Abstract
Cellular heterogeneity poses a significant challenge to understanding tissue level phenotypes and confounds conventional bulk analyses. To facilitate the analysis of signaling at the single-cell level in human tissues, we applied mass cytometry using CyTOF (Cytometry Time-of-Flight) to formalin-fixed paraffin-embedded (FFPE) normal and diseased intestinal specimens. We developed and validated a technique called FFPE-DISSECT (Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue), a single-cell approach for characterizing native signaling states from embedded solid tissue samples. We applied FFPE-DISSECT coupled to mass cytometry and found differential signaling by tumor necrosis factor α (TNF-α) in intestinal enterocytes, goblet cells and enteroendocrine cells, implicating the role of the downstream RAS-RAF-MEK-ERK signaling pathway in dictating goblet cell identity. In addition, application of FFPE-DISSECT, mass cytometry, and data-driven computational analyses to human colon specimens confirmed reduced differentiation in colorectal cancer (CRC) compared to normal colon, and revealed quantitative increases in inter- and intra-tissue heterogeneity in CRC with regards to the modular regulation of signaling pathways. Specifically, modular co-regulation of the kinases P38 and ERK, the translation regulator 4EBP1, and the transcription factor CREB in the proliferative compartment of the normal colon was loss in CRC, as evidenced by their impaired coordination over samplings of single cells in tissue. Our data suggest that this single-cell approach, applied in conjunction with genomic annotation, such as microsatellite instability and mutations in KRAS and BRAF, allows rapid and detailed characterization of cellular heterogeneity from clinical repositories of embedded human tissues. FFPE-DISSECT coupled of mass cytometry can be used for deriving cellular landscapes from archived patient samples, beyond CRC, and as a high resolution tool for disease characterization and subtyping.
Introduction
A distinguishing feature of cancer and other diseases of dysregulated homeostasis is the expanded degree of intra-tissue cellular heterogeneity (1–4). Heterogeneous cell populations arise from an aberrant differentiation process where cells adopt semi-mature or new progenitor states on the Waddington landscape (5). Cellular heterogeneity has been demonstrated to present a significant challenge for treating these diseases, as therapies targeting one cell type may not be effective in another (6). Furthermore, rare cell populations, such as cancer stem cells (7, 8), can adopt specialized, deleterious functions, including therapeutic resistance and metastatic ability (9–13). The phenotypic state of a cell is governed by its genetics and environment; information from these sources are integrated by signaling and transcriptional networks into cellular behaviors. Investigations of cellular heterogeneity immensely benefit from single-cell analysis (14, 15). However, it is not trivial to interrogate multi-pathway signaling activities at single-cell resolution since cellular signaling states can be destabilized outside the native tissue context (16–18).
A tried and true approach for preserving tissue morphology, and even cellular signaling states, is the procedure of formalin fixation coupled to paraffin embedding (FFPE). FFPE has been a standard practice in clinical analysis of tissues for nearly a century, and its ability to preserve tissues at ambient temperatures has been widely demonstrated (19). Due to the effectiveness of FFPE for preserving tissue, large repositories of clinically-annotated patient samples have been collected over the years. These banks are valuable resources for scientific insight when coupled to next-generation analytical approaches (20, 21). Specifically, one of our goals is to conduct single-cell signaling analysis on FFPE tissues to address cellular heterogeneity. In order to achieve this, careful measures must be taken to undo the effects of formalin crosslinking in order to access cells, proteins, and nucleic acids for sophisticated analyses.
To comprehensively assess the phenotypic state of cells, evaluating the activity of a single pathway is not sufficient. Recently, several approaches have been described for measuring protein parameters from FFPE tissue in a multiplex fashion (22). The majority of these advances have been microscopy-based approaches for imaging tissue sections that are ~5 μm in thickness. Approaches that enable multiplexing protein measurements include iterative rounds of fluorescence imaging (23–26) or metal-based detection (27, 28). To achieve single-cell resolution, single or multiple cell border markers are used in conjunction with sophisticated image processing algorithms to extract single cell objects from images (29). Oblique sectioning and imperfect segmentation of partial cells can lead to inaccurate quantification, making these approaches semi-quantitative at best. Furthermore, either due to the iterative nature of cyclic immunofluorescence or rastering of samples for imaging mass-spectrometry, these approaches are low throughput and require multiple days/weeks of analysis to fully sample a given specimen. Given their space-resolving capabilities, we surmise that these techniques will be very powerful when combined with a primary strategy that confers feasibility to analyze a large number of samples with higher quantitative accuracy.
Our lab has recently reported a relatively rapid mass cytometry-based strategy for profiling signaling protein modifications at the single-cell level from solid tissues (16). This strategy, named DISSECT (Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue), involves rapid, short fixation of freshly-isolated tissue to maintain native signaling in intact epithelia, and then a series of coupled procedures for staining and dissociation prior to mass cytometry analysis. The present study examines whether the same approach can be applied to FFPE-preserved tissues, given that FFPE preservation also involves the use of a formaldehyde fixative. In this report, we present an optimized procedure for dissociating single cells from FFPE-preserved solid tissues while maintaining their intact signaling states for mass cytometry analysis. We conducted a proof-of-concept study on a small of cohort of human normal colon and colorectal cancer (CRC) FFPE specimens to sample signaling pathway heterogeneity at the single-cell level. Our results indicate that in normal colonic tissues, signaling pathways are organized into modules according to surface-to-crypt differentiation status. This modular organization is undermined in CRC. In addition, examining tumor samples in combination with genomic markers such as microsatellite instability and mutational status reveals distinct single-cell cancer phenotypes. This hypothesis-generating study demonstrates FFPE-DISSECT coupled to mass cytometry analysis on archival tissues, with the aim to extend to large cohort studies from solid tumor repositories that classify tumors in conjunction with genomic, transcriptomic, epigenomic, and proteomic data.
Results
The FFPE-DISSECT method disaggregates single epithelial cells from archived tissue blocks while preserving cell type and signaling markers
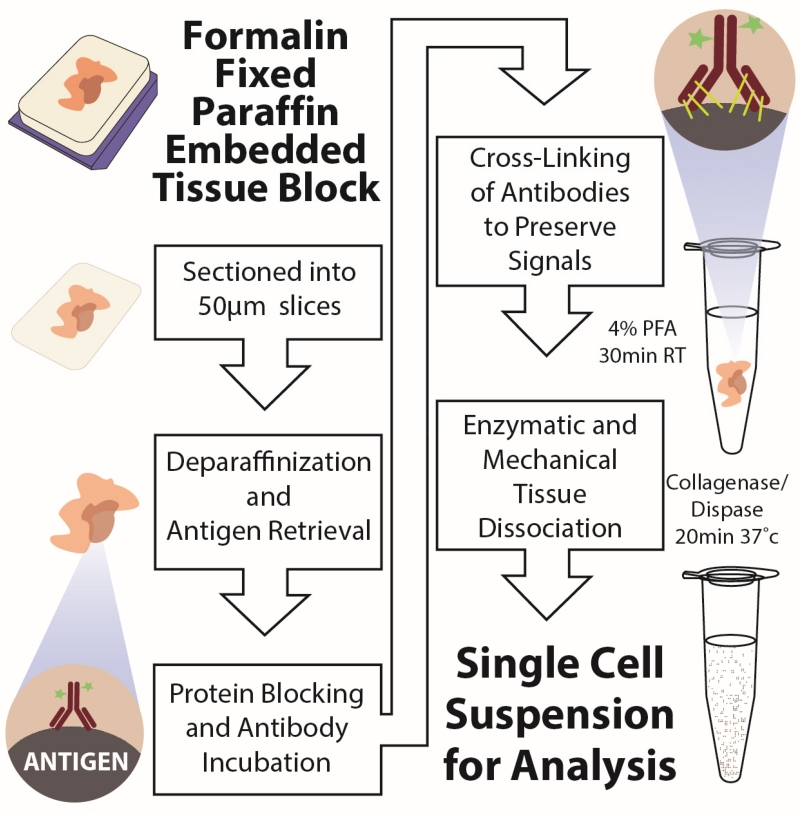
We established a single-cell disaggregation approach for FFPE tissues (FFPE-DISSECT) combining heat-induced antigen retrieval with the whole mount staining and dissociation steps of DISSECT (Fig. 1) (16, 30). The steps of DISSECT were incorporated to enable epithelial signaling state preservation during the disaggregation process. We confirmed single-cell retrieval from FFPE tissues by both brightfield and autofluorescence imaging (fig. S1, A and B). As with DISSECT, because tissue was kept intact until the end of the protocol, cell loss due to pre-analytic processing was minimized. Thus, we routinely yielded 5000-10000 cells (7503 +/− 2830 cells) per square millimeter of tissue from a single 50 μm section. From the approximate area occupied per sample, we estimated that we yielded routinely on the order of a million cells per 50 μm tissue section.
Fig. 1. Schematic of the FFPE-DISSECT process for preserving native epithelial signaling.
Thick (50 μm) tissue slices were sectioned from FFPE blocks, antigen retrieved, and then processed following the steps of the DISSECT procedure. RT, room temperature; PFA, paraformaldehyde.
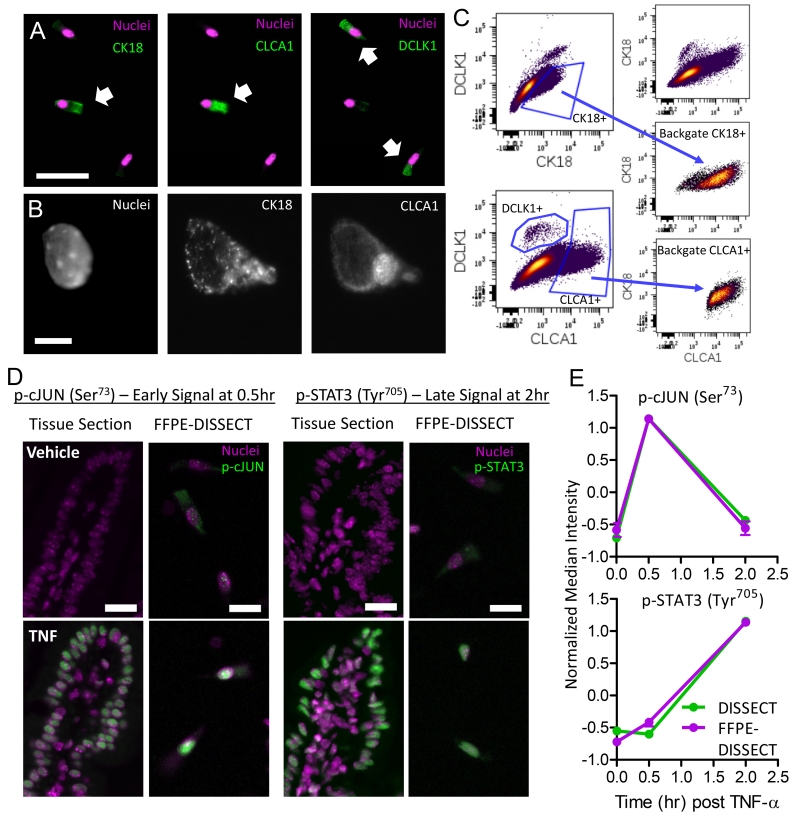
We first determined the preservation of cell identity markers for classifying epithelial cell types using our approach on murine intestinal tissue embedded by FFPE. Up until tissue dissociation, CLCA1 (chloride channel accessory 1) and CK18 (cytokeratin 18), markers for goblet and secretory cells, displayed substantial co-localization in whole-mount immunofluorescent staining, as expected (fig. S2). DCLK1 (doublecortin like kinase 1), a marker of tuft cells, labeled a separate population of CK18−/CLCA1− cells. Upon dissociation, these relationships remained intact in single epithelial cells (Fig. 2A). Furthermore, the correct subcellular localization of proteins within cells can be visualized in the absence of scattered light or convolution from neighboring cells, namely CK18 staining of cytoskeletal structures and CLCA1 staining of mucous granules (Fig. 2B). We then quantitatively verified marker co-expression using multi-parameter flow cytometry. CK18+ cells and CLCA1+ cells were independently gated. Back-gating of CK18+ and CLCA1+ cells revealed that they largely fell within an overlapping population, with CK18 marking a wider population of cells due to its ability to label other cells in the secretory lineage (Fig. 2C, fig. S3A). These results demonstrated that cell types can be discerned in dissociated epithelial cells after FFPE-DISSECT.
Fig. 2. FFPE-DISSECT enables the identification of cell types and quantification of phospho-protein signaling activities.
(A) IF imaging of dissociated cells from FFPE murine intestinal tissues prepared by FFPE-DISSECT, stained for cell type markers CK18, CLCA1, and DCLK1. Scale bar, 50 μm. (B) IF imaging of a single epithelial cell stained for nucleic acid, CK18, and CLCA1. Scale bars, 10 μm. (C) Flow cytometry bi-plots of the mouse ileum prepared by FFPE-DISSECT. Manual gating of goblet cells by CK18 and CLCA1, and tuft cells by DCLK1. CK18 and CLCA1 singular positive cells are back-gated to a bi-axial plot not used for the original gating to demonstrate that the cells comprise an overlapping goblet cell population. (D) IF imaging of intact FFPE intestinal tissues as 5 μm sections, compared to single cells prepared by FFPE-DISSECT, stained for p-cJUN (early signal) and p-STAT3 (late signal) in response to TNF-α at the indicated time points (hr: hours). Scale bars, 20 μm. (E) Quantification of p-cJUN and p-STAT3 from single-cell suspensions generated from murine duodenal tissues, prepared immediately by DISSECT (green), or FFPE-embedded and then by FFPE-DISSECT (magenta), followed by flow cytometry. Median intensities calculated from single cell distributions are displayed for comparisons. Tissues were harvested at specified time points after TNF-α administration. Data are means ± SEM from n=3 animals. Data scales are Z-score values derived from mean centering and variance scaling of each set of time course experiment. Data are representative of n=3 animals.
More importantly, we determined that single cells retained their native signaling states post-dissociation using FFPE-DISSECT. To activate signaling pathways in vivo, we exposed the murine intestinal epithelium to TNF-α (tumor necrosis factor α) via intravenous administration, as we have done previously (31, 32). Duodenal tissues from the same animal were assessed as FFPE tissue sections or as single cells generated by FFPE-DISSECT. Immunofluorescence imaging of tissue sections revealed that the abundance of phosphorylated (p-) cJUN (an early TNF-α-induced signal) was upregulated at 0.5 hours after TNF-α exposure, and p-STAT3 (a late signal) was upregulated at 2 hours after TNF-α exposure (Fig. 2D). Imaging of single cell suspensions prepared by FFPE-DISSECT from serial sectioning of the same tissue block also revealed activation of the two signaling pathways at the appropriate time points compared to vehicle control (Fig. 2D). We quantitatively compared signaling data from single-cell suspensions prepared by the validated DISSECT approach from freshly isolated tissues (16), with those prepared by FFPE-DISSECT from embedded tissues. Using the median intensity calculated from single cell distributions evaluated by flow cytometry (fig. S3B), we confirmed that both DISSECT and FFPE-DISSECT generated comparable signaling data for both p-cJUN and p-STAT3 with similar dynamics (Fig. 2E). These results demonstrated the ability of FFPE-DISSECT in preserving signaling states of p-cJUN and p-STAT3 in single epithelial cells disaggregated from FFPE tissues.
Quantitative single cell-level data is obtained through mass cytometry signaling analysis on FFPE tissue
In clinical practice, excised tissues requiring gross pathological examination may not be immediately fixed. Reports have documented the effects of ischemia and other factors on the degradation of protein signals in other tissues, such as the breast (33). To examine the effects of post-excision time outside of the body on signaling in the intestinal epithelium, we harvested intestinal tissues from mice, and fixed the tissue either immediately, 30 minutes post-excision, or 1 hour post-excision. Following standard FFPE processing, we examined changes in constitutively active signaling pathways at homeostasis, for example, the abundance of p-ERK in the crypt and p-S6 at the tip of the villus. We performed such analysis for markers across a wide breadth of signaling pathways (fig. S4) that we then examined in human patients (figs. S10-S14). For the intestinal epithelium, there was minimal degradation of these signals for up to one hour from the time of harvesting the tissue. We further verified that the length of fixation time, for up to 72 hours, has minimal effect on the detection of representative signaling markers in FFPE (fig. S5).
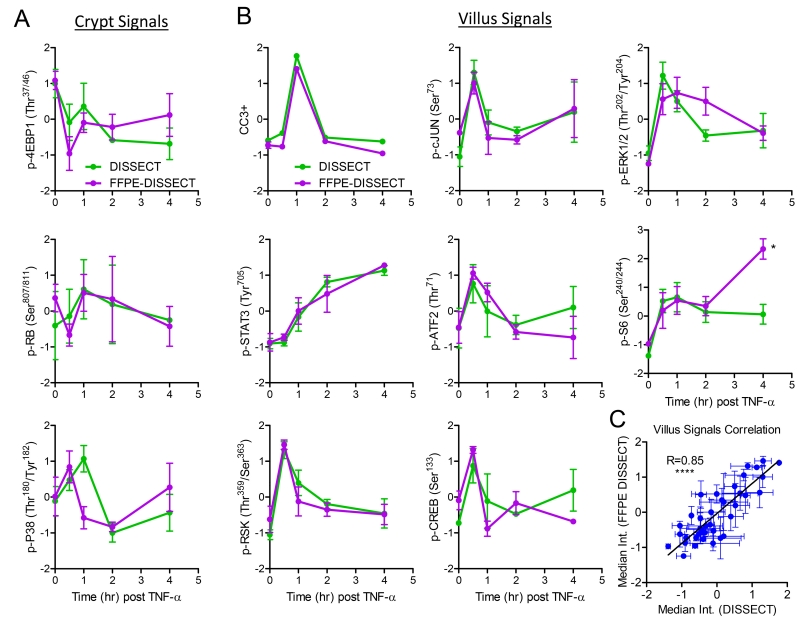
Having successfully assessed the validity of FFPE-DISSECT on selected signaling markers, we next sought to systematically and quantitatively validate our approach over a broad range of signaling pathways. We used mass cytometry as a multiplex technique to quantify a broad range of signaling markers from single cell suspensions, comparing between FFPE-DISSECT preparations from embedded tissues and DISSECT preparations from fresh tissues. Single-cell signaling data obtained by DISSECT have previously been rigorously validated against those generated by conventional bulk approaches such as immunoblotting (16). Mice were stimulated with TNF-α and duodenal tissues were harvested over a time course to sample a quantitative range of signaling activities as a function of phosphorylated protein abundance. Harvested tissues were then divided, either to be freshly processed by DISSECT, or , to be embedded and then processed by FFPE-DISSECT. Mass cytometry analysis was performed on both sets of tissues (isolated from the same animal) using the same panel of metal-conjugated reagents for signaling markers (Table S1). The normalized median intensities of distributions of signaling markers were used as a direct comparison between DISSECT and FFPE-DISSECT preparations (Fig. 3). The DISSECT approach was optimized for scraped mucosa, and thus, the data generated were enriched for villus signals. In contrast, tissue sectioning enabled sampling of the entire epithelium for FFPE-DISSECT. Thus, crypt-enriched signals, such as p-RB, p-4EBP1, and p-P38 (fig. S6A), did not show good concordance between the two methods due to the de-emphasis of crypt signals in DISSECT preps (Fig. 3A). Crypt proliferative signals (p-4EBP1 and p-RB) generated by FFPE-DISSECT showed an initial dip and a subsequent increase after TNF-α exposure, mirroring the proliferative response of the intestinal epithelium to TNF-α (32). Examining villus-enriched signals, a strong correlation between data generated by DISSECT and FFPE-DISSECT was observed (Fig. 3B). Quantitative correlation analyses using villus-enriched signals resulted in a highly significant correlation (R=0.85, p<0.0001) of mass cytometry data generated by DISSECT against FFPE-DISSECT (Fig. 3C). Including crypt-enriched signals resulted in a slightly lower correlation (R=0.76, p<0.0001) (fig. S6B). We further verified FFPE-DISSECT and compared median signals obtained to those obtained by IF imaging (fig. S6, C and D) and quantitative immunoblotting (fig. S6, E and F), comparing across different cohorts of mice similarly stimulated with TNF-α as a time course. Again, FFPE-DISSECT compared favorably. By using FFPE-DISSECT in conjunction with mass cytometry, valid, single-cell level signaling data can be obtained from embedded epithelial tissues.
Fig. 3. Comparison between mass cytometry data generated by FFPE-DISSECT and the validated DISSECT method on the same intestinal tissue.
(A and B) Dynamic signals of TNF-α stimulation time courses, and enriched in either (A) crypts or (B) villus.from murine duodenal harvested from specified time points after TNF-α administration. Tissues were split in two, with one set processed immediately by DISSECT (green), and the other set FFPE-embedded and then processed by FFPE-DISSECT (magenta). Both sets of tissues were analyzed by mass cytometry with the same cross-reacting signaling antibody panel. (C) Correlation analysis combining all villus signaling markers, comparing mass cytometry data generated by DISSECT against FFPE-DISSECT. Quantitative data from different time points were used to generate a range of variation for correlation analysis. Data are means ± SEM from n=3 animals. Data scales are Z-score values derived from mean centering and variance scaling of each set of time course experiment. **** P ≤ 0.0001, by t-test.
Cell type-specific signaling reveals increased secretory cell sensitivity to basal and TNF-α-induced signaling
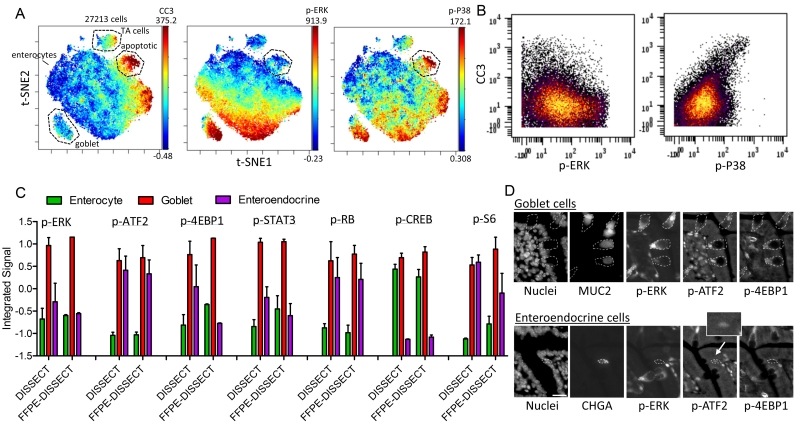
In addition to examining the average over epithelial distributions, we sought to determine how different cell populations in the small intestine respond to TNF-α using our single-cell approach. TNF-α triggers apoptosis and extrusion of duodenal epithelial cells upon hours of induction (31, 32, 34, 35) . Our previous study demonstrated that the onset of apoptosis occurs 1 hour after intravenous administration of exogenous TNF-α in mice, and thus, mass cytometry data enabled by FFPE-DISSECT were obtained from murine duodenal tissues at this time point. t-distributed stochastic neighbor embedding [t-SNE (36)] analysis on 15-channel signaling and cell identity data revealed a CC3+ population of apoptotic epithelial cells (Fig. 4A; for markers see Table S1). This dying cell population has a distinct signaling signature, including the downregulation of p-ERK and upregulation of p-P38 (Fig. 4, A and B, and fig. S7), as reported previously. We previously showed that p-ERK upregulation in neighboring cells surrounding the apoptotic cell is a contact-dependent survival mechanism preventing large-scale barrier defects in the gut (16). We then further evaluated cell-type-specific signaling by integrating signals from the entire TNF-α time course in murine cell populations expressing cell-type-specific markers (CLCA1+ goblet cells, CHGA+ - chromogranin A enteroendocrine cells, CK+/CLCA1−/CHGA− enterocytes). Goblet cells generally have increased signaling across most pathways assayed, whereas enteroendocrine cells selectively upregulate certain pathways when compared to enterocytes (Fig. 4C and fig. S8A). The relative differences in signaling between cell types can be reproduced by DISSECT on freshly isolated tissue, again confirming the validity of our new approach (Fig. 4C and fig. S8A). Furthermore, the upregulation of p-ERK, p-ATF2, and p-4EBP1 in goblet cells, and of only p-ATF2 in enteroendocrine cells, was corroborated by immunofluorescent imaging (Fig. 4D and fig. S8B). These differences were also observed at basal level without TNF-α stimulation, perhaps demonstrating the importance of these signaling pathways in the identity of these cells (fig. S8C). Hereafter, we focused on the role of p-ERK in goblet cell identity.
Fig. 4. Cell-specific signaling in the murine duodenal epithelium.
(A) t-SNE analysis of mass cytometry data from the mouse duodenum exposed to TNF-α for 1 hr, prepared by FFPE-DISSECT. Color overlaid represents the relative quantification of cleaved caspase 3, p-ERK, and p-P38 events, respectively. Labelled cells: apoptotic – CC3+ (3.07%), TA cells – p-4EBP1+ (4.13%), goblet – CLCA1+ (5.44%), enterocytes - CKAE+, CLCA1−, CHGA−. Numbers on right axis represent min and max value of the color scale. (B) Bi-plots of CC3 with p-ERK or p-P38, demonstrating negative correlation in the former and positive correlation in the latter. (C) Signaling specific to epithelial cell types (enterocyte- CKAE+, CLCA1−, CHGA−, goblet – CLCA1+, enteroendocrine – CHGA+) calculated by integrating signal values over the entire TNF-α time course, comparing mass cytometry data generated by DISSECT against FFPE-DISSECT. Data are means ± SEM from n=3 animals. Data scales are Z-score values derived from mean centering and variance scaling over data values for the three cell types for each method. (D) IF imaging to confirm cell type-specific signals (p-ERK, p-ATF2, p-4EBP1) at baseline, unstimulated. Scale bars, 20 μm. Data are representative of n=3 animals.
MEK-ERK signaling is canonically activated by upstream RAS activation. The members of the RAS family of small GTPases (KRAS4A, KRAS4B, NRAS, and HRAS) share N-terminal sequence identity and in vitro effector binding, but have distinct subcellular membrane distribution due to differences in post-transcriptional modifications in their C-terminal hypervariable regions (37). Thus, different RAS isoforms can engage in different signaling effectors, such as RAF (Rapidly accelerated fibrosarcoma), PI3K (Phosphatidylinositol-4,5-bisphosphate 3-kinase), and RAL (RAS-related protein) , which can lead to different phenotypic manifestations. Mutationally activated KRAS in the intestinal epithelium induces hyper-proliferation, whereas activated NRAS does not (38). Given that both activated KRAS(38) and NRAS(34, 39) in the intestine can sensitize downstream MEK-ERK towards activation in different circumstances, we surmise that MAPK-induced goblet cell identity may be a common feature of RAS activation. Villin-Cre driving an activated KRAS (KRasLSL-G12D/+) allele in the murine intestinal epithelium increased the number of goblet cells (fig. S9), but also induced hyperplasia as documented previously (38, 40). The same induction scheme with activated NRAS (NRasLSL-G12D/+) did not result in hyperplastic growth. Surprisingly, NRAS activation led to a similar increase in goblet cells (fig. S9) in the murine intestinal epithelium, a phenotype that has not previously been connected to NRAS activation. Furthermore, in accordance with the role of p-ERK in promoting enterocyte survival, goblet cells have been shown to be resistant to TNF-α-induced apoptosis (16). We have demonstrated here that mass cytometry results from FFPE-DISSECT corroborate with conclusions drawn from fresh tissue assays and produce biological insights, supporting its feasibility for generating meaningful single-cell signaling data from embedded tissues.
Human CRCs present with dysregulated signaling and differentiation
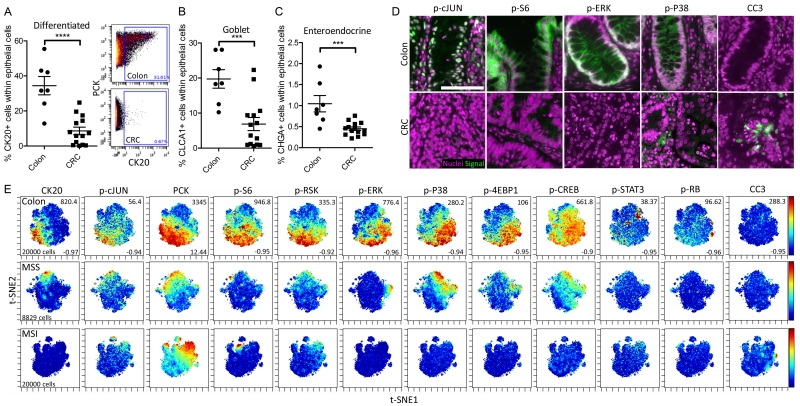
One of the goals for FFPE-DISSECT application to embedded tissue is to enable single-cell signaling analysis on human patient tissue repositories stored as FFPE blocks. To that end, we procured a cohort of clinically-annotated colonic tissue samples from the Western Division of the Cooperative Human Tissue Network (CHTN), situated at the Vanderbilt University Medical Center. After discarding samples with low cellularity (<10%), our cohort included 7 normal colon control samples and 13 (6 MSI – microsatellite instable, and 7 MSS – microsatellite stable) primary CRC samples. Control colon samples were collected from a variety of conditions unrelated to CRC (such as adjacent normal tissue from diverticulitis samples). According to our time-to-fixation optimization, we only selected samples with a post-excision time of <1 hour, a parameter tracked by CHTN. Clinical and pathological attributes of the CRCs, including microsatellite instability and KRAS/BRAF mutational status, were summarized (Table S2). A board-certified pathologist further examined the hematoxylin & eosin stains of these samples to confirm tumor histology (fig. S10). A panel of cross-reacting antibodies against signaling proteins and cell type markers were prepared for mass cytometry analysis (Table S3). These reagents were verified to stain human tissues by immunofluorescence imaging (fig. S11), and to be on-target in a previous report (16). Mass cytometry following FFPE-DISSECT was performed on this cohort of human colon and CRC samples. Because actual tumors comprised only of a minor fraction of tissues resected (fig. S10), we decided to focus specifically on epithelial cells that are marked and can be gated by PCK (pan-cytokeratin) abundance (fig. S12, A and B). From mass cytometry data, we quantitatively assessed the percentage of different epithelial cell types in various differentiation states from normal colon versus CRC tissues within the epithelial compartment. As expected, terminally differentiated cells (CK20+ - cytokeratin 20) were significantly decreased in CRC compared to normal colon (Fig. 5A and fig. S12A). Furthermore, goblet cells (CLCA1+) and enteroendocrine (CHGA+) cells were also significantly decreased (Fig. 5, B and C; and fig. S12C). However, we discovered that a portion of protein markers representing signaling pathway activation were downregulated in CRC (Fig. 5D and fig. S11). This result was paradoxical given that cancer is often driven by mutations that ultimately activate signaling pathways. However, there is evidence from in vivo studies that demonstrate the upregulation of negative feedback mechanisms when MAPK signaling pathways are mutationally activated, only in the context of CRC (41). For instance, mutational activation of KRAS in CRC paradoxically results in the downregulation of p-ERK due to the upregulation of MKP3 ERK phosphatase (38). Furthermore, as shown in our mouse studies, there are substantial signaling activities in differentiated cells and these cells are largely absent in CRC (Fig. 4C, D). To verify that the reduction in signaling of these pathways did not result from poor penetration of fixative, we were able to detect similar stain intensities of multiple signaling markers in the peripheral and central regions of the same tumors, for tumors displaying positive signals (fig. S13). These results, all obtained from one sampling of tissue, suggested that differentiation is impaired in CRC, and these changes are associated with reduced signaling through certain pathways.
Fig. 5. Mass Cytometry analysis of human colorectal cancer specimens prepared by FFPE-DISSECT.
(A to C) Percentage of (A) CK20+ fully differentiated epithelial cells, (B) CLCA+ goblet cells, and (C) CHGA+ enteroendocrine cells in samples of normal human colon compared to samples of human CRC. Data are means ± SEM from n>7 different patient specimens. Inset depicts manual gating of differentiated cells by CK20. *** P ≤ 0.001, **** P ≤ 0.0001, by t-test. (D) IF imaging of signaling markers (p-CJUN, p-S6, p-ERK, p-P38, CC3) comparing normal colon and CRC. Scale bar, 100 μm. (E) t-SNE mapping of mass cytometry data generated from human colon, MSS or MSI CRC specimens, overlaid with signaling and selected differentiation markers. Numbers on right axis represent min and max value of the color scale. The same scales were used between all samples. Proportional downsampling to 20,000 cells was performed for more equivalent representation since some samples have a small representation of actual tumor cells. On average 60,000 cell events were collected per sample. Data are representative of n>6 human patient samples for each group (colon, MSS-CRC, MSI-CRC).
Modular organization of signaling pathways is disrupted in human CRC
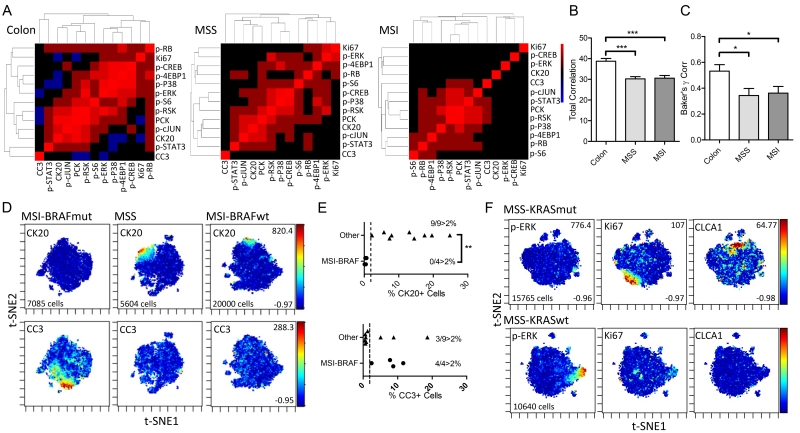
Using t-SNE analysis to visualize multidimensional single-cell data from normal and CRC tissues, we observed defined organization of signaling pathways in normal colon tissues at the single-cell level. The abundances of phosphorylated signaling proteins in different pathways formed distinct patterns on t-SNE maps (Fig. 5E; for markers see Table S3); in one specimen, signaling markers formed a counter-clockwise arrangement in association with surface-to-crypt status marked by cytokeratins. These patterns can be broken down into a modular architecture: p-cJUN correlated with CK20+ differentiated cells; PCK, p-S6, and p-RSK shared similar expression patterns; and p-ERK, p-P38, p-4EBP1, and p-CREB formed another module correlating to less differentiated crypt cells. These modules can also be revealed by calculating the pairwise correlation between signaling markers over individual cells, and using correlative distances for hierarchical clustering per sample (Fig. 6A). Qualitatively, the components within each module were consistent between normal colon samples, signifying robust organization of signaling pathways between cellular populations. The correlation between signaling pathways over single cells was reduced in both MSS and MSI CRC samples, signifying the usage of heterogeneous modes of signaling pathway regulation between individual cells in a tumor (Figs. 5E and 6A). Furthermore, modular organization of signaling pathways from hierarchical clustering was not preserved from sample to sample in CRC, implying significant intertumoral heterogeneity in signaling pathway regulation (fig. S14A). In place of qualitative assessment of signaling heterogeneity on a sample-per-sample basis, we quantitatively assessed intratumoral and intertumoral heterogeneity over all the samples. We evaluated intratumoral signaling regulation by quantifying the magnitude of correlation between all signaling markers measured in a pairwise fashion, with the notion that high correlations signify similar regulatory mechanisms between any pair of pathways utilized by all cells. On a per sample basis, this metric can be represented by the total intensity on a correlative distance heat map (Fig. 6A). Normal colon samples had significantly greater total correlation between signaling markers than did MSS and MSI CRC samples (Fig. 6B), denoting the loss of signaling regulation homogeneity between cells in CRC. We quantitatively evaluated intertumoral signaling heterogeneity by assessing the degree by which signaling modules are similar between samples. For this, we took advantage of tools built previously to assess the similarity between dendrograms to evaluate the degree similarity between the structures of hierarchical clustering trees (42). We used the Baker’s Gamma Correlation coefficient (43), a metric that is insensitive to the height of the branches but is sensitive to the position of each branch, to calculate pairwise similarities between hierarchical trees generated for each sample (fig. S14A). The mean Gamma Correlation pairwise coefficient showed a significant decrease in value in MSS and MSI CRC samples compared to normal colon samples, suggesting that similar signaling modules recurrently exist across different normal colon samples, but less so in CRCs (Fig. 6C). Using tissue-level data to perform the same analysis resulted in different interpretations, again, with normal colon samples having high correlation between sets of signaling markers, MSS samples having less correlation than normal colon tissue, and MSI samples having a correlation between different sets of markers (fig. S14B). This difference may be due to the loss of single-cell resolution where markers expressed in different cells are considered to be in the same compartment as a sample average. These results demonstrated, in a quantitative fashion, that (i) cells within normal colon have shared regulatory mechanisms between pathways but cells in CRC samples do not (intratumoral heterogeneity), and (ii) organized signaling modules recurrently exist between normal colon samples but not between CRC samples (intertumoral heterogeneity).
Fig. 6. Insights into the heterogeneous organization of signaling pathways in CRC from single-cell data.
(A) Heat map and hierarchical clustering derived from pairwise correlative distances between signaling markers calculated over all single cells in a sample. A high pairwise correlation signifies two pathways are regulated in the same way in all cells. Data are representative of n>6 human patient samples for each group (colon, MSS-CRC, MSI-CRC). (B) The mean value of all pairwise correlations between signaling markers calculated per sample, comparing between normal colon, MSS, and MSI. Data are means ± SEM from n>6 different patient specimens. (C) Baker’s γ correlation coefficient comparing the similarity between hierarchical clustering trees computed between all samples within each group (colon, MSS, MSI). Data are means ± SEM from n>6 different patient specimens. (D) t-SNE maps of mass cytometry data generated from a MSI-BRAFV600E mutant tumor compared to MSS or MSI-BRAF wild-type tumors overlaid with the abundance of CK20 and CC3. Numbers on right axis represent min and max value of the color scale. The same scales were used between all samples. Number of cells noted from each single tumor. Data are representative of n=4 MSI-BRAFV600E mutant tumors, n=7 MSS tumors, and n=2 MSI-BRAFWT tumors. (E) Percent CK20+ or CC3+ cells comparing MSI-BRAFV600E mutant tumors (n=4) compared to tumors of other genotypes (n=9). Dotted line represents the 2% threshold, and inset is the number of samples passing the threshold. (F) t-SNE maps of mass cytometry data generated from a MSS-KRASG12C mutant tumor compared to a MSS-KRAS wild-type tumor overlaid with abundances of p-ERK, Ki67, and CLCA1. Data are representative of n=2 KRAS mutant and n=2 KRAS wildtype tumors.* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, by ANOVA followed by Tukey post-test (B,C) or t-test (E).
We next examined whether single-cell signaling properties of tumors are associated with molecular characteristics (Table S2). None of the four microsatellite unstable (MSI) tumors in our set with a BRAFV600E mutation presented with CK20+ differentiated cells, whereas all other nine tumors (MSS or MSI with wild-type BRAF) presented with some degree of differentiation (Fig. 6, D and E). Furthermore, all MSI-BRAF mutant tumors (4/4) had some degree of CC3+ apoptotic cells, whereas only a small proportion (3/9) of other tumors exhibited this phenotype (Fig. 6, D and E). For MSS tumors specifically, a G12 mutation (G12V or G12C) in KRAS downregulated the abundance of p-ERK, increased cell proliferation (Ki67), and upregulated CLCA1+ goblet cell specification compared to tumors with wild-type KRAS (Fig. 6F). These results provided evidence that genetic properties, such as microsatellite instability and mutations, but not pathologic details, such as grade and stage of the tumor, correlate with single-cell signaling phenotypes in CRC.
Discussion
There is an ongoing effort to use next-generation genomic, epigenomic, transcriptomic, and proteomic data to predict tumor outcomes and responses to therapy (44). However, the degree of behavioral diversity within a tumor may be just as important, because different cellular populations may respond to drugs differently and cooperate to produce emergent behaviors. FFPE-DISSECT enables the analysis of single cell signaling activities in archival human tissues. Whereas large academic centers have access to various methods for human tissue preservation, such as flash freezing, most community hospital settings only have access to FFPE. These untapped, large human tissue resources can now be mined at the single-cell level for building appropriately powered models to inform how heterogeneity contribute to tumor behavior and how cellular diversity changes in response to treatment.
There are several caveats to using FFPE-DISSECT, which fall under the same limitations as other FFPE applications. First, the range of antibodies that actually work in FFPE tissues is reduced compare to freshly isolated tissues, because not all conformationally blocked antigens can be retrieved. We somewhat alleviated this problem by only using antibodies that are well-validated (such as by knockdown in human cell lines or mouse tissues) and are widely used in the field for FFPE applications. This problem can perhaps be further addressed in the future by better antibody generation practices. For example, a higher success rate for the generation of antibodies for FFPE applications may be achieved by using fixed proteins as immunogens instead of native peptides. Second, the veracity of a stain of a human FFPE section due to tissue degradation comes into question. The preservation of signals, specifically of signaling proteins, is sensitive to the amount of time the tissue has been outside the body (33). Furthermore, storage conditions of FFPE block, such as temperature and humidity, may introduce variability in the results (45). Standardized operating procedures regarding post-excision time, fixation, and storage, such as those adopted by CHTN, are required to decrease the variability introduced during the tissue preparation step. Third, mass cytometry, although multiplexed, still remains a candidate-based method, and the biological insights derived are only as informative as the biomarker panel allows. A well-known shortcoming of immunohistochemistry techniques is the reliance on cell-type specific markers and morphology to identify cell types, whereas these properties may be altered by concomitant loss of architecture, infiltration of host cells, and dedifferentiation in dysplastic tissue. While we appreciate that cell identities in cancer may not reflect those of normal tissue, the use of multiplex marker panels, specifically those of signaling that represent the functional state of a cell, can allow for the inference of the lineage of origin of cancer cells with unknown identities. Using multiplex single-cell data with comparative algorithms such as Citrus (46), one can determine the similarity of cancer cells to reference signatures of normal cell types in marker space. Furthermore, candidate-based single-cell approaches can be coupled with single-cell RNA sequencing (47), and even other unbiased bulk-based methods to become a powerful discovery tool. There is high potential impact for characterizing unidentified transitional cells in cancer, as they may have altered properties that contribute to malignancy, and more importantly, may be targetable by therapy. All of the above limitations are inherent to FFPE applications in general, and should be considered and be controlled for at the study design phase.
The ability to query signal transduction in a cell-type specific or even at a single-cell level is a defined strength of our approach. The prevalent methods to detect and quantify signaling proteins remain to be bulk approaches such as Western blots and ELISA (32, 48), which assume cellular homogeneity and are not always suitable for tissue analysis. With these approaches, positive signals in small subsets of cell are washed out by larger populations, and the cellular sources of positive signals cannot be determined. For imaging applications in tissues, cell-type specific signaling is usually evaluated with low multiplexity, for instance, looking at one signaling marker with one cell type marker. More recent advances, as mentioned above (23–28, 22), allow for higher multiplexity but at the expense of feasible application on large sample sets. FFPE-DISSECT coupled to mass cytometry is a relatively rapid method for performing multiplex single-cell signaling analysis. It can be used for proposing interesting signaling markers that can be followed up by imaging, as we have done in this study.
The major assumption of FFPE-DISSECT, which begins with archival tissue blocks, is that the tissues are handled properly during the pre-analytical fixation steps. This assumption is widely made in the histopathology field, especially in tissue microarray or cohort studies where hundreds of samples are collected from different sources (49). Improperly fixed tissue will inevitably lead to invalid downstream analyses. To mitigate artifacts arising from this source, a standardized operating procedure was adopted for processing all tissues in this study. First, tissue thickness was limited to 5 mm, which according to common references (50, 51), should allow efficient penetration of fixation within one hour. Second, fixative was incubated with a magnetic stirrer to maximize diffusion. Third, the fixation time was standardized at 24 hours. Last, and most importantly, a board-certified pathologist has reviewed the histology of tissue along with quality assurance-quality control data. Histological characteristics indicating poor fixation quality/inadequate fixative penetration include: (i) processing observations based on nuclear staining and appearance of cytoplasm; (ii) scratches or hatching of the specimen during microtomy; (iii) section disintegrating or pulling apart; (iv) smudging or unusual staining, (v) other unusual artifacts; (vi) stutter; (vii) degree of autolysis; and (viii) cells showing crenation. Samples indicative of fixation problems were not included in this study. Aside from pre-analytical evaluation, additional steps can be taken to identify potential artifacts after data collection. These include: (i) imaging tissue section from the same tissue block to ensure concordance (% of host cell infiltrating, relative intensity of markers) with single-cell data; (ii) imaging single-cell suspensions to ensure disaggregation into single cells; (iii) evaluating proper conjugation of antibodies by staining with both the conjugated and the unconjugated clone coupled to a secondary detection system; and (iv) assessing detection specificity by identifying CyTOF events that are positive for all markers. Many of these artefacts arise from the FFPE process, and we remain hopeful that widespread adoption of standardized procedures and additional technological advances will minimize these issues in the future.
Our approach illuminated differential signaling patterns in different cell types (enterocytes, goblet cells, and enteroendocrine cells), with the conclusion that secretory cells in general are more sensitive to basal and TNF-α-induced signaling. Goblet cells have the highest signaling propensity, with upregulation of many pathways compared to enterocytes. Specifically, goblet cells upregulate the phosphorylation of ERK, which we identified as a survival mechanism against TNF-α-induced apoptosis; accordingly, goblet cells are resistant to TNF-α-induced apoptosis (16). Furthermore, EGFR is a receptor upstream of ERK that plays critical roles in growth, survival, and differentiation in the stem cell niche (52). Following this line of logic, we established a link between RAS activation and goblet cell metaplasia in the intestinal epithelium. This is the first time that NRAS activation has been connected to this phenotype to our knowledge. Demonstrating the casual effect of this pathway, ERK signaling downregulation has been documented to suppress goblet cell specification. Heuberger et al. have shown that epithelial-specific knockout of the phosphatase SHP2 suppresses p-ERK signaling and goblet cell differentiation by modulating transcription factor 4 (TCF4) isoform switching and WNT-dependent transcription (53). This effect on goblet cells can be rescued by gain-of-function in MEK1. A more recent report by De Jong et al. has also shown that the knockout of both ERK1 and ERK2 impairs goblet cell differentiation (54). Goblet cell numbers were reduced but not completely ablated in that study, suggesting that there are compensatory mechanisms to maintain goblet cell number. Using a multiplex cell-type specific approach, we propose other candidate signaling pathways, such as through the phosphorylation of activating transcription factor 2 (ATF2), that may act in synergy with p-ERK to control goblet cell specification.
Cellular heterogeneity is an important topic in cancer biology from both genetic and cell biology perspectives. Differential signaling between cells is a form of heterogeneity that controls cellular behaviors, but is relatively unexplored. Gerdes et al. reported that pathway relationships between signaling components identified from cell lines may not hold true in human tissues when observed at single-cell level (23). Here, we identified signaling pathways that organize into a modular architecture associated with surface-to-crypt identity in the normal colonic epithelium. Consistent correlation between pathways over single cells represent regulatory mechanisms that are recurrently used by all cells in the tissue. Maintenance of modular architecture between samples reflects homogenous organization of signaling pathways. Quantifying these two properties using mass cytometry single-cell data suggest that both intra- and inter- tissue heterogeneity are increased in CRC regarding signaling regulation. Heterogeneity in cancer signaling reflects the relaxation of constraints that allows a cancer cell to sample a wider state space. These constraints can be physical or biochemical, from disorganization of tissue architecture to rewiring of signaling networks. In turn, a cell can adopt novel behaviors and functions outside of normal cellular behaviors, such as epithelial-to-mesenchymal transition (55).
The RAS-RAF-MEK-ERK kinase cascade plays a major role in the pathogenesis of CRC. Activating KRAS and NRAS mutations are found in ~50% of all CRC, and activating BRAF mutations are found in ~10% of CRC (56–58). Mutations in KRAS and downstream BRAF are a biomarker for resistance to upstream EGFR-targeted therapies, as expected (59, 60). However, downstream MEK inhibition has limited efficacy in CRC with KRAS and BRAF mutations (61, 62). While acquired resistance mechanisms, such as upregulation of EGFR family members and BRAF gene amplification (63, 64) are seen in cell lines, an alternative explanation for the lack of efficacy of MEK inhibitors in the clinic may simply be that MAPK signaling downstream of mutant KRAS or BRAF is not upregulated in CRC tissue, as shown in our data here. Unlike cell lines (65), activating KRAS and BRAF in vivo results in negative feedback that upregulates the expression of ERK-targeting phosphatases (41). Work from the Channing Der laboratory has observed that nuclear ERK phosphorylation in human CRC is not correlated to the mutational status of KRAS or, to a lesser extent, BRAF (66). Mouse models also revealed that KRAS activation in normal intestinal epithelium activates ERK, but this effect is inhibited in the context of mouse models of CRC (38). These results support that tissue, cell, and disease contexts strongly govern the influence of genetics on the output of signaling pathways.
Although some argue for redundancy between KRAS and BRAF mutations in MAPK signaling by their mutual exclusion (67), the cancer phenotypes induced by these mutations are vastly different. KRAS mutations mostly occurs in common sporadic CRC that are classified as the CIN (chromosomal instability) phenotype, whereas BRAF mutations mostly occurs in CRC that are classified as the CpG island methylator phenotype (68, 69). Hypermethylation of the MLH1 gene results in diminished DNA repair and induces a MSI-high phenotype distinct from that caused by mismatch repair gene mutations (such as in Lynch’s syndrome) (70, 71). Thus, BRAF mutations are seen in a majority of MSI tumors and rarely in MSS tumors (72). BRAF mutant pathology is also distinct from traditional adenocarcinomas, adopting a serrated morphology (58). Although MSI is often a good prognosis due to the infiltration of cytotoxic T lymphocytes (4, 73), patients with BRAF-mutant tumors have relatively poor overall compared to their BRAF-wild-type counterparts (71, 74). These properties may be due to the lack of differentiation in BRAF-mutant MSI tumors, resulting in a large number of “stem-like cells” , as seen in this study and reported in others (5, 75), which confers resistance to conventional therapies. Perhaps, increased sensitivity to apoptosis in these tumors, as marked by CC3 positivity, can be exploited as a therapeutic option. In our hands, KRAS mutation in MSS tumors is suggested to result in the downregulation of ERK, an increase in the number of goblet cells, and an increase in cell proliferation in distinct populations that may identify with their relative differentiation states. A weakness in our study is the low number of samples in each grouping, especially if we further partition samples by their molecular details. Our intent here is to provide a proof-of-concept application of FFPE-DISSECT on human CRCs, and the hypotheses generated with this small cohort will need to be confirmed in a larger set of tumors. However, given that our approach can be applied to FFPE tissue blocks, one can have access to much larger repositories of retroactively collected samples that can power any study. FFPE-DISSECT coupled to mass cytometry applied to archival samples is a powerful tool to generate large amounts of single-cell data with acceptable throughput. These data are complementary to other precision medicine efforts to molecularly characterize solid tumors for arriving at subtypes that can predict prognosis and therapeutic response.
Materials and Methods
Mouse experiments
All animal experiments were performed under protocols approved by the Vanderbilt University Animal Care and Use Committee and in accordance with NIH guidelines. Mice were stimulated with TNF-α as a time course, and their duodena (proximal small intestine) were collected for analysis as previously described (31, 32). For DISSECT, a previously published protocol was used (16). For FFPE embedding, tissues were fixed in formalin for 24 hours prior and then were subjected to standardized embedding procedures. Tissues were incubated in RPMI when outside of the body for extended time.
Human tissue acquisition
Human normal colon and colorectal cancer tissues were obtained under protocols approved by Vanderbilt University through the Cooperative Human Tissue Network (CHTN). Clinical and pathology reports were attached to each sample prior to de-identification of patient information. An optimized CHTN collection SOP was used. Briefly, specimen sizes were limited to 5 mm in diameter and fixed for 24 hours in magnetically stirred formalin (to facilitate diffusion), after which the specimen was embedded with a standardized FFPE protocol. The time from which the specimen was excised from the patient to the fixative (during which the tissue was examined by the pathologists or their assistants) was recorded as the post-excision time (PET). Specimens with substantial tumor cell content determined by haematoxylin and eosin staining were selected for analysis. Overall, 7 normal colon samples and 13 CRC samples were selected.
DISSECT disaggregation on FFPE tissues
50μm sections were freshly cut from each block and placed in 1.5mL microcentrifuge tubes (Fisher). Samples were heated to 65°c for 25 minutes to melt wax, then washed 3 times with 1mL of histoclear for 8 minutes each. Tissues were then rehydrated in 2 washes each of 100%, 70% and 50% ethanol, then 3 washes of PBS. Samples were washed for 10 minutes in PBS with 0.3% Triton X-100, then washed a final time in PBS before incubation in the HIAR buffer (DAKO). Samples were incubated in the buffer under high heat and pressure for 20 minutes (actively heating for the first 4 minutes), followed by 20 minutes cooling of the bench. Samples were then washed 3 addition times in PBS and stored at 4° until staining.
Tissues were blocked at room temperature (RT) for 30 minutes in 2.5% donkey serum (Jackson ImmunoResearch) in PBS and stained overnight at RT with antibodies diluted in the same buffer. Additional blocking with carefully chosen serum combinations were applied if secondary antibodies were used. After appropriate washing, samples were incubated for 30 minutes in 4% PFA to crosslink antibodies to their targets. Samples were washed, and then incubated for 25 minutes at 37°C in 200μL PBS with 1mg/mL each of collagenase (Calbiotech) and dispase (Life Technologies). Tissues were passaged 5-10 times through a 27ga needle to mechanically dissociate them into single cells. Cells were incubated with a nuclear intercalating agent prior to analysis.
Cytometry analyses
For both fluorescence cytometry and mass cytometry, cells were initially gated using DNA content (Hoescht – fluorescence cytometry) or intercalator (Iridium – mass cytometry) following established procedures to identify intact single cells and eliminate cell doublets and clusters from analysis (16, 76, 77). Single cells were then analyzed for intensity of antibody conjugates. Fluorescence cytometry was performed on a BD LSRII with 5 lasers and mass cytometry was performed on a Fluidigm-DVS CyTOF 1 instrument. Epithelial-specific analysis was achieved by gating cells positive for pan-Cytokeratin (PCK).
Immunofluorescence imaging
FFPE tissues were sectioned at 5 μm and processed using standard immunohistological techniques, stained with appropriate primary or primary/secondary antibodies. DISSECT-processed tissues were also imaged pre- and post- disaggregation in a whole-mount format. Slides were imaged using a Zeiss Axiophot fluorescence/brightfield microscope with a Zeiss Axiocam with 5 channel imaging capabilities. Quantitative analysis of goblet cells in the villus was performed on ImageJ using the particle analysis module. Ratios of areas occupied between the CLCA1 channel and nuclear channel were calculated with a correction factor for the typical size of a goblet granule against the typical size of a nucleus.
Quantitative immunofluorescence imaging and immunoblotting of signaling proteins
The same antibody clones were used for FFPE-DISSECT-CyTOF. Quantifications were performed as described previously (16).
Antibody reagents
Antibodies used in this study are listed tables S1 and S3. All signaling antibodies were previously validated and used in mass cytometry applications (16).
Data analysis and Statistical analysis
t-SNE analysis was performed using the viSNE implementation on Cytobank.org following established single-cell analysis workflows (78–80). Gating for cell types was performed by considering a first decade (101) threshold for cell type specific markers. Unpaired t-tests and correlation analyses were performed using Prism (Graphpad). Multiple comparison tests were performed with ANOVA with Tukey post-hoc test (Graphpad). Correlative distances and heat maps were generated using MATLAB (Mathworks). Hierarchical clustering and dendrogram analysis were performed using the dendextend package in R (42).
Supplementary Material
Acknowledgements
The authors would like to thank Kevin Weller and David Flaherty at the Vanderbilt Flow Cytometry core, Joseph Roland at the Vanderbilt Digital Histology Shared Resource, and Robert Carnahan at Vanderbilt Antibody and Protein Resource for their technical assistance, Ray Mernaugh in the Department of Biochemistry, Kerry Wiles at CHTN, Gregor Ayers at the Vanderbilt Center for Quantitative Sciences, Amrita Banerjee and Sun Wook Kim in the Lau lab for their helpful advice, and Dr. Kevin Haigis at BIDMC for his consultation on RAS mutations.
Funding: K.S.L is funded by R01DK103831, an Innovator Award from the AACR-Landon Foundation (15-20-27-LAUK), a CCFA CDA (308221), VICTR (2UL1TR000445), and pilot project grants from P30CA068485, P30DK058404, P50CA095103, and U24CA159988. A.J.S. is funded by R01DK103831. C.A.H. is funded by a training grant from T32HD007502 and F31GM120940. C.R.S. is funded by a training grant from T32AI007281. ETM is funded by a training grant from R25CA092043. R.J.C. is funded by R01CA174377, and R.J.C. and M.K.W. are funded by P50CA095103. J.M.I. is funded by R00CA143231.
Footnotes
Figure S1: Imaging of single cell suspensions prepared by FFPE-DISSECT from embedded mouse intestinal tissue by brightfield and autofluorescence.
Figure S2: Whole mount staining of embedded mouse intestinal tissue prepared by FFPE-DISSECT before single cell disaggregation.
Figure S3: Fluorescence cytometry of samples prepared by FFPE-DISSECT.
Figure S4: Signals are preserved for up to 1 hour post-excision.
Figure S5: Fixation time does not affect signaling marker detection in FFPE.
Figure S6: Villus signal comparison between FFPE-DISSECT, quantitative imaging, quantitative immunoblotting.
Figure S7: Replicates over multiple animals depicting relationships between apoptosis and signaling pathways using t-SNE and bi-plots.
Figure S8: Cell type-specific signaling in the murine duodenal epithelium.
Figure S9: RAS induction of MEK-ERK signaling induces goblet cell identity.
Figure S10: Hematoxylin and eosin of MSS and MSI CRCs showing tissue areas occupied by tumors.
Figure S11: Signaling in human normal colon and CRC.
Figure S12: Differentiation in human normal colon and CRC.
Figure S13: Evidence of fixative penetration.
Figure S14: Analysis of the organization of signaling pathways in human specimens.
Author contributions: A.J.S. designed and performed all mouse, cytometry, and imaging experiments. C.R.S. extracted and organized human patient data. E.T.M. assisted with imaging experiments. C.A.H. assisted with computational data analysis. M.K.W. facilitated the acquisition of human tissues, performed pathological assessment, and intellectually contributed to the tissue preparation process. J.M.I., and R.J.C. intellectually contributed to the study and the writing of the manuscript. K.S.L. conceived of the study, performed cytometry and imaging analysis, performed computational data analysis, wrote the manuscript, and supervised the research.
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Single-cell data for human tumor and mouse intestine generated by FFPE-DISSECT is shared on Cytobank.org (mouse data: https://community.cytobank.org/cytobank/experiments/57176; human data: https://community.cytobank.org/cytobank/experiments/57155).
References and Notes
- 1.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;6:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagenblast E, Soto M, Gutiérrez-Ángel S, Hartl CA, Gable AL, Maceli AR, Erard N, Williams AM, Kim SY, Dickopf S, Harrell JC, Smith AD, Perou CM, Wilkinson JE, Hannon GJ, Knott SRV. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature. 2015;520:358–362. doi: 10.1038/nature14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smedt L, Lemahieu J, Palmans S, Govaere O, Tousseyn T, Van Cutsem E, Prenen H, Tejpar S, Spaepen M, Matthijs G, Decaestecker C, Moles Lopez X, Demetter P, Salmon I, Sagaert X. Microsatellite instable vs stable colon carcinomas: analysis of tumour heterogeneity, inflammation and angiogenesis. Br. J. Cancer. 2015:1–10. doi: 10.1038/bjc.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat A. a, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton A. a, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011;29:1120–7. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreso A, O’Brien C. a, van Galen P, Gan OI, Notta F, Brown AMK, Ng K, Ma J, Wienholds E, Dunant C, Pollett A, Gallinger S, McPherson J, Mullighan CG, Shibata D, Dick JE. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–8. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton A. a, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozar S, Morrissey E, Nicholson A, van der Heijden M, Zecchini H, Kemp R, Tavaré S, Vermeulen L, Winton D. Continuous Clonal Labeling Reveals Small Numbers of Functional Stem Cells in Intestinal Crypts and Adenomas. Cell Stem Cell. 2013:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin. Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrizi E. Therapeutic implications of colon cancer stem cells. World J. Gastroenterol. 2010;16:3871. doi: 10.3748/wjg.v16.i31.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colak S, Medema JP. Cancer stem cells; important players in tumor therapy resistance. FEBS J. 2014 doi: 10.1111/febs.13023. doi:10.1111/febs.13023. [DOI] [PubMed] [Google Scholar]
- 12.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 13.Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F, Gupta PB. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irish JM, Doxie DB. High-dimensional single-cell cancer biology. Curr. Top. Microbiol. Immunol. 2014;377:1–21. doi: 10.1007/82_2014_367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat. Rev. Cancer. 2006;6:146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 16.Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS, Masuzaki R, Karp SJ, Franklin JL, Gerdes MJ, Irish JM, Coffey RJ, Lau KS. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol. Syst. Biol. 2015;11:835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrahamsen I, Lorens JB. Evaluating extracellular matrix influence on adherent cell signaling by cold trypsin phosphorylation-specific flow cytometry. BMC Cell Biol. 2013;14:36. doi: 10.1186/1471-2121-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng J, Mohammadreza A, Gao W, Merza S, Smith D, Kelbauskas L, Meldrum DR. A minimally invasive method for retrieving single adherent cells of different types from cultures. Sci. Rep. 2014;4:5424. doi: 10.1038/srep05424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv. Biobank. 2013;11:101–6. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Conley A, Zhang H, Kim HL. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics. 2014;15:1087. doi: 10.1186/1471-2164-15-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedegaard J, Thorsen K, Lund MK, Hein A-MK, Hamilton-Dutoit SJ, Vang S, Nordentoft I, Birkenkamp-Demtröder K, Kruhøffer M, Hager H, Knudsen B, Andersen CL, Sørensen KD, Pedersen JS, Ørntoft TF, Dyrskjøt L. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9:e98187. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons J, Herring CA, Banerjee A, Simmons AJ, Lau KS. Multiscale analysis of the murine intestine for modeling human diseases. Integr. Biol. (Camb) 2015;7:740–57. doi: 10.1039/c5ib00030k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11982–7. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J-R, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using CycIF, a high-throughput cyclic immunofluorescence method. Cmpct. 2015;6:1–7. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zrazhevskiy P, Gao X. Quantum dot imaging platform for single-cell molecular profiling. Nat. Commun. 2013;4:1619. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riordan DP, Varma S, West RB, Brown PO. Automated analysis and classification of histological tissue features by multi-dimensional microscopic molecular profiling. PLoS One. 2015;10:1–18. doi: 10.1371/journal.pone.0128975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014;20:436–42. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giesen C, Wang H. a O., Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Günther D, Bodenmiller B. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11:417–22. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 29.Schüffler PJ, Schapiro D, Giesen C, Wang HAO, Bodenmiller B, Buhmann JM. Automatic single cell segmentation on highly multiplexed tissue images. Cytometry. A. 2015;87:936–42. doi: 10.1002/cyto.a.22702. [DOI] [PubMed] [Google Scholar]
- 30.Corver WE, ter Haar NT. High-resolution multiparameter DNA flow cytometry for the detection and sorting of tumor and stromal subpopulations from paraffin-embedded tissues. Curr. Protoc. Cytom. 2011 doi: 10.1002/0471142956.cy0737s55. Chapter 7, Unit 7.37. [DOI] [PubMed] [Google Scholar]
- 31.Lau KS, Cortez-Retamozo V, Philips SR, Pittet MJ, Lauffenburger DA, Haigis KM. Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-α-induced apoptosis in the intestinal epithelium. PLoS Biol. 2012;10:e1001393. doi: 10.1371/journal.pbio.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-α-induced apoptosis and proliferation by MAPKs. Sci. Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinhel IF, Macneill FA, Hills MJ, Salter J, Detre S, A’hern R, Nerurkar A, Osin P, Smith IE, Dowsett M. Extreme loss of immunoreactive p-Akt and p-Erk1/2 during routine fixation of primary breast cancer. Breast Cancer Res. 2010;12:R76. doi: 10.1186/bcr2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau KS, Schrier SB, Gierut J, Lyons J, Lauffenburger DA, Haigis KM. Network analysis of differential Ras isoform mutation effects on intestinal epithelial responses to TNF-α. Integr. Biol. (Camb) 2013;5:1355–65. doi: 10.1039/c3ib40062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gierut JJ, Wood LB, Lau KS, Lin Y-J, Genetti C, Samatar AA, Lauffenburger DA, Haigis KM. Network-level effects of kinase inhibitors modulate TNF-α-induced apoptosis in the intestinal epithelium. Sci. Signal. 2015;8:ra129. doi: 10.1126/scisignal.aac7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 37.Lau KS, Haigis KM. Non-redundancy within the RAS oncogene family: insights into mutational disparities in cancer. Mol. Cells. 2009;28:315–20. doi: 10.1007/s10059-009-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008;40:600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Velho S, Vakiani E, Peng S, Bass AJ, Chu GC, Gierut J, Bugni JM, Der CJ, Philips M, Solit DB, Haigis KM. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer Discov. 2013;3:294–307. doi: 10.1158/2159-8290.CD-12-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, Brown K, Burberry A, Cho KR, Fearon ER. Mutant kras promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology. 2011;141:1003–1013. e10. doi: 10.1053/j.gastro.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cagnol S, Rivard N. Oncogenic KRAS and BRAF activation of the MEK/ERK signaling pathway promotes expression of dual-specificity phosphatase 4 (DUSP4/MKP2) resulting in nuclear ERK1/2 inhibition. Oncogene. 2013;32:564–76. doi: 10.1038/onc.2012.88. [DOI] [PubMed] [Google Scholar]
- 42.Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31:3718–20. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker FB. Stability of Two Hierarchical Grouping Techniques Case 1: Sensitivity to Data Errors. J. Am. Stat. Assoc. 1974;69:440–445. [Google Scholar]
- 44.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJC, Carr S. a, Tabb DL, Coffey RJ, Slebos RJC, Liebler DC. NCI CPTAC, Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie R, Chung J-Y, Ylaya K, Williams RL, Guerrero N, Nakatsuka N, Badie C, Hewitt SM. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J. Histochem. Cytochem. 2011;59:356–65. doi: 10.1369/0022155411398488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2770–7. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciaccio MF, Wagner JP, Chuu C-P, Lauffenburger D. a, Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat. Methods. 2010;7:148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimm DL, Camp RL, Charette LA, Olsen DA, Provost E. Amplification of tissue by construction of tissue microarrays. Exp. Mol. Pathol. 2001;70:255–64. doi: 10.1006/exmp.2001.2363. [DOI] [PubMed] [Google Scholar]
- 50.Bauer DR, Stevens B, Chafin D, Theiss AP, Otter M. Active monitoring of formaldehyde diffusion into histological tissues with digital acoustic interferometry. J. Med. imaging (Bellingham, Wash.) 2016;3:17002. doi: 10.1117/1.JMI.3.1.017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker JR. Principles of Biological Microtechnique; a Study of Fixation and Dyeing. 2015. [Google Scholar]
- 52.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–8. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heuberger J, Kosel F, Qi J, Grossmann KS, Rajewsky K, Birchmeier W. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl. Acad. Sci. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Jong PR, Taniguchi K, Harris AR, Bertin S, Takahashi N, Duong J, Campos AD, Powis G, Corr M, Karin M, Raz E. ERK5 signalling rescues intestinal epithelial turnover and tumour cell proliferation upon ERK1/2 abrogation. Nat. Commun. 2016;7:11551. doi: 10.1038/ncomms11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalluri R, Weinberg RA. Review series The basics of epithelial-mesenchymal transition. 119:2009. doi: 10.1172/JCI39104. doi:10.1172/JCI39104.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 57.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barras D. BRAF Mutation in Colorectal Cancer: An Update. Biomark. Cancer. 2015;7:9–12. doi: 10.4137/BIC.S25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au H-J, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 60.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, Iacovelli R, Bossi I, Lonati V, Ghilardi M, de Braud F, Barni S. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur. J. Cancer. 2015;51:587–94. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 61.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin. Cancer Res. 2008;14:342–6. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 62.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, Maloney L, Gordon G, Simmons H, Marlow A, Litwiler K, Brown S, Poch G, Kane K, Haney J, Eckhardt SG. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol. 2008;26:2139–46. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, Schlicker A, Wessels LFA, Smit EF, Thunnissen E, Halonen P, Lieftink C, Beijersbergen RL, Di Nicolantonio F, Bardelli A, Trusolino L, Bernards R. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 64.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeh JJ, Routh ED, Rubinas T, Peacock J, Martin TD, Shen XJ, Sandler RS, Kim HJ, Keku TO, Der CJ. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol. Cancer Ther. 2009;8:834–43. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahin IH, Kazmi SMA, Yorio JT, Bhadkamkar NA, Kee BK, Garrett CR. Rare Though Not Mutually Exclusive: A Report of Three Cases of Concomitant KRAS and BRAF Mutation and a Review of the Literature. J. Cancer. 2013;4:320–2. doi: 10.7150/jca.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 69.Morkel M, Riemer P, Bläker H, Sers C, Markus PRHBCS. Morkel, Similar but different: distinct roles for KRAS and BRAF oncogenes in colorectal cancer development and therapy resistance. Oncotarget. 2015;6:20785–20800. doi: 10.18632/oncotarget.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin. Cancer Res. 2008;14:3408–15. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, Fuchs CS, Ogino S. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J. Natl. Cancer Inst. 2013;105:1151–6. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fransén K, Klintenäs M, Osterström A, Dimberg J, Monstein H-J, Söderkvist P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–33. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- 73.Xiao Y, Freeman GJ. The Microsatellite Instable Subset of Colorectal Cancer Is a Particularly Good Candidate for Checkpoint Blockade Immunotherapy. Cancer Discov. 2015;5:16–18. doi: 10.1158/2159-8290.CD-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran B, Kopetz S, Tie J, Gibbs P, Jiang Z-Q, Lieu CH, Agarwal A, Maru DM, Sieber O, Desai J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–32. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim JH, Rhee Y-Y, Bae JM, Cho N-Y, Kang GH. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am. J. Surg. Pathol. 2013;37:1532–41. doi: 10.1097/PAS.0b013e31829ab1c1. [DOI] [PubMed] [Google Scholar]
- 76.Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, Irish JM, Kalams SA. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytometry. A. 2016;89:271–80. doi: 10.1002/cyto.a.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leelatian N, Diggins KE, Irish JM. Characterizing Phenotypes and Signaling Networks of Single Human Cells by Mass Cytometry. Methods Mol. Biol. 2015;1346:99–113. doi: 10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amir E-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diggins KE, Ferrell PB, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saeys Y, Van Gassen S, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.