Summary
This study focuses on the genetic history of the Quechua‐Lamistas, inhabitants of the Lamas Province in the San Martin Department, Peru, who speak their own distinct variety of the Quechua family of languages. It has been suggested that different pre‐Columbian ethnic groups from the Peruvian Amazonia, like the Motilones or “shaven heads”, assimilated the Quechua language and then formed the current native population of Lamas. However, many Quechua‐Lamistas claim to be direct descendants of the Chankas, a famous pre‐Columbian indigenous group that escaped from Inca rule in the Andes. To investigate the Quechua‐Lamistas and Chankas’ ancestries, we compared uniparental genetic profiles (17 STRs of Q‐M3 Y‐chromosome and mtDNA complete control region haplotypes) among autochthonous Amazonian and Andean populations from Peru, Bolivia and Ecuador. The phylogeographic and population genetic analyses indicate a fairly heterogeneous ancestry for the Quechua‐Lamistas, while they are closely related to their neighbours who speak Amazonian languages, presenting no direct relationships with populations from the region where the ancient Chankas lived. On the other hand, the genetic profiles of self‐identified Chanka descendants living in Andahuaylas (located in the Apurimac Department, Peru, in the Central Andes) were closely related to those living in Huancavelica and the assumed Chanka Confederation area before the Inca expansion.
Keywords: Quechua‐Lamistas, Chankas, Y‐SNPs, Y‐STRs, mtDNA, Amazonia, Andes, indigenous South Americans, human history
Introduction
Pre‐Columbian cultures from South America did not have writing, and their history started being recorded by the first chroniclers that accompanied the conquistadors. In the absence of a writing system, oral tradition preserves the collective memory, but it becomes less and less accurate as more time passes successively. It is not surprising that more information has been gathered about the Inca Empire, which encompassed millions of inhabitants and a territory that spanned almost all the countries of the Central Andes from the Pacific Ocean to the limit with Western Amazonia. In contrast, very little information exists about the pre‐Columbian history of Amazonian indigenous people.
During the Inca Empire (Tawantinsuyu) and the Hispanic colonial period in Peru, political leaders used Mitma (forced resettlement) and Reducciones (mission reductions) to govern the diverse peoples of the region. Through these actions, people of different ethnicities or regions were mixed together in a locality (mitmaqs, yanakunas, curacazgos or Inca lord governors, and Spanish mission towns; de la Vega, 1609; Ravi Mumford, 2012) for administrative control. In addition, in post‐Columbian times (after 1532), encomiendas and repartimientos (similar to the European feudal system) were introduced into the reduced communities along the Andes, the Pacific Coast and in the Amazon Basin. The Spaniards also exploited the existing sociopolitical model and the infrastructure of the Qhapaq Ñan (Great Inca Road) during the conquest of the Inca Empire. Moreover, the Spanish conquistadors and missionaries, along with native helpers, moved eastwards beyond the Inca borders to colonise some areas of Amazonia, where they also adopted the Quechua language as a lingua franca (San Roman, 1994; García, 1999).
By the 16th century, the Peruvian Cocama and Omagua groups (Tupi‐Guarani language family) dominated regional exchange networks in the Western Amazonia (Amazon, Napo, Ucayali, Marañon Rivers), while an Arawakan trade network covered most of the central Amazon Basin (Diamond & Bellwood, 2003; Hornborg, 2005; Reeve, 2014). Soon after the European conquest came the evangelisation, thus the Franciscan and Jesuit missionaries of Quito introduced the Quichua language (northern Quechua) as a lingua franca to various tribes of the Marañon, Napo and Amazon Rivers (San Roman, 1994; García, 1999). They founded the reduced cities of San Francisco de Borja, Moronas, Pastazas, Jeveros (or Xeveros) and Maynas, inhabited by many local indigenous groups, and commissioned soldiers as well as Jesuits in 1638. Other reducciones resulted also in the mission towns of San Estanislao de Muniches (1652), Presentación de Chayahuitas (1678), Nuestra Señora de las Nieves de Yurimaguas (1689), Santo Tome de Andoas (1707), San Regis de Lamistas (1718), Concepción de Cahuapanas (1726), Corazón de Jesús de Jíbaros (1767), and San Fernando de Mayorunas (1744) among others (García, 1999). It is noteworthy that Jesuit missionaries founded 152 reduced towns from 1638 to 1768 (de Velasco, 1789). Particularly, San Regis de Lamistas or San Regis del Baradero de Lamistas was a reduced village or Baradero (a place of refuge) where people arriving from Lamas suburbs and surrounding areas were settled (de Velasco, 1789).
The Quechua‐Lamistas and Chankas, focal ethnic groups of this study, are Quechua speakers. The term “Quechua” denotes a language family and their varieties are spoken throughout the Andean region of Peru, Bolivia, Chile, Argentina, Ecuador and Colombia (including Putumayo‐Caquetá Rivers). In the Lamas Province, the inhabitants of the Wayku suburb are self‐identified as Quechua‐Lamistas.
On the Pre‐Columbian Settlement of Northern Peruvian Amazonia
According to Riva Herrera (2004), six different ethnic groups were living in the present‐day San Martin Department when the Spanish conquered this area in 1654. They included Tabalosos, Lamas, Amasifuynes, Cascoasoas, Juamuncos and Payanancos (or Payansos). These tribes were also known as Motilones, or “shaven heads”, by the Spaniards (the same term Motilones was used for another ethnic group – the Barí – from Colombia and Venezuela). However, other ethnic groups were recorded being present in that region (Fig. 1) by the Jesuit and Franciscan missionaries (de Velasco, 1789; Scazzocchio, 1981; García, 1999). The Lamas people lived south of the Mayo River, close to Tabalosos villages, while the Amasifuynes, and also Suchiches and Muniches (Scazzocchio, 1981) occupied a region around Tarapoto. The Cascoasoas, Aguanos and Mayorunas were living on the shores of the Huallaga River, the Shawi (Cahuapanas or Chayahuitas) were settled around the Paranapura River, close to Yurimaguas (Loreto Department) and the Awajun were distributed in different areas of Moyobamba (San Martin Department) and Loreto Department. To the south of the Huallaga and Mayo Rivers were also found the Hibitos, Cholones and Payansos, including other minor ethnic groups like the Pativas, Saparinas, Cocamas and Shipibos (Scazzocchio, 1981; García, 1999; Riva Herrera, 2004).
Figure 1.
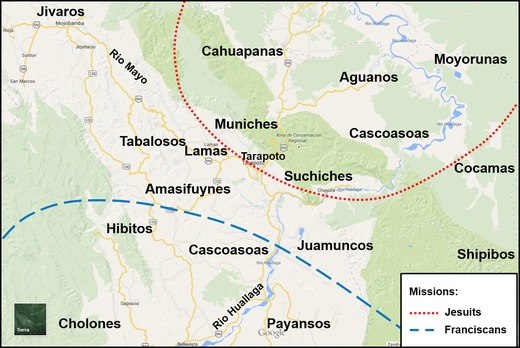
The major ethnic groups mapped by the 17th century in the Department of San Martin, Peru.
By the 17th century, Jesuit and Franciscan missionaries shared the administration of the San Martin region for evangelising purposes. Part of the Central Huallaga Valley fell under Franciscan administration, while the area from Chazuta to the lowlands was granted to the Jesuits (García Jordan, 2001). They formed alliances with the reduced ethnic groups and controlled these areas, as well as salt rock deposits like the “Cerro de la Sal” in Central Amazonia, which was a key element of commerce. Later, by the 19th and 20th centuries, many indigenous communities were displaced from their homelands to profit or run away from slaving and brutal practices of the rubber boom industry (García Jordan, 2001). Throughout this time, most Amazonian populations were affected by displacements, but also by episodes of relative isolation, endogamy and demographic decline that continue to the present day in some areas.
Nowadays, in Peruvian Amazonia, local populations living in 11 departments speak about 51 native languages (of 13 linguistic families). In the Department of San Martin, there are circa 728,800 inhabitants (Census II, 2007, http://www.inei.gob.pe), with three major ethnic groups representing most of these individuals – Quechua‐Lamistas (or Llakwash‐Runa), Awajun (or Aguarunas) and Shawi (or Chayahuitas, Cahuapanas, Paranapuras). However, around the Lamas and San Martin provinces, there are also minor ethnic groups like Shiwilu (or Jeveros). Along the borders of the San Martin and Loreto Departments, there are Cocama‐Cocamilla (Tupi‐Guarani linguistic family), Shipibo (Pano family), Chamicuro (Arawak family), Urarina (unclassified language), among others.
The Awajun live around the Alto Mayo River (circa 2500 inhabitants) and are distributed in 13 communities and four villages in the Moyobamba and Rioja provinces. The Shawi inhabit the districts of Pongo de Cainarachi and Papaplaya, living in two communities, Charapillo (Lamas Province) and Santa Rosa (San Martin Province), and also around Paranapura, Cahuapana and Sillay Rivers (Alto Amazonas Province, Loreto Department). In 1652, some Shawi were integrated into the Xeveros and Nuestra Señora de Loreto de Paranapura missions (Homan, 2014).
The city of Lamas stands at 850 m above sea level in the upper lowlands of Amazonia and was re‐founded in 1656 by Martin de la Riva Herrera with the name “El Triunfo de la Santa Cruz de los Motilones de Lamas” (Schjellerup, 1999). The Spanish conquistadors subdivided Lamas into nine neighbourhoods, including Ankoayllo, Calvario, San Juan, Zaragoza, Muniches, La Plaza, Suchiches, Quilloallpa and Wayku (Scazzocchio, 1981; Schjellerup, 1999). In addition, they forced the Motilones to adopt the use of cotton clothing according to Andean or Inca style, and the Jesuits imposed on them to speak the Quichua (Quechua dialect) from Ecuador (de Velasco, 1789; Schjellerup, 1999).
The Quechua‐Lamista language has been classified as part of the Quechua IIB branch that is related most closely to dialects from Ecuador, Colombia, Chachapoyas and Amazonian Quechuas (Torero, 1964; Taylor, 1979; Adelaar, 2013). The Amazonian Quechua speakers inhabit the areas around the Pastaza, Napo and Bobonaza Rivers and their tributaries in Peru and Ecuador, and Caquetá‐Putumayo region in Colombia (Torero, 1964; Taylor, 1979; Adelaar, 2013; Reeve, 2014). Presently, the Quechua‐Lamista dialect is also dispersed throughout communities of San Martin, Loreto, Amazonas and Madre de Dios Departments.
In the San Martin Department, the self‐identified Quechua‐Lamista‐speaking people are disseminated in nine provinces with around 50,000 inhabitants (Pardo et al., 2001). About 38% of the Quechua‐Lamistas are in the Lamas province. Historically, the Wayku suburb from Lamas is where most of the Quechua‐Lamista‐speaking people were concentrated. They are further divided into clans according to their patrilineal family identified by their last name or “surname”, such as Guerra, Sangama, Shupingahua, Cachique, Tapullima, Amasifuen, Sinarahua; Ishuiza and Salas (Pardo et al., 2001). In our previous study (Sandoval et al., 2013a), the Quechua‐Lamista people from Wayku showed 8.7% genomic ancestry from Eurasia, a finding consistent with the chronicles and the demographic history of the region.
The Highland and Lowland Quechua‐Speaking People from Ecuador
At the time of Inca expansion in the Andes, Tupac Inca Yupanqui and his son Huayna Capac conquered the Cañaris, Quitus, Cayambis, Karankis (Carangues), Otavalos and other Andean populations from Ecuador (Cieza de León, 1553; de la Vega, 1609; Vazques de Espinoza, 1948). Those populations and newly conquered Amazonian regions came under the Spanish encomiendas and reducciones, which were consolidated in 1570 as provinces of the Real Audiencia de Quito by Viceroy Francisco de Toledo (Espinoza Soriano, 1999). Furthermore, those systems also included the Amazonian Quijos (from the Napo River), Canelos (from the Pastaza River), ethnic groups from the Jivaroan linguistic family and other indigenous groups along the Bobonaza, Pastaza, Napo, Tigre, Trompeteros and Barranca Rivers (Reeve, 2014). However, in Ecuador there were about 40 “nations” or main language groups, which included more than 300 different tribes (de Velasco, 1789). On the other hand, the migration of people related to commercial trade networks was widespread around Amazonia and the Andes. For example, many populations from Ecuadorian Amazonia (mostly Quechua‐speaking people) traded salt supplies extracted from the mines in the Huallaga River by the Cocamas and Shipibo‐Conibos who inhabited the Peruvian Marañon and Ucayali Rivers.
What Was the Chanka Confederation?
The archaeological recovery of human remains from the Late Intermediate Period (AD 1000–1400) in the present‐day Huancavelica, Ayacucho and Apurimac regions (Central Andes of Peru) suggests that Andean “Chankas” and their neighbours, frequently assumed to be Quechua speakers, shared many common cultural features, but with their own distinctions (Bauer & Kellett, 2010; Kurin et al., 2014). After the collapse of the Wari Empire at the end of the 11th century, several fragmented small polities or “señorios” (Chanka, Uranmarca, Ankoayllo, Vilca, Hutunsulla, Sula) emerged in this region. They periodically engaged in warfare, although in some situations also formed alliances (de la Vega, 1609; Kurin et al., 2014).
The Chankas were divided into two groups, the Hanan and Hurin (a widespread sociopolitical division in pre‐Hispanic populations from Central Andes), which were distributed in villages with circular towers on mountaintops and governed by sinchis (Rostworowski, 2001). Later, they constituted the Chanka Confederation, and according to several chronicles, circa 1438 they tried to expand south to the Cusco Valley, but were defeated by the Incas and their allies. This was a landmark event considered as the beginning of the Inca state and imperial expansion (Rostworowski, 2001). Afterwards, the Incas occupied the region presently known as Andahuaylas (part of the Chanka heartland), to where several ethnic groups from distant regions were forced to move, including mitmaqs from Chachapoyas and from northeastern Quito (de la Vega, 1609; Espinoza Soriano, 1999; Julien, 2002).
During Spanish colonial times, the Chankas were also part of the encomiendas of “Hanan‐Chankas, Hurin‐Chankas and the Quichuas from Vilcaparo” in Andahuaylas, which were granted to Diego Maldonado in 1539 by conquistador Francisco Pizarro. In addition, several ethnic groups from southeast of Cusco and other regions were also brought into these encomiendas (Julien, 2002).
Part of the Chanka population did not surrender and fled to a still uncertain destination. According to the chronicles of Friar Martin de Murúa (1613): “The Chankas reached the province of Andahuaylas …stole across this and took a large number of people to Chachapoyas, and this is the story among the people here, who say they are in Aricoayllo and Ruparupa. Then, the general Capac Yupanqui sent messages to find out about them, and they continued (northwards) until they reached Cajamarca”. Similar versions were recorded by Cieza de León (1553) and Sarmiento de Gamboa (1572). On the other hand, de la Vega (1609) and Father Cobo (1653) express doubts as to which region or locations the Chankas took refuge in to escape from persecution by the Incas.
To test the different versions of the chronicles and the assumptions about the Chankas’ genetic legacy among Quechua‐Lamista people, we addressed the following questions: (1) Is there a genealogical connection between the Quechua‐Lamista‐speaking people and the self‐identified Chanka group from the Apurimac and Huancavelica areas? (2) Are there genetic affinities between the highland and lowland speakers of Quechua in Ecuador and neighbouring regions of Amazonia and the Quechua‐Lamistas as suggested by their linguistic affinities? (3) Are the Quechua‐Lamistas related to their Amazonian neighbours from Ecuador and Peru as suggested by their geographic vicinity? and (4) Is the genetic pattern of the self‐identified Chankas coherent with the populations of the Chanka Confederation region before the Inca expansion?
To investigate these issues, we compared mtDNA and Y‐chromosome haplotypes of Quechua‐Lamistas and other indigenous populations from the Andes and Amazonia, living in Peru, Bolivia and Ecuador. Based on individual‐ and population‐level genetic analyses, we evaluated hypothetical relationships between Andean and Amazonian ethnic groups from a historical perspective.
Material and Methods
Ethics Statement
Ethical approval for this study (South American Genographic Project) was provided by the Universidade Federal de Minas Gerais (Brazil), Brazilian National Ethics Commission or CONEP (Brazil), the Universidad San Martin de Porres (Peru) and the Universidad de las Américas (Ecuador). The project was explained to the volunteers after previous contact with indigenous confederations, authorities and/or community leaders, in some cases, in their indigenous languages. Signed informed consent forms for all subjects were obtained before the collection of buccal swab samples. Relatives to the third degree were avoided, and men representing unique families were preferentially sampled to maximise analyses with both Y‐chromosome and mtDNA markers.
Sampling
DNA samples were extracted from buccal swabs using standard procedures and according to written informed consent forms from voluntary healthy individuals from 15 different ethnic groups/localities from Peru and Ecuador. Quechua speakers from Peru included Quechua_Lamas (n = 40, from the Wayku suburb and nearby communities, Lamas province, San Martin); Chanka (n = 32, self‐identified Chankas from Andahuaylas, Apurimac), Quechua_HVC (n = 21, Huancavelica); Chopcca (n = 32, Huancavelica); and Kichwa_LO (n = 37, Loreto). Other Amazonian ethnic groups from Peru included Achuar (n = 14, Loreto); Huambisa (n = 12, Loreto); Awajun (n = 24, San Martin); Shawi (n = 8, San Martin); Shipibo (n = 5, Ucayali); Yine (n = 7, Ucayali); and Andoas (n = 70, Loreto). Andean samples from Quechua speakers of Ecuador came from the Province of Imbabura (Quichua, n = 22; and self‐identified Karanki, n = 15; Fig. 2). In addition, for haplotype comparisons, we included published data for Quechua‐speaking people from the Pastaza River, labelled as Kichwa_PTZ (n = 65 for mtDNA, n = 28 for Y‐chromosome) from Ecuador (Baeta et al., 2012; Roewer et al., 2013).
Figure 2.
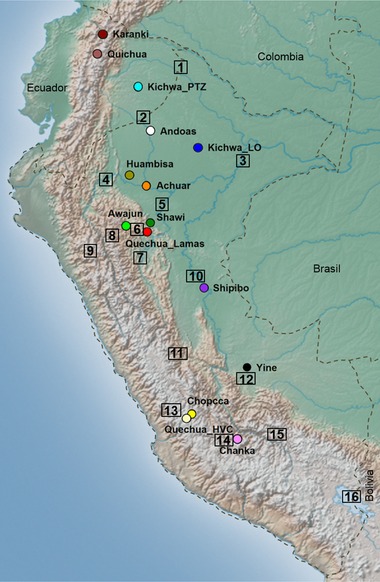
Locations of the 15 Peruvian and Ecuadorian populations under study. Conventional population codes are found in “Material and Methods”. Localities cited in text are also depicted in the numbered squares: 1‐Napo River (Ecuador, Peru); 2‐Pastaza River (Ecuador, Peru); 3‐Amazonas River, Iquitos, Peru; 4‐Marañon River, Peru; 5‐Yurimaguas, Peru; 6‐Moyobamba, Peru; 7‐Huallaga River, Peru; 8‐Chachapoyas, Peru; 9‐Cajamarca, Peru; 10‐Pucallpa, Peru; 11‐Oxapampa, Pasco, Peru; 12‐Nuevo Mundo, Cusco, Peru; 13‐Huancavelica, Peru; 14‐Andahuaylas, Peru; 15‐Cusco, Peru and 16‐Titicaca Lake in the Altiplano region (Peru and Bolivia).
Y‐Chromosome and mtDNA Analyses
The samples were genotyped for five Y‐SNPs found in the South American natives, M130, M242, M346, L54 and M3 (Karafet et al., 2008; Jota et al., 2011), using TaqMan assays (ABI) and a 7900HT Fast Real‐Time PCR System (ABI). Next, the Q‐M3 and Q‐M346* lineages were genotyped with 17 Y‐chromosome short tandem repeats (Y‐STRs) using Y‐filer Kit (ABI). The PCR reactions were performed following Sandoval et al. (2013b) and their products were subjected to capillary electrophoresis using the ABI 3130XL Genetic Analyzer (Applied Biosystems), with the alleles being visualised by the GeneMapper ID v3.2 software (Applied Biosystems, Foster City, California, USA). The DYS389b allele scoring was done by subtracting DYS389I from DYS389II (Zerjal et al., 1997) and the DYS385 marker was not included in the statistical analyses.
The complete mtDNA control region (1122 bp, 16024–576) according to the revised Cambridge Reference Sequence (rCRS; Andrews et al., 1999) was amplified and sequenced following Sandoval et al. (2013b), using a 3130XL Genetic Analyzer (ABI) and Big Dye Terminator v. 3.1. protocol. The sequence alignments were performed in relation to rCRS through SeqScape 2.6 software (Applied Biosystems), and major haplogroup assignment was obtained by MitoTool (Fan & Yao, 2011) or haplogroup prediction tool from the Genographic Project (Behar et al., 2007). Due to phylogenetic uncertainty and alignment controversy, the alignments at nucleotide positions 309.1C, 315.1C, indels at 515–522, 16182C, 16183C, 16193.1C and 16519 were not taken into account in the statistical analyses.
Statistical Analyses
To determine the genetic relationships among Q‐M3 individuals, we used the stepwise mutation model and Median Joining Maximum Parsimony algorithm (Bandelt et al., 1999) through the program Network as described at the Fluxus Engineering website (http://www.fluxus‐engineering.com), and the weighting criteria for Y‐STRs following Sandoval et al. (2013b).
Analysis of Molecular Variance (AMOVA) was performed in Arlequin 3.5 (Excoffier & Lischer, 2010), where Fst indices (Rst for STRs, and ϕst for mtDNA) were obtained to evaluate the genetic differentiation among the 15 Peruvian and Ecuadorian populations. The haplotype diversity indices and neutrality tests for distribution of mtDNA haplotypes were calculated under 10,000 simulated permutations. Furthermore, we considered the genetic distances of Reynolds’ coancestry coefficients, which were used for nonmetric MDS analyses according to Sandoval et al. (2013b). In addition, the Reynolds’ distances were also used in the Mantel test, comparing matrices of genetic and geographic distances, calculated with Geographic Distance Matrix Generator v1.2.3 (http://biodiversityinformatics.amnh.org/open_source/gdmg), with the GenAlEx 6.5 Excel package (http://biology.anu.edu.au/GenAlEx). To identify geographical areas in which probable barriers to gene flow exist, we used the Monmonier's algorithm and Delaunay triangulation with the program Barrier (Manni et al., 2004). Moreover, we applied the Principal Components Analysis (PCA) and Correspondence Analysis methods with the Y‐STR haplotype or mtDNA haplogroup frequencies using the FactoMineR script available in the R project (http://www.r‐project.org).
Results
Y‐Chromosome Results
After SNP and STR genotyping in all 15 Peruvian and Ecuadorian populations, we considered only Q‐M3 Y‐chromosome haplotypes (n = 277) for the analyses because they represent the dominant autochthonous lineage that appeared in all populations. The list of 17 Y‐STR haplotypes of Q‐M3 and Q‐M346* lineages obtained for the studied populations is found in Tables S1a and S1b.
To reveal the genetic relationships among the Y‐STR haplotypes at the individual level, we used them to generate median joining networks (Bandelt et al., 1999). Thus, in Figure 3, we observe shared haplotypes between some Quechua‐Lamistas and individuals from different Amazonian ethnic groups (Achuar, Shipibo, Yine and Andoas locality), including one Cocama‐speaking individual from Iquitos as well as from Loreto, San Martin and Chachapoyas (Fig. S1). In some branches, the Quechua‐Lamistas were clustered according to their clan “relatives” (differentiated by one or two mutation steps), which were identified by patrilineal connections, and a family surname. There were five recognisable male lineage clusters (see also, Table S1a), here named from W1 to W5. Clan W1 (haplotype code = 125, n = 8) is connected to Machiguenga‐speaking individuals (Fig. S2). Another clan was W2 (haplotype code = 26, 27 and 29, n = 4) is derived from haplotypes appearing in Kichwa and Shawi individuals. A third clan was W3 (haplotype codes = 143 and 174, n = 5), which shared a haplotype with Andoas, and was connected by one mutation step to another Quechua‐Lamista, and by four mutation steps to an individual from Andoas. In Figure S1, we show also that the W3 clan shared a haplotype with an individual from the San Martin Department. A fourth clan was W4 (haplotype code = 47 and 48, n = 3), which was linked by one mutation step to a Quechua‐speaking individual from Ecuador (Fig. 3). Also, haplotype 48, which belongs to the same W4 clan, shared a haplotype with an individual from Chachapoyas (Fig. S1). In addition, we observe a shared haplotype (code = 49) between Quechua‐Lamistas (n = 2) and Andoas (n = 2), which is connected by three mutation steps to a Yine‐speaking individual. In another branch, connected to the W4 clan, we observe a shared haplotype between Yine (n = 1) and Quechua‐Lamista (n = 1; Fig. 3, and haplotype code = 79). Clan W5 (haplotype code = 55, n = 2) shared a haplotype with one Achuar‐speaking individual from Rio Tigre (Loreto), a lineage located in a separated branch. In another branch, we observe a shared haplotype (code = 73) between Quechua‐Lamista (n = 1) and a Shipibo (n = 1), a Cocama, and two individuals from Loreto Department (Fig. S1). Finally, haplotype 73 is connected by two mutation steps to a shared haplotype between Achuar and Andoas (Fig. 3; haplotype code 78 in Table S1a). Despite the fact that other haplotypes from Quechua‐Lamistas are heterogeneous and differentiated by several mutations from the five main clans, most of them appeared to be more connected to individuals from Amazonia than to Andean people.
Figure 3.
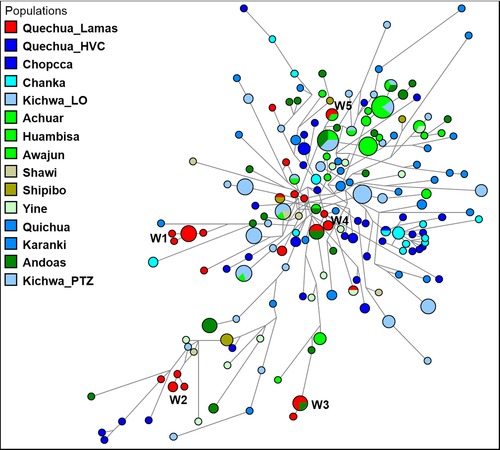
Median joining network for Q‐M3 Y‐STR haplotypes among 15 Peruvian and Ecuadorian populations. The population groups are depicted with distinct colours (red, light blue, blue and its spectrum = Quechuan language family; green spectrum = languages from Amazonian ethnic groups). The haplotypes composed of alleles on 15 Y‐STRs are represented by circles with sizes proportional to numbers of individuals, and branch lengths are proportional to STR mutation steps (one step unit between haplotypes in the W1 branch). W1‐W5 are the specified Quechua‐Lamista clans identified by “surname”.
Following this picture, we observed shared haplotypes among Jivaro‐speaking individuals (Achuar, Huambisa, Awajun) and Amazonian Quechuas from Peru and Ecuador (Kichwa_LO; Kichwa_PTZ). In some branches, the Jivaro and Kichwa share haplotypes between them. On the other hand, there was a similar scenario among the Quechua‐speaking individuals from Peruvian Andes. Most of the haplotypes of the self‐identified Chankas were more related to those of the Quechua_Chopcca‐HVC (Quechua‐speaking people from Huancavelica) than to those from Amazonia.
In the AMOVA analyses of Peruvian and Ecuadorian populations (Table S1c), we observed a complementary picture to the scenario described at the individual level. Without grouping, the genetic variation among the 15 populations showed a moderate differentiation (Rst = 0.0918, p = 0.0000) in comparison to intrapopulational diversity, which was higher (Ris = 0.9082). When all 15 populations were grouped according to different regional zones (Andes vs Amazonia), there was low differentiation among Andean populations (Rst = 0.0625, p = 0.0004) in comparison to Amazonian populations (Rst = 0.0832, p = 0.0000), a general pattern previously noticed among South American autochthonous populations (Tarazona‐Santos et al., 2001). In the MDS plot (Fig. 4) based on the Reynolds's genetic distance estimates, we observed a compact cluster in Andeans, formed by Quichua and Karanki populations from Ecuador. In a similar way, the Peruvian Quechua‐speaking populations from Huancavelica (labelled as Chopcca and HVC) were clustered together, and with the Chanka population, despite the fact that the latter was located distantly in the plot. In contrast, the Amazonian populations are spatially dispersed in the graphic, indicating a high interpopulation differentiation. This picture indicates that Andean populations are less differentiated from each other than Amazonians, as previously reported (Tarazona‐Santos et al., 2001). The levels of expected heterozygosity (He) were in close agreement with the pattern of population diversity in the MDS plot (Table S1d). The spatial configuration of the 15 populations in the PCA plot (Fig. S3) was also in close agreement with that shown by the MDS analysis.
Figure 4.
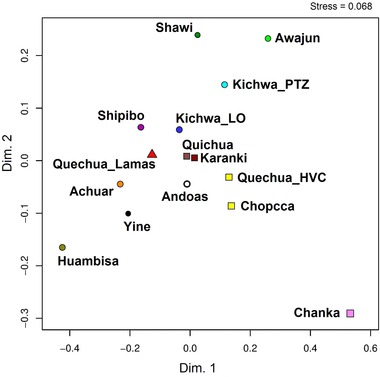
MDS plot for Q‐M3 Y‐STR data among 15 Peruvian and Ecuadorian populations. Reynolds’ Rst genetic distances were used among populations. Amazonian populations are represented by circles, Andean populations by squares and the Quechua‐Lamista population by a triangle.
mtDNA Results
We identified 174 mtDNA CR haplotypes (GenBank accession numbers for 168 new sequences: KT997553‐KT997720). At the individual level, we conducted a phylogenetic analysis of mtDNA CR sequences using the median joining algorithm. The genetic relationship of the mtDNA control region haplotypes among the 404 individuals from 15 populations is shown in Figure 5. There are two interesting features about the Quechua‐Lamistas’ maternal lineages. First, independently of the mtDNA haplogroup, 37 out of 40 individuals from Lamas do not share haplotypes with individuals from other ethnic groups, which could suggest a separation of several generations from other related Amazonians. Another feature is that there is much less genetic differentiation between Quechua‐Lamistas and Amazonians, than between Quechua‐Lamistas and Andeans. The complete set of polymorphic sites of mtDNA sequences or haplotypes relative to rCRS is listed in Table S2a and the most common haplogroup found among the Quechua‐Lamistas was B2. Regarding this lineage, we observed many haplotypes shared among Quechua‐Lamistas, mostly from Wayku and Pamashto, but also from Chumbakiwi and other nearby localities (haplotype code = 157, in Table S2a). In addition, common mtDNA haplotypes were correlated with the self‐declared clans identified by “surname” (W1, W3, W4), appeared only among Quechua‐Lamistas (code = 157). In another branch, two Chopccas and two Chankas shared haplotypes (code = 113), and few other haplotypes were shared between individuals speaking different languages. For example, there was a shared haplotype (code = 50) between Chopcca (n = 1), Jivaro (Awajun, n = 3), Quechuas from Ecuador (Quichua, n = 2; Karanki, n = 2) and Andoas (n = 1). Among other haplotypes, the Jivaroan (n = 6) communities and Andoas (n = 1) shared a haplotype (code = 63); haplotype 34 was also shared between Andoas (n = 10), Huambisa (n = 1) and Kichwa_LO‐PTZ (n = 1); haplotype 163 was shared between Jivaro‐speaking groups (Huambisa, n = 4 and Awajun, n = 2); haplotype 170 was shared among Jivaro (Awajun, n = 1), Arawak (Yine, n = 1) and Andoas (n = 1); haplotype 111 was shared between Kichwa_LO (n = 4) and Jivaro (Achuar, n = 1); and haplotype 58 was shared between Andoas (n = 3) and Awajun (n = 1).
Figure 5.
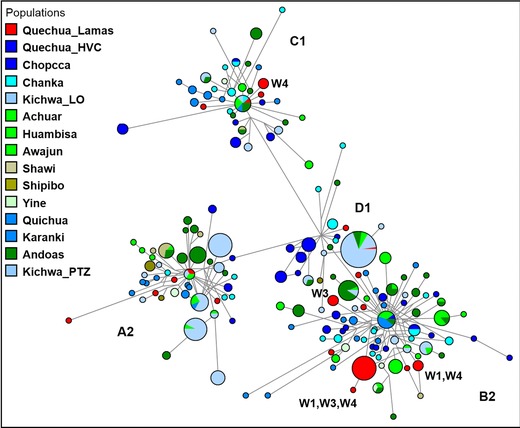
Median Joining network for A2, B2, C1 and D1 control region mtDNA haplotypes among 15 Peruvian and Ecuadorian populations. The population groups are depicted with distinct colours (red, light blue, blue and its spectrum = Quechuan language family; green spectrum = languages from Amazonian ethnic groups). The control region mtDNA haplotypes are represented by circles with sizes proportional to numbers of individuals, and branch lengths are proportional to mutation steps (nucleotide changes). Clusters of haplotypes into four mtDNA haplogroups (A2, B2, C1, D1) are indicated. W1‐W4 are the specified Quechua‐Lamista clans identified by “surname”.
In A2 and D1 haplogroups, we observed shared haplotypes among Kichwa_LO, Jivaro (Achuar) and Quechua‐Lamistas as well as among the Kichwa_LO, Kichwa_PTZ, Andoas, Jivaro (Achuar) and Quechua‐Lamistas, respectively (see also Table S2a). Furthermore, in these haplogroups, most of the shared haplotypes were observed among Quechuas from Pastaza (Ecuador) and Loreto (Peru). However, in the C1 (with haplotype code = 85), one individual (a woman from the Tabalosos locality) from Quechua‐Lamistas (n = 1) shared a haplotype with Kichwa (n = 1), Andoas (n = 2), Jivaro (Achuar, n = 1; Huambisa, n = 1), a Quichua (n = 1) and a Chanka (n = 1). In B2 (haplotype code = 57) and C1 (haplotype code = 12), some haplotypes were shared by Quichua and Karanki from Ecuador, while in the D1, Quechuas and Chopccas from Huancavelica shared a haplotype (haplotype code = 131).
To characterise the genetic variability among the 15 Peruvian and Ecuadorian indigenous communities, we carried out the AMOVA analyses taking into account the pairwise haplotype differences according to Sandoval et al. (2013b) There was considerable variability within populations (Fis = 0.891) and an intermediate level of differentiation among them (Fst = 0.109). When ethnic groups were divided into two regional categories (Andes vs Amazonia), higher interpopulation differentiation was observed among Amazonian subpopulations (Fst = 0.1397, p = 0.000) than among Andeans (Fst = 0.0073, p = 0.229; Table S2b). The haplotype diversity indices and neutrality tests (Tajima's D and Fu's Fs statistics) showed high intrapopulation genetic diversity among Andean populations (Quichua and Karanki, Quechua_Chopcca‐HVC and Chanka) in contrast to Amazonians which had lower values, but with the exceptions of Andoas, Jivaro (Achuar) and Kichwa_LO (Table S2c). The nucleotide diversity indices (π) among the Quechua‐Lamistas were similar to those of Awajun and Huambisa populations. The distribution of mtDNA haplogroup frequencies among the 15 Peruvian and Ecuadorian communities (n = 404) is shown in Table S2d.
The MDS plot based on the Reynolds’ linearised distances of mtDNA's Fst estimates shows that Quechua‐speaking populations from Peru and Ecuador were grouped in a compact cluster in comparison to the Amazonian communities (Fig. 6), and in agreement with previous observations. In addition, the Jivaro populations (Awajun, Huambisa) were associated with each other as expected due to their higher B2 haplotype frequencies, as well as to the Quechua‐Lamista population. However, the Achuar population was located at the opposite side of the two‐dimensional space because of the high frequency of A2 haplotypes. Notwithstanding this configuration in the MDS, the Awajun, Huambisa and Achuar populations belonged to the same Jivaroan language family. Furthermore, there was a close relationship between the Achuar and Quechua speakers from the Loreto and Pastaza River (Kichwa_LO; Kichwa_PTZ), such as Pano (Shipibo) and Cahuapana (Shawi).
Figure 6.
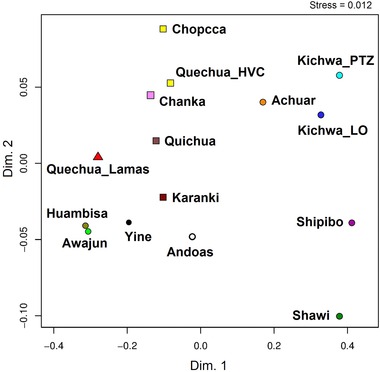
MDS plot for haplogroup A2, B2, C1 and D1 control region mtDNA sequences among 15 Peruvian and Ecuadorian populations. Reynolds’ Φst genetic distances among populations were used. Amazonian populations are represented by circles, Andean populations by squares and the Quechua‐Lamista population by triangles.
In addition, we included the control region mtDNA (16024‐16365 bp) haplotypes of several individuals (n = 51) from lowland Amazonia‐Yurimaguas (Justice et al., 2012) and Chachapoyas (n = 14, from Amazonia‐Andes) in the network analysis. The phylogenetic tree showed that Quechua‐Lamistas and Chankas shared some B2, A2 and C1 haplotypes amongst them and with other individuals from different Amazonian groups (Fig. S4). In the D1 lineage, one Quechua‐Lamista shared a haplotype with the Quechua speakers from Pastaza and Loreto as well as Andoas, Jivaro (Achuar) and Yurimaguas. At the population level, analyses using A2, B2, C1 and D1 haplogroup frequencies, populations of Yurimaguas, Chachapoyas and Andoas showed intermediate levels of genetic diversity relative to Andean and Amazonian populations. Besides, the set of mtDNA haplogroup frequencies among Quechua‐Lamistas ranged between that of Quichua‐Karanki from Ecuador and those of the Huambisa‐Awajun populations (Fig. S5).
Spatial and Geographic Analyses among the 15 Peruvian and Ecuadorian Populations
Statistically, a nonsignificant correlation was observed between Reynolds’ linearised distances of mtDNA (Fst) or Y‐STRs (Rst) data and geographic distances (kilometers) among the 15 populations using the Mantel test (R 2 = 0.048; p = 0.033, for mtDNA; R 2 = 0.026, p = 0.112, for Y‐STRs). However, the spatial analysis using genetic distances and geographic coordinates through Monmonier's algorithm and Delaunay triangulation showed that Jivaroan‐speaking populations (Awajun, Ashuar, Huambisa) were relatively isolated by gene‐flow barriers (Fig. S6), most likely associated with cultural differences, in close agreement with their linguistic affinities. On the other hand, including only Andean populations and Quechua‐Lamistas in the test, we observed that the Ecuadorian and Quechua‐Lamistas communities were also relatively isolated by gene‐flow barriers (figure not shown) from the Peruvian Quechua‐speaking communities (Huancavelica and Apurimac). This phylogeographic pattern is in agreement with the observed linguistic differences between the Quechua IIB (from Ecuador, North Amazonia of Peru, including the Department of San Martin, where Quechuas‐Lamistas live) and the Quechua IIC varieties (from Ayacucho, Huancavelica, Apurimac, Cuzco, Arequipa, Moquegua, Puno, and including Bolivia, Argentina and Chile; Torero, 1964; Adelaar, 2013). On the other hand, these results indicate that the spread of Quechua language into those macroregions was predominantly cultural rather than demographic.
Discussion
Previous genetic studies of HLA‐I and ‐II variation (Arnaiz‐Villena et al., 2006; Moscoso et al., 2006; Rey et al., 2012) among the Quechua‐Lamistas (from Wayku) have shown limited genetic affinities with other Quechua speakers or Andean people, whereas most Native American populations do share HLA haplotypes with each other. Using the correspondence analyses on HLA‐DRB1 and HLA‐DQB1 allele frequencies, Moscoso et al. (2006) and Rey et al. (2012) showed that Quechua‐Lamistas (n = 83) were more related to some Gran Chaco populations than to Bolivian Quechua speakers (n = 80). In contrast, upon comparing ABO and Rh blood group frequencies in some highland populations (Junin, Ancash, Puno) and lowland populations from Amazonia, Frisancho & Klayman (1975) suggested that Quechua‐Lamistas may be descendants of Andean populations. However, it has been shown that the O allele (mostly O01 (01) and O02 (01v) variants) is commonly found throughout the Americas, ranging from 86.5% to 100% (Georges et al., 2012). Thus, the similarity of O blood group frequencies observed by Frisancho & Klayman (1975) among the Quechua‐Lamistas (from Pamashto locality) and other Andean and Amazonian populations may likely be due to genetic drift and to the demographic history of the studied populations (pre‐ and post‐Columbian admixture with Europeans).
Furthermore, in our previous study using a set of 40 ancestry informative markers among 25 Peruvian subpopulations (Sandoval et al., 2013a), we showed that Quechua‐Lamista are genetically more related to populations from Pucallpa and Andoas than to Andean populations. By contrast, the Kaquiabamba and Andahuaylas subpopulations from the Apurimac Department clustered with southern and central Andean populations, respectively. In this study, using uniparental markers (Y, mtDNA), we show closer relationships among the Quechua‐Lamistas and Amazonian ethnic groups, as previously suggested by autosomal markers. Also, it has been shown that Yurimaguas, Chachapoyas and Andoas populations are localities connected by gene flow, suggesting a high interpopulation migration between the Andes and Amazonia (this study). Admixture or migration events also appear to have happened between Arawak linguistic groups (like Machiguenga and Yanesha) and Andean Quechua speakers (including Chankas from Apurimac) and Aymara speakers at the boundaries of Central Andes‐Amazonia (Fig. S2; Sandoval et al., 2013b).
In the post‐Columbian period, after the conquest of the Chachapoyas by Alonso de Alvarado, a series of expeditions were carried out in the Moyobamba region. During his first campaign, the Corregidor of Cajamarca Martin de la Riva Herrera founded the first two reducciones, San Joseph de los Lamas and Virgen del Rosario (currently Tabalosos) in the province of Tabalosos (Schjellerup, 1999). The Spaniards described the presence of the Motilones among several tribes from San Martin region. Although this feature is a representative cultural expression for some Amazonian ethnic groups like the Barí‐Motilones from Colombia‐Venezuela border, the Motilones category and the cultural description of the Chankas of Andahuaylas are quite different: “They (Chankas) had long hair and twisted cord wools which come to fall below the beard” (Cieza de León, 1553). Hence, there is no consistent cultural association between the Motilones and the Chankas, even though the latter were assumed to be Motilones by the European chroniclers or conquistadors. Despite the fact that the Motilones and Tabalosos were devastated by a smallpox epidemic in 1645 and killed during punitive Spanish expeditions, it seems that some of their descendants survived to the present day such as the Quechua‐Lamista clans (Pardo et al., 2001; Riva Herrera, 2004).
From another point of view, Scazzocchio (1981: p. 102) makes reasonable proposals as to the genetic background of the Motilones. “If any time a group of Chankas had arrived in the region, and it was integrated into the system of alliances and hostilities, it is unlikely that it could maintain its genetic material unchanged: no mention is made of a dominant group or one isolated in the tribal mosaic on which all sources are agreed”. Considering this scenario among Quechua‐Lamistas, we showed several clusters or “clans” associated with paternal Q‐M3 lineages that probably reflect an ancient genetic legacy of some Amazonian ethnic groups like the Motilones. However, among B2 lineages of Quechua‐Lamistas, there are three clans (labelled as W1, W3, W4) that share a common haplotype (Fig. 5). This suggests that these clans may have a very recent common maternal ancestor. We can make similar interpretations about the shared haplotypes between W1 and W4 clans (labelled as W1, W4). On the other hand, there is a mosaic of mtDNA lineages (shared haplotypes), which indicates high maternal gene flow around northwestern Amazonia, as well as among Andean populations from the Central Andes (Huancavelica, Apurimac, Cusco, Altiplano region) and Quechua speakers from Ecuador (Sandoval et al., 2013b; this study). This observation is also consistent with the chronicles and registers of the displacement of mitmaqs along the Tawantinsuyu Empire, as described by Espinoza Soriano (1999), Julien (2002) and others. However, based on Y‐chromosome and mtDNA profiles, our results showed that the Quechua‐Lamista‐speaking population is more related to Amazonian populations and to the Quechuas from Ecuador than to the self‐defined Chanka population from Andahuaylas, Apurimac Department (Figs 4 and 6, respectively). Indeed, from our data and the chronicles, it seems that some Chankas, after escaping from the Incas, may have moved to the tropical lowlands of the Huanuco‐Pasco‐Junin regions. After allying with or fighting local Amazonian ethnic groups like the Yanesha, Asheninka, Ashaninka, and Nomatsiguenga (Arawakan language groups that inhabit the “ceja de Selva” from Pasco‐Junin), they were probably incorporated into the local sociocultural system, at least some sinchis like Ankoayllo and their relatives. This suggestion is based on the observation of some shared Y‐STR haplotypes between the Chanka and Machiguenga and Yanesha‐speaking individuals (Fig. S2; Sandoval et al., 2013b). It is noteworthy that the Machiguenga, the Nomatsiguenga and the Asheninka are the closest to each other ethnic groups among the Arawak language family in Peru (Walker & Ribeiro, 2011), and it was expected that their genetic profiles were very similar, as well. Concerning the Chankas, the scenario outlined in the chronicles indicates that they were pursued by the Incas across Chachapoyas, Cajamarca and Moyobamba (including the Huallaga and Marañon Rivers), but they were never found (Sarmiento de Gamboa, 1572; de Velasco, 1789; Vazques de Espinoza, 1948). However, in the post‐Columbian period, Spaniards learned in Cajamarca (northern Andes of Peru) that some Chankas were likely hidden around Moyobamba (San Martin Department). In contrast, our results suggest that they probably hid in the lowlands of the Huanuco‐Pasco‐Junin region, and not in the San Martin region, an area which was inhabited by the Motilones and other Amazonian ethnic groups before the Spanish invasion in the 17th century (Scazzocchio, 1981; García, 1999). On the other hand, it is possible that some Chanka individuals also escaped to the valley of the Apurimac and Ene Rivers, an area that was known to them (de la Vega, 1609; Palma, 2010). Furthermore, during the Inca expansion towards the Altiplano region, some Chankas were probably taken as mitmaqs to the mines of Potosi (Bolivia), and also as part of the Inca legion against the Guarani tribes (Nordenskiold, 1917; Orsúa y Vela, 1965). This is partially supported by a network shown in Figure S2, where some Quechua‐ and Aymara‐speaking individuals from the Altiplano and Titicaca Lake region, including Potosi, shared haplotypes with Chanka people from Andahuaylas. In any case, there was a high gene flow between highland Quechua‐ and Aymara‐speaking populations in the Tawantinsuyu, and also during the Hispanic colonial period, which could have included the Chankas (Sandoval et al., 2013b).
Furthermore, our results indicate a clear genetic kinship between Jivaroan‐speaking groups and Quechua speakers from Amazonia. On the other hand, the observed genetic relationship between populations from Huancavelica and Apurimac (Chankas from Andahuaylas) is coherent with their geographic‐linguistic relationships and reported history since pre‐Columbian times (Rostworowski, 2001). In addition, the Quechua speakers (from Otavalo and Karanki) from Ecuador are closely related, in agreement with the chronicles which indicate that the Quitos‐Otavalos‐Carangues (Karanki) and Cayambis were allies against the Incas (Sarmiento de Gamboa, 1572).
In summary, our results bring important clues concerning the origins of Quechua‐Lamistas, Chankas and northwestern Amazonian people from South America that are consistent with the history described by most chroniclers. The results for Quechua‐Lamistas show a clear evidence of an Amazonian genetic background that shaped their patrilineal clans among inhabitants of the Wayku community, while the genetic footprint of the Chankas appears to lie mostly placed in Central Andes.
Conflict of Interest
The authors state that they have no conflicts of interest.
Author Contributions
Conceived and designed the experiments: JRS, FRS, RF. Collected the samples: JRS, ASG, OA, RF, PPRV, PRR, CPyM, FRS. Performed the laboratory experiments: JRS, DRL, MSJ. Analysed the data: JRS, DRL, FRS. Wrote the paper: JRS, FRS, RF. All authors have reviewed and approved the final version of this manuscript.
Supporting information
Disclaimer: Supplementary materials have been peer‐reviewed but not copyedited.
Table S1: (a) List of 15 Y‐STR haplotypes for Q‐M3 and Q‐M346* lineages; (b) list of DYS385 haplotypes; (c) Y‐STR AMOVA results among 15 Peruvian and Ecuadorian populations; (d) the level of expected heterozygosity (He) among 15 Peruvian and Ecuadorian populations. See attached xls files (MS Excel) in Online.
Table S2: (a) List of mtDNA SNPs according to rCRS; (b) MtDNA AMOVA results among 15 Peruvian and Ecuadorian populations; (c) neutrality Tajima's D and Fu's tests; (d) distribution of A2, B2, C1 and D1 mtDNA haplogroup frequencies. See attached xls files (MS Excel) in Online Resource.
Figure S1 Median Joining network for Q‐M3 Y‐STR haplotypes among Quechua‐Lamista population and individuals from other populations that share haplotypes. Some individuals from Loreto, San Martin and Chachapoyas Departments were included in this analysis.
Figure S2 Median Joining network for Q‐M3 Y‐STR haplotypes among 21 Peruvian, Bolivian and Ecuadorian populations. The population groups are depicted with distinct colours. The haplotypes composed of 15 Y‐STRs are represented by circles with sizes proportional to numbers of individuals, and branch lengths are proportional to STR mutation steps (one step unit between haplotypes in the W1 branch). W1, W2, W3, W4 and W5 are the specified Quechua‐Lamista clans identified by “surname”. Total sample, n = 562; 277 from this study and 285 Y‐STR data from Sandoval et al. (2013b).
Figure S3 PCA scatter plot for Q‐M3 Y‐STR data among the 15 Peruvian and Ecuadorian populations. Reynolds’ Rst genetic distances among populations were used. Amazonian populations are represented by circles, Andean populations by squares and the Quechua‐Lamista population by a triangle.
Figure S4 Median Joining network for A2, B2, C1 and D1 of control region mtDNA haplotype frequencies among 17 Peruvian and Ecuadorian populations. We also included published data from Yurimaguas (n = 51, from Justice et al., 2012) and Chachapoyas (n = 14, our unpublished results) locations (Peru). The population groups are depicted with distinct colours.
Figure S5 Correspondence analyses for the four mtDNA haplogroup frequencies (no control region variation is used) among the 17 Peruvian and Ecuadorian populations. We included published data from Yurimaguas (Justice et al., 2012) and Chachapoyas locations (Peru). Amazonian populations (Chachapoyas is located in the Andes‐Amazonia border) are represented by circles, Andean populations by squares and the Quechua‐Lamista population by a triangle.
Figure S6 Spatial analyses using Monmonier's algorithm among the 15 populations to detect gene‐flow barriers. Barriers shown in red colour are results of Y‐STR data and blue colour barriers are mtDNA results. Green lines are Delaunay triangulation and grey lines the Voronoi tessellation according to geographic locations (GPS of populations).
Acknowledgements
We thank all volunteers who donated their samples to this project, with the support of the Peruvian indigenous confederations FEPIKRESAM, CEPKA, CODEPISAM, FERIAAM, ONDEPIP, Drs. Madaleny Ayre, Luis Roman, Gladys Peralta and all fieldwork helpers, but particularly Prof. Toribio Amasifuen Sangama and Mag. Donaldo Pinedo Macedo. This work received funds from the National Geographic Society, the Waitt Family Foundation and IBM from the United States and from FAPEMIG and CNPq from Brazil for laboratory analysis and fieldwork.
References
- Adelaar, W. F. H. (2013) Quechua I y Quechua II: En defensa de una distinción establecida. Rev Bras Linguist Antropol 5(1), 45–65. [Google Scholar]
- Andrews, R. M. , Kubacka. I. , Chinnery, P. F. , Lightowlers, R. N. , Turnbull, D. M. & Howell, N. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23, 147. [DOI] [PubMed] [Google Scholar]
- Arnaiz‐Villena, A. , Moscoso, J. , Serrano‐Vela, J. I. & Martinez‐Laso, J. (2006) The uniqueness of Amerindians according to HLA genes and the peopling of the Americas. Immunología 25, 13–24. [Google Scholar]
- Baeta, M. , Núñez, C. , Sosa, C. , Bolea, M. , Casalod, Y. , Gonzáles‐Andrade, F. , Roewer, L. & Martínez‐Jarreta, B. (2012) Mitochondrial diversity in Amerindian Kichwa and Mestizo populations from Ecuador. Int J Legal Med 126, 299–302. [DOI] [PubMed] [Google Scholar]
- Bandelt, H. ‐J. , Forster, P. & Röhl, A. (1999) Median‐Joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Bauer, B. S. & Kellett, L. C. (2010) Cultural transformations of the Chanka homeland (Andahuaylas, Peru) during the Late Intermediate Period (A.D. 1000‐1400). Lat Am Antiq 21, 87–111. [Google Scholar]
- Behar, D. M. , Rosset, S. , Blue‐Smith, J. , Balanovsky, O. , Tzur, S. , Comas, D. , Mitchell, R. J. , Quintana‐Murci, L. , Tyler‐Smith, C. , Wells, R. S. & Genographic Consortium (2007) The Genographic Project public participation mitochondrial DNA database. PLoS Genet. 3, e104. doi:10.1371/journal.pgen.0030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobo, B. (1653) Historia del Nuevo Mundo. Publisher: Sevilla, Imp. de E. Rasco, Jimenez de la Espada M, 1892, Spain: New York Public Library. [Google Scholar]
- de la Vega, I. G. (1609) Los comentarios reales de los Incas. Lisboa, Portugal: Impreso en casa de Pedro Crasbeeck. [Google Scholar]
- de León P. C. (1553) Crónica del Perú In: Señorío de los Incas (ed. Franklin Pease G. Y.), 2005, Venezuela: Biblioteca Ayacucho. [Google Scholar]
- de Velasco, J. (1789) Historia del reino de Quito. Edición, prólogo, notas y cronología por Alfredo P. Diez Canseco. pp. XLIX+669 Ecuador. Fundacion Biblioteca Ayacucho, Venezuela. [Google Scholar]
- Diamond, J. & Bellwood, P. (2003) Farmers and their languages: The first expansions. Science 300, 597–603. [DOI] [PubMed] [Google Scholar]
- Espinoza Soriano, W. (1999) Etnohistoria Ecuatoriana. Estudios y documentos. Ediciones Abya‐Yala, 2da. ed Quito, Ecuador. [Google Scholar]
- Excoffier, L. & Lischer, H. E. L. (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10, 564–567. [DOI] [PubMed] [Google Scholar]
- Fan, L. & Yao, Y. ‐G. (2011) MitoTool: A web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 11, 351–356. [DOI] [PubMed] [Google Scholar]
- Frisancho, A. R. & Klayman, J. E. (1975) A‐B‐O and Rh affinities between Highland and Lowland Quechua‐speaking Peruvian populations. Am J Phys Anthropol 43, 285–290. [DOI] [PubMed] [Google Scholar]
- Gamboa, S. de P. (1572) The History of the Incas. Translated and edited by Bauer and Smith, 2007, University of Texas Press, Austin, USA. [Google Scholar]
- García Jordan, P. (2001) En el corazón de las tinieblas… del Putumayo, 1890–1932. Fronteras, caucho, mano de obra indígena y misiones católicas en la nacionalización de la Amazonía. Rev Indias 61, 591–617. [Google Scholar]
- García, O. C. D. L. (1999) Historia de las Misiones en la Amazonia Ecuatoriana. 2da ed, Quito, Ecuador. [Google Scholar]
- Georges, L. , Seidenberg, V. , Hummel, S. & Fehren‐Schmitz, L. (2012) Molecular characterization of ABO blood group frequencies in pre‐Columbian Peruvian highlanders. Am J Phys Anthropol 149, 242–249. [DOI] [PubMed] [Google Scholar]
- Homan, J. (2014) The Cordillera Escalera and the Pueblo Shawi: Ethnohistory. Rapid Biological and Social Inventories Report 26. The Field Museum, Chicago, 167–541.
- Hornborg, A. (2005) Ethnogenesis, regional integration, and ecology in prehistoric Amazonia. Curr Anthropol 46, 589–620. [Google Scholar]
- Jota, M. S. , Lacerda, D. R. , Sandoval, J. R. , Vieira, P. P. R. , Santos‐Lopes, S. S. , Bisso‐Machado, R. , Paixão‐Cortes, V. R. , Revollo, S. , Paz‐Y‐Miño, C. , Fujita, R. , Salzano, F. M. , Bonatto, S. L. , Bortolini, M. C. , Santos, F. R. & The Genographic Consortium (2011) A new subhaplogroup of native American Y‐chromosomes from the Andes. Am J Phys Anthropol 146, 553–559. [DOI] [PubMed] [Google Scholar]
- Julien, C. (2002) Diego Maldonado y los Chancas. Revista Andina 34, 183–197. [Google Scholar]
- Justice, A. , Dean, B. & Crawford, M. H. (2012) Molecular consequences of migration and urbanization in Peruvian Amazonia In: Causes and consequences of human migration. An evolutionary perspective (eds. Crawford M. H. & Campbell) B. C. New York: Cambridge University Press. [Google Scholar]
- Karafet, T. M. , Mendez, F. L. , Meilerman, M. B. , Underhill, P. A. , Zegura, S. L. & Hammer, M. F. (2008) New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurin, D. S. , Lofaro, E. M. , Gómez Choque, D. E. & Krigbaum, J. (2014) A bioarchaeological and biogeochemical study of warfare and mobility in Andahuaylas, Peru (ca. AD 1160–1260). Int J Osteoarchaeol doi:10.1002/oa.2398. [Google Scholar]
- Manni, F. , Guerard, E. & Heyer, E. (2004) Geographic patterns of (genetic, morphological, linguistic) variation: How barriers can be detected by using Monmonier's algorithm. Hum Biol 76, 173–190. [DOI] [PubMed] [Google Scholar]
- Moscoso, J. , Seclen, S. , Serrano‐Vela, J. I. , Villena, A. , Martinez‐Laso, J. , Zamora, J. , Moreno, A. , Ira‐Cachafeiro, J. & Arnaiz‐Villena, A. (2006) HLA genes in Lamas Peruvian‐Amazonian Amerindians. Mol Immunol 43, 1881–1889. [DOI] [PubMed] [Google Scholar]
- Murúa, M. de (1613) Historia general del Perú y descendencia de los Incas. Ed. de Manuel Ballesteros Gabrois, 1986. Serie Crónicas de América: Historia 16; Madrid, Spain. [Google Scholar]
- Nordenskiold, B. E. (1917) The Guarani invasion of the Inca empire in the sixteenth century: An historical Indian migration. Geogr Rev 4, 103–121. [Google Scholar]
- Orsúa y Vela, B. A. de (1965) Historia de la villa imperial de Potosí. Edición de L. Hanke y G. Mendoza. Rhode Island, RI: Brown University Press. [Google Scholar]
- Palma, L. (2010) Socioeconomía, informe temático Proyecto Mesozonificación Ecológica y Económica para el desarrollo sostenible del valle del río Apurímac ‐ VRA. Iquitos, Peru: IIAP. [Google Scholar]
- Pardo, C. M. , Doherty, V. J. & Sangama, S. I. (2001) Los Kechuas Lamistas y la Educación Bilingüe Intercultural: Historia y Razón de un compromiso. Ed. Tarapoto, San Martin, Perú: SAC PT. [Google Scholar]
- Ravi Mumford, J. (2012) Vertical empire: The general resettlement of Indians in the colonial Andes. Durham, NC: Duke University Press. [Google Scholar]
- Reeve, M. ‐E. (2014) Amazonian Quichua in the western Amazon regional interaction sphere. Tipití: SALSA 12, 14–27. [Google Scholar]
- Rey, D. , Areces, C. , Enríquez‐de‐Salamanca, M. , Parga‐Lozano, C. , Abd‐El‐Fatah, S. , Fernández, M. y Arnaiz‐Villena, A. (2012) Los primeros pobladores de América y sus relaciones con poblaciones del Océano Pacífico según los genes HLA. Immunología 31, 83–91. [Google Scholar]
- Riva Herrera, M. de la (2004) La Conquista de los Motilones, Tabalosos, Maynas y Jibaros. Iquitos, Perú: CETA. [Google Scholar]
- Roewer, L. , Nothnagel, M. , Gusmão, L. , Gomes, V. , González, M. , Corach, D. , Sala, A. , Alechine, E. , Palha, T. , Santos, N. , Ribeiro‐dos‐Santos, A. , Geppert, M. , Willuweit, S. , Nagy, M. , Zweynert, S. , Baeta, M. , Núñez, C. , Martínez‐Jarreta, B. , Gonzáles‐Andrade, F. , Fagundes‐de‐Carvalho, E. , Aparecida‐da‐Silva, D. , Builes, J. J. , Turbón, D. , Lopez‐Parra, A. M. , Arroyo‐Pardo, E. , Toscanini, U. , Borjas, L. , Barletta, C. , Ewart, E. , Santos, S. & Krawczak, M. (2013) Continent‐wide decoupling of Y chromosomal genetic variation from language and geography in Native South Americans. PLoS Genet. 9, e1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostworowski, M. (2001) Pachacutec Inca Yupanqui. IEP. Serie: Historia Andina, 23: Lima, Peru. [Google Scholar]
- Sandoval, J. R. , Lacerda, D. R. , Jota, M. S. A. , Salazar‐Granara, A. , Vieira, P. P. R. , Acosta, O. , Cuellar, C. , Revollo, S. , Fujita, R. , Santos, F. R. & The Genographic Project Consortium (2013b) The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: The legacy of the Uros. PLoS One 8, e73006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval, J. R. , Salazar‐Granara, A. , Acosta, O. , Castillo‐Herrera, W. , Fujita, R. , Pena, S. D. J. & Santos, F. R. (2013a) Tracing the genomic ancestry of Peruvians reveals a major legacy of pre‐Columbian ancestors. J Hum Genet 58, 627–634. [DOI] [PubMed] [Google Scholar]
- San Roman, J. V. (1994) Perfiles Históricos de la Amazonia Peruana, 2da. ed Iquitos, Perú: CETA, CAAAP, IIAP. [Google Scholar]
- Scazzocchio, F. (1981) La Conquete des Motilones du Huallaga Central aux XVIIe et XVIIIe siecles. Bull Inst Fr Et And X, 99–111. [Google Scholar]
- Schjellerup, I. (1999) Wayko ‐ Lamas: a Quechua community in the Selva Alta of North Peru under change. Geografisk Tidsskrift, Danish J Geogr Special Issue (I), 199–207. [Google Scholar]
- Tarazona‐Santos, E. , Carvalho‐Silva, D. R. , Pettener, D. , Luiselli, D. , De Stefano, G. F. , Labarga, C. M. , Rickards, O. , Tyler‐Smith, C. , Pena, S. D. & Santos, F. R. (2001) Genetic differentiation in South Amerindians is related to environmental and cultural diversity: Evidence from the Y chromosome. Am J Hum Genet 68, 1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G. (1979) Diccionario normalizado y comparativo quechua: Chachapoyas – Lamas. Paris, France: L'Harmattan. [Google Scholar]
- Torero, A. (1964) Los dialectos Quechuas. Separata de Anales Científicos de la Universidad Agraria 2, 446–478. [Google Scholar]
- Vazques de Espinoza, A. (1948) Compendio y descripción de las Indias occidentales. Smithsonian miscellaneous collections volumen 108. The Smithsonian Institution, Washington, USA. [Google Scholar]
- Walker, R. S. & Ribeiro, L. A. (2011) Bayesian phylogeography of the Arawak expansion in lowland South America. Proc R Soc B 278, 2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal, T. , Dashnyam, B. , Pandya, A. , Kayser, M. , Rower, L. , Santos, F. R. , Schiefenhövel, W. , Fretwell, N. , Jobling, M. A. , Harihara, S. , Shimizu, K. , Semjidmaa, D. , Sajantila, A. , Salo, P. , Crawford, M. H. , Ginter, E. K. , Evgrafov, O. V. & Tyler‐Smith, C. (1997) Genetic relationships of Asians and northern Europeans revealed by Y‐chromosomal DNA analysis. Am J Hum Genet 60, 1174–1183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclaimer: Supplementary materials have been peer‐reviewed but not copyedited.
Table S1: (a) List of 15 Y‐STR haplotypes for Q‐M3 and Q‐M346* lineages; (b) list of DYS385 haplotypes; (c) Y‐STR AMOVA results among 15 Peruvian and Ecuadorian populations; (d) the level of expected heterozygosity (He) among 15 Peruvian and Ecuadorian populations. See attached xls files (MS Excel) in Online.
Table S2: (a) List of mtDNA SNPs according to rCRS; (b) MtDNA AMOVA results among 15 Peruvian and Ecuadorian populations; (c) neutrality Tajima's D and Fu's tests; (d) distribution of A2, B2, C1 and D1 mtDNA haplogroup frequencies. See attached xls files (MS Excel) in Online Resource.
Figure S1 Median Joining network for Q‐M3 Y‐STR haplotypes among Quechua‐Lamista population and individuals from other populations that share haplotypes. Some individuals from Loreto, San Martin and Chachapoyas Departments were included in this analysis.
Figure S2 Median Joining network for Q‐M3 Y‐STR haplotypes among 21 Peruvian, Bolivian and Ecuadorian populations. The population groups are depicted with distinct colours. The haplotypes composed of 15 Y‐STRs are represented by circles with sizes proportional to numbers of individuals, and branch lengths are proportional to STR mutation steps (one step unit between haplotypes in the W1 branch). W1, W2, W3, W4 and W5 are the specified Quechua‐Lamista clans identified by “surname”. Total sample, n = 562; 277 from this study and 285 Y‐STR data from Sandoval et al. (2013b).
Figure S3 PCA scatter plot for Q‐M3 Y‐STR data among the 15 Peruvian and Ecuadorian populations. Reynolds’ Rst genetic distances among populations were used. Amazonian populations are represented by circles, Andean populations by squares and the Quechua‐Lamista population by a triangle.
Figure S4 Median Joining network for A2, B2, C1 and D1 of control region mtDNA haplotype frequencies among 17 Peruvian and Ecuadorian populations. We also included published data from Yurimaguas (n = 51, from Justice et al., 2012) and Chachapoyas (n = 14, our unpublished results) locations (Peru). The population groups are depicted with distinct colours.
Figure S5 Correspondence analyses for the four mtDNA haplogroup frequencies (no control region variation is used) among the 17 Peruvian and Ecuadorian populations. We included published data from Yurimaguas (Justice et al., 2012) and Chachapoyas locations (Peru). Amazonian populations (Chachapoyas is located in the Andes‐Amazonia border) are represented by circles, Andean populations by squares and the Quechua‐Lamista population by a triangle.
Figure S6 Spatial analyses using Monmonier's algorithm among the 15 populations to detect gene‐flow barriers. Barriers shown in red colour are results of Y‐STR data and blue colour barriers are mtDNA results. Green lines are Delaunay triangulation and grey lines the Voronoi tessellation according to geographic locations (GPS of populations).


