Significance
Plants and animals use intracellular immunity receptors, known as nucleotide-binding oligomerization domain-like receptors (NLRs), to defend themselves against invading microbes. In this study, we report the solution structure of the N-terminal coiled-coil (CC) domain from the wheat stem rust resistance protein Sr33. Remarkably, this structure differs substantially from the published crystal structure of the equivalent region from the orthologous barley powdery mildew resistance protein MLA10. Using a structural, biophysical, and functional approach, we compare the Sr33 CC domain with other structurally defined NLR CC domains. Collectively, this work redefines our current understanding of the structure and function of plant NLR CC domains, which has significant implications for future studies into this important class of defense receptors.
Keywords: plant innate immunity, resistance protein, NLR proteins, effector-triggered immunity, nuclear magnetic resonance spectroscopy
Abstract
Plants use intracellular immunity receptors, known as nucleotide-binding oligomerization domain-like receptors (NLRs), to recognize specific pathogen effector proteins and induce immune responses. These proteins provide resistance to many of the world’s most destructive plant pathogens, yet we have a limited understanding of the molecular mechanisms that lead to defense signaling. We examined the wheat NLR protein, Sr33, which is responsible for strain-specific resistance to the wheat stem rust pathogen, Puccinia graminis f. sp. tritici. We present the solution structure of a coiled-coil (CC) fragment from Sr33, which adopts a four-helix bundle conformation. Unexpectedly, this structure differs from the published dimeric crystal structure of the equivalent region from the orthologous barley powdery mildew resistance protein, MLA10, but is similar to the structure of the distantly related potato NLR protein, Rx. We demonstrate that these regions are, in fact, largely monomeric and adopt similar folds in solution in all three proteins, suggesting that the CC domains from plant NLRs adopt a conserved fold. However, larger C-terminal fragments of Sr33 and MLA10 can self-associate both in vitro and in planta, and this self-association correlates with their cell death signaling activity. The minimal region of the CC domain required for both cell death signaling and self-association extends to amino acid 142, thus including 22 residues absent from previous biochemical and structural protein studies. These data suggest that self-association of the minimal CC domain is necessary for signaling but is likely to involve a different structural basis than previously suggested by the MLA10 crystallographic dimer.
Plant diseases constitute a major economic and social burden worldwide, and the appearance of new or more virulent pathogens can pose significant challenges. Plants rely on their innate immunity systems to combat pathogens, an important component of which is the recognition of pathogen effector molecules by resistance proteins, a process commonly referred to as effector-triggered immunity. Resistance protein activation triggers a defense mechanism known as the hypersensitive response, which often culminates in localized cell death at the site of infection, leading to general immunity of the whole plant (1).
One such R protein is encoded by the recently discovered wheat gene Sr33 (2). Sr33 confers resistance to the virulent Ug99 strain of wheat stem rust, Puccinia graminis f. sp. tritici, a pathogen recognized for its potential threat to global food security. Sr33 is orthologous to the barley powdery mildew resistance protein MLA and rye Sr50 genes (3, 4) and encodes a member of the canonical class of plant resistance proteins, consisting of a central nucleotide-binding (NB) domain, a C-terminal leucine-rich repeat (LRR) domain, and an N-terminal coiled-coil (CC) domain (2). Such proteins have a domain arrangement and function similar to the NB oligomerization domain-like receptors (NLRs) from mammals, and are commonly referred to as plant NLRs (5).
Although the molecular details of plant NLR activation and signaling are not fully understood, targeted studies have helped define the roles of their different domains. The central NB domain appears to control the activation of the protein through NB and nucleotide exchange (6–8). The LRR domain plays a role in effector recognition specificity for a number of plant NLRs, and, in some cases, it is implicated in effector binding (9–11). The LRR domain also appears to have a general autoinhibitory role, and structural and biochemical studies of the human NLR proteins NAIP and NLRC4 support this conclusion (12). In plant NLRs, the N-terminal domain generally consists of either a CC domain, as in Sr33, or a Toll/interleukin-1 receptor/resistance protein (TIR) domain. Both CC and TIR domains are implicated in downstream signaling, and have been shown to be necessary and sufficient for cell death responses in a number of systems (6, 13–18).
Oligomerization is key to animal NLR activation and signaling, as demonstrated by the structural characterization of the NAIP2/NLRC4 inflammasome NLR pair (19–21). Our understanding of these processes in plant NLRs is more limited. To date, effector-induced self-association of a full-length plant NLR has been demonstrated for only the TIR-NLRs, including the tobacco mosaic virus resistance protein N (22) and the Arabidopsis protein RECOGNITION OF PERONOSPORA PARASITICA1 (RPP1) (23). Self-association in the absence of effector proteins has been observed in CC-NLR proteins from Arabidopsis RPS5 (24), barley (MLA10) (14), maize (Rp1-D) (18), and wheat (Sr33 and Sr50) (25). However, MLA10, Sr33, and Sr50 also display autoactivity when overexpressed (16, 25), suggesting autoassociation may mimic the activated state.
To date, structure-guided studies of plant NLRs have been restricted to the N-terminal TIR or CC domain (6, 14, 17, 26), with the exception of the noncanonical integrated-sensor heavy metal-associated domain from the rice NLR Pik (27). Although the plant NLR TIR domains have a conserved fold (6, 17), the structures of the two known CC domain fragments from barley MLA10 and potato Rx are strikingly different. The N-terminal amino acids 5–120 of MLA10 (designated MLA105-120) crystallized as an antiparallel homodimer adopting a helix–loop–helix fold. However, the equivalent region from Rx (Rx1-122) adopted a compact, four-helical bundle when crystallized in a 1:1 heteroassociation with the Trp-Pro-Pro (WPP) domain from its cofactor protein RanGAP2 (26). MLA105-120 was also reported to dimerize in vitro, whereas Rx1-122 was monomeric in vitro in the absence of cofactors. A larger CC-containing fragment of MLA10 (MLA101-160) was capable of inducing cell death in planta (14), whereas studies of Rx found no cell death induction by CC domain fragments (28). Although the CC domains from Rx and MLA10 share low sequence identity in this region (<20%), both contain the EDVID motif and are classified within the CCEDVID domain class from plant NLRs (29). These studies suggested that significant structural and mechanistic variation is present among the CCEDVID domains of plant NLRs.
Recently, Cesari et al. (25) found that the CC-containing fragments of MLA10, Sr33, and Sr50 corresponding to MLA10 residues 1–160 induced cell death and self-associated in planta, whereas the smaller 1–120 fragments (equivalent to the MLA10 structure) did not. Thus, to better understand the role of the CC domains in NLR protein signaling, we undertook a structural and functional study of the CC region of the wheat stem rust NLR protein Sr33. Here, we present the solution 3D structure of the Sr33 CC domain (residues 6–120; Sr336-120), determined by NMR spectroscopy. The structure resembles the CC domain of Rx more closely than the CC domain of MLA10. Prompted by this finding, we carried out a detailed biophysical comparison of the CC domains of Sr33, MLA10, and Rx, which suggests that these CC-NLRs all adopt a common fold and are monomeric in solution. We also find that the minimal functional regions for MLA10, Sr33, and Sr50 CC domain self-association and cell death signaling exceed the boundaries used in in vitro and structural studies to date.
Results
The NMR Structure of Sr336-120 Reveals a Compact, Four-Helix Bundle.
Previous structural studies of the Sr33 ortholog MLA10 encompassed residues 5–120 of the CC domain (MLA105-120). For our investigation, we initially focused on an equivalent region within the Sr33 CC domain. Soluble Sr33 CC domain protein was produced by recombinant expression of a construct comprising residues 6–120 in Escherichia coli, and the atomic structure was determined by NMR spectroscopy.
Sr336-120 appears to be monomeric under the conditions used for the NMR structural studies. This conclusion is evidenced by the sharp line widths of the resonances. In addition, the average T2 relaxation rates of the backbone amides yield an overall rotational correlation time of the protein of ∼8.7 ns. This rotational correlation time corresponds to a spherical protein with a molecular mass of ∼13 kDa, compared with the theoretical monomeric molecular mass of 13.1 kDa (30) (SI Appendix, Table S1). Analysis of the assigned chemical shifts (31) revealed four distinctive α-helical regions (α1, residues 7–19; α2, residues 28–51; α3, residues 60–87; and α4, residues 99–115). Other than the termini, two small regions (comprising residues 22–23 and residues 88–91) were found to have near-random-coil chemical shifts, indicating that these regions are highly dynamic. The residues that correspond to 88–91 in the MLA105-120 crystal structure were also poorly defined; however, the residues corresponding to 22–23 appear in an ordered helical region in the MLA105-120 crystal structure. Dihedral angles derived from the chemical shift analysis were used, together with distance restraints from 15N- and 13C-edited NOESY experiments, to calculate the 3D structure of the protein. The structure shows that the protein is folded into a four-helix bundle (Fig. 1A and SI Appendix, Fig. S1). The average root-mean-square distance (RMSD) for the amide backbone atoms (N, Cα, and C′) of residues 6–89 and 98–110 in the ensemble of the 20 lowest energy structures is 0.93 Å (SI Appendix, Table S1).
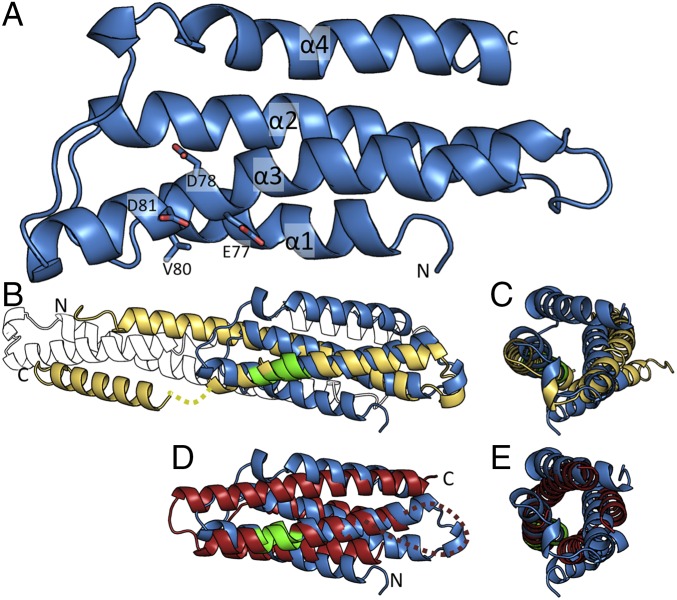
Fig. 1.
Solution structure of Sr33 reveals a four-helix bundle fold. (A) NMR structure of Sr336-120 in cartoon representation, with the individual helices and N and C termini labeled. The conserved EDVID motif (EDAVD in Sr33) is shown in stick representation (colored green in B and D). (B) Superposition of the Sr336-120 structure (blue) and the crystal structure of MLA105-120 (yellow) in cartoon representation. Missing residues in the MLA105-120 structure (amino acids 91–95) are shown by a dotted line. The crystallographic dimer observed for MLA105-120 is shown as a black and white outline. (C) Superposition, as shown in B, rotated 90° around the y axis. (D) Superposition of the Sr336-120 structure (blue) and the crystal structure of Rx1-122 (red) in cartoon representation. Missing residues in the Rx1-122 structure (amino acids 40–50) are shown by a dotted line. (E) Superposition, as shown in D, rotated 90° around the y axis.
The EDVID motif is conserved within this class of CC domains and is important in mediating intramolecular domain interactions in full-length CC-NLR proteins (26, 28). In Sr336-120, the equivalent motif (residues 77–81) encodes residues EDAVD, which reside in the α3 helix, with residues E77, D78, V80, and D81 all surface-exposed (Fig. 1A). Despite the high sequence similarity between Sr336-120 and MLA105-120, these structures differ significantly and the solution structure of Sr336-120 resembles the structure of Rx1-122 more closely (Fig. 1 B–E).
Biophysical Characterization Shows That Sr336-120, MLA105-120, and Rx1-122 Are Predominantly Monomeric and Adopt Compact, Globular Conformations.
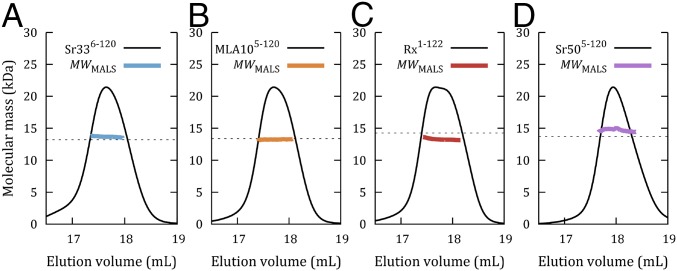
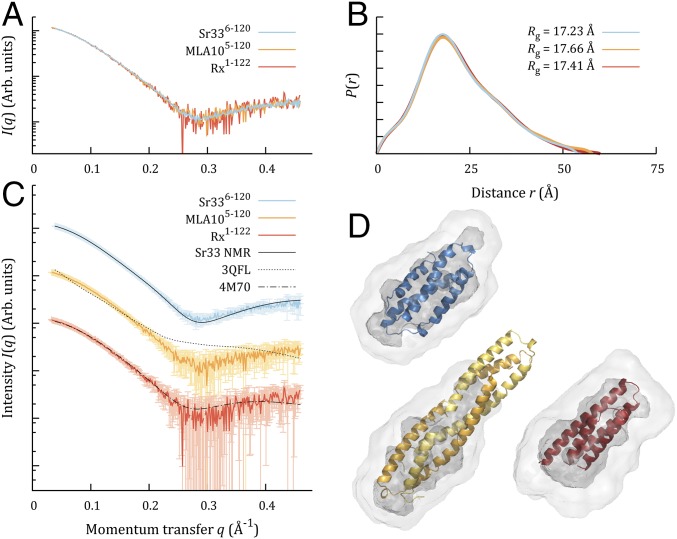
The NMR results suggested that Sr336-120 is monomeric in solution. This result differed from our expectations based on the crystal structure of MLA105-120. We investigated this finding further using in vitro biophysical techniques and also included the MLA105-120 and Rx1-122 constructs used in previous structure determination studies (14, 26), as well as Sr505-120, a rye ortholog of Sr33 (4), in our analysis. Size-exclusion chromatography (SEC)-coupled multiangle light scattering (MALS) experiments on these four proteins found an average molecular mass in solution very close to the predicted monomer sizes, indicating that they were all predominantly monomeric in solution, even at loading concentrations of 30 mg/mL (Fig. 2 A–D). This finding was corroborated by SEC-coupled small-angle X-ray scattering (SAXS) for Sr336-120, MLA105-120, and Rx1-122 (SI Appendix, Fig. S2 A–C and Table S2), which yielded molecular masses between 12 and 14 kDa for Sr336-120 and between 12 and 16 kDa for MLA105-120 and Rx1-122, varying with concentration across the elution. Averaged datasets from different fractions reflect this finding (SI Appendix, Fig. S2 D–I), but the data also suggested that the peak center fractions were suffering from some nonspecific aggregation (SI Appendix, Fig. S2 G–I), which is not present in the tail fractions. SAXS data also contain information about the shape of particles in solution. The scattering from the tail fractions of all three proteins is indistinguishable within experimental error (Fig. 3A), and their calculated properties (SI Appendix, Table S2) and real-space distributions also agree (Fig. 3B). Moreover, the experimental data are consistent with the predicted scattering of the four-helix bundle arrangement observed in the structures of Sr336-120 and Rx1-122. They are not consistent with the structures of either the dimer or individual protomers from the MLA105-120 crystal structure (Fig. 3C and SI Appendix, Table S3). The same conclusion can be drawn if one attempts to superimpose the ab initio reconstructions from the SAXS data onto the corresponding high-resolution structures (Fig. 3D). All three datasets yield compact, globular-shaped envelopes, into which the NMR structure of Sr336-120 and the crystal structure of Rx1-122 can be docked within the proposed envelope. In contrast, the envelopes are clearly smaller than the extended conformation seen in the MLA105-120 crystals.
Fig. 2.
Molecular mass calculations based on SEC-MALS analysis for Sr336-120 (A), MLA105-120 (B), Rx1-122 (C), and Sr505-123 (D). For all proteins, solid black lines represent the normalized refractive index trace (arbitrary units, y axis) for proteins eluted from an in-line Superdex 200 10/300 column. Colored lines under the peaks correspond to the averaged molecular mass (right-hand y axis) distributions across the peak as determined by MALS (MWMALS). Dotted lines indicate the predicted molecular masses of a monomer. The average MWMALS values compared with predicted monomeric molecular mass values are 13.7/13.1 kDa for Sr336-120, 13.3/13.4 kDa for MLA105-120, 13.3/14.3 kDa for Rx1-122, and 14.7/14.1 kDa for Sr505-123.
Fig. 3.
SAXS data from monomeric fractions of Sr336-120, MLA105-120, and Rx1-122 are consistent with compact, globular particles. (A) Datasets from SEC-SAXS are shown as colored lines, with the MLA105-120 and Rx1-122 data scaled to overlay with the Sr336-120 data. Arb., Arbitrary. (B) Normalized distance distribution functions, P(r), are shown as colored lines matching the scattering curve from which they were calculated. All distributions have been scaled to the maxima of the highest peak. (C) SEC-SAXS datasets again plotted as colored lines, now arbitrarily offset in y for clarity. Experimental errors are displayed at 1σ in lighter colors. The theoretical scattering predicted from each 3D structural model is shown as a black line against the corresponding dataset. (D) The first member of the Sr336-120 NMR ensemble (blue), the Rx1-122 crystal structure (red), and the dimeric MLA105-120 crystal structure (yellow) are shown in cartoon representation, docked into ab initio envelopes calculated from their respective scattering datasets. Ab initio models are shown in transparent surface representation, with the average model from 16 independent runs shown in light gray and the filtered model shown in darker gray.
Maekawa et al. (14) found that the MLA105-120 protein coeluted from an analytical SEC column with a 25-kDa protein standard and also that treatment with the amine-to-amine cross-linker bis(sulfosuccinimidyl)suberate (BS3) resulted in covalent dimer formation. We repeated these experiments using our own purified MLA105-120 protein and observed similar results. MLA105-120 indeed elutes at a similar volume to the 25-kDa chymotrypsin marker during SEC (SI Appendix, Fig. S3). However, this behavior was also observed for Sr33, and for Rx, which had been previously found to be monomeric (26). We also observed a similar magnitude of cross-linked dimer to that observed by Maekawa et al. (14) after incubation with BS3 (SI Appendix, Fig. S3). In both experiments, the majority of the MLA105-120 protein remains monomeric even after 2 h of incubation with BS3. In contrast, the flax-rust AvrM effector protein, which forms a stable dimer in solution (32), was immediately cross-linked as a dimer on the addition of BS3 (SI Appendix, Fig. S3). Thus, these data do not support the formation of a stable MLA105-120 dimer in solution. Collectively, biophysical analysis shows that the monomer is the predominant species in solution for Sr336-120, MLA105-120, and Rx1-122, and that this monomer is compact, globular, and consistent with four-helix bundle structures.
Crystal Structure of MLA105-120.
The crystal structure of MLA105-120 reported by Maekawa et al. (14) shows a helix–loop–helix structure that forms a dimer through a large interface. The published MLA105-120 crystal structure was obtained at low pH (4.6) and high salt (2.0 M sodium formate). To investigate whether dimer formation is dependent upon the crystallization conditions, we attempted crystallization of MLA105-120 using alternative precipitants and neutral pH. We found that MLA105-120 crystallized readily in 25% (wt/vol) polyethylene glycol 1500 at pH 7.0. These crystals diffracted X-rays to ∼2.0-Å resolution, and the structure was solved using single-wavelength anomalous diffraction (SI Appendix, Table S4).
The resulting structure resembles the structure described previously for MLA105-120 (14), having a Cα RMSD of 3.7 Å from 110 residues (SI Appendix, Fig. S4), and it forms a similar crystallographic dimer. We observed electron density for all residues (residues 5–120) in the best chain, including the previously undefined residues 91–95. However, there are notable differences between the two structures, particularly with respect to the interactions between the monomers forming the crystallographic dimer. In some regions, the interacting residues in the two protomers are offset by approximately one helical turn relative to the previously published crystallographic dimer (SI Appendix, Fig. S4). Comparison of the dimeric interface using PISA (Protein, Interfaces, Structures, and Assemblies) (33) shows that the structure presented here includes a more extensive hydrogen-bonding network between complementary residues, suggesting that the conformational dynamics of MLA105-120 permit different structural rearrangements during crystallization.
Extended CC Domain Fragments of Sr33 and MLA10 CC Domains Show an Increased Propensity to Self-Associate.
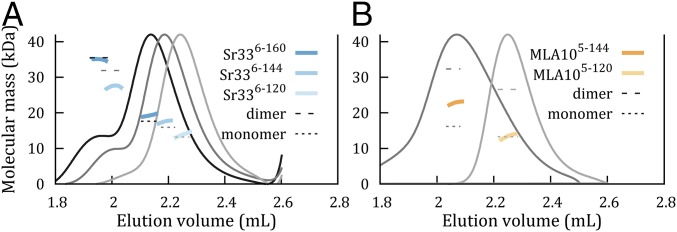
Recent in planta results indicate that the residues between 120 and 160 are required for signaling and self-association of the Sr33, Sr50, and MLA10 N-terminal domains (25). Secondary-structure predictions (34) of these proteins predict a helix extending from residue 98 to residue 138 (SI Appendix, Fig. S5), which would be truncated in the fragments used for structure determination. Using a modified purification buffer system (SI Appendix, SI Methods), we expressed and purified Sr336-144, Sr336-160, and MLA105-144 to homogeneity (SI Appendix, Fig. S5 B and C). SEC-MALS revealed increased self-association in these longer CC domain fragments (Fig. 4). An earlier peak with a molecular mass near the molecular mass expected for the dimer was apparent for Sr336-144 and Sr336-160, while a larger peak at the position expected of the monomer was still present for both. MLA105-144 eluted in a single peak that was more extended and asymmetrical than the single peak of MLA105-120, with a molecular mass 40% higher than the molecular mass expected for the monomer, indicating a polydisperse population of molecules (Fig. 4 and SI Appendix, Table S5). These experiments show that the additional residues promote self-association in both Sr33 and MLA10.
Fig. 4.
Solution studies of CC domains with extended sequences of Sr33 and MLA10. Molecular mass calculations from SEC-MALS analysis for Sr336-120, Sr336-144, and Sr336-160 (A) and MLA105-120 and MLA105-144 (B). Solid gray, dark gray, and black lines represent the refractive index for the three proteins, respectively, when eluted from an in-line Superdex 200 5/150 GL column, normalized to the height of the major peak for clarity. Dotted lines indicate the predicted molecular masses of both monomeric and dimeric species, and colored lines show the experimental molecular mass distributions as determined by MALS (values are shown in SI Appendix, Table S5).
Defining the Minimal CC-Domain Signaling Unit in Sr33, MLA10, and Sr50.
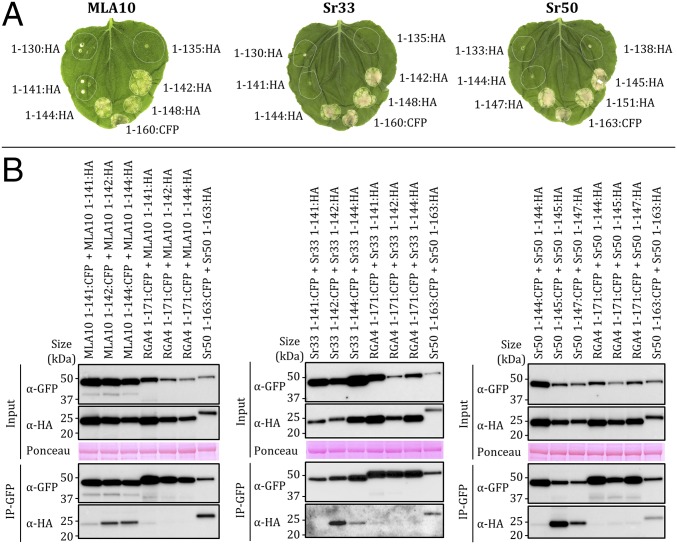
To identify the minimal N-terminal fragment necessary for the signaling function of these proteins, we generated six truncations of the MLA10, Sr33, and Sr50 N-terminal domains at positions surrounding the predicted end of the α4 helix (Fig. 5A). These truncations were transiently expressed in Nicotiana benthamiana under the control of the 35S promoter and fused to a C-terminal HA tag. Fragments truncated at, or beyond, the equivalent of MLA10 residue 142 induced cell death similar to the autoactive 1–160 fragments (16, 25), whereas shorter fragments were inactive (Fig. 5A). Immunoblotting showed that the proteins were stable and accumulated to similar levels (SI Appendix, Fig. S6). These results demonstrate that the minimal N-terminal cell death signaling domains in MLA10, Sr33, and Sr50 extend to the amino acid position equivalent to 142 in MLA10 (SI Appendix, Fig. S5).
Fig. 5.
Minimal autoactive domains of MLA10, Sr33, and Sr50 self-associate in planta. (A) MLA10, Sr33, and Sr50 protein fragments fused to HA or CFP were transiently expressed in N. benthamiana. The autoactive MLA101-160:CFP, Sr331-160:CFP, and Sr501-163:CFP constructs were used as positive controls. Cell death was visualized 5 d after infiltration. Equivalent results were obtained in three independent experiments. (B) Indicated proteins, transiently expressed in N. benthamiana leaves, were extracted 20 h after infiltration and analyzed by immunoblotting with anti-HA (α-HA) and anti-GFP antibodies (α-GFP) (Input). Proteins were immunoprecipitated with anti-GFP beads (IP-GFP) and analyzed by immunoblotting with anti-GFP and anti-HA antibodies. RGA4 (CC domain):CFP fusion was used as a control for specificity. Sr501-163 was used as a positive control (25). Ponceau staining of the RuBisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) large subunit shows equal protein loading.
Autoactive Fragments of MLA10, Sr33, and Sr50 Self-Associate in Planta.
To investigate if cell death induction was correlated with in planta self-association, we performed coimmunoprecipitation (co-IP) experiments using fragments of MLA10, Sr33, and Sr50 equivalent to MLA101-141, MLA101-142, and MLA101-144 fused to CFP or HA tags (Fig. 5B). The CFP-fused CC fragments were all expressed (SI Appendix, Fig. S6B) and displayed equivalent cell death activity as the corresponding HA-tagged fragments (SI Appendix, Fig. S6C). Expression of all proteins in the input was verified by immunoblotting using anti-GFP and anti-HA antibodies (Fig. 5B). CFP-fused proteins were enriched after immunoprecipitation with anti-GFP beads, and HA-fused autoactive fragments of MLA10, Sr33, and Sr50 coprecipitated with their respective CFP-fused fragment. This binding was specific, because they did not coprecipitate, or coprecipitated to a much weaker extent, with a divergent CC domain from the rice RGA4 protein (35) used as a negative control (Fig. 5B). In the case of the inactive CC domains (MLA101-141 and equivalents), these proteins showed much lower levels of self-association. Therefore, these data indicate a clear correlation between self-association and in planta signaling activity.
Discussion
Conservation of Plant NLR CC Domain Structure.
Before this study, the structures of two plant CC domain fragments had been reported. The structure of the CC domain from the potato NLR protein Rx, a four-helix bundle, is “strikingly different” (26) from the structure of the CC domain from the barley NLR protein MLA10, a helix–turn–helix dimer (14). Given the low sequence identity between MLA10 and Rx within the structured region (∼18%) and the fact that Rx1-122 was crystallized bound to the RanGAP2 WPP domain, it has been unclear whether these structural differences may represent divergent types of CC domains or different conformational states. Hao et al. (26) noted that the identification of any representative structure for the class would require characterization of further CCEDVID structures. Interestingly, such variation has not been observed between structures of TIR domains, the other class of plant NLR N-terminal domains, which have a conserved fold despite low sequence identities (36).
The NMR structure of the CC region in Sr336-120, solved here, reveals a compact, four-helix bundle similar to the crystal structure of the Rx1-122 (26), rather than the dimeric arrangement in the crystal structure of MLA105-120 (14), despite the much lower sequence identity (18% versus 82%). Furthermore, biophysical characterization by SEC-MALS showed that these regions of Sr33, MLA10, Rx, and Sr50 were predominantly monomeric in solution (Fig. 2). Similarly, Sr336-120, MLA105-120, and Rx1-122 were indistinguishable by SEC-SAXS shape analysis, and these data were consistent with the four-helix bundle structures of the Sr336-120 NMR structure and Rx1-122 crystal structure, but not with the dimeric MLA105-120 crystal structure (Fig. 3). These results indicate that plant NLR CC domains likely have conserved structures, as is the case for TIR domains.
Self-Association and CC Domain Signaling.
Our findings that neither Sr336-120 nor MLA105-120 self-associate in solution are consistent with recent observations that constructs of Sr33, Sr50, and MLA10 comprising residues 1–120 (or equivalent) of the CC domain do not self-associate or induce a cell death phenotype when transiently expressed in tobacco (25). In contrast, longer constructs comprising residues 1–160 are capable of both in planta self-association (based on co-IP) and cell death activity (25). Building upon these observations, we found that the minimal functional unit for cell death signaling of these proteins extends to a position slightly C-terminal to the predicted end of the last α-helix within the CC domain (Fig. 5). Co-IP experiments demonstrate that CC domain fragments that are capable of causing cell death can also self-associate when expressed in planta, whereas inactive fragments displayed strongly impaired self-association. Solution studies using these longer active constructs showed that the inclusion of additional residues at the C terminus (MLA105-144, Sr336-144, and Sr336-160) also facilitates self-association in vitro (Fig. 4), corroborating the link between self-association and signaling. It is, however, important to note that even for the longest constructs, both monomer and dimer forms of these proteins are present. Collectively, these results establish a correlation between self-association and biological activity, and suggest that CC domain self-association is the switch regulating cell death induction.
The CC Dimer.
Given these findings, it is important to address the tendency of MLA105-120 to crystallize in a dimeric conformation, as well as the previously presented evidence (14) for such a dimer in solution. To interrogate the behavior of this protein in solution, it is necessary both to confirm previous experiments and to relate these experiments to the biophysical results. In their study, Maekawa et al. (14) suggested that MLA105-120 existed as a dimer in solution based on the crystal structure, the slow migration of the protein by SEC, and the appearance of cross-linked bands following incubation with a chemical cross-linking agent, and the fact that the recombinant protein was no longer soluble after the putative dimer interface was disrupted by mutations (14).
We also observed that MLA105-120, Rx1-122 [reported monomer (26)], and Sr336-120 have similar SEC elution times and that these elution times are consistent with the migration rate of a protein approximately double their monomeric molecular masses. Although migration by SEC is often used to estimate molecular masses by comparison with known protein standards, migration rates do not depend solely on molecular mass. The rate of migration is also influenced by particle shape, flexibility, composition, and rates of association and disassociation (37). In contrast, scattering techniques, such as MALS and SAXS, directly relate signal to average mass. These approaches provide a more robust and quantitative means of molecular mass measurement than migration by SEC (37, 38). In the case of MLA105-120, these methods show only a small degree of transient self-association at most. Consistent with these data, MLA105-120 remains predominantly monomeric even after extended incubation with a cross-linking agent. In contrast, the dimeric AvrM protein is rapidly and efficiently cross-linked (SI Appendix, Fig. S3). Chemical cross-linking can be promoted by transient or even nonspecific interactions, and we suggest that the observed behavior is more indicative of weak reversible self-association as opposed to obligate dimer formation.
The process of crystallization selects for states that promote a highly ordered arrangement in the crystal, not necessarily the most prevalent conformation in solution. The comparison of the structure of the MLA105-120 dimer observed in the crystals to the four-helix bundle structures suggests that the monomers have undergone a domain swap during crystallization. SI Appendix, Fig. S7 shows that it is possible to superimpose two copies of the Sr336-120 four-helix bundle side-by-side onto the MLA10 dimer. Repositioning α1 and α4 in Sr336-120 to form a continuous helix would regenerate the helix–loop–helix observed in the MLA105-120 dimer, while retaining the internal hydrophobic contacts. Importantly, this analysis also explains the insolubility of the interface-disrupting mutants reported by Maekawa et al. (14), because the residues that form the dimeric interface in the crystal also form the hydrophobic core of the four-helix bundle monomer, and mutation of these residues may thus be expected to disrupt the protein fold (SI Appendix, Fig. S8).
Our results demonstrate that the region between residues 120 and 142 in both Sr33 and MLA10 is necessary for both self-association and signaling activity, and suggest that the dimeric form is responsible for CC domain signaling. However, the structure and nature of this dimer remain unknown. It remains plausible that the MLA105-120 crystal structures capture some part of the activated signaling dimer. It is noteworthy that domain swapping is common among signaling proteins (39); however, it is not trivial to reconcile this conformation with the importance of the additional C-terminal residues. The C termini are at opposing ends of the rod-shaped dimer, and an extension of the C-terminal helices would project in opposite directions. In the event that these regions folded back onto the body of the domain-swapped CC domain, the C-terminal extensions would be unlikely to interact directly. However, it is possible that these regions may further stabilize a domain-swapped CC domain dimer through interactions with other regions of the protein (SI Appendix, Fig. S9). It is equally plausible that the MLA10 conformation observed in the crystal structures is a product of the crystallization process and is not biologically relevant. In this case, the additional C-terminal residues may promote a thus far uncharacterized self-association event between the monomers in the four-helix bundle conformation. Ultimately, a full structural exploration of the longer, active constructs will be necessary to reveal the structural basis of the self-association and its role in CC-domain signaling.
Mechanism of Signaling by NLR CC Domains.
It has been hypothesized that the self-association of the TIR domains postactivation is positively regulated via self-association of other domains from the full-length plant NLR (40). Structural studies demonstrate self-association of NB and LRR domains in animal NLRs (19–21), and this self-association has been shown by co-IP experiments for the Arabidopsis CC-NLR RPS5 (24) and TIR-NLR RPP1 (23). We propose a similar model of signaling for the Sr33, Sr50, and MLA10 CC-NLRs, in which the transient self-association of the CC domain is stabilized by the full-length NLR to achieve the activated state. These associations would presumably facilitate the recruitment of downstream signaling molecules, as is the case in animal NLRs (19–21, 41) and Toll-like receptors (42), and resemble the mechanism proposed for TIR-NLRs (5, 8).
We demonstrate that both closely and distantly related CC-NLR proteins have structurally similar CC domains, reconciling previously conflicting data and models of activation of this important domain. We show that self-association of the CC domains correlates strongly with cell death activity in the MLA10 and Sr33 CC-NLR proteins, and we define residues comprising the minimal functional unit for these proteins (both biophysically and in planta). Our data redefine the structural understanding of the CC domains from CC-NLR proteins. This work will provide a foundation for further structural studies of the more complete, signaling-competent NLR CC domain.
Materials and Methods
Details of the methods used are provided in SI Appendix, SI Methods, including cloning, protein production and crystallization, structure determination (NMR spectroscopy and X-ray crystallography), and SEC-MALS and SEC-SAXS experiments; constructs for in planta analyses; transient protein expression; and co-IP and cell death assays in N. benthamiana. Primers and construct details are listed in SI Appendix, Tables S6 and S7.
Supplementary Material
Acknowledgments
We thank the Australian Synchrotron scientists for their support and Sambasivam Periyannan and Evans Lagudah for access to the Sr33 cDNA. We acknowledge the University of Queensland Remote Operation Crystallization and X-ray Diffraction Facility, the Queensland NMR Network, and the SAXS-wide-angle X-ray scattering and MX Beamlines at the Australian Synchrotron. This research was supported by Australian Research Council (ARC) Grants DP120100685 and DP160102244, National Health and Medical Research Council Research Fellow Grants 1003325 and 1110971 (to B.K.), ARC Future Fellow Grant FTl10100925 (to M.M.), and ARC Discovery Early Career Researcher Award DE160100893 (to S.J.W.). S.C. is the recipient of Commonwealth Scientific and Industrial Research Organization Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The protein structures and data used to derives these structures have been deposited with the Protein Data Bank, www.pdb.org [PDB ID codes 2NCG (Sr336-120) and 5T1Y (Mla105-120)].
See Commentary on page 12619.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609922113/-/DCSupplemental.
References
- 1.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 2.Periyannan S, et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341(6147):786–788. doi: 10.1126/science.1239028. [DOI] [PubMed] [Google Scholar]
- 3.Seeholzer S, et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant Microbe Interact. 2010;23(4):497–509. doi: 10.1094/MPMI-23-4-0497. [DOI] [PubMed] [Google Scholar]
- 4.Mago R, et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants. 2015;1:15186. doi: 10.1038/nplants.2015.186. [DOI] [PubMed] [Google Scholar]
- 5.Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. Animal NLRs provide structural insights into plant NLR function. Ann Bot. August 25, 2016 doi: 10.1093/aob/mcw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9(3):200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams SJ, et al. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact. 2011;24(8):897–906. doi: 10.1094/MPMI-03-11-0052. [DOI] [PubMed] [Google Scholar]
- 8.Bernoux M, et al. Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell. 2016;28(1):146–159. doi: 10.1105/tpc.15.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodds PN, Lawrence GJ, Ellis JG. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell. 2001;13(1):163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravensdale M, et al. Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 2012;8(11):e1003004. doi: 10.1371/journal.ppat.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22(7):2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341(6142):172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 13.Swiderski MR, Birker D, Jones JD. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22(2):157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 14.Maekawa T, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9(3):187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011;24(8):918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 16.Bai S, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8(6):e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams SJ, et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344(6181):299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- 18.Wang G-F, Balint-Kurti PJ. Cytoplasmic and nuclear localizations are important for the hypersensitive response conferred by maize autoactive Rp1-D21 protein. Mol Plant Microbe Interact. 2015;28(9):1023–1031. doi: 10.1094/MPMI-01-15-0014-R. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350(6259):404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350(6259):399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 21.Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure. 2015;23(12):2349–2357. doi: 10.1016/j.str.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18(2):491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ. Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog. 2016;12(7):e1005769. doi: 10.1371/journal.ppat.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104(7):2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cesari S, et al. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC–NLR proteins. Proc Natl Acad Sci USA. 2016;113(36):10204–10209. doi: 10.1073/pnas.1605483113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao W, Collier SM, Moffett P, Chai J. Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2) J Biol Chem. 2013;288(50):35868–35876. doi: 10.1074/jbc.M113.517417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maqbool A, et al. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife. 2015;4:e08709. doi: 10.7554/eLife.08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rairdan GJ, et al. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20(3):739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collier SM, Moffett P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009;14(10):521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Anglister J, Grzesiek S, Ren H, Klee CB, Bax A. Isotope-edited multidimensional NMR of calcineurin B in the presence of the non-deuterated detergent CHAPS. J Biomol NMR. 1993;3(1):121–126. doi: 10.1007/BF00242480. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ve T, et al. Structures of the flax-rust effector AvrM reveal insights into the molecular basis of plant-cell entry and effector-triggered immunity. Proc Natl Acad Sci USA. 2013;110(43):17594–17599. doi: 10.1073/pnas.1307614110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Buchan DWA, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41(Web Server issue):W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Césari S, et al. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33(17):1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ve T, Williams SJ, Kobe B. Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis. 2015;20(2):250–261. doi: 10.1007/s10495-014-1064-2. [DOI] [PubMed] [Google Scholar]
- 37.Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240(2):155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- 38.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11(6):1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernoux M, Ellis JG, Dodds PN. New insights in plant immunity signaling activation. Curr Opin Plant Biol. 2011;14(5):512–518. doi: 10.1016/j.pbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156(6):1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.