Abstract
Cholesterol accumulates in prostate lesions and has been linked to prostate cancer (PCa) incidence and progression. However, how accumulated cholesterol contributes to PCa development and progression is not completely understood. Cholesterol sulfate (CS), the primary sulfonation product of cholesterol sulfotransferase (SULT2B1b), accumulates in human prostate adenocarcinoma and precancerous prostatic intraepithelial neoplasia (PIN) lesions compared to normal regions of the same tissue sample. Given the enhanced accumulation of CS in these lesions, it was hypothesized that SULT2B1b-mediated production of CS provides a growth advantage to these cells. To address this, PCa cells with RNAi-mediated knockdown (KD) of SULT2B1b were used to assess the impact on cell growth and survival. SULT2B1b is expressed and functional in a variety of prostate cells and the data demonstrate that SULT2B1b KD, in LNCaP and other androgen-responsive (VCaP and C4-2) cells, results in decreased cell growth/viability and induces cell death. SULT2B1b KD also decreases androgen receptor (AR) activity and expression at mRNA and protein levels. While AR overexpression has no impact on SULT2B1b KD-mediated cell death, addition of exogenous androgen is able to partially rescue the growth inhibition induced by SULT2B1b KD in LNCaP cells. These results suggest that SULT2B1b positively regulates the AR either through alterations in ligand availability or by interaction with critical co-regulators that influence AR activity.
Implications
These findings provide evidence that SULT2B1b is a novel regulator of AR activity and cell growth in prostate cancer and should be further investigated for therapeutic potential.
Keywords: SULT2B1b, prostate cancer, androgen receptor, cholesterol, cell death
INTRODUCTION
Prostate cancer (PCa) is the second leading cause of non-cutaneous cancer death among males in the United States with 180,890 new cases and 26,120 deaths estimated in 2016(1). Current treatments for organ-confined disease, including prostatectomy and radiation, have proven to be successful. Unfortunately, the initial treatment for metastatic disease, androgen-deprivation therapy (ADT), is palliative, providing only approximately 11 months of failure-free survival(2). Progression leads to the development of castration resistant prostate cancer (CRPC) for which second generation ADT is available but the response is also short-lived(3,4). Further investigation into the biology of PCa and identification of novel regulators of cellular growth and/or androgen receptor (AR) activity is required for alternate, more curative therapeutic options for men with advanced PCa.
Cholesterol metabolism dysregulation has been investigated in PCa for many years leading to a better understanding of cholesterol’s contribution to disease progression. Through modulation of intracellular signaling and pro-survival pathways, cholesterol metabolism is believed to play a major role in PCa development and progression to the castration non-responsive state, but the mechanisms involved remain elusive(5–9). A number of modified forms of cholesterol are found within cells, namely hydroxysterols, cholesterol esters, and cholesterol sulfate (CS). Previously, desorption electrospray ionization mass spectrometry (DESI-MS) was used to identify unique lipid profiles in human prostate tissue specimens, where CS was observed to be elevated in prostatic intraepithelial neoplasia (PIN) and adenocarcinoma lesions compared to normal tissues(10). This suggested that the enzyme(s) responsible for converting cholesterol to CS were not simply expressed, but functional in these tissues.
Cytosolic sulfotransferases (SULTs) are a class of enzymes that catalyze the sulfonation of various hydroxysteroid substrates. SULT members within this class each have preferential affinity for various substrates. For example, SULT2A1 is expressed in the liver and adrenal gland where it utilizes dehydroepiandrosterone (DHEA) as a substrate to produce and secrete the abundant circulating hormone precursor, DHEA-sulfate; SULT2A1 has been studied in PCa as a result of this function(11). SULT2B1 has two isoforms, SULT2B1a and SULT2B1b, which differ only by 15 amino acids at the N-terminus due to alternate transcriptional start sites of the same gene. SULT2B1a and SULT2B1b each have different substrate affinities.(12) SULT2B1b, the “cholesterol sulfotransferase,” is expressed in a variety of tissues, including the prostate, placenta, and skin, but it is unclear whether SULT2B1a is expressed at the protein level in any human tissues(12–14). SULT2B1b-catalyzed production of cholesterol sulfate (CS) is critical for skin barrier layer formation and proper membrane function of a variety of cell types(13,15). SULT2B1b also is known to influence cell metabolism through regulation of the Liver X Receptor (LXR), a global regulator of cholesterol homeostasis(16).
Studies have shown a potential role of SULT2B1b in the growth and progression of cancer cells(17–20). SULT2B1b was studied in PCa in androgen-free conditions supplemented with DHEA(19,21). However, SULT2B1b has not been previously evaluated in androgen-replete conditions that mimic normal biology and PCa progression, consistent with the conditions in which CS accumulation was observed(10). Here, we investigate the impact of SULT2B1b in PCa cells in the presence of androgen with the hypothesis that enzymatic activity of SULT2B1b supports cell growth and contributes to progression of the disease through modulation of LXR and AR activity. Studies reported herein verify that SULT2B1b is an important metabolic enzyme in multiple PCa cell lines by demonstrating that SULT2B1b knockdown (KD) induces caspase-3 activation and cell death. Additionally, data show that SULT2B1b activity positively correlates with AR activity in PCa cells. This positive regulation of AR by SULT2B1b appears to be LXR-independent and may be due to alterations in AR expression levels or ligand availability, since replenishing testosterone partially overcomes the growth-inhibitory effects of SULT2B1b KD. These studies support a novel mechanism of AR regulation in PCa cells. Thus, therapeutic targeting of SULT2B1b may be a novel approach to inhibiting AR activity in PCa.
MATERIALS AND METHODS
Human Prostate Tissue Source and Immunohistochemistry
A random selection of four unpaired and four paired frozen, human prostate tissue samples were obtained from the Indiana University Simon Cancer Center Tissue Bank. All tissues were handled in accordance with the Indiana University institutional review board and prepared as previously described(10). Frozen sections were prepared on slides and subjected to hematoxylin and eosin (H&E) staining and pathological evaluation.
For SULT2B1b immunohistochemistry (IHC), slides containing frozen tissue specimens were rinsed with ddH20 and subjected to the following treatments: 3% hydrogen peroxide, protein block solution (#X0909, Dako), 1:500 anti-SULT2B1 primary antibody (ab88085, Abcam), peroxidase-linked polymeric anti-mouse antibody (K4006, Dako), and 3,3’-diaminobenzidine (Dako). Samples were washed between each step and Gills II hematoxylin was used as a counterstain.
Desorption Electrospray Ionization (DESI) Mass Spectrometry
A laboratory-built DESI ion source, similar to the commercial 2D source from Prosolia, Inc. (Indianapolis, IN, USA) was coupled to a linear ion trap mass spectrometer (LTQ) controlled by XCalibur 2.0 software (ThermoFisher Scientific, Waltham, MA) and used in all experiments. The negative ionization mode was used with the automatic gain control (AGC) inactivated. Tissues were analyzed as previously described(10). For cell lines, the spray solvent used for DESI-MS was dimethylformamide (DMF)-acetonitrile (ACN) at a 1:1 ratio in volume; both solvents were purchased from Mallinckrodt Baker Inc.(Phillipsburg, NJ) and infused over the cell line material at 1.0 µL/min flow rate through the instrument syringe pump. The DESI source parameters are indicated in Supplemental Materials and Methods. Each cell line was analyzed by manual movement of a 2D moving stage where the slide was held in a fixed position. Full scan mass spectra were acquired in negative ion mode in the mass range m/z 200–1000.
Cell Lines
LNCaP, VCaP, RWPE-1, PC-3, and DU 145 were purchased from the American Tissue Culture Collection (ATCC; Manassas, VA) and maintained in media conditions identical to those recommended by ATCC. C4-2 cells were a generous gift from MD Anderson (Houston, TX) and were maintained in T-medium (Invitrogen) supplemented with 10% FBS and 1% streptomycin/penicillin. Cell lines were verified using cell line authentication testing from DDC Medical (Fairfield, OH) through the generous support of the Prostate Cancer Foundation and results are shown in Supplemental Table 1. All experiments were completed within 20 passages of acquisition from ATCC.
RNA interference (RNAi)
For siRNA KD, SULT2B1 (HSC.RNAI.N004605.12.2), NR1H2 (LXRβ, HSC.RNAI.N007121.12.2), and NR1H3 (LXRα, HSC.RNAI.N005693.12.2) predesigned DsiRNA duplexes were purchased from IDT. Non-targeting siRNA was purchased as a control (Dharmacon Scientific). Transfection using Lipofectamine RNAiMax (Invitrogen) was completed for all siRNA transfections according to the manufacturer’s instructions. For double siRNA KD, combinations of the same amount of siRNA were incubated with double the recommended transfection reagent prior to adding to cells in culture. “Controlx2” indicates twice the amount of non-targeting siRNA and double RNAiMax compared to “Control” samples. For shRNA KD in LNCaP cells, SULT2B1 and control sequences 5’-CCTCTATCATTACTCCAAGAT-3’ (LNKD) and 5’-CCATTAACTCTTTCCCGAAAT-3’ (LNCon), respectively, were inserted into the pLNKO.1 Tet-ON vector (Addgene) and transfected using FuGene HD (Promega). Finally, SULT2B1 shRNA and copGFP Control Lentiviral particles (Santa Cruz Biotechnology, Inc. sc-44399-V and sc-108084, respectively) were transduced in LNCaP cells using Polybrene ® (Santa Cruz sc-134220). The appropriate method of SULT2B1b KD is indicated in figure legends.
RNA isolation, cDNA amplification, and qRT-PCR
Total RNA was isolated using the E.Z.N.A. Total RNA Kit I (Omega Biotek) according to the manufacturer’s instructions. cDNA was prepared by mixing 2–4 µg total RNA, 250µM dNTPs (Amresco), 0.5 µM each of random hexamer and oligo(dT)15 primers (Promega), and 200 units M-MLV reverse transcriptase with included reaction buffer(NEB). qRT-PCR was conducted using PerfeCTa® FastMix® II (Quanta Biosciences) according to the manufacturer’s instructions. PrimeTime ® qRT-PCR gene probes (IDT) used for these studies include: ABCG1 (Hs.PT.56a.20848083.g), AR (Hs.PT.56a.38770693), KLK3 which will be identified herein as “PSA” (Hs.PT.56a.38546086.g), NR1H2 (Hs.PT.56a.45297581.g), NR1H3 (Hs.PT.56a.40638751.g), and SULT2B1 (Hs.PT.56a.25562421.g). “Relative mRNA expression” levels were calculated and normalized to 18s rRNA (Cat#4308329, Applied Biosystems), as described previously.(22)
Antibodies and Reagents
The following antibodies were used for western blots: anti-β-actin clone 8H10D10 ab#3700, anti PARP ab#9542 (Cell Signaling Technologies); anti-SULT2B1 ab88085 (Abcam); anti-AR clone 441, anti-β-tubulin T0198, and goat anti-mouse IgG-HRP (Santa-Cruz); anti-human PSA A0562 (Dako); and goat anti-rabbit IgG-HRP (Vector Laboratories). The approximate molecular weight of each protein is indicated with blots in relevant figures.
Caspase-3 activity was measured after 72 hours of siRNA transfection using the Caspase-Glo® 3/7 assay (Promega, Madison, WI) and analyzed using a Luminoskan Ascent microplate luminometer (Thermo Scientific) according to the manufacturer’s instructions. CellTiter 96® AQueous One Solution Reagent (Promega) was used for MTS cell proliferation assays, and relative absorbance was quantified by a Multiskan FC plate reader (ThermoScientific). For relevant assays, 10µM pan-caspase inhibitor Z-VAD-FMK (Promega) and/or 20µM Necrostatin-1 (Sigma) was added to cells at the time of siRNA transfection. Synthetic androgen, R1881 (Sigma), was used at the indicated concentrations.
Production of DU 145-SULT2B1b and LNCaP-SULT2B1b cells
To produce cell lines with tetracycline-inducible expression of SULT2B1b, consecutive lentiviral transductions were performed. DU 145 or LNCaP cells were transduced with lentivirus produced from the Lenti-X™ Tet-On® Advanced Inducible Expression System (Clontech) and then from the pLVX-Tight-Puro lentiviral vector (Clontech) containing human SULT2B1b cDNA. The SULT2B1b cDNA transcript was obtained from Dr. Charles Falany (University of Alabama). Stable selection of both transductions resulted in DU 145-SULT2B1b or LNCaP-SULT2B1b cells. Additional information is available in Supplemental Materials and Methods.
Soft-agar assay
LNCaP cells were transfected with shRNA plasmids in 60-mm dishes for 24 hours and harvested. Soft agar plates were prepared as described in Supplemental Materials and Methods. Each layer received 1 µg/mL doxycycline for induction of Tet-ON shRNA plasmids. Plates were incubated for 7 days and then cell colonies were counted.
Flow Cytometry: Cell cycle analysis and Annexin-V staining
Treated cells were harvested and then washed twice with PBS, fixed in 75% ethanol, and stored at −20°C for up to 7 days. To prepare for flow cytometry, cells were stained with PI (Biolegend #421301) and, in relevant assays, Annexin-V (Biolegend #640920) according to the manufacturer’s instructions. Staining buffer details are indicated in Supplemental Materials and Methods. Cells were stained at room temperature before analysis on FACSCanto II (BD Biosciences, San Jose, CA). Further analysis of DNA content was completed via FlowJo software version 9 (TreeStar Inc., Ashland, OR).
Luciferase assays and reporter constructs
AR activity was measured by luciferase assay, as described previously(23). SULT2B1b or AR overexpression was conducted transiently under a CMV promoter. Briefly, LNCaP cells were transfected with shRNA/siRNA, pRL-TK (Promega), and a construct controlled by the AR-responsive portion of the PSA promoter driving firefly luciferase(23). Assays with shRNA KD constructs were transfected simultaneously with luciferase plasmids using Fugene HD (Promega). Assays with siRNA KD were first transfected with luciferase plasmids using Fugene HD for 8–16 hours and then media was changed for consecutive transfection with siRNA as described above. After 24 hours RNAi transfection, 1nM R1881 (Sigma) or ethanol control was added to respective wells and incubated for an additional 24 hours. Then, cell lysates were tested for Firefly and Renilla luciferase activity using the Dual Luciferase Reporter Assay kit (Promega)(24) and relative luciferase activity (RLU=Firefly/Renilla) of the AR is shown as mean +/− SEM from at least three independent experiments performed in triplicate.
LXR activity was determined by transfecting LNCaP cells with LXR-RE or Negative Control plasmids (Qiagen CCS-0041L) along with Control or SULT2B1b siRNA. After indicated time of transfection, cells were lysed and analyzed as above. Data was interpreted by determining RLU and then further normalizing samples containing the LXR-RE reporter to samples transfected with the Negative Control reporter.
Statistical analysis
Data were represented as the mean ± standard error of the mean. Statistical analysis was performed using the unpaired two-tailed student’s t test or two-way ANOVA and analyzed by GraphPad Prism 5 (La Jolla, CA) or SAS Enterprise (Cary, NC).
RESULTS
Cholesterol sulfate accumulation correlates with SULT2B1b expression in PCa cells and human tissue specimens
We previously demonstrated that CS accumulation occurs within PIN and PCa tissues compared to normal prostate tissues(10). As an extension of the previous work, a small subset of samples (12 samples ranging from normal to high-grade carcinoma obtained from 8 different patients) was randomly selected from the Indiana University Simon Cancer Center Tissue Bank for comparison of the co-localization of SULT2B1b staining by immunohistochemistry (IHC) and areas of CS accumulation by DESI-MS. These studies showed that areas with CS detection were limited to regions of positive SULT2B1b expression (Figure 1A, Supplemental Figure 1). To study the role of SULT2B1b in vitro, human prostatic epithelial cell lines were analyzed for SULT2B1b expression and activity. Androgen responsive cell lines LNCaP, VCaP, RWPE-1, and the castration non-responsive line C4-2 generally express higher levels of SULT2B1b than AR- cell lines, such as PC-3 and DU 145 cells (Figure 1B–C). In general, SULT2B1b expression in these cell lines corresponds to detection of CS, indicating functional activity of SULT2B1b (Figure 1D). However, it is noteworthy that SULT2B1b can be expressed but not active, as in PC-3 cells (Figure 1B–D). To demonstrate that CS production was the result of SULT2B1b activity specifically, expression of SULT2B1b was induced in cells that normally express a minimal level of SULT2B1b. A stable, doxycycline-inducible DU 145-SULT2B1b cell line was developed to overexpress SULT2B1b. Induction of SULT2B1b expression by doxycycline resulted in a striking increase in the relative abundance of CS (Figure 1E–F). Similarly, inducing SULT2B1b overexpression in doxycycline-inducible LNCaP-SULT2B1b cells resulted in an increase in the relative abundance of CS (Supplemental Figure 2A). Conversely, SULT2B1b knockdown (KD) in LNCaP cells decreased the relative abundance of CS (Supplemental Figure 2B). While not quantitative, these DESI-MS observations provide strong correlative evidence that SULT2B1b activity is responsible for the accumulated CS in PCa.
Figure 1. Cholesterol sulfate accumulation correlates with SULT2B1b expression in PCa cells and human prostate specimens.
A) Analysis of human prostate cancer by IHC for SULT2B1b (left), H&E staining (middle), and DESI-MS detection of cholesterol sulfate at m/z 465 (right). These sections were derived from one representative patient out of a sampling of 8 patients. B–C) Basal SULT2B1b expression was determined in indicated prostate cell lines by qRT-PCR (normalized to 18s rRNA) (B) and western blot (C). Error bars represent the mean +/− SEM of 3 replicate analyses. D) DESI-MS spectra of prostate cell lines from (B–C). Cholesterol sulfate expression is indicated by a peak at m/z 465 (indicated in red). E–F) Stable DU145-SULT2B1b cells were treated with or without 1 µg/mL doxycycline for induction of SULT2B1b cDNA expression and analyzed by DESI-MS. E) A western blot detecting SULT2B1b and β-actin in wildtype DU145 (Ø), DU145-SULT2B1b +Dox, and DU145-SULT2B1b –Dox is shown. F) DESI-MS negative ion spectra in DU145-SULT2B1b +/− Dox cells from (E) indicate the relative abundance of cholesterol sulfate in cells by a peak at m/z 465.
Targeted knockdown of SULT2B1b impairs growth/viability of PCa cells
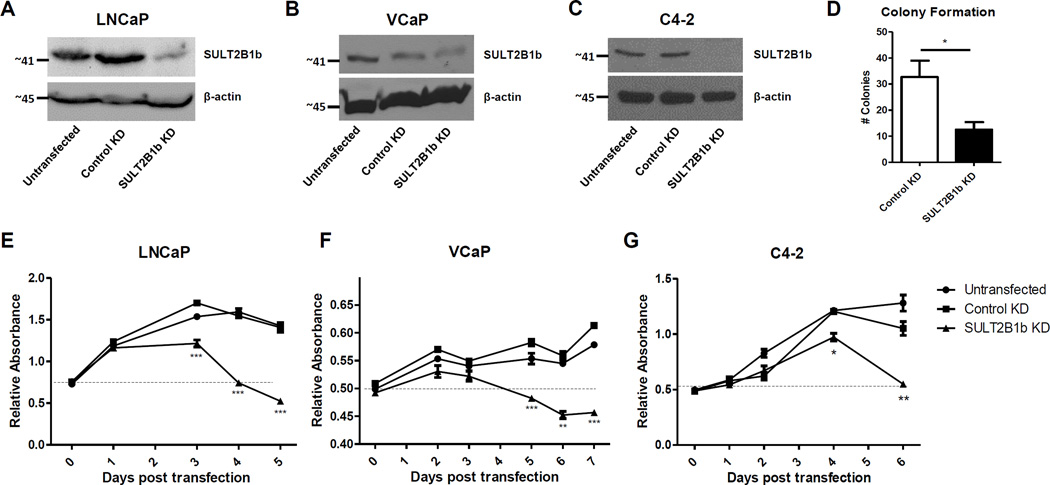
In order to determine if SULT2B1b impacts PCa through production of its product, CS, exogenous CS was added to LNCaP and observed for alterations in phenotype and growth characteristics. Notably, no overt phenotypic changes in growth or cell death were observed (data not shown). Similarly, overexpression of SULT2B1b in LNCaP cells did not enhance growth (data not shown). Thus, RNAi-mediated KD of SULT2B1b was performed to better understand the function of this enzyme in PCa cells. SULT2B1b KD resulted in a decrease in growth/viability of LNCaP, VCaP, C4-2, and RWPE-1 cells as well as decreased soft-agar colony formation in LNCaP cells (Figure 2, Supplemental Figure 3). Importantly, a significant decrease in cell growth/viability was observed using multiple SULT2B1b RNAi sequences in LNCaP cells (Figure 2A,D,E, Supplemental Figure 4A).
Figure 2. Targeted knockdown of SULT2B1b impairs growth/viability of PCa cells.
A–C) Western blot of SULT2B1b protein expression in LNCaP (A), VCaP (B) and C4-2 (C) cells after a 72 hour incubation with non-targeting (control), or SULT2B1b siRNA. D) LNCaP cells were transfected with scrambled control or SULT2B1b shRNA vector. After 24 hours, cells were split into 96-well plates and allowed to grow for 7 days before counting the number of colonies. *indicates p<0.05. Data represents mean +/− SEM of three independent experiments. E–G) Cell viability/growth curves in LNCaP (E), VCaP (F) and C4-2 (G) cells with treatment of control or SULT2B1b siRNA on day 0. Cell viability was measured via MTS assay at indicated time points. Each point represents the mean +/− SEM of 3–4 duplicate wells from each sample and growth curves are representative of three independent experiments. Statistics were completed at each time point using a student’s t-test with Bonferroni correction. *=p-value of <0.05, **=p-value of <0.01, and ***= p-value of <0.001 compared to control KD cells.
SULT2B1b knockdown induces cell death in PCa cells
Further investigations showed SULT2B1b KD increased the percentage of sub-G1 nuclei by cell cycle analysis and significantly increased caspase-3 activity and PARP cleavage in both LNCaP and VCaP cells (Figure 3A–C). Increased sub-G1 nuclei percentages and caspase-3 activation was also observed in C4-2 cells with SULT2B1b KD (Supplemental Figure 5). Furthermore, LNCaP cells with SULT2B1b KD analyzed by flow cytometry showed an increased percentage of Annexin-V+/propidium iodide (PI)- cells compared to control KD cells (Figure 3D–E).
Figure 3. SULT2B1b KD induces cell death in PCa cells.
LNCaP and VCaP cells were transfected with control or SULT2B1b siRNA and harvested after 72 hours. A) Cell cycle analysis of LNCaP (left) and VCaP (right) cells. Cell samples were analyzed by flow cytometry after fixing and staining with propidium iodide (PI). Data is representative of three independent experiments. B) Caspase-3 activity in LNCaP (left) and VCaP (right) cells was determined by luminescence assay after control or SULT2B1b siRNA KD. Results indicate the mean ± SEM of three independent experiments.***indicates p<0.001 C) Western blots of indicated proteins after control or SULT2B1b siRNA transfection in LNCaP (left) and VCaP (right) cells. D) A representative sample of LNCaP cells after incubation with control or SULT2B1b siRNA Annexin-V and PI. Cells were harvested and stained before analysis by flow cytometry. E) Summary of replicate samples shown in (D). Data represents the %Annexin-V+/PI- cells out of the total population of events (excluding doublets) and is representative of three independent experiments performed in duplicate. ****indicates p<0.0001
To determine whether caspase activation was an essential aspect of the induced cell death, abrogation of apoptosis by the addition of a pan-caspase inhibitor, Z-vad-fmk (Z-vad), was tested in SULT2B1b KD LNCaP cells. Results indicated that Z-vad successfully blocked caspase-3 activation and abrogated the enhancement of Annexin-V+/PI- cells after SULT2B1b KD, but LNCaP cell viability and, ultimately, cell death was not altered (Figure 4A–C). While the complete mechanism(s) of cell death are not understood, it is possible that after pan-caspase inhibition SULT2B1b KD cells activated alternative cell death pathways. Additional studies completed in SULT2B1b KD cells with co-treatment of Z-vad and an additional inhibitor of RIP1 kinase, Necrostatin-1, yielded a greater proportion of viable cells compared to Z-vad treatment alone (Figure 4D). It may be of note that although LNCaP cells with SULT2B1b KD respond to Necrostatin-1 treatment with concurrent pan-caspase inhibition, our studies investigating reactive oxygen species production and AnnexinV-/PI+ cells did not show that RIP1 kinase-dependent death pathways (i.e. during necroptosis(25)) are directly induced by SULT2B1b KD (data not shown).
Figure 4. Cell death resulting from SULT2B1b KD persists with pan-caspase inhibition.
A) Caspase-3 activity in LNCaP cells at 72 hours after transfection. Bars indicate the mean±SEM of duplicate experiments. B) Cell viability determined via trypan blue cell counting in LNCaP cells at indicated time points after control or SULT2B1b siRNA KD with and without addition of 20µM pan-caspase inhibitor, Z-vad. C) LNCaP cells were treated with 20µM Z-vad at the time of control or SULT2B1b siRNA transfection. After 72 hours, cells were harvested, incubated with Annexin-V and PI, and analyzed by flow cytometry. Bars represent the mean ± SEM of %Annexin-V+/PI- cells within total events collected (excluding doublets) from three independent experiments. D) Graph of MTS assay endpoint analysis in LNCaP cells after 96 hours of control or SULT2B1b siRNA KD as well as treatment with 20µM Z-vad and/or 10µM Necrostatin-1. Z-vad was added every 24 hours and Necrostatin-1 was added every 48 hours in applicable samples. Bars represent the mean +/− SEM of three independent experiments. Statistical analysis was conducted using two-way ANOVA and Tukey’s post-test. Different letters indicate significant differences among treatments.
SULT2B1b activity modulates AR activity in PCa cells
PCa cells rely on AR activity for growth and stimulation and previous studies have demonstrated that cholesterol can be used as a precursor for androgen synthesis(6,26). Thus, the impact of SULT2B1b modulation on AR expression and activity was evaluated. The data show that SULT2B1b KD by various methods in LNCaP cells decreases AR expression as well as AR activity measured by both prostate specific antigen (PSA) expression and transcription of the AR-response element (AR-RE) within the PSA promotor (Figure 5, Supplemental Figure 4B–C).
Figure 5. SULT2B1b KD decreases AR expression and activity.
A–B) LNCaP cells treated with control or SULT2B1b siRNA were harvested at 72 hours for mRNA expression by qRT-PCR (A) and protein by western blot (B) of indicated genes and proteins, respectively. C) A luciferase reporter construct controlled by the AR-responsive portion of the PSA promoter was transfected into LNCaP cells followed by transfection with control or SULT2B1b siRNA. Cells were harvested and lysed 48 hours after siRNA transfection. Firefly luciferase activity was normalized to Renilla. Relative luciferase units (RLU) are shown for cell conditions with or without addition of 1nM R1881 added 24 hours before harvest. Bars represent the mean ± SEM of three independent experiments. * indicates p<0.05 determined by t-test.
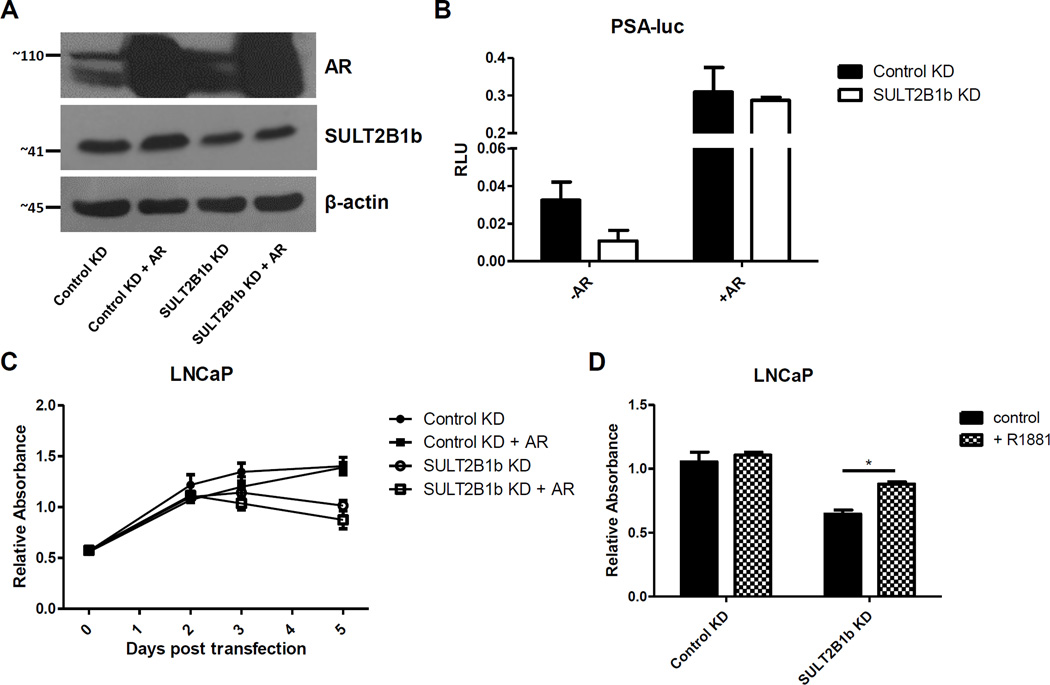
Further investigation showed that the decreased expression of the AR mediated by SULT2B1b KD was not the cause of decreased cell growth, since transient overexpression of the AR in LNCaP cells with SULT2B1b KD showed no increase in cell growth over LNCaP cells with SULT2B1b KD alone (Figure 6A–C). Endpoint analysis of a growth assay in LNCaP cells shows that replenishing the culture medium with synthetic androgen, R1881, partially, yet significantly, rescued the reduced cell growth in SULT2B1b KD LNCaP cells back to Control KD levels (Figure 6D).
Figure 6. AR ligand addition, but not AR overexpression, partially rescues cell growth in LNCaP with SULT2B1b KD.
A) Western blot showing expression levels of indicated proteins with Control or SULT2B1b KD ± AR overexpression. Cells were harvested 60 hours after transfection with Control or SULT2B1b siRNA. B) Luciferase assay showing AR activity after 72 hours of siRNA transfection. C) MTS assay of LNCaP cells with or without AR overexpression followed by Control or SULT2B1b siRNA transfection. The time of siRNA transfection was considered Day 0 and relative absorbance was identified at the indicated time points. Each point represents the mean +/− SEM of 3 replicate wells. D) 72 hour endpoint analysis from an MTS assay of LNCaP cells that were transfected with control or SULT2B1b siRNA with or without addition of 10nM R1881 daily. Each bar represents the mean +/− SEM of 3 replicate wells and similar results were found in duplicate experiments. * indicates p<0.05 determined by two-way ANOVA.
SULT2B1b regulates the AR independently of LXR
Transient overexpression of the human SULT2B1b cDNA was performed to address whether or not SULT2B1b activity regulates LXR activity in PCa cells(27). LNCaP cells with SULT2B1b overexpression (hSULT2B1b vector) showed a significant decrease in LXR activity and a trend of decreased transcription of downstream target gene, ATP-binding cassette (ABC)-G1 (Figure 7A–C).
Figure 7. SULT2B1b regulates the AR independently of LXR.
A–C) LNCaP cells were transiently transfected with a control vector (pcDNA3.1) or a construct containing the human SULT2B1b cDNA under the CMV promoter (hSULT2B1b vector), resulting in SULT2B1b overexpression. Western blot (A) or qRT-PCR (B) was performed 72 hours after transfection. Gene expression was normalized to 18S rRNA. Bars represent the mean +/− SEM of triplicate samples. C) A LXR-responsive luciferase reporter construct under the minimum SV40 promoter was transfected into LNCaP cells with basal expression (control vector) or overexpression (hSULT2B1b vector) of SULT2B1b. Firefly luciferase activity was normalized to Renilla. LXR activity was measured and further normalized to negative control luciferase reporter activity for each sample. D) LNCaP cells were treated with control or LXRβ siRNA and analyzed for LXR activity via luciferase assay after 72 hours. RLU was quantified and normalized as described in (C). T-tests were completed in (B–D) to determine significant differences. *=p-value of <0.05 and ****=p-value of <0.0001 compared to control cells. E–F) Double siRNA KD of LXRβ and SULT2B1b was conducted. Bars represent the mean ± SEM of triplicate samples. E) SULT2B1b and LXRβ expression determined via qRT-PCR. Gene expression was normalized to 18S rRNA. *=p-value of <0.05, **=p-value of <0.01, and ***= p-value of <0.001 compared to control/controlx2 KD cells by One-way ANOVA. F) AR activity was evaluated through PSA expression by qRT-PCR. Gene expression was normalized to 18S rRNA. Statistical analysis was conducted using a one-way ANOVA and Tukey’s post-test. Different letters indicate significant differences among treatments.
Additionally, since LXR activation has been shown to decrease AR activity(28), the question of whether activation of LXR in SULT2B1b KD cells causes decreased AR transcription was addressed. Double siRNA KD of SULT2B1b and LXRβ was performed followed by the assessment of AR activity through PSA expression (Figure 7D–F). LXRα was excluded because LXR activity in LNCaP cells was demonstrated to be due to transcriptional activation of LXRβ (Figure 7D, Supplemental Figure 6). Double KD of SULT2B1b/LXRβ did not impact the siRNA KD efficiency compared to SULT2B1b or LXRβ KD alone (Figure 7E). Regardless of whether or not LXR was present/active, SULT2B1b KD was still able to inhibit AR activity, as shown by a significant reduction in PSA expression (Figure 7F).
DISCUSSION
We previously reported that CS accumulates in precancerous and cancerous human prostate specimens(10). To date, the role of CS in the prostate or the impact of accumulated CS within prostatic cells is not known. Data in this study utilized DESI-MS technology to show CS accumulation is the result of increased SULT2B1b activity, rather than simply expression, within prostate cells (Figure 1, Supplemental Figure 1). Detection of accumulated CS at the macroscopic level reveals either enhanced activity of SULT2B1b or an inability of the tissue to secrete or utilize this product. Nonetheless, the use of DESI-MS in these studies allowed the validation that manipulation of SULT2B1b in cell lines led to functional alterations (Figure 1E–F, Supplemental Figure 2).
Mechanisms of SULT2B1b function other than sulfonation (15,16) have not been well described. In contrast to our findings, two groups independently reported that SULT2B1b KD promotes LNCaP cell growth while maintained in androgen-depleted conditions with DHEA supplementation(19,21). Although we do not fully understand the reason(s) for this discrepancy, culture medium conditions in growth assays may play a role. While their data support the logical hypothesis that SULT2B1b-mediated sulfonation causes “inactivation” of steroid precursors (i.e. DHEA) leading to decreased AR-dependent proliferative signaling, data herein directly show that SULT2B1b activity is positively correlated to AR activity, which implies an alternative mechanism of AR regulation is occurring. While our studies in cell lines support our previous findings that CS accumulates in precancerous and cancerous prostate tissues compared to normal counterparts, we also found that a single benign cell line, RWPE-1, has robust SULT2B1b expression and activity, which likely reflects the heterogeneity of SULT2B1b expression/activity within prostate tissue as has been reported by others(19). In contrast to the findings of Seo et al.(19), recent evidence in cancer cells suggest SULT2B1b plays a role in cancer growth/aggressiveness. It has been demonstrated that SULT2B1b activity increases gastric cancer angiogenesis and tumor volume, SULT2B1b activity promotes hepatocellular carcinoma cell growth in vitro and in vivo, and SULT2B1b expression correlates with poor prognosis and promotes tumor cell growth in colorectal cancer patients.(17,18,20) In fact, the Human Protein Atlas database (http://www.proteinatlas.org/ENSG00000088002-SULT2B1/cancer) of tissue samples supports elevated expression levels of SULT2B1b in PCa compared to normal tissue. As a result of the controversial role of SULT2B1b activity, additional studies in PCa are required to fully understand its function.
We hypothesize that SULT2B1b continues to be essential for PCa cell growth throughout progression of the disease, as our data demonstrate that SULT2B1b KD induces cell death in both androgen-dependent and CRPC cell lines (Figures 2–4, Supplemental Figure 5). However, it remains to be determined whether SULT2B1b activity influences the progression of androgen-dependent PCa cells toward a castration non-responsive state. Notably, our past investigation of human clinical PCa specimens did not find any correlation between CS accumulation and PCa stage/grade(10).
Cell death induced by SULT2B1b KD includes caspase-3 activation, but pan-caspase inhibition does not reduce cell death overall as it appears other death mechanisms, such as RIP1 kinase-related pathways, can become activated (Figure 4B,D). All of these data suggest that KD of SULT2B1b in PCa cells alters cellular metabolism that results in cell death, perhaps by a variety of pathways. Although an exact mechanism of how this cholesterol sulfotransferase could be linked to cell viability was not defined here, the data supporting SULT2B1b-mediated modulation of AR activity likely plays a role.
SULT2B1b activity has been shown to decrease activity of the LXR through inactivation of endogenous oxysterol ligands and we showed similar results in PCa cells (Figure 7A–C)(16). Additionally, chemical activation of the LXR with agonist TO901317 has been shown to induce cell death in LNCaP cells grown in serum-free medium(21). We observed that SULT2B1b KD-induced activation of the LXR was minimal compared to direct activation by TO901317 (data not shown), suggesting that SULT2B1b KD in complete medium conditions may endogenously regulate LXR in addition to alternative pathways that culminate in a cell death phenotype.
A possible alternative would be that SULT2B1b KD induces cell death through its impact on AR expression and activity (Figure 5, Supplemental Figure 4B–C).(29) However, this may not be due to reduced AR expression levels since overexpression of the AR with SULT2B1b KD does not rescue the cell death phenotype (Figure 6A–C). Additionally, even though some of the included cell lines do express AR variants, we do not suspect that SULT2B1b function relies completely on variant forms since SULT2B1b KD decreases viability of a range of cell lines, including benign RWPE-1 cells (Supplemental Figure 3)(30,31). However, it is not known whether SULT2B1b modulation is capable of impacting activity of AR variants. Perhaps SULT2B1b KD influences transcriptional coregulators of the AR, causing this decrease in activity. LXR activation has also been shown to decrease AR activity in LNCaP cells as well as inhibit androgen-dependent proliferation in a SULT2A1-dependent manner(28). Our data suggest that the impact of SULT2B1b on AR activity is independent of LXR activity, since AR activity decreases regardless of the presence of LXR (Figure 7D–F). Due to these findings, it is likely that other novel mechanisms of SULT2B1b-influence on AR activity are at play. Given that de novo androgen synthesis occurs within PCa cells(32), it is also possible that SULT2B1b activity is linked to synthesis of DHT within these cells since addition of AR ligand R1881 abrogates the growth-inhibitory effect of SULT2B1b KD (Figure 6D).
This is the first study to demonstrate that SULT2B1b modulation alters AR activity. Interestingly, in preliminary RNA-sequencing studies, genes modulated by SULT2B1b KD significantly altered 4 out of 5 overrepresented CRPC-related pathways identified by Robinson, et al.(33). Regulation of c-myc, crosstalk with the PI3K/Akt pathway, and impact on a number of other proliferation pathways can lead to the formation of CRPC in an androgen-deprived environment(34–36). It may be important to determine the impact of SULT2B1b modulation on these pathways to better understand whether SULT2B1b aids in progression of PCa toward a castration non-responsive state. Since CRPC retains AR activity in the context of human PCa(26), it is tempting to speculate that SULT2B1b modulation could impact AR activity in this disease stage. Based on our data, it is clear that SULT2B1b is a critical regulator of PCa growth and that SULT2B1b could be the target of a promising anti-cancer therapeutic that would simultaneously activate LXR and inhibit AR activity. These data and further studies may indicate that SULT2B1b is a significant and novel metabolic target for treatment of human PCa at multiple stages of the disease.
Supplementary Material
Acknowledgments
These studies were supported by the Department of Defense Prostate Cancer Research Program grant #W81XWH-14-1-0588, the Purdue University Center for Cancer Research P30CA023168 grant, and the Walther Cancer Foundation.
The authors would like to acknowledge the Prostate Cancer Foundation for generously offering complimentary cell line authentication and DDCMedical for rigorous analysis of our cell lines. We are grateful for the Purdue Genomics Core Facility and support from the Purdue University Center for Cancer Research (PUCCR), NIH grant P30 CA023168, as well as the Walther Cancer Foundation. The authors appreciate the help of Dr. Carol Bain for completing the IHC staining, Dr. James Fleet for his insights in and experience with statistical analysis, Gregory Cresswell and Dr. Bennett Elzey for their expertise in flow cytometry, and Dr. Andrew Thorburn at the University of Colorado for his informed suggestions regarding the cell death components of this project. The authors are also thankful for Xuehong Deng’s technical assistance and continued support of these studies. Finally, the authors wish to acknowledge the staff and faculty members within the PUCCR and the Department of Comparative Pathobiology for their support of this project and for engaging in thought-provoking discussions relating to this work.
Footnotes
There are no conflicts of interest to disclose by the authors of this manuscript.
REFERENCES
- 1.Cancer Facts and Figures 2016 [Internet] [cited 2016 Apr 5]; Available from: http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf. [Google Scholar]
- 2.James ND, Spears MR, Clarke NW, Dearnaley DP, De Bono JS, Gale J, et al. Survival with newly diagnosed metastatic prostate cancer in the docetaxel era: Data from 917 patients in the control arm of the STAMPEDE Trial (MRC PR08, CRUK/06/019) Eur Urol. 2015;67(6):1028–1038. doi: 10.1016/j.eururo.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 3.Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PFA, Sternberg CN, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. [cited 2015 Jan 16];Lancet Oncol [Internet] 2015 Jan 15;16(2):152–160. doi: 10.1016/S1470-2045(14)71205-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25601341. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin M-E, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med [Internet] 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22894553. [DOI] [PubMed] [Google Scholar]
- 5.Locke JA, Nelson CC, Adomat HH, Hendy SC, Gleave ME, Guns EST. Steroidogenesis inhibitors alter but do not eliminate androgen synthesis mechanisms during progression to castration-resistance in LNCaP prostate xenografts. [cited 2014 Jan 30];J Steroid Biochem Mol Biol [Internet] 2009 Jul;115(3–5):126–136. doi: 10.1016/j.jsbmb.2009.03.011. Available from: http://www.sciencedirect.com/science/article/pii/S0960076009001162. [DOI] [PubMed] [Google Scholar]
- 6.Mostaghel Ea, Solomon KR, Pelton K, Freeman MR, Montgomery RB. Impact of circulating cholesterol levels on growth and intratumoral androgen concentration of prostate tumors. [cited 2014 Jan 30];PLoS One [Internet] 2012 Jan;7(1):e30062. doi: 10.1371/journal.pone.0030062. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3261168&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman MR, Solomon KR. Cholesterol and prostate cancer. [cited 2014 Jan 30];J Cell Biochem [Internet] 2004 Jan 1;91(1):54–69. doi: 10.1002/jcb.10724. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14689582. [DOI] [PubMed] [Google Scholar]
- 8.Freeman MR, Cinar B, Lu ML. Membrane rafts as potential sites of nongenomic hormonal signaling in prostate cancer. [cited 2014 Jan 30];Trends Endocrinol Metab [Internet] 2005 Aug;16(6):273–279. doi: 10.1016/j.tem.2005.06.002. Available from: http://www.sciencedirect.com/science/article/pii/S1043276005001244. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang L, Kim J, Adam RM, Solomon KR, Freeman MR. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. 2005;115(4) doi: 10.1172/JCI200519935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, et al. Letters to Analytical Chemistry Cholesterol Sulfate Imaging in Human Prostate Cancer Tissue by Desorption Electrospray Ionization Mass Spectrometry. 2010;82(9):3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falany CN, Comer KA, Dooley TP, Glatt H. Human Dehydroepiandrosterone Sulfotransferase. [cited 2015 Mar 5];Ann N Y Acad Sci [Internet] 1995 Dec;774(1 Dehydroepiand):59–72. doi: 10.1111/j.1749-6632.1995.tb17372.x. Available from: http://doi.wiley.com/10.1111/j.1749-6632.1995.tb17372.x. [DOI] [PubMed] [Google Scholar]
- 12.Falany CN, He D, Dumas N, Frost AR, Falany JL. Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. [cited 2015 Mar 5];J Steroid Biochem Mol Biol [Internet] 2006 Dec;102(1–5):214–221. doi: 10.1016/j.jsbmb.2006.09.011. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1820847&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higashi Y, Fuda H, Yanai H, Lee Y, Fukushige T, Kanzaki T, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J Invest Dermatol. 2004;122:1207–1213. doi: 10.1111/j.0022-202X.2004.22416.x. [DOI] [PubMed] [Google Scholar]
- 14.He D, Meloche CA, Dumas NA, Frost AR, Falany CN. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochem J. 2004;379:533–340. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strott CA. Sulfonation and molecular action. [cited 2014 Mar 26];Endocr Rev [Internet] 2002 Oct;23(5):703–732. doi: 10.1210/er.2001-0040. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12372849. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. Enzymatic Reduction of Oxysterols Impairs LXR Signaling in Cultured Cells and the Livers of Mice. Cell Metab. 2007;5(1):73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Xu Y, Guo F, Ning Y, Zhi X, Yin L, et al. Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo. In: Sun J, editor. PLoS One [Internet]. Public Library of Science. 4. Vol. 8. 2013. Jan, [cited 2014 Mar 6]. p. e60853. Available from: http://dx.plos.org/10.1371/journal.pone.0060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu L, Yang G-Z, Zhang Y, Feng D, Zhai Y-X, Gong H, et al. Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Lab Investig [Internet] 2015 doi: 10.1038/labinvest.2015.84. Available from: http://dx.doi.org/10.1038/labinvest.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo Y-K, Mirkheshti N, Song CS, Kim S, Dodds S, Ahn SC, et al. SULT2B1b sulfotransferase: induction by vitamin D receptor and reduced expression in prostate cancer. [cited 2014 Jan 30];Mol Endocrinol [Internet] 2013 Jun;27(6):925–939. doi: 10.1210/me.2012-1369. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23579488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Zhou H, Ye L, Zhan B. Overexpression of SULT2B1b Promotes Angiogenesis in Human Gastric Cancer. [cited 2016 Apr 15];Cell Physiol Biochem [Internet] 2016 Jan;38(3):1040–1054. doi: 10.1159/000443055. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26937945. [DOI] [PubMed] [Google Scholar]
- 21.He D, Falany CN. Inhibition of SULT2B1b Expression Alters Effects of 3 b -Hydroxysteroids on Cell Proliferation and Steroid Hormone Receptor Expression in Human LNCaP. Prostate Cancer Cells. 2007 Apr;1329:1318–1329. doi: 10.1002/pros.20615. [DOI] [PubMed] [Google Scholar]
- 22.Burcham GN, Cresswell GM, Snyder PW, Chen L, Liu X, Crist SA, et al. Impact of prostate inflammation on lesion development in the POET3(+)Pten(+/−) mouse model of prostate carcinogenesis. [cited 2016 May 25];Am J Pathol [Internet] 2014 Dec;184(12):3176–3191. doi: 10.1016/j.ajpath.2014.08.021. Available from: http://www.sciencedirect.com/science/article/pii/S0002944014005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu C-C, Hu C-D. Transcriptional activity of c-Jun is critical for the suppression of AR function. [cited 2016 Feb 24];Mol Cell Endocrinol [Internet] 2013 Jun 15;372(1–2):12–22. doi: 10.1016/j.mce.2013.03.004. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3646949&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu C-C, Hu C-D. Critical role of N-terminal end-localized nuclear export signal in regulation of activating transcription factor 2 (ATF2) subcellular localization and transcriptional activity. [cited 2016 May 25];J Biol Chem [Internet]. American Society for Biochemistry and Molecular Biology. 2012 Mar 9;287(11):8621–8632. doi: 10.1074/jbc.M111.294272. Available from: /pmc/articles/PMC3318682/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degterev A, Zhou W, Maki JL, Yuan J. Assays for Necroptosis and activity of RIP kinases. Methods Enzymol. 2014;545:1–33. doi: 10.1016/B978-0-12-801430-1.00001-9. [DOI] [PubMed] [Google Scholar]
- 26.Heinlein Ca, Chang C. Androgen receptor in prostate cancer. [cited 2014 Jan 24];Endocr Rev [Internet] 2004 Apr;25(2):276–308. doi: 10.1210/er.2002-0032. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15082523. [DOI] [PubMed] [Google Scholar]
- 27.Bai Q, Xu L, Kakiyama G, Runge-Morris MA, Hylemon PB, Yin L, et al. Atherosclerosis [Internet] 2. Vol. 214. Elsevier; 2011. Feb 1, [cited 2014 Feb 20]. Sulfation of 25-hydroxycholesterol by SULT2B1b decreases cellular lipids via the LXR/SREBP-1c signaling pathway in human aortic endothelial cells; pp. 350–356. Available from: http://www.atherosclerosis-journal.com/article/S0021-9150(10)00959-7/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Gong H, Khadem S, Lu Y, Gao X, Li S, et al. Androgen deprivation by activating the liver X receptor. [cited 2014 Jan 30];Endocrinology [Internet] 2008 Aug;149(8):3778–3788. doi: 10.1210/en.2007-1605. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2488233&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eder IE, Culig Z, Ramoner R, Thurnher M, Putz T, Nessler-Menardi C, et al. Inhibition of LncaP prostate cancer cells by means of androgen receptor antisense oligonucleotides. [cited 2016 Jan 5];Cancer Gene Ther [Internet]. Nature Publishing Group. 2000 Jul 15;7(7):997–1007. doi: 10.1038/sj.cgt.7700202. Available from: http://www.nature.com/cgt/journal/v7/n7/abs/7700202a.html. [DOI] [PubMed] [Google Scholar]
- 30.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. [cited 2016 May 1];Cancer Res [Internet] 2009 Jan 1;69(1):16–22. doi: 10.1158/0008-5472.CAN-08-2764. Available from: http://cancerres.aacrjournals.org/content/69/1/16.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. [cited 2016 May 20];Cancer Res [Internet] 2009 Mar 15;69(6):2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2672822&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke Ja, Guns ES, Lubik Aa, Adomat HH, Hendy SC, Wood Ca, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. [cited 2014 Jan 22];Cancer Res [Internet] 2008 Aug 1;68(15):6407–6415. doi: 10.1158/0008-5472.CAN-07-5997. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18676866. [DOI] [PubMed] [Google Scholar]
- 33.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. [cited 2015 May 21];Cell [Internet] 2015 May;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. Available from: http://www.sciencedirect.com/science/article/pii/S0092867415005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. [cited 2016 Jan 5];J Clin Invest [Internet] 2003 Dec;112(11):1724–1731. doi: 10.1172/JCI19035. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=281646&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karantanos T, Corn PG, Thompson TC. Oncogene [Internet] 49. Vol. 32. Macmillan Publishers Limited; 2013. Dec 5, [cited 2015 Oct 2]. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches; pp. 5501–5511. Available from: http://dx.doi.org/10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. [cited 2016 Jan 5];Cancer Cell [Internet] 2011 May 17;19(5):575–586. doi: 10.1016/j.ccr.2011.04.008. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3142785&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.