SUMMARY
Bacteria employ surveillance complexes guided by CRISPR (clustered, regularly interspaced, short palindromic repeats) RNAs (crRNAs) to target foreign nucleic acids for destruction. Although most type I and type III CRISPR systems require four or more distinct proteins to form multi-subunit surveillance complexes, the type I-C systems use just three proteins to achieve crRNA maturation and double-stranded DNA target recognition. We show that each protein plays multiple functional and structural roles: Cas5c cleaves pre-crRNAs and recruits Cas7 to position the RNA guide for DNA binding and unwinding by Cas8c. Cryoelectron microscopy reconstructions of free and DNA-bound forms of the Cascade/I-C surveillance complex reveal conformational changes that enable R-loop formation with distinct positioning of each DNA strand. This streamlined type I-C system explains how CRISPR pathways can evolve compact structures that retain full functionality as RNA-guided DNA capture platforms.
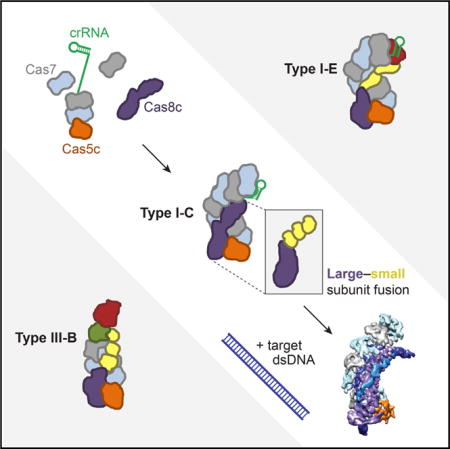
Graphical abstract
INTRODUCTION
Bacteria and archaea defend themselves against infection using adaptive immune systems comprising clustered, regularly interspaced, short palindromic repeats (CRISPR) arrays and CRISPR-associated (cas) genes. CRISPR interference occurs in three stages. First, the Cas1-Cas2 CRISPR integrase inserts foreign DNA segments called spacers into the CRISPR locus of the host genome (Amitai and Sorek, 2016). Next, CRISPR array transcription produces precursor crRNAs that are processed into mature crRNAs containing a single spacer and part of the flanking repeat sequences (Charpentier et al., 2015; Hochstrasser and Doudna, 2015; Li, 2015). Finally, crRNAs assemble with one or more Cas proteins to form RNA-guided surveillance complexes that can base pair with complementary foreign nucleic acid target sequences and trigger degradation (van der Oost et al., 2014; Wright et al., 2016). CRISPR-Cas systems have undergone extensive diversification and have been classified into two broad classes, six types, and many subtypes based on the architecture of their cas gene loci (Abudayyeh et al., 2016; Makarova et al., 2015; Shmakov et al., 2015). Despite requiring multiple proteins to form crRNA-guided surveillance complexes, type I CRISPR loci are the most abundant and are found in the genomes of both cultured and uncultivated bacteria and archaea (Burstein et al., 2016; Makarova et al., 2015). How type I systems can differ in composition yet retain similar DNA recognition and unwinding activities is not known.
All type I surveillance complexes share the ability to bind double-stranded DNA sequences at sites that can base pair with the guide sequence in the crRNA. In the Escherichia coli type I-E system, the 400-kDa surveillance complex Cascade/I-E (CRISPR-associated complex for anti-viral defense) includes the crRNA and five unique proteins: a “large subunit” (Cse1), a dimer of small subunits (Cse2), six copies of a backbone subunit (Cas7), and subunits that cap the 5′ end of the crRNA (Cas5e) and the 3′ stem-loop (Cas6e) (Hayes et al., 2016; Jore et al., 2011; Wiedenheft et al., 2011). Double-stranded DNA (dsDNA) target recognition by Cascade/I-E requires the protospacer adjacent motif (PAM), a short sequence upstream of the target site that is critical for distinguishing foreign DNA from the CRISPR locus in the host genome (Mojica et al., 2009). When a bona fide target is identified, base pairing and protein binding induce DNA unwinding to form an “R-loop” structure in which RNA-DNA hybridization occurs within Cascade and the noncomplementary or “non-target” strand is displaced. Stable R-loop formation and conformational changes within the complex recruit the Cas3 effector nuclease-helicase to cleave the target DNA (Blosser et al., 2015; Hochstrasser et al., 2014; Mulepati and Bailey, 2013; Redding et al., 2015; Rutkauskas et al., 2015; Sinkunas et al., 2013; Szczelkun et al., 2014; Westra et al., 2012).
Although Cascade/I-E has been studied extensively, the type I-C system occurs more often in nature and requires only three unique proteins to form its DNA targeting complex, Cascade/I-C (Makarova et al., 2011b, 2015; Nam et al., 2012). One reason for the streamlined type I-C composition is the absence of Cas6, the endoribonuclease employed by most type I systems for pre-crRNA processing (Carte et al., 2008; Charpentier et al., 2015; Haurwitz et al., 2010; Hochstrasser and Doudna, 2015; Li, 2015). I-C systems enlist Cas5 (referred to here as Cas5c) for this purpose, a protein that is noncatalytic in other type I systems and observed in Cascade/I-E crystal structures to make sequence-specific contacts to the conserved crRNA 5′ end (Jackson et al., 2014; Mulepati et al., 2014; Zhao et al., 2014). Another reason for the streamlined type I-C composition is the apparent fusion of the large and small subunit proteins into a single protein encoded by the cas8c gene (formerly csd1) (Makarova et al., 2011a). In Cascade/I-E, the large subunit protein Cse1 recognizes the PAM sequence and recruits the Cas3 nuclease-helicase for target degradation, while two copies of the small subunit protein Cse2 stabilize base pairing between the crRNA and target DNA strand (Hayes et al., 2016; Hochstrasser et al., 2014; Sashital et al., 2012). However, this arrangement is only found in the I-A and I-E CRISPR subtypes, and it is unclear how these activities are performed in the majority of type I subtypes, which lack a separately encoded small subunit (Makarova et al., 2011b, 2015).
To determine how the three proteins of Cascade/I-C are sufficient to assemble into a crRNA-guided complex capable of site-specific DNA capture, we investigated the binding interactions, assembly pathway, and molecular architecture of Desulfovibrio vulgaris Cascade/I-C. Biochemical studies and cryoelectron microscopy (cryo-EM) reconstructions show that Cas5c remains bound to the conserved crRNA 5′ end after cleavage through low-affinity interactions with the RNA sequence and other subunits. The crRNA 3′ stem-loop structure caps Cas7 (formerly Csd2) oligomerization along the crRNA, allowing seven Cas7 protomers to bind the length of the crRNA. The architecture of the Cas8c subunit, which associates using both RNA-protein and protein-protein interactions, resembles a large subunit fused to three small subunit domains. Cryo-EM reconstructions of Cascade/I-C both alone and bound to a target dsDNA reveal conformational changes that trigger R-loop formation in which the complete path of the non-target DNA strand is visible. Cascade/I-C induces a pronounced 60° bend angle in the target DNA that traverses the complex, suggesting a mechanism of DNA unwinding that involves sequential protein-DNA and RNA-DNA interactions. The compact composition of Cascade/I-C may lend itself to biotechnology applications, as exemplified by its recent use for gene repression in bacteria (Leenay et al., 2016).
RESULTS
Cas5c Binds the 5′ Handle Sequence of the Pre-crRNA
This study focused on the type I-C CRISPR system found on the pDV megaplasmid of D. vulgaris str. Hildenborough, in which slightly overlapping genes encode the Cas5c, Cas8c, and Cas7 subunits of Cascade/I-C (Nam et al., 2012) (Figure 1A). As observed in other I-C systems (Garside et al., 2012; Koo et al., 2013; Nam et al., 2012; Punetha et al., 2014), the catalytic Cas5c protein cleaves the precursor CRISPR RNA (pre-crRNA) repeat directly 3′ of the hairpin (Figure S1A) to generate 64–68 nucleotide (nt) mature crRNAs, depending on the length of the spacer.
Figure 1. Tight Binding of Cas5c to the Pre-crRNA Repeat Is Mediated by the First 4–5 Nucleotides of the 5′ Handle Region.
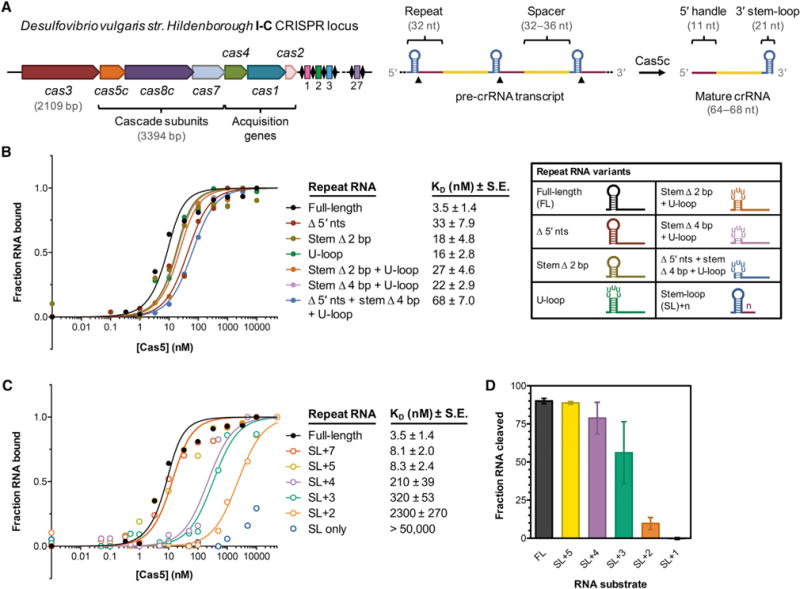
(A) Overview of the type I-C cas genes and CRISPR array found on the Desulfovibrio vulgaris str. Hildenborough megaplasmid. The length of each cas gene is drawn approximately to scale, whereas the repeat-spacer array is drawn much larger for clarity.
(B) At left, normalized florescence polarization (FP) measurements of Cas5c binding to the full-length pre-crRNA repeat and several variants in which the 5′ end and stem-loop have been altered. Cas5c was incubated with each 5′ fluorescein-labeled RNA for 30 min at 37°C prior to FP measurement. Each point represents the average of at least three independent replicates and calculated dissociation constant values (KD) are listed along with the standard error of the fit parameter (SE). Error bars representing ±1 SD are omitted for clarity but can be found in Figure S1C. At right, graphical depictions of the modified repeat sequences tested.
(C) Normalized fluorescence polarization binding data for Cas5c binding to pre-crRNA repeats in which the 5′ handle is progressively truncated. Data represent at least three independent replicates. Error bars representing ±1 SD are omitted for clarity but can be found in Figure S1C.
(D) Quantified cleavage efficiencies of several pre-crRNA variants by Cas5c after incubation for 1 hr at 37°C. Each bar is the average of at least three independent replicates; error bars represent ±1 SD.
See also Figure S1 and Tables S1 and S2.
We wondered how recognition of the pre-crRNA repeat by Cas5c might differ from that by Cas6, the endoribonuclease responsible for crRNA maturation in all other type I and III systems. Using fluorescently labeled variants of the pre-crRNA repeat with a deoxyguanosine 5′ of the cleavage site to prevent cleavage (Figure S1B), we measured fluorescence polarization to quantify binding by Cas5c (Figures 1A, 1B and S1C). We observed high-affinity binding (3.5 ± 1.4 nM) of Cas5c to the full-length repeat, somewhat weaker than the 50 pM to 3 nM affinities observed between Cas6 proteins and their cognate RNA substrates (Niewoehner et al., 2014; Sashital et al., 2011; Sternberg et al., 2012). Unlike Cas6 (Sternberg et al., 2012), Cas5c tolerated drastic mutations and/or deletions of the 5′ nucleotides and stem-loop, with only modest defects in binding affinity observed. Next, we systematically truncated the repeat from the 3′ end. We found that this 11-nt segment, known as the 5′ handle for its eventual position in the mature crRNA, was highly sensitive to truncation (Figures 1C and S1C), in agreement with previous work (Nam et al., 2012). Trimming down to 5 nts after the stem-loop (SL+5) was tolerated, but each additional nucleotide deletion caused substantial losses in binding affinity. The same RNA substrates that were bound poorly by Cas5c were also cleaved less efficiently (Figure 1D) (Nam et al., 2012). Thus, Cas5c functions conversely to Cas6, primarily recognizing the 5′ handle rather than the 3′ stem-loop.
Cascade/I-C Subunit Binding to Different crRNA Segments Directs Assembly
Since Cas5c was sensitive to deletion of bases in the 5′ handle but had no apparent affinity for the 3′ stem-loop of the crRNA, we quantified the binding affinity of Cas5c for its crRNA cleavage products using fluorescence polarization. Surprisingly, Cas5c has a dissociation constant (KD) of only 2–4 μM for the crRNA 5′ handle, the full-length mature crRNA, or a crRNA lacking the 3′ stem-loop, affinities that are 500-fold weaker than those measured for the pre-crRNA repeat (Figures 2A and 2B). To ensure that the 5′ fluorescein label did not interfere with Cas5c binding, we also quantified Cas5c affinity for a 5′ handle RNA containing a 5′ hydroxyl end (the end chemistry that results from Cas5c cleavage) (Garside et al., 2012; Nam et al., 2012) and a 3′ fluorescein, and observed similar binding affinity (Figure S2A). Deletion of the 5′ handle reduces binding affinity to 13 ± 1.1 μM, similar to that observed for a random 34-nt protospacer sequence (Figure 2B). Hence, Cas5c binds preferentially to the 5′ handle region of the mature crRNA.
Figure 2. Sequence-Specific Interactions with the 5′ Handle, Single-Stranded RNA Affinity, and Cooperativity Drive Complex Assembly, Which Is Capped by the 3′ Stem-Loop.
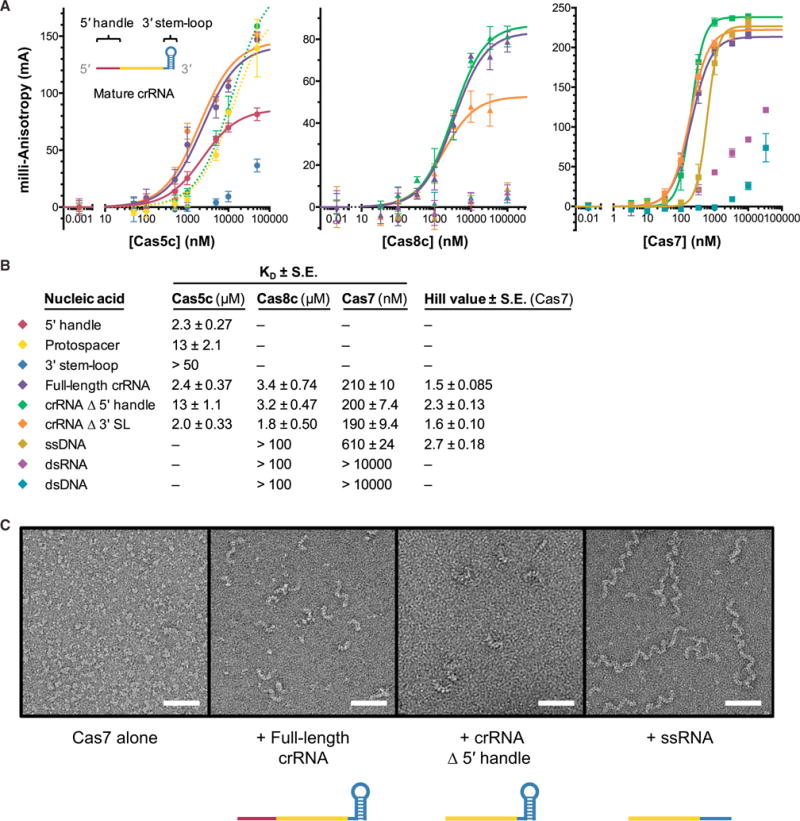
(A) Fluorescence anisotropy data showing binding of Cas5c (left), Cas8c (middle), and Cas7 (right) to different regions of a crRNA and various types of nucleic acids. Dotted lines indicate that binding has not reached saturation, making the curve fitting unreliable. Cas7 data were fit with a Hill equation to account for apparent cooperativity. Each point is the average of at least three independent replicates; error bars represent ±1 SD.
(B) Legend for plots in (A) and summary of calculated dissociation constants (KD values) for all three proteins and Hill coefficients for Cas7 data.
(C) Representative electron micrographs from negative stain electron microscopy analysis of RNA-induced Cas7 oligomerization. Scale bar, 500 Å.
See also Figure S2 and Tables S1 and S2.
We next investigated whether Cas8c and Cas7, the other two subunits of Cascade/I-C, bind preferentially to any region of the crRNA. We found that Cas8c, the larger subunit, has low micromolar affinity for ssRNA and does not bind detectably to DNA or double-stranded RNA (dsRNA) (Figures 2A and 2B). This result suggests that although Cas8c interacts with the crRNA, other contacts are responsible for its positioning in Cascade/I-C. The backbone protein Cas7 binds single-stranded RNA with a KD of ~200 nM. We noticed nonrandom residuals when the data were fit with a standard binding equation, so we included the Hill coefficient, yielding nH values greater than 1.5 for all variants of the crRNA. An nH value greater than 1 indicates positive cooperativity in the assembly of Cas7 subunits, consistent with their ability to oligomerize (Jore et al., 2011; Lintner et al., 2011) and reminiscent of RecA filaments (Chen et al., 2008; Sorek et al., 2013).
We hypothesized that because none of the Cascade/I-C subunits have high binding affinity for dsRNA, the 3′ stem-loop may serve as a way to “cap” the complex, preventing addition of Cas7 subunits beyond the 3′ end of the crRNA (Figures 2A and 2B). The Cas6 subunit accomplishes this function in Cascade/I-E (Gesner et al., 2011; Sashital et al., 2011; Wiedenheft et al., 2011), but the lack of Cas6 in I-C systems implies that another element must play this role. To test this possibility, we used negative stain electron microscopy to assess the oligomerization state of Cas7 alone and incubated with various RNA constructs. In the absence of a crRNA, Cas7 is monomeric (Figure 2C), consistent with its behavior during size exclusion chromatography (Figure S2B). When incubated with either a full-length 66-nt crRNA or a 55-nt crRNA missing the 5′ handle, we observed filaments of up to ~8 Cas7 subunits (Figures 2C and S2C). However, when we mixed Cas7 with a 44-nt ssRNA lacking the 3′ hairpin, we observed the formation of many long, helical filaments (Figures 2C and S2C), presumably due to Cas7 oligomerization along the ssRNA and bridging adjacent RNA molecules. Together, these data suggest that the 3′ stem-loop serves to cap Cas7 oligomerization at the 3′ end of the crRNA, determining the maximum length of Cascade/I-C.
Protein-Protein Contacts Interlock the Components of Cascade/I-C
Unlike the high-affinity, cooperative crRNA binding measured for Cas7 subunits, Cas5c and Cas8c exhibited only weak binding to the crRNA. To understand how these subunits assemble into the Cascade/I-C complex, we tested binding between each pair of Cascade/I-C subunits using isothermal titration calorimetry (ITC). The results indicate that all three proteins have low micromolar affinity for each other (Figure 3A) with binding that is both enthalpically and entropically driven. Similar heats were not detected when buffer was substituted for either component in control experiments (Figure S2D). We compiled the individually quantified protein-protein and protein-RNA affinities to map the network of interactions that govern assembly and stability of Cascade/I-C (Figure 3B). This web of individually weak binding interactions suggests that the complex is stabilized by avidity. The sequence of the 5′ handle and structure of the 3′ stem-loop in the crRNA direct the localization and incorporation of subunits, which form hydrophobic and electrostatic interactions with each other that facilitate assembly of a stable targeting complex.
Figure 3. Cascade/I-C Is Connected by a Network of Protein-Protein and Protein-RNA Interactions.
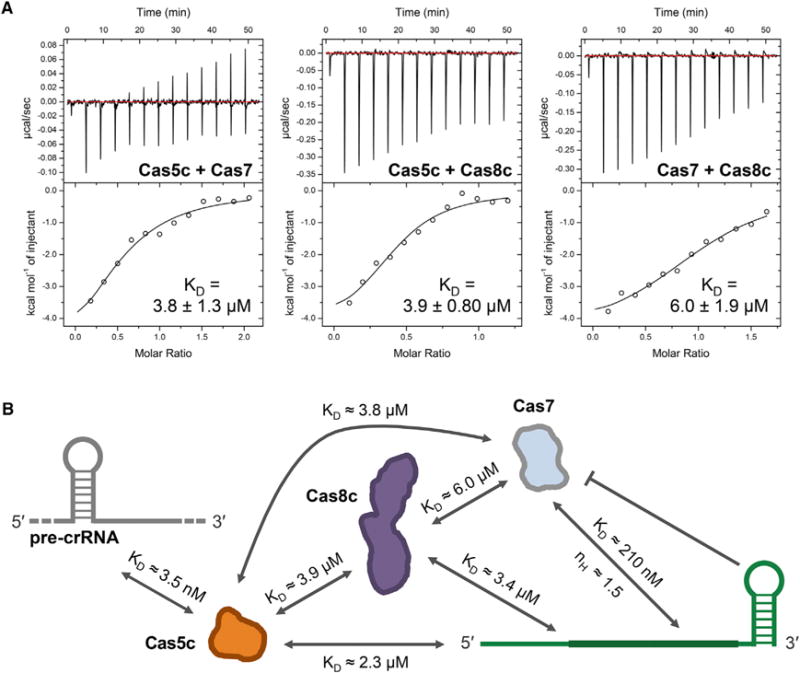
(A) Representative isothermal titration calorimetry (ITC) thermograms showing that all three Cascade/I-C subunits exhibit low micromolar affinity for each other in the absence of crRNA. Affinities are reported as KD ± standard error of the fit parameter (SE). Three independent experiments for each interaction yielded the following values for Cas5c-Cas7: KD = 3.8 ± 1.3 μM, n = 0.59 ± 0.082, ΔH = 5,100 ± 2,200 cal/mol, ΔS = 8.6 ± 7.5 cal/mol/deg; Cas5c-Cas7: KD = 3.9 ± 0.80 μM, n = 0.57 ± 0.14, ΔH = 4,800 ± 520 cal/mol, ΔS = 9.4 ± 1.7 cal/mol/deg; and Cas7-Cas8c: KD = 6.0 ± 1.9 μM, n = 1.0 ± 0.30, ΔH = 5,200 ± 610 cal/mol, ΔS = 7.4 ± 2.3 cal/mol/deg.
(B) Map of all quantified interactions contributing to assembly and stability of the Cascade/I-C complex.
See also Figure S2 and Tables S1 and S2.
Cryo-EM Reveals Superhelical Cascade/I-C Architecture
To further understand how this minimal system assembles into a fully functional RNA-guided surveillance complex, we used cryo-EM for structural analysis. We purified Cascade/I-C by expressing His6-MBP-tagged Cas5c along with Cas8c, Cas7, and a repeat-spacer array containing three copies of the same spacer (Figure S3, top; Tables S1 and S2). Aligned and averaged micrographs of apo-Cascade/I-C complexes (containing a crRNA guide) showed caterpillar-like structures measuring ~200 Å in the longest dimension (Figures S4A and S4B, left). Using three-dimensional (3D) classification and refinement in RELION (Scheres, 2012), we obtained a final 3D structure of apo-Cascade/I-C at 6.7 Å resolution (using the 0.143 Fourier Shell Correlation [FSC] criterion) from a final set of ~24,000 particles (Figures 4A, S4C, S4D; Movie S1). Identifying individual subunits by visual inspection and segmentation was straightforward, revealing a repeating backbone of seven Cas7 subunits (Cas7.1–7.7, Figure 4A) cradling the entire length of the crRNA. A single Cas5c subunit caps the 5′ end. The 3′ stem-loop is not resolved in our structure, likely because it is flexible. Accordingly, the Cas7.1 subunit, located near the 3′ end of the crRNA, has regions of low resolution (Figures S4D and S4E). In the “belly” of the complex opposite the Cas7 backbone, there is a large density for Cas8c that occupies a position comparable to the densities for both the Cse1 and two Cse2 subunits of Cascade/I-E (Jackson and Wiedenheft, 2015). This proposed stoichiometry implies a molecular weight of ~342 kDa, consistent with the retention volume measured by size exclusion chromatography (Figure S3, top left).
Figure 4. Architecture of crRNA-Guided and dsDNA Target-Bound Cascade/I-C Complexes.
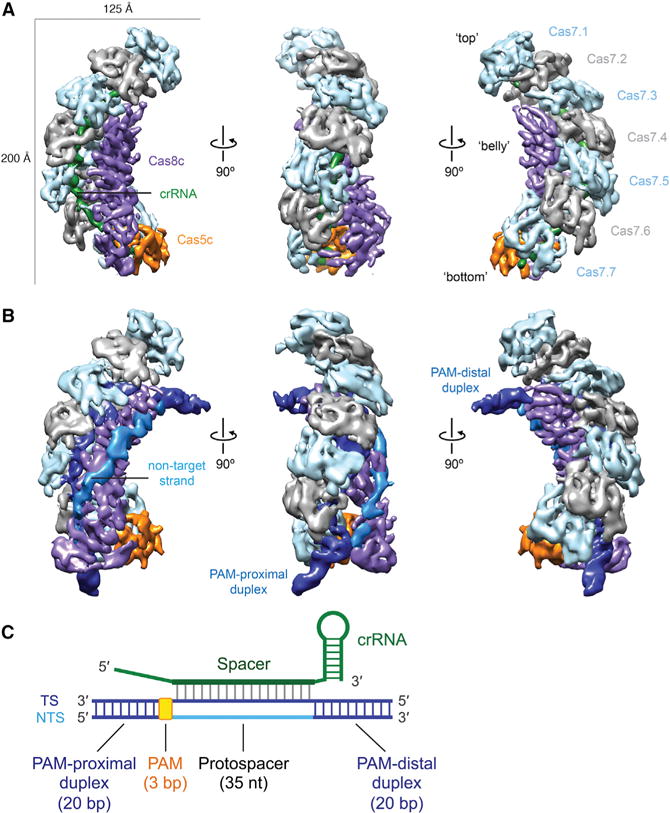
(A and B) Cryo-EM reconstructions of intact apo-Cascade/I-C (crRNA-bound) (A) and “2× duplex” dsDNA target-bound Cascade/I-C (B) at 6.7- and 8.8-Å resolution, respectively. Subunits are segmented and colored as indicated. Details of these structures can be seen in Movies S1 and S2.
(C) Schematic of crRNA and target dsDNA used for cryo-EM analysis. TS, target strand; NTS, non-target strand; PAM, protospacer-adjacent motif.
See also Figures S3 and S4 and Tables S1 and S2.
Structural Rearrangements upon Target dsDNA Binding by Cascade/I-C
To explore the structural basis for dsDNA target capture and R-loop formation by this minimal complex, we bound Cascade/I-C to a 75-base pair (bp) dsDNA target bearing complementary protospacer and functional PAM sequences (Leenay et al., 2016) and determined its structure using cryo-EM (Figures 4B, 4C, S3, and S4A, right). Two-dimensional (2D) alignment and classification of the target-bound complex showed particles with additional density projecting out from the top and bottom in an arc configuration (Figure S4B). We obtained two distinct structures of the dsDNA target-bound Cascade at 7.6- and 8.8-Å resolution (using the 0.143 FSC criterion) after one and three rounds of 3D classification, respectively (Figures S4C, S4E–S4G, and S5). In the lower-resolution structure (2× duplex), we observed two rod-like densities for the ends of the dsDNA duplex and a tube-like density connecting these two ends that likely corresponds to the non-target DNA strand (Figure 4B; Movie S2). The PAM-proximal duplex is anchored between the bottom two Cas7 subunits (Cas7.6 and 7.7) and the bottom of the Cas8c subunit, while the PAM-distal duplex extends from the α-helical domain of Cas8c. The non-target strand emerges from the duplexed PAM and snakes along the entire length of the Cas8c subunit, running prominently on the outside of the complex toward the top of Cas8c before rejoining with the target strand to create the other end of the duplex. In the higher-resolution structure (1× duplex), the PAM-distal end of the target duplex is not visible, suggesting that this end of the duplex is held less rigidly (Figure S4G).
We next turned to dissecting the architecture of the large, elongated Cas8c subunit. In both the apo- and target-bound Cascade structures, the top of Cas8c, located in the “belly” of the complex, can be easily segmented into three identical (cross-correlation coefficient of 0.97) sets of five alpha-helices (Figure 5A, left; Movie S2). This distinctive helical arrangement is structurally conserved in the small subunits of type I and type III surveillance complexes (Venclovas, 2016), confirming the bioinformatic prediction that Cas8c is a large subunit fused to a small subunit domain (Makarova et al., 2011a). The type III targeting complexes have three copies of the small subunit (Csm2 or Cmr5), while the type I-E targeting complex has two copies of its slightly larger small subunit (Cse2). In this regard, the I-C system appears to resemble the type III interference complex architecture (Figures S6A and S6B) (Osawa et al., 2015; Staals et al., 2014; Taylor et al., 2015). We refer to the three small subunit domains (displayed in Figure 5A in peach, gold, and yellow) as Cas8cSS1–3. The apparent Cas8c N-terminal domain, which we term Cas8cLS (large subunit), is flexible, making it difficult to identify unambiguously any secondary structural features (Figure 5A, middle). The Cas8cLS is poorly resolved in the apo structure (Figure S4D), but it rearranges and becomes ordered upon dsDNA binding (Figure 5A, right).
Figure 5. Details of Cas8c Subunit Architecture, Conformational Changes Induced by Target Binding, and R-Loop Structure.
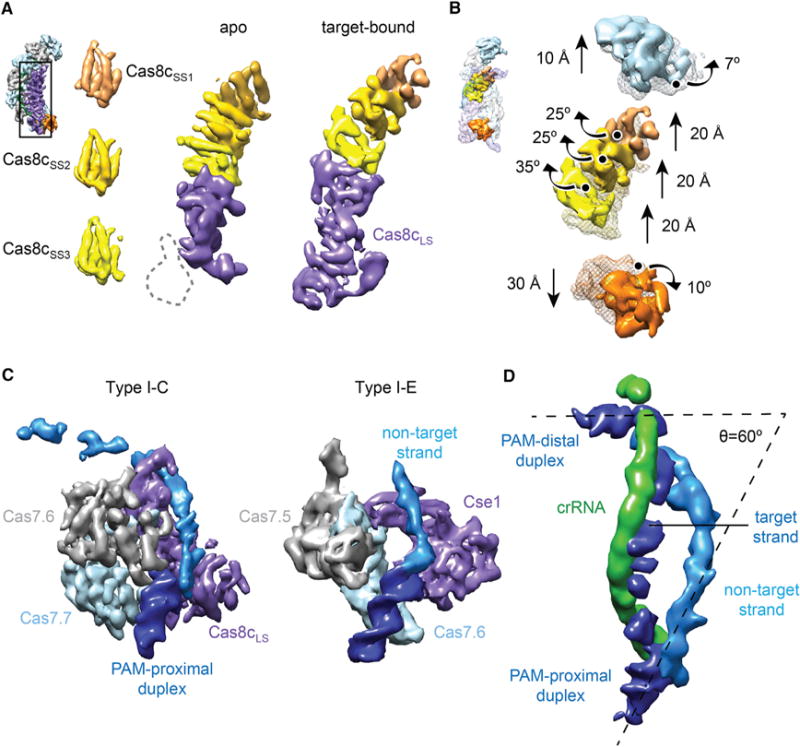
(A) The Cas8c subunit of the Cascade/I-C complex (inset) contains three indistinguishable sets of five helices (Cas8cSS1, Cas8cSS2, and Cas8cSS3) that are characteristic of small subunits (left). The remaining density of Cas8c in the apo structure (middle) shows similarities to a large subunit (Cas8cLS, purple). In the target-bound structure, the bottom region of Cas8cLS rearranges and becomes ordered (right). Inset: view of the apo structure used for this analysis.
(B) Aligning apo-Cascade/I-C (transparent mesh) and the target-bound complex (surface) based on the Cas7.2–7.6 backbone (removed for clarity), reveals that the complex undergoes structural rearrangements involving the Cas5c, Cas7.1, and the Cas8c small subunit domains upon dsDNA binding. Inset: view of the target-bound structure used for this analysis. Details of these movements can be seen in Movie S3.
(C) Comparison of the PAM-proximal DNA interaction with the bottom two Cas7 subunits and Cas8cLS or Cse1 from the “1× duplex” type I-C (left) and type I-E Cascade (right, PDB: 5H9F) structures, respectively.
(D) Architecture of the R-loop from the “2× duplex” dsDNA target-bound structure. The target DNA is bent to form a 60° angle between DNA ends. Subunits are colored as in Figure 1A or as indicated. A detailed view of the R-loop can be seen in Movie S4.
See also Figures S3–S5 and Tables S1 and S2.
To better describe the conformational changes that occur during target binding, we aligned both structures by superposition of the middle five Cas7 subunits, which remain relatively unchanged (cross-correlation coefficient of 0.95), and then compared the positions and orientations of all other subunits. This comparison shows a concerted structural rearrangement upon DNA substrate binding (Movie S3). The Cas7.1 subunit at the top of the complex rotates by ~7° and moves an additional 10 Å upward (Figure 5B). The three Cas8c small subunit domains swing (~25°–35° rotation) toward the center of the complex and move ~20 Å upward (Figure 5B), creating a tight cleft to hold the crRNA:target strand heteroduplex. Cas5c translates by ~30 Å toward the bottom of the complex, presumably driven by the conformational changes in Cas8c.
The Cascade/I-C Cas8c Subunit Binds the PAM and Stabilizes R-Loop Formation
We compared the PAM-proximal DNA end in our 1× duplex Cascade/I-C structure to the recently published crystal structure of E. coli Cascade bound to a target strand and a portion of the non-target strand (Hayes et al., 2016). In Cascade/I-C, the PAM-proximal DNA end contacts the inside edge of Cas7.6, Cas7.7, and the Cas8cLS (Figures 4B and 5C, left), whereas in Cascade/I-E, the PAM-proximal duplex sits between the bottom two Cas7 subunits (Figure 5C, right) (Hayes et al., 2016; Hochstrasser et al., 2014; van Erp et al., 2015). Interestingly, all Cas7 subunits except Cas7.7 are identical. Cas7.7 appears to be in either an alternative conformation or disordered (Figure S6C). The conformational ordering of Cas8cLS upon binding to the region of the target DNA duplex containing the PAM and the similarity between the overall orientation of the duplex and non-target strand in the type I-C and I-E structures suggest that Cas8c recognizes the PAM sequence, presumably in the same manner as Cse1, in duplexed form on the target strand via contacts in the minor groove (Hayes et al., 2016). By calculating the 3D difference map between the apo- and target-bound (containing both visible DNA ends) structures, we observed density for nearly the entire R-loop (Figures 5D, S6D, and S6E; Movie S4). Cas8c contacts both ends of the duplex and is the only subunit to contact the non-target strand (Figure 4B), which appears to run along the same surface of each small subunit domain. These results indicate that in addition to PAM recognition, Cas8c is the primary protein responsible for maintaining a stable, locked R-loop structure (Blosser et al., 2015; Rutkauskas et al., 2015; Szczelkun et al., 2014). Notably, the bound DNA is bent by 120° relative to a B-form DNA helical axis to a final angle of 60° (Figure 5D), consistent with the propensity of Cascade/I-E to bend DNA (Westra et al., 2012). This structural distortion may facilitate unwinding of the target dsDNA and help to hold apart the target and non-target strands after R-loop formation.
DISCUSSION
Although many type I and III CRISPR systems recognize foreign DNA using a crRNA-guided surveillance complex comprising four or more unique Cas proteins, type I-C systems include just three such proteins (Makarova et al., 2015). Biochemical and structural data presented here show how the three-protein surveillance complex Cascade/I-C from D. vulgaris assembles and targets double-stranded DNA (Figure 6). This naturally streamlined system helps explain evolutionary connections between diverse CRISPR immunity subtypes.
Figure 6. Comparison Between Type I-C and I-E Cascade Structures and Division of Labor Between Subunits.
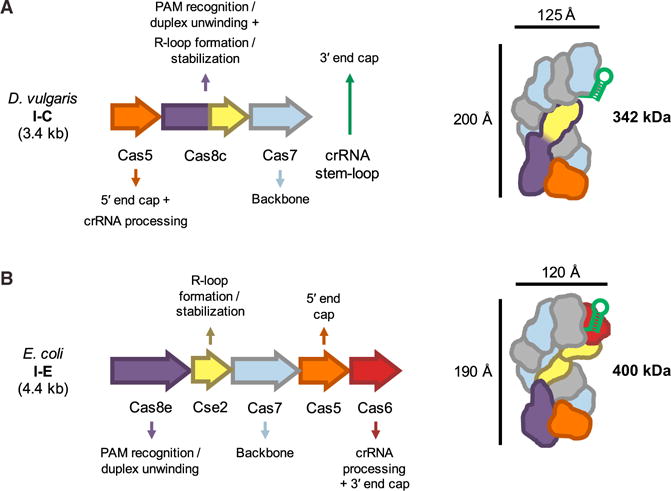
(A) Overview of Desulfovibrio vulgaris type I-C cas genes that form Cascade/I-C, the function of each subunit, and overall complex architecture. Length of the cas gene operon and molecular weight of the protein complex are indicated in kilobases (kb) and kilodaltons (kDa), respectively.
(B) Overview of the Escherichia coli type I-E Cascade complex architecture and subunit functions, as in (A).
See also Figures S6A and S6B.
Taken together, the biochemical and structural data suggest a cooperative model for Cascade/I-C assembly. Cas5c binds to the pre-crRNA repeat with high affinity (KD ≈3 nM) and cleaves at the base of the stem-loop to generate a mature crRNA. Specific binding to the 5′ crRNA repeat sequence and interactions with Cas7 position Cas5c at one end of the assembling complex. Cooperative oligomerization of Cas7 (nH = 1.5–2.3) along the crRNA stops at the RNA 3′ end stem-loop, accommodating seven Cas7 protomers. RNA binding by Cas7 is the highest affinity individual interaction (200 nM) involved in complex assembly after crRNA maturation. The Cas8c subunit associates with the complex through a set of weak protein-protein interactions with Cas5c (4 μM) and Cas7 (6 μM), as well as association with the single-stranded region of the crRNA (3 μM). The lower affinity of the Cas8c “large subunit” for the rest of the interference complex is consistent with observations that Cse1—the large subunit of Cascade/I-E—readily dissociates from Cascade/I-E (Sashital et al., 2012).
The Cas8c subunit appears to be the workhorse of the complex. It recognizes the dsDNA PAM sequence and stabilizes the R-loop through contacts to the displaced non-target DNA strand, two functions that are divided between the large (Cse1) and small (Cse2) subunit proteins of Cascade/I-E (Figure 6) (Hayes et al., 2016). By analogy to the type I-E system, Cas8c likely also recruits Cas3 for target degradation (Hochstrasser et al., 2014; Mulepati and Bailey, 2013; Sinkunas et al., 2013). The three distinct C-terminal domains of Cas8c bear structural similarity to a five-helix motif conserved in type I and III small subunits, confirming the hypothesis that Cas8c represents a functional fusion of small and large subunits. This natural fusion protein, observed in the I-B, I-U, I-D, and I-F CRISPR subtypes (Makarova et al., 2015), is the most common gene arrangement in type I systems. It will be interesting to see how many copies of the individual small subunit or fused small subunit domains exist in other systems, as it is not straightforward to predict this feature from sequence alone.
Although several crystal and cryo-EM structures of Cascade/I-E are available, none have revealed the structure of the entire R-loop formed when a dsDNA target sequence is engaged with the surveillance complex. In the D. vulgaris Cascade/I-C R-loop structure presented here, the non-target DNA strand follows a similar path to that observed in the E. coli Cascade complex (Hayes et al., 2016), guided along the top of the Cas8c large subunit domain and emerging near the small subunit domains. We find that the non-target strand continues along the outside of a ridge created by the small subunit domain of Cas8c. We hypothesize that the non-target strand is positioned similarly in all type I Cascade complexes, and that actively guiding this strand is critical for recruitment of and cleavage by the Cas3 nuclease-helicase, as observed in the E. coli system (Blosser et al., 2015; Hayes et al., 2016; Hochstrasser et al., 2014; Mulepati and Bailey, 2013; Rutkauskas et al., 2015; Sinkunas et al., 2013; Szczelkun et al., 2014). It remains to be seen how a recently described Cascade/I-F from Shewanella putrefaciens CN-32 is able to function despite its apparent lack of a Cas8 subunit (Gleditzsch et al., 2016). The similarity between the orientation of the PAM-proximal duplex DNA end observed in the Cascade/I-C and Cascade/I-E structures suggests that in both complexes, the large subunits recognize the PAM in a similar manner. Additionally, the variability in PAM sequences tolerated by Cascade/I-C (Leenay et al., 2016) suggests a plasticity in PAM recognition similar to that determined for the I-E system, which is thought to be a consequence of reading the minor groove (Hayes et al., 2016). Hence, we propose that minor groove recognition of a PAM in double-stranded form is likely a feature common to all type I systems.
It was observed previously that Cascade/I-E induces DNA bending, but the bend angle was not quantified (Westra et al., 2012). We find that the type I-C Cascade surveillance complex causes a pronounced bend in target dsDNA, distorting it from 180° to 60°. Structural studies showed that the type II Cas9 protein also distorts its target dsDNA during recognition, albeit to a much less acute DNA bend angle of 150° (Jiang et al., 2016). Thus, target bending is a conserved feature of DNA binding by both type I and type II systems and may contribute to DNA unwinding. Cascade complexes contain a crRNA with a longer spacer (~30–35 nts) than the 20-nt spacer used by Cas9 (Deltcheva et al., 2011). This fact suggests a possible relationship between the extent of DNA bending and the length of the DNA that must be unwound for base-pairing. It remains unclear whether DNA bending facilitates unwinding and crRNA base-pairing with the target strand, or if target distortion is instead a consequence of R-loop formation. Further studies are needed to more thoroughly understand the mode of DNA recognition by Cascade prior to unwinding, RNA-DNA hybridization and R-loop locking (Blosser et al., 2015; Rutkauskas et al., 2015; Szczelkun et al., 2014). This may in turn guide the use of Cascade/I-C for applications in genome manipulation in both bacteria (Leenay et al., 2016) and eukaryotes.
EXPERIMENTAL PROCEDURES
Cloning
Genomic DNA from Desulfovibrio vulgaris str. Hildenborough was obtained from the Joint Bioenergy Institute (JBEI). The genes encoding Cas5c (NCBI: YP_009170.1), Cas8c (NCBI: YP_009171.1), and Cas7 (NCBI: YP_009172.1) were PCR amplified as a single operon for Cascade/I-C expression as well as individually and cloned into the pHMGWA destination vector (Invitrogen). The pHMGWA expression vector encodes an N-terminal His6-MBP (maltose binding protein) tag followed by a tobacco etch virus (TEV) protease recognition site.
To generate a plasmid for expressing pre-crRNAs, pACYCDuet-1 (Novagen) was digested with EcoNI and KpnI and a series of partially overlapping oligos (Table S2) were mixed, annealed, and ligated into this vector. Several clones were sequenced and we chose a final construct containing a T7 promoter sequence followed by three copies of the spacer sequence, each flanked by repeats. For further details, see Supplemental Experimental Procedures.
Protein Purification
Cas5c, Cas8c, and Cas7 were overexpressed in BL21(DE3) Rosetta pLysS cells. For purification of Cascade/I-C, the protein subunits (Cas5c-Cas8c-Cas7) and a synthetic CRISPR array containing three copies of D. vulgaris spacer #2 were co-expressed in BL21(DE3) cells. Cells were grown at 37°C prior to induction of protein expression by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After overnight growth at 16°C, cells were harvested and lysed by sonication in lysis buffer. Lysate was clarified by centrifugation at 27,000 × g and purified using Ni-NTA affinity resin (QIAGEN). The resin was washed with wash buffer and eluted with buffer containing 300 mM imidazole. TEV protease was added, and the protein-TEV mixture was dialyzed at 4°C overnight against wash buffer. Cleaved His6-MBP tag, uncut protein, and His-TEV were removed by an additional Ni purification.
Cas5c and Cas7 were further purified by size exclusion chromatography using a Superdex 75 16/60 or Superdex 200 10/300 GL column (GE Healthcare), respectively, pre-equilibrated in gel filtration buffer. Cascade/I-C was purified by size exclusion chromatography using a pre-equilibrated Superose 6 prep grade or 10/300 GL column. All proteins and the Cascade/I-C complex were analyzed for purity by SDS-PAGE and either buffer exchanged or dialyzed into storage buffer. Final stocks were concentrated, flash frozen in liquid nitrogen, and stored at −80°C. For further details, see Supplemental Experimental Procedures.
Oligonucleotide Preparation
RNA and DNA oligonucleotides used in cleavage assays, gel shifts, and electron microscopy were purchased from Integrated DNA Technologies (IDT) (Table S1). Oligonucleotides were purified on denaturing gels containing 15% (v/v) 29:1 polyacrylamide, 7M urea, and 1 × Tris/Borate/EDTA (TBE). RNA or DNA bands were visualized by UV light, excised, and eluted by soaking gel pieces in deionized H2O. Gel pieces were filtered out, and DNA was ethanol-precipitated and resuspended in deionized H2O. DNA duplexes were formed by mixing equimolar amounts of each DNA strand in 40 mM Tris (pH 8.0), 38 mM MgCl2, and 1 mM spermidine, heating at 95°C for 2 min, and slow cooling at room temperature for at least 10 min. Duplexes were resolved on a native gel containing 6% (v/v) 29:1 polyacrylamide and 1 × TBE and purified as previously described. RNA and DNA samples were 5′ end labeled with [γ-32P] ATP using polynucleotide kinase (PNK, New England Biolabs) for 30 min at 37°C. PNK was heat-denatured at 65°C for 20 min, and excess ATP was removed using a G-25 spin column (GE Healthcare).
RNA Cleavage Assays
All cleavage assays were performed in 1 × cleavage buffer [20 mM HEPES–NaOH (pH 7.5), 100 mM KCl, 5% glycerol, and 1 mM TCEP]; 1 μM Cas5c was incubated with 0.2–2.5 nM 5′ radiolabeled RNA at 37°C for 30 min and resolved on denaturing gels containing 15%–20% (v/v) 29:1 polyacrylamide, 7 M urea, and 1 × TBE. Gels were dried, RNA was visualized by phosphorimaging, and images were quantified using ImageQuant software (GE Healthcare). Reported values are the average of at least three independent replicates, and error bars represent the SD. For further details about RNA cleavage assays, including Cas5c cleavage site mapping, see Supplemental Experimental Procedures.
Fluorescence Polarization
Fluorescence polarization assays were performed in 1 × binding buffer [20 mM HEPES–NaOH (pH 7.5), 100 mM KCl, 5% (v/v) glycerol, 1 mM TCEP, 10 ug/ml heparin, and 0.01% Igepal CA-630]. Cas8c assays were performed in 1 × binding buffer containing 150 mM KCl. The 5′ fluorescein-labeled RNAs (IDT) were added to protein solution at a final concentration of 10–20 nM, and the RNA-protein mixture was incubated at 37°C for 30 min. Measurements were made by excitation at 485 nm and monitoring emission at 535 nm. All data analysis was performed using Prism software (GraphPad). Each experiment was performed in at least triplicate and error bars, when included, represent the SD. For further details, see Supplemental Experimental Procedures.
Isothermal Titration Calorimetry
All isothermal titration calorimetry (ITC) experiments were conducted on a MicroCal Auto-iTC200 system (GE Healthcare). Purified Cas5c and Cas7 were dialyzed at 4°C against 20 mM HEPES–NaOH (pH 7.5), 100 mM KCl, 5% (v/v) glycerol, and 1 mM TCEP; 100–150 μM Cas7 was titrated into the cell containing 10–30 μM Cas5c using one 0.5-μl injection followed by twelve 3.1-μl injections at 37°C. For Cas8c ITC experiments, all proteins were dialyzed at 4°C against 20 mM HEPES–NaOH (pH 7.5), 300 mM KCl, 10% (v/v) glycerol, and 1 mM TCEP; 175 μM Cas8c was titrated into the cell containing 17.5–30 μM Cas5c, and 200 μM Cas8c was titrated into 20–35 μM Cas7 using one 0.5-μl injection followed by twelve 3.1-μl injections at 37°C. Reported values for each interaction are the average of three independent experiments. Modified Origin software (GE Healthcare) was used for analysis. For further details, see Supplemental Experimental Procedures.
Electrophoretic Mobility Shift Assays
Gel shift assays were performed in 1 × binding buffer [20 mM HEPES–NaOH (pH 7.5), 100 mM KCl, 5% (v/v) glycerol, 1 mM TCEP, 10 μg/ml heparin, and 0.01% Igepal CA-630] with radiolabeled DNA added to a final concentration of 0.15–0.5 nM. Samples were incubated at 37°C for 30 min and resolved at 4°C on 6% (v/v) 29:1 polyacrylamide gels containing 1 × TBE. Gels were quantified using ImageQuant software (GE Healthcare) and the data were fit to standard one-site binding isotherm using Prism (GraphPad). Reported KD values are the average of at least three independent experiments, and error bars represent the SD. For further details, see Supplemental Experimental Procedures.
Negative-Stain Electron Microscopy
Cas7 samples were applied to glow-discharged 400-mesh continuous carbon grids and stained with 2% (w/v) uranyl acetate solution. Data were acquired using a Tecnai F20 Twin transmission electron microscope operated at 120 keV. For further details, see Supplemental Experimental Procedures.
Cryo-EM
4-μl droplets of the sample (~0.25 mg·ml−1) were placed onto C-flat grids and rapidly plunged into liquid ethane. Data were acquired using a FEI Titan Krios transmission electron microscope (at Janelia Research Campus Cryo-EM Shared Resource), operated at 300 keV on a Gatan K2 Summit direct electron detector. Three-dimensional reconstruction was performed using RELION (Scheres, 2012). For further details about cryo-EM, 3D reconstruction, and docking and analysis, see Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Desulfovibrio vulgaris Cas5c cleaves pre-crRNAs and stays bound to the 5′ handle
The Cas7 backbone assembles cooperatively, capped by the crRNA stem-loop
Cascade/I-C resembles type I and III complexes; large and small subunits are fused
Cryo-EM reveals R-loop stabilization by Cas8c and pronounced target DNA bending
In Brief.
Hochstrasser et al. describe the assembly and architecture of a minimal CRISPR interference complex. Type I-C Cascade’s three subunits each play multiple functional roles. Cryo-EM structures reveal the importance of the Cas8c subunit in PAM recognition and R-loop formation.
Acknowledgments
We thank R. Louder, A. Patel, E. Kellogg, P. Grob, T. Houweling, Z. Yu and C. Hong for expert electron microscopy assistance and S. Floor, P. Kranzusch, T. Liu, J. Nuñez, S. Sternberg, S. Strutt, and R. Wilson for helpful discussions and critical reading of the manuscript. D.W.T. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2218-15). J.A.D and E.N. are Howard Hughes Medical Institute Investigators. J.A.D. is a co-founder of Editas Medicine, Intellia Therapeutics, and Caribou Biosciences and a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics, and Driver.
Footnotes
ACCESSION NUMBERS
The accession numbers for the structures of apo-Cascade/I-C, 2× dsDNA target-bound Cascade/I-C, and 1× dsDNA target-bound Cascade/I-C reported in this paper are EMDataBank: EMD: 8294, EMD: 8295, and EMD: 8296, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, two tables, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.07.027.
AUTHOR CONTRIBUTIONS
M.L.H., D.W.T., J.E.K., E.N., and J.A.D. designed research. M.L.H. conducted the experiments and prepared the EM samples. D.W.T. and J.E.K. performed electron microscopy and determined 3D structures. M.L.H., D.W.T., J.E.K., E.N., and J.A.D. analyzed the data. M.L.H., D.W.T, E.N., and J.A.D. wrote the manuscript.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- Blosser TR, Loeff L, Westra ER, Vlot M, Künne T, Sobota M, Dekker C, Brouns SJJ, Joo C. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell. 2015;58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Sun CL, Brown CT, Sharon I, Anantharaman K, Probst AJ, Thomas BC, Banfield JF. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 2016;7:10613. doi: 10.1038/ncomms10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, Almo SC, Mehta G, MacMillan AM. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–2028. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L. Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucleic Acids Res. 2016;44:5872–5882. doi: 10.1093/nar/gkw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RP, Xiao Y, Ding F, van Erp PBG, Rajashankar K, Bailey S, Wiedenheft B, Ke A. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530:499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Wiedenheft B. A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PBG, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Koo Y, Ka D, Kim EJ, Suh N, Bae E. Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system. J Mol Biol. 2013;425:3799–3810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Structural Principles of CRISPR RNA Processing. Structure. 2015;23:13–20. doi: 10.1016/j.str.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copié V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Héroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Punetha A, Sivathanu R, Anand B. Active site plasticity enables metal-dependent tuning of Cas5d nuclease activity in CRISPR-Cas type I-C system. Nucleic Acids Res. 2014;42:3846–3856. doi: 10.1093/nar/gkt1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell. 2015;163:854–865. doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 2015;10:1534–1543. doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and functional fharacterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Staals RHJ, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DW, Zhu Y, Staals RHJ, Kornfeld JE, Shinkai A, van der Oost J, Nogales E, Doudna JA. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp PBG, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. 2015;43:8381–8391. doi: 10.1093/nar/gkv793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venclovas Č. Structure of Csm2 elucidates the relationship between small subunits of CRISPR-Cas effector complexes. FEBS Lett. 2016;590:1521–1529. doi: 10.1002/1873-3468.12179. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


