Abstract
DNA damage interferes with the progression of transcription machineries. A tight coordination of transcription with signaling and repair of DNA damage is thus critical for safeguarding genome function. This coordination involves modulations of chromatin organization. Here, we focus on the central role of chromatin dynamics, in conjunction with DNA Damage Response (DDR) factors, in controlling transcription inhibition and restart at sites of DNA damage in mammalian cells. Recent work has identified chromatin modifiers and histone chaperones as key regulators of transcriptional activity in damaged chromatin regions. Conversely, the transcriptional state of chromatin before DNA damage influences both DNA damage signaling and repair. We discuss the importance of chromatin plasticity in coordinating the interplay between the DDR and transcription, with major implications for cell fate maintenance.
Keywords: DNA damage, transcription regulation, transcription coupled repair, histone dynamics, chromatin modifications, genome integrity
Introduction
Our genome is constantly assaulted by genotoxic agents, which not only compromise the integrity of DNA and its packaging into chromatin [1] but also perturb cell transcriptional programs by interfering with the progression and function of transcription machineries [2,3]. If left unrepaired, DNA damage can lead to the production of aberrant transcripts or to prolonged transcription inhibition, with adverse cellular outcomes including cell death and disease. A tight control of transcription, in close coordination with signaling and repair of DNA damage, is thus critical for safeguarding cellular functions.
In this review, we focus on the central role of chromatin dynamics, in conjunction with DNA Damage Response (DDR) factors, in regulating transcription at sites of DNA damage in mammalian cells.
Role of DDR factors in transcriptional regulation of damaged genes
In addition to regulating the expression of stress-responsive genes, DNA lesions result in transcriptional inhibition in damaged chromatin regions, which is critical to avoid interference between transcription and repair machineries [2]. DNA damage-induced transcriptional arrest is rapid and transient, as initially revealed by analyzing the incorporation of radiolabeled uridine analogs into nascent transcripts in UltraViolet (UV)-irradiated human fibroblasts [4]. Such control of transcriptional activity is localized to sites of DNA lesions, as shown by irradiating cells through micropore filters [5], and recovery of RNA synthesis after UV damage is dependent on proficient UV damage repair [4,5]. This response is not restricted to UV damage as RNA synthesis is also excluded from γH2A.X foci forming upon exposure of cells to ionizing radiation (IR), a DNA Double-Strand Break (DSB) inducing agent, suggesting that transcription is also suppressed at DSB sites [6]. Profiling RNA polymerase II (RNAPII) occupancy and nascent transcript production in the vicinity of the breaks showed that transcription is inhibited at damaged genes or at genes immediately adjacent to DSBs induced by site specific endonucleases [7–9]. In addition to impacting RNAPII transcription, DNA damage also inhibits the synthesis of ribosomal RNA by RNAPI in mammalian cells [10,11] but the underlying mechanisms deserve further investigation.
Identifying the molecular players regulating transcription in damaged chromatin has been the focus of intense research, providing evidence for the existence of repair factors dedicated to the repair of DNA lesions in transcribed genes. Cells have indeed developed specific pathways for rapid removal of DNA lesions on the transcribed DNA strand, called Transcription Coupled Repair (TCR), primarily characterized in the context of Nucleotide Excision Repair of UV damage (reviewed in [2,12,13]), and recently described after DSB induction in yeast [14]. UV damage and more generally bulky adducts are RNAPII blocking lesions, leading to the accumulation of stalled RNAPII molecules at sites of DNA damage and thus transcription inhibition. TCR factors, Cockayne Syndrome (CS) proteins and UV-stimulated scaffold protein A (UVSSA) in particular, are required both for efficient repair of these lesions and for transcription recovery, which involves displacement of stalled RNAPII and/or its targeting to ubiquitin-dependent proteasome degradation (reviewed in [2,13]). In addition to repair factors, recent studies have implicated transcription factors in regulating transcription recovery post UVC damage. For instance, Eleven-nineteen Lysine-rich Leukemia (ELL) has been proposed to serve as a docking platform for factors that would stimulate RNAPII restart [15], and transcription factor IIS (TFIIS) would facilitate the backtracking of RNAPII stalled at DNA lesions [16]. Unlike the response to UV damage, which directly blocks RNAPII progression, transcription arrest at sites of DSBs is controlled by DNA damage sensor proteins. Indeed, the signaling kinase Ataxia Telangectasia Mutated (ATM) and the DSB repair factor DNA-dependent Protein Kinase (DNA-PK) were identified as key players of break-mediated inhibition of transcription in response to FokI and I-PpoI induced breaks, respectively [8,9]. It was also suggested that Poly(ADP-Ribose) Polymerase (PARP) facilitates transcriptional silencing at DSBs [17]. It is conceivable that the localization of the breaks, their nature and their number may influence which DSB sensor is required for transcriptional arrest. Furthermore, it is still to be determined whether turning off the enzymatic pathways and reversing the modifications involved in transcription arrest is sufficient for transcription restart in response to DNA breaks or if additional mechanisms are at play.
Importantly, DDR factors identified as regulators of transcriptional arrest or recovery at damaged sites act on a chromatinized template, where DNA is wrapped around histone proteins to form nucleosomes. It is thus crucial to integrate chromatin dynamics in the control of damaged gene expression.
Importance of chromatin dynamics in the transcriptional regulation of damaged genes
Transcription and DNA damage repair both require a transient disorganization of chromatin, underlying the importance of chromatin plasticity in regulating these processes.
Chromatin modifications
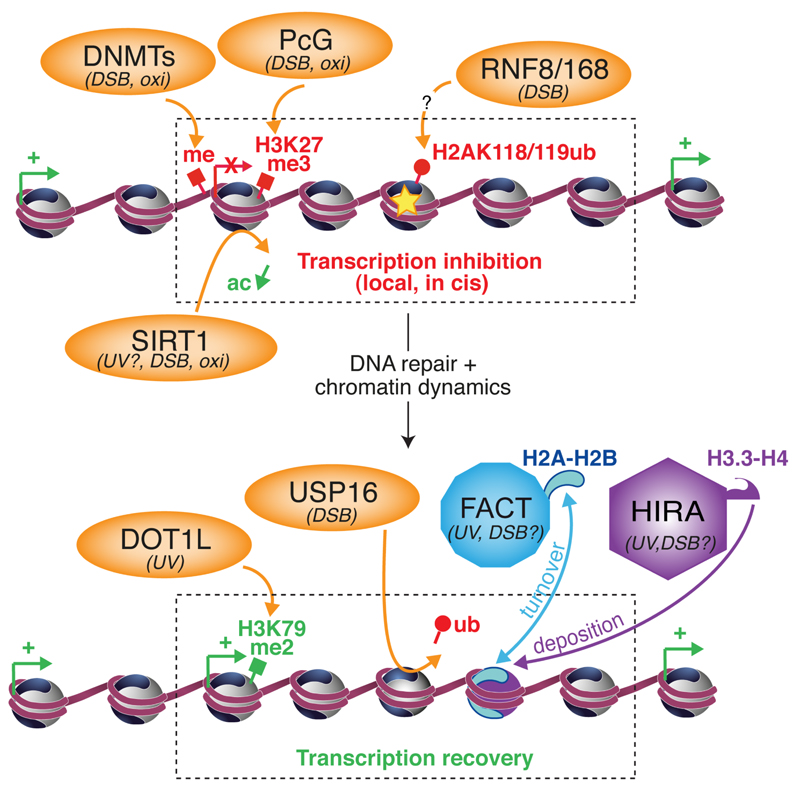
Chromatin modifications - affecting both the DNA and histone proteins - have been involved in the transcriptional regulation of damaged genes. Indeed, ubiquitylation of H2A histones, a modification implicated in transcription repression, was found enriched at DSBs and associated with the ATM-dependent transcription silencing in response to FokI-induced breaks [8] (Figure 1). However, ubiquitylated H2A was not involved in response to I-PpoI-mediated cuts [9], again suggesting that how and where DSBs are induced in the genome may influence the underlying mechanism of transcriptional arrest. DSBs and oxidative damage, when introduced in CpG island-rich promoters, also trigger the accumulation of chromatin modifiers, including DNA methyltransferases (DNMT 1 and 3a), Polycomb group proteins (PcG) along with histone deacetylases (SIRT1, Sirtuin 1) [18,19] (Figure 1). The targeting of these well-characterized mediators of transcription repression correlates with the enrichment of histone marks associated with transcription silencing (hypoacetylation, H3K9 and K27 trimethylation). Similarly, trimethylated H3K27 was observed at DNA damage foci induced by ion micro-irradiation, with a concomitant exclusion of H3K4 trimethylation, a histone modification typical of actively transcribed chromatin [20]. SIRT1 deacetylase activity has also been involved in suppressing RNA synthesis in response to UV damage in repair deficient cells [21], but whether it operates similarly in repair proficient cells deserves further investigation. In addition to these repressive chromatin modifiers, several heterochromatin components and marks, generally associated with transcription repression, are found enriched at damaged sites [1,22,23], but it is not yet clear if they actually contribute to the transcriptional silencing of damaged chromatin.
Figure 1. Importance of chromatin dynamics in the transcriptional regulation of damaged genes.
Chromatin modifiers (orange) and histone chaperones (blue, purple) involved in regulating transcription inhibition (top) and recovery (bottom) in damaged chromatin regions in response to DSBs, UV lesions, oxidative damage (DNA damage is represented by a star). DNA-damage-responsive chromatin modifications associated with active transcription (green) and silencing (red) are represented. ac : acetylation ; me : methylation ; ub : ubiquitylation. Given that RNF8/168 ubiquitin ligases are not known to target K118/119 residues on H2A, an intermediate factor is likely involved as indicated by a question mark.
DNA damage-induced histone modifications also impact on the reactivation of gene expression after repair of DNA lesions. Indeed, Ubiquitin Specific Protein 16 (USP16) relieves ATM-mediated transcription silencing at DSB sites, most likely via its capacity to deubiquitylate H2A [8] (Figure 1). H3K79me2, a histone mark enriched in actively transcribed regions, accumulates transiently at gene promoters upon UVC irradiation, and its down-regulation by knockout of the methyltransferase Disruptor of Telomeric silencing-1 homolog (DOT1L) prevents transcription recovery after UV damage in mouse fibroblasts [24]. Treating cells with the histone deacetylase inhibitor Trichostatin A, which results in chromatin decompaction, rescues the DOT1L phenotype, suggesting that DOT1L-dependent methylation of H3K79 may favor an open chromatin structure around the UV-repressed gene promoters, which facilitates transcription re-initiation after repair (Figure 1). In light of these data, it would be of major interest to perform in depth analysis of the dynamic changes of activating and repressive histone marks after DNA damage in order to identify additional chromatin modifying factors involved in transcription regulation after genotoxic stress.
Histone mobilization
Not only does genotoxic stress alter histone modification profiles at gene promoters, but it also leads to the mobilization of histone proteins [25]. Chromatin remodeling complexes are key players in histone dynamics and they participate in signaling and repair of DNA lesions [26]. However, their contribution to DNA damage-induced transcription regulation is far from being completely understood. It has been proposed that PARP-dependent remodelers could mediate transcription silencing at DNA breaks [17], but there is still no direct evidence in the literature supporting such model. The remodeling activity of the repair factor CSB is important for UV resistance and not for recruiting downstream TCR factors to chromatin, it is thus tempting to speculate that this activity plays a role in transcription recovery [27]. Besides chromatin remodeling factors, histone chaperones can mobilize histones in and out of chromatin. Recent studies have unveiled the importance of two histone chaperone complexes in transcription restart following UV damage in human cells, the H3.3-specific chaperone Histone Regulator A (HIRA) and the H2A-H2B chaperone Facilitating Chromatin Transcription (FACT) (reviewed in [28]) (Figure 1). HIRA is targeted to UV-damaged chromatin regions where it deposits newly synthesized H3.3 histones in a manner coupled to DNA damage detection and it facilitates transcription recovery after repair [29]. Because H3.3 histones generally bear modifications associated with active transcription and tend to make nucleosomes more labile [30], these incorporated H3.3 histones have been proposed to constitute a chromatin bookmark that renders damaged chromatin prone for transcription restart upon completion of DNA repair by maintaining a plastic and transcription-permissive chromatin structure at sites of DNA lesions. In line with this hypothesis, it has been recently demonstrated that transcription induction in mouse embryonic stem cells is primed by H3.3 histone deposition, via an opening of chromatin at enhancers and promoters [31]. Given that HIRA and H3.3 also accumulate at DSB sites [29,32], it is tempting to speculate that HIRA-mediated deposition of H3.3 promotes transcription resumption at DSBs by a similar mechanism. In addition to its contribution to transcription recovery, it will be interesting to investigate whether the function of HIRA in stimulating transcriptional activation of stress-responsive genes described in yeast [33] is conserved in mammalian cells. Noteworthy, point mutations have been recently identified in H3.3 in several human cancers, some of them affecting H3 modification profiles [34,35]. In light of these findings, it will be important to assess the functional consequences of such mutations on transcription restart after UV damage, which could contribute to their oncogenic potential.
Similar to HIRA, the histone chaperone FACT was shown to promote transcription recovery after UV damage repair [36]. FACT is recruited to UV-damaged chromatin independently of DNA repair and accelerates H2A-H2B turnover in UV-damaged regions [36]. Such increased histone turnover may destabilize nucleosomes, facilitating backtracking of stalled RNAPII away form the DNA lesion, and ultimately transcription resumption. Stimulating histone exchange at sites of DNA damage may also be a way to dilute repressive histone marks associated with transcription silencing in damaged regions, thus favoring the restoration of a transcriptionally competent chromatin landscape. FACT also accumulates at IR-induced foci [37], suggesting a potential role for FACT in controlling transcription restart after DSB repair as well. Notably, FACT is also important for protecting cells against transcription-induced DNA damage by promoting the resolution of R-loops resulting from transcription-replication conflicts [38]. A detailed analysis of the underlying mechanism should bring new insights into how FACT controls the crosstalk between the DDR and transcription. Moreover, both HIRA and FACT could also indirectly regulate transcription recovery after UV damage, by promoting TCR of UV lesions, which deserves to be closely examined in future studies.
Altogether, significant advances have been made in identifying chromatin modifiers and histone chaperones as key regulators of transcriptional activity in damaged chromatin regions, highlighting the importance of chromatin dynamics in coordinating the interplay between the DDR and transcription.
Impact of chromatin transcriptional state on damage signaling and repair
Conversely, the transcriptional state of chromatin before DNA damage influences both DNA damage signaling and repair. For instance, heterochromatin, which is mostly transcriptionally silent, is known to be refractory to late steps of the DSB response, which are restricted to the periphery of heterochromatin domains (reviewed in [1]). Nevertheless, depletion of heterochromatin components that are targeted to sites of DNA breaks, such as heterochromatin protein 1 (HP1), impairs DSB signaling and repair, suggesting an active contribution of heterochromatin proteins to the DSB response.
In addition to regulating DNA damage signaling and repair, the pre-existing transcriptional state of chromatin may be instrumental for fine-tuning the repair process as recently demonstrated in the context of DSB repair. DSBs are processed by two major pathways, homologous recombination, which uses a homologous sequence for repairing the damaged DNA, and non homologous end-joining, which repairs DSBs by direct ligation of the two broken ends. A higher frequency of incorrect end use during end-joining of tandem DSBs was observed in an active transcription context in human cells [39]. Furthermore, DSBs located in actively transcribed chromatin regions, marked by the transcription elongation-associated histone modification H3K36me3, are preferentially repaired by homologous recombination because they facilitate the recruitment of factors involved in DNA-end resection [40,41]. However, studies in yeast reported an opposite function for H3K36me3 in promoting the end-joining process [42,43], suggesting that the role of H3K36me3 in determining DSB repair pathway choice is not strictly conserved during evolution.
Finally, recent data revealed that, unexpectedly, transcriptional activity is not completely inhibited in damaged chromatin as small non-coding RNA species are produced at DSB sites with a major role in controlling initial DDR steps [44–46]. This pathway is evolutionarily conserved but it has only been reported in the response to DNA breaks so far and future studies should examine whether it also occurs in response to other types of DNA lesions. Furthermore, it is still unclear how these DNA Damage Response RNAs (DDRNA) control DNA damage signaling and repair. It would be of major interest to investigate whether they cooperate with factors regulating chromatin plasticity to induce chromatin changes at sites of DNA breaks, which would stimulate the recruitment of DDR factors.
Conclusions
Recent work has shed light on the key regulatory role of chromatin dynamics, together with DDR factors, for stopping and re-establishing transcriptional programs after DNA damage. Chromatin can thus be viewed as an integration platform that coordinates DNA damage signaling and repair with transcription (Figure 2). It will be important to further dissect the molecular bases of this interplay between the DDR, chromatin and transcription and to understand how DDR factors work in concert with chromatin changes to orchestrate DNA repair and transcription control. This will help us to fully appreciate the consequences of disease-associated alterations in the chromatin landscape on genome integrity and function.
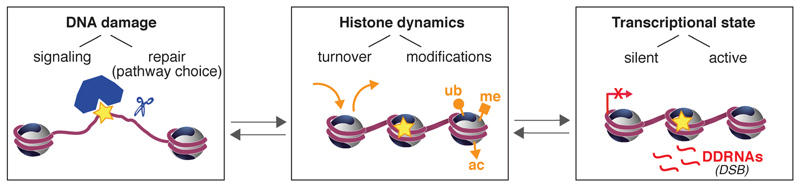
Figure 2. Histone dynamics at the core of the interplay between the DDR and transcription.
DNA damage-induced histone dynamics, including histone exchange and post-translational modifications, govern local changes in chromatin transcriptional activity. Conversely, chromatin transcriptional state, via histone modifications, modulates DNA damage signaling and repair.
Highlights.
DNA damage signaling and repair regulate transcription of damaged chromatin
Chromatin dynamics control transcription inhibition and recovery in damaged regions
Chromatin transcriptional state influences DNA damage signaling and repair
Acknowledgements
We thank Evi Soutoglou for critical reading of the manuscript. We apologize to colleagues whose work could not be cited due to space constraints. Research in the laboratory is supported by the European Research Council (ERC-2013-StG-336427 “EpIn”), the French National Research Agency (ANR-12-JSV6-0002-01), the “Who am I?” laboratory of excellence, EDF Radiobiology program and the Fondation ARC. S.A. is recipient of a PhD fellowship from University Pierre & Marie Curie.
References
- [1].Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- [2].Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- [3].Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- [5].Moné MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, et al. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Solovjeva LV, Svetlova MP, Chagin VO, Tomilin NV. Inhibition of transcription at radiation-induced nuclear foci of phosphorylated histone H2AX in mammalian cells. Chromosome Res. 2007;15:787–797. doi: 10.1007/s10577-007-1162-x. [DOI] [PubMed] [Google Scholar]
- [7].Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. Embo J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- [10].Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- [11].Calkins AS, Iglehart JD, Lazaro J-B. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Research. 2013;41:7378–7386. doi: 10.1093/nar/gkt502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- [13].Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harbor Perspectives in Biology. 2013;5:a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chaurasia P, Sen R, Pandita TK, Bhaumik SR. Preferential repair of DNA double-strand break at the active gene in vivo. J Biol Chem. 2012;287:36414–36422. doi: 10.1074/jbc.M112.364661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mourgues S, Gautier V, Lagarou A, Bordier C, Mourcet A, Slingerland J, et al. ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc Natl Acad Sci U S A. 2013;110:17927–17932. doi: 10.1073/pnas.1305009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jensen A, Mullenders LHF. Transcription factor IIS impacts UV-inhibited transcription. 2010;9:1142–1150. doi: 10.1016/j.dnarep.2010.08.002. [DOI] [PubMed] [Google Scholar]
- [17].Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].O'hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genetics. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].O'hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Seiler DM, Rouquette J, Schmid VJ, Strickfaden H, Ottmann C, Drexler GA, et al. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. 2011;19:883–899. doi: 10.1007/s10577-011-9244-1. [DOI] [PubMed] [Google Scholar]
- [21].Vélez-Cruz R, Zadorin AS, Coin F, Egly J-M. Sirt1 suppresses RNA synthesis after UV irradiation in combined xeroderma pigmentosum group D/Cockayne syndrome (XP-D/CS) cells. Proc Natl Acad Sci USA. 2013;110:E212–20. doi: 10.1073/pnas.1213076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vissers JHA, van Lohuizen M, Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci. 2012;125:3939–3948. doi: 10.1242/jcs.107375. [DOI] [PubMed] [Google Scholar]
- [23].Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oksenych V, Zhovmer A, Ziani S, Mari P-O, Eberova J, Nardo T, et al. Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack. PLoS Genetics. 2013;9:e1003611. doi: 10.1371/journal.pgen.1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Polo SE. Reshaping chromatin after DNA damage: the choreography of histone proteins. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cho I, Tsai P-F, Lake RJ, Basheer A, Fan H-Y. ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair. PLoS Genetics. 2013;9:e1003407. doi: 10.1371/journal.pgen.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mandemaker I, Vermeulen W, Marteijn J. Role of chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus. 2014;5 doi: 10.4161/nucl.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Adam S, Polo SE, Almouzni G. Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- [30].Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011:1–14. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang X, Li L, Liang J, Shi L, Yang J, Yi X, et al. Histone acetyltransferase 1 promotes homologous recombination in DNA repair by facilitating histone turnover. J Biol Chem. 2013;288:18271–18282. doi: 10.1074/jbc.M113.473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chujo M, Tarumoto Y, Miyatake K, Nishida E, Ishikawa F. HIRA, a conserved histone chaperone plays an essential role in low-dose stress response via transcriptional stimulation in fission yeast. J Biol Chem. 2012;287:23440–23450. doi: 10.1074/jbc.M112.349944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yuen BTK, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer Cell. 2013;24:567–574. doi: 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- [37].Oliveira DV, Kato A, Nakamura K, Ikura T, Okada M, Kobayashi J, et al. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci. 2014;127:763–772. doi: 10.1242/jcs.135855. [DOI] [PubMed] [Google Scholar]
- [38].Herrera-Moyano E, Mergui X, García-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014 doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. Journal of Biological Chemistry. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pfister SX, Ahrabi S, Zalmas L-P, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter ACG, et al. SETD2-Dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell Rep. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pai C-C, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ, et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nature Communications. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jha DK, Strahl BD. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nature Communications. 2014;5:3965. doi: 10.1038/ncomms4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- [46].Michalik KM, Böttcher R, Förstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Research. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]