Abstract
IL-6 is a cytokine that is involved in various physiological and pathological conditions, and approaches using gain-of-function transgenic animals have contributed in elucidating IL-6 function. However, studies of the multiple functions of IL-6 in vivo are very time consuming because they require the generation of transgenic mice that harbor the gene encoding IL-6 under the control of specific promoters to mimic different pathologies. Here, we report the establishment of a conditional human IL-6 transgenic mouse, LGL-IL6, which conditionally expresses human IL-6 by taking advantage of the well-characterized Cre recombinase drivers.
Keywords: conditional transgenic mouse, Cre-loxP system, IL-6
Introduction
IL-6 is among the major cytokines that mediate inflammation. IL-6 plays important roles in acute inflammation and in various physiological and pathological conditions, such as aging, obesity, diabetes, osteoporosis, rheumatoid arthritis, osteoarthritis, sepsis, cognitive function, and fibrosis [1, 2, 6,7,8, 10, 20]. The IL-6 signal is transduced by its receptors IL-6R and gp130 and modulates downstream gene expression, such as Socs3, by phosphorylating STAT3. Of clinical importance, the intervention of the IL-6 signaling has been already applied to treatments for rheumatoid arthritis and Castleman’s disease [9, 13].
Transgenic approaches for studying the in vivo function of IL-6 have been conducted during the last three decades. The earliest reports found that transgenic expression driven by the immunoglobulin heavy-chain enhancer or the major histocompatibility complex class-I promoter induces IgG1 plasmacytosis or plasmacytomas, respectively [17, 18]. The overexpression of IL-6 in skin by the keratin 14 promoter induces the thickening of the stratum corneum but not inflammation of the skin [19]. Moreover, multiple pathogenic alterations occur in mice that express IL-6 through glial fibrillary acidic protein or neuron-specific enolase gene promoter to target glial cells or neurons, respectively [3, 5]. Humanized IL-6/IL-6 receptor system in mice has been more recently developed via the transgenic approach to demonstrate IL-6 pathology [21].
However, the establishment of a transgenic line with a defined promoter activity is labor intensive. Constitutive overexpression of a transgene typically occurs when the promoter is active, for example, during embryogenesis and/or perinatally. Moreover, because IL-6 is often pathogenic, the overexpression of IL-6 during embryogenesis may be lethal and, even they survived, it may result in selecting founders that keep the expression levels of the transgene as low as avoiding the lethality and maintaining the fertility for propagation. It is difficult to generate individual gain-of-function transgenic lines to study the multiple in vivo functions of IL-6 for each pathology. Here, we report the establishment of a transgenic line that conditionally expressed human IL-6. It enables to drive spatiotemporal expression of human IL-6 by appropriate Cre-dependent recombination.
Materials and Methods
Materials
The mouse hybridoma cell line 7-TD-1 was obtained from RIKEN Cell Bank (resource number RCB1190). Recombinant human IL-6 and tamoxifen were purchased from Wako (Osaka, Japan) and Sigma-Aldrich (St. Louis, MO, USA), respectively.
Construction of IL6-T2A-mCherry
To monitor the transgene expression by fluorescence in vivo, the human IL-6 coding sequence was fused to T2A-mCherry from pUbC-rLoxP-mChe [16]. The mCherry sequence was modified adding a signal sequence from the mouse proacrosin at the N-terminus for extracellular sorting and a consensus sequence for attaching a glycosylphosphatidylinositol (GPI)-anchor at the C-terminus to detect the fluorescence associated with the plasma membrane [16]. Because the self-cleaving T2A peptide was digested at the C-terminal site of the peptide, the resulting IL-6 peptide was extended with a 21-amino acid stretch (GRAGEGRGSLLTCGDVEENPG) by the cleavage. DNA fragments were fused using PCR and the InFusion HD Kit (Takara Bio, Shiga, Japan) combined with pEF4-mycHisA (Invitrogen, Carlsbad, CA, USA) to construct the expression plasmid pEF4-hIL6-T2A-mCherry. The sequence was confirmed by sequencing.
Transfection and fluorescence microscopy
HEK293T cells do not express detectable endogenous IL-6 or IL-6 receptor. HEK293T cells were transfected without (mock) or with pEF4-hIL6-T2A-mCherry using FuGENE HD Transfection Reagent (Promega, Madison, WI, USA), according to manufacturer’s protocol. Cells and cell culture supernatants were assayed 48 h after transfection. To estimate the protein level of the human IL-6, ELISA was performed as described bellow. To detect the expression of mCherry, the transfected cells were photographed using a BIOREVO fluorescence microscope (Keyence, Osaka, Japan).
Generation of LGL-IL6 mice
The CAG promoter [14] was used to drive the ubiquitous expression of the transgene. EGFP and SV40 polyadenylation signal sequences were inserted between the two loxP sites to block transcription through the IL6-T2A-mCherry sequence in the absence of Cre activity. DNA fragments were generated using PCR and combined using the InFusion Kit that are described above. Nucleotide sequence analysis was conducted to confirm the structure of the construct. The vector sequence will be provided on request.
A 5.4 kbp Dra III-Xho I fragment was isolated to remove the vector region and the purified DNA fragment was injected into fertilized pronuclear-stage eggs of C57BL/6NCrSlc (SLC, Shizuoka, Japan), and then healthy eggs were transplanted into recipients. Genomic PCR was performed to identify founders, and PCR was used to determine the genotypes of mice harboring the transgene using the primers as follows: LGL-EGFP-1F, 5′-CGACAAGCAGAAGAACGGCATCAAGGTG-3′ and LGL-EGFP-1R, 5′- CAAGCTGACCCTGAAGTTCATCTGCACC-3′.
Tamoxifen administration
Rosa-CreERT2 mice (B6.129-Gt (ROSA) 26Sortm1 (cre/ERT2) Tyj/J, Stock 008463) were obtained from the Jackson Laboratory [23]. Mice were mated to produce offspring that carried LGL-IL6 and Rosa-CreERT2 (Cre/tg), genotyped, and treated with tamoxifen that was dissolved in canola oil (20 mg/ml). Tamoxifen (100 mg/kg) was intraperitoneally injected into 4-week-old mice on days 0 and 7 and were sacrificed on day 10. Littermates of the Cre/tg mice harboring either the LGL-IL-6 or RosaCreERT2 transgene served as controls. Mice were housed in specific pathogen-free condition in the animal facility of National Center for Geriatrics and Gerontology (NCGG). All animal experiments were performed according to the protocols approved by the Ethics Committee for Animal Experimentation of NCGG.
Human IL-6 ELISA
An IL-6 ELISA kit (Human) was purchased from Diaclone (Besançon, France). Conditioned media prepared from supernatants of cultures of HEK293T cells were stored at −80°C until assay. The frozen medium was thawed and diluted 1:100,000 in PBS. The concentrations of human IL-6 in the diluted samples were measured using the ELISA kit, according to the manufacturer’s protocol. Mouse serum samples were diluted 1:5,000 in PBS for analysis. Each sample was tested at least in triplicate, and the values were obtained from mice of each genotype (n=4 per group).
IL-6 bioassay
To measure the specific activity of human IL-6 that was expressed by hIL6-T2A-mCherry, we performed a bioassay using the IL-6 dependent 7-TD-1 cell line [22]. Serial dilutions of the conditioned medium from mock, pEF4-hIL6-T2A-mCherry-transfectants, or recombinant hIL-6 were added to flat-bottom microwells. The supernatant from mock-transfected cells served as the negative control. 7-TD-1 cells (5 × 103) were added to each well and incubated for 72 h at 37°C. Cell proliferation was determined using a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan), and specific activity was determined by the relative values to the protein content. One unit was calculated as 106/ED50 of the culture media or commercially available recombinant IL-6, in which a half-maximal proliferation of 7-TD-1 cells was adopted.
Confocal imaging
Mouse kidney sections were visualized using an LSM 5 EXCITER confocal laser microscope (CarlZeiss, Oberkochen, Germany). Dual-color images were captured in a sequential manner. All images were obtained using a ×63 oil-immersion objective. Scanning was performed with a pinhole size of 1.0 airy unit and seventeen times line averaging. The images were stored in a 574 × 574-pixel, 12-bit TIFF file format.
SDS-PAGE and immunoblot analyses
Mouse serum proteins (0.3-µl serum equivalent) were subjected to SDS-PAGE under reducing conditions [11] and detected using CBB. For immunoblotting, serum (0.2-µl serum equivalent) or tissue proteins (10 µg) that were separated using SDS-PAGE were electrophoretically transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA) that was first immersed in the StartingBlock Blocking Buffer in TBS (Thermo Fisher Scientific, Waltham, MA, USA). Blots were probed with the following antibodies: pSTAT3 mAb (diluted 1:100,000; Abcam, Cambridge, UK), Stat3 pAb (diluted 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), β-actin DirecT (diluted 1:30,000; MBL, Aichi, Japan), or mouse IgG (diluted 1:20,000; GE Healthcare, Chicago, IL, USA) using ImmunoStar LD or Zeta (Wako). Blots were imaged using the LAS 4000 Imaging System (Fujifilm, Tokyo, Japan).
Real-time PCR
Total RNA was isolated using an miRNeasy Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. Total RNA was treated with RNase-free DNase (QIAGEN) to eliminate possible contamination with genomic DNA. The template was synthesized using Superscript VILO (Thermo Fisher Scientific), and real-time PCR was performed using SYBR Premix Ex Taq II reagents (TOYOBO, Osaka, Japan) and a CFX96 Real-Time PCR Detection System (Bio-Rad). For the tissue distribution of the transgene expression, the copy number of the RNA was calculated using serial dilution of the amplicon and arbitrary units were given by the copy number of the transgene-specific mRNA per 1 ng of RNA. For the expression of the downstream genes for IL-6 signaling, the mean values of the expression in relative to those The primers (P1 and P2) specific for the human IL6 transgene were as follows: P1, 5′-GTAGACTCGACTAGCTTGGGCTG-3′ and P2, 5′-TGGGGCGGCTACATCTTTGGAATC-3′. Other primers (Supplementary Table I) were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast).
Statistical analysis
All data are expressed as mean ± SD. To compare mRNA levels expressed by the control vs. Cre/tg mice, statistical analysis was performed using a two-tailed unpaired Student’s t-test (StatFlex 6 software; Artech, Osaka, Japan).
Results
In vitro expression of the hIL-6-mCherry construct
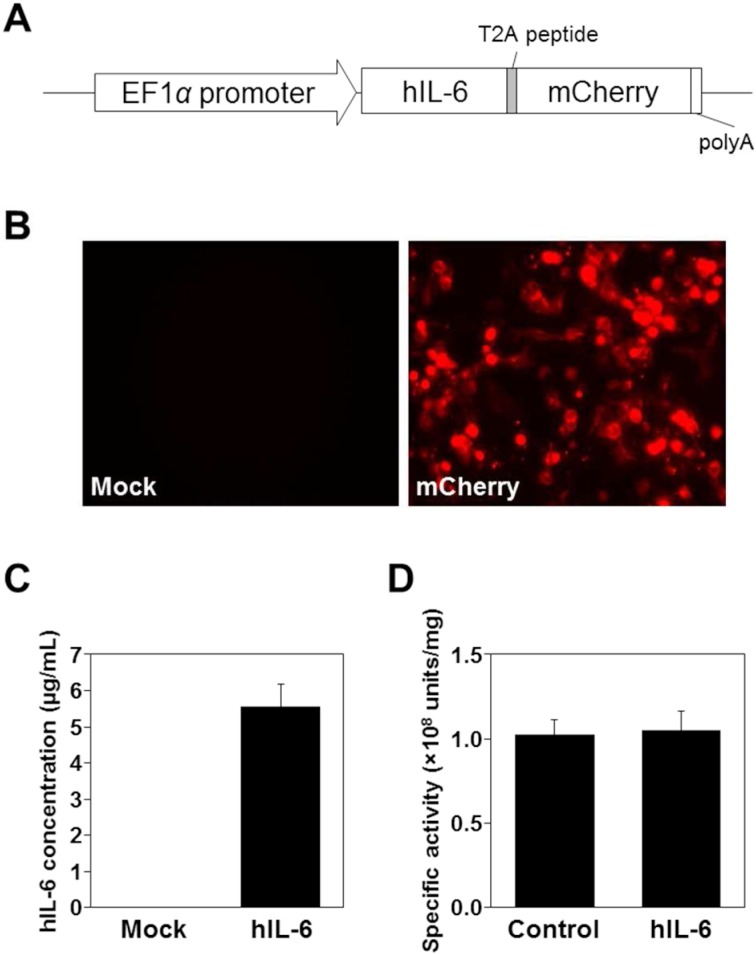
To enable monitoring transgene expression by visualizing the expressing cells with red fluorescence, human IL6 cDNA was fused to mCherry mediated by T2A, an autocleavable peptide in the cells (Fig. 1A). To distinguish the transgene product from the endogenous IL-6, human IL-6, which can activate mouse IL-6 signaling pathway, was used for this study. The mCherry was modified by addition of signal peptide and GPI-anchoring sequences at the N-terminal and C-terminal ends, respectively, to achieve cell surface expression. To confirm the validity of the construct, the IL6-T2A-mCherry unit was expressed in HEK293T cells. When pEF4-hIL6-T2A-mCherry was transfected in HEK293T cells, the red fluorescence was detected on the cells but not on mock-transfected cells (Fig. 1B). Using ELISA, the concentration of human IL-6 in the culture medium of the transfected cells was 5.6 µg/ml (Fig. 1C) but was undetectable in a medium that was harvested from cultures of the mock-transfected cells.
Fig. 1.
Expression of IL-6-T2A-mCherry. A, Structure of the expression unit in pEF4-hIL6-T2A-mCherry. B, Fluorescent images of mCherry in HEK293T cells transfected with empty vector (left panel, mock) or pEF4-hIL6-T2A-mCherry (right panel, mCherry). C, Expression of IL-6 proteins in the culture medium of the transfected cells. Concentration of IL-6 expressed in the culture medium was measured by ELISA. D, Specific activities of IL-6 proteins. By bioassay for IL-6 using a mouse hybridoma cell line, 7-TD-1, the specific activities of recombinant human IL-6 were compared (n=3). Control, biologically active recombinant human IL-6 (Wako); hIL-6, human IL-6 detected in the culture media of cells transfected with pEF4-hIL6-T2A-mCherry.
It has been reported that the C-terminal sequence of IL-6 was important for its activity [12]. Therefore, it was crucial to verify the activity of the present human IL-6 construct containing C-terminal extension of 21 amino acid residues that are derived from T2A sequence (Fig. 1A). The culture medium from the transfected 293T cells was subjected to bioassay of IL-6 using 7-TD-1 cells. As shown in Fig. 1D, the specific activity of the culture medium was comparable with that of a biologically active recombinant IL-6 (1.02 ± 0.09 vs. 1.05 ± 0.10 (108 units/mg)), suggesting that the human IL-6 expressed by IL6-T2A-mCherry unit possessed biological activity that was sufficient for use as a transgene in vivo.
Generation of transgenic mice that conditionally express hIL-6
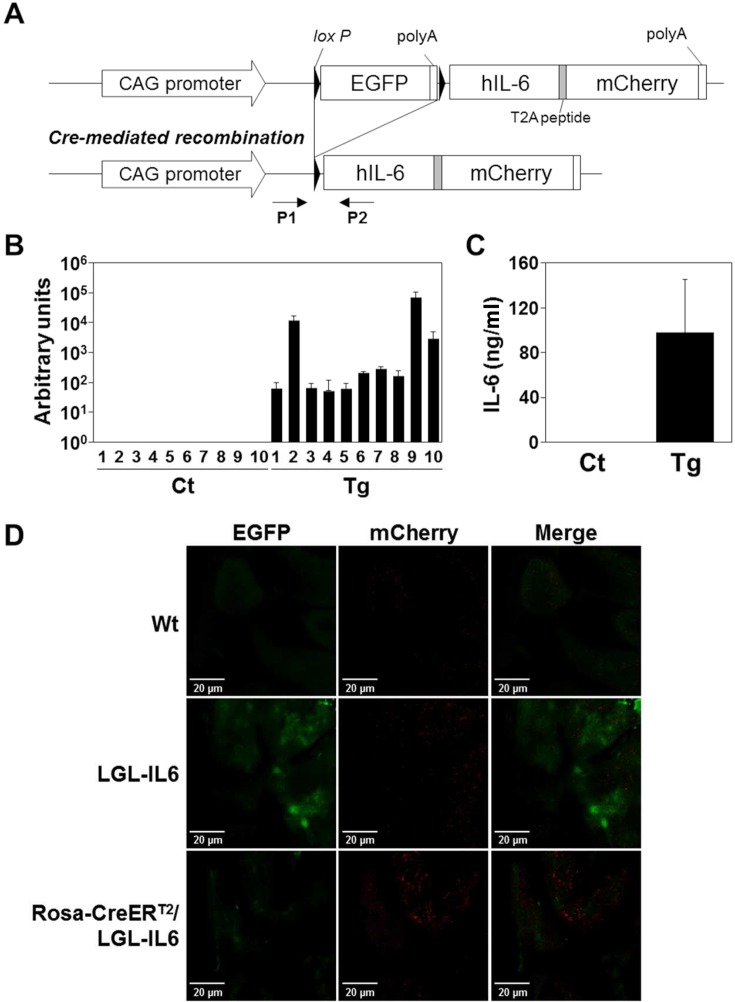
To achieve Cre-dependent conditional expression, a sequence encoding EGFP-polyA with flanking loxP elements was introduced upstream of the hIL-6-mCherry sequence (Fig. 2A). Of eight transgenic founders, two failed to propagate. Transgene expression, monitored by EGFP expression, was detected in the progeny of three lines. The expression of the transgene declined during the propagation in one of the lines, and the other two lines designated Li29 and Li56 were obtained as the conditional transgenic mice, LGL-IL6. Li29 expressed the transgene in all tissues tested, and thus, was subjected to further analysis.
Fig. 2.
Systemic expression of IL-6 in the LGL-IL6 transgenic mouse line. A, LGL-IL6 transgene (top). In the absence of Cre activity, only EGFP protein is expressed. Cre-mediated recombination deletes the floxed EGFP-polyA sequence, allowing the expression of IL6-T2A-mCherry. P1 and P2, PCR primers. B, Expression of IL6 mRNA in tissues. Mating LGL-IL6 with Rosa-CreERT2 mice generated progeny (Rosa-CreERT2/LGL-IL6) that systematically expressed IL-6 after treatment with tamoxifen. Arbitrary units were calculated as the estimated copy numbers of transgene-specific mRNA per 1 ng of total RNA. Ct, control mice including both Rosa-CreERT2 and transgenic (LGL-IL6) littermates; Tg, Rosa-CreERT2/LGL-IL6 mice. Both mice were treated with tamoxifen as described in the Materials and Methods. 1, brain; 2, heart; 3, lung; 4, liver; 5, kidney; 6, spleen; 7, stomach; 8, intestine; 9, skeletal muscle; and 10, skin. C, Serum IL-6 levels of Cre/tg mice. ELISA analysis of serum human IL-6 levels of control (Ct) and Cre/tg mice (Tg) treated with tamoxifen (n>4). D, Recombination mediated fluorescent protein expression.
Next, we mated Li29 with a Rosa-CreERT2 mouse to generate a progeny with tamoxifen-dependent systemic expression of the transgene. Following an intraperitoneal injection of tamoxifen, transgene expression was detected only in mice with the Rosa-CreERT2/LGL-IL6 (Cre/tg) genotype (Fig. 2B), and IL-6 expression was detected in all tissues tested. The administration of tamoxifen increased the serum IL-6 levels (~92 ng/ml) of the Cre/tg mice, whereas those of the control mice were undetectable (Fig. 2C), suggesting that the loxP-EGFP-polyA-loxP sequence prevented IL-6 expression by the transgene. With the same protocol, the massive death has been observed in Li56 mice that were compound heterozygotes of Rosa-CreERT2 (not shown).
To validate the system, fluorescent images of the tissues from the mice were analyzed. In Fig. 2D, green fluorescent originated by EGFP was observed in LGL-IL6 and Rosa-CreERT2/LGL-IL6 mice, but not in wild-type mice. The red fluorescence originated by mCherry was seen only in Rosa-CreERT2/LGL-IL6 mice, but not wild-type or LGL-IL6 transgenic mice, suggesting that the fluorescence switch by Cre mediated recombination has been confirmed in vivo.
Protein patterns in serum
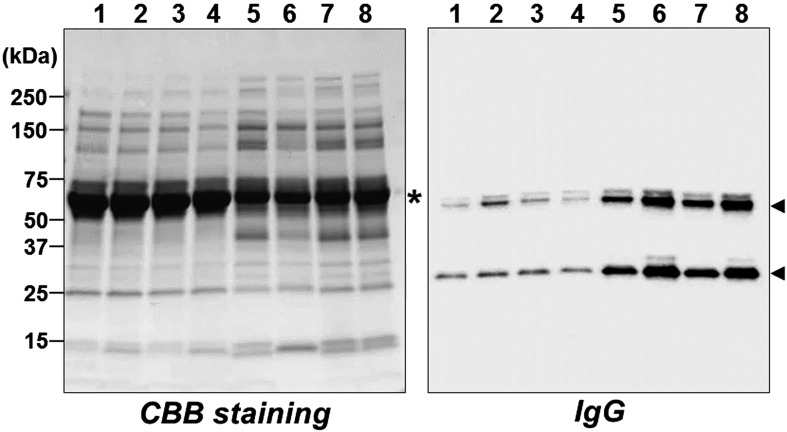
The marked elevation of IL-6 levels in the serum led to presume alteration of serum protein profile. As expected, significant alteration of protein patterns in serum was evident (left panel, Fig. 3). The level of serum albumin was also significantly decreased in Cre/tg mice, suggesting an abnormality in the physical condition of the transgenic mice that was presumably caused by the overexpression of IL-6. In contrast, IgG levels increased in Cre/tg mice (right panel, Fig. 3). When Li29 mice were injected with tamoxifen once every 2 weeks, some phenotypes, including splenomegaly, became pronounced (the detailed phenotype will be described elsewhere).
Fig. 3.
Serum proteins. Sera were prepared from tamoxifen-treated control and Rosa-CreERT2/LGL-IL6 (Cre/tg) mice. Left panel, SDS-PAGE analysis (CBB staining); right panel, immunoblot analysis to detect IgG using an anti-mouse IgG. Lanes 1–4, sera from control mice (1–2, Rosa-CreERT2; 3–4, LGL-IL6 mice); lanes 5-8, sera from Cre/tg mice. Arrowheads, the moieties corresponding to IgG heavy and light chains; asterisk, serum albumin.
IL-6 signaling in transgenic mice
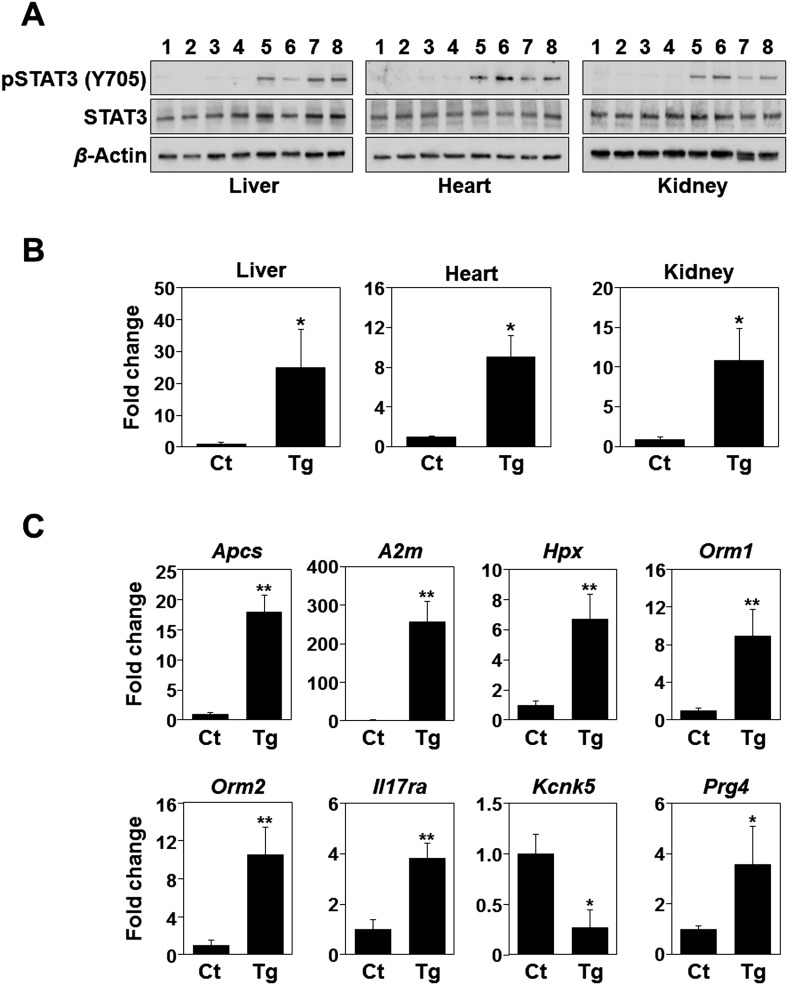
To verify the action of transgenic IL-6, phosphorylation levels of STAT3 were determined in the liver, heart, and kidney. As shown in Fig. 4A, the levels of STAT3 phosphorylation at Tyr705 in the tissues of Cre/tg mice were significantly higher compared with those of control mice. Next the expression of Socs3, a common target gene of the IL-6 signaling pathway, was analyzed. The expression of Socs3 was markedly increased in the tissues of Cre/tg mice comparing to those of control mice, suggesting that the tamoxifen-induced transgene expression evoked the expected responses in the tissues (Fig. 4B). In addition to the common signaling responses of IL-6, gene expression profile in liver was determined. Several IL-6 responsive genes in liver have been previously described [4, 15] and used as targets of real-time PCR. As shown in Fig. 4C, the changes in expression of the genes were consistent with the previous findings, well reflecting IL-6 actions in mouse liver [4, 15].
Fig. 4.
IL-6 responses in the tissues from Cre/tg mice. A, Phosphorylation of STAT3 proteins in mouse tissues. Phosphorylated STAT3 (pSTAT3 [Y705]), total STAT3 (STAT3), and β-actin proteins were detected by immunoblotting. Lanes 1–4, control mice (1–2, Rosa-CreERT2; 3–4, LGL-IL6 mice); lanes 5–8, Cre/tg mice. Tissue sources were as indicated. B, Real-time PCR analysis of Socs3 expression. Socs3 expression levels are shown as fold-changes of Cre/tg mice (Tg) compared with those of control mice (Ct) consist of two Rosa-CreERT2 and two LGL-IL6 genotypes. C, Expression of IL-6-responsive genes in the mouse liver. *P<0.05; **P<0.001 (t-test, n=4).
Discussion
Here, we report the systemic and tamoxifen-dependent expression of human IL-6 by the progeny of an IL-6 transgenic mouse line mated with a Rosa-CreERT2 transgenic mouse. Even additional 21 amino acid stretch was appended at the C-terminal end, the IL-6 originated from the transgene unit was biologically active and well reproduced the responses previously reported.
The construct was designed to undergo Cre-mediated recombination leading to the expression of mCherry instead of EGFP [16]. The transgene seemed to present in tandem with multiple copies in the chromosome of the line, Li29 (not shown). Theoretically, at least one copy of the IL-6-mCherry unit tends to be activated by Cre mediated recombination, as evidenced by the gene and protein expression, even existed as tandem, although variation could be existed. In fact, some EGFP expression was still remained in the tamoxifen-treated Cre/tg mice and not uniformly detected in the tissues. Although the appearance of mCherry signal corresponds to the transgene expression in the LGL-IL6 mice, the disappearance of EGFP signal was not essential to be evidenced for transgene inexpression.
Elevation of IL-6 levels in mouse sera was seen within 2 days after a single injection of tamoxifen (not shown). In the previous study, serum IgG levels were significantly elevated in IL-6 overexpressing mice [17]. The difference in the degree of the increase may be explained by the differences in the duration of the IL-6 overexpression and/or the expression levels of IL-6 transgene. Although serum IL-6 levels could not be simply compared between studies, the serum levels in tamoxifen-treated Cre/tg mice reported here were even higher compared with those of the transgenic mice with plasmacytosis, suggesting that the difference in duration may account for the difference between the two phenotypes. However, detailed analysis is required to explain the phenotype.
The broad distribution of tissues that expressed the transgene harbored by the LGL-IL6 line makes possible a wide range of applications of conditional overexpression of human IL-6 in mice using various Cre transgenic lines. Compared with injection, infusion, or both of recombinant IL-6, it may be possible to achieve a stable and more distinct IL-6-induced pathology if a suitable Cre driver is available. The inducibility of the expression achieved using the Cre driver is advantageous as well. Although the limitations in the availability of an appropriate Cre driver may be problematic, the number of Cre mice with useful distinct genotypes is increasing. Therefore, we expect that the LGL-IL6 line will serve as a useful tool for studying the function of IL-6 in vivo and the pathogenesis of disease mediated by the inappropriate expression of IL-6.
Conflict of Interest
All authors have no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was supported by The Research Funding for Longevity Sciences (24–12, 27–16) from National Center for Geriatrics and Gerontology (NCGG), Japan and the Program for Promotion of Fundamental Studies in Health Sciences conducted by the National Institute of Biomedical Innovation of Japan (10–43 and 10–44).
References
- 1.Blanchard F., Duplomb L., Baud’huin M., Brounais B.2009. The dual role of IL-6-type cytokines on bone remodeling and bone tumors. Cytokine Growth Factor Rev. 20: 19–28. doi: 10.1016/j.cytogfr.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 2.Calabrese L.H., Rose-John S.2014. IL-6 biology: implications for clinical targeting in rheumatic disease. Nat. Rev. Rheumatol. 10: 720–727. doi: 10.1038/nrrheum.2014.127 [DOI] [PubMed] [Google Scholar]
- 3.Campbell I.L., Abraham C.R., Masliah E., Kemper P., Inglis J.D., Oldstone M.B., Mucke L.1993. Neurologic disease induced in transgenic mice by cerebral overexpression of interleukin 6. Proc. Natl. Acad. Sci. USA 90: 10061–10065. doi: 10.1073/pnas.90.21.10061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fattori E., Della Rocca C., Costa P., Giorgio M., Dente B., Pozzi L., Ciliberto G.1994. Development of progressive kidney damage and myeloma kidney in interleukin-6 transgenic mice. Blood 83: 2570–2579. [PubMed] [Google Scholar]
- 5.Fattori E., Lazzaro D., Musiani P., Modesti A., Alonzi T., Ciliberto G.1995. IL-6 expression in neurons of transgenic mice causes reactive astrocytosis and increase in ramified microglial cells but no neuronal damage. Eur. J. Neurosci. 7: 2441–2449. doi: 10.1111/j.1460-9568.1995.tb01042.x [DOI] [PubMed] [Google Scholar]
- 6.Fielding C.A., Jones G.W., McLoughlin R.M., McLeod L., Hammond V.J., Uceda J., Williams A.S., Lambie M., Foster T.L., Liao C.T., Rice C.M., Greenhill C.J., Colmont C.S., Hams E., Coles B., Kift-Morgan A., Newton Z., Craig K.J., Williams J.D., Williams G.T., Davies S.J., Humphreys I.R., O’Donnell V.B., Taylor P.R., Jenkins B.J., Topley N., Jones S.A.2014. Interleukin-6 signaling drives fibrosis in unresolved inflammation. Immunity 40: 40–50. doi: 10.1016/j.immuni.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter C.A., Jones S.A.2015. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16: 448–457. doi: 10.1038/ni.3153 [DOI] [PubMed] [Google Scholar]
- 8.Kishimoto T.2005. Interleukin-6: from basic science to medicine--40 years in immunology. Annu. Rev. Immunol. 23: 1–21. doi: 10.1146/annurev.immunol.23.021704.115806 [DOI] [PubMed] [Google Scholar]
- 9.Kishimoto T.2010. IL-6: from its discovery to clinical applications. Int. Immunol. 22: 347–352. doi: 10.1093/intimm/dxq030 [DOI] [PubMed] [Google Scholar]
- 10.Kraakman M.J., Kammoun H.L., Allen T.L., Deswaerte V., Henstridge D.C., Estevez E., Matthews V.B., Neill B., White D.A., Murphy A.J., Peijs L., Yang C., Risis S., Bruce C.R., Du X.J., Bobik A., Lee-Young R.S., Kingwell B.A., Vasanthakumar A., Shi W., Kallies A., Lancaster G.I., Rose-John S., Febbraio M.A.2015. Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell Metab. 21: 403–416. doi: 10.1016/j.cmet.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U.K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. doi: 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 12.Li X., Rock F., Chong P., Cockle S., Keating A., Ziltener H., Klein M.1993. Structure-function analysis of the C-terminal segment of human interleukin-6. J. Biol. Chem. 268: 22377–22384. [PubMed] [Google Scholar]
- 13.Nishimoto N., Kishimoto T.2004. Inhibition of IL-6 for the treatment of inflammatory diseases. Curr. Opin. Pharmacol. 4: 386–391. doi: 10.1016/j.coph.2004.03.005 [DOI] [PubMed] [Google Scholar]
- 14.Niwa H., Yamamura K., Miyazaki J.1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193–199. doi: 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- 15.Ramadoss P., Chiappini F., Bilban M., Hollenberg A.N.2010. Regulation of hepatic six transmembrane epithelial antigen of prostate 4 (STEAP4) expression by STAT3 and CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 285: 16453–16466. doi: 10.1074/jbc.M109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart M.D., Jang C.W., Hong N.W., Austin A.P., Behringer R.R.2009. Dual fluorescent protein reporters for studying cell behaviors in vivo. Genesis 47: 708–717. doi: 10.1002/dvg.20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suematsu S., Matsuda T., Aozasa K., Akira S., Nakano N., Ohno S., Miyazaki J., Yamamura K., Hirano T., Kishimoto T.1989. IgG1 plasmacytosis in interleukin 6 transgenic mice. Proc. Natl. Acad. Sci. USA 86: 7547–7551. doi: 10.1073/pnas.86.19.7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suematsu S., Matsusaka T., Matsuda T., Ohno S., Miyazaki J., Yamamura K., Hirano T., Kishimoto T.1992. Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Proc. Natl. Acad. Sci. USA 89: 232–235. doi: 10.1073/pnas.89.1.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turksen K., Kupper T., Degenstein L., Williams I., Fuchs E.1992. Interleukin 6: insights to its function in skin by overexpression in transgenic mice. Proc. Natl. Acad. Sci. USA 89: 5068–5072. doi: 10.1073/pnas.89.11.5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udagawa N., Takahashi N., Katagiri T., Tamura T., Wada S., Findlay D.M., Martin T.J., Hirota H., Taga T., Kishimoto T., Suda T.1995. Interleukin (IL)-6 induction of osteoclast differentiation depends on IL-6 receptors expressed on osteoblastic cells but not on osteoclast progenitors. J. Exp. Med. 182: 1461–1468. doi: 10.1084/jem.182.5.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda O., Tateishi H., Higuchi Y., Fujii E., Kato A., Kawase Y., Wada N.A., Tachibe T., Kakefuda M., Goto C., Kawaharada M., Shimaoka S., Hattori K., Jishage K.2013. Novel genetically-humanized mouse model established to evaluate efficacy of therapeutic agents to human interleukin-6 receptor. Sci. Rep. 3: 1196. doi: 10.1038/srep01196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Snick J., Cayphas S., Vink A., Uyttenhove C., Coulie P.G., Rubira M.R., Simpson R.J.1986. Purification and NH2-terminal amino acid sequence of a T-cell-derived lymphokine with growth factor activity for B-cell hybridomas. Proc. Natl. Acad. Sci. USA 83: 9679–9683. doi: 10.1073/pnas.83.24.9679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T.2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665. doi: 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.