ABSTRACT
The mammalian nucleus has a distinct substructure that cannot be visualized directly by conventional microscopy. In this study, the organization of the DNA within the nucleus of multiple myeloma (MM) cells, their precursor cells (monoclonal gammopathy of undetermined significance; MGUS) and control lymphocytes of the representative patients is visualized and quantified by superresolution microscopy. Three‐dimensional structured illumination microscopy (3D‐SIM) increases the spatial resolution beyond the limits of conventional widefield fluorescence microscopy. 3D‐SIM reveals new insights into the nuclear architecture of cancer as we show for the first time that it resolves organizational differences in intranuclear DNA organization of myeloma cells in MGUS and in MM patients. In addition, we report a significant increase in nuclear submicron DNA structure and structure of the DNA‐free space in myeloma nuclei compared to normal lymphocyte nuclei. Our study provides previously unknown details of the nanoscopic DNA architecture of interphase nuclei of the normal lymphocytes, MGUS and MM cells. This study opens new avenues to understanding the disease progression from MGUS to MM. J. Cell. Biochem. 116: 704–710, 2015. © 2014 The Authors. Journal of Cellular Biochemistry published by Wiley Periodicals, Inc.
Keywords: 3D STRUCTURED ILLUMINATION MICROSCOPY, DNA NANO STRUCTURE, MULTIPLE MYELOMA, MONOCLONAL GAMMOPATHY OF UNDETERMINED SIGNIFICANCE
Plasma cell disorders are a spectrum of diseases characterized by the proliferation of neoplastic plasma cells of B‐cell lineage that produce monoclonal immunoglobulin [Rajkumar et al., 2006; Dimopoulos and Terpos, 2010]. This spectrum includes asymptomatic conditions such as monoclonal gammopathy of unknown significance (MGUS) as well as the symptomatic malignant condition, multiple myeloma (MM) [Rajkumar et al., 2006; Dimopoulos and Terpos, 2010]. The risk of progression from MGUS to symptomatic MM is approximately 1% per year [Rajkumar, 2005; Korde et al., 2011].
Abnormal plasma cells in MGUS and MM are thought to be morphologically identical [Kastritis and Dimopoulos, 2014]. These cells also share common cytogenetic features as well as genetic and epigenetic alterations [Klewes et al., 2013; Kastritis and Dimopoulos, 2014]. Although MGUS and MM cells can be distinguished from normal plasma cells by genetic and phenotypic markers, there is no single marker that distinguishes between MGUS and MM cells [Zingone and Kuehl, 2011]. To our knowledge, no previous study has provided a detailed description of the subnuclear architecture of these abnormal plasma cells in MGUS and MM.
The mammalian cell nucleus has a unique structural and functional organization [Raska et al., 1992; Cooper, 2000]. It contains morphologically distinct chromatin domains and protein subcompartments that fit into a limited space [Qumsiyeh, 1999; Cremer and Cremer, 2001]. Several studies have shown that a specific nuclear architecture is related to transcriptional activity [van Driel and Verschure, 2001; Rajapakse and Groudine, 2011]. A better understanding of nuclear structure of the myeloma cell might reveal underlying molecular mechanisms in the pathogenesis of the disease.
Conventional light microscopy, with a resolution limited by the diffraction limit of the objective lens, has been widely used in modern cell and cancer biology. The recent development of superresolution fluorescence microscopy techniques allows us to evaluate spatial relationships within subcellular and suborganelle structures beyond the diffraction limit [Hell, 2007; Heilemann, 2010; Schermelleh et al., 2010; Leung and Chou, 2011]. Such optical nanoscopy techniques provide the ability of accurate measurements of subcellular structures at a level previously achieved only by electron microscopy [Baddeley et al., 2010].
Three‐dimensional structured illumination microscopy (3D‐SIM) is a super‐resolution method, which provides a higher image resolution than conventional widefield microscopy [Gustafsson, 2008; Schermelleh et al., 2010]. In short, a periodic illumination pattern results in heterodyne detection of high frequency information that would otherwise be lost. Images are acquired for multiple pattern orientations and phases and computationally recombined as a superresolution image [Gustafsson et al., 2008; Shroff et al., 2009; Wicker et al., 2013]. 3D‐SIM has revealed the subcellular localization of key proteins in cells [Sonnen et al., 2012; Strauss et al., 2012], the fine details of nuclear envelope [Schermelleh et al., 2008], chromosome structure [Carlton, 2008; Flors and Earnshaw, 2011; Green et al., 2011], or even the specialized cellular structure such as endothelial cell fenestrations [Cogger et al., 2010] and the cytokinetic Z ring in live bacteria [Turnbull et al., 2014]. The application of this technique is compatible with both fixed and live cells [Hirvonen et al., 2009]. Using 3D‐SIM, Righolt et al. (2014) had measured the DNA organization in the interphase nuclei of Hodgkin's lymphoma and revealed a significant increase in submicron DNA structures of Hodgkin cells and Reed‐Sternberg cells compared to normal lymphocytes that clearly distinguish the three cell types from each other.
In our current study, we have used 3D‐SIM to examine the three‐ dimensional ultrastructure of the interphase nucleus of myeloma cells from untreated MM patients and compared them to malignant plasma cells of untreated MGUS patients and normal lymphocytes of both patient groups. The aim of this study was to determine whether or not 3D‐SIM allowed us to differentiate between MGUS and MM nuclei based on their superresolution DNA structure.
MATERIALS AND METHODS
PATIENTS
This study was approved by CancerCare Manitoba Research Resource Impact Committee and Research Ethics Board on human studies at University of Manitoba, Manitoba, Canada (Ethics Reference No. H2010:170) and the Ethics Review Committee on Human Research of the University of Tartu (Protocol No. 194T‐11). The study population consisted of a total of 20 patients, which where subdivided into two groups: MM (N = 10) and MGUS (N = 10). Informed consent was obtained from all patients. All patients conformed to the diagnostic criteria according to the International Myeloma Working Group (IMWG) [Kyle and Rajkumar, 2009]. All blood samples were collected before the start of any treatment. All patients were treatment naive. Control lymphocytes were examined from the identical patients (i.e., from patients presenting with MM or MGUS).
ISOLATION OF LYMPHOCYTES AND MYELOMA CELLS
Ten milliliters peripheral blood from each patient was collected in EDTA‐treated tubes. Mononuclear cells were overlaid in Ficoll‐Paque (GE Healthcare Life Sciences, Baie d'Urfe, Quebec, Canada) and separated by centrifugation at 200 g for 30 min. The remove buffy coat was washed with 10 ml of a 1 x phosphate buffered saline (PBS) solution.
DAPI STAINING
The isolated cells were subsequently placed onto slides. The slides were incubated in 3.7% formaldehyde (Sigma‐Aldrich, Oakville, Ontario, Canada) for 30 min and washed three times in 1xPBS for 5 min each while shaking at room temperature. Slides were stained with 4′,6‐diamidino‐2‐ phenylindole (DAPI) (0.1 µl/ml) and incubated in the dark for 3 min. Excess DAPI was removed with ddH2O. The slides were then mounted with Vestashield (Vector Laboratories, Burlington, Ontario, Canada). The slides were covered with a coverslip (No. 1 1/2, Schott, Mainz, Germany) and sealed with nail polish. The slides were stored at 4 °C until imaging.
IDENTIFICATION OF MYELOMA CELLS
Our previous work had described the identification of myeloma cells based on the intensity of green fluorescence signals emitted by the fluorescein isothiocyanate–labeled CD138 antibody and on the size and intensity of the DAPI counterstained nucleus. In comparison, normal lymphocyte nuclei had a smaller, rounder shape and emitted a brighter DAPI signal than myeloma nuclei [Klewes et al., 2013]. In this study, we have identified myeloma and lymphocyte nuclei based on size and intensity of the DAPI staining.
IMAGE ACQUISITION
All images from isolated cells were captured using a Zeiss Elyra PS1 SIM equipped with a Zeiss Plan Apochromat inverted 63x/1.40 oil immersion objective lens using an Andor EM‐CCD iXon 885 camera and a 1.6 x tube lens at room temperature. The DAPI channel was obtained with 405 nm laser excitation, 23 µm diffraction grating and filter cube SR Cube 07. The lateral pixel size, Δx and Δy, was 79 nm in the recorded images and 40 nm in the reconstructed image. The z‐stacks were acquired by capturing slices taken at 91 nm intervals through each nucleus, and consisted of 60–85 slices collected sequentially. Cell nuclei were chosen by the operator. A field of view was selected and the z‐stack boundaries were defined manually. The 3D‐SIM and widefield images were reconstructed using ZEN 2012 black edition (Carl Zeiss, Jena, Germany). Image stacks were exported as 16‐bit tiff image sequences.
The image processing was performed in Matlab (MathWorks, Natick, MA) with the toolbox DIP image [Luengo Hendriks et al., 1999]. A central z‐plane was manually selected. The nucleus was automatically detected by isodata thresholding. The granulometry of the DNA structure was measured with a morphological sieve applied to the error‐function clipped images [Duin et al., 2007; Luengo Hendriks et al., 2007]. The coefficient of variation and the skewness of the intensity histogram over the detected region were also calculated. See [Righolt et al., 2014] for full details of the methodology.
STATISTICAL ANALYSIS
Group data were expressed as mean ± SD. For 3D‐SIM imaging data, the distributions were compared using two‐sided, two‐sample Kolmogorov–Smirnov (KS) tests to determine the significance of difference. P‐values of <0.05 were considered statistically significant.
RESULTS
Clinical characteristics of all patients included in this study are described in Table I. The two patient groups were similar in age. The average age of the MM and MGUS groups is 67.4 ± 14.7 and 67.2 ± 14.9 years, respectively. The MM group was composed of 3 cases at stage I, 5 cases at stage II and 2 cases at stage III according to the International Staging System (ISS) [Greipp et al., 2005]. The majority of the patients in both groups were classified in the IgG group. The percentage of bone marrow plasma cells (BMPC) and the level of secreted monoclonal protein (M‐protein) increased with disease progression to symptomatic MM (Table I).
Table I.
Clinical Characteristics of Patient Included in This Study
| Clinical characteristic | MGUS patients | MM patients |
|---|---|---|
| Mean age (year) | 67.2 ± 14.9 | 67.4 ± 14.7 |
| BMPC (%) | 4.2 ± 2.5 | 38.8 ± 33.5 |
| Immunoglobulin isotype (mg/dL) | ||
| IgG | 16.1 ± 8.6 | 34.5 ± 25.9 |
| IgA | 3.9 ± 2.8 | 2.6 ±5.3 |
| IgM | 2.3± 4.4 | 0.4 ± 0.2 |
| M protein (g/L) | 9.2 ± 7.2 | 29.1 ± 18.8 |
BMPC indicates bone marrow plasma cells.
Lymphocytes and myeloma nuclei from MM and MGUS samples were identified and then imaged using 3D‐SIM. After acquisition and image reconstruction (see Materials and Methods), we determined the intranuclear DNA structure. We analyzed a total of 534 lymphocytes, 259 MGUS and 279 MM nuclei. Figure 1 illustrates the nuclear DNA structure of normal lymphocyte as well as MGUS and MM nuclei. Nuclear DNA structures were well defined and clearly visible in 3D‐SIM images compared to widefield images. In normal lymphocytes, the DNA structure within the nucleus generally appeared as a fine‐grained texture and exhibited uniform distribution. On the contrary, myeloma cells had a relatively coarse texture and uneven distribution of their nuclear DNA. The full z‐stacks for these cells are shown in Movies 1–3 (Supplemental Information). Additionally, we observed numerous well‐defined areas void of DAPI staining (“holes”) in 3D‐SIM images of the myeloma nuclei, which were difficult to observe in the corresponding widefield images (Fig.1). “Holes” refer to DNA poor or DNA‐free nuclear space [Righolt et al., 2014]. While most of the myeloma nuclei have the large scale “holes” within their nuclei, we have hardly detected these structures in lymphocyte nuclei.
Figure 1.
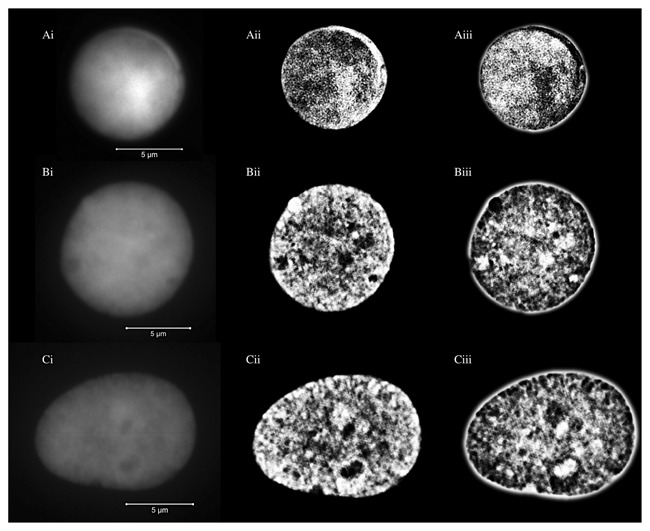
Representative images from DAPI‐stained nuclei of a normal lymphocyte (A), MGUS nucleus (B) and MM nucleus (C). Widefield images are shown in images (i), unclipped SIM images in images (ii), and negative unclipped SIM images in images (iii). The scale bars are 5 μm in length. The SIM images showed the DNA structures in greater detail compared to the widefield images. Note the difference in the DNA structures and also the size of the nuclei between the different cell types.
To quantify whether there are differences between normal lymphocytes and myeloma nuclei, we have used granulometry to evaluate the size distribution of the DNA structure and the DNA‐free space [Righolt et al., 2014]. The granulometry shows the differences in nuclear size in the microns, whereas the differences in the DNA‐structure occur at submicron size. Normal lymphocytes have the smallest amount of submicron DNA structure. The two‐sided, two‐sample Kolmogorov‐Smirnov (KS) test showed that the amount of the intranuclear submicron DNA structure in myeloma nuclei was significantly increased compared to normal lymphocyte nuclei (P = 10−88). The KS test also showed significant alterations in the granule size distribution of the DNA‐free space of myeloma nuclei compared to lymphocyte nuclei (P = 10−168 for MM nuclei vs lymphocytes and P = 10−231 for MGUS nuclei vs lymphocytes), as described in Table II. Among myeloma nuclei, the DNA‐free space of MM nuclei and MGUS nuclei was significantly different (P = 10−8) as measured by dark granulometry and shown in Figure 2. However, there was no significant difference of the DNA submicron structure between MM and MGUS nuclei (P = 0.68) when light granulometry was measured. There was no difference of nuclear DNA structure and DNA‐free space between normal lymphocyte nuclei in MM and MGUS patients (P = 0.99). Note that all samples examined were from treatment naïve patients (Materials and Methods).
Table II.
The Differences of Intranuclear Organization Between MM, MGUS and Lymphocyte Nuclei Using the Two‐Sided, Two‐sample Kolmogorov–Smirnov (KS) Analysis
| Differences in intranuclear organization | P value |
|---|---|
| DNA submicron structure | |
| Lymphocyte vs MGUS nuclei | 2.0 × 10−88 |
| Lymphocyte vs MM nuclei | 2.5 × 10−88 |
| MGUS vs MM nuclei | 0.68 |
| Intranuclear DNA‐free space | |
| Lymphocyte vs MGUS nuclei | 4.1 × 10−231 |
| Lymphocyte vs MM nuclei | 1.1 × 10−168 |
| MGUS vs MM nuclei | 1.0 × 10−8 |
Figure 2.
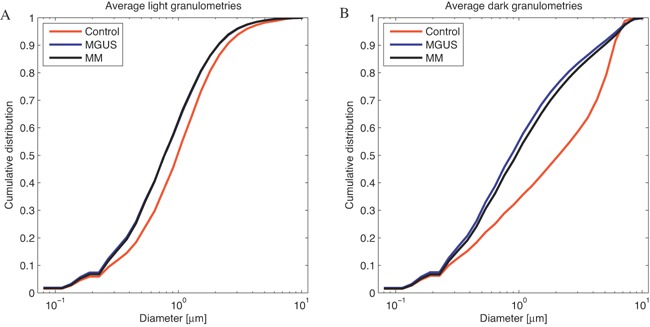
Measurement of the size distribution of the DNA structure of normal lymphocyte (red lines), MGUS (blue lines) and MM (black lines) using granulometry. While the differences in nucleus size of each cell type might reach up to several μm, the differences in the DNA structure size were represented in the sub‐micron size range. Granulometries revealed a significant increase in the amount of DNA submicron structure in MGUS and MM nuclei compared to control lymphocyte (P = 10−88) (A). The differences between MGUS and MM are significant for DNA‐free space (P = 10−8) but not for DNA submicron structure (P = 0.68) (B). Note that both MGUS and MM nuclei were significantly larger in size than normal lymphocytes.
In summary, the application of 3D‐SIM microscopy revealed details of nuclear DNA organization in MM and MGUS nuclei. Our data showed that myeloma nuclei have significantly increased submicron DNA structure and an increase in DNA‐free space compared to normal lymphocyte nuclei. Moreover, MGUS and MM nuclei differ significantly in their dark granulometries (“empty nuclear space”) indicating that MGUS and MM represent two distinct types of plasma cell malignancies. This is the first time that differences between MGUS and MM nuclei can be visualized and quantified at the organizational DNA level.
DISCUSSION
The eukaryotic interphase nucleus is a highly compartmentalized structure [Kumaran et al., 2008]. Chromosomes and other nuclear components are non randomly organized within the nucleus [Kumaran et al., 2008; Cremer and Cremer, 2010]. Each chromatin territory influences gene expression and nuclear function [Sproul et al., 2005; Kumaran et al., 2008; Solovei et al., 2009]. In the present study, we have used 3D‐SIM to provide a quantitative evaluation of the size distribution of nuclear DNA in abnormal myeloma nuclei at a level of accuracy beyond the conventional optical diffraction limit of light microscopes. 3D‐SIM allows increased resolution in all three directions, thus opening new possibilities to study the nuclear architecture at ultrastructure level. Our study showed a significant change of the size distribution of nuclear DNA of MM nuclei compared to MGUS and normal lymphocytes. This alteration reflects the structural changes of the cell nucleus and the distribution of nuclear DNA. These changes may be associated with a series of genetic and epigenetic alterations in the transformation process of a normal cell into a malignant cell [Liu et al., 2014]. Malignant cells generally gain multiple types of chromosomal aberrations, including rearrangements, translocation, deletion and duplications [Lobo, 2008]. Previous studies have shown that chromosome size and gene density have a significant impact on the nuclear arrangement of chromosome territories [Tanabe et al., 2002; Sproul et al., 2005; Kumaran et al., 2008]. 3D‐SIM, therefore, yields the information of the alterations of DNA organization that reflect genetic changes in interphase nucleus.
In this study, we observed that MGUS nuclei have significantly higher DNA‐free space than MM nuclei, whereas no difference in nuclear DNA submicron structure was measured between the two types of nuclei. The increase in the nuclear DNA submicron structure in MGUS and MM nuclei might be the result of the accumulation of genetic and epigenetic alterations. This finding is consistent with the notion that genetic alterations occur early in the premalignant stage of myeloma and continue throughout the malignant stage in MM [Morgan GJ et al., 2012]. Cell transformation is associated with rearrangements in the nuclear DNA. The difference in DNA‐free space between MGUS and MM nuclei might be resulting from changes in the condensation and the rearrangement of the DNA. These alterations are likely associated with changes in DNA organization as revealed by 3D‐SIM.
This is the first study that demonstrates differences in the nuclear DNA organization between MGUS and MM nuclei. The characteristic morphological changes between malignant and premalignant cells are not visible by conventional light microscopy due to mostly nanoscale changes, for example, nuclear chromatin texture [Liu et al., 2014]. Our findings provide crucial information to differentiate myeloma nuclei between MGUS and MM patients. Our data also show the potential role of 3D‐SIM to become a new powerful tool that can visualize morphological changes enabling the identification of premalignant cells. This will also enhance one's ability to predict the risk of progression to a more malignant stage of the disease.
Altogether, this study showed significantly altered nuclear DNA organization of MM nuclei compared to MGUS and normal lymphocyte nuclei. Righolt et al.(2014) had used superresolution microscopy to demonstrate a significant and progressive change of DNA organization from control lymphocytes through Hodgkin cells to Reed‐Sternberg cells. Their study also hypothesized that nuclear DNA structure might be related to disease aggressiveness in patients with Hodgkin's lymphoma.
Our current findings in myeloma are in line with the previous study in Hodgkin's lymphoma and indicate significant alterations of DNA structure during both the premalignant and malignant stages. These alterations might be common in cancer nuclei in general. Further study is warranted to support our finding of superresolution nuclear DNA alteration in different clinical staging and patient groups. The alteration of nuclear DNA structure might be one of the common changes during tumorigenesis. Further studies should include various types of malignancies to uncover the organization of the nuclear DNA in interphase nuclei, i.e., whether there might be common changes in various types of cancer or rather organizational changes in a cancer type‐specific manner.
A better understanding of organizational changes in nuclear DNA of myeloma cells in the premalignant and malignant stage of disease may lead to the integration of advances in cancer biology and therapy for a better treatment of myeloma. This method could provide a new tool for the individualized risk evaluation of disease progression from MGUS to MM. In conclusion, our study demonstrates that 3D‐SIM is a new tool for a better understanding of the nuclear ultrastructure of DNA in cancer.
Supporting information
Supplementary Data S1.
Supplementary Data S2.
Supplementary Data S3.
ACKNOWLEDGEMENT
This study is dedicated to Dr. Adebayo Olujohungbe (1963‐2013). The authors thank the patients who contributed blood samples to this study. We thank Donna Hewitt for obtaining patient consent. Samples from Estonia were provided by the Hematology and Oncology clinic, Tartu University Hospital.
Conflict of interest: The authors declare no potential conflict of interest.
REFERENCES
- Baddeley D, Chagin VO, Schermelleh L. 2010. Measurement of replication structures at the nanometer scale using super‐resolution light microscopy. Nucl Acids Res 38(2):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM. 2008. Three‐dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res 16(3):351–365. [DOI] [PubMed] [Google Scholar]
- Cogger VC, McNerney GP, Nyunt T, DeLeve LD, McCourt P, Smedsrød B, Le Couteur DG, Huser TR. 2010. Three‐dimensional structured illumination microscopy of liver sinusoidal endothelial cell fenestrations. J Struct Biol 171(3):382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM. 2000. The cell: A molecular approach. 2nd edition Sunderland (MA): Sinauer Associates Internal organization of the nucleus. [Google Scholar]
- Cremer T, Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev Genet 2:292–301. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2(3):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos MA, Terpos E. 2010. Multiple myeloma. Ann Oncol 21(7):vii143–vii 150. [DOI] [PubMed] [Google Scholar]
- Duin RPW, Juszczak P, Paclik P, Pekalska E, de Ridder D, Tax DMJ, Verzakov S. 2007. PRTools, a Matlab toolbox for pattern recognition. Delft, The Netherlands: Delft University of Technology.
- Flors C, Earnshaw WC. 2011. Super‐resolution fluorescence microscopy as a tool to study the nanoscale organization of chromosomes. Curr Opin Chem Biol 15(6):838–844. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. 2008. Three‐Dimensional Resolution Doubling in Wide‐Field Fluorescence Microscopy by Structured Illumination. Biophys J 94(12):4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LC, Kalitsis P, Chang TM, Cipetic M, Kim JH, Marshall O, Turnbull L, Whitchurch CB, Vagnarelli P, Samejima K, Earnshaw WC, Choo KHA, Hudson DF. 2011. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci 125(6):1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greipp PR, Miguel JS, Durie BGM, Crowley JJ, Barlogie B, Bladé J, Boccadoro M, Child JA, Avet‐ Loiseau H, Kyle RA, Lahuerta JJ, Ludwig H, Morgan G, Powles R, Shimizu K, Shustik C, Sonneveld P, Tosi P, Turesson I, Westin J. 2005. International staging system for multiple myeloma. J Clin Oncol 23:3412–3420. [DOI] [PubMed] [Google Scholar]
- Heilemann M. 2010. Fluorescence microscopy beyond the diffraction limit. J Biotechnol 149(4):243–251. [DOI] [PubMed] [Google Scholar]
- Hell SW. 2007. Far‐field optical nanoscopy. Science 316:1153–1158. [DOI] [PubMed] [Google Scholar]
- Hirvonen LM, Wicker K, Mandula O, Heintzmann R. 2009. Structured illumination microscopy of a living cell. Eur Biophys J 38:807–812. [DOI] [PubMed] [Google Scholar]
- Kastritis E, Dimopoulos MA. 2014. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma In: Schey SA. Yong KL. Marcus R. Anderson KC. editors. Myeloma: Pathology, diagnosis and treatment. New York: Cambridge University Press; p 121–133. [Google Scholar]
- Klewes L, Vallente R, Dupas E, Brand C, Grün D, Guffei A, Sathitruangsak C, Awe JA, Kuzyk A, Lichtensztejn D, Tammur P, Ilus T, Tamm A, Punab M, Rubinger M, Olujohungbe A, Mai S. 2013. Three‐dimensional nuclear telomere organization in multiple myeloma. Transl Oncol 6:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde N, Kristinsson SY, Landgren O. 2011. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM): novel biological insights and development of early treatment strategies. Blood 17(21):5573–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Thakar R, Spector DL. 2008. Chromatin dynamics and gene positioning. Cell 132(6):929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle RA, Rajkumar SV. 2009. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia 23(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung BO, Chou KC. 2011. Review of super‐resolution fluorescence microscopy for biology. Appl Spectrosc 65(9):967–980. [DOI] [PubMed] [Google Scholar]
- Liu Y, Uttam S, Alexandrov S, Bista RK. 2014. Investigation of nanoscale structural alterations of cell nucleus as an early sign of cancer. BMC Biophys 7(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo I. 2008. Chromosome abnormalities and cancer cytogenetics. Nature Education 1(1):35–43. [Google Scholar]
- Luengo Hendriks CL, van Vliet LJ, Rieger B, van Ginkel M. 1999. DIPimage: A scientific image processing toolbox for MATLAB. Delft: Quantitative Imaging Group, The Netherlands: Delft University of Technology.
- Luengo Hendriks CL, van Kempen GMP, van Vliet LJ. 2007. Improving the accuracy of isotropic granulometries. Pattern Recognit Lett 28(7):865–872. [Google Scholar]
- Morgan GJ, Walker BA, Davies FE. 2012. The genetic architecture of multiple myeloma. Nat Rev Cancer 12(5):335–348. [DOI] [PubMed] [Google Scholar]
- Qumsiyeh MB. 1999. Structure and function of the nucleus: Anatomy and physiology of chromatin. Cell Mol Life Sci 55(8–9):1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I, Groudine M. 2011. On emerging nuclear order. J Cell Biol 192(5):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar SV. 2005. MGUS and smoldering multiple myeloma: Update on pathogenesis, natural history, and management. Hematology Am Soc Hemalol Educ Program 1:340–345. [DOI] [PubMed] [Google Scholar]
- Rajkumar SV, Dispenzieri A, Kyle RA. 2006. Monoclonal gammopathy of undetermined significance, Waldenström macroglobulinemia, AL amyloidosis, and related plasma cell disorders: diagnosis and treatment. Mayo Clin Proc 81(5):693–703. [DOI] [PubMed] [Google Scholar]
- Raska I, Dundr M, Koberna K. 1992. Structure‐function subcompartments of the mammalian cell nucleus as revealed by the electron microscopic affinity cytochemistry. Cell Biol Int Rep 16(8):771–789. [DOI] [PubMed] [Google Scholar]
- Righolt CH, Guffei A, Knecht H, Young IT, Stallinga S, van Vliet LJ, Mai S. 2014. Differences in nuclear DNA organization between lymphocytes, Hodgkin and Reed–Sternberg cells revealed by structured illumination microscopy. J Cell Biochem 115:1441–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, Leonhardt H, Sedat JW. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320:1332–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. 2010. A guide to super‐resolution fluorescence microscopy. J Cell Biol 190(2):165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff SA, Fienup JR, Williams DR. 2009. Phase‐shift estimation in sinusoidally illuminated images for lateral superresolution. J Opt Soc Am A Opt Image Sci Vis 26(2):413–424. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, LanctÔt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137(2):356–368. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D‐structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1(10):965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA. 2005. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6:775–781. [DOI] [PubMed] [Google Scholar]
- Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D‐SIM super resolution microscopy reveals a bead‐like arrangement for FtsZ and the division machinery: Implications for triggering cytokinesis. PLoS Biol 10(9):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Müller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. 2002. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci 99:4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull L, Strauss MP, Liew AT, Monahan LG, Whitchurch CB, Harry EJ. 2014. Super‐resolution imaging of the cytokinetic Z ring in live bacteria using fast 3D‐structured illumination microscopy (f3D‐SIM). J Vis Exp 29(91):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel R, Verschure PJ. 2001. Nuclear organization and gene expression: Visualization of transcription and higher order chromatin structure. In: Mapping protein/DNA interactions by cross‐linking [Internet]. Paris: Institut national de la sant& #233; et de la recherche m& #233;dicale. [PubMed]
- Wicker K, Mandula O, Best G, Fiolka R, Heintzmann R. 2013. Phase optimisation for structured illumination microscopy. Opt Express 21(2):2032–2049. [DOI] [PubMed] [Google Scholar]
- Zingone A, Kuehl WM. 2011. Pathogenesis of monoclonal gammopathy of undetermined significance (MGUS) and progression to multiple myeloma. Semin Hematol 48(1):4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data S1.
Supplementary Data S2.
Supplementary Data S3.


