Abstract
Helicases unwind double-stranded nucleic acids, remove secondary structures from single-stranded nucleic acids, and remove proteins bound to nucleic acids. For many helicases, the mechanisms for these different functions share the ability to translocate with a directional bias as a result of ATP binding and hydrolysis. The nonstructural protein 3 (NS3) is an essential enzyme expressed by the hepatitis C virus (HCV) and is known to catalyze the unwinding of both DNA and RNA substrates in a 3′-to-5′ direction. We investigated the role of nucleic acid binding in the unwinding mechanism by examining ATP-independent unwinding. We observed that even in the absence of ATP, NS3 helicase domain (NS3h) unwound duplexes only when they contained a 3′-tail (i.e., 3′-to-5′ directionality). Blunt-ended duplexes and 5′-tailed duplexes were not melted even in the presence of a large excess concentration of the protein. NS3h was found to diffuse rapidly along single-stranded DNA at a rate of 30 nt2·s−1. Upon encountering an appropriate single-strand/double-strand (ss/ds) junction, NS3h slowly melted the duplex under conditions with excess protein concentration relative to DNA concentration. When a biotin-streptavidin block was placed into the ssDNA region, no melting of DNA was observed, suggesting that NS3h must diffuse along the ssDNA, and that the streptavidin blocked the diffusion. We conclude that the specific interaction between NS3h and the ss/dsDNA junction, coupled with diffusion allows binding energy to melt duplex DNA with a directional bias. Alternatively, we found that the full-length NS3 protein did not exhibit strict directionality and was dependent on duplex DNA length. NS3 was able to unwind the duplex even in the presence of the biotin-streptavidin block. We propose a non-canonical model of unwinding for NS3 in which the enzyme binds directly to the duplex via protein-protein interactions to melt the substrate.
Helicases are enzymes that manipulate nucleic acids by coupling the energy of ATP hydrolysis to protein conformational changes that unwind and/or translocate DNA or RNA.1–4 The mechanisms for these enzymes have received intense study over the past two decades. Different families of helicases have been identified based on sequence analysis.5 Some helicases readily oligomerize into hexameric rings, while other helicases appear to function as monomers or dimers.6 The non-structural protein 3 (NS3) from the hepatitis C virus (HCV) is one of the most studied helicases from superfamily 2. It is of medical interest as a target for anti-viral drug development, but it also serves as an excellent model system for studying helicase mechanisms.7 This enzyme is a multifunctional protein that contains a N-terminal protease domain and a C-terminal helicase domain. The domains can be separated while retaining each individual function, but helicase and protease activity are allosterically linked.8 NS3 can interact with itself, but it also reportedly can function as a monomer. HCV is an RNA virus, so the biologically relevant substrate for NS3 is RNA. However, this enzyme readily functions on DNA as well, and some evidence indicates that NS3 can be observed in the nucleus of patients with HCV infection.9 While nuclear DNA helicase II (SF2 helicase), vaccinia NPH II (SF2 helicase), and human coronavirus 229E (SF1 helicases) are also known to function on both nucleic acids,10–12 most helicases, even very closely related SF2 enzymes (e.g. DENV NS3), typically only function on either DNA or RNA.13
Studies on this enzyme have been conducted on both RNA and DNA substrates, and structural studies have demonstrated that the interactions made between NS3 and the two different nucleic acid structures are highly similar.14–16 It is due to this lack of strict substrate specificity that NS3 is able to work on both substrates with similar but not identical kinetic parameters.17,18
The ability of helicases to unwind dsNA can be broken down into two functions: translocation and base pair melting. Translocation has been examined in depth through many approaches, resulting in models for stepwise movement of the enzymes along the nucleic acid lattice.4,6,19 The helicase domain of NS3 (NS3h) has served as a model enzyme for studying translocation and base pair melting. In the case of NS3h, movement has been suggested to occur in one nt steps, driven by hydrolysis of one ATP.20 An alternative model based on single molecule FRET experiments supports a “spring-loaded” mechanism in which the enzyme hydrolyzes ATP, building up strain through movement of subdomains along the DNA, then the enzyme releases the strain by springing forward while melting 3 base pairs.21
The chemical mechanism for helicase-catalyzed base pair melting has received less attention than translocation. In previous studies, it has been shown that NS3 and NS3h bind tightly to the ss/ds junction causing destabilization of the duplex.22 In the current study, we have examined unwinding of dsDNA in the absence of ATP in order to investigate the role of protein-DNA binding during the base pair melting process. Results indicate that NS3h readily diffuses along ssDNA, but interacts tightly with ss/dsDNA junction. A directional bias for unwinding of DNA is observed in the absence of ATP, which emphasizes the importance of the specific interaction between the enzyme and the ss/dsDNA junction. In the case of full-length NS3, directionally-biased melting is partially lost for ATP-independent melting of short duplexes which appears to be due to protein-protein interactions that are much stronger compared to the truncated NS3h protein.
EXPERIMENTAL PROCEDURES
Materials
Nucleotide radiolabeling was achieved using [β-32P] ATP purchased from PerkinElmer Life Sciences and T4 polynucleotide kinase obtained from New England Biolabs. HEPES, EDTA, βME, SDS, MOPS, Tris, NaCl, MgCl2, KOH, ATP, KMnO4, glycerol, formamide, arylamide, bisacrylamide, xylene cyanole, bromophenol blue, and urea were all purchased from Fisher. Poly(U) was acquired from Amersham Biosciences, and Sephadex G-25 was from Sigma. Streptavidin Dynabeads were obtained from Invitrogen.
Oligonucleotides and Proteins
DNA oligonucleotides were obtained from Integrated DNA Technologies (Table 1); all nucleic acids were purified via preparative gel electrophoresis prior to use in experimental assays and radiolabeled as previously described.23 DNA substrates are shown in Table 1. For each substrate, the top strand (10 pmols) was radiolabeled. Duplexes were prepared by adding 1.2 equivalents of unlabeled oligonucleotide (bottom strand) to the radiolabeled oligonucleotide, followed by heating to 95 °C, then allowing slow cooling to room temperature. DNA substrates were stored in a solution buffer consisting of 10 mM HEPES (pH 7.5) and 0.1 mM EDTA. Recombinant full-length NS3 was derived from the HCV replicon 1b replicon consensus sequence and expressed and purified as previously described.23–25
Table 1.
DNA Substrates
| Name | Sequence | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T15-22 bp | 3′- | TTT | TTT | TTT | TTT | TTT | AGG | ACA | GTC | GGA | TCG | CAG | TCA | G | ||
| 5′- | TCC | TGT | CAG | CCT | AGC | GTC | AGT | C | ||||||||
| T15(bio·dT-12)-22 bp | 3′- | TTT | TTT | TTT | TT/Bio·dT/ | TTT | AGG | ACA | GTC | GGA | TCG | CAG | TCA | G | ||
| 5′- | TCC | TGT | CAG | CCT | AGC | GTC | AGT | C | ||||||||
| 22 bp-T15 | 3′- | AGG | ACA | GTC | GGA | TCG | CAG | TCA | GTT | TTT | TTT | TTT | TTT | T | -5′ | |
| 5′- | TCC | TGT | CAG | CCT | AGC | GTC | AGT | C | ||||||||
| 22 bp | 3′- | AGG | ACA | GTC | GGA | TCG | CAG | TCA | G | |||||||
| 5′- | TCC | TGT | CAG | CCT | AGC | GTC | AGT | C | ||||||||
| T15-30 bp | 3′- | TTT | TTT | TTT | TTT | TTT | CAT | CAT | GCA | GGA | CAG | TCG | GAT | CGC | AGT | CAG |
| 5′- | GTA | GTA | CGT | CCT | GTC | AGC | CTA | GCG | TCA | GTC | ||||||
ATP-Independent, Multiple Turnover Unwinding
DNA unwinding assays are conducted by incubating a helicase with a radiolabeled duplex substrate and then initiating the reaction upon addition of ATP leading to formation of ssDNA. The presence of a nucleic acid (NA) trap that is complementary to the unlabeled strand of the substrate prevents reannealing of separated ssDNA products. Detection of these products is by autoradiography after native gel electrophoresis separates the ssDNA product from the dsDNA. The ATP-independent unwinding reactions were initiated by mixing 2 nM radiolabeled substrate with 500 nM NS3 or NS3h (final concentrations reported) at 37 °C after an initial pre-incubation period of 5 min to allow the solutions to reach the desired temperature prior to the reaction. The reaction was performed in 25 mM MOPS (pH 7.0), 50 mM NaCl, 10 mM MgCl2, 2 mM βME, 0.1 mg/mL BSA, and 0.1 mM EDTA. The multiple turnover unwinding reaction proceeded at 37 °C for the desired time, followed by mixing with a quench solution containing 100 μM nt poly(U), 60 nM annealing trap, 0.2 M EDTA, and 0.7% SDS. An aliquot of the quenched reaction mixture (20 μl) was added to 4 μl loading buffer (0.1% bromophenol blue, 0.1% xylene cyanol, and 30% glycerol) and resolved on a native 20% polyacrylamide gel. The radiolabeled products were detected using a PhosphorImager (GE Healthcare Life Sciences), and quantitation of the radioactivity in each band was performed using ImageQuant software (GE Healthcare, Piscataway, NJ). The ratio of single-stranded to double-stranded DNA was determined and plotted as a function of time using Kaleidagraph (Synergy Software, Reading, PA).
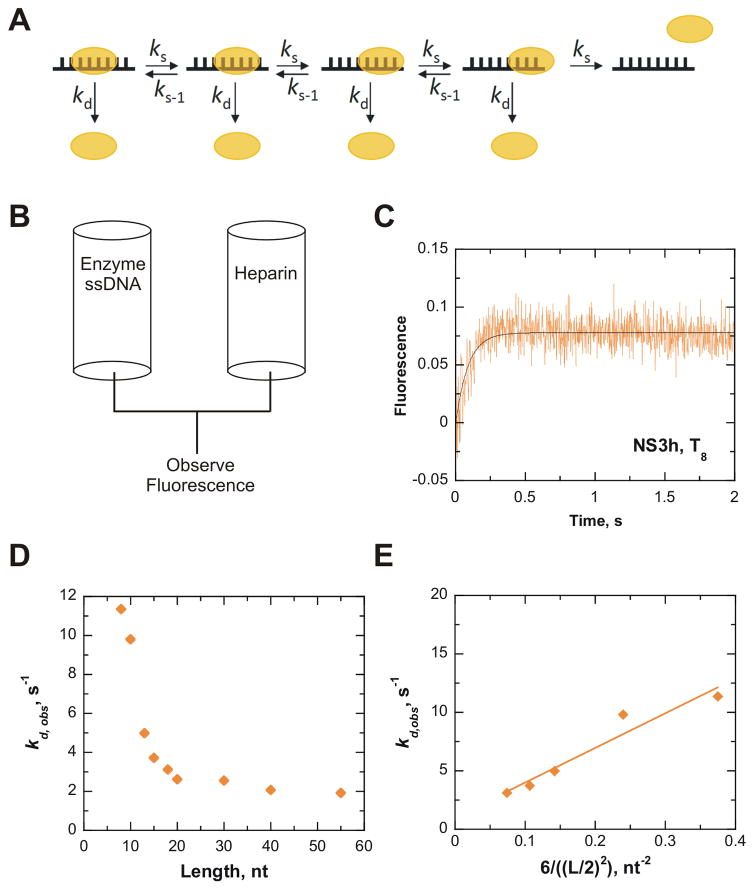
Stopped-flow Translocation Assay
All concentrations indicated are after mixing. The dissociation of NS3h from ssDNA in the absence of ATP was measured by using a SX.18MV stopped-flow instrument (Applied Photophysics) maintained at 37 °C. NS3h was incubated with ssDNA, followed by rapid mixing with heparin (4 mg/ml). The change in intrinsic tryptophan fluorescence over time was monitored with excitation wavelength of 280 nm using 1 mm slit width. Emission was measured using an Oriel 51980 filter with a 340–600 nm bandpass. 100 nM NS3h was pre-incubated with 1.2 μM ssDNA (T8, T10, T13, T15, T18, T20, T30, T40, and T55) in a buffer containing 25 mM MOPS (pH 7.0), 50 mM NaCl, 0.1 mM EDTA, and 2 mM βME in the presence and absence of 10 mM MgCl2, where indicated. The reaction was initiated by rapid mixing with 4 mg/mL heparin and 10 mM Mg2+ (Figure 2B). For each enzyme-substrate pair, five to six kinetic traces were collected and averaged. The data were then fit to a single exponential to obtain the observed dissociation rate constant (kd,obs). The rate of translocation (kt) or diffusional sliding (ks) along ssDNA was determined as described by Young et al., 1994 (see text for details).26
Figure 2.
NS3h slides along ssDNA in an ATP-independent manner. Diffusional sliding of NS3h along ssDNA was determined from dissociation of the enzyme from oligonucleotides of increasing length (L). All concentrations reported are final after mixing. A. A model for diffusion of NS3h along a 8mer oligonucleotide. NS3h binds 6 nt based on the crystal structure. Each step of diffusional sliding (ks) is shown in one direction, but actually can occur with equal probability in both directions. Sliding is assumed to occur in increments of one nt. Dissociation from the end of the DNA was estimated to occur in three individual steps, which allows half the six sites of interaction to dissociate. NS3h can also dissociate from internal positions, kd. B. Protocol for rapid mixing of a solution containing a pre-bound NS3h-ssDNA complex with a solution of heparin, which will sequester any unbound helicase and prevent rebinding of NS3h to the substrate. C. A representative progress curve from dissociation of 200 nM NS3h from 1.2 μM strands T8 upon rapid mixing with 4 mg/mL heparin. The data was fit to a single exponential to obtain an observed dissociation rate (kd,obs) of 11.4 ± 0.8 s−1. D. The kd,obs rates obtained for different lengths of ssDNA are plotted (
 ). E. The rates of dissociation from oligonucleotide that display a length-dependence (T8, T10, T13, T15, T18) were used to determine the rate of sliding. The sliding rate (ks) was determined via analysis of the linear regression fit of the observed dissociation rates plotted versus the equation describing diffusion along a linear polymer.26 The ks of NS3h along ssDNA in the absence of ATP binding and hydrolysis was determined to be 30 nt2/s (
). E. The rates of dissociation from oligonucleotide that display a length-dependence (T8, T10, T13, T15, T18) were used to determine the rate of sliding. The sliding rate (ks) was determined via analysis of the linear regression fit of the observed dissociation rates plotted versus the equation describing diffusion along a linear polymer.26 The ks of NS3h along ssDNA in the absence of ATP binding and hydrolysis was determined to be 30 nt2/s (
 ).
).
RESULTS
NS3h can unwind DNA substrates containing a properly oriented ss/dsDNA junction in the absence of ATP
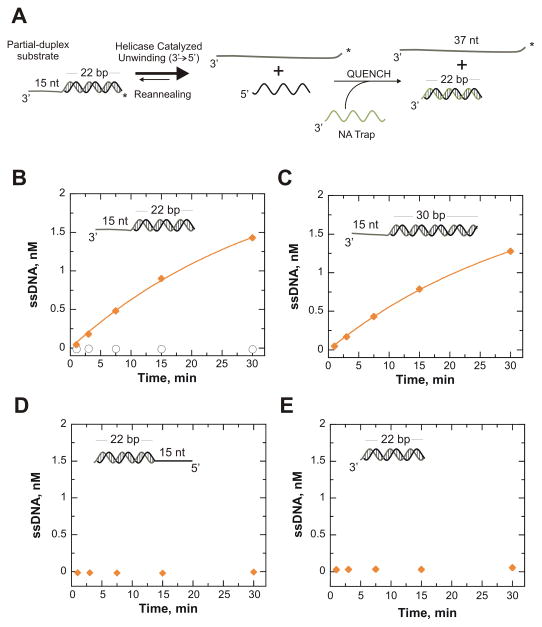
Binding of the enzyme to the substrate is the first step in translocation and unwinding activities of a helicase. Helicase ATPase activity may be needed for directional movement, base pair melting, or both. In order to understand the specific role of nucleic acid binding to base pair melting, ATP-independent unwinding of NS3h on a variety of DNA substrates was investigated. Under excess enzyme concentration relative to substrate concentration (500 nM NS3h and 2 nM substrate) and multiple turnover conditions, ATP-independent unwinding of a substrate with a 15 thymidine 3′-tail adjacent to a 22 bp mixed sequence duplex (T15-22 bp; see Table 1 for sequences) was observed for NS3h at 37 °C (Figure 1B). NS3h was capable of separating nearly 75% of the substrate in 30 minutes.
Figure 1.
ATP-independent unwinding of DNA substrates by NS3h requires interaction with a properly-oriented junction. ATP-independent unwinding was initiated by mixing 500 nM NS3h with 2 nM DNA in the presence of 10 mM MgCl2 at 37 °C (
 ). A. An unwinding assay is performed on a radiolabeled duplex substrate with or without a single-stranded extension. The action of a helicase unwinds the duplex resulting in two separate DNA strands. A trapping strand that is complimentary to the unlabeled displaced strand is placed in the quench of an ATP independent unwinding assay to prevent reannealing. B. The DNA substrate, T15-22 bp, featuring a ssDNA 3′-ssDNA tail made up of 15 thymidine residues was unwound by NS3h (
). A. An unwinding assay is performed on a radiolabeled duplex substrate with or without a single-stranded extension. The action of a helicase unwinds the duplex resulting in two separate DNA strands. A trapping strand that is complimentary to the unlabeled displaced strand is placed in the quench of an ATP independent unwinding assay to prevent reannealing. B. The DNA substrate, T15-22 bp, featuring a ssDNA 3′-ssDNA tail made up of 15 thymidine residues was unwound by NS3h (
 ). The data was fit to a single exponential resulting in an observed rate of unwinding of 0.033 ± 0.004 min−1. To ensure the efficiency of the protein trap, 100 μM nt poly(U) was pre-incubated with the DNA then mixed with enzyme (○) to begin the unwinding reaction. C. NS3h was also able to unwind a DNA substrate of 30 bp, T15-30 bp (
). The data was fit to a single exponential resulting in an observed rate of unwinding of 0.033 ± 0.004 min−1. To ensure the efficiency of the protein trap, 100 μM nt poly(U) was pre-incubated with the DNA then mixed with enzyme (○) to begin the unwinding reaction. C. NS3h was also able to unwind a DNA substrate of 30 bp, T15-30 bp (
 ). The data was fit to a single exponential to obtain the observed unwinding rate of 0.029 ± 0.003 min−1. D. NS3h was unable to unwind a DNA substrate containing a 5′-ssDNA tail adjacent to the 22 bp duplex (22 bp-T15) (
). The data was fit to a single exponential to obtain the observed unwinding rate of 0.029 ± 0.003 min−1. D. NS3h was unable to unwind a DNA substrate containing a 5′-ssDNA tail adjacent to the 22 bp duplex (22 bp-T15) (
 ). E. NS3h was unable to unwind a blunt-ended DNA substrate comprised of only the 22 bp duplex.
). E. NS3h was unable to unwind a blunt-ended DNA substrate comprised of only the 22 bp duplex.
These experiments were performed under multiple-turnover conditions in the absence of a protein trap so that the enzyme could bind, dissociate, and re-associate with the DNA throughout the duration of the unwinding reaction. The protein trap (100 μM nt poly(U)) was added at the designated times to sequester the enzyme and stop the unwinding reaction. To ensure that the protein trap was efficient, the poly(U) and the duplex DNA substrate were pre-incubated together prior to the initiation of unwinding by the addition of the enzyme. NS3h did not unwind the substrate in the presence of excess poly(U), confirming the protein trap was effective (Figure 1B, open circles).
To determine whether this reaction was dependent on the duplex length, the assay was repeated on a DNA substrate with the same 15 thymidine ssDNA tail attached to a slightly longer duplex of 30 bp, T15-30 bp (Figure 1C). NS3h unwound T15-30 bp in with a similar rate and amplitude as the 22 bp substrate.
NS3h ATP-independent unwinding displays apparent directional bias
During ATP-dependent processes, NS3 and NS3h are known to have an inherent 3′ → 5′ translocase and helicase activities.27 To determine if this directionality is also observed in ATP-independent unwinding, a 15 thymidine 5′-tail attached to a 22 bp duplex identical in sequence to the previous experiments (22 bp-T15) was utilized. NS3h was unable to unwind the substrate with the 5′-tail (Figure 1D). The requirement for a 3′-oriented tail in this unwinding reaction is evidence for a directional bias inherent in the unwinding activity of NS3h, independent of ATP binding and hydrolysis. This also suggests that the helicase domain itself exhibits this property, even in the absence of the protease domain.
NS3h did not unwind a blunt-ended DNA duplex substrate consisting of 22 bp in the absence of a ssDNA tail (Figure 1E). This data supports similar reports of the requirement of a ss region for these enzymes to load onto before unwinding can occur.23,28
It is possible that diffusional sliding on ssDNA may play a part in the observed ATP-independent unwinding. The finding that NS3h requires the 3′-ssDNA tail to melt substrates but is inactive on the 5′-ssDNA tail in the absence of ATP suggests that the polarity of binding to ssDNA and recognition of the ss/ds junction is important in the unwinding reaction. Hence, a specific interaction between NS3h and the ss/dsDNA junction appears to mediate the melting reaction. Previously, Levine and Patel reported a junction specific interaction between NS3h and DNA substrates.22 Results here suggest that binding to ssDNA alone is insufficient to melt the duplex, but if the binding to ssDNA is on the appropriate side of the ss/ds junction, duplex melting can occur.
NS3h diffusion on ssDNA in the absence of ATP
Methods to determine ATP-dependent translocation of helicases on ssDNA were developed by several labs.18,26,29,30. NS3h binds to the ssDNA and presumably it may diffuse or slide randomly along the DNA (Figure 2A). To examine diffusional sliding of NS3h, a series of stopped-flow experiments were performed to measure dissociation of NS3h from oligonucleotides of increasing lengths (1.2 μM: T8, T10, T13, T15, T18, T20, T30, T40, and T55). NS3h was incubated at 37 °C with the ssDNA followed by rapid mixing with a solution containing heparin (4 mg/mL), and the intrinsic tryptophan fluorescence of NS3h was measured over time (Figure 2B). Heparin serves as a protein trap to prevent the helicase from rebinding the ssDNA after the dissociation event. It also prevents any unbound NS3h in solution at the start of the reaction from binding to the ssDNA after the reaction begins. The observed rate of dissociation (kd,obs), which encompasses the dissociation constant from internal positions along the ssDNA and the dissociation of the enzyme from the end of the oligonucleotide, is obtained by fitting the observed change in fluorescence to a single exponential (Figure 2C). If the enzyme diffuses along ssDNA, then it will dissociate from the end of the shorter DNA sequences faster than it dissociates from longer DNA sequences, which is exactly what we observed (Supplemental Figure 1).
The observed rate of dissociation (kd,obs) decreased as the length of DNA increased up to ~ 20 nt (Figure 2D). The relationship between the diffusion rate and observed dissociation rate has been described by others26 and can be evaluated according to the following equation:
| Equation 1 |
In this equation, kd,obs is the observed dissociation rate, ks is the rate of diffusional sliding along the ssDNA, and L is the length of the oligonucleotide. This equation describes the relationship between the kd,obs and the ks as a function the distance the enzyme travels along the substrate adjusted for a random initial binding site (Figure 2A). The model in Figure 2A shows diffusional sliding from only one end of the ssDNA, but diffusion can occur in either direction, according to the rate constants ks and ks−1.
The rate of diffusional sliding, ks, was determined by plotting the kd,obs according to Equation 1 and fitting the data to a linear regression (Figure 2E). The observed dissociation rates for oligonucleotide of lengths 8, 10, 13, 15, and 18 were used in the analysis (Figure 2D), resulting in a ks of 30 nt/s for NS3h. Longer oligonucleotides were not used in this analysis because dissociation was essentially independent of length for > 18 nt. For comparison, rates of 1–46 nt/s have been reported for ATP-dependent translocation by NS3h.18,31,32
The more rapid dissociation constant from shorter oligonucleotides could be due simply to weaker binding to the shorter oligos, as reported previously, rather than diffusional sliding from the ends of the ssDNA. Levin and Patel33 measured the affinity for oligonucleotides dT7, dT10, dT12, dT15, and dT20, with Kd values of 47, 7.4, 0.4, 0.5, and 0.9 nM, respectively. Hence, the shortest oligonucleotide binds with weaker affinity, but the others bind with similar affinity. Therefore, we further tested the idea that NS3h might be sliding on ssDNA by placing a protein block into the ssDNA path.
NS3h ATP-independent unwinding requires diffusional sliding
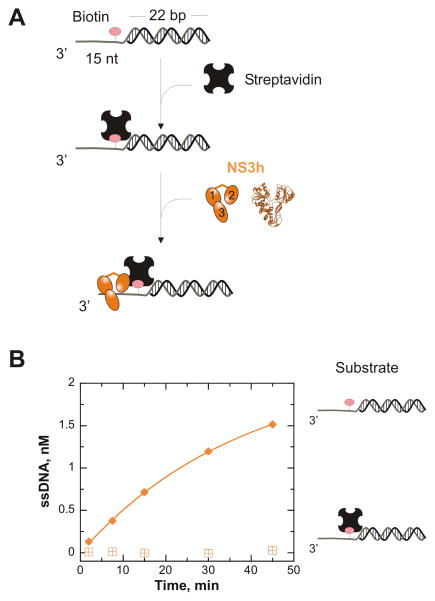
The previous results lead us to hypothesize that NS3h utilizes the ssDNA tail to bind and then slide into the ss/dsDNA junction. Once the enzyme reaches the ss/dsDNA junction, the specific interaction with the junction may lead to melting of a few base pairs.22 To test this idea, a substrate was designed that could block diffusion along the ssDNA, but not necessarily block direct binding to the ss/dsDNA junction. A biotin label was placed into the ssDNA so that a streptavidin block could be placed on the ssDNA tail (Figure 3A). A substrate (2 nM) with a biotin-dT analogue at position 12 of a 3′-T15 tail (3 nt 3′ to the ss/ds junction) attached to a 22 bp substrate (T15(bio·dT-12)-22 bp; see Table 1) was pre-incubated with streptavidin (120 nM) to place a protein block just ahead of the ss/ds junction of the 3′-tailed DNA substrate.
Figure 3.
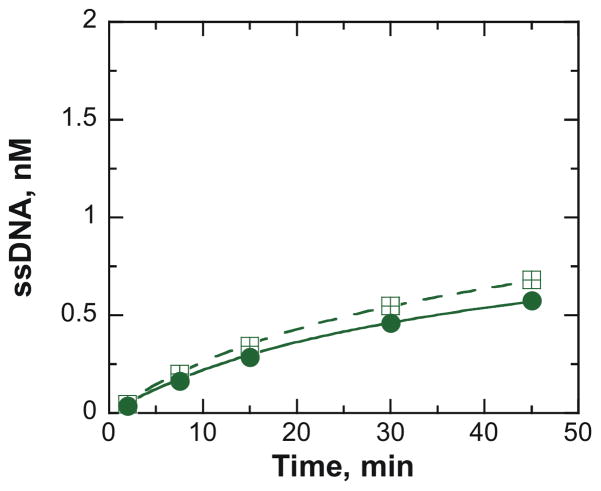
ATP-independent unwinding by NS3h is halted by the presence of a protein block that prevents sliding into the duplex. A. The streptavidin block was created by incubating 2 nM DNA substrate containing a biotin-thymidine analogue (pink) at position 12 with 120 nM streptavidin (black tetramer) at 37°C prior to initiating the ATP-independent unwinding reaction. The streptavidin-blocked DNA substrate was mixed with 500 nM NS3h (orange) to initiate the ATP-independent unwinding reaction. B. The presence of streptavidin (
 ) halted NS3h unwinding of the biotinylated substrate, T15(bio·dT-12)-22 bp. NS3h was able to unwind this substrate in the absence of streptavidin (
) halted NS3h unwinding of the biotinylated substrate, T15(bio·dT-12)-22 bp. NS3h was able to unwind this substrate in the absence of streptavidin (
 ).
).
The streptavidin-blocked substrate was then subjected to ATP-independent unwinding by NS3h (500 nM) to determine the reliance of diffusional sliding into the duplex on the unwinding mechanism. The presence of the streptavidin block completely eliminates ATP-independent unwinding of this substrate by NS3h (Figure 3B). The presence of excess streptavidin in the unwinding reaction does not interfere with the ability of NS3h to unwind the substrate in the absence of the biotin-thymidine analogue (Supplemental Figure 2). It is possible that streptavidin might prevent binding of NS3h to the DNA substrate. However, KMnO4 footprinting of NS3h on T15(bio·dT-12)-22 bp in the presence and absence of streptavidin indicates that the enzyme does bind to the substrate (Supplemental Figure 3). Therefore, the observed cessation of DNA melting in the presence of the protein block is not due to the lack of bound enzyme molecules. Results here illuminate the possibility of a previously unknown feature for this enzyme: diffusional sliding along ssDNA into the duplex in the absence of nucleotide binding and hydrolysis. Directionally biased binding to the ss/ds DNA junction can then stabilize melted base pairs at the junction.
NS3 unwinding does not display strict polarity in the absence of ATP
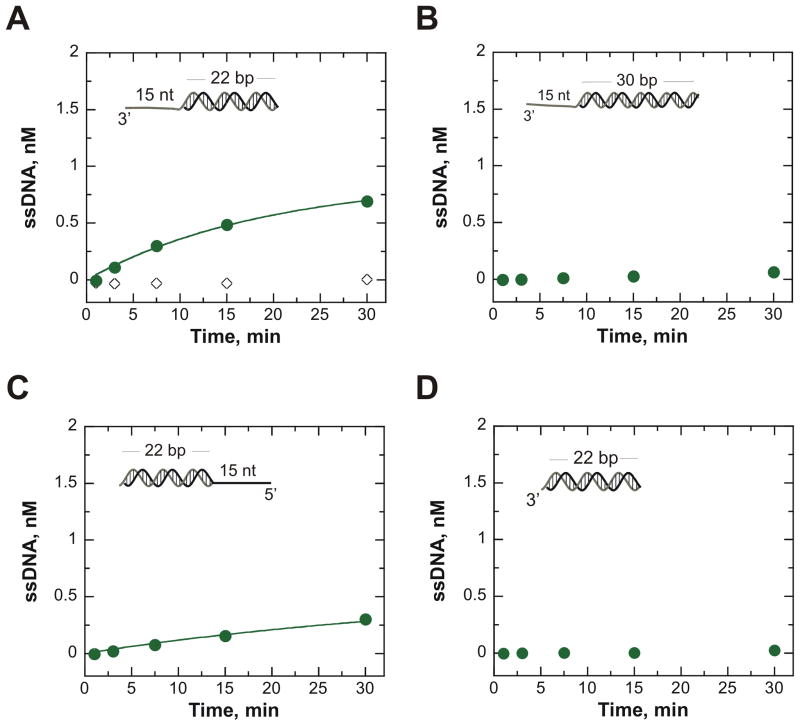
It is well-established that full-length NS3 does not always behave as NS3h.18,32,34 To investigate the effect of nucleic acid binding on the mechanism of unwinding by NS3, a series of similar experiments were conducted using the same conditions as for NS3h. Under conditions of excess enzyme concentration (500 nM NS3 and 2 nM DNA), ATP-independent unwinding of a substrate with a 15 thymidine 3′-tail adjacent to the 22 bp mixed base duplex region was observed for NS3 (Figure 4A). NS3 did not unwind the substrate in the presence of poly(U) confirming that the protein trap is effective (Figure 4A).
Figure 4.
ATP-independent unwinding of DNA substrates by NS3 does not display strict directional bias and is reduced by increasing the length of the duplex. ATP-independent unwinding was initiated by mixing 500 nM NS3 with 2 nM DNA at 37 °C. A. The DNA substrate, T15-22 bp, featuring a ssDNA 3′-tail was unwound by NS3 (
 ). The data was fit to a single exponential resulting in an observed rate constant of 0.05 ± 0.01 min−1. To ensure the efficiency of the protein trap, 100 μM nt poly(U) was pre-incubated with the DNA then mixed with enzyme (◇) to begin the unwinding reaction. B. NS3 was unable to unwind a DNA substrate featuring a duplex length of 30 bp (
). The data was fit to a single exponential resulting in an observed rate constant of 0.05 ± 0.01 min−1. To ensure the efficiency of the protein trap, 100 μM nt poly(U) was pre-incubated with the DNA then mixed with enzyme (◇) to begin the unwinding reaction. B. NS3 was unable to unwind a DNA substrate featuring a duplex length of 30 bp (
 ). C. NS3 was able to unwind a DNA substrate containing a 5′-tail adjacent to the 22 bp duplex (22 bp-T15) (
). C. NS3 was able to unwind a DNA substrate containing a 5′-tail adjacent to the 22 bp duplex (22 bp-T15) (
 ). This data was fit to a single exponential resulting in an observed rate constant of 0.02 ± 0.01 min−1. D. NS3 was unable to unwind a blunt-ended DNA substrate comprised of only the 22 bp duplex (
). This data was fit to a single exponential resulting in an observed rate constant of 0.02 ± 0.01 min−1. D. NS3 was unable to unwind a blunt-ended DNA substrate comprised of only the 22 bp duplex (
 ).
).
To determine if the directionality exhibited by NS3h is also present for NS3, the substrate featuring a 5′-tail attached to a 22 bp duplex (22 bp-T15) was investigated. Surprisingly, NS3 was able to separate this 5′-tailed substrate (Figure 4C), albeit at a slower rate and with a lower unwinding amplitude than what was observed on a comparable substrate containing a 3′-tail (Figure 4A). This result indicates that NS3 exhibits a different mechanism than NS3h for ATP-independent unwinding of duplex DNA. Interestingly, the addition of 5 mM ATP with the presence of 10 mM MgCl2 completely abolishes this apparent 5′ → 3′ unwinding activity by NS3, and no product formation can be detected (Supplemental Figure 4). Hence, the 3′ → 5′ directionality for translocation that is driven by ATP hydrolysis prevents the melting reaction with the 5′-tailed substrate.
Similar to NS3h, NS3 could not unwind a substrate made up of a blunt-ended 22 mixed base duplex in the absence of a ss tail region during a one hour reaction time (Figure 4D). This data supports prior reports of the requirement of a ss region for these enzymes to load onto before unwinding can occur.23,28
We suggest that NS3 binding directly to the duplex, directed by protein-protein interactions, can account for the observed “reverse” in directionality in the ATP-independent unwinding reaction. This conclusion is similar to the reported unwinding mechanism for other DExD/H-box and DEAD-box proteins in which strict directionality is also not observed.35–39
NS3 ATP-independent unwinding demonstrates a length dependency
Length-dependence of this ATP-independent activity for NS3 was examined by measuring unwinding of a substrate with an extended duplex region of 30 bp adjacent to the 15 thymidine 3′-tail region (T15-30 bp). In contrast to NS3h, NS3 was unable to unwind a measurable amount of the DNA substrate containing the 30 bp duplex in the absence of ATP (Figure 4B).
It has been reported that unwinding of this substrate requires only two kinetic steps by NS3 in the presence of ATP.23,40 The T15-30 bp is long enough to remain at least partially in native duplex form even if first 18–20 bp are unwound, which is the reported size of a single kinetic step size for NS3.23,40. It is possible that large kinetic step size relates in some manner to the binding site size of the oligomeric form of NS3.
NS3 ATP-independent unwinding of DNA substrates is not blocked by a streptavidin bound to biotin in the ssDNA tail
To explore the mechanism of NS3 mediated melting of the duplex, the substrate containing biotin-streptavidin block was utilized. Surprisingly, NS3 melted the T15(bio·dT-12)-22 bp similarly in the presence and absence of the streptavidin block with little or no observed decline in product formation (Figure 5). NS3 is able to bind to this substrate in the presence or absence of streptavidin (Supplemental Figure 6). This result indicates that diffusion does not play an important role in the ATP-independent unwinding mechanism of full-length NS3. Furthermore, it suggests that the observed ATP-independent unwinding reaction may be mediated by NS3 oligomers that can bind to the ssDNA and then extend past the protein block and interact directly with the duplex due to NS3-NS3 interactions (see discussion). This result is consistent with the model whereby NS3 is capable of interacting with both the tail region and the duplex portion of a substrate to locally unwind the duplex.41,42
Figure 5.
ATP-independent unwinding by NS3 is unaffected by the presence of a large protein block. The reaction was conducted as shown in Figure 3A. The ATP-independent unwinding reaction was initiated by mixing the streptavidin-blocked DNA substrate with 500 nM NS3. The presence of streptavidin (
 ) did not stop NS3 unwinding T15(bio·dT-12)-22 bp. In the absence of the streptavidin block, NS3 unwinds this substrate (
) did not stop NS3 unwinding T15(bio·dT-12)-22 bp. In the absence of the streptavidin block, NS3 unwinds this substrate (
 ) similarly to the substrate in the absence of the biotin-thymidine analogue (Figure 4A).
) similarly to the substrate in the absence of the biotin-thymidine analogue (Figure 4A).
DISCUSSION
Experiments reported here were designed to probe the interaction between these enzymes and nucleic acids and the role of protein-protein interactions in the overall unwinding mechanism. NS3h was found to diffuse along ssDNA, then engage the ss/dsDNA junction from the 3′-direction. In the presence of excess enzyme concentration, NS3h melted the duplex in a directionally biased manner, even in the absence of ATP. Conversely, full-length NS3 melting was not completely biased in the 3′-to-5′ direction. We propose that the oligomeric structure of NS3 can bind directly to the ssDNA then guide additional protein molecules to the duplex to melt and separate the strands.
NS3h
While the ATP-driven directional movement on ssDNA is certainly a key feature of helicase activity, the protein-nucleic acid interactions are also critical. ATP-independent unwinding of DNA emphasized the role of protein-nucleic acid interactions in the melting step of the reaction. The simple idea for DNA melting under these conditions is that ssDNA forms at the ss/dsDNA junction due to thermal fraying and can be bound by NS3h. The surprising event in this reaction is that NS3h unwinds the duplex with a directional bias. A previous report showed that NS3h binds more tightly to a ss/dsDNA junction compared to ssDNA alone.22 The same study suggested that the energy required for duplex separation is provided by NS3h binding to the nucleic acid substrate and not by ATP binding and hydrolysis.
The inability for NS3h or NS3 to melt the blunt-ended duplex indicated that simple sequestration of thermally melted duplex was insufficient for unwinding under the conditions examined here. The requirement of a 3′-ssDNA tail led to the idea that NS3h binds to the ssDNA tail, and then slides into the ss/dsDNA junction. This idea was tested by placing a biotin label on the ssDNA so that in the presence of streptavidin, a block was present that prevents diffusional sliding into the junction. The presence of this protein block completely eliminated ATP-independent melting of the DNA substrate (Figure 3). The presence of streptavidin did not prevent binding of NS3 to the substrate. Therefore, the protein block likely prevents diffusion into the duplex region.
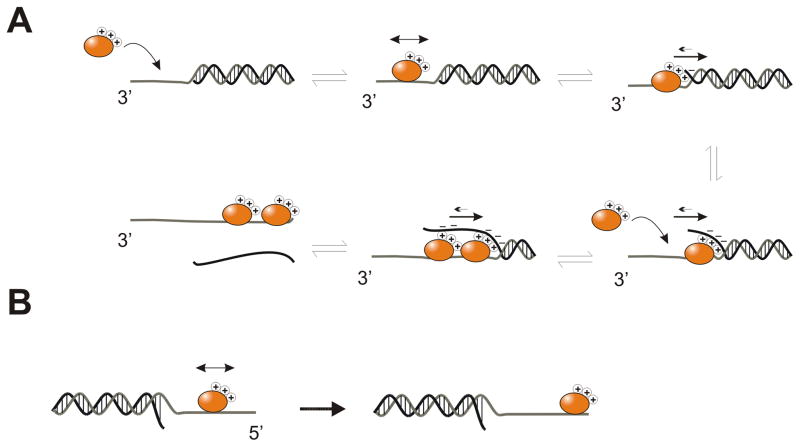
The strong directional bias for DNA unwinding in the absence of ATP, with the known interaction between NS3h and a ss/dsDNA junction, leads us to propose the model in Figure 6. NS3h binds to the ssDNA tail, and the protein can diffuse off of the 3′-end of the ssDNA or it can slide into the ss/dsDNA junction. Specific interactions between the displaced strand of the duplex strengthen the interaction between NS3h and the DNA. These interactions are depicted by the charge symbols in the diagram, but could be stacking or other interactions. Due to this new interaction, the direction of diffusion is now biased due to the additional interaction between one face of the enzyme and the displaced strand. Additional molecules of NS3h can bind to the ssDNA. These molecules can further add to the directional bias in diffusion by preventing the lead molecule from sliding back towards the 3′-end. Thus, upon encountering the junction from the 3′-side (Figure 6A), NS3h interacts specifically with the junction, melting 1–2 base pairs at a time, as reported previously 22. The asymmetric interactions at the junction lead to a 3′-to-5′ biased DNA unwinding reaction, even in the absence of ATP.
Figure 6.
Model for DNA unwinding by NS3h in the absence of ATP. A. NS3h binds a substrate and can diffuse along the ssDNA tail region (indicated with a black arrow). Upon engaging the ss/ds junction, additional interactions occur that add stability to the complex, thereby favoring this species. Additional NS3h molecules can also bind to the cleared ssDNA tail. The trailing protein can also favorably interact with the displaced strand, leading to diffusion with an apparent directional bias. This results in the separating of base pairs in the 3′ → 5′ direction and eventually full strand separation. B. NS3h bound the 5′-tail does not form specific interactions with the ss/ds junction, therefore it is unable to unwind a DNA substrate containing a 5′-tail.
Alternatively, if NS3h binds to a substrate containing only a 5′-ssDNA tail (Figure 6B), the favorable interaction between the protein and the ss/dsDNA junction cannot occur because the incorrect surface of NS3h faces the junction. Hence, the protein simply dissociates rather than sequestering ssDNA to separate the duplex. This mechanism emphasizes the importance of the specific interaction between NS3h and a properly-oriented ss/dsDNA junction, which is also likely to be important in the overall ATP-dependent DNA unwinding reaction.
Identification of the specific residues of NS3h that are responsible for the interaction at the junction begins with examination of the structure of the enzyme. A large beta hairpin is believed to serve as a wedge that helps separate the two DNA strands by forming favorable interactions with the DNA.22,43,44 Indeed, mutagenesis of the Phe444 within this wedge uncouples ATP driven DNA unwinding activity.44
The data for diffusion on ssDNA (30 nt2/s) can be compared to the data for ATP-dependent translocation on ssDNA. Several reports have measured ATP-dependent translocation. The short range of oligonucleotide lengths over which a change in kd,obs is measured has been shown in the presence of ATP and is likely a result of limited processivity per binding event.18 Rates of 1–46 nt/s have been reported for NS3h for ATP-dependent translocation.18,31,32
NS3
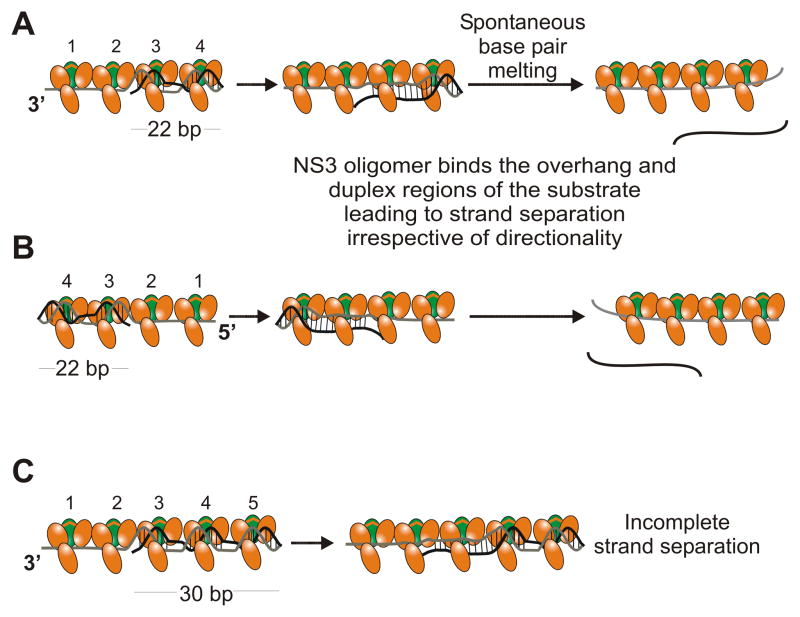
In the absence of ATP, NS3 appears to unwind DNA substrates by a mechanism that does not utilize diffusional sliding and is therefore translocation-independent. NS3 unwinds DNA duplexes via local strand separation in the absence of ATP (Figure 7). This mechanism is similar to the unwinding mechanism utilized by a number of DEAD-box proteins, which is unlike the translocation-dependent canonical mechanism of unwinding.35–39 We propose a model in which NS3 binds the ss extension portion of a partial-duplex substrate and loads additional molecules directly onto the duplex region due to protein-protein interactions, leading to separation of the strands in an ATP-independent manner. This protein binding and alignment between NS3 and DNA substrates was indicated by DNA footprinting experiments.41 This interaction in the duplex not only leads to the separation of 3′-tail-containing substrates, but can also lead to the unwinding of 5′-tail-containing substrates. This suggests that once a molecule of NS3 binds to ssDNA, near a ss/dsDNA junction, the other NS3 molecules can interact with the duplex correctly and separate the strands (Figure 7). The decrease in unwinding efficiency observed on the DNA substrate with the 5′-tail (Figure 4C) is likely due to the binding polarity displayed by NS3 and the lack of appropriate interaction with the ss/dsDNA junction. Strand separation by NS3 is limited by the duplex length further indicating a different mechanism for NS3 compared to NS3h. We suggest that diffusion of NS3 is limited due to the oligomeric nature of the enzyme, which limits the length of duplex that can be melted in the absence of ATP. Diffusional sliding was recently investigated and found to be important for NS3.45 However, these investigators examined RNA rather than DNA. NS3 is capable of melting both RNA and DNA, but clearly differences can be observed for each substrate.
Figure 7.
ATP-independent unwinding of DNA substrates is mediated by an oligomeric form of NS3. A. Multiple molecules of NS3 (protease domain shown in green, helicase domain in orange) are capable of binding both the ssDNA tail and duplex regions of a substrate.41 Preferential binding of NS3 to the junction properly aligns the oligomer to optimally interact with the duplex leading to direct separation of base pairs in the duplex. NS3 is capable of disrupting a sufficient number of base pairs such that duplex lengths of 22 bp can separate at 37 °C. B. On a substrate containing a 5′-tail, protein-protein interactions guide NS3 to bind to the duplex region, after initial binding to the ssDNA, which leads to melting of the duplex. C. ATP-independent unwinding of DNA substrates by NS3 oligomer is length-dependent. Slightly longer duplexes are sufficiently stable to remain intact despite binding and melting of some of the dsDNA.
Our results point to an intriguing diffusion property shared by some helicases, but not others. Helicase unwinding assays are typically performed by incubating the helicase with the DNA or RNA substrate prior to addition of ATP. During the incubation time, some helicases assemble into a functional complex.46 Other helicases appear to achieve functional status more rapidly such as the case with Dda, 47 Pif1,48 and PcrA,49 for example. For those enzymes that assemble rapidly, it is curious that little or no melting of short duplexes occurs during the incubation period. This observation implies that these enzymes may not diffuse or may not recognize the ss/ds junction in the same manner as NS3h. Lack of diffusion may be due to the nature of the interactions between helicases and nucleic acid. We suggest that enzymes that do not diffuse readily contain binding sites in which one or more bases are “flipped” out of the normal, stacked position, into a specific helicase binding site. In the case of PcrA, base flipping clearly occurs.50 Base flipping is also proposed to occur in Dda.51 No evidence for base flipping exists in the case of NS3h, despite numerous x-ray crystallographic structural studies.15,16 Use of ATP-independent unwinding assays may be a simple way to distinguish enzymes that readily diffuse from those that do not. Recent work from the Lohman laboratory characterized diffusion of the human SSB, hRPA. Interestingly, hRPA uses diffusion to unwind duplex DNA more efficiently from ssDNA on the 5′ side of a hairpin.52
In summary, we report a surprising property of NS3h: directionally biased melting of dsDNA in the absence of ATP. This result emphasizes the importance of specific interactions between the enzyme and the ss/dsDNA junction. Bi-directional diffusion of NS3h on ssDNA appears to be rapid, and contributes to the observed melting.
Supplementary Material
Acknowledgments
Funding
Funding for this work was provided by National Institutes of Health grant R01 GM089001 to K.D.R and C.E.C.
ABBREVIATIONS
- HCV
hepatitis C virus
- NS3
nonstructural protein 3
- NA
nucleic acids
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA
- β-ME
β-mercaptoethanol
- BSA
bovine serum albumin
- SDS
sodium dodecyl sulfate
Footnotes
The authors declare no competing financial interest.
SUPPORTING INFORMATION FOR PUBLICATION
Supporting experiments include ATP-dependent unwinding of a DNA substrate with a 5′-ssDNA overhang, ATP-independent unwinding of both NS3 and NS3h on a DNA substrate not featuring a biotinylated analog to gauge the effect of excess streptavidin on the unwinding process, and KMnO4 footprinting of the biotinylated DNA substrate in the presence and absence of streptavidin to show NS3h binding to the substrate under unwinding conditions. This material is available free of charge via the internet at htts://pubs.acs.org.
References
- 1.Jarmoskaite I, Bhaskaran H, Seifert S, Russell R. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc Natl Acad Sci U S A. 2014;111:E2928–E2936. doi: 10.1073/pnas.1404307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci (Landmark Ed) 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 7.Raney KD, Sharma SD, Moustafa IM, Cameron CE. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beran RK, Serebrov V, Pyle AM. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 9.Errington W, Wardell AD, McDonald S, Goldin RD, McGarvey MJ. Subcellular localisation of NS3 in HCV-infected hepatocytes. J Med Virol. 1999;59:456–462. doi: 10.1002/(sici)1096-9071(199912)59:4<456::aid-jmv6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 11.Bayliss CD, Smith GL. Vaccinia virion protein I8R has both DNA and RNA helicase activities: implications for vaccinia virus transcription. J Virol. 1996;70:794–800. doi: 10.1128/jvi.70.2.794-800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seybert A, Hegyi A, Siddell SG, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CC, Huang ZS, Chiang PL, Chen CT, Wu HN. Analysis of the nucleoside triphosphatase, RNA triphosphatase, and unwinding activities of the helicase domain of dengue virus NS3 protein. FEBS Lett. 2009;583:691–696. doi: 10.1016/j.febslet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, Brendza KM, Somoza JR. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 16.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopal V, Patel SS. Single strand binding proteins increase the processivity of DNA unwinding by the hepatitis C virus helicase. J Mol Biol. 2008;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 20.Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Jr, Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat Struct Mol Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 23.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 24.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 26.Young MC, Kuhl SB, von Hippel PH. Kinetic theory of ATP-driven translocases on one-dimensional polymer lattices. J Mol Biol. 1994;235:1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- 27.Morris PD, Byrd AK, Tackett AJ, Cameron CE, Tanega P, Ott R, Fanning E, Raney KD. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5′-biotinylated oligonucleotides but not from 3′-biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 28.Beran RK, Bruno MM, Bowers HA, Jankowsky E, Pyle AM. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J Mol Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 29.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 30.Tomko EJ, Fischer CJ, Lohman TM. Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods. 2010;51:269–276. doi: 10.1016/j.ymeth.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khaki AR, Field C, Malik S, Niedziela-Majka A, Leavitt SA, Wang R, Hung M, Sakowicz R, Brendza KM, Fischer CJ. The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties. J Mol Biol. 2010;400:354–378. doi: 10.1016/j.jmb.2010.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matlock DL, Yeruva L, Byrd AK, Mackintosh SG, Langston C, Brown C, Cameron CE, Fischer CJ, Raney KD. Investigation of translocation, DNA unwinding, and protein displacement by NS3h, the helicase domain from the hepatitis C virus helicase. Biochemistry. 2010;49:2097–2109. doi: 10.1021/bi901977k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levin MK, Patel SS. Helicase from hepatitis C virus, energetics of DNA binding. J Biol Chem. 2002;277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- 34.Frick DN, Rypma RS, Lam AM, Gu B. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem. 2004;279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Potratz JP, Tijerina P, Del CM, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jankowsky E. RNA helicases at work: binding and rearranging. Trends in Biochemical Sciences. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q, Del CM, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Serebrov V, Beran RK, Pyle AM. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J Biol Chem. 2009;284:2512–2521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raney VM, Reynolds KA, Harrison MK, Harrison DK, Cameron CE, Raney KD. Binding by the hepatitis C virus NS3 helicase partially melts duplex DNA. Biochemistry. 2012;51:7596–7607. doi: 10.1021/bi300654v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng W, Dumont S, Tinoco I, Jr, Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci U S A. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 44.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Cheng W, Bustamante C, Oster G. Coupling translocation with nucleic acid unwinding by NS3 helicase. J Mol Biol. 2010;404:439–455. doi: 10.1016/j.jmb.2010.09.047. [DOI] [PubMed] [Google Scholar]
- 46.Ding SC, Kohlway AS, Pyle AM. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J Virol. 2011;85:4343–4353. doi: 10.1128/JVI.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci U S A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J Biol Chem. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chisty LT, Toseland CP, Fili N, Mashanov GI, Dillingham MS, Molloy JE, Webb MR. Monomeric PcrA helicase processively unwinds plasmid lengths of DNA in the presence of the initiator protein RepD. Nucleic Acids Res. 2013;41:5010–5023. doi: 10.1093/nar/gkt194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 51.He X, Byrd AK, Yun MK, Pemble CW, Harrison D, Yeruva L, Dahl C, Kreuzer KN, Raney KD, White SW. The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation. Structure. 2012;20:1189–1200. doi: 10.1016/j.str.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen B, Sokoloski J, Galletto R, Elson EL, Wold MS, Lohman TM. Diffusion of Human Replication Protein A along Single-Stranded DNA. J Mol Biol. 2014;426:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.