Summary
Background
Polymorphic variants within human melanocortin‐3 receptor gene (MC3R) gene have been associated with obesity. However, its influence on infancy and early childhood adiposity has not been reported before.
Objectives
We assessed associations between genotype at polymorphic sites within MC3R with early childhood adiposity and interaction with early childhood appetitive traits.
Methods
We studied 1090 singletons in an Asian mother–offspring cohort genotyped for MC3R and in a subgroup (n = 422) who had completed Child Eating Behaviour Questionnaires (CEBQ) at 12 months. Children were followed from birth to 48 months, and up to 10 measurements of body mass index and five measures of triceps and subscapular skin‐folds were obtained.
Results
Independent of potential confounders, each additional MC3R minor allele copy was associated with greater body mass index standard deviation score [B{95% confidence interval}: 0.004 units/month {0.001,0.007}; p = 0.007], triceps [0.009 mm/month {0.001,0.02}; p = 0.021] and subscapular skin‐fold [0.008 mm/month {0.002,0.01}; p = 0.011] gain velocity in the first 48 months. Each additional MC3R minor allele copy was also associated with increased odds of overweight [odds ratio {95% confidence interval}: 1.48{1.17–1.88}] and obesity [1.58{1.10–2.28}] in the first 48 months. Every additional copy of MC3R minor allele was positively associated with ‘slowness‐in‐eating’ appetitive trait [0.24{0.06,0.39}, p = 0.006]; however, the relationship between ‘slowness‐in‐eating’ with adiposity gain was not statistically significant.
Conclusions
Our findings support the role of MC3R genetic variants in adiposity gain during early childhood.
Keywords: Childhood adiposity, MC3R, single nucleotide polymorphism
Introduction
Overweight and obesity are commonly associated with increased risk of chronic diseases including type 2 diabetes and cardiovascular disease and present a massive public health challenge as they rapidly become a worldwide epidemic 1, 2. Non‐syndromic or common obesity is often viewed as a complex, multifactorial condition where exposure to an ‘obesogenic’ environment, coupled with an underlying genetic susceptibility to excessive weight gain, causes an obese phenotype 3, 4, 5. Linkage studies in humans and mice 6, 7 have reported that polymorphisms within melanocortin‐3‐receptor (MC3R) locus are associated with susceptibility for obesity. Mouse models with genetic alterations that disrupt MC3R exhibited hypophagia, higher energy efficiency, hyperleptinemia and reduced linear growth accompanied by higher percentage of body fat without increased body weight compared with wild‐type mice 8, 9.
In humans, two case–control studies have independently demonstrated two missense MC3R variants, Thr6Lys (rs3746619) and Val81Ile (rs3827103), which are in near complete linkage disequilibrium (LD), were significantly associated with increased adiposity in childhood and exhibited reduced in vitro activity compared with wild‐type MC3R 10, 11. We have previously described that Singaporean obese children with Thr6Lys/Val81Ile variants exhibited significantly higher leptin levels, percentage body fat and insulin sensitivity 11. Thus, there is evidence for the role of MC3R polymorphism in variation of human adiposity, and its effects on childhood adiposity appear to be mediated by altered energy intake and possibly altered sensitivity to satiety 12, 13. However, the age at which the variants start to alter phenotype is unknown. In this study, we assessed the association between MC3R coding variants rs3746619 and rs3827103 with adiposity and appetitive traits during early childhood in an Asian mother–offspring prospective cohort.
Materials and Methods
Study population
This study is embedded in Growing Up in Singapore Towards Healthy Outcomes (GUSTO) cohort study, designed to test specific hypotheses related to developmental pathways to cardio‐metabolic disorders in Chinese, Malay and Indian participants in Singapore. The study has previously been described in detail elsewhere 14. Pregnant women in their first trimester were recruited from two major public hospitals with obstetric services, KK Women's and Children's Hospital and National University Hospital, between June 2009 and September 2010. Of 3751 screened, 2034 met eligibility criteria, and 1247 women were recruited, of which there were 1176 deliveries, and 1090 with complete umbilical cord genotype data available and 422 with completed Child Eating Behaviour Questionnaire (CEBQ) (Figure S1). The women gave informed consent to participate in the study, which was approved by both National Healthcare Group Domain Specific Review Board and SingHealth Centralized Institutional Review Board.
Antenatal data
Socio‐demographic and health data (e.g. age, education and gestational diabetes) were captured during recruitment visits and at 26–28 weeks of gestation. Maternal ethnicity was self‐reported, and homogenous paternal ethnic background was an eligibility criteria; ethnicity was later confirmed by genotype analysis. Gestational age (GA) was determined by dating ultrasonography in first trimester and reported in completed weeks.
Infant anthropometry measurements
Measurements of weight and length, adopted from standardized protocols 15, were obtained at birth and at 3, 6, 9, 12, 15, 18, 24, 36 and 48 months of age. Weight was measured to the nearest gramme using calibrated scales [SECA 334 Weighing Scale {birth to 18 months}; SECA 803 Weighing Scale {24 to 48 months}, SECA Corp, Hamburg, Germany]. Recumbent length (from birth to 18 months) was measured from the top of the head to soles of feet using an infant mat (SECA 210 Mobile Measuring Mat) to the nearest 0.1 cm. Standing height (between 18 and 48 months) was measured using a stadiometer (SECA stadiometer 213) from the top of participant's head to his or her heels. For reliability, all measurements were taken in duplicates and averaged. Two skin‐folds (triceps and subscapular) were measured using standardized protocols 15 at birth and at 18, 24, 36 and 48 months in triplicates using Holtain skin‐fold callipers (Holtain Ltd, Crymych, UK) on the right side of the body, recorded to the nearest 0.2 mm. Anthropometric training and standardization sessions were conducted quarterly (once every 3 months), and observers were trained to obtain anthropometric measurements that, on average, were closest to the values measured by a master anthropometrist. Assessment of reliability was estimated by inter‐observer technical error of measurement and coefficient of variation 16 (Table S1).
Genotyping of melanocortin‐3‐receptor variants
Genotyping was performed on DNA extracted from frozen umbilical cords using Illumina omniexpress + exome array platform, which type the most frequent single nucleotide polymorphisms (SNPs) genome‐wide from all three HapMap phases 17, capturing the greatest amount of common SNP variation. DNA hybridization to arrays and scanning was performed by Expression Analysis Inc. Data were processed in GenomeStudio Genotyping Module™. Genotyping calls were made by the GenCall software, which incorporates a clustering (GenTrain) and calling algorithm (Bayesian model). GenCall score of each SNP probe and call rate of each sample are generated. Genotypes with GenCall scores less than 0.15 were considered missing. There were no poorly performing samples as defined by low sample call rates or GenCall Scores.
Only nine variants within MC3R were genotyped in the array, and the frequencies and positions of these SNPs are described in Table S2. Two variants (rs3746619 and rs3827103) were polymorphic in our study population, representing missense polymorphisms 17C > A (Thr6Lys) and 241G > A (Val81Ile), respectively. LD, in the form of r 2, was estimated using Haploview 18. The two variants exhibited strong LD in each ethnic group (r 2 > 0.9) (Figure S2), indicating no big difference across ethnicities. Thus, we discuss results for rs3746619 as the representative variant for MC3R, as it exhibited the highest minor allele frequency (Table S2).
Infant feeding and appetitive traits
Mothers were asked about infant milk‐feeding using questionnaires, based on 24‐h recall, at house visits when infants were 3, 6, 9 and 12 months of age. In accordance with World Health Organization guidelines 19, feeding practices were classified into exclusive, predominant, partial breastfeeding and formula‐feeding. In our data collection, both direct breastfeeding and expressed breastmilk intake were classified as breastfeeding.
Appetitive traits were measured using CEBQ 20, which is a self‐report completed by mothers. This was sent to all mothers by postal service prior to 12‐month visit. Questionnaires distributed were in English unless a preference for another language (Mandarin, Malay or Tamil) was expressed, in which translated versions were distributed. CEBQ relates to a period when the child was predominantly fed solid food, with each questionnaire item answered using a 5‐point Likert frequency scale (1 = never, 2 = rarely, 3 = sometimes, 4 = often and 5 = always). Only 422 children had questionnaires that were completed and returned by mothers.
Principal component analysis with Varimax normalized rotation was used to reduce the 35‐item CEBQ to seven subscales, which were the same as the original CEBQ 20. Questions with reverse scales were first reverse‐scored, and a factor loading cut‐off of 0.5 was applied. Four subscales measured food‐approach (food responsiveness, enjoyment of food, emotional over‐eating and desire to drink), whilst three measured food‐avoidant behaviours (emotional under‐eating, satiety responsiveness and slowness in eating) 21. Internal reliability for each derived subscale was calculated using Cronbach's alpha reliability coefficients, with coefficients ranging from 0.6 to 0.9, indicating a moderate‐to‐good internal reliability of the subscales in our study cohort (Table S3).
Statistical analyses
Descriptive statistics were reported as means and standard deviations for continuous variables and percentages for categorical variables. Infant body mass index (BMI) was calculated as infant weight/(infant height)2. Age‐specific and gender‐specific standard deviation scores (SDS) were calculated for BMI, referencing the local Singapore population 22. Overweight and obesity status was defined as having a BMI above 85th (i.e. BMI SDS > 1.036) and 95th percentile (i.e. BMI SDS > 1.645), respectively 22. We examined the longitudinal effect of MC3R on BMI SDS, triceps and subscapular skin‐folds trajectory using linear mixed effects models, which accounts for correlation between repeated measures on the same individual and allows for incomplete outcome data, assuming that they are missing at random 23. Maximum likelihood was the method of estimation, and an unstructured working covariance matrix for random effects parameters was chosen. The Akaike information criterion statistic facilitated model selection, and final models included linear, quadratic and cubic terms for children's ages and age–allele interaction to estimate change in BMI SDS, triceps and subscapular skin‐folds over time associated with each additional copy of MC3R minor allele. Besides the fixed effect of age, we allowed for a random intercept and linear slope for age. There was no evidence of significant differences in maternal health and socio‐demographic factors across offspring MC3R genotype (Table S4); hence, only breastfeeding duration, ethnicity and birthweight‐for‐GA were included as covariates.
Generalized estimating equations (GEE) were used to estimate the association between MC3R and odds of overweight or obesity in the first 48 months longitudinally, adjusting for ethnicity, breastfeeding duration and birthweight‐for‐GA. Similar with linear mixed effects models, GEE models are often used for repeated measures data as it account for within‐subject correlation between observations across time and allows for missing outcome data. In all models, potential effect modifications by ethnicity were investigated by adding the interaction term of MC3R with ethnicity to the fully adjusted model. We also analysed associations between MC3R with the seven CEBQ subscales at 12 months using multivariable linear regression with Bonferroni‐corrected multiple testing. In all analyses, an additive genetic model was used, where genotypes were coded as having 0, 1 or 2 copies of the minor allele. All analyses were performed using Stata version 13.0 (Statacorp College Station, TX, USA).
Results
Offspring clinical characteristics are shown in Table 1. No significant differences in birthweight, gender, skin‐folds at birth and MC3R genotype were observed across the three ethnic groups. We noted significant differences in GA (p = 0.038), breastfeeding duration (p < 0.001) and offspring BMI SDS at birth (p = 0.001) across the three ethnic groups.
Table 1.
Clinical characteristics of offspring
| Offspring | Chinese (n = 617) | Malay (n = 276) | Indian (n = 197) | Total (n = 1090) | p value |
|---|---|---|---|---|---|
| Gestational age (weeks) | 38.4 ± 1.4 | 38.1 ± 1.3 | 38.2 ± 1.6 | 38.3 ± 1.4 | 0.038 |
| Gender | 0.917 | ||||
| Male | 324 (52.5) | 149 (54.0) | 105 (53.3) | 578 (53.0) | |
| Female | 293 (47.5) | 127 (46.0) | 92 (46.7) | 512 (47.0) | |
| Birthweight (kg) | 3.1 ± 0.4 | 3.1 ± 0.4 | 3.0 ± 0.5 | 3.1 ± 0.4 | 0.081 |
| Birth adiposity measures | |||||
| Body mass index standard deviation score | −0.28 ± 1.37 | −0.03 ± 1.32 | −0.49 ± 1.38 | −0.25 ± 1.37 | 0.001 |
| Triceps skin‐folds (mm) | 5.4 ± 1.2 | 5.5 ± 1.3 | 5.2 ± 1.2 | 5.4 ± 1.2 | 0.052 |
| Subscapular skin‐folds (mm) | 5.0 ± 1.2 | 5.0 ± 1.2 | 4.8 ± 1.1 | 4.9 ± 1.2 | 0.141 |
| Any breastfeeding duration | <0.001 | ||||
| <6 months | 254 (53.8) | 153 (77.3) | 86 (60.6) | 493 (60.7) | |
| ≥6 months | 218 (46.2) | 45 (22.7) | 56 (39.4) | 319 (39.1) | |
| MC3R rs3746619 | 0.069 | ||||
| CC | 385 (62.4) | 150 (54.3) | 104 (52.8) | 639 (58.6) | |
| AC | 199 (32.3) | 108 (34.1) | 78 (39.6) | 385 (35.3) | |
| AA | 33 (5.3) | 18 (6.5) | 15 (7.6) | 66 (6.1) |
Numbers represent mean ± SD or n (%) # P value for continuous variables by one‐way analysis of variance; for categorical variables by chi‐square analysis
Figures 1A–C illustrates the modelled BMI SDS, triceps and subscapular skin‐folds trajectory, respectively, in the first 48 months of life for MC3R. We observed an increase in BMI SDS, triceps and subscapular skin‐folds velocity across age for each additional copy of MC3R minor allele (p = 0.007, 0.021 and 0.011, respectively), with an estimated rate of increase of 0.004 units/month (95% CI: 0.001, 0.007) for BMI (Fig. 1A), 0.009 mm/month (95% CI: 0.001,0.02) for triceps (Fig. 1B) and 0.008 mm/month (95% CI: 0.002,0.01) for subscapular skin‐folds, respectively (Fig. 1C), independent of potential confounders (i.e. breastfeeding duration, ethnicity and birthweight‐for‐GA). These differences in adiposity across MC3R genotype began to manifest significantly from 24 months of age (BMI: [B{95% CI}: 0.09{0.01,0.17}; p = 0.025], triceps: [0.18{0.05,0.32}; p = 0.007] and subscapular skin‐folds: [0.19{0.07,0.32}; p = 0.002]). In addition, we noted no significant interactions between MC3R and ethnicity, indicating that there were no ethnic‐specific associations in the relationship between MC3R and adiposity.
Figure 1.
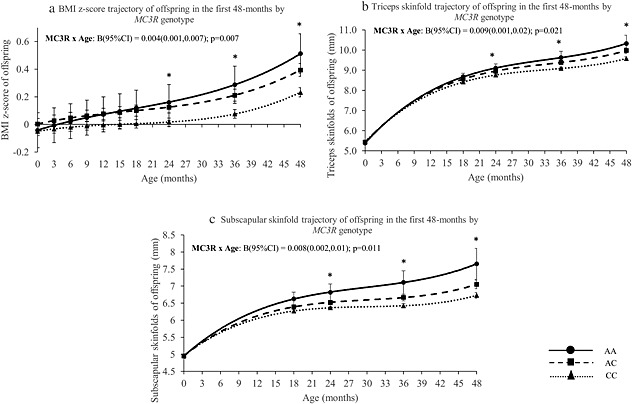
(A) Body mass index standard deviation score, (B) triceps skin‐folds and (C) subscapular skin‐folds trajectory in the first 48 months by offspring MC3R genotype. Adjusted for ethnicity, breastfeeding duration and birthweight‐for‐gestational age. Dotted line = CC genotype; Dashed line = AC genotype; Solid line = AA genotype. *p < 0.05 for every additional copy of minor allele; MC3R, melanocortin‐3‐receptor.
Additionally, we observed that every additional copy of MC3R minor allele contributed to an increase in proportion of overweight (Figure S3) and obesity (Figure S4) at 12, 24, 36 and 48 months of age. The longitudinal GEE model illustrated that MC3R increased the risk of being overweight (p = 0.001) or obese (p = 0.014) in the first 48 months, with each additional copy of MC3R minor allele increasing odds of overweight by 1.48 times (95%CI: 1.17–1.88) and obesity by 1.58 times (95%CI: 1.10–2.28) after adjustment for potential confounders (Table 2). Similarly, we noted no significant interactions between MC3R and ethnicity, indicating no ethnic‐specific associations in the relationship between MC3R with overweight or obesity in children.
Table 2.
Estimated odds of child overweight and obesity in the first 48 months by longitudinal logistic regression using GEE model, according to offspring MC3R minor allele and other characteristics
| Odds ratio | 95% Confidence interval | P value | ||
|---|---|---|---|---|
| Lower bound | Upper bound | |||
| Overweight | ||||
| rs3746619 (per A allele) | 1.48 | 1.17 | 1.88 | 0.001 |
| Birthweight‐for‐GA (per unit z‐score increase) | 1.62 | 1.39 | 1.88 | <0.001 |
| Malay (vs. Chinese) | 2.99 | 1.60 | 5.57 | 0.001 |
| Indian (vs. Chinese) | 2.69 | 1.31 | 5.53 | 0.007 |
| Breastfeeding duration (per month increase) | 0.98 | 0.95 | 1.01 | 0.126 |
| Age (per year increase) | 1.03 | 0.94 | 1.13 | 0.531 |
| Obesity | ||||
| rs3746619 (per A allele) | 1.58 | 1.10 | 2.28 | 0.014 |
| Birthweight‐for‐GA (per unit z‐score increase) | 1.68 | 1.36 | 2.08 | <0.001 |
| Malay (vs. Chinese) | 2.85 | 1.65 | 4.93 | <0.001 |
| Indian (vs. Chinese) | 2.63 | 1.48 | 4.67 | 0.001 |
| Breastfeeding duration (per month increase) | 0.96 | 0.92 | 1.01 | 0.106 |
| Age (per year increase) | 1.00 | 0.99 | 1.01 | 0.808 |
Odds ratios represent the odds of being overweight or obese in the first 48 months.
GA, gestational age; GEE, generalized estimating equations; MC3R, melanocortin‐3‐receptor.
We additionally analysed association of MC3R with each CEBQ subscale for those with completed CEBQ questionnaires (n = 422). This subgroup tended to be Chinese with at least 12 years of education, longer breastfeeding duration, lower BMI, longer gestational duration and higher birthweight compared with those who did not complete the questionnaire (Table S5); thus, these variables were included as potential confounders in subsequent analyses. Independent of these potential confounders, we observed an association between MC3R with the ‘slowness‐in‐eating’ subscale only [B{95% CI}: 0.24{0.06,0.39}; p = 0.006] (Table S6). However, within the sample size studied, ‘slowness‐in‐eating’ was not associated with child's gain in adiposity in the first 48 months [−0.0001 SDS units/month {−0.003, 0.003}; p = 0.966] (data not shown).
Discussion
In this study, we have identified novel associations between MC3R with early childhood adiposity gain in the first 48 months of life, with the differences in adiposity across MC3R genotype manifesting as early as 24 months of age. To our knowledge, this is the first study to assess the relationship between MC3R and adiposity at such an early age. Additionally, we demonstrated that every additional MC3R minor allele was associated with increasing odds of child overweight and obesity in the first 48 months. Earlier reports 10, 11, 24 have documented associations of the same MC3R polymorphism with adiposity in older, obese children, whilst other studies 24, 25, 26 identified similar relationships in adult populations. We have thus extended these findings to show that MC3R associated with adiposity in early childhood, further supporting the role of MC3R minor allele variants in determining early childhood adiposity.
Our findings are consistent with other studies from different populations that have independently described associations between MC3R variants with childhood adiposity. A recent GWAS study conducted on Hispanic children had reported nominal association of MC3R variants with childhood obesity 27. Feng et al. 10 showed that children who were homozygous for Thr6Lys and Val81Ile variants were significantly heavier, had more body fat, greater plasma leptin and insulin concentrations and greater insulin resistance than children who were wild‐type or heterozygous for these variants. These homozygous variants were also only observed amongst children who were at risk of overweight (BMI ≥ 85th percentile), or obesity (BMI ≥ 95th percentile). We also described similar observations in a population of obese Singaporean children, whereby obese children with Thr6Lys and Val81Ile variants exhibited significantly higher leptin levels and percentage body fat, highlighting that MC3R may be a predisposing factor for excessive body weight gain in children 11. However, other studies have reported null findings between MC3R risk variants with childhood obesity. A recent study 12 found insufficient evidence of significant association between childhood obesity and common MC3R variants, including the rs3827103 variant. Another study on a group of Polish children 28 showed that the common MC3R rs3827103 polymorphism was widely distributed amongst obese and control cohorts, suggesting that its predisposing effect to obesity may be rather unlikely. Potential causes for the discrepancy between studies could be epistatic interactions between MC3R alleles and other loci, which may be differential between populations. Studies that have shown an effect of MC3R have been conducted in those of African 10, Asian 11 or Hispanic 27 populations, whilst studies which failed to show a significant effect have been mostly conducted in European populations 28, 29. Data from HapMap have identified that MC3R minor allele frequencies are more frequent in Hispanics, Asians and Africans (HapMap Data Rel 28 Phase II + III, dbSNP b126). Thus, the contribution of these MC3R variants in predisposing to increased adiposity may vary significantly from population to population, and the alleles may be differentially penetrant in different populations or cultures with different food customs.
Interestingly, we also reported positive associations between MC3R with ‘slowness‐in‐eating’ at 12 months, consistent with findings from a Chilean cohort study, where obese boys carrying risk MC3R variants exhibited higher scores for ‘slowness‐in‐eating’ compared with those without 12, indicating that children with risk MC3R variants were perceived by their mothers to be eating more slowly compared with those without the risk allele. This is in contrast with current literature that have reported children with risk MC3R variants having higher energy intake 13 and children with food‐approach behaviours exhibiting greater weight gain 30. Our findings, however, appear to be consistent with phenotypes observed in MC3R knockout mice, which exhibited hypophagia compared with wild‐type littermates and were unusually susceptible to high fat diet‐induced obesity, partly explained by physical inactivity 8, 9. These mouse studies indicated that the mechanism in which MC3R variation leads to increased body fat was not increased food intake, but increased feed efficiency. Taken together, we postulate that children with risk MC3R variants may be eating more but taking a longer time to complete meals, which mothers ultimately perceive as ‘slowness‐in‐eating’. Unfortunately, our study lacks data on child energy intake; thus, we were unable to test this hypothesis.
Strengths of our study include the prospective design with high follow‐up rate in an Asian population. To date, there are no published studies relating MC3R genetic variants with adiposity in the first 48months of life; thus, our study provides useful and informative data on this relationship. There are, however, limitations to consider. Firstly, GUSTO was not designed to detect associations between gene variants and adiposity, and thus may not be sufficiently powered to detect the association. However, analyses between MC3R with various measures of adiposity were conducted using longitudinal models, making use of repeated measurements for each individual over a period of time, which would subsequently result in increased power. Our study genotyped only nine of the most common MC3R variants, but many more variants exist and not all were genotyped. Secondly, although all analyses were adjusted for potential confounders of adiposity, we could not rule out possible confounding by population stratification at higher resolution than ethnicity. Thirdly, this study used anthropometry as indicators of adiposity, but lacks more detailed measures of body composition, such as dual x‐ray absorptiometry or air displacement plethysmography, thus unable to further distinguish if these MC3R genetic variants were associated with fat mass or fat‐free mass. Lastly, CEBQ was only a self‐report measure by mothers on child's appetite, which may not be a direct reflection of the child's energy intake. Furthermore, the relatively low response rate of CEBQ completers (those with completed CEBQ questionnaire), which represented only 40% of the sample, may lack statistical power to answer the hypothesis of relation between ‘slowness‐in‐eating’ and adiposity. The results thus need to be taken as exploratory, with a need for further replication and research.
In summary, this study has provided evidence of significant association between MC3R variants with early childhood adiposity in an Asian population. Our study provided clues for the relationship of MC3R on early childhood adiposity and highlighted the plausible role of MC3R susceptibility alleles on weight regulation pathways at an early age. Follow‐up studies with larger numbers would be necessary to examine if these variants would continue to influence adiposity at later ages with more detailed measures of body composition and to address the role of MC3R in human weight regulation and pathogenesis of obesity.
Conflict of Interest statement
IMA, MTT, ALT, JDH, PLQ, CFFM, SES, SMS, KK, NL, FY, and YSL have nothing to declare. PDG, KMG and YSC have received reimbursement for speaking at conferences sponsored by companies selling nutritional products. They are part of an academic consortium that has received research funding from Abbot Nutrition, Nestec and Danone.
Funding
This study is under Translational Clinical Research (TCR) Flagship Programme on Developmental Pathways to Metabolic Disease, NMRC/TCR/004‐NUS/2008; NMRC/TCR/012‐NUHS/2014 funded by the National Research Foundation (NRF) and administered by the National Medical Research Council (NMRC), Singapore. KMG is supported by the National Institute for Health Research through the NIHR Southampton Biomedical Research Centre and by the European Union's Seventh Framework Programme (FP7/2007‐2013), project EarlyNutrition under grant agreement n°289346.
Clinical Trial Registration
This study is registered under the Clinical Trials identifier NCT01174875; http://www.clinicaltrials.gov/ct2/show/NCT01174875?term=GUSTO&rank=2
Supporting information
Supporting info item
Acknowledgements
The co‐authors acknowledge the contribution of the rest of the GUSTO study group that includes Dennis Bier, Arijit Biswas, Cai Shirong, Helen Chan, Jerry Chan, Yiong Huak Chan, Cornelia Chee, Audrey Chia, Chiang Wen Chin, Chng Chai Kiat, Mary Chong, Chong Shang Chee, Chua Mei Chien, Mary Daniel, Ding Chun Ming, Anne Ferguson‐Smith, Eric Andrew Finkelstein, Marielle Fortier, Doris Fok, Anne Goh, Daniel Goh, Joshua J Gooley, Han Wee Meng, Mark Hanson, Mikael Hartman, Michael Heymann, Stephen Hsu Chin‐Ying, Hazel Inskip, Jeevesh Kapur, Lee Bee Wah, B. F. P. Leutscher‐Broekman, Lim Sok Bee, Loh Seong Feei, Low Yen Ling, Iliana Magiati, Krishnamoorthy N, Cheryl Ngo, Pang Wei Wei, Prathiba Agarwal, Qiu Anqi, Quah Boon Long, Jen Richmond, Anne Rifkin‐Graboi, Allan Sheppard, Lynette Pei‐Chi Shek, Borys Shuter, Leher Singh, So Wing Chee, Walter Stunkel, Su Lin Lin, Tan Kok Hian, Tan Soek Hui, Teoh Oon Hoe, Terry Yoke Yin Tong, Hugo Van Bever, Rob Van Dam, Sudhakar Venkatesh, Helena Marieke Verkooijen, Inez By Wong, P. C. Wong, George S. H. Yeo. We thank Dr Mabel Yap (Ministry of Health, Singapore) and the National Healthcare Group Polyclinics (Singapore) for the BMI‐for‐age charts and SDS. K. M. G. is supported by the National Institute for Health Research through the NIHR Southampton Biomedical Research Centre. I. M. A. researched the data, wrote, edited and reviewed the manuscript. M. T. T., A. L. T., J. D. H., Q. P. L., C. F. F. M., X. L. and S. E. S. researched the data, reviewed the manuscript and contributed to the discussion. S. M. S., K. K., K. M. G., P. D. G., Y. S. C., N. L. and F. Y. reviewed the manuscript and contributed to the discussion. Y. S. L. edited and reviewed the manuscript and contributed to the discussion
Aris, I. M. , Tint, M. T. , Teh, A. L. , Holbrook, J. D. , Quah, P. L. , Chong, M. F.‐F. , Lin, X. , Soh, S. E. , Saw, S.‐M. , Kwek, K. , Godfrey, K. M. , Gluckman, P. D. , Chong, Y. S. , Lek, N. , Yap, F. , and Lee, Y. S. (2016) MC3R gene polymorphisms are associated with early childhood adiposity gain and infant appetite in an Asian population. Pediatric Obesity, 11: 450–458. doi: 10.1111/ijpo.12086.
The copyright line for this article was changed on 08 November 2016 after original online publication.
References
- 1. Dans A, Ng N, Varghese C et al. The rise of chronic non‐communicable diseases in Southeast Asia: time for action. Lancet . 2011; 377(9766): 680–689. doi: 610.1016/S0140-6736(1010)61506-61501. Epub 62011 Jan 61525. [DOI] [PubMed] [Google Scholar]
- 2. Kopelman P. Health risks associated with overweight and obesity. Obes Rev Mar 2007; 8(Suppl 1): 13–17. [DOI] [PubMed] [Google Scholar]
- 3. Gonzalez JR, Estevez MN, Giralt PS, et al. Genetic risk profiles for a childhood with severe overweight. Pediatr Obes . 2014; 9(4): 272–280. doi: 210.1111/j.2047-6310.2013.00166.x. Epub 02013 Apr 00129. [DOI] [PubMed] [Google Scholar]
- 4. Kotanidou EP, Kalinderi K, Kyrgios I, et al. Apelin and G212A apelin receptor gene polymorphism in obese and diabese youth. Pediatr Obes . 2015; 10(3): 213–219. doi: 210.1111/ijpo.1251. Epub 2014 Jul 1124. [DOI] [PubMed] [Google Scholar]
- 5. Lee YS. Genetics of nonsyndromic obesity. Curr Opin Pediatr . 2013; 25(6): 666–673. doi: 610.1097/MOP.1090b1013e3283658fba. [DOI] [PubMed] [Google Scholar]
- 6. Fox CS, Heard‐Costa NL, Vasan RS, et al. Genomewide linkage analysis of weight change in the Framingham Heart Study. J Clin Endocrinol Metab 2005; 90(6): 3197–3201. Epub 2005 Mar 3115. [DOI] [PubMed] [Google Scholar]
- 7. Lembertas AV, Perusse L, Chagnon YC, et al. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest 1997; 100(5): 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler AA, Kesterson RA, Khong K, et al. A unique metabolic syndrome causes obesity in the melanocortin‐3 receptor‐deficient mouse. Endocrinology 2000; 141(9): 3518–3521. [DOI] [PubMed] [Google Scholar]
- 9. Chen AS, Marsh DJ, Trumbauer ME, et al. Inactivation of the mouse melanocortin‐3 receptor results in increased fat mass and reduced lean body mass. Nat Genet 2000; 26(1): 97–102. [DOI] [PubMed] [Google Scholar]
- 10. Feng N, Young SF, Aguilera G, et al. Co‐occurrence of two partially inactivating polymorphisms of MC3R is associated with pediatric‐onset obesity. Diabetes 2005; 54(9): 2663–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YS, Poh LK, Kek BL, Loke KY. The role of melanocortin 3 receptor gene in childhood obesity. Diabetes 2007; 56(10): 2622–2630 Epub 2007 Jul 2616. [DOI] [PubMed] [Google Scholar]
- 12. Obregon AM, Amador P, Valladares M et al. Melanocortin‐3 receptor gene variants: association with childhood obesity and eating behavior in Chilean families. Nutrition . 2010; 26(7–8): 760–765. doi: 710.1016/j.nut.2009.1007.1005. Epub 2010 Feb 1019. [DOI] [PubMed] [Google Scholar]
- 13. Savastano DM, Tanofsky‐Kraff M, Han JC, et al. Energy intake and energy expenditure among children with polymorphisms of the melanocortin‐3 receptor. Am J Clin Nutr . 2009; 90(4): 912–920. doi: 910.3945/ajcn.2009.27537. Epub 22009 Aug 27535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soh SE, Tint MT, Gluckman PD, et al. Cohort profile: Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study. Int J Epidemiol 2013; 2: 2. [DOI] [PubMed] [Google Scholar]
- 15. Hamilton CM, Strader LC, Pratt JG, et al. The PhenX Toolkit: get the most from your measures. Am J Epidemiol . 2011; 174(3): 253–260. doi: 210.1093/aje/kwr1193. Epub 2011 Jul 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaefer F, Georgi M, Zieger A, Scharer K. Usefulness of bioelectric impedance and skinfold measurements in predicting fat‐free mass derived from total body potassium in children. Pediatr Res 1994; 35(5): 617–624. [PubMed] [Google Scholar]
- 17. The International HapMap C . A haplotype map of the human genome. Nature . 10/27/print 2005;437(7063): 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21(2): 263–265 Epub 2004 Aug 2005. [DOI] [PubMed] [Google Scholar]
- 19. World Health Organization . Indicators for assessing infant and young child feeding practices, in Conclusions of a consensus meeting held 6–8 November 2007 in WashingtonD.C., USA.2008.
- 20. Wardle J, Guthrie CA, Sanderson S, Rapoport L. Development of the Children's Eating Behaviour Questionnaire. J Child Psychol Psychiatry Oct 2001; 42(7): 963–970. [DOI] [PubMed] [Google Scholar]
- 21. Quah PL, Chan YH, Aris IM, et al. Prospective associations of appetitive traits at 3 and 12 months of age with body mass index and weight gain in the first 2 years of life. BMC Pediatr 2015; 15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. National Healthcare Group Polyclinics . Age and gender specific national BMI cut‐offs (Singapore). 2000.
- 23. Finucane MM, Samet JH, Horton NJ. Translational methods in biostatistics: linear mixed effect regression models of alcohol consumption and HIV disease progression over time. Epidemiol Perspect Innov 2007; 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zegers D, Beckers S, Mertens IL, Van Gaal LF, Van Hul W. Common melanocortin‐3 receptor variants are not associated with obesity, although rs3746619 does influence weight in obese individuals. Endocrine . 2010; 38(2): 289–293. doi: 210.1007/s12020-12010-19386-12025. Epub 12010 Oct 12023. [DOI] [PubMed] [Google Scholar]
- 25. Mencarelli M, Dubern B, Alili R, et al. Rare melanocortin‐3 receptor mutations with in vitro functional consequences are associated with human obesity. Hum Mol Genet . 2011; 20(2): 392–399. doi: 310.1093/hmg/ddq1472. Epub 2010 Nov 1093. [DOI] [PubMed] [Google Scholar]
- 26. Mencarelli M, Walker GE, Maestrini S, et al. Sporadic mutations in melanocortin receptor 3 in morbid obese individuals. Eur J Hum Genet . 2008; 16(5): 581–586. doi: 510.1038/sj.ejhg.5202005. Epub 5202008 Jan 5202030. [DOI] [PubMed] [Google Scholar]
- 27. Comuzzie AG, Cole SA, Laston SL, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One . 2012; 7(12): e51954. doi: 51910.51371/journal.pone.0051954. Epub 0052012 Dec 0051914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cieslak J, Majewska KA, Tomaszewska A et al. Common polymorphism (81Val > Ile) and rare mutations (257Arg > Ser and 335Ile > Ser) of the MC3R gene in obese Polish children and adolescents. Mol Biol Rep . 2013; 40(12): 6893–6898. doi: 6810.1007/s11033-11013-12808-11038. Epub 12013 Oct 11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schalin‐Jantti C, Valli‐Jaakola K, Oksanen L, et al. Melanocortin‐3‐receptor gene variants in morbid obesity. Int J Obes Relat Metab Disord 2003; 27(1): 70–74. [DOI] [PubMed] [Google Scholar]
- 30. McCarthy EK, Chaoimh CN, Murray DM, et al. Eating behaviour and weight status at 2 years of age: data from the Cork BASELINE Birth Cohort Study. Eur J Clin Nutr 2015; 12(10): –130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item


