Abstract
DNA damage signaling and repair machineries operate in a nuclear environment, where DNA is wrapped around histone proteins and packaged into chromatin. Understanding how chromatin structure is restored together with the DNA sequence during DNA damage repair has been a topic of intense research. Indeed, chromatin integrity is central to cell functions and identity. Yet, chromatin shows remarkable plasticity in response to DNA damage. This review presents our current knowledge of chromatin dynamics in the mammalian cell nucleus in response to DNA-double strand breaks and UV lesions. I provide an overview of the key players involved in regulating histone dynamics in damaged chromatin regions, focusing on histone chaperones and their concerted action with histone modifiers, chromatin remodelers and repair factors. I also discuss how these dynamics contribute to reshaping chromatin and, by altering the chromatin landscape, may affect the maintenance of epigenetic information.
Keywords: Chromatin remodeling, genotoxic stress, histone chaperones, histone variants, histone modifications
Introduction
The cell nucleus is a critical organelle that is central to cell functions as it stores genetic information within the DNA sequence. However, DNA molecules need to be highly compacted to fit the small nuclear volume. This compaction is achieved by the incorporation of DNA into chromatin [1], a repeated nucleoproteic structure whose basic unit is the nucleosome [2]. In the nucleosome core particle, DNA wraps around an octamer of histone proteins, composed of two H2A-H2B dimers flanking a (H3-H4)2 tetramer. Interactions between nucleosomes, stabilized by the association of linker histones, non-histone proteins and structural RNA components, drive further compaction of chromatin fibers [3,4].
Furthermore, histone proteins, through their post-translational modifications [5] and the existence of sequence variants [6], confer additional variations to chromatin structure and contribute to encoding epigenetic information, which regulates gene expression without changes to the DNA sequence. The maintenance of chromatin integrity with the inheritance of epigenetic information through cell generations is thus key for preserving cell functions and identity.
While chromatin is the physiological substrate for all DNA metabolic reactions, access of replication, repair and transcription factors is impeded by chromatin compaction. One of the key features of chromatin function is thus its exquisitely dynamic nature. Chromatin dynamics involve the concerted action of histone chaperones [7,8], histone modifying enzymes [5], and remodeling factors [9,10] and play a crucial role in response to DNA damage [11–14]. Arising from endogenous products of cell metabolism and from exposure to environmental mutagens [15], DNA damage indeed poses a major threat to genome stability and to the maintenance of chromatin organization. DNA damage detection and repair are accompanied by profound rearrangements of the nucleosomal fiber, as described in the Access/Prime-Repair-Restore model [16,17]. This model, based on seminal work by M. Smerdon and colleagues, predicts a transient disorganization of damaged chromatin, which primes chromatin for repair of DNA lesions, followed by restoration of chromatin structure. Chromatin thus serves as an integration platform that coordinates DNA repair with the maintenance of cellular functions.
In this review, I present our current knowledge of chromatin dynamics in response to DNA damage in the mammalian cell nucleus, focusing on the mobilization of histone proteins. I also discuss how studying damaged chromatin dynamics has provided exciting insights into the plasticity of epigenetic information in response to genotoxic stress, and I highlight future challenges in this rapidly expanding field.
Monitoring histone dynamics in response to DNA damage in human cells
Over the past decades, series of experimental approaches have been designed to monitor chromatin dynamics in response to DNA damage and to address how this may affect the maintenance of chromatin integrity.
Chromatin rearrangements: transient disorganization of nucleosomal fibers
Measuring DNA accessibility to nucleases within damaged chromatin in vivo was instrumental for revealing the transient disorganization of the nucleosomal fiber in response to DNA damage. Partial digestion of UV-damaged chromatin with Micrococcal Nuclease (MNase) or DNA Nuclease 1 (DNase I) indeed showed a transient increase in nuclease sensitivity of chromatin regions undergoing repair of UltraViolet C (UVC) lesions in confluent human fibroblasts [18,19]. These pioneering experiments have led to the Access/Prime-Repair-Restore model (Fig. 1), which is still the prominent view of the chromatin rearrangements triggered in response to DNA damage [16,17].
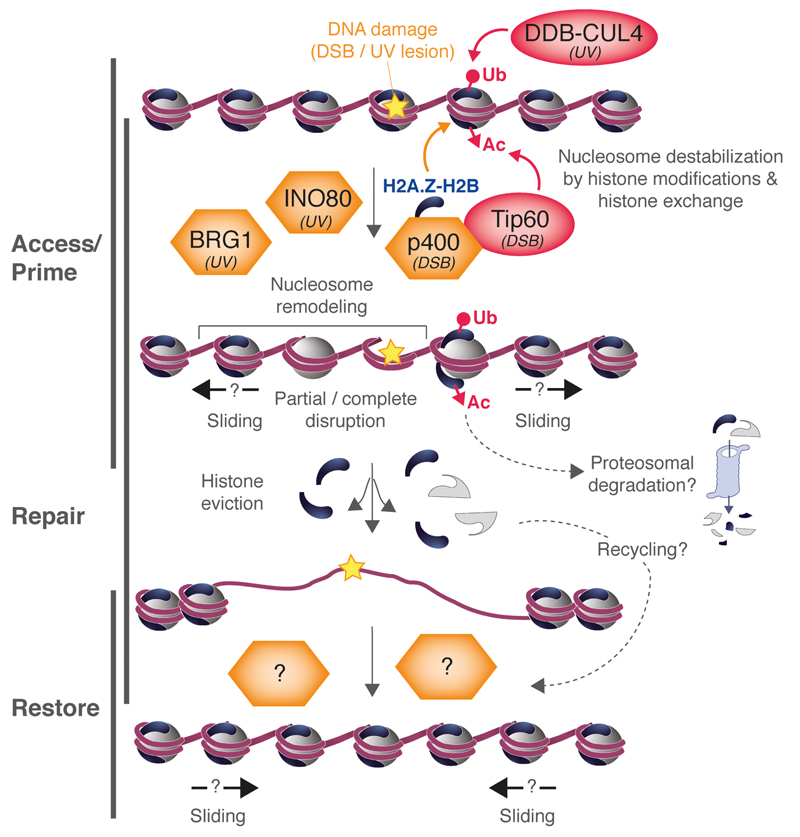
Fig. 1.
Role of histone modifying enzymes and remodeling factors in histone dynamics in response to DNA damage (DSBs or UV lesions). DNA damage-induced histone modifications (red) by acetylation (Ac) and ubiquitylation (Ub) promote nucleosome destabilization, and acetylation may drive histones to proteosomal degradation. The indicated nucleosome remodelers (orange) are involved in histone exchange, nucleosome sliding and/or disruption with histone eviction from damaged chromatin. Displaced histones may be re-positioned/re-deposited after repair of DNA damage. The contribution of remodelers to chromatin restoration is still to be determined.
Remarkably, these rearrangements can span several kilobases on chromatin [20] despite the small size the repair patch, which is around 30 nucleotides for Nucleotide Excision Repair (NER) of UVC damage in human cells [21]. Furthermore, a global relaxation of chromatin affecting the whole nucleus has been reported in response to local UVC irradiation, as highlighted by an increased sensitivity of chromatin to denaturation by hydrochloric acid [22]. This is in apparent contrast to the local expansion of chromatin observed at sites of DNA breaks induced by laser micro-irradiation in human cells expressing H2A or H2B histones tagged with a photoactivatable version of GFP (PA-GFP). The expansion of damaged chromatin at these sites is an active process that requires adenosine triphosphate (ATP) and Poly(ADP-ribose) Polymerase (PARP) activity [23,24]. However, alterations of chromatin compaction were not examined outside damaged areas in these studies. Thus, it is still unclear how far away from damaged regions chromatin disorganization actually spreads. Determining the extent of chromatin alterations will be critical to evaluate the impact of genotoxic stress on the epigenome overall. It will be particularly interesting to examine whether chromatin organization into nuclear domains regulates the spreading of chromatin disruption in response to DNA damage by imposing structural barriers to chromatin disorganization.
Histone mobility/displacement
Histone proteins are mostly incorporated into chromatin [25] and thus poorly mobile in human cells, as measured by Fluorescence Recovery After Photobleaching on cells expressing GFP-tagged histones [26]. However, a slight increase in core histone mobility, most pronounced for the H2A.X variant, has been observed after DNA break induction in nuclear regions exposed to laser micro-irradiation [27]. Consistent with these observations, histones are more readily extracted from chromatin in cells treated with DNA break-inducing agents like ionizing radiation (IR) or the radiomimetic drug bleomycin compared to undamaged cells [28,29]. IR-induced release of histones to the soluble fraction has also been reported [30]. Such nucleosome destabilization is an early and transient response to DNA damage, occurring around 30 min after acute genotoxic treatment.
Nucleosome destabilization in response to genotoxic stress results in histone removal from damaged DNA. Indeed, chromatin immunoprecipitation experiments at site specific DNA double strand breaks (DSBs) induced by the homing endonuclease I-PpoI in the human genome have revealed a loss of core histones over a region of 3 kilobases around DSBs [31,32]. Similar analyses in cells expressing the AsiSI restriction enzyme have also shown reduced histone H3 occupancy in the vicinity of DSBs [33]. Interestingly, nucleosomes are partially disassembled in G1 cells, with the displacement of H2A-H2B only, whereas all core histones are removed in asynchronous cells, most likely as a result of DSB repair by homologous recombination in S-G2 while DSBs are repaired by non-homologous end-joining in G1 [32]. Similar to the response to DSBs, a local reduction in core and linker histone density has also been observed at sites of UVC irradiation in human cells [34]. It is still to be determined whether this actually reflects histone eviction from chromatin due to the disruption of damaged nucleosomes and/or histone sliding away from the lesions as a result of nucleosome remodeling.
Histone accumulation and nucleosome restoration
The transient increase in nuclease sensitivity of repaired DNA observed in UV-irradiated human fibroblasts indicates that nucleosome disruption after DNA damage is only temporary. Nucleosome restoration after UV is revealed by the re-establishment of a canonical DNase I footprint [18] and is complete with deposition of linker histone H1 [35]. Nucleosome reassembly also occurs in response to DSBs. Indeed, the use of I-PpoI endonuclease fused to a destabilization domain to allow DSB repair showed that core histone removal from DSBs was transient. Histone levels around DSB sites are back to normal within 2 hours post damage [32].
Several in vitro assays have been devised for recapitulating nucleosome assembly coupled to DNA repair, including damaged plasmid supercoiling and histone deposition onto damaged DNA immobilized on magnetic beads [36,37]. More recently, the development of in vivo methods that discriminate between parental and newly synthesized histones was instrumental for investigating histone deposition during chromatin re-assembly after DNA damage. In particular, local UVC irradiation of human cells short term after transient transfection of tagged histones, H3.1-Flag-HA [38] or H2A-GFP [39], revealed an incorporation of these newly synthesized histones in damaged chromatin regions. Note that this incorporation represents higher histone exchange and does not reflect an accumulation of histone proteins in damaged regions as no local increase is detected in total histone levels. Given that post-translational modifications on new soluble histones are distinct from parental histone marks as shown for histone H3 [25], the replacement of parental histones by new histones is likely to dilute the original information conveyed by chromatin. Chromatin re-assembly after DNA damage thus represents a window of opportunity for modulating epigenetic information. New histone deposition may be accompanied by a recycling of parental histones displaced from damaged regions at the onset of the damage response, but the contribution of parental histones to the composition of repaired chromatin is still to be determined.
The dynamics of incorporation of newly synthesized H3 histones in damaged chromatin regions has been further explored by combining local UVC irradiation with SNAP-tag based imaging of new histones. Thus, it was shown that not only H3.1 but also H3.3 variants are deposited de novo at sites of UVC damage [40]. Interestingly, H3.3 has also been found to accumulate at DSBs [41]. The centromeric H3 variant Centromeric Protein A (CENPA) by contrast does not show de novo accumulation at UV sites, and although CENPA accumulation was initially reported at DSBs induced by laser micro-irradiation and by the endonuclease I-SceI [42], it has not been reproduced in recent studies [43].
Regarding outer core histones H2A and H2B, their incorporation rate is enhanced by approximately two folds at sites of local UVC irradiation in human cells, reflecting higher histone exchange in damaged chromatin [39]. Indeed, accelerated incorporation of fluorescently labeled H2A-H2B histones was observed at sites of UVC irradiation upon cell fusion or after photobleaching half of the nucleus. Interestingly, H3.1 and H4 did not show such a response in this experimental setting, which may reflect their lower mobility compared to H2A-H2B histones [26].
Concerning H2A variants, while no specific accumulation of H2A.X at damage sites has been reported so far, an enrichment of H2A.Z at DSBs was revealed by chromatin immunoprecipitation upon DNA cutting with a zinc-finger nuclease in silent chromatin regions [28]. However, similar experiments using the AsiSI restriction enzyme to induce site specific DSBs did not reveal H2A.Z enrichment [44]. It is possible that H2A.Z enrichment at DSBs is only detectable in poorly or non-transcribed regions of the genome, where H2A.Z basal levels are low.
The macroH2A1.1 variant, known for its ability to bind ADP-ribose via its macrodomain, is recruited to DNA breaks in human cells in a PARP-dependent manner [45–47]. Intriguingly however, macroH2A1.1 is not stably incorporated into nucleosomes at DSBs, as highlighted by the necessity to use crosslinking agents to reveal its binding to chromatin unlike intrinsic chromatin components [47].
Altogether, the studies presented in this section reveal significant alterations of the chromatin landscape in response to DNA damage, with a prominent role of histone dynamics. Recent work has shed light on the complex network of regulatory factors that ensure fine-tuning of histone mobilization in damaged chromatin as described below.
Regulatory factors controlling histone dynamics in response to DNA damage
Histone modifications and nucleosome remodeling
Histone proteins are targets of a wide range of post-translational modifications that regulate chromatin functions [5]. Among them, acetylation and ubiquitylation have been associated with histone dynamics in response to DNA damage (Fig. 1). In particular, H2A ubiquitylation by the UV damage detection complex comprising DNA Damage Binding Proteins 1 and 2 and Cullin 4 (DDB2-DDB1-CUL4) has been shown to destabilize damaged nucleosomes with eviction of ubiquitylated H2A in vitro [48], but it remains to be established whether it operates similarly in vivo. UV-induced ubiquitylation of H3 and H4 by the same complex has also been proposed to weaken the association of histones with DNA in human cells, facilitating histone release from nucleosomes [49]. Likewise, the increased mobility of H2A.X observed at laser-induced breaks has been linked to histone modifications, namely H2A.X acetylation by Tat-interacting protein 60 (Tip60, also known as KAT5) and subsequent ubiquitylation [27]. How such histone modifications govern histone mobility in response to DNA damage is a complex and still unresolved issue. Histone acetylation was recently shown to drive core histones to proteosomal degradation [50]. Histone modifications can also impair nucleosome stability directly by affecting histone-histone interactions and histone-DNA contacts, or through the recruitment of chromatin remodelers.
Chromatin remodelers use the energy from ATP hydrolysis to disrupt histone-DNA interactions, thus promoting nucleosome sliding, eviction or histone exchange [9,10]. The ATP-dependency of chromatin expansion in response to laser damage [23] and of histone displacement from UVC damage sites [34] is indicative of a possible involvement of remodelers in the dynamics of damaged chromatin. All families of remodelers have been implicated in the DNA damage response as they are recruited to damaged chromatin and/or contribute to damage repair and cell survival after damage [51]. However, only a few of them have been formally shown to stimulate histone dynamics in response to genotoxic stress (Fig. 1). The catalytic subunit of Tip60 remodeling complex - p400 - promotes histone H3 displacement from DSBs induced by the restriction enzyme AsiSI [33]. The remodeler brahma-related gene 1 (BRG1, also known as SMARCA4), and to a lesser extent inositol requiring mutant 80 (INO80), increase chromatin accessibility upon global UV irradiation in human cells as revealed by MNase digestion profiles [52,53]. However, it is unclear whether their nucleosome remodeling activity is restricted around DNA damage sites or affects the whole nucleus.
Current studies mainly support a role for remodelers in promoting chromatin accessibility by moving histones or nucleosomes away from the damage site. Nevertheless, this does not exclude a possible function in histone deposition, as recently shown for p400, which promotes H2A.Z enrichment at DSBs, thus facilitating subsequent chromatin alterations [29]. In addition to its ATP-dependent remodeling activity, a conserved amino-terminal region in p400 could mediate a histone chaperone function towards H2A.Z-H2B based on crystal structure studies of the budding yeast ortholog in complex with histones [54].
Histone chaperones
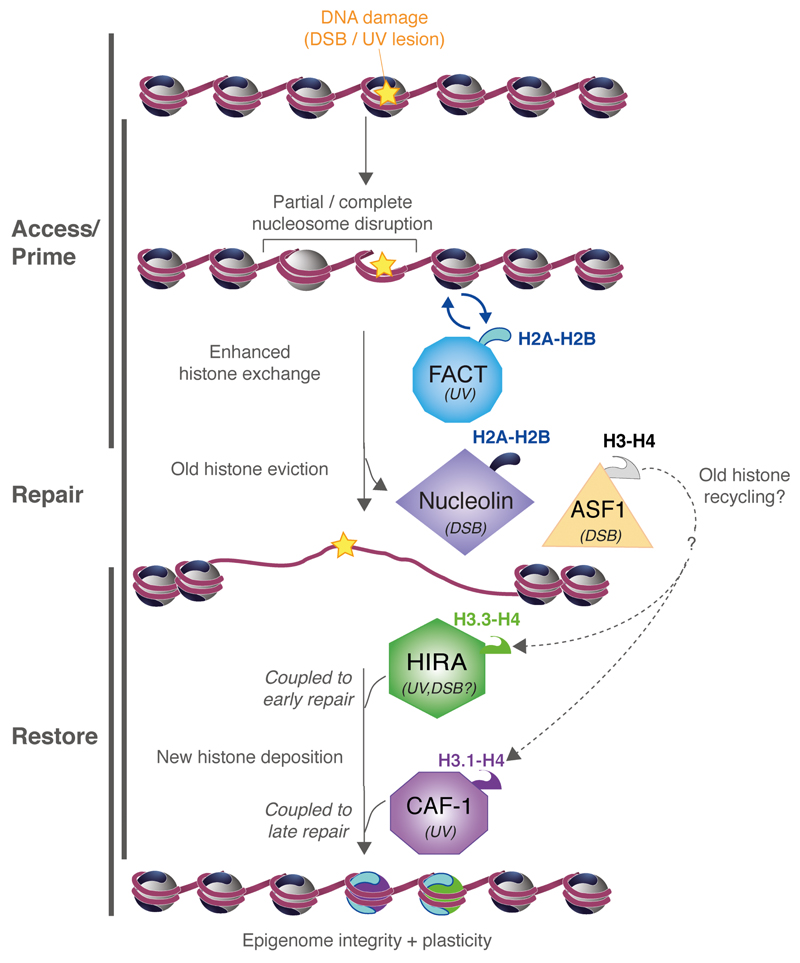
Histone chaperones escort histone proteins from their point of synthesis up to their deposition onto DNA [7,8]. Histone H3 chaperones [55] were first to be involved in regulating histone dynamics in response to DNA damage, in particular the H3.1-specific chaperone Chromatin Assembly factor -1 (CAF-1) [56,57], which stimulates histone deposition coupled to DNA synthesis [58,59]. CAF-1 indeed promotes chromatin assembly in response to UV damage in vitro [60], acting in synergy with another histone H3 chaperone named Anti-Silencing Factor 1 (ASF-1)[61]. In human cells, CAF-1 is recruited to UV-damaged chromatin [62,63] and is key for restoring the integrity of chromatin structure by mediating de novo incorporation of H3.1 histones after UV damage repair [38]. Recent work has shown that CAF-1 is also a chaperone for the H3.2 variant [64]. It is thus likely that CAF-1 also mediates de novo deposition of H3.2 at UV sites.
Another H3 chaperone that has recently been involved in restoring UV-damaged chromatin in human cells is Histone Regulator A (HIRA) [40]. As opposed to CAF-1, HIRA specifically associates with the H3.3 variant [57] and promotes nucleosome assembly independently of DNA synthesis in vitro [57,65] and in vivo [59]. HIRA is targeted to UV-damaged chromatin in human cells, where it promotes deposition of newly synthesized H3.3 variants [40]. Similarly, HIRA is recruited to DSBs, where it stimulates H3.3 accumulation [40,41,66]. Intriguingly, HIRA is also required for the de novo incorporation of H3.1 in UV-damaged chromatin [40], revealing unanticipated cross-talk between H3 variant deposition pathways in response to DNA damage in vivo, which deserves to be closely examined in future studies. Not only does HIRA contribute to restoring chromatin structure in response to genotoxic stress, but it is also critical for proper reactivation of transcription once repair is complete. HIRA-dependent H3.3 incorporation coupled to DNA damage recognition could thus serve as a chromatin bookmark that licenses damaged chromatin for transcription restart upon completion of DNA repair [40]. Importantly, recurrent somatic point mutations of histone H3.3 were shown to alter H3 post-translational modifications and to be dominant driver events in several human cancers [6,67–69]. In light of recent data showing that some of these mutations affect H3.3 incorporation into chromatin upon transcription activation [70,71], it will be interesting to examine the how they impact H3.3 deposition by HIRA and transcription restart at sites of DNA damage, which could contribute to their oncogenic potential.
While HIRA function in the DNA damage response is now clearly established, whether the other H3.3-specific chaperone Death domain associated protein (DAXX) [72,73] also contributes to H3.3 dynamics in response to DNA damage is still to be determined. Nevertheless, a role for DAXX in response to genotoxic stress has been recently revealed in cells overexpressing the centromeric H3 variant CENPA. DAXX indeed promotes deposition of overexpressed CENPA on chromosome arms and this increases cell viability after DNA damage [74].
Besides histone chaperones mobilizing H3 variants in damaged chromatin, other chaperones stimulate H2A histone dynamics, as exemplified by Facilitates Chromatin Transcription (FACT) and Aprataxin and PNPK-like factor (APLF). APLF promotes the accumulation of macroH2A.1 macrodomain at sites of laser-induced breaks in human cells [46] but its function in damaged chromatin regions deserves more extensive characterization. Recent studies directly implicate FACT in regulating histone turnover in response to DNA damage. Initially identified as a histone chaperone promoting H2A-H2B dynamics during transcription [75], FACT was later shown to stimulate H2A/H2A.X exchange in nucleosomes [76]. FACT is recruited to IR-induced breaks and to UV–damaged chromatin with, similar to HIRA, a role in transcription recovery after UV damage repair [39]. Furthermore, FACT enhances H2A-H2B turnover at UV sites. The Suppressor of Ty 16 homolog (SUPT16H) subunit of the FACT heterodimer appears to be the driving force for its recruitment to UV-damaged chromatin and for stimulating histone dynamics at UV sites. Interestingly, FACT activity is regulated after DNA damage: SUPT16H poly(ADP-ribosyl)ation in response to DSBs impairs FACT ability to interact with nucleosomes and the resulting histone exchange reactions [76,77].
As described above, histone chaperones are usually put forward as factors stimulating chromatin assembly and thus nucleosome restoration after DNA damage (Fig. 2; Table 1). However, some chaperones have the opposite effect, as recently shown for nucleolin and ASF1, which both contribute to histone eviction from nucleosomes around DSBs [30,32]. Because AFS1 binding to histones occludes the H3-H4 tetramerization interface [78], it is likely that H3 and H4 histones are evicted from damaged chromatin as dimers. These data should prompt us to further examine potential roles for histone chaperones not only in nucleosome re-formation after repair but also in chromatin destabilization at the earliest stages of the damage response. It also raises the possibility that some chaperones are involved at both stages, acting as histone acceptors and donors, thus promoting recycling of displaced histones as proposed for ASF1 at the replication fork [79].
Fig. 2.
Role of histone chaperones in histone dynamics in response to DNA damage (DSBs or UV lesions). Nucleosome disorganization after DNA damage is followed by nucleosome re-assembly with de novo histone deposition and potential recycling of displaced histones. The indicated histone chaperones promote histone exchange, histone eviction and de novo deposition in damaged chromatin. ASF1, known to act both as a histone donor and acceptor, may facilitate the recycling of displaced histones by coupling nucleosome disassembly and re-assembly.
Table 1. Human histone chaperones promoting histone dynamics in response to DNA damage.
| Histone chaperone | Histone chaperone function | Role in histone dynamics at damage sites | Mode of recruitment to damaged chromatin | References |
|---|---|---|---|---|
| APLF | H3-H4 & macroH2A dynamics | Accumulation of macroH2A.1 macrodomain at laser-induced breaks | Recruited to DNA breaks by repair factors (Ku, XRCC4, XRCC1, PARP) | [46,87,88] |
| ASF1 | H3-H4 donor/acceptor | H3-H4 removal from DSBs | n.d. | [32] |
| CAF-1 | H3.1/2-H4 deposition (sites of DNA synthesis) | New H3.1 deposition coupled to NER synthesis | Direct binding to PCNA (sites of repair synthesis) | [38,60,62,63,86] |
| DAXX | -H3.3-H4 deposition (silent chromatin) - Deposition of overexpressed CENPA (chromosome arms) |
Deposition of overexpressed CENPA (chromosome arms) increases damage tolerance | n.d. | [74] |
| FACT | H2A-H2B eviction/deposition | - H2A.X/H2A replacement (facilitated by H2A.X phosphorylation) - H2A-H2B turnover at UV sites |
n.d. | [39,76] |
| HIRA | H3.3-H4 deposition (active chromatin) | - New H3.3 deposition at NER sites (coupled to UV damage detection) - H3.3 enrichment at DSBs |
Recruited to UV-damaged DNA by the ubiquitylation activity of DDB-CUL4 complex | [40,41] |
| Nucleolin | H2A-H2B eviction during transcription | Core histone removal from DSBs | Recruited to DSBs by MRN | [30,32] |
| p400 | - Putative H2A.Z-H2B chaperone - Nucleosome remodeler |
- H2A.Z deposition at DSBs (silent chromatin?) - H3 displacement from DSBs |
Targeted to DSBs as part of Tip60 complex by MRN | [28,33,93] |
Cross-talks between histone modifications, histone chaperones and remodelers
Histone modifications, histone chaperones and remodeling factors all contribute to histone dynamics in response to DNA damage in a coordinated and inter-dependent manner. Indeed, there is growing evidence that histone modifications regulate nucleosome assembly/disassembly by histone chaperones. For instance, phosphorylation and acetylation on histone H4 determine nucleosome assembly of H3 variants by regulating the association of H3.1 and H3.3 with their respective chaperones CAF-1 and HIRA [80,81]. Regarding inner core histones, DNA damage–induced phosphorylation of H2A.X facilitates the exchange of nucleosomal H2A.X by the histone chaperone FACT [76]. In turn, histone chaperones can influence histone modifications with a role in the DNA damage response. Indeed, FACT promotes H2B ubiquitylation at DSBs by recruiting the ubiquitin ligase Ring Finger protein 20, which is important to stimulate DSB repair by homologous recombination [82]. Finally, while nucleosome remodeling complexes are often associated with histone modifying activities, they can also be targeted to nucleosomes by histone modifications. For example, binding of the BRG1 remodeler to IR-damaged nucleosomes in human cells is mediated by its interaction with acetylated H3 on γH2AX-containing nucleosomes [83]. Similarly, UV-induced acetylation on H3 stabilizes nucleosome association of the yeast ortholog to BRG1 complex, facilitating UV damage repair in vitro [84]. Altogether, the concerted actions of histone modifiers, chaperones and remodelers contribute to fine-tuning histone dynamics for an efficient and timely response to DNA damage.
Role of repair factors
The dynamics of damaged chromatin are tightly coordinated with repair pathways as highlighted by the contribution of repair factors to damaged chromatin rearrangements (Table 1).
Repair factors can have a direct effect on nucleosome stability when binding to damaged DNA within chromatin. Indeed, reconstitution of base excision repair reactions with DNA damage-containing nucleosomes in vitro has shown that binding of the Ligase3-X-Ray Cross-Complementing 1 (XRCC1) repair factor complex disrupts gap- and nick-containing nucleosomes [85].
Repair factors also indirectly affect damaged chromatin reorganization by recruiting histone chaperones and chromatin remodelers. In particular, the histone chaperone CAF-1 is recruited to sites of repair synthesis through its direct binding to the polymerase sliding clamp Proliferating Cell Nuclear Antigen (PCNA) [86], while the association of the APLF histone chaperone to DNA break repair factors helps targeting APLF to DNA breaks [87]. In some cases, the enzymatic activity of repair factors is necessary for the recruitment of histone chaperones, as exemplified by APLF binding to DNA breaks that is also mediated by poly(ADP-ribosyl)ation [88], and as demonstrated for HIRA targeting to UV damaged chromatin, which requires the ubiquitylation activity of the DDB2-DDB1-CUL4 complex involved in UV damage detection [40]. In addition to contributing to HIRA recruitment, the UV damage response protein DDB2 also promotes chromatin decompaction and ATP-dependent histone displacement from sites of local UVC irradiation [34]. How DDB2 stimulates such chromatin rearrangements is currently unclear. This function appears to be independent of the E3-ubiquitin ligase activity of the DDB2 complex, and is most likely indirect, via the recruitment of ATP-dependent chromatin remodelers still to be identified. A role of PARP has been found in promoting chromatin expansion at sites of UVC irradiation and at laser-induced breaks [24,34]. It is thus tempting to speculate that PAR-dependent chromatin remodelers could be involved in this response [89–92]. Finally, the transient loss of H2B histones observed around I-PpoI-induced DSBs requires the DSB sensor complex MRE11-RAD50-NBS1 (MRN) [31]. Recent work has shed light into how MRN can promote histone dynamics around DNA breaks as this repair factor recruits both the histone chaperone nucleolin and Tip60 remodeling complex to DSBs [32,93]. Altogether, these studies reveal intimate relationships between repair pathways and factors involved in chromatin dynamics, providing a molecular basis for the close coordination of chromatin reorganization with DNA damage repair.
Conclusions and Future directions
How epigenome integrity is preserved together with the DNA sequence during DNA damage repair is a fascinating question that has raised increasing interest over the past decades. As discussed throughout this review, chromatin displays remarkable plasticity after DNA damage, with profound changes at the level of histone proteins. Whether histone dynamics at damage sites effectively contribute to restoring chromatin integrity or rather compromise epigenome stability, leaving a scar on chromatin, is still a matter of debate. Investigating the fate of parental histones that are displaced from damaged chromatin will be essential to determine whether and how the original information is preserved. Is there recycling of parental histones during the repair process or are they targeted to proteasome degradation due to protein damage? Although much effort has been invested in deciphering histone dynamics in response to DNA damage, we are only beginning to understand the underlying mechanisms and how they operate in a concerted manner. How are H2A-H2B and H3-H4 dynamics coordinated at damage sites? Is there evidence for combinatorial histone variant deposition within repaired nucleosomes? Is there a mechanistic coupling between nucleosome disorganization and reorganization? These important questions remain open for future studies.
Beyond histone dynamics, we still know very little about larger scale chromatin plasticity in human cells exposed to genotoxic stress. New technologies, such as super-resolution microscopy and chromosome conformation capture techniques to map preferential chromatin interactions genome-wide, should greatly advance our understanding of large-scale chromatin reorganization in response to DNA damage. In this respect, future challenges include deciphering the mobilization of non-histone chromatin proteins at damage sites [17,94], the influence of chromatin organization into specialized nuclear domains [95], and the importance of DNA damage-induced chromosome mobility [96,97]. Finally, it will be critical to determine if reshaping of damaged chromatin operates similarly throughout development, in stem cells and aging cells in particular, characterized by an exacerbated DNA damage response and specific chromatin organization.
Acknowledgements
I thank Salomé Adam for critical reading of the manuscript. Research in the laboratory is supported by the European Research Council (ERC-2013-StG-336427 “EpIn”), the French National Research Agency (ANR-12-JSV6-0002-01), the “Who am I?” laboratory of excellence, EDF Radiobiology program and the Fondation ARC.
Abbreviations used
- APLF
Aprataxin and PNPK-Like Factor
- ASF-1
Anti-Silencing Factor 1
- ATP
adenosine triphosphate
- BRG1
brahma-related gene 1
- CAF-1
Chromatin Assembly Factor 1
- CENPA
Centromeric Protein A
- CUL4A
Cullin 4A
- DAXX
Death domain associated protein
- DDB1 & DDB2
DNA Damage Binding Protein 1 & 2
- DNase I
DNA Nuclease 1
- DSB
Double Strand Break
- FACT
Facilitates Chromatin Transcription
- FRAP
fluorescence recovery after photobleaching
- HIRA
Histone Regulator A
- INO80
inositol requiring mutant 80
- IR
ionizing radiation
- MNase
micrococcal nuclease
- MRN
MRE11-RAD50-NBS1
- NER
Nucleotide Excision Repair
- PA-GFP
photoactivatable-green fluorescent protein
- PARP
Poly(ADP-ribose) Polymerase
- PCNA
Proliferating Cell Nuclear Antigen
- SUPT16H
Suppressor of Ty 16 homolog
- Tip60
Tat-interacting protein 60
- UV
UltraViolet
- XRCC
X-Ray Cross-Complementing
References
- [1].Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- [2].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [3].Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–86. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- [5].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maze I, Noh K-M, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014;15:259–71. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- [8].Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: the means to the end. Febs J. 2011;278:3579–95. doi: 10.1111/j.1742-4658.2011.08281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2013;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peterson CL, Almouzni G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Price BD, D'Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–54. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- [15].Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smerdon MJ. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991;3:422–8. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- [17].Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–34. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- [18].Smerdon MJ, et al. Distribution within chromatin of deoxyribonucleic acid repair synthesis occurring at different times after ultraviolet radiation. Biochemistry. 1980;19:2992–3000. doi: 10.1021/bi00554a025. [DOI] [PubMed] [Google Scholar]
- [19].Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proc Natl Acad Sci USA. 1978;75:4238–41. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mathis GA, Althaus FR. Isolation of 8-methoxypsoralen accessible DNA domains from chromatin of intact cells. Cell Biol Toxicol. 1990;6:35–45. doi: 10.1007/BF00135025. [DOI] [PubMed] [Google Scholar]
- [21].Hu J, Choi J-H, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide Excision Repair in Human Cells: FATE OF THE EXCISED OLIGONUCLEOTIDE CARRYING DNA DAMAGE IN VIVO. J Biol Chem. 2013;288:20918–26. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. Embo J. 2003;22:975–86. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–34. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J Cell Sci. 2013;126:889–903. doi: 10.1242/jcs.109413. [DOI] [PubMed] [Google Scholar]
- [25].Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–16. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- [26].Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–53. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Molecular and Cellular Biology. 2007;27:7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–33. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, et al. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kobayashi J, Fujimoto H, Sato J, Hayashi I, Burma S, Matsuura S, et al. Nucleolin Participates in DNA Double-Strand Break-Induced Damage Response through MDC1-Dependent Pathway. PLoS ONE. 2011;7:e49245–5. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Berkovich E, Monnat RJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- [32].Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci U S A. 2013;110:16874–9. doi: 10.1073/pnas.1306160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, et al. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J Cell Biol. 2012;199:1067–81. doi: 10.1083/jcb.201205059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol. 2012;197:267–81. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smerdon MJ, Watkins JF, Lieberman MW. Effect of histone H1 removal on the distribution of ultraviolet-induced deoxyribonucleic acid repair synthesis within chromatin. Biochemistry. 1982;21:3879–85. doi: 10.1021/bi00259a024. [DOI] [PubMed] [Google Scholar]
- [36].Gérard A, Polo SE, Roche D, Almouzni G. Methods for studying chromatin assembly coupled to DNA repair. Meth Enzymol. 2006;409:358–74. doi: 10.1016/S0076-6879(05)09021-X. [DOI] [PubMed] [Google Scholar]
- [37].Adam S, Polo SE. Chromatin Dynamics during Nucleotide Excision Repair: Histones on the Move. Ijms. 2012;13:11895–911. doi: 10.3390/ijms130911895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–93. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- [39].Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51:469–79. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- [40].Adam S, Polo SE, Almouzni G. Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- [41].Yang X, Li L, Liang J, Shi L, Yang J, Yi X, et al. Histone acetyltransferase 1 promotes homologous recombination in DNA repair by facilitating histone turnover. J Biol Chem. 2013;288:18271–82. doi: 10.1074/jbc.M113.473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JYJ, Berns MW, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–7. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle. 2013;12:3070–82. doi: 10.4161/cc.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Taty-Taty G-C, Courilleau C, Quaranta M, Carayon A, Chailleux C, Aymard F, et al. H2A.Z depletion impairs proliferation and viability but not DNA double-strand breaks repair in human immortalized and tumoral cell lines. Cell Cycle. 2014;13:399–407. doi: 10.4161/cc.27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–9. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- [46].Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, et al. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012;586:3920–5. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lan L, Nakajima S, Kapetanaki MG, Hsieh CL, Fagerburg M, Thickman K, et al. Monoubiquitinated histone H2A destabilizes photolesion-containing nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase. Journal of Biological Chemistry. 2012;287:12036–49. doi: 10.1074/jbc.M111.307058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang H, Zhai L, Xu J, Joo H-Y, Jackson S, Erdjument-Bromage H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–94. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- [50].Qian M-X, Pang Y, Liu CH, Haratake K, Du B-Y, Ji D-Y, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–24. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, et al. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2010;107:17274–9. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhao Q, Wang Q-E, Ray A, Wani G, Han C, Milum K, et al. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. Journal of Biological Chemistry. 2009;284:30424–32. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, et al. The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer. Mol Cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hamiche A, Shuaib M. Chaperoning the histone H3 family. Biochim Biophys Acta. 2013;1819:230–7. doi: 10.1016/j.bbagrm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- [56].Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- [57].Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- [58].Stillman B. Chromatin assembly during SV40 DNA replication in vitro. 1986;45:555–65. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- [59].Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, et al. Dynamics of histone h3 deposition in vivo reveal a nucleosome gap-filling mechanism for h3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–41. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- [60].Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–96. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- [61].Mello JA, Silljé HHW, Roche DMJ, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–34. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–75. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. Embo J. 2003;22:5163–74. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Latreille D, Bluy L, Benkirane M, Kiernan RE. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Research. 2014 doi: 10.1093/nar/gkt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ray-Gallet D, Quivy J-P, Scamps C, Martini EM-D, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- [66].Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–28. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fontebasso AM, Liu X-Y, Sturm D, Jabado N. Chromatin remodeling defects in pediatric and young adult glioblastoma: a tale of a variant histone 3 tail. Brain Pathol. 2013;23:210–6. doi: 10.1111/bpa.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yuen BTK, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer Cell. 2013;24:567–74. doi: 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sarai N, Nimura K, Tamura T, Kanno T, Patel MC, Heightman TD, et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. Embo J. 2013;32:2392–406. doi: 10.1038/emboj.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Newhart A, Rafalska-Metcalf IU, Yang T, Joo LM, Powers SL, Kossenkov AV, et al. Single Cell Analysis of RNA-mediated Histone H3.3 Recruitment to a Cytomegalovirus Promoter-regulated Transcription Site. J Biol Chem. 2013;288:19882–99. doi: 10.1074/jbc.M113.473181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–65. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, et al. Mislocalization of the Centromeric Histone Variant CenH3/CENP-A in Human Cells Depends on the Chaperone DAXX. Mol Cell. 2014;53:631–44. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- [75].Belotserkovskaya R, Oh S, Bondarenko V, Orphanides G, Studitsky V, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–3. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- [76].Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- [77].Huang J, Chen W, Chang Y, Wang H, Chuang W, Lee S. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006;34:2398–407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- [79].Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–31. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- [80].Zhang H, Han J, Bin Kang, Burgess R, Zhang Z. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J Biol Chem. 2012;287:6573–81. doi: 10.1074/jbc.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kang B, Pu M, Hu G, Wen W, Dong Z, Zhao K, et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 2011;25:1359–64. doi: 10.1101/gad.2055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Oliveira DV, Kato A, Nakamura K, Ikura T, Okada M, Kobayashi J, et al. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci. 2014;127:763–72. doi: 10.1242/jcs.135855. [DOI] [PubMed] [Google Scholar]
- [83].Lee H-S, Park J-H, Kim SJ, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. Embo J. 2010;29:1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Duan M-R, Smerdon MJ. Histone H3 Lysine 14 (H3K14) Acetylation Facilitates DNA Repair in a Positioned Nucleosome by Stabilizing the Binding of the Chromatin Remodeler RSC (Remodels Structure of Chromatin) Journal of Biological Chemistry. 2014;289:8353–63. doi: 10.1074/jbc.M113.540732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Odell ID, Barbour J-E, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, et al. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol Cell Biol. 2011;31:4623–32. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Moggs JG, Grandi P, Quivy JP, Jónsson ZO, Hübscher U, Becker PB, et al. A CAF- 1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Molecular and Cellular Biology. 2000;20:1206–18. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Grundy GJ, Moulding HA, Caldecott KW, Rulten SL. One ring to bring them all-The role of Ku in mammalian non-homologous end joining. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.02.019. [DOI] [PubMed] [Google Scholar]
- [88].Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- [89].Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. Embo J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–4. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Vissers JHA, van Lohuizen M, Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci. 2012;125:3939–48. doi: 10.1242/jcs.107375. [DOI] [PubMed] [Google Scholar]
- [95].Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–54. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Miné-Hattab J, Rothstein R. DNA in motion during double-strand break repair. Trends Cell Biol. 2013;23:529–36. doi: 10.1016/j.tcb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–64. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]