Abstract
The malaria parasite Plasmodium spp. varies the expression profile of its genes depending on the host it resides in and its developmental stage. Virtually all messenger RNA (mRNA) is expressed in a monocistronic manner, with transcriptional activation regulated at the epigenetic level and by specialized transcription factors. Furthermore, recent systems‐wide studies have identified distinct mechanisms of post‐transcriptional and translational control at various points of the parasite lifecycle. Taken together, it is evident that ‘just‐in‐time’ transcription and translation strategies coexist and coordinate protein expression during Plasmodium development, some of which we review here. In particular, we discuss global and specific mechanisms that control protein translation in blood stages of the human malaria parasite Plasmodium falciparum, once a cytoplasmic mRNA has been generated, and its crosstalk with mRNA decay and storage. We also focus on the widespread translational delay observed during the 48‐hour blood stage lifecycle of P. falciparum—for over 30% of transcribed genes, including virulence factors required to invade erythrocytes—and its regulation by cis‐elements in the mRNA, RNA‐processing enzymes and RNA‐binding proteins; the first‐characterized amongst these are the DNA‐ and RNA‐binding Alba proteins. More generally, we conclude that translational regulation is an emerging research field in malaria parasites and propose that its elucidation will not only shed light on the complex developmental program of this parasite, but may also reveal mechanisms contributing to drug resistance and define new targets for malaria intervention strategies. WIREs RNA 2016, 7:772–792. doi: 10.1002/wrna.1365
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Plasmodium spp., the causative agents of malaria, are unicellular protozoan parasites belonging to the phylum Apicomplexa that use a multi‐stage developmental program to transition between their mammalian host and mosquito vector: while replicative stages in the host are strictly haploid and intracellular, cell division in the vector occurs in an extracellular milieu, within a cyst‐like structure. Each developmental stage (Figure 1) has a distinct morphology and physiology that is determined by its gene expression profile, as revealed by stage‐specific transcriptomic and proteomic analyses of human and rodent malaria parasites.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 The best studied amongst these is the 48‐hour intraerythrocytic developmental cycle (IDC) of the lethal human malaria parasite, Plasmodium falciparum, when a single parasite replicates within mature erythrocytes to produce 8–32 daughter cells (i.e., merozoites; Figure 1). Indeed, systems‐wide studies have shown that the P. falciparum IDC is characterized by a cyclic pattern of steady‐state messenger RNA (mRNA) expression, with more than 75% of the genes attaining peak mRNA levels at only one time‐point of the 48‐h cycle.3, 11, 19 Moreover, for approximately 30% of genes, a delay is observed between peak mRNA and protein levels;11, 21 in select cases, this corresponds to a delay in ribosome association.5, 6
Figure 1.
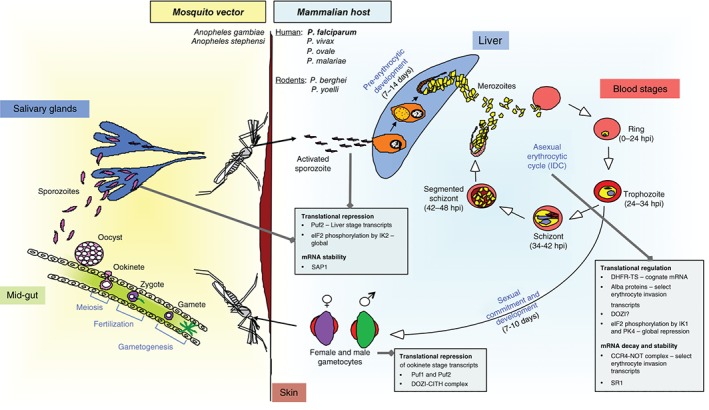
Lifecycle of Plasmodium spp. parasites in the mammalian host and mosquito vector. The timing of different stages of P. falciparum in the human host is indicated. Instances of translational regulation and mRNA decay are also indicated. Note that the monkey malaria parasite P. knowlesi has recently been identified to infect humans.
Research in the past decade has shown that gene expression in malaria parasites is governed by epigenetic, transcriptional, post‐transcriptional, and post‐translational regulatory mechanisms.36, 37, 38, 39, 40 Because protein production is the primary outcome of gene expression, mRNA translation, in particular, is coordinated in time and space to fine‐tune protein levels and respond to a variety of environmental cues:41 in Plasmodium spp., this occurs not just during the IDC, but also facilitates developmental stage transitions. The prevalence of post‐transcriptional control is also supported by the apparent lack of specialized transcription factors encoded by the parasite genome, relative to genes encoding RNA‐binding and ‐regulatory proteins.42, 43, 44
Similar to other eukaryotes, the life of a Plasmodium spp. mRNA begins in the nucleus, when the transcriptional machinery is recruited to the promoter of a gene to synthesize pre‐mRNA (or nascent mRNA) that corresponds to the 5′‐untranslated region (UTR), coding sequence (CDS), introns, and 3′UTR of the gene (Figure 2). Regulatory events include the synthesis of cryptic RNAs from cognate promoters of select P. falciparum genes, which may be cotranscriptionally degraded by exoribonucleases;45 antisense transcription from a downstream promoter27, 46 (Figure 2); transcription from intronic promoters for select P. falciparum virulence gene families;47, 48 and stage‐dependent regulation of specific genes by the 27‐member specialized transcription factor family ApiAP2 (Apicomplexan Aptela‐2).49 Subsequent processing of the pre‐mRNA via splicing (for intron‐containing genes), accompanied by the addition of the m7GpppN cap structure to the 5′ end and the poly(A) tail to the 3′ end, results in a mature mRNA that is competent for cytoplasmic export (Figure 2). In the cytoplasm, the mature mRNA faces three fates: (1) decoding of its message by the 80S ribosome and tRNAs charged with amino acids to synthesize specific polypeptides, i.e., translation, (2) degradation by exo‐ or endo‐ribonucleases, i.e., decay, and (3) sequestration within specific ribonucleoprotein (RNP) complexes to inhibit translation and/or decay, i.e., repression (Figure 2). The equilibrium amongst these processes is a key determinant of the proteomic signature at any given stage of parasite growth, although the steady‐state protein levels also depend on protein degradation rates.
Figure 2.

Stages of mRNA maturation and cytoplasmic outcomes. Once a pre‐mRNA is transcribed in the nucleus from a given GOI (gene of interest), it is co‐ and post‐transcriptionally processed by splicing, 5′ cap modification, and 3′ polyadenylation to yield a mature mRNA. For select GOIs in Plasmodium falciparum, so‐called cryptic RNAs and antisense RNAs have been described in asexual blood stages, which may regulate transcription and/or translation. The mature mRNA is then bound by nuclear ribonucleoprotein (RNP) complexes and transported to the cytoplasm where it faces three outcomes: (1) Translation, mediated by the 80S ribosome to synthesize a polypeptide chain; (2) Decay, mediated by ribonucleases; and (3), Repression, which may occur in the context of mRNA‐RNP (mRNP) granules composed of different classes of proteins such as RNA helicases, RNA‐binding proteins, and translation‐associated factors.
Here, we summarize the current knowledge of translational regulation in malaria parasites, both at the global and mRNA‐specific levels, with an emphasis on mechanisms that are prevalent during the P. falciparum IDC. We discuss how insights from transcriptome‐wide studies of ribosome occupancy and mRNA decay, the identification of cis‐acting elements in the UTRs of select mRNAs, and the characterization of the mRNA interactomes of trans‐acting RNA‐binding proteins (RBPs), have provided a complex, but as yet incomplete, picture of translational control in this unicellular pathogen. Because translational regulation during Plasmodium spp. stage transitions has recently been reviewed elsewhere,36 we will discuss these findings in brief, where relevant.
CORE TRANSLATION APPARATUS AND ITS REGULATION IN PLASMODIUM
The core translation machinery is highly conserved in Plasmodium spp. with regard to its repertoire of ribosome components, translation factors, tRNA molecules, and aminoacyl‐tRNA synthetases (reviewed in Jackson et al.).50 However, certain unique features have been noted as the malaria parasite progresses from mammalian development (consisting of liver stages, the IDC, and sexual stages, i.e., gametocytes; Figure 1) to mosquito stages (consisting of gamete, zygote, ookinete, oocyst, and sporozoite stages; Figure 1).
Translation Machinery
In contrast to most eukaryotes, which have ribosomal DNA (rDNA) clusters comprising hundreds of tandemly repeated units, Plasmodium genomes feature 4–8 single‐copy rDNA units on different chromosomes: these differ in sequence as well as expression profile during lifecycle progression, and are broadly classified into A‐type (liver and blood asexual stage‐specific), O‐type (ookinete‐specific), and S‐type (sporozoite‐specific) rDNA.51, 52, 53, 54, 55, 56, 57, 58, 59 Moreover, the different rRNAs are functionally divergent. For example, heterologous complementation studies in yeast showed that P. falciparum S‐Type rRNA were not functionally equivalent to A‐Type rRNA.58 This hinted at the presence of regulatory elements within the different rRNAs that could contribute to translational control, either globally or for a subset of mRNAs. Additionally, in the rodent malaria parasite P. berghei, S‐type rDNA loci appear to be dispensable for lifecycle completion and only affect parasite fitness,60 whereas in a second rodent parasite P. yoelii, an S‐type rRNA gene was associated with oocyst development defect (ODD) by quantitative trait locus analysis.61 Not only was this phenotype verified by targeted genetic knockout of the rDNA locus, ODD was also observed upon episomal complementation of the mutant strain,61 emphasizing the importance of tight regulation of rRNA dose and timing of expression. The concept of specialized ribosomes with stage‐specific functions is enticing, but due to alterations in rDNA sequence and locus numbers across different Plasmodium species, a generalized interpretation requires further research.
Recently, high‐resolution cryo‐electron microscopy (cryo‐EM) reconstructions of the P. falciparum blood stage, cytoplasmic 80S ribosome either bound to the antimalarial drug emetine62 or bound to tRNAs63 were published. While the core structure was well conserved, some P. falciparum‐specific differences were identified including large rRNA expansion segments (ESs) at the periphery of the 18s rRNA; these ESs may be targeted by the translational regulation machinery in vivo and present as potential targets for drug intervention.62, 63 Additionally, both studies noted the striking absence of PfRACK1 (receptor for activated C kinase protein)64 from the conserved binding site near the mRNA exit channel; RACK1 has been shown to associate with eukaryotic ribosomes in stoichiometric quantities and is thought to be involved in translational regulation, especially internal ribosomal entry site (IRES)‐mediated, cap‐independent translation initiation.65 Absence of RACK1 from the Plasmodium blood stage ribosome could either be due to loss during purification or could point to important mechanistic differences in P. falciparum translational regulation; of note, PfRACK1 copurified with blood stage polysomes.5 Moreover, RACK1‐ribosome association may be regulated in a stage‐ and rRNA‐dependent manner.
Another notable aspect is the moonlighting, extra‐ribosomal function of Plasmodium 60S‐ribosomal stalk proteins P0 and P2. P0 has been found on the merozoite and gametocyte surface and has been implicated in the invasion process66 while P2 is found on the infected erythrocyte surface during early schizogony and is thought to be involved in regulation of nuclear division.67 Extra‐ribosomal functions of eukaryotic ribosomal proteins, such as regulation of ribosome biogenesis and stress sensing, have been previously reported,68 and it appears that Plasmodium spp. ribosomal proteins may also have evolved alternate functions. Overall, the heterogeneity in Plasmodium rRNAs, ribosome structure and the function of ribosomal proteins need to be better understood, especially to shed light on translational regulation and parasite development.
Global Regulation of Translation
Translation in eukaryotes can be divided into three phases, initiation, elongation, and termination, all of which are subject to regulation (Figure 3). In Plasmodium spp., mechanistic conservation has been experimentally demonstrated for some of the phases. For example, in model eukaryotes, during cap‐dependent translation initiation, the 5′ m7GpppN cap structure of an mRNA is bound by eIF4E (eukaryotic translation Initiation Factor 4E), which is part of the heterotrimeric translation initiation complex eIF4F consisting of the helicase eIF4A, the scaffold protein eIF4G, and eIF4E69 (Figure 3). Subsequently, interaction of eIF4F with poly(A)‐binding protein (PABP), decorating the 3′ poly(A)‐tail, results in the formation of a closed‐loop mRNA‐RNP (mRNP) structure that both promotes efficient translation and deters mRNA decay (Figure 2). The interactions of P. falciparum eIF4E,70, 71 eIF4F,72, 73 and PABP72, 73 have been demonstrated in vitro. Next, eukaryotic translation begins when the 80S ribosome and the methionine‐loaded initiator tRNA are assembled on the start codon with the help of eIF269 (Figure 3); Plasmodium eIF2 has been annotated, but not functionally characterized. However, phosphorylation of the alpha subunit of eIF2, eIF2α, under conditions of stress, one of the most common modes of global mRNA translational repression in eukaryotes, has been demonstrated in Plasmodium (Figure 3).40 The three eIF2α serine/threonine kinases encoded by the parasite genome, eIK1, eIK2, and PK4, are differentially expressed during its lifecycle and exhibit specialized function for stress response and stage‐specific repression of translation in sporozoites and schizonts74, 75, 76, 77, 78 (Table 1). Additionally, the translation elongation complex, which contains four subunits EF1α, β, γ, and δ, was purified from P. falciparum blood stages under native conditions79 (Figure 3), although its functionality remains uncharacterized. Given that new modes of regulation of elongation are being discovered in model organisms,80 and that translational accuracy and efficiency of the AT‐rich mRNAs of P. falciparum may be determined by ribosome stalling at low complexity regions such as asparagine‐encoding AAT tracts,81 translation elongation in malaria parasites needs to be further elucidated. This is especially important because the GTP‐dependent elongation factor PfeEF2 was recently described as a target of the small molecule inhibitor DDD10749882 (discussed below), although PfeEF2 function in translation elongation has not been demonstrated in vitro or in vivo. Finally, while eukaryotic peptide chain release factors (eRFs; Figure 3) that facilitate translation termination have been annotated in Plasmodium spp. (http://plasmodb.org), their function is not yet characterized.
Figure 3.

Molecular basis of translational regulation in malaria parasites. Translational regulation in the cytoplasm can occur during the initiation, elongation, and termination phases. This can be co‐regulated by factors that mediate decay and repression. eIF, eukaryotic Initiation Factor; tRNAmet, tRNA charged with methionine; eEF, eukaryotic Elongation Factor; eRF, eukaryotic Release Factor; eIF4F complex, eIF4E + eIF4G + eIF4A; PABP, Poly(A)‐Binding Protein; UTR, Untranslated Region; CDS, Coding Sequence; uORF, upstream Open Reading Frame; PBE, Puf‐Binding Element.
Table 1.
Molecular Regulators of Translation, Decay, and Repression That Have Been Characterized in Plasmodium spp.
| Biological Process | Protein/Protein Complex | Species | Lifecycle Stage | Expressed in Asexual Stages? | Remarks | References |
|---|---|---|---|---|---|---|
| Translation | eIF2 α Kinases: IK1, IK2, PK4 | P. falciparum P. berghei | IDC, Sporozoite, Gametocytes | Yes | IK1 is primarily transcribed in asexual blood stages, IK2 in sporozoites and PK4 during the IDC and in gametocytes; IK1 and IK2 are not essential to parasite growth (all stages of development) whereas the PK4 gene cannot be deleted in blood stages | 73, 74, 75, 76, 77 |
| PfDZ50 | P. falciparum | IDC | Yes | DDX6/Dhh1 RNA helicase and homolog of DOZI; RNA‐binding and RNA helicase activities of PfDZ50 have been demonstrated in vitro as has its ability to bind to PfeIF4E and repress translation | 112 | |
| PfAlba1 | P. falciparum | IDC | Yes | PfAlba1 is essential to blood stage development; The in vivo RNA interactome of PfAlba1 consists of >105 mRNAs, including mRNAs encoding erythrocyte invasion proteins such as Rap1, AMA1, RhopH3 and CDPK1 | 120, 122 | |
| mRNA decay | PfCaf1 | P. falciparum | IDC | Yes | Deadenylase subunit of the CCR4‐NOT complex; Of the 1031 mRNAs that are misregulated upon PfCaf1 depletion, mRNAs encoding erythrocyte egress and invasion proteins are over‐represented | 125 |
| RNA exosome (RRP6, DIS3) | P. falciparum | IDC | Yes | PfRRP6 and PfDIS3 coimmunoprecipitate with the exosome and localize to the nucleus and cytoplasm, respectively, of ring stage parasites | 45 | |
| Translational repression | PbDOZI‐CITH complex | P. berghei | Female gametocytes | Yes | PbDOZI and PbCITH localize to the cytoplasm in P‐granule‐like structures, with the PbDOZI‐CITH complex composed of 11 proteins; Targets include ~730 mRNAs, several of which are repressed in sexual stages and translated only in the zygote and ookinete | 130, 131, 132 |
| Puf2 | P. falciparum P. berghei P. yoelii | Gametocytes, Sporozoites | Yes | PfPuf2 binds to the Puf‐binding element in the 5' and 3′UTRs of two mRNAs, Pfs25 and Pfs28, in gametocytes, and translationally represses them; PbPuf2 and PyPuf2 translationally regulate the UIS mRNAs in sporozoites | 138, 139, 140, 141, 142 | |
| Bruno/CELF | P. falciparum P. berghei | IDC, Female gametocytes | Yes | PfCELF1 localizes to punctate structures in the cytoplasm; ~1100 mRNA targets of PfCELF1 have been identified using in vitro RNA‐binding assays; PbCELF2 associates with the DOZI‐CITH complex | 132, 144 | |
| Alba1, 2 and 3 | P. berghei P. falciparum | Female gametocytes, IDC | Yes | PbAlba1, 2 and 3 associate with the DOZI‐CITH complex; PfAlba1, 2 and 3 bind to RNA in vitro; The cytoplasmic localization of PfAlba1 and 2 in trophozoites and schizonts is punctate and reminiscent of P‐granules | 120, 132 |
Translation in the Apicoplast and Mitochondria
There is evidence that, in addition to bulk translation in the cytoplasm, both Plasmodium organelles of endosymbiotic origin, the apicoplast and the mitochondrion, are translationally active.83, 84, 85 The apicoplast, which is a nonphotosynthetic remnant of the plastid, is a peculiarity of several organisms belonging to the phylum Apicomplexa and is required for fatty acid and isoprenoid biosynthesis, and parasite survival.86 The 35 kb apicoplast genome encodes its own set of rRNAs, tRNAs, some ribosomal proteins and the bacterial translation elongation factor EF‐Tu,87 while the highly reduced 6 kb genome of the mitochondrion encodes only three proteins and fragmented rRNAs.88, 89 To achieve functional translation in each organelle, many factors including ribosomal proteins, translation factors, and tRNA synthetases, have to be encoded by the nuclear genome, with some factors shared between the cytoplasm and these organelles.50, 90, 91, 92 Furthermore, nuclear‐encoded, divergent P. falciparum homologues of the bacterial initiation factors IF1, 2, and 3 were recently characterized, and specific isoforms localized to either the apicoplast or mitochondrion.93 Organellar translation is of particular interest, in view of being the predicted target of inhibitors of prokaryotic translation, which are also active against malaria parasites94, 95, 96 (discussed below).
LESSONS FROM SYSTEMS‐WIDE STUDIES OF P. FALCIPARUM ASEXUAL BLOOD STAGES
The first instance of translational regulation during the P. falciparum IDC was observed in 2002 for dihydrofolate reductase–thymidylate synthase (DHFR‐TS), a bifunctional enzyme that is essential for parasite growth.97 Binding of PfDHFR‐TS to its cognate mRNA repressed translation,98 which could be alleviated by antifolate treatment; importantly, such a treatment did not affect steady‐state PfDHFR‐TS mRNA levels.99 These findings supplemented previous studies in P. berghei, where researchers had reported translational repression in gametocyte stages (discussed below). Nonetheless, the widespread prevalence of translational control in blood stages became evident only in the postgenomic area.
Transcriptomics and Proteomics
The draft genome of P. falciparum was first published in 200243 and was closely followed by the transcriptome and proteome of several lifecycle stages.3, 9, 19 The transcriptome of the 48‐h IDC was measured independently by two groups in 2003: Le Roch et al. used 25 base‐long oligonucleotide arrays to analyze the transcriptomes of rings [8–16 hours post erythrocyte invasion (hpi)], trophozoites (22–30 hpi), schizonts (36–42 hpi), and merozoites,19 while Bozdech et al. performed a higher time‐resolution study, where they used 70 base‐long oligonucleotide arrays to measure the IDC transcriptome at 1‐h intervals.3 Together, it was revealed that at least 3240 of the then‐annotated approximately 5400 P. falciparum genes (the latest annotation being ~5770 genes) were transcribed during this stage and exhibited a cyclic pattern of expression, with >75% of the mRNAs reaching peak steady‐state levels at only one time‐point; a study in 2011 extended this analysis to 4670 IDC mRNAs.11 This resulted in the assignment of a ‘Fourier phase’ to several genes, which corresponded to peak timing of mRNA expression. Notably, transcripts that encoded proteins belonging to the same biological process such as DNA replication, protein translation (peak mRNA at 8–12 hpi), proteasomal degradation, and so on, were co‐expressed, leading to a model of ‘just‐in‐time’ transcription, i.e., induction of gene expression occurred only at one specific point of the lifecycle, exactly when protein function was required. Moreover, when the proteome of select lifecycle stages including a few blood stage time‐points was measured by Multidimensional Protein Identification Technology (MudPIT)9 and mass spectrometry (MS),25 and of the IDC at 2‐h intervals by two‐dimensional DIfferential Gel Electrophoresis (2D‐DIGE),11 a periodic pattern of protein expression was observed for the IDC. However, this pattern was more discontinuous than the transcriptional cascade, with two distinct breaks at 6–14 and 26–34 hpi, indicative of replenishment/restocking of proteins at these time‐points.11
Thereafter, when the transcriptomic profiles of seven different P. falciparum stages—including four IDC time‐points19—were compared to their protein profiles, it became evident that there was a delay of 11–18 h between peak mRNA and protein levels for approximately 30% of the 2584 analyzed genes.21 This hinted at widespread translational regulation, especially during the transition from merozoite to ring stages, and from ring to trophozoite stages,21 supporting a model of ‘just‐in‐time’ translation for several mRNAs. A subsequent study revisited this question, but for a reduced set of genes and at 2‐h intervals of the IDC.11 Using microarrays for transcriptome analysis, and 2D‐DIGE followed by MS for proteomic analysis, the steady‐state expression patterns of 125 proteins (with 2.9 isoforms per protein on average) were compared to their mRNA profiles. Again, a delay of 11–15 h was observed between peak mRNA and protein levels for a majority of the full‐length protein isoforms. However, using a mathematical model, the authors attributed this delay to rates of protein translation (k transl) and degradation (k deg) alone, and ruled out checkpoints between transcription and translation, lending support to the ‘just‐in‐time’ transcription model. Nevertheless, given the relatively small number of proteins analyzed in this study and the assumptions of k transl and k deg made by the researchers, it remains unclear if the model can fully explain the widespread translational delay observed during this stage. In general, the genome‐wide picture of translation regulation that has emerged from these studies is incomplete and needs to be further elucidated by evaluating the blood stage proteome at high time‐resolution, using quantitative proteomic analysis techniques such as SILAC (Stable Isotope Labeling by Amino acids in Cell culture) and iTRAQ (Isobaric Tagging reagents for Relative and Absolute Quantification), both of which have been adapted to P. falciparum in vitro culture.25, 100
Ribosome Occupancy Studies
To globally identify instances of translational regulation, genome‐wide approaches that correlate ribosomal occupancy of mRNAs to their steady‐state levels have been developed,101 with the premise that ribosome–mRNA association is an indicator of active protein production. One such study by Bunnik et al. utilized sucrose gradients to isolate polysome‐mRNA complexes from three time‐points of the P. falciparum IDC—0, 18, and 36 hpi—and analyzed protein and RNA composition by MudPIT and directional RNA‐seq, respectively.5 Upon comparing polysome profiles to the trancriptome at each time‐point, the authors found that for 1749 mRNAs, peak steady‐state levels and polysome association peaks were coincident, while for 738 mRNAs, there was a partial delay, with polysome occupancy sustained into the next time‐point, even in the absence of detectable steady‐state mRNA levels. Finally, for 1280 mRNAs, there was a marked delay in polysome occupancy (of up to 18 h) indicating that these mRNAs were maintained (or possibly stored) in a translationally inactive state until the time when protein synthesis was required, supporting the model of ‘just‐in‐time’ translation. Such a mechanism was especially evident for proteins that are required during or immediately after erythrocyte invasion, and are involved in remodeling the erythrocyte as the parasite establishes its niche.
A second study by Caro et al. utilized ribosome profiling to evaluate the diverse pool of mRNA footprints (28–30 nt long) generated by actively translating 80S ribosomes in five IDC stages—11, 21, 31, and 45 hpi and merozoites (considered as the 2 h time‐point); the ribosome footprints were identified by directional RNA‐seq.6 When the ribosome footprint density of 3605 genes was compared to mRNA abundance at each time‐point, a strong correlation was apparent for 3110 genes, all of which had a single mRNA peak; this indicated that there was no apparent delay between mRNA production and ribosome association. This was in contrast to the observations by Bunnik et al. and could be partly explained by: (1) the different time‐points analyzed in the two studies, and (2) the fact that Bunnik et al. analyzed polysomes (i.e., two or more ribosomes per mRNA), whereas Caro et al. assessed both monosomes and polysomes. Next, the authors calculated the translational efficiency (TE) of each mRNA as: ribosome footprint rpkM/mRNA rpkM; where rpkM is the reads per kilobase of exon model per million sequencing reads mapped; and identified 177 mRNAs with high TE and 124 mRNAs with low TE at each time‐point of the IDC tested. These corresponded, respectively, to translationally up‐ or downregulated mRNAs, although the authors did not rule out translational regulation of mRNAs that did not exhibit extreme TE patterns. Notably, 73 of the 177 mRNAs with elevated TE encode proteins involved in merozoite egress and/or erythrocyte invasion, key parasite virulence factors.
Furthermore, the two studies identified potential cis‐regulators of translation. Bunnik et al. mapped polysome‐associated sequencing reads to the 5′UTR and 3′UTR of select mRNAs as well as to intronic and intergenic sequences: e.g., 409 mRNAs showed twofold higher coverage in the 5′UTR as compared to the CDS. The authors correlated this higher coverage to the presence of upstream Open Reading Frames (uORFs) in the 5′UTR (Figure 3) that could ‘trap’ the 80S ribosome and inhibit translation of the coding sequence, as has been observed in yeast and mammals;102 however, they did not further validate this hypothesis. In contrast, Caro et al., who also mapped ribosomal footprints to the 5′UTR of a subset of genes, found that ribosome occupancy in the 5′UTR did not correlate with the presence of predicted uORFs. The exceptions were var2csa, which encodes the surface virulence antigen PfEMP1‐CSA (Erythrocyte Membrane Protein 1‐Chondroitin Sulphate A), and PF3D7_0531000, which encodes a conserved Plasmodium spp. protein of unknown function. Indeed, it has been demonstrated that the 360‐base uORF in the 5′UTR sequence of var2csa represses translation of the CDS and that this effect can be transiently reversed in vitro. 103, 104, 105 An independent bioinformatic study found 22860 uORFs in the −350 to −1 position of 5211 P. falciparum genes, with an over‐representation in select virulence genes such as var. 106 Moreover, the uORFs from the 5′UTR of three genes were able to repress translation of a downstream reporter gene.106 Overall, it is possible that uORF translation may not only inhibit translation initiation of the CDS, but may also result in protein products such as functional small peptides.107 Additionally, the presence of ribosomes in the 5′UTR may be dependent on IRESs and other sequences,108 which remain undescribed in P. falciparum, but, if they exist, might have effects on translation elongation and termination. It is also important to note that in the ribosome profiling study by Caro et al.,6 some of the footprints might have originated from RNP complexes that migrate at the same size as the 80S ribosome in sucrose gradients109 and should be cautiously interpreted. Overall, systems‐wide studies support translational control in blood stages of P. falciparum, although the extent of translational delay and its contribution to steady‐state protein levels still remains unclear.
mRNA Decay during the IDC
Studies in yeast and mammals have shown that the half‐life (t 1/2) of an mRNA is closely related to its biological role and can be altered in response to different environmental stimuli and during development; this in turn affects translational capacity.110 Therefore, an in‐depth analysis of mRNA decay in P. falciparum may shed light on instances of translational regulation. To this end, DeRisi and colleagues measured the t 1/2 of >4700 P. falciparum mRNAs at four time‐points—10, 20, 30, and 44 hpi—using Actinomycin‐D treatment (to block transcription) and 70‐nt microarrays.111 While mRNA t 1/2 varied from as little as 1 min to >138 min and was not correlated to abundance or CDS length, the mRNAs of proteins that participate in the same biological pathway exhibited similar patterns of decay, as has been observed in yeast.112, 113 Moreover, for 2744 mRNAs, t 1/2 increased during the IDC with a mean of 9.5 min at 10 hpi to a mean of 65.4 min at 44 hpi. The lengthening of t 1/2 during a single developmental cycle appears to be unique to Plasmodium spp. One explanation for this may be the phasic expression of mRNA decay components, with most profiles showing peak mRNA abundance at 10 and 20 hpi (http://plasmodb.org), although it is unclear if the corresponding proteins exhibit a similar profile. Finally, the correlation between ribosomal occupancy of an mRNA, rate of translation, and its decay rate remains to be measured, as does the contribution of other regulatory factors, including mRNA sequestration, to maintain this developmentally regulated pattern of decay.
MOLECULAR REGULATORS OF TRANSLATION
In silico analyses of the Plasmodium genome have identified conserved RNA metabolism pathways,38, 44 and homologues of the major classes of RNA‐binding and ‐processing proteins;44 however, experimental validation of these proteins, and their contribution to translational control in Plasmodium spp. is still lagging behind. In the following sections, we delineate the few cis‐ and trans‐acting molecular players that have been characterized thus far.
Translational Regulation: Timing and Ribosome Association
Besides global repression of translation initiation via eIF2α phosphorylation, in higher eukaryotes, translation initiation can also be regulated by eIF4E‐binding proteins, which inhibit eIF4E–eIF4G interaction, in turn inhibiting binding of PABP to the eIF4F complex and preventing the cap‐dependent loading of the ribosome onto an mRNA (Figure 3).114 One potential eIF4E‐binding protein PfDZ50, a DDX6/DHH1‐like RNA helicase, and the homolog of DOZI (Development of Zygote Inhibited; see below), was recently described in P. falciparum blood stages115 (Table 1). Immunofluorescence studies showed that PfDZ50 localized to the cytoplasm during the IDC, though without an obvious pattern. Next, in vitro assays demonstrated that recombinant PfDZ50 binds to both DNA and RNA, hydrolyzes ATP, unwinds RNA, and represses translation in a reticulocyte lysate system. Finally, recombinant PfDZ50 interacted with PfeIF4E in vitro and this interaction relieved PfDZ50‐mediated translational repression, most likely by titrating away PfDZ50 from reticulocyte eIF4E. Nonetheless, the impact of PfDZ50‐PfeIF4E interaction in vivo, and PfDZ50's mRNA targets are not known. Also, whether signaling pathways such as MAPK (mitogen‐activated protein kinase) can regulate translation initiation116 in Plasmodium spp. is an open question.
Alternatively, secondary structure elements or cis‐acting sequences in the 5′UTR can induce and regulate cap‐independent initiation of translation,108 all of which remain unexplored in Plasmodium spp. For example, the 5′UTR of select var genes contains a cis‐element that putatively inhibits the translation of the CDS;117 whether this element mediates cap‐independent translation of a uORF or prevents ribosomal loading at the AUG of the CDS is unclear. Another mode of regulating translation at 5′UTRs was recently reported in P. falciparum that infected erythrocytes expressing the sickle‐cell variant of hemoglobin, HbS.118 Lamonte et al. found that the human miRNAs miR‐451 and let‐7i were overexpressed in HbS‐erythrocytes, and that these miRNAs translocated into the parasite cell and were trans‐spliced to the 5′ end of the mRNAs encoding essential P. falciparum genes. This resulted in translational repression by preventing ribosomal loading and consequently, defects in parasite growth: nevertheless, how widespread this phenomenon of host resistance is, remains to be seen. It is also unclear whether such a regulatory mechanism has any fitness benefits or evolutionary significance for Plasmodium spp. given the lack of success in detecting small (<25 nt) parasite‐derived RNAs119 and the absence of a functional RNAi pathway.120
Trans‐acting regulators of translation include RBPs of the Alba (Acetylation lowers binding affinity) family121, 122, 123, 124 (Figure 3; Table 1); Alba proteins were initially characterized in archaea as DNA‐binding proteins,121, 125 but a divergence of their function in other protozoan parasites such as Toxoplasma gondii,126 Trypanasoma cruzi,127 and Leishmania infantum 128 toward RNA regulation has been described as has their role in stress adaptation in plants.129 In P. falciparum, it was recently shown that PfAlba1 binds to a subset of IDC mRNAs including those encoding erythrocyte invasion proteins such as RAP1, AMA1, RhopH3, and CDPK1, and may regulate their association with the ribosome in a stage‐dependent manner, thus determining the timing of protein expression.124 In particular, this transcriptome‐wide analysis evidenced the presence of translational RNA regulons in P. falciparum, which are most likely composed of mRNAs encoding proteins of the same biological process, e.g., erythrocyte invasion. Given that Plasmodium spp. encode up to six Alba family proteins (PfAlba1‐6),44 that the PfAlbas appear to have different cellular localization and DNA/RNA specificity patterns,122, 123 and that PfAlba1‐4 coprecipitate with polysomes of blood stages,5 the roles of these proteins in regulating translation needs to be evaluated in greater detail. Furthermore, the diversification of Plasmodium Alba1 and Alba2 by the addition of an RGG domain and of Alba4 by the addition of a membrane‐tethering ENTH/VHS module,122 indicates that these proteins may perform distinct functions. Lastly, the evolutionary proximity of the Albas to RNA‐binding components of RNase P/RNase MRP RNP complexes121 also has implications for the crosstalk between translation regulation and mRNA decay. It is interesting to conjecture that the Alba proteins, which are of archaeal origin, may be master regulators of Plasmodium translation.130
mRNA Decay and Stabilization
mRNA decay can take place in a 5′–3′ or 3′–5′ manner, and typically involves decapping, removal of the poly(A)‐tail, i.e., deadenylation, and degradation by ribonucleases; this is different from nonsense‐mediated mRNA decay which is mediated by Upf1 and involved endonucleolytic cleavage.131 In Plasmodium spp., mRNA decapping enzymes such as DCP1 and DCP2, and members of the CCR4/NOT complex, implicated in transcription initiation and elongation and mRNA deadenylation, are conserved38, 44, 111 (Figure 3). Preliminary studies in P. falciparum showed that disruption of the deadenylase subunit Caf1 of the CCR4/NOT complex altered gene expression during the IDC, in particular, mistimed expression of proteins involved in erythrocyte invasion and egress, resulting in a proliferation defect132 (Table 1). Upon deadenylation, a transcript is vulnerable to 3′–5′ degradation by the RNA exosome, a complex of nine noncatalytic core proteins and associated exoribonucleases (exoRNases) which is involved in RNA processing, quality control and decay, both in the nucleus and cytoplasm.133, 134 Putative homologues of some of the core proteins, as well as the exoRNases DIS3/RRP44 (exoRNaseII‐related family) and RRP6 (RNase D family) are found in the Plasmodium genome,38, 44, 45, 111 but the exact composition of the complex still remains undefined (Figure 3). Using coimmunoprecipitation assays, Zhang et al. demonstrated that PfDis3 and PfRRP6 associated with the exosome core, and using immunofluorescence assays, localized PfDIS3 and PfRRP6 to the cytoplasm and the nucleus, respectively, of ring stage parasites45 (Table 1). Furthermore, they showed that PfRNaseII, a noncanonical exoribonuclease, post‐transcriptionally regulates ~200 genes—including a subset of var genes and noncoding RNAs (ncRNAs)—in an RNA exosome‐independent fashion, and identified, for the first time, the presence of cryptic RNAs in the malaria parasite. Nonetheless, the regulation of mRNA decay and its correlation to translation is not well understood in Plasmodium spp.
Besides mRNA decay components, other trans‐acting factors may regulate mRNA stability. These include splicing regulators such as P. falciparum SR1 (Serine/Arginine‐rich protein 1), which shows both nuclear and cytoplasmic localization.135 Indeed, it was recently shown that PfSR1 binds to 64 transcripts via two purine‐rich RNA motifs, similar to its human counterpart SRSF1, and that PfSR1 overexpression resulted in the early detection of gametocyte‐specific intron‐less transcripts.136 However, the molecular basis of PfSR1‐mediated post‐transcriptional regulation is not known. Also, it remains to be seen whether the other splicing factors encoded by the parasite genome perform similar post‐transcriptional functions, as has been observed for several mammalian SR proteins.137
Translational Repression
The presence of translational repression in malaria parasites was first evidenced by the analysis of a P. berghei surface antigen P28 (previously called Pbs21). While p28 mRNA was abundantly transcribed in mature gametocytes in the host, the protein was only expressed after transmission to mosquitoes, in zygotes and ookinetes.138, 139 Subsequent studies identified a similar regulatory pattern for another P. berghei mRNA that encodes a mosquito‐stage surface antigen P25 and 7 other mRNAs.14 Thereafter, Waters, Mair, and colleagues described PbDOZI, and its interacting partner CITH (CAR‐I/Trailer Hitch Homolog) as central regulators of translational repression in P. berghei female gametocytes140, 141, 142 (Figure 3; Table 1). PbDOZI/CITH‐containing mRNP complexes appeared as punctate P granule‐like structures in the cytoplasm and bound to 731 transcripts (488 for PbDOZI and 551 for PbCITH, with an overlap of 154) including p25 and p28; 211 of these were downregulated in gametocytes upon PbDOZI/CITH depletion.140, 141, 142 The authors also determined the composition of PbDOZI‐CITH‐containing mRNA‐protein (mRNP) complexes using tandem affinity purification and identified RBPs such as PbBRUNO/CELF and PbAlbas, and translation factors such as PbeIF4E and PbPABP, as components of the complex; notably, the complex did not contain RNases.142 Taken together, these studies validated the presence of a so‐called ‘maternal mRNA repressome’ in P. berghei, which is coordinated by PbDOZI‐CITH at the level of mRNA stability and translational repression and is essential for developmental stage transitions. The question that remains is whether a similar complex exists in P. falciparum gametocytes.
In addition to regulation by the PbDOZI‐CITH complex, the mRNAs of P25 and P28 are regulated by cis‐acting elements in the 5′UTR and 3′UTR sequences. A 47‐base cis‐acting regulatory element in the 3′UTR of translationally repressed mRNAs of P. berghei gametocytes was first identified by in silico analysis.14 Thereafter, using a GFP reporter system, the impact of the 47‐base element on TE was measured in P. berghei gametocytes, as was the impact of the 5′ and 3′UTRs of p25, p28 and pb000245.02.0, another translationally repressed gametocyte mRNA.143 This led to the identification of a U‐rich cis‐regulatory motif in the 5′UTR of p25 that was homologous to the 47‐base element, and was necessary and sufficient to confer translational repression. Nonetheless, it remains unclear if the binding of PbDOZI, PbCITH, or other members of the complex to p25, p28, and so on, depends on these cis‐elements.
Adding further complexity to translational regulation in sexual stages is the observation that pfs25 and pfs28, P. falciparum homologs of p25 and p28, are regulated by the Puf (Pumilio and fem‐3 binding factor homolog) family of RBPs144, 145, 146, 147 (Figure 3; Table 1); Puf proteins regulate diverse processes in eukaryotes by binding to the 3′UTRs of their mRNA targets and repressing translation and/or promoting degradation.148 In P. falciparum, PfPuf1 and PfPuf2 were shown to be differentially expressed in gametocytes, and to bind in vitro to the RNA sequence of the Nanos‐responsive element from the hunchback mRNA of Drosophila melanogaster. 144, 145 Subsequent analysis demonstrated an essential role for PfPuf2 in repressing gametocytogenesis, in particular male differentiation,146 and translational repression of pfs25 and pfs28 mRNAs via binding to Puf‐binding elements (PBEs) in the 5′ and 3′UTRs of its target mRNAs147 (Figure 2). Simultaneously, several groups assessed the contribution of P. berghei and P. yoelli Puf proteins to translational repression and parasite development.149, 150, 151, 152, 153 A key conclusion from these studies was that Puf2 binds to and inhibits the translation of UIS (Upregulated in Infectious Sporozoites) mRNAs in sporozoite stages, with the proteins being translated only after liver cell invasion. Notably, the UIS mRNAs are also post‐transcriptionally regulated at the level of stability by sporozoite asparagine‐rich protein 1 (SAP1).154
Although systems‐wide studies of P. falciparum have hinted at pervasive, temporal translational repression during the IDC, other than PfAlba1, molecular regulators of translation are poorly described; indeed, the impact of the DOZI‐CITH complex and Puf proteins during this stage in P. falciparum is a black box. A putative component of the P. falciparum DOZI‐CITH complex, Bruno/CELF, was recently characterized155 (Figure 3 and Table 1). Wongsombat et al. identified two Bruno/CELF homologs in the P. falciparum genome, CELF1 and CELF2, and showed that GFP‐tagged PfCELF1 localized to both the nucleus and cytoplasm of IDC stages. Next, using an in vitro UV crosslinking‐based assay, they demonstrated that, PfCELF1, but not PfCELF2, bound to 12‐mer RNA sequences. Moreover, using in vitro RNA immunoprecipitation‐microarray analysis, they found that PfCELF1 bound to 1376 features from 1040 mRNAs, including the CELF1 mRNA, while PfCELF2 bound to 26 features from 22 mRNAs. Bioinformatic analysis of PfCELF1's mRNA targets led to the identification of a UG‐rich motif that the authors predict is the binding site of this protein. One of the proposed functions of PfCELF1 was the regulation of splicing, although it was not validated. Moreover, the contribution of the 180‐odd Plasmodium RBPs44 to the translational delay observed in blood stages, which could be partially achieved by the storage of mRNAs in structures similar to the PbDOZI‐CITH‐containing maternal granules, is not explored. Again, the PfAlba proteins and PfCELF1 are front‐runners for such a storage mechanism given their punctate localization in the cytoplasm of trophozoite and schizont stages,122, 155 which is reminiscent of P‐granules (Table 1). Overall, the mechanism of translational repression in malaria parasites is complex, but as yet in its nascent stages of clarification. Future work could focus on specific sets of mRNAs that are subject to translational control at different stages of the parasite lifecycle as well as RBPs that exhibit features similar to P‐granule components.44
TRANSLATION INHIBITORS AS DRUGS
Translation is an essential, conserved process: therefore, inhibitor design inherently faces the challenge of selectivity for the parasite over host enzymes, which is not easily predicted. Nevertheless, antimalarial activities of antibiotics like chloramphenicol and tetracyclines that inhibit prokaryotic translation were described more than 60 years ago.156, 157, 158, 159 These antibiotics were thereafter shown to target the translation machinery of the apicoplast; as described above, this organelle was obtained by secondary endosymbiosis and, like the mitochondrion, possesses a prokaryotic‐type 70S ribosome, albeit with a divergent composition. Indeed, in silico analysis revealed that the Plasmodium spp. apicoplast and mitochondrial ribosomes have a reduced number of ribosomal proteins with differences in the rRNA sequence and structure.95 Furthermore, the effect of disrupting apicoplast translation on parasite proliferation is consistently observed only in the second cycle, after cell division and reinvasion, and has been termed the ‘delayed death phenotype’ (i.e., the effect of the antibiotic against apicoplast translation and/or transcription begins in the parental generation, and accumulates in the progeny, leading to the irreversible loss of apicoplast function and parasite death). This presents a problem with regards to the therapeutic potential of these antibiotics and is a much debated intervention strategy.160 However, apicoplast‐targeted aminoacyl‐tRNA synthetases (aaRSs), enzymes that couple cognate tRNAs with amino acids, have attracted attention as therapeutic targets and specific inhibitors against apicoplastic isoleucyl‐tRNA synthetase161 and lysyl‐tRNA synthetase162 have been described.
In recent years, the cytoplasmic translation apparatus has also emerged as a potential target for antimalarials. Firstly, small molecule inhibitors of cytoplasmic aaRSs have been characterized. One such molecule, Cladosporin, was identified in a cell‐based high throughput screen of natural compounds against both liver and blood stage proliferation of P. falciparum, and was demonstrated to block de novo protein synthesis by directly targeting cytoplasmic lysyl tRNA syntethase (KRS).163 In silico docking studies revealed differences in the ATP‐binding pocket between the parasite and human enzymes as the cause of Cladosporin's 100‐fold higher cytotoxicity against the parasite as compared to human cells.163 Another drug, borrelidin, was shown to be active against threonyl‐tRNA synthetase from bacteria, Plasmodium spp., yeast and humans; in Plasmodium spp., threonyl‐tRNA synthetase is shared between the cytoplasm and apicoplast.92, 164 In a separate study, analogues of borrelidin were identified that show higher selectivity and lower cytotoxicity against human cells165 compared to the original molecule.166 A further screen of a library of borrelidin analogs identified two compounds with improved features that were able to clear parasites from P. yoelii infected mice.167 Secondly, small molecule inhibitors of ribosome‐associated factors have been described. In 2015, Baragan et al. identified PfeEF2 as the target of the antimalarial compound, DDD107498.82 DDD107498, which is not toxic to mammalian cells and is now in the advanced nonclinical development stage as a therapeutic, inhibited de novo protein synthesis and displayed antimalarial activity against multiple lifecycle stages of P. falciparum. Therefore, it is tempting to speculate that the inhibition of protein synthesis indeed presents as an effective intervention strategy for attaining multi‐stage activity against malaria parasites.
CONCLUSIONS AND FUTURE PERSPECTIVES
The discovery that steady‐state mRNA levels do not strictly correlate to steady‐state protein levels is not unprecedented and different models have been proposed to describe how eukaryotic mRNA‐protein equilibrium is maintained.168, 169 In malaria parasites, the steady‐state transcriptome and proteome of different developmental stages, each of which has distinct morphological and physiological properties, appears to be maintained by ‘just‐in‐time’ transcriptional and translational regulatory mechanisms. This results in phenotypes such as the transcriptional cascade of the 48‐h P. falciparum IDC, translational delay to fine‐tune protein expression, and translational repression of maternal mRNAs in sexual stages and UIS mRNAs in sporozoite stages. All of these phenomena are likely regulated by Plasmodium DNA‐binding proteins and/or RBPs and their co‐factors. In recent years, one such class of proteins, the Albas, has emerged as an important regulator of translation in blood stages and potentially, in sexual stages. Therefore, in‐depth analysis of Plasmodium RBPs will be key to gain further insights into translational control mechanisms in this parasite. We anticipate that the utilization of CRISPR/Cas9‐based genome editing techniques,170 and in the future, CRISPR‐based mRNA degradation techniques,171 will expedite this process. Alternately, cis‐regulatory sequences can be bioinformatically predicted in the 5′ and 3′UTRs of co‐regulated sets of mRNAs and tested for their contribution to translational regulation; of note, Caro et al. described the 5′ and 3′UTRs of 3569 P. falciparum IDC mRNAs (average length of 607–1140 nt and 518–622 nt, respectively).6 Additionally, given that more than 60% of the annotated Plasmodium proteins are of unknown function, several of these may contribute to mRNA‐binding and translational regulation as illustrated in a recent study on African trypanosomes.172
In general, the field of translational regulation has burgeoned in recent years due to the application of techniques such as ribosome profiling,109 transcriptome‐wide detection of mRNA ‘epigenetic’ modifications,173, 174 and quantitative high‐throughput proteomics,175, 176 to name a few. In particular, RNA modifications have emerged as important biological regulators of tRNA, rRNA, mRNA, and ncRNA function. For example, base modifications in human tRNAs have a direct impact on translation rates and disease,177 while in bacteria, changes in tRNA modifications regulate the TE of stress response proteins.178 Furthermore, N6‐methyl‐adenosine (m6A) modifications in mRNA and long ncRNAs regulate RNA tertiary structure and/or recruitment of m6A‐binding proteins.179 Therefore, future studies could focus on whether Plasmodium RNAs are subject to post‐transcriptional regulation at the level of base modifications. Indeed, such a mechanism may contribute to various parasite phenotypes that are yet to be explored at the molecular level and may even explain the observation that clinical isolates of P. falciparum which are resistant to the drug artemisinin exhibit an extended ring phase transcriptomic profile.180
To conclude, we would like to emphasize that there are many outstanding questions in the field of Plasmodium translational regulation that need to be resolved. These include: Do blood stages simply use translational regulation as a mechanism to fine‐tune gene expression (hence its widespread occurrence), whereas the transition stages between the host and vector—which do not have the same precise timing as asexual blood stages—use translational control to rapidly respond to environmental changes by repressing specific sets of mRNAs? Indeed, does mRNA‐specific translational regulation occur at all during the IDC, and if yes, how can these specific events be identified? Moreover, are multivariant gene families such as var and erythrocyte invasion components subject to both epigenetic and translational control in order to better adapt to host responses? Do changes in tRNA modifications play a role in parasite development and drug resistance by altering blood stage translation outcomes, as has been observed in human disease states177? Given that phosphoproteomic studies have identified post‐translational modifications (PTMs; such as phosphorylation) of Plasmodium eIF, eEF, and eRF proteins (http://plasmodb.org), do these PTMs modulate their translational regulatory function at different stages of the parasite lifecycle? Is ribosomal profiling combined with proteomics the way forward to identify true instances of translational control? Investigating the answers to these questions will reveal the where, when, what, who, how, and how much, of translational control occurs in malaria parasites and may lead to the establishment of new paradigms in RNA biology. Finally, it is noteworthy to mention that all of the systems‐wide studies described here were performed using laboratory‐adapted P. falciparum strains such as 3D7, HB3, DD2, and W2. While they have highlighted important physiological phenomena, their extrapolation to growth of the parasite within humans is not always straightforward. Therefore, in the future, transcriptomic studies of field isolates should be coupled with proteomic studies to reveal true instances of translational regulation in malaria parasites and their contribution to disease severity.
ACKNOWLEDGMENTS
Research in the Scherf laboratory is supported by the European Research Council Advanced Grant (ERC PlasmoSilencing), ANR‐13‐ISV3‐0003‐01 NSFC (n° 81361130411), and the French Parasitology consortium ParaFrap (ANR‐11‐LABX0024) awarded to A. S. S.S.V. was supported by the European Molecular Biology Organization Long‐Term Fellowship, the Marie Sklodowska‐Curie International Incoming Fellowship (FP7‐MC‐IIF‐302451), and the Pasteur Roux Postdoctoral Fellowship. D.D. is supported by the ParaFrap Postdoctoral Fellowship. We also acknowledge PlasmoDB, a member of pathogen‐databases that are housed under the NIAID‐funded EuPathDB Bioinformatics Resource Center (BRC) umbrella, for regular updates and ready access to Plasmodium genome information, including gene annotations.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Acharya P, Pallavi R, Chandran S, Chakravarti H, Middha S, Acharya J, Kochar S, Kochar D, Subudhi A, Boopathi AP, et al. A glimpse into the clinical proteome of human malaria parasites Plasmodium falciparum and Plasmodium vivax . Proteomics Clin Appl 2009, 3:1314–1325. [DOI] [PubMed] [Google Scholar]
- 2. Anderson DC, Lapp SA, Akinyi S, Meyer EV, Barnwell JW, Korir‐Morrison C, Galinski MR. Plasmodium vivax trophozoite‐stage proteomes. J Proteomics 2015, 115:157–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol 2003, 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA 2008, 105:16290–16295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bunnik EM, Chung DW, Hamilton M, Ponts N, Saraf A, Prudhomme J, Florens L, Le Roch KG. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum . Genome Biol 2013, 14:R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caro F, Ahyong V, Betegon M, DeRisi JL. Genome‐wide regulatory dynamics of translation in the asexual blood stages. eLife 2014, 3:e04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui L, Fan Q, Hu Y, Karamycheva SA, Quackenbush J, Khuntirat B, Sattabongkot J, Carlton JM. Gene discovery in Plasmodium vivax through sequencing of ESTs from mixed blood stages. Mol Biochem Parasitol 2005, 144:1–9. [DOI] [PubMed] [Google Scholar]
- 8. Daily JP, Le Roch KG, Sarr O, Ndiaye D, Lukens A, Zhou Y, Ndir O, Mboup S, Sultan A, Winzeler EA, et al. In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. J Infect Dis 2005, 191:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 2002, 419:520–526. [DOI] [PubMed] [Google Scholar]
- 10. Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. Quantitative protein expression profiling reveals extensive post‐transcriptional regulation and post‐translational modifications in schizont‐stage malaria parasites. Genome Biol 2008, 9:R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foth BJ, Zhang N, Chaal BK, Sze SK, Preiser PR, Bozdech Z. Quantitative time‐course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum . Mol Cell Proteomics 2011, 10:M110 006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grüner AC, Hez‐Deroubaix S, Snounou G, Hall N, Bouchier C, Letourneur F, Landau I, Druilhe P. Insights into the P. y. yoelii hepatic stage transcriptome reveal complex transcriptional patterns. Mol Biochem Parasitol 2005, 142:184–192. [DOI] [PubMed] [Google Scholar]
- 13. Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol Biochem Parasitol 2004, 136:35–42. [DOI] [PubMed] [Google Scholar]
- 14. Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 2005, 307:82–86. [DOI] [PubMed] [Google Scholar]
- 15. Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH. Differential transcriptome profiling identifies Plasmodium genes encoding pre‐erythrocytic stage‐specific proteins. Mol Microbiol 2004, 51:1221–1232. [DOI] [PubMed] [Google Scholar]
- 16. Kappe SH, Gardner MJ, Brown SM, Ross J, Matuschewski K, Ribeiro JM, Adams JH, Quackenbush J, Cho J, Carucci DJ, et al. Exploring the transcriptome of the malaria sporozoite stage. Proc Natl Acad Sci USA 2001, 98:9895–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khan SM, Franke‐Fayard B, Mair GR, Lasonder E, Janse CJ, Mann M, Waters AP. Proteome analysis of separated male and female gametocytes reveals novel sex‐specific Plasmodium biology. Cell 2005, 121:675–687. [DOI] [PubMed] [Google Scholar]
- 18. Lasonder E, Janse CJ, van Gemert GJ, Mair GR, Vermunt AM, Douradinha BG, van Noort V, Huynen MA, Luty AJ, Kroeze H, et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog 2008, 4:e1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 2003, 301:1503–1508. [DOI] [PubMed] [Google Scholar]
- 20. Le Roch KG, Zhou Y, Batalov S, Winzeler EA. Monitoring the chromosome 2 intraerythrocytic transcriptome of Plasmodium falciparum using oligonucleotide arrays. Am J Trop Med Hyg 2002, 67:233–243. [DOI] [PubMed] [Google Scholar]
- 21. Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res 2004, 14:2308–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Llinás M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 2006, 34:1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno‐Pérez DA, Dégano R, Ibarrola N, Muro A, Patarroyo MA. Determining the Plasmodium vivax VCG‐1 strain blood stage proteome. J Proteomics 2014, 113C:268–280. [DOI] [PubMed] [Google Scholar]
- 24. Ngwa CJ, Scheuermayer M, Mair GR, Kern S, Brügl T, Wirth CC, Aminake MN, Wiesner J, Fischer R, Vilcinskas A, et al. Changes in the transcriptome of the malaria parasite Plasmodium falciparum during the initial phase of transmission from the human to the mosquito. BMC Genomics 2013, 14:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol Microbiol 2004, 52:1187–1199. [DOI] [PubMed] [Google Scholar]
- 26. Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, Barrell B, Pain A, Berriman M, et al. New insights into the blood‐stage transcriptome of Plasmodium falciparum using RNA‐Seq. Mol Microbiol 2010, 76:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, Kube M, Reinhardt R, Ng GH, Manickam R, et al. A global view of the nonprotein‐coding transcriptome in Plasmodium falciparum . Nucleic Acids Res 2010, 38:608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roobsoong W, Roytrakul S, Sattabongkot J, Li J, Udomsangpetch R, Cui L. Determination of the Plasmodium vivax schizont stage proteome. J Proteomics 2011, 74:1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siau A, Toure FS, Ouwe‐Missi‐Oukem‐Boyer O, Ciceron L, Mahmoudi N, Vaquero C, Froissard P, Bisvigou U, Bisser S, Coppee JY, et al. Whole‐transcriptome analysis of Plasmodium falciparum field isolates: identification of new pathogenicity factors. J Infect Dis 2007, 196:1603–1612. [DOI] [PubMed] [Google Scholar]
- 30. Sorber K, Dimon MT, DeRisi JL. RNA‐Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res 2011, 39:3820–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva‐Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SH. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci USA 2008, 105:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 2011, 10:410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Q, Brown S, Roos DS, Nussenzweig V, Bhanot P. Transcriptome of axenic liver stages of Plasmodium yoelii . Mol Biochem Parasitol 2004, 137:161–168. [DOI] [PubMed] [Google Scholar]
- 34. Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, Collins WE, Hoffman SL, Zhou Y, Vinetz JM, et al. A systems‐based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis 2010, 4:e653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Young JA, Fivelman QL, Blair PL, de la Vega P, Le Roch KG, Zhou Y, Carucci DJ, Baker DA, Winzeler EA. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology‐based pattern identification. Mol Biochem Parasitol 2005, 143:67–79. [DOI] [PubMed] [Google Scholar]
- 36. Cui L, Lindner S, Miao J. Translational regulation during stage transitions in malaria parasites. Ann N Y Acad Sci 2015, 1342:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doerig C, Rayner JC, Scherf A, Tobin AB. Post‐translational protein modifications in malaria parasites. Nat Rev Microbiol 2015, 13:160–172. [DOI] [PubMed] [Google Scholar]
- 38. Hughes KR, Philip N, Starnes GL, Taylor S, Waters AP. From cradle to grave: RNA biology in malaria parasites. Wiley Interdiscip Rev RNA 2010, 1:287–303. [DOI] [PubMed] [Google Scholar]
- 39. Voss TS, Bozdech Z, Bártfai R. Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol 2014, 20:88–95. [DOI] [PubMed] [Google Scholar]
- 40. Zhang M, Joyce BR, Sullivan WJ, Nussenzweig V. Translational control in Plasmodium and toxoplasma parasites. Eukaryot Cell 2013, 12:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van Der Kelen K, Beyaert R, Inzé D, De Veylder L. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol 2009, 44:143–168. [DOI] [PubMed] [Google Scholar]
- 42. Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum . Genome Res 2004, 14:1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature 2002, 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reddy BP, Shrestha S, Hart KJ, Liang X, Kemirembe K, Cui L, Lindner SE. A bioinformatic survey of RNA‐binding proteins in Plasmodium. BMC Genomics 2015, 16:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Q, Siegel TN, Martins RM, Wang F, Cao J, Gao Q, Cheng X, Jiang L, Hon CC, Scheidig‐Benatar C, et al. Exonuclease‐mediated degradation of nascent RNA silences genes linked to severe malaria. Nature 2014, 513:431–435. [DOI] [PubMed] [Google Scholar]
- 46. Siegel TN, Hon CC, Zhang Q, Lopez‐Rubio JJ, Scheidig‐Benatar C, Martins RM, Sismeiro O, Coppée JY, Scherf A. Strand‐specific RNA‐Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum . BMC Genomics 2014, 15:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum . RNA 2009, 15:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum‐infected erythrocytes. Cell 1995, 82:89–100. [DOI] [PubMed] [Google Scholar]
- 49. Painter HJ, Campbell TL, Llinás M. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol 2011, 176:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson KE, Habib S, Frugier M, Hoen R, Khan S, Pham JS, Ribas de Pouplana L, Royo M, Santos MA, Sharma A, et al. Protein translation in Plasmodium parasites. Trends Parasitol 2011, 27:467–476. [DOI] [PubMed] [Google Scholar]
- 51. Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP, McCutchan TF. Structurally distinct, stage‐specific ribosomes occur in Plasmodium. Science 1987, 238:933–937. [DOI] [PubMed] [Google Scholar]
- 52. Li J, McConkey GA, Rogers MJ, Waters AP, McCutchan TR. Plasmodium: the developmentally regulated ribosome. Exp Parasitol 1994, 78:437–441. [DOI] [PubMed] [Google Scholar]
- 53. Li J, Wirtz RA, McConkey GA, Sattabongkot J, McCutchan TF. Transition of Plasmodium vivax ribosome types corresponds to sporozoite differentiation in the mosquito. Mol Biochem Parasitol 1994, 65:283–289. [DOI] [PubMed] [Google Scholar]
- 54. Li J, Gutell RR, Damberger SH, Wirtz RA, Kissinger JC, Rogers MJ, Sattabongkot J, McCutchan TF. Regulation and trafficking of three distinct 18 S ribosomal RNAs during development of the malaria parasite. J Mol Biol 1997, 269:203–213. [DOI] [PubMed] [Google Scholar]
- 55. McCutchan TF, Li J, McConkey GA, Rogers MJ, Waters AP. The cytoplasmic ribosomal RNAs of Plasmodium spp. Parasitol Today 1995, 11:134–138. [DOI] [PubMed] [Google Scholar]
- 56. Rogers MJ, Gutell RR, Damberger SH, Li J, McConkey GA, Waters AP, McCutchan TF. Structural features of the large subunit rRNA expressed in Plasmodium falciparum sporozoites that distinguish it from the asexually expressed subunit rRNA. RNA 1996, 2:134–145. [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson J, van Spaendonk RM, Choudhuri R, Sinden RE, Janse CJ, Waters AP. Heterogeneous ribosome populations are present in Plasmodium berghei during development in its vector. Mol Microbiol 1999, 31:253–260. [DOI] [PubMed] [Google Scholar]
- 58. Velichutina IV, Rogers MJ, McCutchan TF, Liebman SW. Chimeric rRNAs containing the GTPase centers of the developmentally regulated ribosomal rRNAs of Plasmodium falciparum are functionally distinct. RNA 1998, 4:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waters AP, Syin C, McCutchan TF. Developmental regulation of stage‐specific ribosome populations in Plasmodium. Nature 1989, 342:438–440. [DOI] [PubMed] [Google Scholar]
- 60. van Spaendonk RM, Ramesar J, van Wigcheren A, Eling W, Beetsma AL, van Gemert GJ, Hooghof J, Janse CJ, Waters AP. Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei . J Biol Chem 2001, 276:22638–22647. [DOI] [PubMed] [Google Scholar]
- 61. Qi Y, Zhu F, Eastman RT, Fu Y, Zilversmit M, Pattaradilokrat S, Hong L, Liu S, McCutchan TF, Pan W, et al. Regulation of Plasmodium yoelii oocyst development by strain‐ and stage‐specific small‐subunit rRNA. mBio 2015, 6:e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. Cryo‐EM structure of the Plasmodium falciparum 80S ribosome bound to the anti‐protozoan drug emetine. eLife 2014, 3:e03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun M, Li W, Blomqvist K, Das S, Hashem Y, Dvorin JD, Frank J. Dynamical features of the Plasmodium falciparum ribosome during translation. Nucleic Acids Res 2015, 43:10515–10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sartorello R, Amaya MJ, Nathanson MH, Garcia CR. The plasmodium receptor for activated C kinase protein inhibits Ca(2+) signaling in mammalian cells. Biochem Biophys Res Commun 2009, 389:586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gallo S, Manfrini N. Working hard at the nexus between cell signaling and the ribosomal machinery: an insight into the roles of RACK1 in translational regulation. Translation (Austin) 2015, 3:e1120382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh S, Sehgal A, Waghmare S, Chakraborty T, Goswami A, Sharma S. Surface expression of the conserved ribosomal protein P0 on parasite and other cells. Mol Biochem Parasitol 2002, 119:121–124. [DOI] [PubMed] [Google Scholar]
- 67. Das S, Basu H, Korde R, Tewari R, Sharma S. Arrest of nuclear division in Plasmodium through blockage of erythrocyte surface exposed ribosomal protein P2. PLoS Pathog 2012, 8:e1002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins. Mol Cell 2009, 34:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004, 5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shaw PJ, Ponmee N, Karoonuthaisiri N, Kamchonwongpaisan S, Yuthavong Y. Characterization of human malaria parasite Plasmodium falciparum eIF4E homologue and mRNA 5’ cap status. Mol Biochem Parasitol 2007, 155:146–155. [DOI] [PubMed] [Google Scholar]
- 71. Tuteja R, Pradhan A. PfeIF4E and PfeIF4A colocalize and their double‐stranded RNA inhibits Plasmodium falciparum proliferation. Commun Integr Biol 2010, 3:611–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tuteja R. Identification and bioinformatics characterization of translation initiation complex eIF4F components and poly(A)‐binding protein from Plasmodium falciparum . Commun Integr Biol 2009, 2:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tuteja R, Pradhan A. Isolation and functional characterization of eIF4F components and poly(A)‐binding protein from Plasmodium falciparum . Parasitol Int 2009, 58:481–485. [DOI] [PubMed] [Google Scholar]
- 74. Fennell C, Babbitt S, Russo I, Wilkes J, Ranford‐Cartwright L, Goldberg DE, Doerig C. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress‐response to amino‐acid starvation. Malar J 2009, 8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Möhrle JJ, Zhao Y, Wernli B, Franklin RM, Kappes B. Molecular cloning, characterization and localization of PfPK4, an eIF‐2alpha kinase‐related enzyme from the malarial parasite Plasmodium falciparum . Biochem J 1997, 328:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Solyakov L, Halbert J, Alam MM, Semblat JP, Dorin‐Semblat D, Reininger L, Bottrill AR, Mistry S, Abdi A, Fennell C, et al. Global kinomic and phospho‐proteomic analyses of the human malaria parasite Plasmodium falciparum . Nat Commun 2011, 2:565. [DOI] [PubMed] [Google Scholar]
- 77. Zhang M, Fennell C, Ranford‐Cartwright L, Sakthivel R, Gueirard P, Meister S, Caspi A, Doerig C, Nussenzweig RS, Tuteja R, et al. The Plasmodium eukaryotic initiation factor‐2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J Exp Med 2010, 207:1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang M, Mishra S, Sakthivel R, Rojas M, Ranjan R, Sullivan WJ, Fontoura BM, Ménard R, Dever TE, Nussenzweig V. PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc Natl Acad Sci USA 2012, 109:3956–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Takebe S, Witola WH, Schimanski B, Günzl A, Ben MC. Purification of components of the translation elongation factor complex of Plasmodium falciparum by tandem affinity purification. Eukaryot Cell 2007, 6:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Richter JD, Coller J. Pausing on polyribosomes: make way for elongation in translational control. Cell 2015, 163:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Frugier M, Bour T, Ayach M, Santos MA, Rudinger‐Thirion J, Théobald‐Dietrich A, Pizzi E. Low complexity regions behave as tRNA sponges to help co‐translational folding of plasmodial proteins. FEBS Lett 2010, 584:448–454. [DOI] [PubMed] [Google Scholar]
- 82. Baragaña B, Hallyburton I, Lee MC, Norcross NR, Grimaldi R, Otto TD, Proto WR, Blagborough AM, Meister S, Wirjanata G, et al. A novel multiple‐stage antimalarial agent that inhibits protein synthesis. Nature 2015, 522:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Afonso A, Neto Z, Castro H, Lopes D, Alves AC, Tomás AM, Rosário VD. Plasmodium chabaudi chabaudi malaria parasites can develop stable resistance to atovaquone with a mutation in the cytochrome b gene. Malar J 2010, 9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chaubey S, Kumar A, Singh D, Habib S. The apicoplast of Plasmodium falciparum is translationally active. Mol Microbiol 2005, 56:81–89. [DOI] [PubMed] [Google Scholar]
- 85. Roy A, Cox RA, Williamson DH, Wilson RJ. Protein synthesis in the plastid of Plasmodium falciparum . Protist 1999, 150:183–188. [DOI] [PubMed] [Google Scholar]
- 86. Kalanon M, McFadden GI. Malaria, Plasmodium falciparum and its apicoplast. Biochem Soc Trans 2010, 38:775–782. [DOI] [PubMed] [Google Scholar]
- 87. Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, et al. Complete gene map of the plastid‐like DNA of the malaria parasite Plasmodium falciparum . J Mol Biol 1996, 261:155–172. [DOI] [PubMed] [Google Scholar]
- 88. Feagin JE, Gardner MJ, Williamson DH, Wilson RJ. The putative mitochondrial genome of Plasmodium falciparum . J Protozool 1991, 38:243–245. [DOI] [PubMed] [Google Scholar]
- 89. Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum . PLoS One 2012, 7:e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Biswas S, Lim EE, Gupta A, Saqib U, Mir SS, Siddiqi MI, Ralph SA, Habib S. Interaction of apicoplast‐encoded elongation factor (EF) EF‐Tu with nuclear‐encoded EF‐Ts mediates translation in the Plasmodium falciparum plastid. Int J Parasitol 2011, 41:417–427. [DOI] [PubMed] [Google Scholar]
- 91. Gupta A, Mir SS, Jackson KE, Lim EE, Shah P, Sinha A, Siddiqi MI, Ralph SA, Habib S. Recycling factors for ribosome disassembly in the apicoplast and mitochondrion of Plasmodium falciparum . Mol Microbiol 2013, 88:891–905. [DOI] [PubMed] [Google Scholar]
- 92. Jackson KE, Pham JS, Kwek M, De Silva NS, Allen SM, Goodman CD, McFadden GI, Ribas de Pouplana L, Ralph SA. Dual targeting of aminoacyl‐tRNA synthetases to the apicoplast and cytosol in Plasmodium falciparum . Int J Parasitol. 2012, 42:177–186. [DOI] [PubMed] [Google Scholar]
- 93. Haider A, Allen SM, Jackson KE, Ralph SA, Habib S. Targeting and function of proteins mediating translation initiation in organelles of Plasmodium falciparum . Mol Microbiol 2015, 96:796–814. [DOI] [PubMed] [Google Scholar]
- 94. Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol 2008, 24:279–284. [DOI] [PubMed] [Google Scholar]
- 95. Gupta A, Shah P, Haider A, Gupta K, Siddiqi MI, Ralph SA, Habib S. Reduced ribosomes of the apicoplast and mitochondrion of Plasmodium spp and predicted interactions with antibiotics. Open Biol 2014, 4:140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vaidya AB, Painter HJ, Morrisey JM, Mather MW. The validity of mitochondrial dehydrogenases as antimalarial drug targets. Trends Parasitol 2008, 24:8–9. [DOI] [PubMed] [Google Scholar]
- 97. Garrett CE, Coderre JA, Meek TD, Garvey EP, Claman DM, Beverley SM, Santi DV. A bifunctional thymidylate synthetase‐dihydrofolate reductase in protozoa. Mol Biochem Parasitol 1984, 11:257–265. [DOI] [PubMed] [Google Scholar]
- 98. Zhang K, Rathod PK. Divergent regulation of dihydrofolate reductase between malaria parasite and human host. Science 2002, 296:545–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nirmalan N, Sims PF, Hyde JE. Translational up‐regulation of antifolate drug targets in the human malaria parasite Plasmodium falciparum upon challenge with inhibitors. Mol Biochem Parasitol 2004, 136:63–70. [DOI] [PubMed] [Google Scholar]
- 100. Briolant S, Almeras L, Belghazi M, Boucomont‐Chapeaublanc E, Wurtz N, Fontaine A, Granjeaud S, Fusaï T, Rogier C, Pradines B. Plasmodium falciparum proteome changes in response to doxycycline treatment. Malar J 2010, 9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kuersten S, Radek A, Vogel C, Penalva LO. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip Rev RNA 2013, 4:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA 2014, 5:765–778. [DOI] [PubMed] [Google Scholar]
- 103. Amulic B, Salanti A, Lavstsen T, Nielsen MA, Deitsch KW. An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog 2009, 5:e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bancells C, Deitsch KW. A molecular switch in the efficiency of translation reinitiation controls expression of var2csa, a gene implicated in pregnancy‐associated malaria. Mol Microbiol 2013, 90:472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mok BW, Ribacke U, Rasti N, Kironde F, Chen Q, Nilsson P, Wahlgren M. Default Pathway of var2csa switching and translational repression in Plasmodium falciparum . PLoS One 2008, 3:e1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kumar M, Srinivas V, Patankar S. Upstream AUGs and upstream ORFs can regulate the downstream ORF in Plasmodium falciparum . Malar J 2015, 14:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 2014, 15:193–204. [DOI] [PubMed] [Google Scholar]
- 108. Mitchell SF, Parker R. Modifications on translation initiation. Cell 2015, 163:796–798. [DOI] [PubMed] [Google Scholar]
- 109. Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 2015, 16:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet 2013, 29:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shock JL, Fischer KF, DeRisi JL. Whole‐genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half‐life during the intra‐erythrocytic development cycle. Genome Biol 2007, 8:R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. García‐Martínez J, Aranda A, Pérez‐Ortín JE. Genomic run‐on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 2004, 15:303–313. [DOI] [PubMed] [Google Scholar]
- 113. Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 2002, 99:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Merrick WC. eIF4F: a retrospective. J Biol Chem 2015, 290:24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tarique M, Ahmad M, Ansari A, Tuteja R. Plasmodium falciparum DOZI, an RNA helicase interacts with eIF4E. Gene 2013, 522:46–59. [DOI] [PubMed] [Google Scholar]
- 116. Mahoney SJ, Dempsey JM, Blenis J. Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog Mol Biol Transl Sci 2009, 90:53–107. [DOI] [PubMed] [Google Scholar]
- 117. Brancucci NM, Witmer K, Schmid C, Voss TS. A var gene upstream element controls protein synthesis at the level of translation initiation in Plasmodium falciparum . PLoS One 2014, 9:e100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, Thornburg CD, Telen MJ, Ohler U, Nicchitta CV, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012, 12:187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett 2006, 580:5185–5188. [DOI] [PubMed] [Google Scholar]
- 120. Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters AP, Cowman AF, Crabb BS, de Koning‐Ward TF. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res 2009, 37:3788–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Aravind L, Iyer LM, Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol 2003, 4:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chene A, Vembar SS, Riviere L, Lopez‐Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig‐Benatar C, Hernandez‐Rivas R, Scherf A. PfAlbas constitute a new eukaryotic DNA/RNA‐binding protein family in malaria parasites. Nucleic Acids Res 2012, 40:3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Goyal M, Alam A, Iqbal MS, Dey S, Bindu S, Pal C, Banerjee A, Chakrabarti S, Bandyopadhyay U. Identification and molecular characterization of an Alba‐family protein from human malaria parasite Plasmodium falciparum . Nucleic Acids Res 2012, 40:1174–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Vembar SS, Macpherson CR, Sismeiro O, Coppée JY, Scherf A. The PfAlba1 RNA‐binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol 2015, 16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Peeters E, Driessen RP, Werner F, Dame RT. The interplay between nucleoid organization and transcription in archaeal genomes. Nat Rev Microbiol 2015, 13:333–341. [DOI] [PubMed] [Google Scholar]
- 126. Gissot M, Walker R, Delhaye S, Alayi TD, Huot L, Hot D, Callebaut I, Schaeffer‐Reiss C, Dorsselaer AV, Tomavo S. Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J Mol Biol 2013, 425:1287–1301. [DOI] [PubMed] [Google Scholar]
- 127. Mani J, Güttinger A, Schimanski B, Heller M, Acosta‐Serrano A, Pescher P, Späth G, Roditi I. Alba‐domain proteins of Trypanosoma brucei are cytoplasmic RNA‐binding proteins that interact with the translation machinery. PLoS One 2011, 6:e22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dupé A, Dumas C, Papadopoulou B. An Alba‐domain protein contributes to the stage‐regulated stability of amastin transcripts in Leishmania. Mol Microbiol 2014, 91:548–561. [DOI] [PubMed] [Google Scholar]
- 129. Verma JK, Gayali S, Dass S, Kumar A, Parveen S, Chakraborty S, Chakraborty N. OsAlba1, a dehydration‐responsive nuclear protein of rice (Oryza sativa L. ssp. indica), participates in stress adaptation. Phytochemistry 2014, 100:16–25. [DOI] [PubMed] [Google Scholar]
- 130. Bunnik EM, Le Roch KG. PfAlba1: master regulator of translation in the malaria parasite. Genome Biol 2015, 16:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense‐mediated mRNA decay pathway. Eukaryot Cell 2014, 13:1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, Andrews RM, Ellis PD, Khan SM, Lin JW, Janse CJ, et al. CCR4‐associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot Cell 2011, 10:1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Januszyk K, Lima CD. The eukaryotic RNA exosome. Curr Opin Struct Biol 2014, 24:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol 2016, 17:227–239. [DOI] [PubMed] [Google Scholar]
- 135. Eshar S, Allemand E, Sebag A, Glaser F, Muchardt C, Mandel‐Gutfreund Y, Karni R, Dzikowski R. A novel Plasmodium falciparum SR protein is an alternative splicing factor required for the parasites’ proliferation in human erythrocytes. Nucleic Acids Res 2012, 40:9903–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Eshar S, Altenhofen L, Rabner A, Ross P, Fastman Y, Mandel‐Gutfreund Y, Karni R, Llinás M, Dzikowski R. PfSR1 controls alternative splicing and steady‐state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs. Mol Microbiol 2015, 96:1283–1297. [DOI] [PubMed] [Google Scholar]
- 137. Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 2015, 6:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Paton MG, Barker GC, Matsuoka H, Ramesar J, Janse CJ, Waters AP, Sinden RE. Structure and expression of a post‐transcriptionally regulated malaria gene encoding a surface protein from the sexual stages of Plasmodium berghei . Mol Biochem Parasitol 1993, 59:263–275. [DOI] [PubMed] [Google Scholar]
- 139. Shaw MK, Thompson J, Sinden RE. Localization of ribosomal RNA and Pbs21‐mRNA in the sexual stages of Plasmodium berghei using electron microscope in situ hybridization. Eur J Cell Biol 1996, 71:270–276. [PubMed] [Google Scholar]
- 140. Guerreiro A, Deligianni E, Santos JM, Silva PA, Louis C, Pain A, Janse CJ, Franke‐Fayard B, Carret CK, Siden‐Kiamos I, et al. Genome‐wide RIP‐Chip analysis of translational repressor‐bound mRNAs in the Plasmodium gametocyte. Genome Biol 2014, 15:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, Khan SM, Dimopoulos G, Janse CJ, Waters AP. Regulation of sexual development of Plasmodium by translational repression. Science 2006, 313:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mair GR, Lasonder E, Garver LS, Franke‐Fayard BM, Carret CK, Wiegant JC, Dirks RW, Dimopoulos G, Janse CJ, Waters AP. Universal features of post‐transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog 2010, 6:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Braks JA, Mair GR, Franke‐Fayard B, Janse CJ, Waters AP. A conserved U‐rich RNA region implicated in regulation of translation in Plasmodium female gametocytes. Nucleic Acids Res 2008, 36:1176–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Cui L, Fan Q, Li J. The malaria parasite Plasmodium falciparum encodes members of the Puf RNA‐binding protein family with conserved RNA binding activity. Nucleic Acids Res 2002, 30:4607–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Fan Q, Li J, Kariuki M, Cui L. Characterization of PfPuf2, member of the Puf family RNA‐binding proteins from the malaria parasite Plasmodium falciparum . DNA Cell Biol 2004, 23:753–760. [DOI] [PubMed] [Google Scholar]
- 146. Miao J, Li J, Fan Q, Li X, Li X, Cui L. The Puf‐family RNA‐binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum . J Cell Sci 2010, 123:1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Miao J, Fan Q, Parker D, Li X, Li J, Cui L. Puf mediates translation repression of transmission‐blocking vaccine candidates in malaria parasites. PLoS Pathog 2013, 9:e1003268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA 2011, 2:471–492. [DOI] [PubMed] [Google Scholar]
- 149. Müller K, Matuschewski K, Silvie O. The Puf‐family RNA‐binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite. PLoS One 2011, 6:e19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Gomes‐Santos CS, Braks J, Prudêncio M, Carret C, Gomes AR, Pain A, Feltwell T, Khan S, Waters A, Janse C, et al. Transition of Plasmodium sporozoites into liver stage‐like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog 2011, 7:e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJJ, Kappe SH. Perturbations of Plasmodium Puf2 expression and RNA‐seq of Puf2‐deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol 2013, 15:1266–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Silva PA, Guerreiro A, Santos JM, Braks JA, Janse CJ, Mair GR. Translational control of UIS4 protein of the host‐parasite interface is mediated by the RNA binding protein Puf2 in Plasmodium berghei sporozoites. PLoS One 2016, 11:e0147940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zhang M, Mishra S, Sakthivel R, Fontoura BM, Nussenzweig V. UIS2: a unique phosphatase required for the development of plasmodium liver stages. PLoS Pathog 2016, 12:e1005370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Aly AS, Lindner SE, MacKellar DC, Peng X, Kappe SH. SAP1 is a critical post‐transcriptional regulator of infectivity in malaria parasite sporozoite stages. Mol Microbiol 2011, 79:929–939. [DOI] [PubMed] [Google Scholar]
- 155. Wongsombat C, Aroonsri A, Kamchonwongpaisan S, Morgan HP, Walkinshaw MD, Yuthavong Y, Shaw PJ. Molecular characterization of Plasmodium falciparum Bruno/CELF RNA binding proteins. Mol Biochem Parasitol 2014, 198:1–10. [DOI] [PubMed] [Google Scholar]
- 156. Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum . Antimicrob Agents Chemother 2006, 50:3124–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Dahl EL, Rosenthal PJ. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother 2007, 51:3485–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Goodman CD, Su V, McFadden GI. The effects of anti‐bacterials on the malaria parasite Plasmodium falciparum . Mol Biochem Parasitol 2007, 152:181–191. [DOI] [PubMed] [Google Scholar]
- 159. Sidhu AB, Sun Q, Nkrumah LJ, Dunne MW, Sacchettini JC, Fidock DA. In vitro efficacy, resistance selection, and structural modeling studies implicate the malarial parasite apicoplast as the target of azithromycin. J Biol Chem 2007, 282:2494–2504. [DOI] [PubMed] [Google Scholar]
- 160. Pradel G, Schlitzer M. Antibiotics in malaria therapy and their effect on the parasite apicoplast. Curr Mol Med 2010, 10:335–349. [DOI] [PubMed] [Google Scholar]
- 161. Istvan ES, Dharia NV, Bopp SE, Gluzman I, Winzeler EA, Goldberg DE. Validation of isoleucine utilization targets in Plasmodium falciparum . Proc Natl Acad Sci USA 2011, 108:1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Hoen R, Novoa EM, López A, Camacho N, Cubells L, Vieira P, Santos M, Marin‐Garcia P, Bautista JM, Cortés A, et al. Selective inhibition of an apicoplastic aminoacyl‐tRNA synthetase from Plasmodium falciparum . Chembiochem 2013, 14:499–509. [DOI] [PubMed] [Google Scholar]
- 163. Hoepfner D, McNamara CW, Lim CS, Studer C, Riedl R, Aust T, McCormack SL, Plouffe DM, Meister S, Schuierer S, et al. Selective and specific inhibition of the plasmodium falciparum lysyl‐tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 2012, 11:654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Nass G, Hasenbank R. Effect of Borrelidin on the threonyl‐tRNA‐synthetase activity and the regulation of threonine‐biosynthetic enzymes in Saccharomyces cerivisiae . Mol Gen Genet 1970, 108:28–32. [DOI] [PubMed] [Google Scholar]
- 165. Sugawara A, Tanaka T, Hirose T, Ishiyama A, Iwatsuki M, Takahashi Y, Otoguro K, Ōmura S, Sunazuka T. Borrelidin analogues with antimalarial activity: design, synthesis and biological evaluation against Plasmodium falciparum parasites. Bioorg Med Chem Lett 2013, 23:2302–2305. [DOI] [PubMed] [Google Scholar]
- 166. Otoguro K, Ui H, Ishiyama A, Kobayashi M, Togashi H, Takahashi Y, Masuma R, Tanaka H, Tomoda H, Yamada H, et al. In vitro and in vivo antimalarial activities of a non‐glycosidic 18‐membered macrolide antibiotic, borrelidin, against drug‐resistant strains of Plasmodia. J Antibiot (Tokyo) 2003, 56:727–729. [DOI] [PubMed] [Google Scholar]
- 167. Novoa EM, Camacho N, Tor A, Wilkinson B, Moss S, Marín‐García P, Azcárate IG, Bautista JM, Mirando AC, Francklyn CS, et al. Analogs of natural aminoacyl‐tRNA synthetase inhibitors clear malaria in vivo. Proc Natl Acad Sci USA 2014, 111:E5508–E5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. de Sousa AR, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009, 5:1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012, 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez‐Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR‐Cas9 system. Nat Biotechnol 2014, 32:819–821. [DOI] [PubMed] [Google Scholar]
- 171. Plagens A, Richter H, Charpentier E, Randau L. DNA and RNA interference mechanisms by CRISPR‐Cas surveillance complexes. FEMS Microbiol Rev 2015, 39:442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Erben ED, Fadda A, Lueong S, Hoheisel JD, Clayton C. A genome‐wide tethering screen reveals novel potential post‐transcriptional regulators in Trypanosoma brucei . PLoS Pathog 2014, 10:e1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Linder B, Grozhik AV, Olarerin‐George AO, Meydan C, Mason CE, Jaffrey SR. Single‐nucleotide‐resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 2015, 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Breker M, Schuldiner M. The emergence of proteome‐wide technologies: systematic analysis of proteins comes of age. Nat Rev Mol Cell Biol 2014, 15:453–464. [DOI] [PubMed] [Google Scholar]
- 176. Larance M, Lamond AI. Multidimensional proteomics for cell biology. Nat Rev Mol Cell Biol 2015, 16:269–280. [DOI] [PubMed] [Google Scholar]
- 177. Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 2015, 16:98–112. [DOI] [PubMed] [Google Scholar]
- 178. Zhong J, Xiao C, Gu W, Du G, Sun X, He QY, Zhang G. Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet 2015, 11:e1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6‐methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014, 15:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, et al. Drug resistance: population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 2015, 347:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]


