Abstract
Stargardt type 3 macular degeneration is dependent on a dominant defect in a single gene, ELOVL4 (elongase of very long chain fatty acids 4). The encoded enzyme, ELOVL4, is required for the synthesis of very long chain polyunsaturated fatty acids (VLC-PUFAs), a rare class of > C24 lipids. In vitro expression studies suggest that mutated ELOVL4STGD3 proteins fold improperly, resulting in ER stress and formation of cytosolic aggresomes of wild type and mutant ELOVL4. Although a number of mouse models have been developed to determine whether photoreceptor cell loss in STGD3 results from depletion of VLC-PUFAs, aggresome-dependent cell stress or a combination of these two factors, none of these models adequately recapitulates the disease phenotype in humans. Thus, the precise molecular mechanism by which ELOVL4 mutation causes photoreceptor degeneration in mice and in human patients remains to be characterized. This mini review compares and evaluates current STGD3 mouse models and determines what conclusions can be drawn from past work.
Keywords: ELOVL4, STGD3, Very long chain polyunsaturated fatty acids (VLC-PUFAs), Transgenic mice, Knock-in mice, Knock-out mice, pLox, Cre, Phenotype
19.1 Introduction
Stargardt 3 is an early onset macular degeneration characterized by a progressive loss of central vision (McMahon and Kedzierski 2010; Zhang et al. 2001; Bernstein et al. 2001). Similarly to the more prevalent Stargardt 1 disease, the STGD3 phenotype has been associated with defects in a single gene (Bernstein et al. 2001; Zhang et al. 2001), the elongase of very long chain fatty acids 4 (ELOVL4). Mutations found in STGD3 patients affect the C-terminal end of the ELOVL4 protein that contains a di-lysine motif thought to regulate protein retention within the endoplasmic reticulum (ER) (Vasireddy et al. 2010). This is believed to derail proper localization of the protein to the ER, where very long chain fatty acid (VLC-FA) synthesis takes place and to suppress the biosynthetic capacity of the wild type ELOVL4 enzyme by removing it as well from the ER (Agbaga et al. 2008; Guillou et al. 2010; Logan et al. 2013). Because ELOVL4 expression in adult vertebrate eyes is limited to the photoreceptor layer (Zhang et al. 2003; Agbaga et al. 2008), its VLC-PUFA products are likely to play specific but yet to be defined, functions in cones and rods (Zemski Berry et al. 2014). It has been hypothesized that these lipids provide superior fluidity and stabilizing highly curved regions of cell membranes and might therefore play a function in phototransduction, outer segment maintenance and/or formation and release of synaptic vesicles (SanGiovanni and Chew 2005; Guillou et al. 2010; McMahon and Kedzierski 2010; Bennett et al. 2014b).
19.2 Cell Culture Studies
The leading hypotheses of STGD3 pathomechanisms are based on studies of transgenic cell cultures where ELOVL4 was expressed alone and/or in combination with the STGD3-causing mutant ELOVL4 gene (Karan et al. 2005; Logan et al. 2013). These studies showed that the mutant protein aggregates with the wild type version and translocates it from the ER to the cytoplasm, possibly forming aggresomes (Ambasudhan et al. 2004; Karan et al. 2005; Grayson and Molday 2005). The impaired trafficking hypothesis provides a plausible mechanism for the dominant inheritance of the disease in STGD3 patients. It also predicts that photoreceptor cells expressing mutated ELOVL4 face ER stress and unfolded protein response (Lin et al. 2008) in parallel to the lost ELOVL4 function and depletion of VLC-PUFAs. Thus, it would be important to determine whether STDG3 is primarily mediated by loss of function due to mutated ELOVL4 or, as observed in other degenerative diseases of photoreceptor cells (Lin and Lavail 2010), as a result of protein misfolding and ER stress.
While misrouting is sufficient to induce loss of enzyme function (Logan et al. 2014), recent studies also established that the mutant ELOVL4 protein’s loss of function and dominant negative effect is not necessarily driven by insufficient ER retention (Logan et al. 2013). In vivo analysis in the transgenic Xenopus model showed that the mutant protein is trafficked to the photoreceptor outer segment, but it does not impede the normal compartmentalization of wild type ELOVL4 (Agbaga et al. 2014). Thus, whether and to what extent information from in vitro studies can be applied to understand the human STGD3 disease process remains an open question.
19.3 Mouse Models: Knock-IN and Knock-OUT Strains
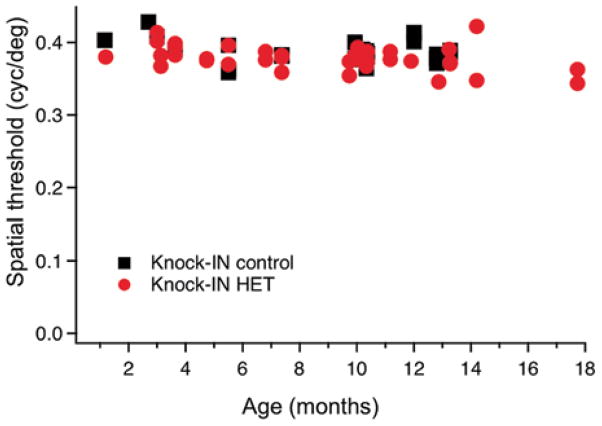
An overarching aim of STGD3 animal model development has been to unveil the causal connection between the genotype and early-onset progressive cone degeneration observed in humans. The early studies were stymied by the discovery that global homozygous Elovl4 knockout and knock-in of the human mutation into the mouse Elovl4 gene are perinatally lethal due to loss of skin acylceramides, required to maintain the water barrier function (Vasireddy et al. 2007). Heterozygotes of both strains are viable but Elovl4+/− mice show no detectable phenotype (Raz-Prag et al. 2006; Li et al. 2007), suggesting that Elovl4 haploinsufficiency and decreased function does not result in degeneration in the mouse. Further questions were raised by the observation that knock-in (KI) heterozygote mice, which represent the closest genetic approximation to the human condition, do not exhibit early-onset cone degeneration. Rather, late onset (8–15 months) and conflicting physiological changes were reported for KI animals: maximal scotopic ERG b-wave amplitudes were increased (Vasireddy et al. 2006) in one, decreased in a different study (McMahon et al. 2007). Consistent with other reports (McMahon et al. 2007; Mandal et al. 2014), our own analysis shows ~ 50 % decrease of C30-C36 VLC-PUFA levels in retinas of KI mice (53.7 ± 8.8 % of control). However, this decrease in VLC-PUFA content was not sufficient to induce a behavioral phenotype. As shown in Fig. 19.1, the KI strain exhibits no visual acuity defect, as measured by their optomotor tracking behavior.
Fig. 19.1.
Visual performance was measured in Knock-IN heterozygous mice and wild type strain controls using their optomotor reflex to establish a spatial frequency threshold (OptoMotry, Cerebral Mechanics). Each symbol represents the visual acuity for the test of a single mouse at the specified age. No significant difference between the groups was detected
19.4 Mouse Models: Transgenic and Cell Specific Knockout Mice
Other STGD3 mouse models include transgenic mice that express either human wild type or mutant ELOVL4 (Karan et al. 2005; Kuny et al. 2010, 2012; Barabas et al. 2013) or cell specific knockdowns where mouse Elovl4 was knocked out specifically from cones (Harkewicz et al. 2012; Barabas et al. 2013) or rods (Harkewicz et al. 2012; Barabas et al. 2013; Marchette et al. 2014) or the entire retina (Bennett et al. 2014a, 2014b). Unexpectedly, only the transgenic strains show early onset photoreceptor degeneration (Karan et al. 2005; Kuny et al. 2010, 2012) with onset time and severity depending on transgene expression level. A major discrepancy with regard to the human disease is that degeneration in transgenic animals starts with a massive loss of rods only secondarily followed by cone degeneration (Kuny et al. 2012; Barabas et al. 2013).
Pan-retinal Elovl4 KOs were characterized by decreased synaptic vesicle size & number in rod terminals, formation of ectopic rod-bipolar synapses associated with sprouting of bipolar dendrites and a reduction in scotopic ERG causing late (after 12 months) degeneration of rods (Bennett et al. 2014a, 2014b). Interestingly, the phenotype was not associated with changes in the postsynaptic excitatory response (Bennett et al. 2014b).
Conditional elimination of ELOVL4 from a single photoreceptor cell class gave discrepant results. The first study indicated a reduction in VLC-PUFA content and loss of rod and cone function in rod and cone conditional knockout (cKO) animals, respectively (Harkewicz et al. 2012). However, the subsequent two studies found no effect on rod (Barabas et al. 2013; Marchette et al. 2014) or cone function and survival (Barabas et al. 2013). Differences between cre expression and knockdown efficiency do not account for these discrepancies as the same cre system with approximately 60–80 % efficiency (Le et al. 2006; Barabas et al. 2013) was used in all of these studies. The latter studies (Barabas et al. 2013; Marchette et al. 2014) observed no effect on scotopic or photopic ERGs or visual behavior even when the highly efficient iCre-75 was used to cause a massive (98 %) reduction in retinal VLC-PUFA content (Barabas et al. 2013). Major distinguishing factors in these studies were the use of different controls (C57B/6 mice in the Harkewicz et al. study, and congenic controls in the Barabas et al and Marchette et al studies), as well as the ages of the mice varied.
The important conclusion from knockdown studies is that deletion of Elovl4 from photoreceptor cells does indeed deplete retinal > C30 VLC-PUFA levels (Barabas et al. 2013; Bennett et al. 2014a). Selective and highly efficacious elimination of the gene (together with near total loss of VLC-PUFAs from the mouse retina) shows a late-onset rod phenotype but no single KO or cKO strains has so far replicated the early cone loss phenotype seen in STGD3 patients.
19.5 Open Questions
Taken together, many open questions remain with respect to the pathophysiology of STGD3. Among the fundamental unresolved issues are (1) what is the function of VLC-PUFAs in photoreceptors? (2) What is the actual cause of the autosomal dominance of ELOVL4? And (3) Why does STGD3 affect cones in humans, are macular cones more sensitive to loss of VLC-PUFAs? The mild-to-none phenotypes of knock-in heterozygotes (McMahon et al. 2007; Vasireddy et al. 2006), knock-out heterozygotes (Raz-Prag et al. 2006; Li et al. 2007), and cell-specific homozygote knockout mice (Barabas et al. 2013; Marchette et al. 2014) and the late-onset rod-specific phenotype of total retinal knockdowns (Bennett et al. 2014a, 2014b) suggest that significant loss of VLC-PUFA levels is not sufficient to cause early onset cone degeneration in the mouse retina. It is possible that residual VLC-PUFAs (~ 2 % of control) are sufficient to maintain mouse photoreceptors, especially given that normal levels of VLC-PUFAs in mice are approximately 10 times higher compared to human (post mortem) retinal tissue (Liu et al. 2013). This may confer resistance to mouse photoreceptors in the form of VLC-PUFA “buffering”. Indeed, clinical and biochemical studies indicate that the human retina may be more sensitive to VLC-PUFA depletion. An inverse association was found between the severity of STGD3 and dietary intake of VLC-PUFA precursors (Hubbard et al. 2006) and loss of VLC-PUFAs was exacerbated in AMD patient eyes compared to age matched controls (Liu et al. 2010). The above data give an impetus to mouse studies, which will need to endow the mouse retina with at least some features of the human macula, establish the relative importance of loss of VLC-PUFAs and presence of the mutated protein and unveil the function of VLC-PUFAs in the healthy retina.
Contributor Information
Peter Barabas, Email: peter.barabas@utah.edu, Department of Ophthalmology and Visual Sciences, John A. Moran Eye Institute, University of Utah School of Medicine, Salt Lake City, UT 84132, USA.
Aruna Gorusupudi, Email: aruna.gorusupudi@utah.edu, Department of Ophthalmology and Visual Sciences, John A. Moran Eye Institute, University of Utah School of Medicine, Salt Lake City, UT 84132, USA.
Paul S Bernstein, Email: paul.bernstein@hsc.utah.edu, Department of Ophthalmology and Visual Sciences, John A. Moran Eye Institute, University of Utah School of Medicine, Salt Lake City, UT 84132, USA.
David Krizaj, Email: david.krizaj@hsc.utah.edu, Department of Ophthalmology and Visual Sciences, John A. Moran Eye Institute, University of Utah School of Medicine, Salt Lake City, UT 84132, USA.
References
- Agbaga MP, Brush RS, Mandal MN, et al. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc Natl Acad Sci U S A. 2008;105:12843–12848. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbaga MP, Tam BM, Wong JS. Mutant ELOVL4 that causes autosomal dominant stargardt-3 macular dystrophy is misrouted to rod outer segment disks. Invest Ophthalmol Vis Sci. 2014;55:3669–3680. doi: 10.1167/iovs.13-13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R, Wang X, Jablonski MM, et al. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004;83:615–625. doi: 10.1016/j.ygeno.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Barabas P, Liu A, Xing W, et al. Role of ELOVL4 and very long-chain polyunsaturated fatty acids in mouse models of Stargardt type 3 retinal degeneration. Proc Natl Acad Sci U S A. 2013;110:5181–5186. doi: 10.1073/pnas.1214707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LD, Brush RS, Chan M, et al. Effect of reduced retinal VLC-PUFA on rod and cone photoreceptors. Invest Ophthalmol Vis Sci. 2014a;55:3150–3157. doi: 10.1167/iovs.14-13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LD, Hopiavuori BR, Brush RS, et al. Examination of VLC-PUFA-deficient photo-receptor terminals. Invest Ophthalmol Vis Sci. 2014b;55:4063–4072. doi: 10.1167/iovs.14-13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PS, Tammur J, Singh N, et al. Diverse macular dystrophy phenotype caused by a novel complex mutation in the ELOVL4 gene. Invest Ophthalmol Vis Sci. 2001;42:3331–3336. [PubMed] [Google Scholar]
- Grayson C, Molday RS. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J Biol Chem. 2005;280:32521–32530. doi: 10.1074/jbc.M503411200. [DOI] [PubMed] [Google Scholar]
- Guillou H, Zadravec D, Martin PG, et al. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Harkewicz R, Du H, Tong Z, et al. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem. 2012;287:11469–11480. doi: 10.1074/jbc.M111.256073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard AF, Askew EW, Singh N, et al. Association of adipose and red blood cell lipids with severity of dominant Stargardt macular dystrophy (STGD3) secondary to an ELOVL4 mutation. Arch Ophthalmol. 2006;124:257–263. doi: 10.1001/archopht.124.2.257. [DOI] [PubMed] [Google Scholar]
- Karan G, Lillo C, Yang Z, et al. Lipofuscin accumulation, abnormal electrophysiology, and photoreceptor degeneration in mutant ELOVL4 transgenic mice: a model for macular degeneration. Proc Natl Acad Sci U S A. 2005;102:4164–4169. doi: 10.1073/pnas.0407698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuny S, Gaillard F, Mema SC, et al. Inner retina remodeling in a mouse model of stargardt-like macular dystrophy (STGD3) Invest Ophthalmol Vis Sci. 2010;51:2248–2262. doi: 10.1167/iovs.09-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuny S, Gaillard F, Sauvé Y. Differential gene expression in eyecup and retina of a mouse model of Stargardt-like macular dystrophy (STGD3) Invest Ophthalmol Vis Sci. 2012;53:664–675. doi: 10.1167/iovs.11-8418. [DOI] [PubMed] [Google Scholar]
- Le YZ, Zheng L, Zheng W, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006;12:389–398. [PubMed] [Google Scholar]
- Li W, Chen Y, Cameron DJ, et al. Elovl4 haploinsufficiency does not induce early onset retinal degeneration in mice. Vis Res. 2007;47:714–722. doi: 10.1016/j.visres.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Lavail MM. Misfolded proteins and retinal dystrophies. Adv Exp Med Biol. 2010;664:115–121. doi: 10.1007/978-1-4419-1399-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Chang J, Lin Y, et al. Long-chain and very long-chain polyunsaturated fatty acids in ocular aging and age-related macular degeneration. J Lipid Res. 2010;51:3217–3229. doi: 10.1194/jlr.M007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Terry R, Lin Y, et al. Comprehensive and sensitive quantification of long-chain and very long-chain polyunsaturated fatty acids in small samples of human and mouse retina. J Chromatogr A. 2013;1307:191–200. doi: 10.1016/j.chroma.2013.07.103. [DOI] [PubMed] [Google Scholar]
- Logan S, Agbaga MP, Chan MD, et al. Deciphering mutant ELOVL4 activity in autosomal-dominant Stargardt macular dystrophy. Proc Natl Acad Sci U S A. 2013;110:5446–5451. doi: 10.1073/pnas.1217251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan S, Agbaga MP, Chan MD, et al. Endoplasmic reticulum microenvironment and conserved histidines govern ELOVL4 fatty acid elongase activity. J Lipid Res. 2014;55:698–708. doi: 10.1194/jlr.M045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal NA, Tran JT, Zheng L, et al. In vivo effect of mutant ELOVL4 on the expression and function of wild-type ELOVL4. Invest Ophthalmol Vis Sci. 2014;55:2705–2713. doi: 10.1167/iovs.13-13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchette LD, Sherry DM, Brush RS, et al. Very long chain polyunsaturated fatty acids and rod cell structure and function. Adv Exp Med Biol. 2014;801:637–645. doi: 10.1007/978-1-4614-3209-8_80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, Kedzierski W. Polyunsaturated very-long-chain C28-C36 fatty acids and retinal physiology. Br J Ophthalmol. 2010;94:1127–1132. doi: 10.1136/bjo.2008.149286. [DOI] [PubMed] [Google Scholar]
- McMahon A, Jackson SN, Woods AS, et al. A Stargardt disease-3 mutation in the mouse Elovl4 gene causes retinal deficiency of C32-C36 acyl phosphatidylcholines. FEBS Lett. 2007;581:5459–5463. doi: 10.1016/j.febslet.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz-Prag D, Ayyagari R, Fariss RN, et al. Haploinsufficiency is not the key mechanism of pathogenesis in a heterozygous Elovl4 knockout mouse model of STGD3 disease. Invest Ophthalmol Vis Sci. 2006;47:3603–3611. doi: 10.1167/iovs.05-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanGiovanni JP, Chew EY. The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog Retin Eye Res. 2005;24:87–138. doi: 10.1016/j.preteyeres.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Vasireddy V, Jablonski MM, Mandal MN, et al. Elovl4 5-bp-deletion knock-in mice develop progressive photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2006;47:4558–4568. doi: 10.1167/iovs.06-0353. [DOI] [PubMed] [Google Scholar]
- Vasireddy V, Uchida Y, Salem N, Jr, et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (> or = C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum Mol Genet. 2007;16:471–482. doi: 10.1093/hmg/ddl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasireddy V, Wong P, Ayyagari R. Genetics and molecular pathology of Stargardt-like macular degeneration. Prog Retin Eye Res. 2010;29:191–207. doi: 10.1016/j.preteyeres.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemski Berry KA, Gordon WC, Murphy RC, et al. Spatial organization of lipids in the human retina and optic nerve by MALDI imaging mass spectrometry. J Lipid Res. 2014;55:504–515. doi: 10.1194/jlr.M044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Han M, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Yang Z, Karan G, et al. Elovl4 mRNA distribution in the developing mouse retina and phylogenetic conservation of Elovl4 genes. Mol Vis. 2003;9:301–307. [PMC free article] [PubMed] [Google Scholar]