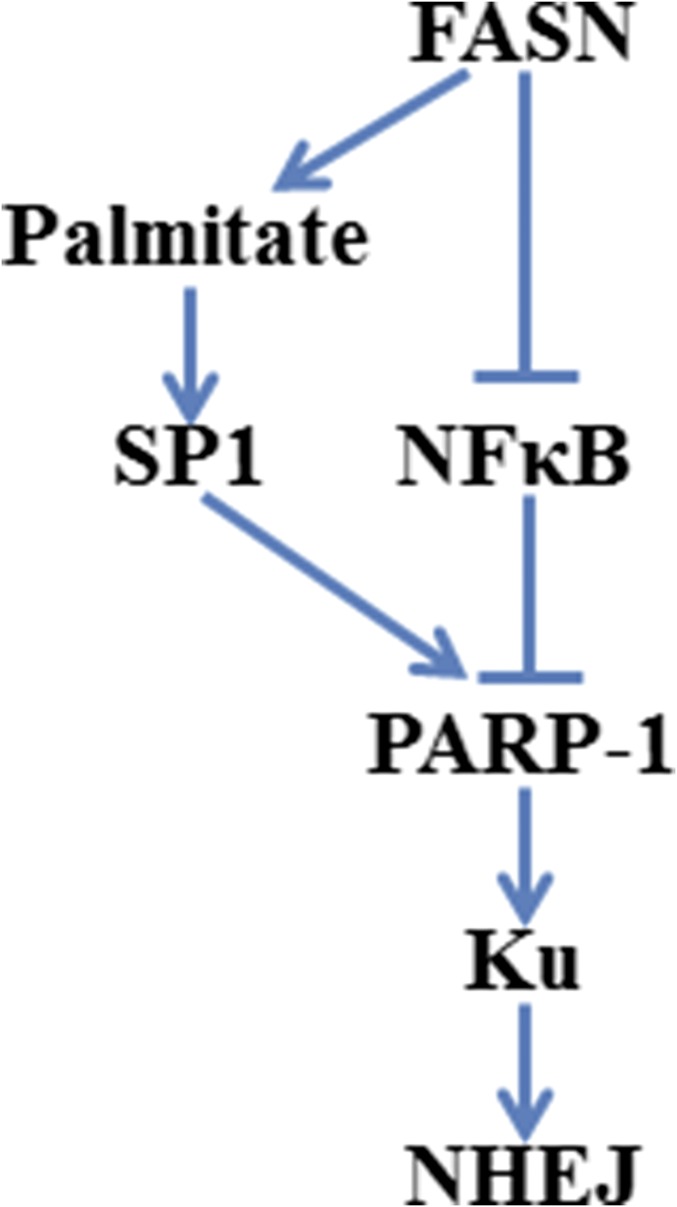
Accumulating evidence shows that FASN expression is up-regulated in human cancers and provides survival advantages for cancer cells. FASN overexpression has also been associated with resistance to anticancer treatments (1). However, the molecular mechanisms of FASN function in drug resistance are largely unknown. In the present study, we show that FASN overexpression suppresses NF-κB whereas up-regulates SP1, which in turn increases PARP-1 expression and facilitates recruitment of Ku proteins to IR- or drug-induced DSBs for NHEJ repair. This cascade of events results in resistance to treatments, including IR and doxorubicin, that elicit DSBs (Fig. S7).
Fig. S7.
Schematic representation of FASN regulation of NHEJ repair of DSBs.
In addition to IR, doxorubicin, and bleomycin, which all cause DSBs, we also found that FASN overexpression contributes to resistance to cisplatin, H2O2, and UVB, which primarily cause other types of DNA lesions repaired by the nucleotide excision repair (NER) and base excision repair (BER) pathways. Because PARP-1 is known to play important roles in both the BER and NER pathways (19–22), FASN may also contribute to resistance to cisplatin, H2O2, and UVB by up-regulating NER and BER activities via increasing PARP-1 expression and activity. In line with our findings, overexpression and constitutive hyperactivation of PARP-1 have been associated with cisplatin resistance in non–small-cell lung cancer (23).
As a nuclear enzyme, PARP-1 is activated by DNA damage. Activated PARP-1 posttranslationally modifies many nuclear proteins such as histones and DNA repair proteins by adding ADP-ribose polymers onto lysine residues, a process termed poly(ADP-ribosyl)ation (15), and has been implicated in playing important roles in different DNA repair pathways (24, 25). For NHEJ repair of DSBs, PARP-1 was thought to exercise its role via both a DNA-PK–dependent (26) and –independent manner (27). Two other members of the PARP family, PARP-2 and PARP-3, also possess poly(ADP-ribose) polymerase activity (28). However, PARP-1 is thought to be the major contributor and responsible for over 90% of cellular PARylation (29, 30). Thus, it is possible that PARP-1 is the major downstream mediator of FASN in the NHEJ repair of DSBs.
It is currently unknown how increased PARP-1 enhances Ku recruitment to chromatin for NHEJ repair of DSBs. Recently, it was found that PARP in Dictyostelium discoideum promotes Ku recruitment/retention to chromatin via PARylating nuclear proteins that bind Ku proteins at its C-terminal PAR-binding zinc-finger (PBZ) domain (14). However, human Ku70 and Ku80 do not possess a PBZ domain. Thus, it is unlikely that human Ku proteins bind directly to PARylated chromatin via a PBZ domain. APLF, also known as Xip1, is a recently discovered DNA repair protein with two C-terminal PBZ domains, which are required for the recruitment of APLF to sites of DNA damage (31, 32). Interestingly, APLF is known to associate with core NHEJ components such as XRCC4-DNA ligase IV and Ku proteins (33–35). It is thus tempting to speculate that the augmented Ku protein recruitment to chromatin by increased PARP-1 expression in human cancer cells may be via association with APLF that binds to the abundant PARylated chromatin via its PBZ domain carrying Ku.
It is noteworthy that PARP-1 has also been suggested to compete with Ku proteins in binding to DNA ends and, thus, perhaps inhibits classical NHEJ repair activity and may contribute to DSB repair via an alternative NHEJ (36). However, we clearly showed that increased PARP-1 expression and activity up-regulate instead of inhibit Ku protein recruitment and DNA-PK activity. Although the reason is currently unclear for the difference between these studies, the results from these studies indicate that PARP-1 may contribute to NHEJ repair of DSBs via different pathways.
Our finding that FASN may reduce HR repair of DSBs via up-regulating PARP-1 is interesting but consistent with previous observations. Initially, PARP-1 was thought to have a protective role against HR via binding to DNA breaks and preventing the HR machinery from recognizing and processing DNA lesions (37). Indeed, it was found later that inhibition or loss of PARP-1 increased HR activity in cells (38) or in vivo (39). PARP-1 may inhibit HR activity by PARylating BRCA1 and inhibiting its activity (40) or by inhibiting the expression of BRCA1 (41) and BRCA2 (42). It has also been observed that overaccumulation of PARylation also interferes with HR activity (43). Clearly, FASN regulates both NHEJ and HR via up-regulation of PARP-1. The promotion of the NHEJ pathway over HR may be a unique solution adopted by FASN-overexpressing cancer cells to achieve efficient repair of DSBs, as HR was associated with a slower rate of repair compared with NHEJ (44, 45). Our finding suggests that inhibiting FASN in HR-deficient cells may provide a novel approach of eliminating cancer cells in combination with DNA-damaging treatments or with PARP inhibitors.
Although it has been shown that the rat PARP-1 promoter is regulated by a composite binding site for the transcription factors SP1 and NF1, we found a similar composite binding site for SP1 and p65 in the promoter of human PARP-1. Whereas SP1 activates, p65 inhibits PARP-1 transcription by binding to the same composite site in a mutually exclusive manner. Composite elements are regulatory sequences commonly found in vertebrate gene promoters (46) that contain two or more closely positioned binding sites for distinct transcription factors and provide a way for fine-tuning gene expression. Binding of transcription factors to composite elements can either synergistically or antagonistically regulate gene expression. Similar to our findings, NF-κB antagonizes SP1 activation of the P-selectin promoter via an NF-κB/SP1 composite site (47).
Previously, we found that FASN suppresses TNF-α production (4), which is consistent with our current finding of reduced NF-κB expression by FASN. However, it is unknown how FASN inhibits NF-κB expression. Although the end product of FASN catalysis, palmitate, may mediate FASN regulation of SP1 expression, it does not appear to affect NF-κB. Previously, we found that supplementation of palmitate increased drug resistance of breast cancer cells (2), which is consistent with the up-regulation of SP1, which in turn increases NHEJ. Thus, lipid metabolism may play an important role in cancer cell survival against genotoxic insults. However, it remains to be determined how palmitate regulates SP1 and how FASN regulates NF-κB. We are currently working in this direction.
Experimental Procedures
All experiments involving human cell lines and recombinant DNA were approved by the Institutional Biosafety Committee at Indiana University.
Construct Engineering.
A pCβA-SP1 construct was engineered for ectopic SP1 overexpression. Briefly, cDNA encoding SP1 was purchased from Open Biosystems and released from the vector, pOTB7, by double digestion with EcoRI and XhoI. The cDNA fragment was inserted into the mammalian cell expression vector, pCβA (48), digested with EcoRI and XhoI, resulting in pCβA-SP1, and confirmed by DNA sequencing.
Cell Lines and Transient Transfections.
Human breast cancer cell line MCF7 and pancreatic cancer cell line Panc-1 were cultured at 37 °C with 5% (vol/vol) CO2 in DMEM supplemented with 10% (vol/vol) FBS, 100 units per mL penicillin, and 100 μg/mL streptomycin. FASN-overexpressing MCF7 (M/FASN) and Panc-1 (P/FASN) cells and their respective vector-transfected control clones (M/Vec, P/Vec), as well as stable MCF7/AdVp3000 cells with FASN knockdown (M3K/Sh) and its control scrambled shRNA-transfected clone (M3K/Scr) were generated previously (2, 8) and maintained in DMEM supplemented with 10% (vol/vol) FBS, 100 units per mL penicillin, 100 mg/mL streptomycin, and 400 µg/mL G418.
For transient transfection of pCβA-SP1, pcDNA3-p65-flag (49), or p65 shRNA (49) constructs and their respective control plasmids, cells were seeded at 3 × 105 cells per well in six-well plates and cultured for 24 h before transfection with 2 μg plasmids using Metafectene Pro (Biontex) according to the manufacturer’s instructions. For transient transfection of PARP-1 siRNA (Santa Cruz; sc-29437) or FASN siRNA (Santa Cruz; sc-43758), 6 × 105 cells were plated in six-well plates for 24 h, followed by transfection with siRNAs using Metafectene Pro according to the supplier’s instructions. The final siRNA concentrations for PARP-1 and FASN were 50 and 100 nM, respectively. At 48 h after transfection, the cells were collected or seeded for different assays.
Western Blot Analysis.
Western blot analysis was performed as we previously described (2, 4) using primary antibodies against FASN (BD Biosciences; 610962), PARP-1 (Cell Signaling; 9542), histone H3 (Cell Signaling; 2650), NF-κB p65 (Santa Cruz; sc372), Ku70 (Santa Cruz; sc-1487), γ-H2AX (Millipore; 05636), and anti-PAR (Trevigen; 4335). Final images were captured using FluorChem HD2 imager, and staining intensity was quantified using the AlphaEaseFC program associated with the FluorChem HD2 imager (both from Alpha Innotech).
Thiazolyl Blue Tetrazolium Bromide and Colony-Formation Survival Assays.
A thiazolyl blue tetrazolium bromide (MTT) survival assay was performed as previously described (50). Briefly, cells in 96-well plates were cultured for 24 h and treated with various concentrations of doxorubicin, bleomycin, cisplatin, or H2O2 continuously for 3 d followed by addition of MTT to a final concentration of 0.5 mg/mL and incubation at 37 °C for 4 h. The medium was then removed and formazan was solubilized in dimethyl sulfoxide, and the OD570 was measured by a Synergy H1 hybrid reader (BioTek). IC50s were obtained from the fitted curves generated by Prism 5.0 (GraphPad Software). The relative resistance factor (RRF) was calculated using the formula RRF = IC50(test)/IC50(control) with the control normalized as 1. For MCF7/AdVp3000 (M3K)-derived cell lines, 10 µM fumitremorgin C (FTC) was preincubated for 30 min before addition of doxorubicin to inhibit high ABCG2 activities in these cell lines. For the MTT assay following PARP-1 knockdown, MCF7/FASN cells were transiently transfected with control scramble or PARP-1 siRNA. At 24 h after transfection, cells were replated in 96-well plates and treated with doxorubicin for survival analysis.
The colony-formation assay was performed as previously described (51) for ionizing radiation and UVB treatments. Briefly, 100 cells were plated in 60-mm dishes and incubated for 24 h followed by treatment with IR or UVB and continuous culture for 10 d before removing the medium and washing the cells with PBS. Colonies were fixed and stained with a solution containing 0.05% (wt/vol) crystal violet and 20% (vol/vol) methanol in PBS.
Immunofluorescence Staining and Imaging.
Immunofluorescence staining and imaging were performed as previously described (52). Briefly, cells were cultured on coverslips in 60-mm culture dishes and treated with IR. At different times following IR treatment, the cells on coverslips were washed with PBS, fixed with 50:50 (vol/vol) acetone/methanol at room temperature for 10 min, blocked with 1% BSA in PBS for 30 min, and probed with anti–γ-H2AX antibody (1:100; Millipore; 05636) at room temperature for 1 h, followed by washing and incubation with FITC-conjugated goat anti-mouse IgG at room temperature for 30 min. The coverslips were washed again and incubated with 4′,6-diamidine-2-phenylindole dihydrochloride (DAPI) (1 μg/mL in PBS) for 10 min in the dark. The coverslips were then mounted on slides before viewing with an Olympus 2 confocal microscope.
Neutral Comet Assay.
The neutral comet assay was performed using a kit from Trevigen according to the manufacturer’s instructions. Briefly, cells following IR were harvested and mixed with low-temperature-melting agarose as single-cell suspensions at 37 °C. The resulting cell/agarose mixture was immediately layered onto CometSlides (Trevigen). The agarose was allowed to set for 1 h at 4 °C and cells on the slides were then lysed at 4 °C for 30 min in the dark. After lysis, the slides were subjected to electrophoresis and then immersed twice in distilled water for 10 min and once in 70% (vol/vol) ethanol for 5 min. The slides were then dried completely at room temperature and stained with SYBR Green I (Trevigen). Comets were observed and recorded by a Zeiss Axiovert 25 fluorescence microscope equipped with a Zeiss CCD camera and analyzed with CometScore version 1.5 (TriTek). The olive tail moment was determined by scoring 100 cells in each sample as previously described (53).
Host Cell Reactivation Assay.
The host cell reactivation NHEJ assay was performed as previously described (54) with minor modifications. Briefly, 5 × 104 cells per well were seeded in 24-well plates and cultured overnight before transfection using Lipofectamine Plus of either 400 ng intact (control) or HindIII-linearized (test) pGL3-encoding firefly luciferase (FL) together with 20 ng pRL-TK (Promega)–encoding renilla luciferase (RL) as a control for transfection efficiency. Cells were harvested at 8 h following transfection, and both FL and RL activities were determined using a Dual-Luciferase Reporter Assay System (Promega). FL activities from both control and test groups were first normalized to that of RL activities before calculating NHEJ activity using the formula NHEJ activity = (normalized FL activity in the test group) × 100/(normalized FL activity in the control group).
The host cell reactivation HR assay was performed as previously described (55) with modifications. Briefly, the pGL3 plasmid was digested with NcoI and XcmI to generate a fragment containing the SV40 promoter and the 5′ portion of the FL gene as well as with BsrGI and BamHI to generate another fragment containing the 3′ portion of the FL gene and the polyA signal. These two DNA fragments, containing a 245-bp-overlapping region, were purified and cotransfected along with the pRL-TK control plasmid into 7 × 104 cells per well in a 24-well plate using Metafectene Pro transfection reagent. Transfections with either the NcoI-XcmI or BsrGI-BamHI fragment alone were used as negative controls. At 24 h after transfection, cells were harvested for the Dual-Luciferase Reporter Assay as described above. FL activities were first normalized to RL activities for transfection efficiency before deriving relative HR activity using the formula HR activity (fold) = (normalized FL activity in P/FASN or M3K/Sh cells)/(normalized FL activity in P/Vec or M3K/Scr cells).
DNA-PK Activity.
DNA-PK activity was analyzed using the SignaTECT DNA-Dependent Protein Kinase Assay System (Promega) according to the manufacturer’s instructions. Briefly, 10 μg nuclear extract with endogenous DNA removed by DEAE-Sepharose filtration was incubated with biotinylated peptide substrate, [γ-32P]ATP, and either DNA-PK activation buffer or DNA-PK control buffer for 5 min at 30 °C. Termination buffer was added, and 10 μL of each reaction sample was spotted onto a SAM2TM biotin-capture membrane. The SAM2TM membrane were then washed and dried before analysis by scintillation counting. DNA-PK activity is expressed as specific activity in pmol ATP/min per μg protein.
Real-Time Quantitative PCR Analysis.
Real-time quantitative PCR analysis was performed as previously described (4, 56). Briefly, cells were harvested, and total RNA was extracted using an RNeasy Mini Kit (Qiagen) followed by real-time RT-PCR using the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems). Data were normalized to an internal control gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer pairs used were 5′-CCCAGGGTCTTCGGATAG-3′ (forward) and 5′-AGCGTGCTTCAGTTCATACA-3′ (reverse) for PARP-1. Amplification of a single PCR product was confirmed by melt-curve analysis.
NF-κB Activity Assay.
For the NF-κB activity assay, P/FASN, M3K/Sh, and their control P/Vec and M3K/Scr cells were seeded at 5 × 104 cells per well in 24-well plates and transfected with a PathDetect NF-κB Cis-Reporting plasmid (Agilent Technologies). pRL-TK–encoding renilla luciferase was cotransfected as a control for transfection efficiency. At 48 h after transfection, cells were harvested for luciferase assay using the Dual-Luciferase Reporter Assay System as described above.
Cell-Cycle Analysis.
Cells seeded in 100-mm dishes at 6 × 105 cells per dish were cultured for 3 d before treatment and cell-cycle analysis. For serum starvation, the cells were washed with serum-free media and cultured in the same media for 48 h before fixation in 70% (vol/vol) ethanol at room temperature for 30 min. For thymidine block, the cells were treated with 2 mM thymidine for 28 h followed by thymidine removal and culture for 10 h in complete media without thymidine. The cells were treated again with 2 mM thymidine for 24 h followed by fixation with ethanol as described above. For nocodazole treatment, cells were cultured in media containing 100 ng/mL nocodazole for 18 h and followed by fixation with ethanol as described above. The fixed cells were then washed with PBS, stained with 50 μg/mL propidium iodide, and digested with 100 μg/mL RNase at 37 °C for 30 min before analysis using a FACScan flow cytometer (BD Bioscience). Cell-cycle distribution was analyzed with the ModFit LT program (Verity Software House).
Chromatin Immunoprecipitation.
ChIP was performed as previously described (57) using a Millipore ChIP Assay Kit. Briefly, Panc-1 cells were seeded at 1 × 106 in 100-mm dishes and cultured for 24 h before transfection with Flag-p65 cDNA or vector control. At 48 h post transfection, cells were fixed with 1% formaldehyde and subjected to ChIP analysis. Primers for the PARP-1 promoter were 5′-CCGGGTCCTCCAAAGAGCTA-3′ (forward) and 5′-GCCGTTCCCTGATAGATTGCT-3′ (reverse). Data were analyzed as the percentage of input of the total samples and calculated as previously described (58).
Statistical Analysis.
Student’s t test was used for all statistical analysis, with P < 0.05 considered significant. All experiments were performed independently at least three times for statistical analysis.
Acknowledgments
The authors thank Dr. Tao Lu for the p65 shRNA and overexpression constructs, and Dr. John Turchi and Dr. Derek Woods for discussions. This work was supported in part by a Department of Defense (DOD) Predoctoral Fellowship (to X.W.), NIH NRSA F31 CA165603 (to V.F.), NIH Grant R01 CA140582 (to J.-T.Z.), and DOD Grant BC150290 (to J.-T.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609934113/-/DCSupplemental.
References
- 1.Liu H, Liu JY, Wu X, Zhang JT. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int J Biochem Mol Biol. 2010;1(1):69–89. [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Liu Y, Zhang JT. A new mechanism of drug resistance in breast cancer cells: Fatty acid synthase overexpression-mediated palmitate overproduction. Mol Cancer Ther. 2008;7(2):263–270. doi: 10.1158/1535-7163.MCT-07-0445. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Qin L, Fako V, Zhang JT. Molecular mechanisms of fatty acid synthase (FASN)-mediated resistance to anti-cancer treatments. Adv Biol Regul. 2014;54:214–221. doi: 10.1016/j.jbior.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, et al. Fatty acid synthase causes drug resistance by inhibiting TNF-α and ceramide production. J Lipid Res. 2013;54(3):776–785. doi: 10.1194/jlr.M033811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5(9–10):1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang M, Wang H, Böcker W, Iliakis G. Complex H2AX phosphorylation patterns by multiple kinases including ATM and DNA-PK in human cells exposed to ionizing radiation and treated with kinase inhibitors. J Cell Physiol. 2005;202(2):492–502. doi: 10.1002/jcp.20141. [DOI] [PubMed] [Google Scholar]
- 7.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, et al. Role of fatty acid synthase in gemcitabine and radiation resistance of pancreatic cancers. Int J Biochem Mol Biol. 2011;2(1):89–98. [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, Schultz M, Kruh GD, Tew KD. Increased expression of DNA-dependent protein kinase confers resistance to adriamycin. Biochim Biophys Acta. 1998;1381(2):131–138. doi: 10.1016/s0304-4165(98)00020-8. [DOI] [PubMed] [Google Scholar]
- 10.Shintani S, et al. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci. 2003;94(10):894–900. doi: 10.1111/j.1349-7006.2003.tb01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luzhna L, Golubov A, Ilnytskyy S, Chekhun VF, Kovalchuk O. Molecular mechanisms of radiation resistance in doxorubicin-resistant breast adenocarcinoma cells. Int J Oncol. 2013;42(5):1692–1708. doi: 10.3892/ijo.2013.1845. [DOI] [PubMed] [Google Scholar]
- 12.Redon CE, et al. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16(18):4532–4542. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo X, Kraus WL. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26(5):417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couto CA, et al. PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J Cell Biol. 2011;194(3):367–375. doi: 10.1083/jcb.201012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38(19):6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopperton KE, Duncan RE, Bazinet RP, Archer MC. Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity. Exp Cell Res. 2014;320(2):302–310. doi: 10.1016/j.yexcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Bergeron MJ, Leclerc S, Laniel MA, Poirier GG, Guérin SL. Transcriptional regulation of the rat poly(ADP-ribose) polymerase gene by Sp1. Eur J Biochem. 1997;250(2):342–353. doi: 10.1111/j.1432-1033.1997.0342a.x. [DOI] [PubMed] [Google Scholar]
- 18.Laniel MA, Poirier GG, Guérin SL. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J Biol Chem. 2001;276(23):20766–20773. doi: 10.1074/jbc.M010360200. [DOI] [PubMed] [Google Scholar]
- 19.Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7(3):165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 20.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robu M, et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci USA. 2013;110(5):1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adham M, et al. Nasopharyngeal carcinoma in Indonesia: Epidemiology, incidence, signs, and symptoms at presentation. Chin J Cancer. 2012;31(4):185–196. doi: 10.5732/cjc.011.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michels J, et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 2013;73(7):2271–2280. doi: 10.1158/0008-5472.CAN-12-3000. [DOI] [PubMed] [Google Scholar]
- 24.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 25.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1(3):328–346. [PMC free article] [PubMed] [Google Scholar]
- 26.Ruscetti T, et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273(23):14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell J, Smith GC, Curtin NJ. Poly(ADP-ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int J Radiat Oncol Biol Phys. 2009;75(5):1520–1527. doi: 10.1016/j.ijrobp.2009.07.1722. [DOI] [PubMed] [Google Scholar]
- 28.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Yamanaka H, Penning CA, Willis EH, Wasson DB, Carson DA. Characterization of human poly(ADP-ribose) polymerase with autoantibodies. J Biol Chem. 1988;263(8):3879–3883. [PubMed] [Google Scholar]
- 30.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 31.Kanno S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26(8):2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bekker-Jensen S, et al. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem. 2007;282(27):19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- 33.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 34.Shirodkar P, Fenton AL, Meng L, Koch CA. Identification and functional characterization of a Ku-binding motif in aprataxin polynucleotide kinase/phosphatase-like factor (APLF) J Biol Chem. 2013;288(27):19604–19613. doi: 10.1074/jbc.M112.440388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grundy GJ, et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32(1):112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20(10):405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 38.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31(17):4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claybon A, Karia B, Bruce C, Bishop AJ. PARP1 suppresses homologous recombination events in mice in vivo. Nucleic Acids Res. 2010;38(21):7538–7545. doi: 10.1093/nar/gkq624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, et al. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4(12):1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, et al. A novel crosstalk between BRCA1 and poly (ADP-ribose) polymerase 1 in breast cancer. Cell Cycle. 2014;13(21):3442–3449. doi: 10.4161/15384101.2014.956507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, et al. Poly(ADP-ribose) polymerase-1 down-regulates BRCA2 expression through the BRCA2 promoter. J Biol Chem. 2008;283(52):36249–36256. doi: 10.1074/jbc.M803693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Illuzzi G, et al. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res. 2014;42(12):7776–7792. doi: 10.1093/nar/gku505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30(6):1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47(2):320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kel OV, Romaschenko AG, Kel AE, Wingender E, Kolchanov NA. A compilation of composite regulatory elements affecting gene transcription in vertebrates. Nucleic Acids Res. 1995;23(20):4097–4103. doi: 10.1093/nar/23.20.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano F, et al. Functional interference of Sp1 and NF-kappaB through the same DNA binding site. Mol Cell Biol. 1998;18(3):1266–1274. doi: 10.1128/mcb.18.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong Z, Zhang JT. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Mol Biol Cell. 2003;14(9):3942–3951. doi: 10.1091/mbc.E02-12-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Chen Q, Zhang JT. Structural and functional consequences of mutating cysteine residues in the amino terminus of human multidrug resistance-associated protein 1. J Biol Chem. 2002;277(46):44268–44277. doi: 10.1074/jbc.M207003200. [DOI] [PubMed] [Google Scholar]
- 51.Li Z, et al. Role of 14-3-3σ in poor prognosis and in radiation and drug resistance of human pancreatic cancers. BMC Cancer. 2010;10:598. doi: 10.1186/1471-2407-10-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Q, Yang Y, Li L, Zhang JT. The amino terminus of the human multidrug resistance transporter ABCC1 has a U-shaped folding with a gating function. J Biol Chem. 2006;281(41):31152–31163. doi: 10.1074/jbc.M603529200. [DOI] [PubMed] [Google Scholar]
- 53.Olive PL, Wlodek D, Banáth JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 1991;51(17):4671–4676. [PubMed] [Google Scholar]
- 54.Boeckman HJ, Trego KS, Turchi JJ. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol Cancer Res. 2005;3(5):277–285. doi: 10.1158/1541-7786.MCR-04-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai YS, Huang JL, Lin CS. Application of host cell reactivation in evaluating the effects of anticancer drugs and environmental toxicants on cellular DNA repair activity in head and neck cancer. In: Chen CC, editor. Selected Topics in DNA Repair. INTECH Open Access; Rijeka, Croatia: 2011. pp. 465–482. [Google Scholar]
- 56.Liu RY, et al. Role of eIF3a in regulating cisplatin sensitivity and in translational control of nucleotide excision repair of nasopharyngeal carcinoma. Oncogene. 2011;30(48):4814–4823. doi: 10.1038/onc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang W, et al. A small molecule compound targeting STAT3 DNA-binding domain inhibits cancer cell proliferation, migration, and invasion. ACS Chem Biol. 2014;9(5):1188–1196. doi: 10.1021/cb500071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zampieri M, et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4(3):e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]