Significance
Genomes are organized into domains of different structure and activity, yet our understanding of their formation and regulation is poor. We show that Caenorhabditis elegans chromatin domain organization in early embryos and third-larval stage animals is remarkably similar despite the two developmental stages containing very different cell types. Chromatin domains separate genes into those with stable versus developmentally regulated expression. Analyses of chromatin domain structure suggest that transcription regulation and germ-line chromatin regulation play roles in separating chromatin domains. Our results further our understanding of genome domain organization.
Keywords: chromatin states, chromatin domains, C. elegans, boundary
Abstract
Eukaryotic genomes are organized into domains of differing structure and activity. There is evidence that the domain organization of the genome regulates its activity, yet our understanding of domain properties and the factors that influence their formation is poor. Here, we use chromatin state analyses in early embryos and third-larval stage (L3) animals to investigate genome domain organization and its regulation in Caenorhabditis elegans. At both stages we find that the genome is organized into extended chromatin domains of high or low gene activity defined by different subsets of states, and enriched for H3K36me3 or H3K27me3, respectively. The border regions between domains contain large intergenic regions and a high density of transcription factor binding, suggesting a role for transcription regulation in separating chromatin domains. Despite the differences in cell types, overall domain organization is remarkably similar in early embryos and L3 larvae, with conservation of 85% of domain border positions. Most genes in high-activity domains are expressed in the germ line and broadly across cell types, whereas low-activity domains are enriched for genes that are developmentally regulated. We find that domains are regulated by the germ-line H3K36 methyltransferase MES-4 and that border regions show striking remodeling of H3K27me1, supporting roles for H3K36 and H3K27 methylation in regulating domain structure. Our analyses of C. elegans chromatin domain structure show that genes are organized by type into domains that have differing modes of regulation.
The complete genome sequence, which provides the information necessary for constructing an organism, is interpreted in the context of chromatin. Covalent modifications of histone tails and histone variants can regulate or reflect genome function, and so are markers of chromatin state and genomic activity (1). For example, H3K4me3 often marks active promoters, H3K36me3 transcription elongation, and H3K27me3 Polycomb-silenced regions. Previous studies have shown that genomic regions of similar activity harbor shared combinations of modifications, termed chromatin states, and that subdividing the genome according to these combinations is a powerful method for annotation and uncovering novel functional regions (2–5). Here, we apply chromatin state mapping to two developmental stages of the model organism Caenorhabditis elegans and use the resulting maps to investigate genome domain organization and its regulation.
C. elegans is highly amenable for global studies of chromatin structure and function because it has a small, well-annotated genome (30× smaller than human), and work of the modENCODE consortium has provided a large number of datasets mapping the locations of chromatin-associated factors, such as histone modifications and transcription factors (TFs) (6–10). C. elegans chromatin shows features in common with those of other organisms, such as the type of marking at regulatory regions and at active or inactive genes (7, 10–12). Additionally, the derivation of a single set of chromatin states for C. elegans, Drosophila, and human using a single joint genome analysis and data from eight histone modifications highlighted the common properties of chromatin in the three organisms (10). However, because chromatin differences also exist, the jointly derived chromatin states are not ideal for C. elegans-specific analyses, and no other C. elegans chromatin state maps have been published.
Previous studies have described broad properties of C. elegans genome organization. The distal “arms” and central regions of the autosomal chromosomes show differences in transcriptional activity, chromatin composition, and recombination rate (6, 7, 11, 13, 14). Central regions have higher average gene expression, moderate enrichment of histone modifications associated with active transcription, and lower meiotic recombination than distal arm regions. In contrast, most features associated with heterochromatin, such as H3K9 methylation and nuclear envelope association, are found on the chromosome arms (7, 15). However, the chromosome arms are not purely heterochromatic. Actively transcribed genes reside on the chromosome arms and these genes are marked by histone modifications associated with gene activity, as in the central regions (7). In addition, the X chromosome shows extensive chromatin differences compared with autosomes because of dosage compensation (16). These previous studies have provided a large-scale picture of C. elegans chromosome organization.
Here, we investigate C. elegans chromatin and genome organization and its regulation through the generation and analyses of C. elegans-specific chromatin state maps for early embryos (EE) and third larval (L3) stages. As in other organisms, chromatin states correlate with many biological features, including enhancers, promoters, transcription elongation, gene ends, repeat regions, and inactive genes. Analyzing patterns of states revealed that chromatin domains of differing activity separate germ-line and broadly expressed genes from developmentally regulated genes. The properties of domains and the border regions between them point to roles for transcription regulatory regions and germ-line chromatin marking in domain separation. Our results provide a framework for future studies of chromatin structure and function in C. elegans.
Results
Twenty State Models of C. elegans Chromatin.
To investigate features and domain organization of C. elegans chromatin, we derived 20 state EE and L3 chromatin state maps, using hidden Markov models (HMM) and ChIP data for 17 histones or modifications (Materials and Methods). Patterns and levels of enrichment of many histone modifications differ on C. elegans autosomes compared with the X chromosome, reflecting dosage compensation (7, 16–18), which caused whole-genome chromatin state maps to subdivide into separate autosomal and X-chromosome–specific states. Therefore, for each stage we generated a separate map for autosomes and the X chromosome (Fig. 1A, Fig. S1, and Dataset S1). The EE and L3 autosomal chromatin states show much greater similarity to each other than do the EE and L3 chromosome X states (Figs. S1 and S2), consistent with alterations in chromatin structure and marking induced by dosage compensation after the EE stage (16). The chromatin states were annotated by analyzing the associations of states with a range of different genomic features (Fig. 1 and Figs. S1, S3, and S4). As well as differences in enrichment levels for histone modifications, the states show differences in median length (250–1,250 bp), genomic coverage (2.2–9%), and GC content (25–44%) (Fig. S1). Below, we briefly describe chromatin state annotation and properties using L3 autosomes as an example. We then use the states to investigate autosomal chromatin domain organization and its regulation.
Fig. 1.
Chromatin states and domains. (A) L3 state key and annotation. (Left) State numbers and annotations; (Right) relative enrichment or underenrichment of the indicated histones or histone modifications in each state. The scale bar shows the average z-score of the mark. (B) IGV screenshot of 340-kb region on chromosome I (chr I: 7,996,451–8,337,598) showing genes, domains, chromatin states, H3K36me3, and H3K27me3 in EE and L3.
States Associated with Active Genes and Enhancers.
We found that states 1–8 predominantly mark high and moderately expressed genes, with states 1–5 most associated with genes in the highest quintile of expression (Figs. S3 and S4G). These chromatin states mark different types of genic regions: promoter (state 1), 5′ proximal (state 2), transcription elongation (states 3–7), and enhancer (state 8) (Figs. S1 and S4 G and H). The different transcription elongation states are associated with different expression levels and genic regions. For example, state 3 marks highly expressed exons, whereas state 6 marks transcribed regions of lower expression, and state 5 typically marks gene ends (Figs. S3 and S4G).
We define states 8–10 as likely enhancer regions based on their enrichment for chromatin modifications typical of enhancers (high H3K4me1, high H3K27ac, low H3K4me3) and their association with annotated enhancers (Figs. S1 and S4H). Annotated noncoding RNA genes that are unclassified in Wormbase are also frequently associated with states 8–10, suggesting that some of these may be enhancer transcripts (Fig. S4I). Other noncoding RNA genes (e.g., microRNAs, transfer RNAs, Piwi-interacting RNAs) show different state enrichments (Fig. S4I).
States Associated with Inactive Genes.
We found that inactive and lowly expressed genes are associated with states 16–20 (Figs. S3 and S4G). In contrast to states associated with active transcription, inactive states usually do not mark particular gene regions but instead are more uniformly distributed across genes (Figs. S3 and S4G). However, inactive states do show differential enrichment in genic versus intergenic regions (Fig. S3). Consistent with known associations of histone modifications with silenced genes (19), inactive states are enriched for H3K27me3 (a mark of Polycomb-mediated silencing; states 17–19) or both H3K27me3 and H3K9me3 (a mark of heterochromatin; states 16 and 20) (Fig. S1). Co-occurrence of H3K27me3 and H3K9me3 in C. elegans has been noted previously (10).
Inactive states 16–20 show different chromosomal distribution patterns and genic associations (Fig. S4L). For example, inactive state 17, which is enriched for H3K27me3, is highly prevalent on chromosome V (Fig. S4L). Chr V is unique in harboring a large fraction (68%) of the 1,383 odorant receptors annotated in C. elegans (20). These receptors are transcriptionally inactive in most cells, usually being expressed in only one or a few neurons (21). We found that state 17 is highly associated with odorant receptor genes, marking 62% of them genome-wide. In addition, state 17 marks 57% of odorant receptor pseudogenes (of 290) and 18% of pseudogenes of other classes (Fig. S4L). The finding of a chromatin state associated with both pseudogenes and a class of widely silenced genes (odorant receptors) suggests that that these loci may be repressed by a shared mechanism.
Mixed States.
Because the histone modification mapping was conducted in whole animals, it was expected that states associated with tissue-specific gene expression might display enrichment for histone modifications of both active and inactive genes. Indeed, states 13 and 14 display this pattern, and genes marked by these states are enriched for having high gene expression combined with high H3K27me3 (Fig. S4K). States 13 and 14 are also enriched for tissue-specific genes identified by gene-expression profiling (Fig. S4K).
Repeats.
The C. elegans genome harbors about 100,000 annotated repeat elements, which fall into 163 families (22). We found that six chromatin states are highly associated with repeats (states 6, 7, 12, 13, 15, and 16) (Fig. S4J). Different repeat classes and individual repeats are associated with different chromatin states (Fig. S4J). The variation of chromatin states on different repeat types may reflect differences in their regulation or function.
Chromatin States on Autosomes Demarcate Chromatin Domains of Different Activities.
We next used the chromatin states to investigate chromatin domain structure and its regulation. We focused on autosomes because the chromatin states are highly similar in the EE and L3 maps, facilitating comparative analyses. In browsing, we observed that states associated with the highest or lowest quintiles of gene expression (states 1–5 and states 16–20, respectively) (Fig. S3) were located in extended genomic domains interspersed with the other 10 states (Fig. 1B). Based on these patterns, we defined high activity domains, low activity domains, and border regions in EE and L3 autosomal chromatin. In brief, high activity domains were defined as regions containing states 1–5 but not 16–20, starting and ending with a state from 1–5. Similarly, low activity domains were defined as regions containing states 16–20 but not 1–5, starting and ending with a state from 16–20. The genomic regions between high and low activity domains were defined as borders. The remaining states (states 6–15) were not used in domain definitions and form parts of all three region types. Table S1 gives statistics on domain sizes and numbers of genes in domains. Domains are larger and contain more genes than expected by chance (Materials and Methods and Table S1). For example, in EE the median active domain is 13,054 bp (4,506 bp expected) and contains four genes (one expected), whereas the median regulated domain is 23,874 bp (13,647 bp expected) and contains four genes (two expected). Fig. S5 shows the distribution of the 20 chromatin states and Fig. S6 the distribution of histone modifications in the different regions; we note that chromatin state 11 is highly associated with domain borders in both EE and L3 domain maps.
Table S1.
Domain statistics and simulation of expected domain size and gene number
| Stage | Domains (n) | Genes (n) | Median domain length (bp) | Expected median domain length (bp) | Median genes per domain | Expected median genes per domain | Mean domain length | Expected mean domain length | Mean genes per domain | Expected mean genes per domain |
| EE active | 1,046 | 6,047 | 13,054 | 4,506 | 4 | 1 | 25,318 | 9,492 | 5.8 | 2.0 |
| EE border | 2,013 | 1,914 | 2,461 | 600 | 1 | 0 | 5,272 | 1,865 | 1.0 | 0.4 |
| EE regulated | 1,041 | 9,424 | 23,874 | 13,647 | 4 | 2 | 42,358 | 21,696 | 9.1 | 4.6 |
| L3 active | 1,274 | 6,198 | 13,608 | 4,328 | 3 | 1 | 22,184 | 8,587 | 4.9 | 1.8 |
| L3 border | 2,471 | 1,996 | 2,405 | 500 | 1 | 0 | 4,343 | 1,794 | 0.8 | 0.4 |
| L3 regulated | 1,257 | 9,220 | 18,670 | 11,567 | 3 | 2 | 33,619 | 18,503 | 7.3 | 3.8 |
The domains were defined based on chromatin states associated with high- and low-gene expression; however, they are not uniform in activity. For example in L3 larvae, 20% of genes in the top quintile of expression lie in low activity domains, and 11% of genes in the bottom 40% of expression lie in high activity domains. Based on the analyses below, we will refer to the high activity domains as “active,” the low activity domains as “regulated,” and the regions in between them as “border.” Across each autosome, the distribution of active and regulated domains is relatively uniform, although chromosomes vary in the relative proportions of active and regulated domains (Fig. S7).
Chromatin Domain Structure of Early Embryos and L3 Larvae Is Strikingly Similar.
To investigate the developmental regulation of chromatin domains, we compared the positions of domains and borders in EE and L3 larvae. These two samples represent very different populations of cells. The profiled EE samples contained undifferentiated cells undergoing cell division (1–300 cell-stage embryos), whereas the L3 larval samples contained ∼85% differentiated somatic cells and ∼15% mitotic germ cells (7). Surprisingly, the autosomal chromatin domain structure of EE and L3 larvae is strikingly similar, with 85% of EE border regions overlapping an L3 border. Additionally, 91% of bases in active domains and 89% of bases in regulated domains are in common between EE and L3 stages. The overall consistency in domain structure between EE and L3 larvae suggests that mechanisms determining shared organization are largely independent of cell fate.
Properties of Domains and Borders.
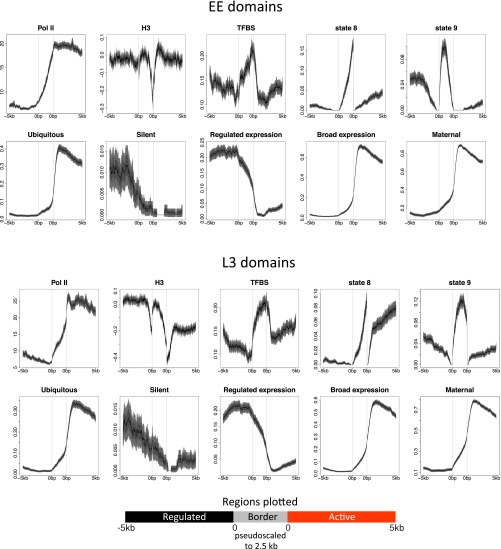
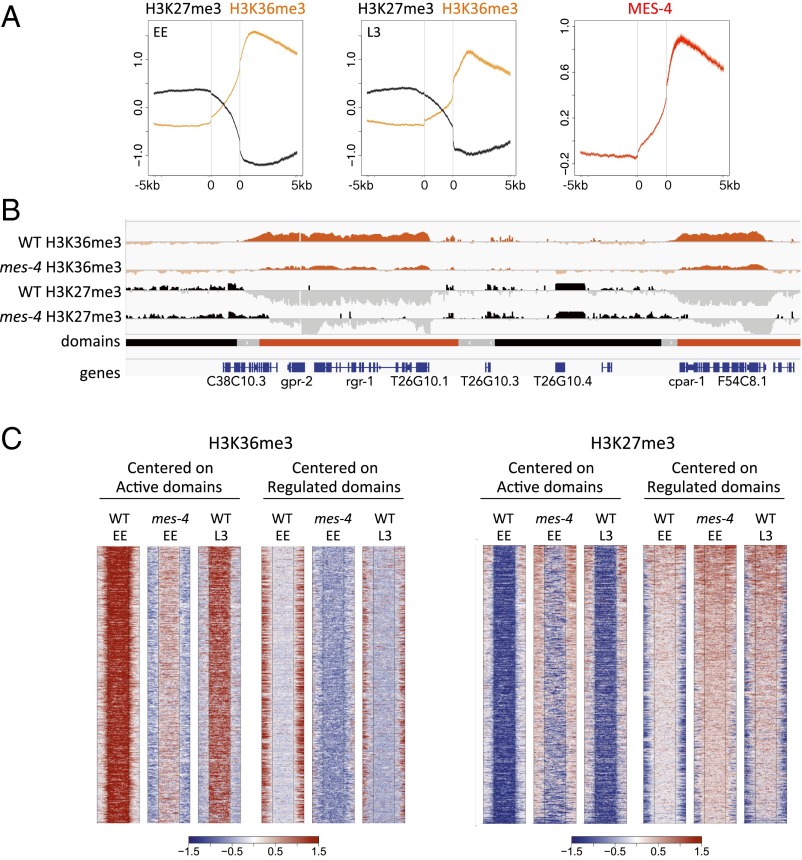
We next investigated chromatin domain properties. As expected, RNA polymerase II levels sharply increase at the transitions from borders to active domains (Fig. 2). The transitions from regulated domains to border regions and from border regions to active domains have low levels of histone H3, indicative of nucleosome depletion, suggesting that these regions are more accessible than the neighboring chromatin (Fig. 2). Intriguingly, two families of repeat elements (CELE1 and CELE2) are particularly associated with borders (Fig. S8). Additionally, border regions are enriched for enhancer chromatin states and distal TF binding sites, typical of enhancers, suggesting that these regions have transcription regulatory activity (Fig. 2). We also observed that active domains contain high levels of H3K36me3 and that regulated domains contain high levels of H3K27me3 (Figs. 1B and 3A). These correspond with alternating blocks of H3K36me3 and H3K27me3 that were previously noted in EE chromatin (23).
Fig. 2.
Properties of EE and L3 autosomal active, border, and regulated domains. Plots are centered and anchored at borders pseudoscaled at 2.5 kb, and show 5 kb into regulated domains (Left) and 5 kb into active domains (Right). Lines show mean signal, darker-filled areas show standard error, and lighter-filled areas are 95% confidence intervals. Gray vertical lines indicate edges of the border region.
Fig. 3.
MES-4 regulates chromatin marking at domain edges. (A) Plots of H3K36me3 and H3K27me3 across regulated, border, and active domains in EE and L3, and of MES-4 in EE. Plots are centered at borders pseudoscaled at 2.5 kb, and show 5 kb into regulated domains (Left) and 5 kb into active domains (Right). Lines show mean signal, darker-filled areas show SE, and lighter-filled areas are 95% confidence intervals. Gray vertical lines indicate edges of the border region. (B) IGV screenshot showing H3K36me3 (−2.0 to 4.0 log2 z-score) and H3K27me3 (−2.0 to 1.0 log2 z-score) signal tracks in wild-type and mes-4 RNAi early embryos. (C) Heatmaps comparing H3K36me3 and H3K27me3 signals across EE active domains and EE regulated domains in wild-type and mes-4 RNAi early embryos. Signals are centered on active domains or regulated domains (pseudoscaled at 5 kb) as indicated, and plot 2.5 kb into borders on either side.
We found that the genes that reside in active and regulated domains have different properties. Not surprisingly, ubiquitously expressed genes lie predominantly in active domains, whereas genes with low or no detectable expression [depth of coverage per million reads (dcpm) < 0.005 in all stages; n = 637] are usually found in regulated domains (Fig. 2). However, the majority of genes in regulated domains are detectably expressed at one or both stages (54% are in the top 60% of expression).
To further investigate the properties of genes in the different domains, we annotated genes as broadly expressed or developmentally regulated using the coefficient of variation (CV) of gene expression across 35 developmental stages and cell types (24). Broadly expressed genes have a similar expression level across the 35 conditions, leading to low CV values, whereas developmentally regulated genes have high CV values because of differential expression across conditions. We considered genes in the bottom third of CV values as broadly expressed and genes in the top third of CV values as developmentally regulated. Gene-expression variation (CV score) shows a remarkable association with domain type. Genes with broad expression across development and cell types (low CV) lie primarily in active domains. Furthermore, most genes (86%) in active domains have maternally contributed mRNA, indicating that they are expressed in the germ line (Fig. 2). Consistent with this, clustering of germ line-expressed genes has been previously noted (25). In contrast, genes with developmentally regulated expression (high CV) are predominantly found in regulated domains (Fig. 2). Conditionally regulated genes (e.g., those induced upon bacterial infection) are also associated with regulated domains (Table S2).
Table S2.
Association of condition-induced gene expression with EE domains
| Condition | n | Active (%) | Border (%) | Regulated (%) | Source |
| Induced by heat | 240 | 11.7 | 11.3 | 77.1 | Brunquell et al. (60) |
| Induced by copper | 235 | 8.5 | 9.4 | 82.1 | Hattori et al. (59) |
| Common induction by three bacterial pathogens (Serratia marcescens, Enterococcus faecalis, Photorhabdus luminescens) | 540 | 6.3 | 9.3 | 84.4 | Engelmann et al. (61) |
To summarize, the genome is organized into two types of domains: H3K36me3-rich “active” domains containing genes that are broadly and germ line-expressed, and H3K27me3-rich “regulated” domains containing genes with regulated expression. The correspondence between domain type and H3K36me3 or H3K27me3 levels suggests that these modifications may play roles in defining active and regulated domains.
A Role for MES-4 in Domain Definition.
The association of active domains with maternal gene expression and high levels of H3K36me3 prompted us to investigate a possible relationship with maternal-effect sterile 4 (MES-4), a germ-line H3K36 histone methyltransferase. Two H3K36me3 methyltransferases have been studied in C. elegans. Histone methyltransferase-like 1 (MET-1) is a Set2 family transcription coupled H3K36 methyltransferase active in most cells (26–28). MES-4 is a germ-line–specific nuclear-receptor–binding SET domain family H3K36 histone methyltransferase with transcription-independent activity (27–29). In the germ line, MES-4 marks expressed genes with H3K36me3, and this germ-line marking is inherited and maintained in EE by maternally contributed MES-4 (27–29). Following knockdown of mes-4, EE show reduced H3K36me3 on germ-line–expressed genes, which is accompanied by increased H3K27me3 (23). As expected, we found that MES-4 is enriched in active domains (Fig. 3A).
The previous studies of mes-4 focused on patterns of chromatin marking and regulation of individual germ-line genes (23, 27). To investigate whether MES-4 is important for domain definition, we analyzed patterns of H3K36me3 and H3K27me3 in wild-type and mes-4(RNAi) early embryos at the level of domains (using data from ref. 23). We found that H3K36me3 still marks active domains in mes-4(RNA) embryos, but both the level and extent of H3K36me3 coverage over active domains is reduced (Fig. 3 B and C). Complementing the reduction of H3K36me3 over active domains, we observed that H3K27me3 coverage at regulated domains is expanded (Fig. 3 B and C). Because MES-4 is a germ-line H3K36 methyltransferase, these results suggest that chromatin regulation in the germ line contributes to the definition of active and regulated domains.
Developmental Remodeling of H3K27 and H3K36 Methylation at Domain Borders.
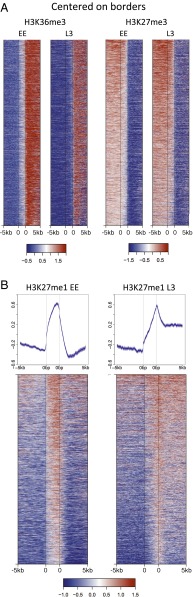
We next investigated whether patterns of H3K36 and H3K27 methylations in domains or borders were developmentally regulated. We observed H3K36me3 marking at borders is altered between EE and L3. In contrast to the sharp rise in H3K36me3 levels across EE borders, in L3 H3K36me3 levels are relatively low and constant in border regions (Fig. 4A). Although H3K27me3 patterns at borders are not obviously changed between EE and L3 (Fig. 4A), there is a striking remodeling of H3K27me1 patterns. In EE, borders have a strong peak of H3K27me1 enrichment, with lower levels in neighboring active and regulated domains (Fig. 4B). At the L3 stage, the H3K27me1 pattern is dramatically altered, with high levels at active domain edges and within active domains (Fig. 4B). The remodeling of H3K36 and H3K27 methylation patterns at border regions suggest that this regulation may play a role in domain definition.
Fig. 4.
Remodeling of H3K36me3 and H3K27me1 marking from EE to L3. (A) Border H3K36me3 is reduced from EE to L3. Plots show heatmaps of H3K36me3 and H3K27me3 signals centered at borders pseudoscaled at 2.5 kb, and show 5 kb into regulated domains (Left) and 5 kb into active domains (Right). (B) H3K27me1 at borders is remodeled from EE to L3. Upper plots average signals centered at borders pseudoscaled at 2.5 kb, with 5 kb into regulated domains (Left) and 5 kb into active domains (Right). Lines show mean signal, darker-filled areas show SE, and lighter-filled areas are 95% confidence intervals. Gray vertical lines indicate edges of the border region. Lower panels show heatmaps of the same regions. Plots at bottom show heatmaps of H3K27me1 signals centered at borders pseudoscaled at 2.5 kb, and show 5 kb into regulated regions (Left) and 5 kb into active regions (Right). Heatmap rows are sorted by decreasing mean signal and show the same regions and order.
Long Intergenic Regions and Enrichment of TF Binding in Borders.
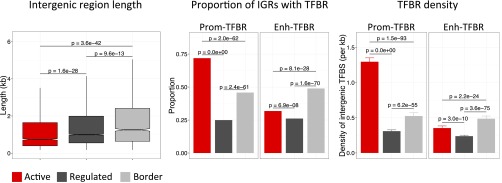
The finding that enhancer chromatin states and TF binding are enriched at border regions suggested that borders may harbor unusual regulatory regions. Indeed, we found that properties of intergenic regions in borders are different from those in active or regulated domains. First, we observed that intergenic regions in borders are long compared with those that lie in active or regulated domains (Fig. 5). We also found that regulated domains harbor longer intergenic regions than active domains (Fig. 5). Therefore, intergenic regions vary in length according to chromatin domain-type across the genome. Borders, which mark the transitions between active and regulated chromatin domains, have the longest intergenic regions.
Fig. 5.
Intergenic regions at borders are long and enriched for enhancer TF binding sites. (A) Boxplots of intergenic region lengths in the specified regions; P values show significance of distribution differences tested using the Mann–Whitney U test. (B) Proportion of intergenic regions overlapping at least one TFBR of the indicated type; P values show differences in proportions tested using a z-test. (C) Density of Prom-TFBR, and Enh-TFBR per kilobase of intergenic region; P values show significance of distribution differences tested using the Mann–Whitney U test.
We next investigated the density of regulatory elements in intergenic regions in different domains. For this analysis, we separated 35,062 modENCODE TF binding regions (TFBRs) (8, 9, 11) into two classes: those containing a promoter (Prom-TFBR; n = 8,388) and those not containing a promoter, which are likely enhancers (Enh-TFBR, n = 26,674). We found that border intergenic regions more often contain an Enh-TFBR and have a higher density of Enh-TFBRs than those in active or regulated domains (Fig. 5). In contrast, intergenic regions within active domains more often have a Prom-TFBR and have a higher density of Prom-TFBRs than intergenic regions in borders or regulated domains (Fig. 5). Therefore, the properties of intergenic regions in borders are different from those in domains, as they are longer and more enriched for Enh-TFBRs. The location of extended transcriptional regulatory regions between active and regulated chromatin suggests that transcriptional activity may play a role in separating chromatin domains.
Discussion
Here we derived chromatin state maps for two developmental stages of C. elegans and used the resulting chromatin states to investigate chromatin domain organization. We show that C. elegans autosomes are subdivided into extended chromatin domains of differing activity separated by border regions containing long, regulatory element-rich, intergenic regions. Chromatin domain positions are remarkably consistent between EE and L3 larvae, despite the two stages being comprised of nearly nonoverlapping cell types. Therefore, chromatin domain organization appears to be a basal property of the genome.
Fig. 6 shows a simple model to explain our observations. The two types of chromatin domains contain different types of genes and are subject to different modifications. “Active” domains contain broadly expressed genes and “regulated” domains contain genes that have regulated expression. Genes in active domains are expressed in the germ line and are subject to H3K36me3 marking by MES-4 there (27–29). This modification is inherited and maintained in EE by maternally provided MES-4. When gene expression is zygotically activated in the embryo, these genes would be marked by the transcription-coupled H3K36 histone methyltransferase MET-1 (26–28), preserving this pattern somatically. Genes with developmentally or conditionally regulated expression lie in regulated domains that are marked by H3K27me3. These genes show low or no H3K36me3 modification. Although the profiling in mixed tissues done here may have limited the ability to detect tissue-specific H3K36me3, the results suggest that regulated genes acquire no or only low levels of H3K36me3 when they are transcribed. In support of this possibility, a recent study showed that regulated genes are not marked by H3K36me3 when they are expressed (30). We propose that H3K36me3 marking may be specifically relevant to genes with stable expression across development, possibly aiding the stability of their expression. Consistent with this idea, it was recently demonstrated that H3K36me3 marking plays a role in gene-expression stability during aging in C. elegans (31). H3K36me3 could also play a role in the preservation of chromatin domain structure.
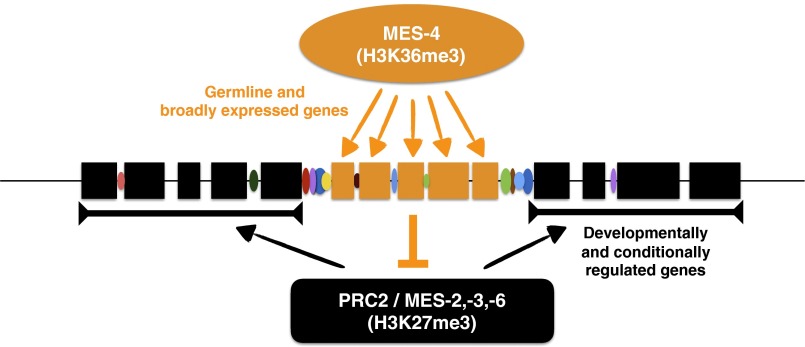
Fig. 6.
Model for regulation of chromatin domains by MES-4, PRC2, and transcription regulatory regions. The genome is subdivided into domains of germ-line and broadly expressed genes (orange boxes), and domains of genes with developmentally or conditionally regulated expression (black boxes). MES-4 marks germ-line–expressed genes with H3K36me3, which inhibits deposition of H3K27me3 by PRC2 (23, 27), leading to demarcation of chromatin domains. Borders separating domains contain long intergenic regions enriched for TF binding (colored ovals); these regulatory regions may play a role in domain separation.
The Polycomb-repressive complex PRC2 can generate H3K27me3, H3K27me2, and H3K27me1, although it is not well understood how the different levels of modification are regulated (32–36). A large body of work has shown that PRC2 functions to maintain and propagate transcriptional repression, but it can also be permissive for transcriptional activation (37–39). Similar to the patterns analyzed here for C. elegans, broad domains of H3K27me3 or H3K27me2 laid down by PRC2 that anticorrelate with high gene activity or H3K36me3 have been observed in other organisms (23, 34, 40–43). This anticorrelation is consistent with the inhibition of PRC2 activity by H3K36me3 and other histone modifications associated with gene activity (32, 33, 35, 36, 44, 45). These patterns suggest that PRC2, together with features of active chromatin such as H3K36me3, and interactions between them, may play a conserved role in the formation of domains of differing chromatin activity.
Although the experiments performed here on whole animals (EE and larvae) captured the high similarity of chromatin domains in development, the mixed tissues precluded our ability to study how chromatin domains might be regulated in individual cell types. Performing similar studies using purified cell types would be needed to investigate this question.
The features of the border regions between active and regulated domains suggest a role for transcription regulation in separating domains. We observed that intergenic regions at borders are generally longer than those in active or regulated domains and are more enriched for TF binding sites distal from promoters, which are likely to define enhancers or other regulatory elements, such as insulators. These properties of borders, combined with their location between active and regulated chromatin domains, suggest that transcription regulatory regions may be involved in domain separation. For example, the binding of factors to borders might generate a blocking structure or a platform for interactions. It is also possible that transcriptional activity or the generation of chromatin accessibility is important. Border regions show high chromatin accessibility, as do functional boundaries in other organisms even when neighboring genes are not active (46–50). Mechanisms operating at border regions could also act in conjunction with those actively specifying domains.
A future goal would be to understand the relationship between chromatin domains, which reflect chromatin activity, and 3D structure. Using techniques to measure chromatin interaction frequencies (“C” methods), it has been shown that chromatin has different levels of 3D organization within the nucleus, from broad chromosome territories to smaller scaled topologically associating domains (49, 51–54). Chromatin interaction mapping gives information about overall genomic structure, but not underlying genomic activity, whereas chromatin state mapping provides information on chromatin composition, but not on physical interactions. A Hi-C chromatin interaction map for C. elegans was recently published, defining 10–20 large interaction domains per chromosome, each containing an average of ∼200 genes (54). This is much higher than the 7–12 genes in human, mouse, and Drosophila topologically associating domains (49, 51–53), suggesting that these C. elegans domains may be functionally different. Because the chromatin domains defined herein contain an average of five to nine genes (median of four), the large C. elegans Hi-C–derived domains would each harbor many chromatin domains. Unfortunately, the resolution of the C. elegans Hi-C domain boundaries (10 kb) (54) is currently not sufficient for a comparison with the border locations defined here. Future higher-resolution chromatin interaction studies will be needed to determine how C. elegans chromatin domains relate to 3D structure.
In summary, our results point to roles for germ-line chromatin marking and transcription regulatory regions at chromatin domain borders in organizing the genome into functional domains and provide a framework for studies of chromatin structure and function in C. elegans. The future identification and functional analyses of sequences and factors that control chromatin structure will allow a better understanding of the mechanisms and functions of genome domain organization.
Materials and Methods
Datasets and Processing.
The datasets used for generating the chromatin state maps were EE and L3 ChIP-chip or ChIP-seq histone and histone modification data (7, 10). Data are available from the Gene Expression Omnibus (GEO) and data.modencode.org/ (Table S3 gives GEO accession numbers). For ChIP-chip data, log ratios of experiment signal over input signal were normalized, z-scored, and then averaged over replicates. Probes assigned to repeat regions were omitted. ChIP-seq data were processed using BEADS (55) at 1-bp resolution and averaged for the matching 50 bases of the ChIP-chip probes, then logged and z-scored. The data were then corrected for outliers by considering a moving window of nine probes: the central value was replaced by the average of the adjacent values, if (| x − m |)/s > 3, where x is the central value and m and s are the mean and sample variance of the remaining eight values. The data were reduced to the set of probes for which data were available for all 17 marks.
Table S3.
Datasets and GEO accession numbers
| Description | Stage | GEO accession no. | Used for chromatin state maps |
| H3 | EE | GSE22722 | x |
| H3 | L3 | GSE22734 | x |
| H3K27ac | EE | GSE22748 | x |
| H3K27ac | L3 | GSE25355 | x |
| H3K27me1 | EE | GSE26178 | x |
| H3K27me1 | L3 | GSE26179 | x |
| H3K27me3 | EE | GSE26180 | x |
| H3K27me3 | L3 | GSE22730 | x |
| H3K36me1 | EE | GSE22744 | x |
| H3K36me1 | L3 | GSE26181 | x |
| H3K36me2 | EE | GSE22717 | x |
| H3K36me2 | L3 | GSE22729 | x |
| H3K36me3 | EE | GSE22719 | x |
| H3K36me3 | L3 | GSE22731 | x |
| H3K4me1 | EE | GSE22747 | x |
| H3K4me1 | L3 | GSE25357 | x |
| H3K4me2 | EE | GSE22741 | x |
| H3K4me2 | L3 | GSE22732 | x |
| H3K4me3 | EE | GSE49739 | x |
| H3K4me3 | L3 | GSE28770 | x |
| H3K79me2 | EE | GSE22738 | x |
| H3K79me2 | L3 | GSE26182 | x |
| H3K79me3 | EE | GSE22739 | x |
| H3K79me3 | L3 | GSE26183 | x |
| H3K9me2 | EE | GSE22740 | x |
| H3K9me2 | L3 | GSE22733 | x |
| H3K9me3 | EE | GSE22720 | x |
| H3K9me3 | L3 | GSE22728 | x |
| H4K20me1 | EE | GSE22754 | x |
| H4K20me1 | L3 | GSE25358 | x |
| H4K8ac | EE | GSE26184 | x |
| H4K8ac | L3 | GSE25356 | x |
| HTZ-1 | MxE | GSE50302 | x |
| HTZ-1 | L3 | GSE49717 | x |
| Pol II | EE | GSE25788 | |
| Pol II | L3 | GSE25792 | |
| H3K36me3 | EE | GSE38180 | |
| H3K27me3 | EE | GSE38180 | |
| MES-4 | EE | GSE38180 | |
| H3K36me3 | mes-4 RNAi EE | GSE38159 | |
| H3K27me3 | mes-4 RNAi EE | GSE38159 |
Throughout, genome coordinates used were WS220. Other data used: C. elegans WS224 gene positions lifted over to WS220 coordinates (www.wormbase.org); operon annotations from Wormbase WS220; definition of arm and center chromosomal regions (14); categories of noncoding genes from WS220; odorant receptors (hand curated list from C. Bargmann, Rockefeller University, New York, based on ref. 20); tissue-specific genes (n = 748), genes core enriched in neurons, intestine, hypodermis, body wall muscle, or coelomocytes from (56); repeats from Dfam2.0 (22); ubiquitous genes (n = 2,575) (27); promoter and enhancer annotations (57); RNA Polymerase II ChIP (11) (EE: GEO accession no. GSE25788, L3: GEO accession no. GSE25792); MES-4 ChIP (23) (GEO accession no. GSE38180); H3K36me3 and H3K27me3 ChIP in wild-type and mes-4 RNAi EE (23) (GEO accession nos. GSE38180 and GSE38159); stage-specific RNA-seq data (24). Silent genes were defined as those with <0.005 dcpm in EE, LE, L1, L2, L3, L4, and YA hermaphrodite RNA-seq from ref. 24) (n = 1,921). Maternally expressed genes were defined using Cel-seq data profiling the AB and P1 blastomeres of two cell-stage embryos (58). RNA in AB and P1 blastomeres is maternally contributed (and therefore germ line-expressed) because these blastomeres have negligible gene expression. Genes with rpkm (reads per kilobase of transcript per million mapped reads) > 0 in both AB and P1 were classified as maternal (n = 7,980). Genes with broad or regulated expression were defined based on gene-expression variability scores, which are the CV in gene expression (ratio of the SD and mean expression) across 35 samples of different stages and cell types; CV values are from ref. 24. A low CV value indicates a gene has similar expression across all samples (broad expression), whereas a high CV indicates a gene has high variation in expression across samples (regulated expression). Because genes with very low expression often have high CV values, we only considered those with moderate to high expression in at least one developmental stage (dcpm >0.2, n = 13,739). Genes in the bottom third of these CV values were defined as having broad expression and genes in the top third of CV values were defined as having regulated expression. Metagene plots and heatmaps were generated using Seqplots (przemol.github.io/seqplots/). Condition-specific gene expression was assessed using copper-induced genes from table S4 of ref. 59, heat-induced genes with log2(fold change) > 1.5 from table S1 in ref. 60, and genes induced by three bacterial pathogens from table S10 in ref. 61. The IGV Integrative Genome Viewer was used to visualize data (62, 63).
Generating the Chromatin State Models.
We chose 20 states as a practical compromise between capturing the complexity of biological features and ease of interpretation; models with a larger number of states contained states that were superficially similar to each other. The states were found using a standard HMM for multivariate gaussians (each state having parameters for mean and covariance of the marks), with v1.0.4 of RHMM (crantastic.org/packages/RHmm) (64), with runtime parameters: nStates = 20, dis = “NORMAL”, and control = list (verbose = 2, init = “KMEANS”, iter = 500). Because whole-genome state maps separated into autosomal and X chromosome-specific states, we generated autosomal and chromosome X chromatin state maps for each stage: EE autosomes, EE Chr X, L3 autosomes, and L3 Chr X. For each, 40 replicate chromatin state HMMs were generated from different random number seeds. To assess the consistency of the 40 replicates, we first matched states between every pair of replicates so that they are comparable. The Jaccard Index (ratio of the length of intersection between two regions to the union of genomic coverage of the two regions) was calculated between every state in one replicate and every state in the other, forming a 20 × 20 similarity matrix from which the pair of states with highest similarity was matched iteratively until all states were matched. Then, an overall Jaccard Index was calculated between every pair of replicates, either from the same stage (e.g., between matched states in the 40 EE autosome replicates) or between stages (e.g., EE autosome to L3 autosome). Within a stage, the replicate 20 state models were very similar to each other (Fig. S2A); one from each set was chosen for analysis (EE autosome, EE Chr X, L3 autosome, L3 Chr X). Chromatin states were matched between the chosen EE autosome and L3 autosome map and between the EE Chr X and L3 Chr X maps by maximizing the Jaccard Index. Fig. S2 B and C shows the similarity between individual EE and L3 autosomal chromatin states in the chromatin state maps analyzed in this report.
Because the chip probes are 50-bases long and 147 bases wrap around one nucleosome, chromatin states of one or two probes were considered too short to be biologically meaningful and so reassigned to adjacent states. Segments of exactly one probe were assigned alternately to the left or right state: segments of exactly two probes were split, the first probe assigned to the left state, the second to the right. Where a continuous instance of a state was defined, for example for statistics on length of states, a run of consecutive probes of the same state including gaps between probes up to a gap of 500 bases was used. Probes on either side of larger gaps were treated as being in different individual states. Dataset S1 gives coordinates of the 20 states in each of the four maps. In feature charts, the cells show fold-enrichment on a log2 scale. Cells were colored gray if there were too few data points for statistical confidence.
Definition of High and Low Activity Domains.
Autosomal domains were defined from the chromatin states as follows: states 1–5 (the most strongly associated with the highest quintile of gene expression) were defined as active; states 16–20 (the most strongly associated with the lowest quintile of gene expression) were defined as inactive; and states 6–15 were defined as neutral. Regions containing active states and the neutral states among them without interruption by states 16–20 were defined as high activity domains (later renamed “active domains”), ending with an active state. Regions containing inactive states and the neutral states among them without interruption by states 1–5 were defined as low activity domains (later renamed “regulated domains”), ending with an inactive state. Regions between active and regulated domains were defined as borders. Single active states between two inactive states and single inactive states between two active states were considered neutral to prevent singleton states breaking up domains; this occurred in only a small number (∼10%) of domains. Domains with less than 50% of their length covered by states were removed. Dataset S2 gives coordinates for regulated, border, and active regions in EE and L3. Table S1 gives the number of domains of each type, lengths, and number of genes per domain, along with expected numbers based on the simulation described below. To count the number of genes per domain, protein-coding genes were assigned to the domain that overlaps their midpoints. The expected number of genes per domain was obtained by permutation, in which domains were called from randomly shuffled chromatin states (Table S1). To control for the positive effect of genic organization on domain length (e.g., association of promoter, transcription elongation, gene end states, and operons), states associated with the same gene were shuffled as a single unit and operon genes were shuffled together and considered single genes. The permutation was repeated 100 times.
Intergenic Region Length and TF Binding Analyses.
Intergenic regions were defined as regions on autosomes between annotated protein coding genes, excluding those where genes overlap, and those separating genes in operons (n = 13,705). To avoid biases caused by outliers, we excluded the top and bottom 10% of intergenic region lengths. Intergenic regions were assigned to the domain which had the largest value of reciprocal overlap as defined as (length_overlap)2/(length_domain × length_IGR). TF binding regions (n = 35,062) were merged modENCODE TF peak calls for 90 C. elegans factors (Supplemental File S1 in ref. 65); data are from refs. 8, 9, and 11. Promoter annotations were protein coding transcription start sites (TSSs) from refs. 57 and 66. For genes with no TSS annotation in either set Wormbase TSSs were used. TF binding regions were annotated as promoters if they overlapped a TSS annotation (Prom-TFBR, n = 8,388). TF binding regions that did not overlap the TSS set were annotated as likely enhancers (Enh-TFBR, n = 26,674). TF binding regions were assigned to intergenic regions on the basis of simple overlap. Differences in the distributions of intergenic lengths and TF binding site densities in different chromosome domains were tested using the Mann–Whitney U test. We tested if the proportions of intergenic regions hosting TFBRs are the same in different chromosome domains using a z-test.
Supplementary Material
Acknowledgments
We thank R. Chen, T. Gaarenstroom, C. Gal, and J. Janes for helpful comments on the manuscript; P. Kharchenko for advice in the early stages of the project; and C. Bargmann, J. Ho, B. Alver, and L. Hillier for providing processed or curated data. N.H., P.S., M.A.C., and J.A. were supported by Wellcome Trust Senior Research Fellowship 054523 or 101863 (to J.A.); K.J.E. and T.A.D. were supported by Wellcome Trust Research Career Development Fellowship 083563 (to T.A.D.); and J.A. and T.A.D. received core funding from the Wellcome Trust (092096) and Cancer Research UK (C6946/A14492).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608162113/-/DCSupplemental.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 7.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21(2):227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu W, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21(2):245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle AP, et al. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512(7515):453–456. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho JW, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512(7515):449–452. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerstein MB, et al. modENCODE Consortium Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41(3):376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141(1):159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5(3):e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119(1):73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau AC, Csankovszki G. Balancing up and downregulation of the C. elegans X chromosomes. Curr Opin Genet Dev. 2015;31:50–56. doi: 10.1016/j.gde.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vielle A, et al. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 2012;8(9):e1002933. doi: 10.1371/journal.pgen.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells MB, Snyder MJ, Custer LM, Csankovszki G. Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol Cell Biol. 2012;32(9):1710–1719. doi: 10.1128/MCB.06546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beisel C, Paro R. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet. 2011;12(2):123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 20.Robertson HM, Thomas JH. The putative chemoreceptor families of C. elegans. Wormbook. 2006;6:1–12. doi: 10.1895/wormbook.1.66.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83(2):207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 22.Hubley R, et al. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44(D1):D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Reports. 2012;2(5):1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512(7515):445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinke V, Cutter AD. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics. 2009;181(4):1219–1228. doi: 10.1534/genetics.108.099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134(16):2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 27.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6(9):e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3(1):15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bender LB, et al. MES-4: An autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133(19):3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Lluch S, et al. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat Genet. 2015;47(10):1158–1167. doi: 10.1038/ng.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pu M, et al. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29(7):718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 33.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 34.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53(1):49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz YB, et al. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet. 2010;6(1):e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8(1):9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20(3):266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Pauler FM, et al. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19(2):221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38(6):700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 42.Heger P, Marin B, Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HG, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 2015;25(8):1170–1181. doi: 10.1101/gr.188920.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14(18):1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- 45.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 46.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315(5817):1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 47.Fourel G, et al. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2(2):124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48(3):471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Bortle K, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15(6):R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148(3):458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523(7559):240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011;39(15):e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21(2):325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen RA, et al. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res. 2013;23(8):1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Reports. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Hattori A, Mizuno T, Akamatsu M, Hisamoto N, Matsumoto K. The Caenorhabditis elegans JNK signaling pathway activates expression of stress response genes by derepressing the Fos/HDAC repressor complex. PLoS Genet. 2013;9(2):e1003315. doi: 10.1371/journal.pgen.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engelmann I, et al. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One. 2011;6(5):e19055. doi: 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taramasco O, Bauer S. 2013. RHmm: Discrete, univariate or multivariate gaussian, mixture of univariate or multivariate gaussian HMM functions for simulation and estimation. Available at crantastic.org/packages/RHmm. Version 1.0.4.
- 65.Chen RA, et al. Extreme HOT regions are CpG-dense promoters in C. elegans and humans. Genome Res. 2014;24(7):1138–1146. doi: 10.1101/gr.161992.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.