Significance
Horizontal gene transfer (HGT) is the nonsexual transfer and genomic integration of genetic materials between organisms. In eukaryotes, HGT appears rare, but parasitic plants may be exceptions, as haustorial feeding connections between parasites and their hosts provide intimate cellular contacts that could facilitate DNA transfer between unrelated species. Through analysis of genome-scale data, we identified >50 expressed and likely functional HGT events in one family of parasitic plants. HGT reflected parasite preferences for different host plants and was much more frequent in plants with increasing parasitic dependency. HGT was strongly biased toward expression and protein types likely to contribute to haustorial function, suggesting that functional HGT of host genes may play an important role in adaptive evolution of parasites.
Keywords: HGT, phylogenomics, validation pipeline, genomic transfer, parasitism
Abstract
Horizontal gene transfer (HGT) is the transfer of genetic material across species boundaries and has been a driving force in prokaryotic evolution. HGT involving eukaryotes appears to be much less frequent, and the functional implications of HGT in eukaryotes are poorly understood. We test the hypothesis that parasitic plants, because of their intimate feeding contacts with host plant tissues, are especially prone to horizontal gene acquisition. We sought evidence of HGTs in transcriptomes of three parasitic members of Orobanchaceae, a plant family containing species spanning the full spectrum of parasitic capabilities, plus the free-living Lindenbergia. Following initial phylogenetic detection and an extensive validation procedure, 52 high-confidence horizontal transfer events were detected, often from lineages of known host plants and with an increasing number of HGT events in species with the greatest parasitic dependence. Analyses of intron sequences in putative donor and recipient lineages provide evidence for integration of genomic fragments far more often than retro-processed RNA sequences. Purifying selection predominates in functionally transferred sequences, with a small fraction of adaptively evolving sites. HGT-acquired genes are preferentially expressed in the haustorium—the organ of parasitic plants—and are strongly biased in predicted gene functions, suggesting that expression products of horizontally acquired genes are contributing to the unique adaptive feeding structure of parasitic plants.
Horizontal gene transfer (HGT) is the movement and genomic integration of genetic material across strong species boundaries. HGT involving prokaryotes (1) has been repeatedly associated with adaptive evolution (2), such as the acquisition of antibiotic resistance (3), resistance to heavy metal (4), and pesticide degradation (5). Although there was a massive endosymbiotic transfer of genes into the nuclear genome from eubacterial ancestors of plastids (6) and mitochondria (7), relatively few cases of functional eukaryote-to-eukaryote HGTs have been detected or studied in detail (8).
As HGT is detected in eukaryotic genomes with greater frequency, it is becoming increasingly possible to address questions of the mechanism, function, and potentially adaptive significance of HGTs. In plants, the horizontal acquisition of genes from microbial sources has been hypothesized to play an important role in early land plant evolution (9). HGT among plant lineages may also occur; a notable example involves the adaptive transfer of a photoreceptor gene from bryophytes that enabled ferns to adapt to low-light conditions (10).
Reported HGT events in plants most commonly involve mitochondrial sequences (11, 12)—for instance, the repeated invasion of mitochondrial coxI by a group I homing intron in diverse angiosperm lineages (13, 14) and widespread incorporation of fragments or entire mitochondrial genomes from algae or moss sources by the giant Amborella mitochondrial genome (15, 16). Active recombination processes and an absence of genomic downsizing pressures to remove excess sequences are probably important factors in mitochondrial HGT in plants (11, 16, 17), but additional steps are required to increase the likelihood of integrated sequences being propagated through sexual reproduction (16).
A second apparent concentration of horizontally acquired sequences in plant genomes is associated with parasitic plants, where both mitochondrial (14, 17–23) and nuclear horizontal transfers (24–29) have been identified. Repeated horizontal acquisitions of mitochondrial atpI by parasitic flowering plants occurred in four extreme parasite lineages where the parasite lives inside the host for much of its lifespan (14). The intimate contact between parasitic plants and their host plants, facilitated by the haustorium, a conspicuous adaptation of parasitic plants (30), may increase the likelihood of genetic exchange, especially in parasites with direct phloem–hloem connections (14, 18). The observation of massive mRNA movement between parasitic Cuscuta and its host (31) suggests a likely route of HGT via mRNA intermediates. We thus hypothesize that parasitic plants act as nexus points for HGT and that HGT will be more frequent in more nutritionally dependent parasite species.
Among all of the parasitic lineages (at least 11 independent origins) of flowering plants (14), Orobanchaceae is the only family that has a complete spectrum of parasitic capabilities (30) and thus is ideal for testing these predictions. Four cases of nuclear gene HGT have been identified in this family to date (26–29), but this is likely to be a small fraction of the number of events. This is due to the challenges in HGT discovery and to the lack of a well-established approach for use with complex plant genomes. The previous three cases were discovered by using a BLAST-based detection approach (26–28), which impeded discovery on a large scale because BLAST-based predictions are subject to high false positive errors from incomplete sampling in the database as well as extensive follow-up work to identify erroneous results. Inspired by Xi et al. (25), who used a phylogenomic approach for HGT inferences in parasitic Rafflesia, we developed a robust phylogenomic pipeline for HGT detection followed by a comprehensive validation procedure to reveal the extent of HGT in three parasitic Orobanchaceae with different degrees of parasitic dependence. However, HGT inferences with gene trees can also suffer from high error rates due to factors including misidentification of contaminant sequences, insufficient taxon sampling, complex gene birth and death processes, gene tree errors, inappropriate rooting, and frame-shift errors. We thus took these factors into account in our pipeline and carefully evaluated each potential HGT candidate with additional data to identify and reduce potential sources of error. We focus on potentially functional genes acquired by parasitic plants by concentrating our search on extensive transcriptome evidence for members of Orobanchaceae grown on host plants with known genome sequences (30, 32, 33); in addition to greatly improving the accuracy of HGT identification, it offers the opportunity to identify the expression patterns associated with genes derived from HGT events (28).
Results and Discussion
Analytical Schema for Detection of HGT.
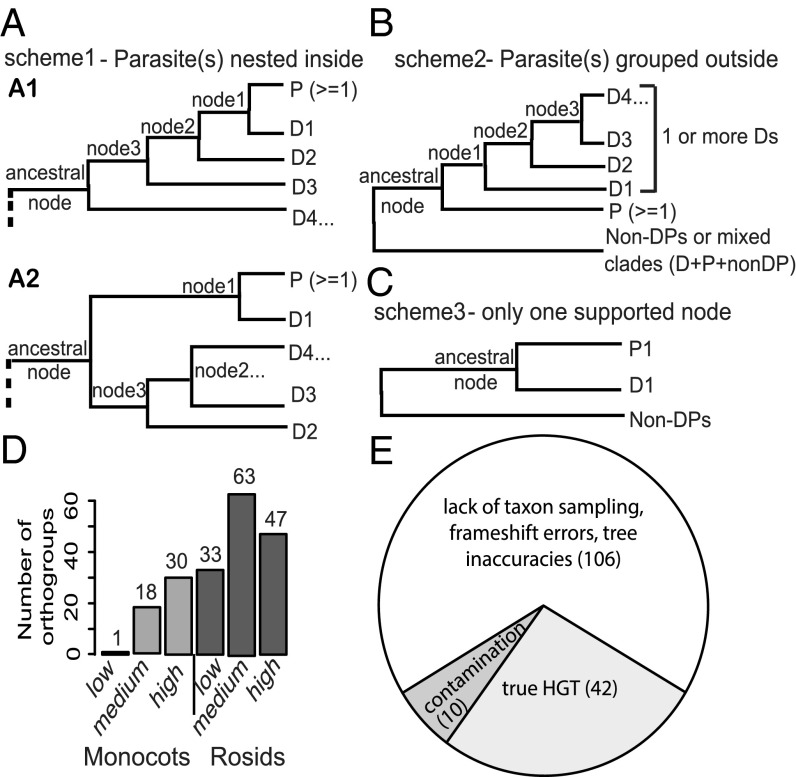
An initial phylogenomic screen was used to identify putative HGTs from transcriptome assemblies of parasitic plants. This approach used an automated pipeline to build phylogenetic trees for each approximate gene family (orthogroup), containing genes from 22 representative sequenced plant genomes and six transcriptomes (32). Four species of Orobanchaceae were tested, including the nonparasitic Lindenbergia, and three parasitic plants with increasing degrees of parasitic dependence—Triphysaria versicolor (facultative hemiparasite—partly heterotrophic when attached to host), Striga hermonthica (obligate hemiparasite—photosynthetic but most carbon derived from host), and Phelipanche aegyptiaca (Syn. Orobanche aegyptiaca) (obligate holoparasite—nonphotosynthetic and fully heterotrophic) (30). Three models illustrate topologies indicative of HGTs we sought to detect (Fig. 1). With a goal of identifying unambiguous HGTs, we focused our search on rosid and monocot donors because of the relatively large number of finished genomes in these groups and the relatively large genetic distance from Orobanchaceae (as all the parasites are asterids) (SI Appendix, Fig. S1). In all three models, we identified “ancestral” nodes, defined here as the node containing exclusively parasite genes and genes in the donor clade. To allow for incomplete sampling of all donor lineages and complexities of gene family evolution, we used a loose initial bootstrap support (BS) cutoff of 50 in the initial phylogenomic screening. The first model describes a scenario where genes from the parasitic plant or nonparasitic relative are supported (BS ≥ 50) as nested within a donor clade (Fig. 1A). The second model (Fig. 1B) describes a case where the parasite’s genes are placed outside of the donor clade. In both cases, two nodes supporting (BS ≥ 50) the grouping of parasitic genes with donor clades were required. The third model (Fig. 1C) requires only one node supporting the placement of the parasite’s gene(s) as sisters of donor clades.
Fig. 1.
Three models for phylogenomic identification of HGTs and further examination of the preliminary-screened HGT candidates. (Scheme 1) Parasitic genes (P) are nested inside donor clades (D). (Scheme 2) Parasitic gene group outside of the donor clade. (Scheme 3) Only one node of donor sequence is sister to parasite genes. In this study, donor refers to distantly related monocot and rosid sequences. Ancestral node is defined to be composed of exclusively parasitic and donor sequences. In A1, at least two nodes within the ancestral node (including the ancestral) are required to have BS ≥ 50; in A2, both the ancestral node and node 1 are required to have BS ≥ 50. In scheme 2 (B), the ancestral node and at least one node within the ancestral node are required to have BS ≥ 50. In scheme 3 (C), only the node that supports the grouping of the parasitic gene and donor sequence is required to have BS ≥ 50. “Non-DPs” refers to nonparasitic, nondonor sequences. (D) A total of 192 HGT orthogroup trees from the initial screening were classified into low-, medium-, and high-confidence categories based on a scoring scheme (SI Appendix, Table S1). Gray colors represent the HGT orthogroups identified in the monocots; darker gray colors represent the rosids. (E) The number of HGT candidate orthogroups manually curated as true HGTs (light gray); artifacts resulting from insufficient taxon sampling, frame shift errors, or tree inaccuracies (white); or fungal or host contamination (dark gray). The 42 “true” HGT orthogroups all fit scheme 1 of A.
High-Confidence HGT Events.
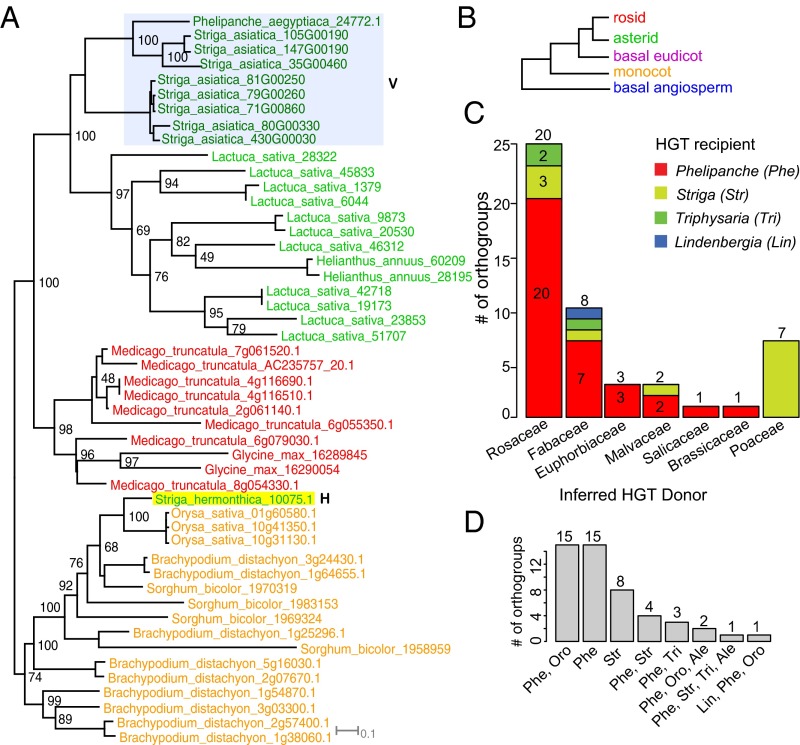
Custom scripts searching for topologies consistent with these three models resulted in the identification of a set of 192 gene trees with preliminary evidence for HGT (143 orthogroups with potential HGTs from rosids and 49 with potential HGTs from monocots) in the three focal species. Only one orthogroup (3861) was identified as including a potential HGT in the nonparasitic “control” species Lindenbergia; all others involved the parasitic species only. We then applied a scoring scheme to assign these 192 candidate HGT trees to low-, medium-, and high-confidence groups based on tree characteristics including the BS for key nodes surrounding the inferred HGT event, sampling of the donor clades grouped with the HGT genes, and branch length heterogeneity (Fig. 1D; see SI Appendix, Table S1 for detailed criteria). The authenticity of the 158 HGT trees in the medium- and high-confidence groups was then validated with follow-up analyses, including manual examination of branch lengths and sequence alignments, correcting any translation or alignment errors, and examining the phylogenetic stability with increased taxon sampling. We examined low-coverage transcriptome sequences from eight more parasitic Orobanchaceae species and also from 10 related nonparasitic Lamiales taxa from the 1kp transcriptome project (34), four sequenced asterid genomes (Phytozome) (35), and the Striga asiatica genome (under github “dePamphilis/HGT_PNAS_2016”) (see Methods, HGT Validation by Increased Taxon Sampling). This resulted in a final set of 42 HGT orthogroups (SI Appendix, Fig. S2), with the remaining 150 putative HGT orthogroups determined to be artifactual or merely low confidence (Fig. 1E and SI Appendix, Fig. S4). The primary source of artifacts (106 out of 150) was insufficient taxon sampling (SI Appendix, Fig. S4 A and B), especially of the order (Lamiales) that contains Orobanchaceae (36). Other artifacts came from frame-shift errors (SI Appendix, Fig. S4 D and E) and contamination, either from the experimental host (nine orthogroups) (SI Appendix, Fig. S4C) or from fungal contamination (one orthogroup). The topology and BS values for the 42 HGT orthogroup trees strongly support (gene trees all fit model 1) the placement of parasitic genes within the donor clade, indicating a clear HGT origin (Figs. 2A and 3A and SI Appendix, Fig. S2). For instance, in Fig. 2A, StHeBC3_10075.1 from S. hermonthica is placed within a grass clade with the proximal node supported with BS 100, and two additional deeper nodes with BS 100, supporting a strong case of HGT. Eleven out of 42 trees suggested a polyphyletic origin of HGT genes (multiple distinct transfers within a gene family). Thus, we used the Shimodaira–Hasegawa (SH) test (37) to evaluate whether more than one transfer was most likely based on the available data. A single transfer could not be rejected for orthogroup 3861 (SI Appendix, Fig. S2.11). Interestingly, the other 10 trees each supported more than one transfer (SI Appendix, Table S2), suggesting a propensity for certain gene families to include successful horizontal transfers. A minimum of 52 horizontal transfer events were thus inferred from these 42 gene families. That a majority, fully 78%, of the candidates (67% of initial medium and high confidence) were (i) excluded due to low support, (ii) identified as artifacts (via increased taxon sampling), or (iii) identified as contamination from host or other organisms illustrates the challenge of accurate HGT discovery.
Fig. 2.
RAxML-based maximum likelihood (ML) trees supporting HGT, donor families, and recipient taxa inferred from the 42 HGT set. Orthogroup tree (12835) supports a grass-derived Pong-like TE in S. hermonthica. HGT sequence is labeled with “H” and vertically inherited sequences with “V.” The species abbreviations are shown in SI Appendix, Fig. S1. (B) A hypothetical tree illustrates the color-coding system for each angiosperm lineage represented in A and Fig. 3. (C) Mapping of parasitic recipient taxa onto inferred donor family (x axis). Each genus in HGT recipient is followed with a three-letter code used in D. Total number of HGT orthogroups inferred from each donor family is placed on top of each bar. Numbers within each bar represent number of orthogroups; the number of singletons is not shown due to space limitations (SI Appendix, Table S9). (D) Number of HGT orthogroups supports transfers from shared and unique parasitic genera. Ale, Alectra; Lin, Lindenbergia; Oro, Orobanche; Phe, Phelipanche; Str, Striga; Tri, Triphysaria.
Fig. 3.
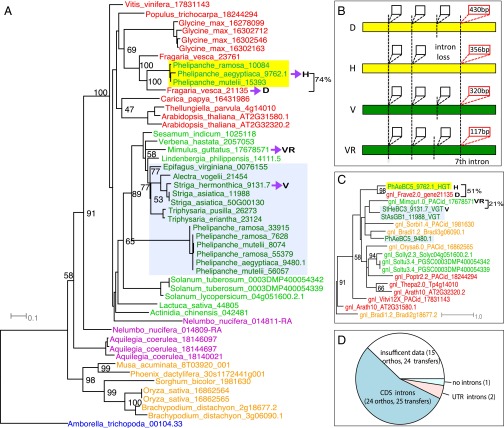
Genomic horizontal transfer of a tRNAHis guanylyltransferase from Frave (Fragaria) (or its ancestor) to Phelipanche. (A) A coding-sequence (CDS) tree by RAxML from represented species across angiosperm lineages. D, inferred donor (in Fragaria); H, the parasitic HGT gene; V, vertical parasitic gene; VR, related sequence of the vertical parasitic gene (in Mimulus). Seventy-four percent represents the CDS similarity between the HGT gene and its inferred donor. (B) Gene structure with four selected introns for the four sequences (H, D, V, and VR). Yellow and green bars represent coding sequence; the vertical dashed lines represent the intron positions; the boxes represent introns. At least four conserved intron positions were shown on the gene structure; the third intron was lost in the HGT gene, and the fourth intron on the graph (which is the seventh intron of the Mimulus gene) showed strong sequence similarity between the HGT gene and its donor (marked by red intron boxes with length within). (C) The phylogeny of the seventh intron (marked red in B) from genes on the CDS tree: the HGT gene groups with its donor supported by 98% BS, whereas the vertically inherited gene groups with a close relative (Mimulus sequence). The intron sequence similarity between the HGT (HGT) gene and its donor (D) is 51%, and the intron sequence similarity between the vertical gene (V) and its vertical relative (VR) is 21%. (D) The number of HGT orthogroups that support 25 genomic transfers in 24 HGT orthogroups containing introns in the CDS region (dark blue) and 2 HGT orthogroups containing introns in the UTR region (pink). The remaining orthogroups contain 15 HGT orthogroups (24 transfers) with insufficient genomic data to infer presence of introns (white) and one orthogroup containing HGT gene without introns (light blue).
Our analyses with explicit phylogenetic schema and stringent evaluation by the use of increased taxon sampling represent a robust approach for HGT identification. In addition, it includes two of the published high-confidence HGT cases (26, 28). Two other published HGTs in Orobanchaceae (27, 29) were detected in the stage 4-specific assembly (28) but not in the (generally more complete) combined assembly we examined in this study. In addition, these 42 orthogroups include six orthogroups encoding transposable elements (TEs), which is consistent with their invasive nature. Four TE-related sequences encode ORFs with abundant transcripts in haustorial tissues (orthogroup 1021, 5002, 14230, and 15149), suggesting a potentially active role in the parasites, a scenario similar to the recently identified Brassicaceae-derived hobo-Ac-Tam3 transposon (hAT) in P. aegyptiaca (29). We also provided multiple lines of evidence for cross-validation of HGT sequences. This includes sequences that were confirmed by RT-PCR (22 events) or PCR amplification with genomic DNA (three events), genomic sequence data (27 events), or present in more than one parasitic species (11 events) (SI Appendix, Table S3). All these HGT genes were verified with at least one additional line of evidence, providing additional validation for HGT not due to assembly errors. In addition, the genomic contigs of HGT sequences had read depths equivalent to those of genomic contigs of the vertically transmitted sequences (SI Appendix, Fig. S3), rejecting the possibility that they may represent sample contamination. Finally, the use of increased taxon sampling sometimes helped to identify a more likely donor lineage, as seen in orthogroup 17, where an initial donor lineage (Populus, a rosid) gave way to a more likely donor lineage (Beta, a caryophyllid) after additional taxon sampling (SI Appendix, Fig. S2.1).
Mechanism of HGT
Transfers from Ancestral Host Lineages.
A majority of these HGTs could be assigned to ancestral donors from known host lineages. All of the HGTs from grass donors (Poaceae) were discovered in Striga (Table 1; SI Appendix, Table S9, and Fig. 2C), which (except for Striga gesnerioides) are specialized parasites of Poaceae (38). In Phelipanche, inferred donors reflected a wide range of dicot families with the majority from Rosaceae and Fabaceae, consistent with the host range of this parasite and its congeners (30) (Fig. 2C and SI Appendix, Table S9). In 38 orthogroups, the transfer was inferred to be unique to one genus (15 are unique in Phelipanche, eight are unique in Striga) or in two closely related genera (15 occurred both in Phelipanche and Orobanche) (Fig. 2D and SI Appendix, Table S9).
Table 1.
Information of the 42 HGT orthogroups including the HGT recipient, donor, expression, Dn/Ds, functional category, and homology-based annotation
| Ortho group | Recipient | Donor | Intron | Expression | Dn/Ds | Functional category | Annotation based on homology |
| 226 | P | Poptr | Y/Y | 1, 4.2 | P | Defense | Cytochrome P450 |
| 1685 | P | Gyma + Medtr | Y/Y | >2 | P | Defense | Cysteine-rich receptor-like kinase |
| 2376 | P | Poptr + Theca | Y/Y | >2 | RP | Defense | Proteasome subunit alpha type |
| 14624 | S | Sorbi + Zea | N/N-5′UI | NA | P | Defense (disease resistance) | BTB/POZ |
| 23343 | P | Theca | — | 5.2 | P | Defense (disease resistance) | Disease resistance protein |
| 11841 | P | Frave | Y/Y | 5.1 | SP | Defense | Hyoscyamine 6-dioxygenase-like |
| 1886 | P | Frave | — | 6.2 | RP | Defense (immunity) | Ankyrin repeat family protein |
| 11437 | P | Frave | — | >2 | RP | Defense and nodule development | Kelch modif related to galactose oxidase |
| 8888 | P | Frave | — | 2, 4.1 | RP | Transcription | Poly(A) polymerase |
| 18709 | P | Arath | Y/Y | Int | POS | Transcription | Nucleolin 2-like |
| 806 | P | Theca | Y/Y | Int | RP | Translation | Valyl-tRNA synthetase |
| 2270 | P, S | Theca | Y/Y | >2 | RP | Translation | Methionyl-tRNA synthetase |
| 4067 | P | Frave | Y/Y | 41 | RP | Translation | tRNAHis guanylyltransferase |
| 10050 | P | Frave | — | 42 | RP | Translation | Histidine-tRNA ligase |
| 13892 | P | Medtr | Y/Y | Int | P | Translation | Ribosomal protein S13 |
| 17 | P | Betvu | Y/Y | 4.2, 5.1, 6.2 | P | Nutrient transport | ABC transporter C family member 3 |
| 9613 | P | Glyma | Y/Y | 0 | RP | Nodule development and cytokinin biosynthesis | Cytosolic purine 5-nucleotidase |
| 15246 | P | Medtr | — | 6.2 | P | Defense-related (insect toxin) | Albumin I (28) |
| 1226 | P | Poptr | Y/Y | 3, int | RP | Diverse | Alpha/beta-Hydrolases |
| 10143 | P | Frave | — | 4.2 | RP | Diverse | Tubulin-specific chaperone D |
| 3861 | P + L | Glyma | Y/Y | >2 | POS | Diverse | Poly(ADP ribose) glycohydrolase |
| 4598 | P | Medtr | — | Int | RP | Diverse | Nuclear pore complex protein |
| 19696 | P | Poptr | Y/Y | 4.1, 6.2 | SP | Diverse | Ubiquitin-like–specific protease 1 |
| 16703 | S | Orysa | Y/Y | 52 | P | Diverse | Zinc finger, GRF-type |
| 4572 | P | Frave | — | 3, 4.1 | P | Diverse | FBD-associated F-box protein |
| 5896 | S | Glyma | Y/Y | 5.2, 6.1 | P | Plastid-to-nucleus signaling | Uroporphyrinogen-III synthase |
| 218 | P, S, T | Prunus | Y/Y (2) | >2 | P | TE | hAT transposon |
| 1021 | P, T | Frave + Malus | Y/Y | >2 | P | TE | hAT transposon |
| 5002 | P | Prunus | Y/Y | Int | P | TE | hAT transposon |
| 12835 | S | Sorbi | N/N | 0 | P | TE | Putative harbinger Transposase-derived nuclease |
| 14230 | P | Prunus | Y/Y | 42 | P | TE | MULE transposase |
| 15149 | P | Frave | Y/Y | Int | P | TE | hAT transposon |
| 13512 | P | Frave | — | 51 | POS | Unknown | Unknown |
| 14233 | S | Sorbi + Orysa | Y/Y | 6.1 | SP | Unknown | Unknown |
| 14675 | P | Frave | — | 6.2 | P | Unknown | Unknown |
| 18354 | P | Frave | — | Int | P | Unknown | Unknown |
| 20190 | P | Frave | — | int, 2 | P | Unknown | Unknown |
| 23480 | P | Frave | — | Int | P | Unknown | Unknown |
| 18774 | S | Orysa | Y/Y | 0 | P | Unknown | Unknown |
| 13656 | S | Sorbi + Orysa | — | Int | SP | Unknown | Hypothetical protein |
| 19297 | S | Bradi | N/N-3′UI | NA | P | Unknown | Unknown (26) |
| 20188 | P | Frave | Y/Y | Int | P | Unknown | Unknown |
Recipient column: L, Lindenbergia; P, Phelipanche; S, Striga; T, Triphysaria. Intron column: “—”, not determined; 3′UI and 5′UI mean 3′/5′-UTR introns; N/N, absence of introns in both donor and recipient; Y/Y, presence of intron in both donor and recipient gene (orthogroup 218 has two genomic transfers with introns). Expression: >2, means highly expressed in more than two stages; int, interface. Dn/Ds: P, purifying selection; POS, positive selection; RP, relaxed purifying selection; SP, stronger purifying selection. Functional category: TE, transposable element. Donor is represented by the five-letter code of species abbreviations in SI Appendix, Fig. S1. Note that the donor is the indicated taxon or an ancestor. Expression column represents the stages with primary expression for the HGT genes. Haustorial stages are 3, 4 (4.1 and 4.2), and int. For detailed information about these stages, please refer to ref. 32.
Any HGT events coincident with the origin of parasitism would have occurred in a common ancestor of the parasites. Previously reported cases of HGT to microbial parasites or pathogens of plants often encode cell wall-degrading enzymes (39) and thus are implicated in host invasion. Surprisingly, although cell wall-modifying enzymes are well-represented in haustorial tissues (24), no such proteins were identified in our HGT search. Instead, numerous proteins involved in cell wall modification processes in the haustorium were attributed to gene duplications that occurred in an ancestor of all parasitic lineages of Orobanchaceae (32). Our HGT phylogenies, however, supported predominantly recent occurrences that were restricted to individual genera. In only one case (i.e., orthogroup 218, SI Appendix, Fig. S2.2), the transfer was detected in almost all of the parasitic taxa (Phelipanche, Striga, Triphysaria, and Alectra) (Fig. 2D and SI Appendix, Table S9), but the SH test indicated that this likely involved at least two (more recent) transfers instead of a single ancestral HGT event (SI Appendix, Table S2). Therefore, although gene duplications often preceded and underpin the origin of parasitism in Orobanchaceae (32), HGT events are more recent and are likely to have been facilitated by parasite connections.
Increased Numbers of HGT with Increased Heterotrophic Dependence.
The absence of HGT events involving the nonparasitic common ancestor of the Orobanchaceae supports the hypothesis that parasitic (heterotrophic) interactions lead to more HGT events than occurs between free-living organisms. The number of HGT events also appears to increase in parasites with greater host dependence. We detected only one likely HGT in Lindenbergia, the free-living sister lineage to all parasitic Orobanchaceae. In T. versicolor, the facultative hemiparasite, two HGT events were found (Table 1 and SI Appendix, Fig. S2.2 and S2.5). In Striga, the obligate hemiparasite, 10 orthogroup trees support HGTs and seven were from grasses (Poaceae). A majority (34 orthogroups) of the HGTs were detected in Phelipanche, the obligate holoparasite with the strongest host dependence (Figs. 2D and 3A and SI Appendix, Fig. S2).
Several factors could account for the increasing number of HGTs in parasites with increased host dependence. First, the lifestyles of facultative and obligate parasites are quite different with respect to the timing of host invasion and parasite gamete formation. The seedlings of the obligate parasites, which require host plant-induced germination stimulation, are in contact with host plants from a very young developmental stage, thus increasing the chances that cells that experience HGT events will develop into germ-line tissues (40). In contrast, facultative parasites like Triphysaria develop roots and aboveground parts before parasitism occurs. In these plants, host-derived gene fragments that cross the haustorium need to be subsequently transported into developing flowers to be captured in germ-line cells. There is also clear evidence for phloem connections between host and P. aegyptiaca (41), allowing for more HGTs along with the genetic exchange of nucleic acids via phloem (31). Similar phloem connections have not been observed in Triphysaria (42).
Integration of Genomic Fragments.
Signatures of the donor molecule should persist in the genome, giving clues to the mechanism of transfer. For instance, a nuclear HGT reported in Striga supports a possible mRNA-mediated transfer, as the HGT lacked introns and seemed to contain a remnant poly-A tail, whereas the donor Sorghum gene lacked a poly-A tail (26). Documented translocation of host RNA into Triphysaria (43) and Phelipanche (44) as well as the massive movement of host RNA into Cuscuta would support an RNA-based mechanism for HGT in parasitic plants (31). In contrast, a horizontally acquired albumin 1 gene in Phelipanche and related taxa (28) and horizontally acquired Brassicaceae-specific strictosidine synthase-like (SSL) genes contained introns in genomic sequences of both donor and the parasites (Phelipanche and Cuscuta), all consistent with direct genomic transfers without an RNA intermediate (27). To test the hypothesis of mRNA-mediated transfer, we examined coding sequence structure (exon–intron boundaries) in the 42 HGT orthogroups. We had sufficient genomic data to examine 28 genes from 52 horizontal transfer events (27 orthogroups) (Table 1), although three HGT genes lacked coding sequence (CDS) introns in both donor and recipient (Fig. 3D). Although these genes lacked CDS introns, two of these had introns in their UTRs (Fig. 3D). A gene in Orthogroup 14624 (BTB/POZ domain containing protein) was transferred from an ancestor of Sorghum bicolor into S. hermonthica, and the 5′-UTR intron shows 87% sequence identity between the donor and recipient gene (CDS, 91%; 5′-UTR, 87%; 3′-UTR, 68%) (SI Appendix, Fig. S5). In the other case, a gene in orthogroup 19297, a conserved 3′-UTR intron (3′-UTR intron, 78%; CDS, 85%; 5′-UTR, 54%; 3′-UTR, 82%), is present in both the donor and recipient (SI Appendix, Fig. S6). It is noteworthy that this HGT event was previously identified by Yoshida et al. (26), who speculated, based in part on the presence of a remnant poly-A tail in the cDNA, that this HGT event may have been mediated by integration of a mature mRNA rather than a genomic fragment. Our analyses identified the presence of a 284-bp high-identity intron in the 3′-UTR, suggesting that this event (like the majority of cases reported here) was mediated by a genomic fragment rather than an mRNA. Only one orthogroup (12835—a Pong-like TE) lacked introns in both the donor and recipient gene, and the nonconserved flanking region failed to inform whether genomic or mRNA-mediated transfer was supported (Table 1). The remaining 24 HGT orthogroups contained 25 genes whose donor and recipient contained CDS introns. We further reduced the list to 13 orthogroups with full-length gene assemblies, allowing us to examine similarities and differences in intron positions and sequences between donor and recipient.
All 13 orthogroups showed congruence of CDS structure between donor and recipient, suggesting a transfer of a genomic fragment containing the gene, rather than an mRNA intermediate. Intron positions are highly conserved (SI Appendix, Tables S4 and S5 and Fig. S7) (although with occasional intron loss; Fig. 3B), suggesting maintenance of intron structure for functional transcription. These sequences provide support for genomic fragments as HGT intermediates but do not help to diagnose the source of the horizontally acquired sequence. We constructed phylogenies using the intron sequences only and compared them to phylogenies constructed with exons only, finding that three of the orthogroup phylogenies were well-resolved (orthogroup 4067, 806, and 2270) (Table 1). The intron phylogenies were congruent with the CDS phylogenies, indicating the same donor lineage as inferred from exon sequence and providing strong support of a genomic fragment-mediated HGT (Fig. 3 A–C and SI Appendix, Fig. S7). The strongest example, orthogroup 4067 [tRNAHis guanylyltransferase—required for translation (45)], not only exhibits strong CDS similarity with its inferred Fragaria donor (∼74%) (Fig. 3A), but the intron sequences maintain ∼51% similarity (Fig. 3 B and C), even higher than that between the vertically inherited parasite gene and its close relative in Mimulus (∼21%) (Fig. 3 B and C). These results show that all of the resolvable HGT events were likely mediated by genomic fragments containing the donor genes rather than by RT-mediated transfer.
Functional HGT
Tissue-Specific HGT Expression.
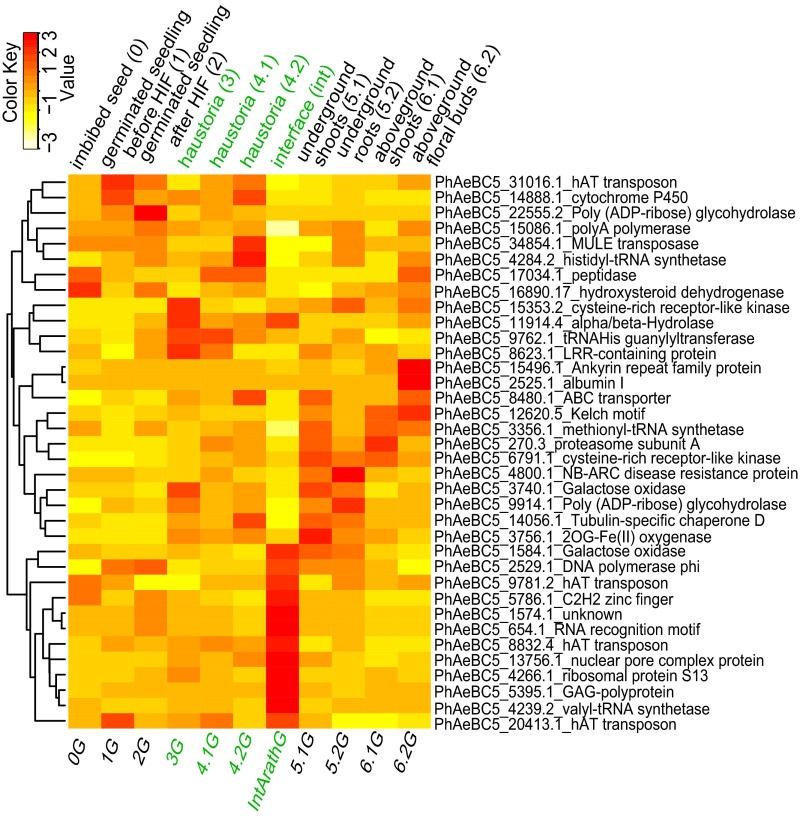
A total of 37/49 HGT genes show expression with maximum Fragments Per Kilobase of Exon Per Million Fragments Mapped (FPKM) greater than 5 in at least one developmental stage, indicating that most are actively transcribed. In addition, 36/42 HGT orthogroups contain HGT genes from more than one parasitic taxon (SI Appendix, Fig. S2), suggesting evolutionary conservation in the parasites. The species with the most HGTs is P. aegyptiaca, and a majority of the candidate genes show tissue-specific expression (Fig. 4; for Striga and Triphysaria, see SI Appendix, Fig. S9). The expression profiles of P. aegyptiaca HGT genes (Table 1) revealed a distinctive cluster of interface-specific expression (Fig. 4) and an equal number with abundant expression in haustorial tissues. A subset of these genes encodes functions related to transcription and protein synthesis (Table 1), and in each of these gene trees, HGT has added an extra gene along with the vertically inherited gene family member. This additional gene may increase the rate of transcription and protein synthesis specifically at the haustorium interface, where such processes go on at elevated levels (46, 47).
Fig. 4.
Heat map showing the expression of HGT transgenes in P. aegyptiaca. Expression is shown with FPKM-transformed z scores to ensure even signal intensity across stages. Rows represent HGT genes, with their identified domain shown on the right; columns represent stages (below) or tissues (above). Haustorial and interface tissues are colored in green. Genes were clustered on the left to show similarity.
HGTs Are Evolving Under Constraint and Are Likely Functional.
For each of the HGT orthogroup phylogenies, we estimated Ds (the frequency of synonymous substitutions per synonymous site) and Dn (the frequency of nonsynonymous substitutions per nonsynonymous site) and ω (the ratio Dn/Ds; values less than 1 indicate purifying selection) for each of the HGT protein-coding sequences and related genes. A branch test, implemented in PAML (Phylogenetic Analysis by Maxiumum Likelihood) (48), compares the Dn/Ds estimate for the foreground (HGT genes) to the background (non-HGT orthogroup members). The same or even stronger levels of purifying selection in parasitic HGT genes were observed in 27 orthogroups (Table 1 and SI Appendix, Table S6). An additional test—RELAX (49)—was implemented to identify whether HGT sequences experienced stronger levels of selection (purifying or adaptive) or relaxed selective constraint (compared with the background). The same levels of selection were observed between HGT sequences and the background genes in 23 orthogroups, whereas stronger levels of selection were observed for HGT sequences of 12 orthogroups (SI Appendix, Table S8). These results show that HGT-encoded proteins are largely evolving under strong constraint, indicating a likely functional role in parasitic plants. Additional evidence comes from conservation of a predicted 3D structure for HGT proteins in comparison with their nonparasitic orthologs in Arabidopsis thaliana (SI Appendix, Fig. S8).
In summary, three primary lines of evidence support a functional role for these horizontally acquired sequences in parasitic Orobanchaceae: (i) HGT sequences are detected and commonly conserved across species boundaries; (ii) the sequences are actively and differentially transcribed, frequently with a bias toward haustorial expression; and (iii) all of the high-confidence HGT genes are evolving under purifying selection, consistent with the conservation of functional protein structures. Notably, a group of HGTs related to transcription and translation are highly expressed in haustoria of P. aegyptiaca (Table 1), the species with the greatest host dependence. As haustoria have a high metabolic rate associated with loading host nutrients (46, 47), it is possible that horizontal transfers of such gene functions help Phelipanche efficiently mobilize host resources transported through haustorial tissues. Noteworthy, although not serving as direct evidence, no parasite-to-host transfers were identified using the same approach. If the intimate contact between the parasite and the host—the haustorium—provides a mechanism for the exchange of genetic elements facilitating HGT, we expect that horizontal transfers also occurred from parasite to host. A lack of nuclear transfers from parasite to host suggests that such transfers are generally not functional.
Evidence of Adaptive Evolution of HGTs.
Our observation of haustorial expression in a majority of the HGT genes suggests a likely contribution of HGT to parasitic adaptation. To corroborate this idea, we examined the possibility of adaptive signatures on protein sequences of these HGTs (see Methods, Selective Constraint Analyses). Out of 15 HGT orthogroups (SI Appendix, Table S6), 13 contain potentially adaptive sites present in HGT genes of parasites. Interestingly, 9 out of 13 orthogroups were identified by RELAX (49) that show a different level of selective constraint in HGT sequences compared with the background. Five orthogroups show stronger levels of selection, and four show relaxed constraint; both patterns could be associated with the presence of adaptive sites (SI Appendix, Tables S6 and S8). These sites are unchanged in nonparasitic species, including Mimulus, the close nonparasitic relative of Orobanchaceae (SI Appendix, Table S7). Of these, six orthogroups have genes encoding functions related to transcription and translation (orthogroup 8888, 18709, 806, 4067, 10050, and 13512), and four orthogroups contain genes with abundant haustorial expression (orthogroup 1226, 8888, 18709, and 806) (Table 1). The signatures of adaptive sites and their retention as haustorial genes in the genome suggest that these changes in HGT proteins are under positive selection and may have provided novel functions contributing to increased parasite fitness.
HGT of Defense-Related Genes.
Our list of HGT events contains a group of genes in orthogroups related to defense responses—orthogroup 226 (50), 1685 (51), 2376 (52), 14624 (53), 23343 (54), 11841 (55), 1886 (56), and 11437 (57) (Table 1). For instance, orthogroup 23343 contains an ortholog of an Arabidopsis gene encoding an NB-ARC domain-containing disease resistance protein (SI Appendix, Table S10) (58, 59). Orthogroup 1685 encodes a cysteine-rich receptor-like protein kinase, and the ortholog in Arabidopsis (AT1G70520) is up-regulated during pathogen infection and rapid cell death (51). Orthogroups 2376 and 14624 contain genes in Arabidopsis and rice that are up-regulated in response to pathogens and parasite attack, respectively [based on analyses with PLEXdb (60)].
Genes involved in defense that have been obtained by horizontal transfer could have been co-opted by the parasites for defense against pathogens that attack it as well as its host. However, six of the HGT genes in defense-related orthogroups show elevated expression in haustoria, suggesting that these genes may play a role in the parasite–host interactions. HGTs related to defense responses may provide a mechanism to attenuate the attack of the host plant immune system against the parasite during haustorial formation. It is also possible that parasite invasion sites are more susceptible to microbial pathogens, in which case enhanced defense responses may reduce the risk of infection. This model is also a potential explanation for the haustorial up-regulation of the putative defense-related HGTs. The specific role of defense-related genes in this potentially multitrophic interaction remains to be discovered.
Conclusion
In this study, we developed a phylogenomic pipeline that parses large-scale phylogenetic trees for preliminary HGT identification, followed by careful validation with further analyses and increased taxon sampling. Our final 42 HGT trees (52 high-confidence HGT events in three parasites of Orobanchaceae) support the placement of focal HGT gene(s) being nested within donor clades with at least two strong nodes, instead of just appearing as a sister to a putative donor lineage. This approach proves to be stringent but also robust to the challenges of genome-scale HGT discovery. Our analyses of intron sequences and structure support genomic fragment integration of HGTs rather than RNA-mediated retroprocessing events. Although unexpected, considering the well-documented mRNA transfer that occurs between parasitic plants and their hosts (31), we hypothesize that compared with mRNA, transfers of genomic fragments will more often result in functional transfers because genomic regions can contain intact promoters that may be recognized by the recipient plant species. Cross-species promoter recognition is common in experimental transformation studies (61, 62), even among very distantly related plant species (63). These hypotheses could be tested experimentally by comparing the capacity of Orobanchaceae parasites to recognize and transcribe sequences with foreign promoters (from other eudicots and from monocots) versus the likelihood of substantial transcription of a randomly inserted cDNA.
Functional roles conferred by these HGT genes have identified HGT as a mechanism contributing to the adaptive evolution of parasitic plants. Our methods likely have underestimated the number of horizontally transferred genes because (i) the phylogenomic approach in this study relies on an fairly complete and accurate construction of gene family phylogenies, (ii) large and complex gene families do not always produce well-resolved trees, and (iii) we restricted our search of possible donor lineages to distantly related monocot and rosid groups for enhanced signal-to-noise ratio. With the increasing availability of genome sequences and other genomic-scale data, along with increasingly rigorous standards for discovery and evaluation, many more examples of functional HGT are likely to be revealed.
Similar to the “you are what you eat” model in explaining the eubacterial origin of nuclear genes of phagotrophic protists (64), the massive HGT we identified in parasitic plants from their hosts again reflect the feeding habit of parasitic organisms. The hypothesis of increased HGT frequency in endoparasites compared with exoparasites was proposed in studies of HGT in the parasitic plants Rafflesia (20, 25) and Cynomorium (22). Our study revealed increased numbers of HGT among related species with increased heterotrophic dependence, a pattern that could be corroborated with rigorous HGT identification in a much larger sampling of parasitic taxa from Orobanchaceae and other parasitic lineages (65) of varying ages and degrees of nutritional dependence.
Methods
Transcriptome sequencing, de novo assembly (including read cleaning and adapter filtering), postprocessing, expression quantification (using CLC workbench), and annotation [predicted protein sequences were used to search against Swissprot, TAIR10 (The Arabidopsis Information Resource 10), tremble, and Pfam domain databases] followed Yang et al. (32).
Removal of Contamination.
Sequences were cleaned by removing nonplant transcripts and transcripts of the host plants used for growing the parasites [Medicago or Zea for Triphysaria, Sorghum for Striga, and Arabidopsis for Phelipanche (30)] with BLASTN (nucleotide BLAST) (E-value of 1e-10).
Phylogenomic Construction of Parasite Gene Trees.
ORFs and protein sequences encoded by assembled transcripts were predicted with ESTScan version 2.0 (66). A total of 586,228 protein coding genes of 22 representatives of sequenced land plant genomes were classified into 53,136 orthogroups using OrthoMCL (67). The selected taxa include nine rosids (A. thaliana, Thellungiella parvula, Carica papaya, Theobroma cacao, Populus trichocarpa, Fragaria vesca, Glycine max, Medicago truncatula, and Vitis vinifera), three asterids (Solanum lycopersicum, Solanum tuberosum, and Mimulus guttatus), two basal eudicots (Nelumbo nucifera and Aquilegia coerulea), five monocots (Oryza sativa, Brachypodium distachyon, S. bicolor, Musa acuminate, and Phoenix dactylifera), one basal angiosperm (Amborella trichopoda) (68), one lycophyte (Selaginella moellendorffii), and one moss (Physcomitrella patens). Unigenes from Lindenbergia, Triphysaria, Striga, Phelipanche, and two Asteraceae species, Lactuca sativa and Helianthus annuus, were assigned into the 22-genome orthogroup classifications by BLASTP (69) with e-value ≤ 1e-5 and Hidden Markov Models (HMMs) (70). This resulted in 13,125 orthogroup phylogenetic trees containing at least one parasitic species in the phylogeny. Orthogroup phylogenies were generated with an automated approach (ref. 32 and https://github.com/dePamphilis/PlantTribes) where codon alignments were used to estimate a maximum likelihood tree using RAxML version 7.2.7 with the GTRGAMMA model (71).
HGT Screening on Phylogenetic Trees.
Customized Python scripts were developed to screen incongruent phylogenies. The python script used the tree-parsing functions available in the ete2 libraries (72) to traverse one node at a time and extract members above each node. To decrease the false positive rate for HGT discovery, the script searched for donors in distantly related rosid and monocot groups rather than more closely related asterid lineages, which would be more prone to false-positive HGT. An ancestral node was determined when traversing to a node whose left and right branches were exclusively composed of parasite and donor sequences. The script then examined all of the inner nodes within the ancestral node for BS values that support the grouping of parasite and donor sequences. Three models of topology (Fig. 1) represent HGTs with decreasing degrees of confidence. The script reported orthogroups that match any of them. After the automated screening, the HGT candidate orthogroups were further classified into three categories: low-confidence, medium-confidence, and high-confidence trees. The classification criteria were based on a scoring scheme that considered whether the donor clade contained at least two donor sequences, bootstrap values supporting the grouping of the parasite gene and donor sequences, and the presence of long-branch clades. Each of these three factors was assigned a score, and the summed score was used to assign trees to each of the confidence levels (SI Appendix, Table S1). The medium- and high-confidence orthogroup trees were then examined carefully for possible sources of errors, including contamination, potential for long-branch artifacts, and insufficient taxon sampling. Frame-shift errors were fixed by manually introducing 1–2 bp to achieve translations that were much more conserved in comparison with other species.
HGT Validation by Increased Taxon Sampling.
For HGT validation, we added more taxa from related species, including five sequenced asterid genomes and 10 transcriptomes from 1kp in the Lamiales order (34). The genomes include the following: Beta vulgaris (beet), Actinidia chinensis (kiwifruit), Utricularia gibba, Sesamum indicum, and S. asiatica (parasite in Orobanchaceae). The transcriptomes include the following: Strobilanthes dyeriana (Acanthaceae), Mansoa alliacea (Bignoniaceae), Sinningia tuberosa (Gesneriaceae), Salvia spp. (Lamiaceae), Olea europaea (Oleaceae), Epifagus virginiana (Orobanchaceae), Paulownia fargesii (Paulowniaceae), Antirrhinum majus (Plantaginaceae), Rehmannia glutinosa (Rehmanniaceae), and Verbena hastata (Verbenaceae). Also, we added genes from transcriptomes of above-ground tissues derived from eight additional parasitic Orobanchaceae: Alectra vogelii, Myzorrhiza californica, Orobanche minor, Phelipanche mutelii, Phelipanche ramosa, Striga gesneroides, Triphysaria eriantha, and Triphysaria pusilla. To make sure that all of the HGTs were captured from these added taxa, we used HMMs (70) (hmmsearch with 1e-5). For lineage-specific HGT orthogroups, a superorthogroup tree (68) was constructed to ensure the inclusion of all homologous sequences.
Validation of HGT Sequences.
RT-PCR was used to verify transcriptome assembly for HGT sequences. HGT sequences (from combined builds) were aligned with BLASTN against haustoria stage-specific assemblies to identify corresponding transcripts; these were then used as templates to design primers for PCR amplification with haustorial cDNA. Genome PCR used HGT transcriptome sequences as templates to design primers for subsequent PCR with genomic DNA. Verification was confirmed when the sequenced genomic PCR product matched the assembled transcript, and occasionally introns were revealed (SI Appendix, Table S3).
Intron Analyses.
Intron positions were extracted to examine if they were conserved in multiple sequence alignments (MSAs). For each orthogroup, the peptide sequences were aligned using MAFFT version 7 (73), which were then forced onto coding sequences (CDS) to generate the CDS alignment. A customized Perl script was used to extract the intron positions in each coding sequence, and the corresponding positions were mapped onto the CDS alignment. To identify intron positions in Orobanchaceae genes, we generated and assembled shallow genomic sequences from S. hermonthica and P. aegyptiaca. Coding sequence was predicted for each transcript using ESTScan (66) and was then aligned to genomic sequences using BLASTN with an e-value cutoff of 1e-05. Manual curation was then performed for each CDS-genomic DNA alignment to make sure introns start with GT, and end with AG. To extract intron sequences for each gene from fully sequenced plant genomes, we used the gff file for the intron regions of each gene. Intron sequences of genes in sequenced genomes were extracted from genomic sequences in Phytozome 10 (35) using samtools (74) and betools [index command “samtools faidx,” “fastaFromBed” in BEDTools (75) was used by indicating the genome reference using “-fi” and the gff file using “-bed,” which generated an output file using “-fo”]. Intron sequences for parasite genes were obtained by blasting the coding sequence onto genomic sequences. For intron phylogenies, introns were concatenated to increase the number of informative sites for tree reconstruction with the same approach as building the tree of CDS.
Selective Constraint Analyses.
To identify signatures of adaptive or purifying selection, we conducted two likelihood ratio tests using the branch model and branch-site model in PAML (48). In each test, we identified the HGT genes on a phylogenetic tree as the foreground branches, which were compared with the remaining background sequences on the tree. The branch model tested if the foreground HGT branches had the same level of protein sequence constraint as the background nonparasitic sequences. Levels of sequence constraint were measured by estimating a Dn/Ds ratio (omega, ω) for both foreground and background. The null model (a one-ratio model) estimated a single ω for all sequences. A likelihood ratio test was performed to infer if the branch model fit the data significantly better than the one-ratio model. A nonsignificant result from the test indicated the same level of protein constraint between the HGT genes and nonparasitic background; a significant result can reflect either (i) a higher ω in the HGT branch than the background, indicating either relaxed constraint (if ω < 1) or adaptive evolution (if ω > 1) or (ii) a lower ω in the HGT branch than the background, indicating stronger levels of purifying selection. HGT sequences that had relaxed constraint were further tested with a branch-site model to identify the presence of adaptive sites for the indicated foreground lineages. To perform the branch-site test, parameters “model =2, NSsites=2, fix_omega=0” were indicated in the codeml files for both the branch-site model and null model in PAML. The former model used an additional parameter “fix_omega=1,” whereas the latter model used “fix_omega=0.” Sites with probability greater than 0.95 from a Bayes Empirical Bayes analysis represented likely adaptive sites. RELAX analyses (49) were also performed to evaluate whether selective constraints are stronger in foreground (HGT) branches compared with background (non-HGT) branches. The analyses was performed using the RELAX tool on datamonkey server (test.datamonkey.org/relax/). An input file that contains the codon sequence alignment and the RAxML tree was provided, and the foreground braches (test branches) and background branches (reference branches) were then indicated before running. A likelihood ratio test was also performed to evaluate if selection varied between test and reference branches. A significant result (P value < 0.05) indicates a stronger level of selection if K is greater than 1 or a weaker selection or relaxed constraint if K is less than 1.
Genome Assembly of Parasite Species.
Illumina data for S. hermonthica and P. aegyptiaca were de novo assembled using CLC Assembly Cell v 4.1 (https://www.qiagenbioinformatics.com/products/clc-assembly-cell/):
“novo_assemble -o contigs.fasta -p fb ss 180 250 -q -i reads1.fq reads2.fq.”
Estimation of Number Of Transfers.
We used a SH test (37) to estimate the number of transfer events from 42 HGT orthogroup trees. Trees in which HGT genes did not form a monophyletic clade were constrained to represent one event, and a RAxML tree with constrained HGT clade was produced. An SH test was performed in RAxML version 7.2.7 (71) to test if the likelihood of the constrained tree was significantly worse than that of the original tree.
Supplementary Material
Acknowledgments
We thank Dr. Craig Praul and the Huck Genomics Core Facility for transcriptome sequencing and the gift of Striga genome sequences generated on a trial run of the Illumina HiSeq2500 sequencer that was purchased as NSF Equipment Grant MRI-1229046 (to C.W.d.); Tony Omeis for growing M. californica (with Grindelia host) in the Biology Greenhouse at Penn State University from material originally provided by Alison Colwell; K. Shirasu and S. Yoshida for access to the S. asiatica genome sequence and annotation; D. E. Soltis, M. K. Deyholos, M. W. Chase, and C. Wang for collecting nine of the 10 1KP samples used for HGT validation in this study; Ning Jiang (Michigan State University) for discussion of Pong-like TEs; and J. Naumann, J. Der, J. Palmer, M. Axtell, D. Cosgrove, S. Maximova, and M. Guiltinan for helpful suggestions. This research was supported by NSF Plant Genome Research Program Awards DBI-0701748 and IOS-1238057 (to J.H.W., C.W.d., M.P.T., and J.I.Y.), with additional support from the Plant Biology graduate program (Z.Y., L.A.H., S.J., and H.Z.) and from the Genetics graduate program as well as the Biology Department (Y.Z.) at Penn State University; National Institute of Food and Agriculture Project 131997 (to J.H.W.); and NSF Grant IOS-1213059 (to M.P.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data are archived at National Center for Biotechnology Information BioProject (ID codes SRP001053 and SRP083761) and at ppgp.huck.psu.edu. Scripts and alignments were submitted to github user “dePamphilis” under directory “HGT_PNAS_2016.”
1Z.Y. and Y.Z. share senior authorship.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608765113/-/DCSupplemental.
References
- 1.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102(40):14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 3.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombs JM, Barkay T. Molecular evidence for the evolution of metal homeostasis genes by lateral gene transfer in bacteria from the deep terrestrial subsurface. Appl Environ Microbiol. 2004;70(3):1698–1707. doi: 10.1128/AEM.70.3.1698-1707.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGowan C, Fulthorpe R, Wright A, Tiedje JM. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol. 1998;64(10):4089–4092. doi: 10.1128/aem.64.10.4089-4092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFadden GI. Chloroplast origin and integration. Plant Physiol. 2001;125(1):50–53. doi: 10.1104/pp.125.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyall SD, Johnson PJ. Origins of hydrogenosomes and mitochondria: Evolution and organelle biogenesis. Curr Opin Microbiol. 2000;3(4):404–411. doi: 10.1016/s1369-5274(00)00112-0. [DOI] [PubMed] [Google Scholar]
- 8.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: Building the web of life. Nat Rev Genet. 2015;16(8):472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 9.Yue J, Hu X, Sun H, Yang Y, Huang J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun. 2012;3:1152. doi: 10.1038/ncomms2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li FW, et al. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc Natl Acad Sci USA. 2014;111(18):6672–6677. doi: 10.1073/pnas.1319929111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archibald JM, Richards TA. Gene transfer: Anything goes in plant mitochondria. BMC Biol. 2010;8:147. doi: 10.1186/1741-7007-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 13.Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA. 1998;95(24):14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101(51):17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 17.Mower JP, et al. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010;8:150. doi: 10.1186/1741-7007-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science. 2004;305(5684):676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- 19.Mower JP, Stefanović S, Young GJ, Palmer JD. Plant genetics: Gene transfer from parasitic to host plants. Nature. 2004;432(7014):165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- 20.Xi Z, et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9(2):e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nickrent DL, Blarer A, Qiu YL, Vidal-Russell R, Anderson FE. Phylogenetic inference in Rafflesiales: The influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol. 2004;4:40. doi: 10.1186/1471-2148-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellot S, et al. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol Evol. 2016;8(7):2214–2230. doi: 10.1093/gbe/evw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci USA. 2015;112(27):E3515–E3524. doi: 10.1073/pnas.1504491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis CC, Xi Z. Horizontal gene transfer in parasitic plants. Curr Opin Plant Biol. 2015;26:14–19. doi: 10.1016/j.pbi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Xi Z, et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida S, Maruyama S, Nozaki H, Shirasu K. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science. 2010;328(5982):1128. doi: 10.1126/science.1187145. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D, et al. Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer. BMC Plant Biol. 2014;14:19. doi: 10.1186/1471-2229-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol Biol. 2013;13:48. doi: 10.1186/1471-2148-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun T, et al. Two hAT transposon genes were transferred from Brassicaceae to broomrapes and are actively expressed in some recipients. Sci Rep. 2016;6:30192. doi: 10.1038/srep30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15(4):227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science. 2014;345(6198):808–811. doi: 10.1126/science.1253122. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol. 2015;32(3):767–790. doi: 10.1093/molbev/msu343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honaas LA, et al. Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 2013;13:9. doi: 10.1186/1471-2229-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matasci N, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96(1):336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 37.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 38.Musselman LJ. The biology of Striga, Orobanche, and other root-parasitic weeds. Annu Rev Phytopathol. 1980;18(1):463–489. [Google Scholar]
- 39.Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19(6):613–619. doi: 10.1016/j.gde.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Huang J. Horizontal gene transfer in eukaryotes: The weak-link model. BioEssays. 2013;35(10):868–875. doi: 10.1002/bies.201300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aly R, et al. Movement of protein and macromolecules between host plants and the parasitic weed Phelipanche aegyptiaca Pers. Plant Cell Rep. 2011;30(12):2233–2241. doi: 10.1007/s00299-011-1128-5. [DOI] [PubMed] [Google Scholar]
- 42.Heide-Jørgensen HS, Kuijt J. The haustorium of the root parasite Triphysaria (Scrophulariaceae), with special reference to xylem bridge ultrastructure. Am J Bot. 1995;82(6):782–797. [Google Scholar]
- 43.Tomilov AA, Tomilova NB, Wroblewski T, Michelmore R, Yoder JI. Trans-specific gene silencing between host and parasitic plants. Plant J. 2008;56(3):389–397. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- 44.Aly R, et al. Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol J. 2009;7(6):487–498. doi: 10.1111/j.1467-7652.2009.00418.x. [DOI] [PubMed] [Google Scholar]
- 45.Heinemann IU, Nakamura A, O’Donoghue P, Eiler D, Söll D. tRNAHis-guanylyltransferase establishes tRNAHis identity. Nucleic Acids Res. 2012;40(1):333–344. doi: 10.1093/nar/gkr696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pielach A, Leroux O, Domozych DS, Knox JP, Popper ZA. Arabinogalactan protein-rich cell walls, paramural deposits and ergastic globules define the hyaline bodies of rhinanthoid Orobanchaceae haustoria. Ann Bot (Lond) 2014;114(6):1359–1373. doi: 10.1093/aob/mcu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visser JH, Dörr I, Kollmann R. The “hyaline body” of the root parasite Alectra orobanchoides Benth. (Scrophulariaceae), its anatomy, ultrastructure and histochemistry. Protoplasma. 1984;121:146–156. [Google Scholar]
- 48.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 49.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 2015;32(3):820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang IS, Hwang BK. Role of the pepper cytochrome P450 gene CaCYP450A in defense responses against microbial pathogens. Planta. 2010;232(6):1409–1421. doi: 10.1007/s00425-010-1266-y. [DOI] [PubMed] [Google Scholar]
- 51.Chen K, Fan B, Du L, Chen Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol. 2004;56(2):271–283. doi: 10.1007/s11103-004-3381-2. [DOI] [PubMed] [Google Scholar]
- 52.Marino D, Peeters N, Rivas S. Ubiquitination during plant immune signaling. Plant Physiol. 2012;160(1):15–27. doi: 10.1104/pp.112.199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyle P, et al. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell. 2009;21(11):3700–3713. doi: 10.1105/tpc.109.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto T, Yamada Y. Hyoscyamine 6beta-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, in alkaloid-producing root cultures. Plant Physiol. 1986;81(2):619–625. doi: 10.1104/pp.81.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, et al. The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol. 2012;159(4):1857–1865. doi: 10.1104/pp.112.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Curtis RH, Pankaj, Powers SJ, Napier J, Matthes MC (2013) The Arabidopsis F-box/Kelch-repeat protein At2g44130 is upregulated in giant cells and promotes nematode susceptibility. Mol Plant Microbe Interact 26(1):36–43. [DOI] [PubMed]
- 58.Ascencio-Ibáñez JT, et al. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148(1):436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization and signaling. Front Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. PLEXdb: Gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012;40(Database issue):D1194–D1201. doi: 10.1093/nar/gkr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oo MM, et al. Evaluation of rice promoters conferring pollen-specific expression in a heterologous system, Arabidopsis. Plant Reprod. 2014;27(1):47–58. doi: 10.1007/s00497-014-0239-x. [DOI] [PubMed] [Google Scholar]
- 62.Atkinson TJ, Halfon MS. Regulation of gene expression in the genomic context. Comput Struct Biotechnol J. 2014;9:e201401001. doi: 10.5936/csbj.201401001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu B, et al. Contribution of NAC transcription factors to plant adaptation to land. Science. 2014;343(6178):1505–1508. doi: 10.1126/science.1248417. [DOI] [PubMed] [Google Scholar]
- 64.Doolittle WF. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14(8):307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 65.Naumann J, et al. Single-copy nuclear genes place haustorial Hydnoraceae within piperales and reveal a cretaceous origin of multiple parasitic angiosperm lineages. PLoS One. 2013;8(11):e79204. doi: 10.1371/journal.pone.0079204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iseli C, Jongeneel CV, Bucher P. 1999 ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol, 138–148. [PubMed] [Google Scholar]
- 67.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342(6165):1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 69.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 72.Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: A python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.