Abstract
Pancreatic islet failure is a characteristic feature of impaired glucose control in diabetes mellitus. Circadian control of islet function is essential for maintaining proper glucose homeostasis. Circadian variations in transcriptional pathways have been described in diverse cell types and shown to be critical for optimization of cellular function in vivo. In the current study, we utilized Short Time Series Expression Miner (STEM) analysis to identify diurnally expressed transcripts and biological pathways from mouse islets isolated at 4 h intervals throughout the 24 h light-dark cycle. STEM analysis identified 19 distinct chronological model profiles, and genes belonging to each profile were subsequently annotated to significantly enriched Kyoto Encyclopedia of Genes and Genomes biological pathways. Several transcriptional pathways essential for proper islet function (e.g., insulin secretion, oxidative phosphorylation), cell survival (e.g., insulin signaling, apoptosis) and cell proliferation (DNA replication, homologous recombination) demonstrated significant time-dependent variations. Notably, KEGG pathway analysis revealed “protein processing in endoplasmic reticulum - mmu04141” as one of the most enriched time-dependent pathways in islets. This study provides unique data set on time-dependent diurnal profiles of islet gene expression and biological pathways, and suggests that diurnal variation of the islet transcriptome is an important feature of islet homeostasis, which should be taken into consideration for optimal experimental design and interpretation of future islet studies.
Keywords: islet, circadian, transcriptome, endoplasmic reticulum, circadian rhythms
type 2 diabetes mellitus (T2DM) is a complex polygenic disease. Pancreatic islet failure constitutes the primary pathophysiological event in T2DM driven by the loss of glucose-stimulated insulin secretion, decline in beta cell numbers, and beta cell dedifferentiation, partly attributed to the induction of endoplasmic reticulum (ER) and oxidative stress (5, 26, 41, 46). A number of genetic and environmental susceptibilities have been proposed to initiate/promote islet failure in T2DM (42). More recently, environmental and genetic factors associated with circadian rhythm disruption have been shown to increase susceptibility to T2DM in humans, mediated in part through deleterious effects on the pancreatic islet (6, 30, 40). Thus, increased understanding of molecular mechanisms driving circadian regulation of pancreatic islets may provide important insights into the pathophysiology of islet failure in T2DM.
The circadian system provides an adaptive advantage by organizing cellular transcriptional and translational cycles to anticipate and respond to environmental changes due to shifts in the light-dark (LD) cycle (33). Thus, circadian rhythms in behavioral, physiological, and cellular processes have been widely observed, and disruption of these rhythms is associated with diverse pathologies (38). Circadian regulation of insulin secretion is particularly critical for normal islet function, given its significance to restrain insulin secretion during the inactive/sleep phase and optimize insulin production and release during the active/feeding phase of the circadian cycle (3). Evidence suggests that disruption of circadian rhythms results in impaired islet function and consequent predisposition to T2DM (6, 24, 34, 35). Moreover, metabolic abnormalities associated with T2DM have also been shown to impair circadian control of insulin secretion (22).
The molecular mechanism of the circadian clock is driven by core “clock genes” that form transcriptional-translational feedback loops (45). The positive limb of this circuit includes genes Clock and Bmal1, which encode basic helix-loop-helix Per-Arnt-Single-minded proteins that initiate transcription through conserved promoter regions of target genes (15). Indeed, deletion of Bmal1 in beta cells leads to impaired insulin secretion, diminished cell proliferation, survival, and glucose intolerance (23, 24, 36). Recent studies demonstrated that CLOCK/BMAL1 complexes regulate time-dependent transcription of key beta cell exocytosis and metabolic genes via binding to distinct beta cell enhancers, thus providing molecular mechanism underlying circadian control of insulin secretion (32). However, despite increased insights into mechanisms guiding circadian control of islet function, circadian patterns of islet gene expression throughout the 24 h LD cycle in vivo remain unexplored. Insights into temporal regulation of distinct genome-wide pathways in islets in vivo will 1) add to the understanding of molecular mechanisms driving circadian control of islet homeostasis and 2) provide a much-needed reference for islet researchers examining transcriptional control of islets, to date typically performed at a single time point during the 24 h LD cycle.
MATERIALS AND METHODS
Study design.
All procedures were approved by the Institutional Animal Care and Use Committee. A total of 40 male C57BL6 mice were purchased from the same vendor (Jackson Labs) and housed in identical cages and environmental conditions (humidity, temperate, intensity of light) throughout the duration of the study. Mice were fed irradiated chow diet (24.7 kcal% protein, 62.1 kcal% carbohydrates, 13.2 kcal% fat; #5053; LabDiet, St. Louis, MO) and housed at University of California, Los Angeles (UCLA) and Mayo Clinic, Rochester, animal facilities under standard 12 h light/12 h dark (LD) cycle. By convention, time of lights on (0600) is denoted as Zeitgeber time (ZT) 0 and lights off (1800) as ZT 12. For determination of diurnal patterns of gene expression, mice were housed in groups of four per cage (matched by body weight), and one cage was demarcated for each ZT time-point to minimize disturbance to other animals in vivarium and avoid stress associated with cage separation. For islet isolation during “dark” time points (ZT 12, 16, and 20), mice were euthanized under red light. All mice had ad libitum access to food and water throughout the study to recapitulate physiological in vivo conditions. In short, 22 mice were euthanized within the 24 h timeframe at ZT 0 (0600), 4 (1000), 8 (1400), 12 (1800), 16 (2200), and 20 (0200) time points for isolation of islets to assess circadian mRNA expression (n = 3–4 per time point). A separate cohort of 18 mice was euthanized at the identical six time points, and whole pancreas was immediately harvested and fixed in 4% paraformaldehyde for subsequent immunofluorescence analysis (n = 3 per time point). Prior to euthanasia, blood was drawn from all 40 mice for subsequent determination of plasma glucose and insulin levels (n = 6–7 per time point).
Behavioral monitoring of circadian activity.
For behavioral monitoring, mice were housed individually in cages outfitted with an optical beam sensor system (Respironics, Murrysville, PA) to monitor circadian behavioral rhythms in activity for 10 days in LD cycle.
Analysis of circadian metabolic profiles.
Plasma was sampled from the heart at ZT 4, 8, 12, 16, and 20 and immediately stored at −80°C for subsequent glucose and insulin measurements. Plasma glucose was measured by glucose oxidase method (Analox instruments). Plasma insulin was measured using ELISA (Alpco Diagnostics, Salem, NH).
Circadian mRNA expression in isolated islets.
Pancreatic islets were isolated by the collagenase method as previously described in detail (18). In short, following ∼15 min collagenase infusion-digestion, digested pancreas containing islets plus separated exocrine tissue was placed on ice and immediately washed three times in working buffer (HBSS, NaHCO3, HEPES, pH 7.4). The homogenate was then transferred to a Petri dish containing cold RPMI medium and kept inside an ice bucket. Endocrine islets were identified in the inverted ZEISS microscope (Carl Zeiss Microscopy, Thornwood, NY), and all islets in the dish were handpicked directly into the Eppendorf tube placed on ice. Collected islets were washed with PBS to remove RPMI medium and immediately transferred to RNeasy RLT Lysis Buffer (Qiagen, Valencia, CA) and stored at −80°C for subsequent RNA isolation. Total RNA was extracted using RNeasy Mini Kit (Qiagen), quantified by NanoDrop ND-100 (BioTek, Winooski, VT) and assessed for overall integrity using agarose gel electrophoresis. Microarray analysis was performed utilizing Agilent Whole Genome Expression Array platform according to manufacturer's instructions (Mus musculus) (Arraystar, Rockville, MD). In short, total RNA was amplified and converted to fluorescent cRNA (version 5.7, Agilent Technologies, Santa Clara, CA) and subsequently hybridized for Whole Genome Oligo Microarray (4X44K, Agilent Technologies). Array images were acquired (Agilent Scanner G2505C), analyzed (Agilent Extraction Software, version 11), and quantified by the quantile normalization method for microarray data (GeneSpring GX, Agilent Technologies) that normalizes fluorescence intensity distributions from scanned array images across samples (4).
Short time-series expression miner analysis.
Short time-series expression miner (STEM) analysis is a validated and commonly used bioinformatics approach designed to determine statistically significant time-dependent gene expression profiles and allows for determination of significantly enriched biological pathways within elucidated profiles (12, 13). STEM assumes gene expression values represent log ratios relative to the expression of the first time point (i.e., ZT 0); thus the first value of the time series will always be 0. STEM first selects a distinct set of predetermined temporal model profiles representing potential time-dependent gene expression patterns. Importantly, temporal profiles are predetermined and thus independent of mRNA expression data, which enables STEM to determine the statistical significance of the number of genes assigned to each profile compared with what is expected based on chance as determined by a permutation test on the time points (13). The set of potential temporal model profiles to be tested is determined by the number of time points (n), which is 6 in our case (samples collected at ZT 0, 4, 8, 12, 16, 20), and a parameter c, which specifies the maximum unit change between the successive time points, which means that between consecutive sampling intervals a profile can either increase or decrease by 1, . . . . to up to c units or remain unchanged. We used the default value of c as 2. The profile that is unchanged at all time-points is excluded from the set of potential profiles. In addition, a parameter m determines the number of model profiles are actually selected for testing, which was set to its default value of 50. The set of profiles selected by STEM for testing are designed to be both representative of all potential temporal profiles and distinct from each other.
STEM then filters transcripts that did not change sufficiently; we used the default setting of filtering genes for which the difference between the maximum and minimum expression value for the gene was <1. For the remaining genes, STEM assigns transcripts to the particular model profile for which it has the highest correlation coefficient (13). STEM subsequently determines profiles statistically significantly enriched by comparing the number of genes assigned with what would be expected based on permutation of the time points with Bonferroni correction for multiple comparisons (13). Following determination and segregation of genes into statistically significant and distinct temporal model profiles, genes included in the 19 model chronological profiles were exported for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the standard enrichment computation method by R script with P values corrected for multiple testing as false discovery rate.
Immunofluorescence.
Paraffin-embedded pancreatic sections were stained for insulin (1:100, ab7842; Abcam, Cambridge, MA) and BMAL1 (1:500, ab3350; Abcam) utilizing standard immunofluorescence protocol (36). For quantification of BMAL1 expression, 10 islets from each animal (n = 3 per time-point) were imaged at same exposure time and acquisitions settings. Slides were viewed and imaged under a Zeiss microscope (Carl Zeiss Microscopy), and images were acquired and quantified with ZEN Pro software (Carl Zeiss Microscopy).
Statistical analysis and calculations.
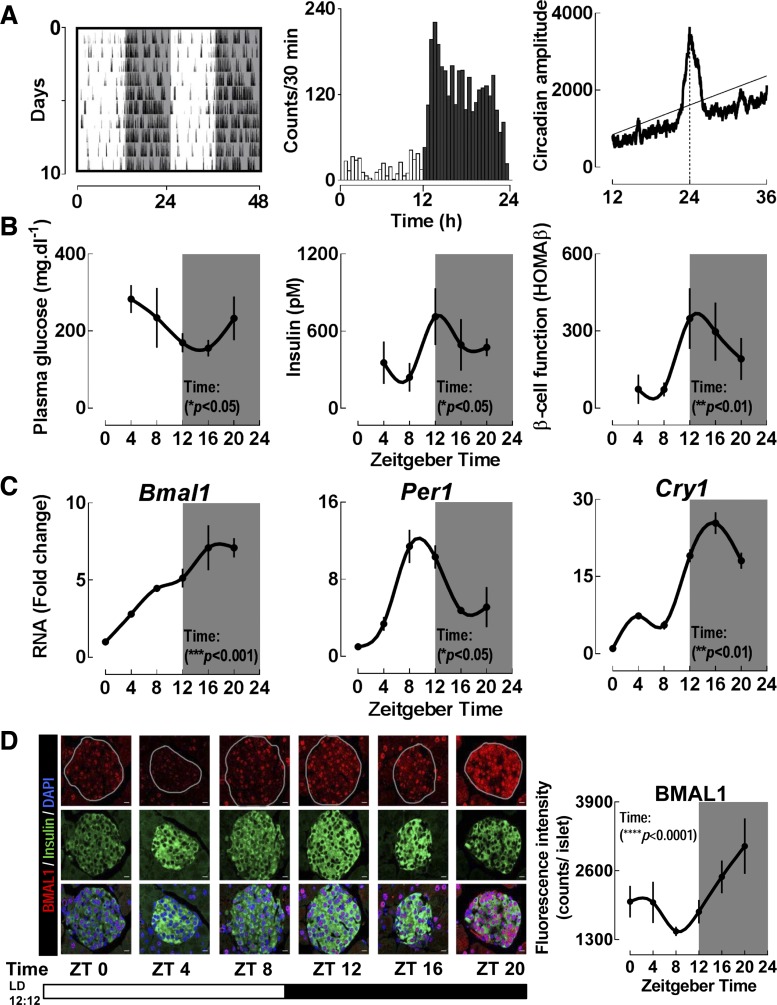
To quantitatively assess dominant circadian period in activity, we utilized χ2 periodogram algorithm analysis (ClockLab, Actimetrics, Evanston, IL). In short, the χ2 periodogram method estimates circadian periodicity by examining data at different period lengths and provides quantification of the variance of data at each time period relative to the defined confidence interval (99%) (43). Statistical analysis for diurnal changes in metabolic profiles and clock gene/protein expression (Fig. 1) was performed by repeated-measures one-way ANOVA with significance for “time” variable taken as evidence of diurnal rhythmicity (GraphPad Prism v.6.0, San Diego, CA). Beta cell function was estimated by using homeostasis model assessment index of beta cell function [HOMAβ = (20 × insulin)/(glucose − 3.5); insulin in mU/l and glucose in mmol/l] (25). Heat maps were generated using free software R console (v.3.2.2, Vienna, Austria).
Fig. 1.
C57BL6 mice display significant diurnal variations in behavioral, physiological, and molecular parameters. A: representative activity recordings in a C57BL6 mouse (n = 9 examined) monitored for 10 days under light-dark (LD) cycle. Shaded areas represent periods of dark (left). Mean circadian activity (expressed as activity counts/30 min) across the 24 h circadian day (center). χ2 periodogram analysis demonstrating the dominant circadian period of exactly 24 h in a representative mouse under LD cycle (right). Solid horizontal line represents statistically significant threshold in determination of the dominant circadian period. Broken vertical black line represents dominant circadian period of exactly 24 h. B: assessment of diurnal plasma glucose, insulin, and homeostatic model assessment of beta cell function (HOMA-β). Each time point represents mean ± SE (n = 6–7 per group). Statistical significance was determined by repeated-measures 1-way ANOVA and shows significant effects of “Time” (P < 0.05) for each metabolic parameter. C: assessment of diurnal mRNA expression of selected key clock genes (Bmal1, Per1, and Cry1) from islet lysates isolated across the 24 h circadian day (n = 3–4 per time point). Statistical significance was determined by repeated-measures 1-way ANOVA and shows significant effects of “Time” (P < 0.05) for every clock gene. Quantitative PCR analysis was used to determine ΔΔCT based fold-changes in expression of clock genes utilizing GAPDH as the endogenous control gene. D: representative examples and corresponding quantification of pancreatic islets stained by immunofluorescence for BMAL1 in red and insulin in green imaged at ×63 magnification from C57BL6 mice collected at 4 h intervals (n = 3 at each time point) across the 24 h day (scale bars, 25 μm). Statistical significance was determined by repeated-measures 1-way ANOVA and shows significant effects of “Time” (P < 0.05). ZT, Zeitgeber time.
RESULTS
As expected, C57BL6 mice displayed significant diurnal variations in behavioral, physiological, and molecular parameters (Fig. 1, A–D). Under the standard LD cycle, mice displayed robust behavioral circadian rhythms and significant diurnal variations in plasma glucose and insulin, as well as a measure of beta cell function (HOMA-β) (P < 0.05 for all parameters, Fig. 1B). Diurnal mRNA expression patterns of key clock genes (Bmal1, Per1, and Cry1) in isolated islets were also consistent with the proposed model of mammalian circadian clockwork (Fig. 1C). Furthermore, immunofluorescence analysis confirmed significant diurnal patterns of clock protein (BMAL1) expression, which was colocalized to beta cells (Fig. 1D).
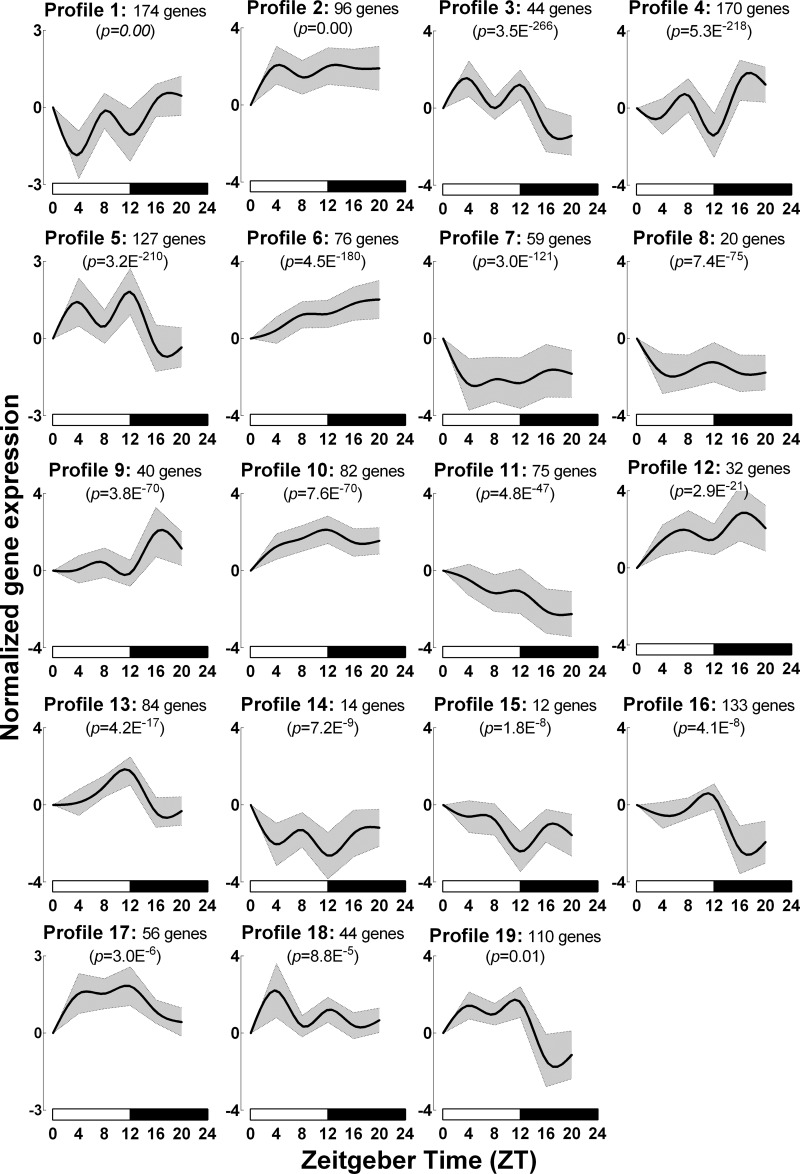
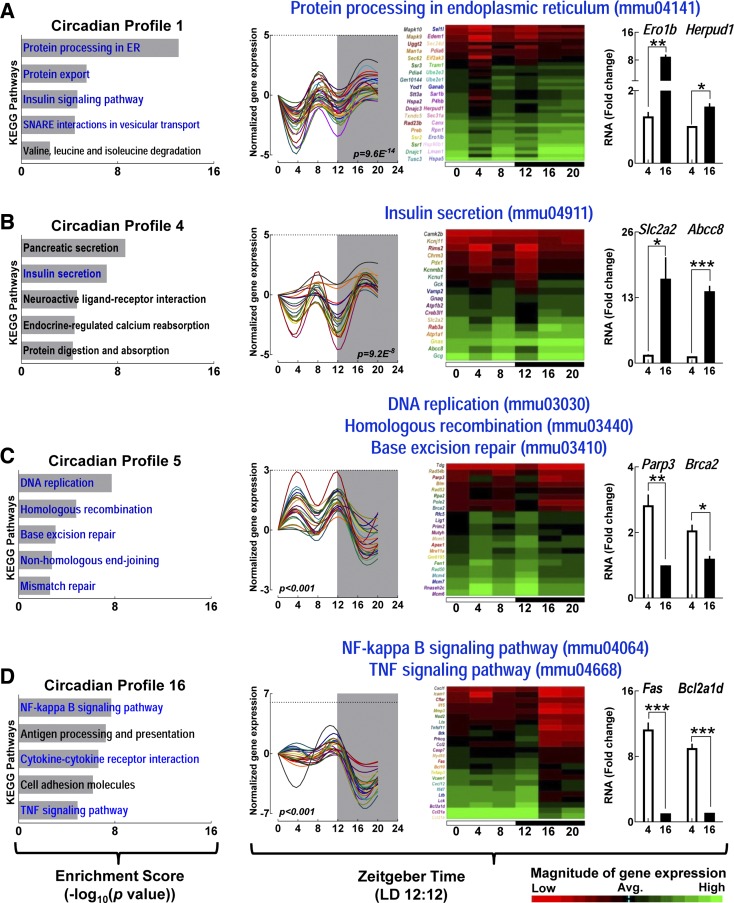
Next, we utilized STEM to identify time-dependent expressed transcriptional profiles from mouse islets isolated at 4 h intervals (ZT 0, 4, 8, 12, 16, and 20). STEM identified 19 distinct time-dependent model profiles that demonstrated statistically significant overenrichment of islet genes (P < 0.01, Figs. 2 and 3). Seven out of 19 profiles (#1, 2, 4, 6, 9, 10, and 12) displayed increased mRNA expression during the active/dark phase of the circadian cycle (ZT 16–20), eight (#3, 7, 8, 11, 14, 15, 18, and 19) profiles displayed a tendency for enhanced gene expression during the inactive/light phase (ZT 0–8), and four profiles (#5, 13, 16, and 17) demonstrated elevated mRNA expression at the onset of dark cycle (ZT 12) (Fig. 3, Table 1). To gain insights into time-dependent expression patterns of distinct biological pathways in islets in vivo, we further analyzed each of the 19 model profiles to quantify significant enrichment of specific regulatory pathways and individual genes utilizing KEGG pathway analysis (Figs. 4 and 5 and Table 1). The analysis revealed “protein processing in endoplasmic reticulum (mmu04141)” as the most significantly enriched biological pathway (P = 9.58E−14, KEGG enrichment score = 13.02, Figs. 4 and 5A) represented in STEM-identified profile #1. The ER protein processing pathway displayed increased expression during the dark/active phase (ZT 16-20) and was enriched with transcripts essential for regulation of 1) ER chaperone function [e.g., Hspa5 (Bip), Hsp40, Grp94], 2) protein folding (e.g., Ero1β, Edem10), 3) unfolded protein response [e.g., Eif2Ak3 (Perk)], ER-associated protein degradation (e.g., Hsp70, Dsk2, Rad23b), and ubiquitination (e.g., Ube2e1, Ube2e3, Herpud1). In line with these observations, biological pathways involved in regulation of “insulin secretion - mmu04911” [P = 9.23E−8, e.g., Slc2a2 (Glut2), Gck (Glucokinase), Creb3L1 (Creb), Pdx1, Kcnj11 (Kir6.2)], “insulin signaling - mmu04910” (P = 2.5E−5, e.g., Insr, Irs1), “SNARE interactions in vesicular transport - mmu04130” (P = 4.0E−5, e.g., Bet1, Snap29, Vamp4), and “oxidative phosphorylation - mmu00190” (P = 6.8E−5 e.g., Atp5c1, Cox4l1, Ndufa2) all displayed similar enriched expression profile during the dark/active phase of the circadian cycle (Fig. 5B, Table 1).
Fig. 2.
C57BL6 mice display circadian variations in islet gene expression. A: heat map representing genome-wide mRNA expression in pancreatic islets of C57BL6 mice isolated at 4 h intervals across the 24 h circadian cycle. B: short time-series expression miner (STEM) analysis identified 19 distinct time-dependent model profiles that demonstrated statistically significant overenrichment of islet genes, of which representative 6 profiles are depicted. For each of the 6 depicted profiles, genes annotated to significantly enriched KEGG biological pathways are represented as a graph (left) and heat map (right). The graph depicts mean (solid line) ± SE (shaded region) pattern of all significantly KEGG-enriched genes across the 24 h circadian cycles within each profile (Permutation test P values included in graphs indicate statistical significance for the number of transcripts assigned to that pattern vs. number expected by chance).
Fig. 3.
Graphical depiction of 19 statistically significant chronological profiles in islet gene expression identified by STEM analysis. Diurnally expressed islet genes are clustered into 19 unique chronological model profiles arranged by significance as obtained from STEM analysis. Each profile graph depicts mean (solid line) ± SE (shaded region) expression pattern of all significantly Kyoto Encyclopedia of Genes and Genomes (KEGG)-enriched genes across the 24 h circadian cycles within each profile (Permutation test P values included in graphs indicate statistical significance for the number of transcripts assigned to that pattern vs. number expected by chance).
Table 1.
Model chronological profiles and enriched KEGG pathways within profiles
| Model STEM Profiles |
KEGG Pathway Enrichment Within Model STEM Chronological Profiles |
|||||
|---|---|---|---|---|---|---|
| Profile # | P Value | Annotated Genes, n | Pathway ID | Definition - Mus musculus (mouse) | Fisher P Value | Enrichment Score (-log10 P value) |
| 1 | 0.00 | 174 | mmu04141 | protein processing in endoplasmic reticulum | 9.58E−14 | 13.02 |
| mmu03060 | protein export | 4.32E−06 | 5.36 | |||
| mmu04910 | insulin signaling pathway | 2.47E−05 | 4.61 | |||
| mmu04130 | SNARE interactions in vesicular transport | 4.04E−05 | 4.39 | |||
| 2 | 0.00 | 96 | mmu00190 | oxidative phosphorylation | 6.79E−05 | 4.17 |
| 3 | 3.50E−266 | 44 | mmu04060 | cytokine-cytokine receptor interaction | 4.41E−05 | 4.36 |
| mmu04115 | p53 signaling pathway | 0.0160 | 1.80 | |||
| 4 | 5.30E−218 | 170 | mmu04972 | pancreatic secretion | 2.60E−09 | 8.59 |
| mmu04911 | insulin secretion | 9.23E−08 | 7.03 | |||
| mmu04080 | neuroactive ligand-receptor interaction | 2.69E−05 | 4.57 | |||
| 3.20E−210 | 127 | mmu03030 | DNA replication | 2.08E−08 | 7.68 | |
| mmu03440 | homologous recombination | 1.85E−05 | 4.73 | |||
| mmu03410 | base excision repair | 0.0010 | 3.01 | |||
| 6 | 4.50E−180 | 76 | mmu01040 | biosynthesis of unsaturated fatty acids | 0.0012 | 2.94 |
| mmu01212 | fatty acid metabolism | 0.0057 | 2.24 | |||
| 7 | 3.00E−121 | 59 | mmu00532 | glycosaminoglycan biosynthesis | 0.0094 | 2.03 |
| mmu00620 | pyruvate metabolism | 0.0149 | 1.83 | |||
| 8 | 7.40E−75 | 20 | mmu00310 | lysine degradation | 0.0030 | 2.53 |
| 9 | 3.80E−70 | 40 | mmu00020 | citrate cycle (TCA cycle) | 0.0053 | 2.28 |
| mmu04151 | PI3K-Akt signaling pathway | 0.0392 | 1.41 | |||
| 10 | 7.60E−70 | 82 | mmu03020 | RNA polymerase | 3.23E−05 | 4.49 |
| mmu03050 | proteasome | 2.91E−03 | 2.54 | |||
| mmu04310 | Wnt signaling pathway | 1.34E−02 | 1.87 | |||
| 11 | 4.80E−47 | 75 | mmu04015 | Rap1 signaling pathway | 6.44E−05 | 4.19 |
| mmu04014 | Ras signaling pathway | 0.0145 | 1.84 | |||
| mmu04350 | TGF-beta signaling pathway | 0.0490 | 1.31 | |||
| 12 | 2.90E−21 | 32 | mmu04015 | Rap1 signaling pathway | 0.0090 | 2.05 |
| mmu00520 | amino sugar and nucleotide sugar metabolism | 0.0439 | 1.36 | |||
| 13 | 4.20E−17 | 84 | mmu04660 | T cell receptor signaling pathway | 1.61E−09 | 8.79 |
| mmu04210 | apoptosis | 9.11E−07 | 6.04 | |||
| 14 | 7.20E−09 | 14 | mmu00514 | other types of O-glycan biosynthesis | 0.0085 | 2.07 |
| 15 | 1.80E−08 | 12 | mmu04915 | estrogen signaling pathway | 0.0180 | 1.74 |
| 16 | 4.10E−08 | 133 | mmu04064 | NF-kappa B signaling pathway | 2.28E−08 | 7.64 |
| mmu04060 | cytokine-cytokine receptor interaction | 2.66E−07 | 6.58 | |||
| mmu04668 | TNF signaling pathway | 1.45E−05 | 4.84 | |||
| 17 | 3.00E−06 | 56 | mmu04142 | lysosome | 0.0024 | 2.62 |
| mmu04330 | Notch signaling pathway | 0.0366 | 1.44 | |||
| 18 | 8.80E−05 | 44 | mmu00190 | oxidative phosphorylation | 0.0009 | 3.04 |
| 19 | 0.01 | 110 | mmu04662 | B cell receptor signaling pathway | 0.0002 | 3.72 |
Left: table depicts 19 distinct significant time-dependent model profiles (numbered 1–19) obtained by short time-series expression miner (STEM) analysis with corresponding P values for statistically significant overenrichment of genes. Right: number of annotated genes belonging to each of the 19 chronological profiles significantly enriched for specific KEGG biological pathways depicted with their respective Fisher P values and enrichment scores.
Fig. 4.
Distinct circadian gene expression profiles in islets are enriched with biological pathways essential for maintenance of islet homeostasis. KEGG pathway analysis was conducted for genes enriched within each of the 19 unique chronological model profiles previously identified by STEM. Bar graphs represent notable KEGG biological pathways significantly enriched within each chronological profile.
Fig. 5.
Biological pathways essential for maintenance of islet homeostasis exhibit circadian variations. A–D: KEGG pathways analysis for enrichment of genes within 4 representative chronological model profiles (#1, 4, 5, and 16) in mouse islets in vivo. Bar graphs on the left represent the top 5 significantly enriched KEGG biological pathways within each profile. Selected biological pathways shown in blue font are highlighted given their critical importance for islet function. Significantly enriched genes within highlighted KEGG pathways are further depicted as individual graphs and heat maps on the right. Graphs represent quantile normalized mRNA expression levels with each significantly rhythmic gene shown in different color trace (Fisher P value included). Shaded area inside graphs represents period of dark. Data on far right of each row represent quantitative PCR validation of selected key transcripts from each of the 4 depicted biological pathways performed (n = 3 per gene) at ZT 4 and ZT 16. Statistical significance was determined by unpaired t-test and denoted by ***P < 0.001, **P < 0.01, and *P < 0.05.
In contrast, KEGG-identified biological pathways regulating cellular proliferation displayed a tendency for enriched gene expression during the light/inactive phase (ZT 0–12) of the circadian cycle (Fig. 5C, Table 1). Notably, key biological pathways related to positive regulation of the cell cycle, clustered in the same circadian profile (#5), which included: “DNA replication - mmu03030” (P = 2.08E−8), “homologous recombination - mmu03440” (P = 2.08E−8), and “base excision repair - mmu03410” (P = 0.001) (Fig. 5C). Similarly, biological pathways regulating inflammatory response clustered in profile #16 with increased expression during the dark phase (ZT 20) and attenuated expression at the end of light phase (ZT 12) including: “NF-κB signaling - mmu04064” (P = 2.3E−8), “cytokine-cytokine interaction - mmu04060” (P = 2.7E−7), and “TNF signaling - mmu04668” (P = 1.5E−5) (Fig. 5D). Other notable diurnally expressed pathways shown to be important for regulation of islet function include “fatty acid metabolism - mmu01212” (P = 0.0057), “apoptosis - mmu04210” (P = 9.1E−7), and “proteasome function - mmu03050” (P = 2.9E−3) (Table 1, Fig. 4).
DISCUSSION
Understanding of mechanisms underlying failed islet function and survival in T2DM is essential for development of novel therapeutic and preventative approaches (10). Robust circadian rhythms in insulin secretion have been documented in humans and animal models in vivo, as well as in isolated islets in vitro (3, 32, 37). It was recently demonstrated that the circadian clock regulates insulin secretion through transcriptional control of key genes regulating beta cell exocytosis machinery and metabolism (32). In addition, a functional circadian clock is required for proper regulation of beta cell proliferation, response to oxidative stress, and mitochondrial function and, when disrupted, compromises regulation of glucose homeostasis (23, 24, 36). Our current results are consistent with previous observations and provide detailed characterization of diurnal variations of key transcripts and biological pathways in mouse islets in vivo in relation to changes in the LD cycle. Specifically, the current study provides data on distinct diurnal patterns of islet gene expression and KEGG-identified biological pathways and emphasizes the complex time-dependent nature of transcriptional control of islet homeostasis in vivo.
Consistent with previous studies in metabolically active tissues (e.g., liver) (31), a significant fraction of the mouse islet transcriptome exhibited distinct diurnal expression patterns. STEM analysis identified 19 distinct time-dependent profiles of gene expression that were subsequently annotated for enrichment of genes belonging to KEGG-defined biological pathways. Importantly, several key biological pathways essential for regulation of islet cell function, survival, and proliferation demonstrated robust (and often antiphase) diurnal expression patterns. These pathways encompass diverse biological functions including regulation of insulin secretion, insulin signaling, DNA replication, apoptosis, oxidative phosphorylation, cytokine function, and fatty acid metabolism.
Protein processing in the ER was identified as one of the most enriched diurnally regulated biological pathways in mouse islets. It has been estimated that beta cells are capable of synthesizing, processing, degrading, and secreting over 10,000 molecules (e.g., insulin and islet amyloid polypeptide) per minute in the basal state alone (9). This capacity for protein processing and degradation undoubtedly rises with the increased insulin demand associated with obesity, aging, and various insulin-resistant states (39). Thus, beta cells are known to possess highly complex ERs that orchestrate protein production, folding, as well as disposal and degradation of misfolding proteins (20). Efficient degradation of misfolded proteins is essential for prevention of detrimental effects consequent of ER and oxidative stress (41), which can be partly attributed to accumulation of reactive oxygen species (ROS) (21). Indeed, pancreatic beta cells are particularly vulnerable to the deleterious effects of accumulating ROS, given very low beta cell expression of key antioxidant enzymes (21). Importantly, induction of ER and oxidative stress is a characteristic feature of islet failure in patients with T2DM (16). Subsequently, our studies suggest that circadian organization of ER protein processing provides a mechanism by which to optimize ER function and thus avoid increased protein misfolding and consequent ROS accumulation. This is achieved by upregulating ER processing and degradation machinery during the time of increased nutrient availability and increased secretory demand (i.e., active/feeding cycle). Indeed, genetic deletion of key ER transcripts that displayed rhythmic expression in our study [e.g., Grp78 (Bip), Ero1β, Eif2Ak3 (Perk)] is associated with induction of beta cell dysfunction, apoptosis, and T2DM (1, 17, 29). Moreover, disruption of circadian rhythms has been previously shown to accelerate development of ER stress-induced beta cell dysfunction and apoptosis in animal models of T2DM (14, 36).
Another notable observation in our study relates to temporal distribution of genes regulating various aspects of cell cycle regulation. Interestingly we observed significant enrichment in genes regulating DNA replication and repair pathways in temporal profiles exhibiting peak gene expression during the light/inactive phase of the circadian cycle. In addition, genes regulating other aspects of cell cycle regulation (e.g., DNA damage response) clustered in temporal profiles with peak expression at the end of the dark/active phase of the circadian day, thus highlighting the complexity of temporal transcriptional control of cell cycle progression. Indeed, the circadian clock has been long been appreciated as exerting transcriptional control of the cell cycle and DNA damage checkpoints (19, 27). Consistent with this, impaired beta cell proliferative potential has been reported in beta cell-specific Bmal1 null mice (36). Studies in yeast have demonstrated restriction of DNA replication to reductive/glycolytic phase of the metabolic cycle in order to maintain genome stability and avoid DNA damage (7). Indeed, the evolution of circadian clocks has been attributed to the “escape from light” hypothesis, which suggests clock-mediated gating of cell division and DNA damage as the driving force for the evolution of the circadian clock mechanism (33). It is thus intriguing to hypothesize that the same mechanism may be conserved in pancreatic islets in vivo, thus clarifying the observed restriction of key islet DNA replication and repair transcripts to the inactive/light phase of the animal's circadian cycle. Furthermore, recent studies demonstrated that the reduction in insulin biosynthesis and ER function enhances beta cell proliferative capacity (44), an observation consistent with antiphase expression of genes regulating cell proliferation and ER protein processing observed in our study. Indeed, environmental or genetic disruption of the circadian clock increases beta cells' vulnerability to DNA damage-induced beta cell apoptosis (35, 36) and restricts beta cell proliferative capacity in vivo (36).
Consistent with previous studies, we report robust diurnal expression of key clock genes in pancreatic mouse islets isolated at distinct phases of 24 h circadian cycle (24). Notably, the temporal expression pattern of a key circadian transcription factor, BMAL1 (enriched during the dark/active phase), was consistent with the essential role of this transcription factor in regulation of insulin secretion. Recent work utilized cultured entrained human and mouse islets to demonstrate that BMAL1 colocalizes with PDX-1 and binds to distinct beta cell enhancers to control transcription of genes regulating insulin secretion and exocytosis (32). Consistent with these results, we observed parallel patterns in temporal expression of BMAL1 protein and insulin secretion genes previously shown to be direct targets of BMAL1 (e.g., Slc2a2) (32). In addition, previously shown induction of beta cell dysfunction in Bmal1-deficient beta cells further underscores the critical importance of the beta cell circadian clock in regulation of insulin secretion (23, 24).
It is important to note that diurnal changes in islet gene expression observed in our study may be driven by changes in feeding cycles as well as diurnal fluctuations in hormonal secretion (e.g., melatonin and corticosterone). In tissues exposed to changes in nutrient availability (e.g., pancreas and liver), timing of feeding contributes to circadian regulation of gene expression (47). Consistent with these studies, we have previously reported that timed feeding is capable of entraining the phase of islet circadian clocks in vivo (37). Also, diurnal secretion of key clock-driven hormones (e.g., melatonin and corticosterone) may be another mechanism responsible for temporal regulation of islet gene expression. For example, melatonin and corticosterone have been shown to modulate cellular clock gene expression and impart significant changes in the islet transcriptome (2, 8, 28). Further studies are required to elucidate exact physiological and molecular mechanisms underlying observed diurnal changes in islet gene expression.
It is important to acknowledge the limitations of our study. First, the necessity for islet isolation from the pancreas raises the question whether the isolation procedure per se might have influenced our results. However, we previously demonstrated that the islet isolation procedure does not appear to reset or alter the phase of clock gene expression in pancreatic islets (34). In addition, the increased induction of cellular stress is evident in cultured isolated islets only after a relatively prolonged incubation period (days to weeks) (11), whereas in our study islets were immediately preserved (within minutes) upon completion of the isolation procedure. Our study also does not provide data on potential physiological consequences (i.e., insulin secretion, cell proliferation) associated with various diurnal patterns of islet gene expression. This is particularly relevant for patterns with analogous temporal expression profiles (e.g., profiles 3 and 19). Although our current study design precludes us from assessing physiological consequences attributed to changes in temporal gene expression, it is important to note that each of the 19 significant chronological model profiles displayed enrichment for distinct KEGG biological pathways. Furthermore, studies have shown that even modest changes in the amplitude of circadian clock gene expression are associated with significant biological effects in islets (34).
In conclusion, diurnal variations in islet gene expression optimize islet function and homeostasis to periods of nutrient availability and fasting and when disrupted, result in a predisposition to islet failure and T2DM. To date, the majority of the studies examining transcriptional control of islet function in vivo rely on data obtained from a single chronological time point, typically collected during the light/inactive phase of animals' circadian cycle. Our data underscore distinct temporal patterns of gene expression in diverse biological pathways critical for regulation of islet homeostasis (e.g., ER function, insulin secretion, cell proliferation, cell apoptosis, mitochondrial function, fatty acid metabolism etc.). Thus, circadian regulation of islet gene expression should be taken into consideration for optimal experimental design and interpretation of islet work. In addition, future studies will be needed to delineate whether induction of beta cell failure in T2DM compromises circadian control of islet gene expression, thereby accelerating the decline in beta cell function and survival.
GRANTS
We acknowledge funding support from National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-098468 (to A. V. Matveyenko) and the Center for Regenerative Medicine (Mayo Clinic, Rochester, MN). The dataset is publically available and deposited into the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database repository of high throughput gene expression (accession number GSE80692).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
K.R. performed experiments; K.R., J.Q., and J.E. analyzed data; K.R., J.E., and A.V.M. interpreted results of experiments; K.R. prepared figures; K.R., J.Q., and J.E. edited and revised manuscript; K.R., J.Q., J.E., and A.V.M. approved final version of manuscript; K.R., J.Q., and A.V.M. conception and design of research; A.V.M. drafted manuscript.
REFERENCES
- 1.Awazawa M, Futami T, Sakada M, Kaneko K, Ohsugi M, Nakaya K, Terai A, Suzuki R, Koike M, Uchiyama Y, Kadowaki T, Ueki K. Deregulation of pancreas-specific oxidoreductin ERO1beta in the pathogenesis of diabetes mellitus. Mol Cell Biol 34: 1290–1299, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Boden G, Ruiz J, Urbain JL, Chen X. Evidence for a circadian rhythm of insulin secretion. Am J Physiol Endocrinol Metab 271: E246–E252, 1996. [DOI] [PubMed] [Google Scholar]
- 4.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 52: 102–110, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Buxton OM, Cain SW, O'Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA, Shea SA. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med 4: 129ra143, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science 316: 1916–1919, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Costes S, Boss M, Thomas AP, Matveyenko AV. Activation of melatonin signaling promotes beta-cell survival and function. Mol Endocrinol 29: 682–692, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costes S, Langen R, Gurlo T, Matveyenko AV, Butler PC. beta-Cell failure in type 2 diabetes: a case of asking too much of too few? Diabetes 62: 327–335, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFronzo RA, Abdul-Ghani MA. Preservation of beta-cell function: the key to diabetes prevention. J Clin Endocrinol Metab 96: 2354–2366, 2011. [DOI] [PubMed] [Google Scholar]
- 11.Erbel S, Reers C, Nawroth PP, Ritzel RA. Prolonged culture of human islets induces ER stress. Exp Clin Endocrinol Diabetes 118: 81–86, 2010. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J, Nau GJ, Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics 21, Suppl 1: i159–i168, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms 26: 423–433, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–1569, 1998. [DOI] [PubMed] [Google Scholar]
- 16.Haataja L, Gurlo T, Huang CJ, Butler PC. Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr Rev 29: 303–316, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell 7: 1153–1163, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Huang CJ, Haataja L, Gurlo T, Butler AE, Wu X, Soeller WC, Butler PC. Induction of endoplasmic reticulum stress-induced beta-cell apoptosis and accumulation of polyubiquitinated proteins by human islet amyloid polypeptide. Am J Physiol Endocrinol Metab 293: E1656–E1662, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Johnson CH. Circadian clocks and cell division: what's the pacemaker? Cell Cycle 9: 3864–3873, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest 110: 1389–1398, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman RJ, Back SH, Song B, Han J, Hassler J. The unfolded protein response is required to maintain the integrity of the endoplasmic reticulum, prevent oxidative stress and preserve differentiation in beta-cells. Diabetes Obes Metab 12, Suppl 2: 99–107, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee A, Ader M, Bray GA, Bergman RN. Diurnal variation in glucose tolerance. Cyclic suppression of insulin action and insulin secretion in normal-weight, but not obese, subjects. Diabetes 41: 750–759, 1992. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Moulik M, Fang Z, Saha P, Zou F, Xu Y, Nelson DL, Ma K, Moore DD, Yechoor VK. Bmal1 and beta-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced beta-cell failure in mice. Mol Cell Biol 33: 2327–2338, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419, 1985. [DOI] [PubMed] [Google Scholar]
- 26.Meier JJ, Butler PC. Insulin secretion. In: Endocrinology (5th ed), edited by DeGroot LJ, Jameson JL. Philadelphia, PA: Elsevier Saunders, 2005, p. 961–973. [Google Scholar]
- 27.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA 104: 3342–3347, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama K, Hirai K. The melatonin agonist ramelteon induces duration-dependent clock gene expression through camp signaling in pancreatic INS-1 beta-cells. PLoS One 9: e102073, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313: 1137–1140, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med 8: e1001141, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002. [DOI] [PubMed] [Google Scholar]
- 32.Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, Allred AL, Bradfield CA, Dinner AR, Barish GD, Bass J. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350: aac4250, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 55: 16–54, 1993. [DOI] [PubMed] [Google Scholar]
- 34.Qian J, Block GD, Colwell CS, Matveyenko AV. Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes 62: 3469–3478, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian J, Yeh B, Rakshit K, Colwell CS, Matveyenko AV. Circadian disruption and diet-induced obesity synergize to promote development of beta-cell failure and diabetes in male rats. Endocrinology 156: 4426–4436, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakshit K, Hsu TW, Matveyenko AV. Bmal1 is required for beta cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice. Diabetologia 59: 734–743, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rakshit K, Qian J, Colwell CS, Matveyenko AV. The islet circadian clock: entrainment mechanisms, function and role in glucose homeostasis. Diabetes Obes Metab 17, Suppl 1: 115–122, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy AB, O'Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol 20: 36–44, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera JF, Costes S, Gurlo T, Glabe CG, Butler PC. Autophagy defends pancreatic beta cells from human islet amyloid polypeptide-induced toxicity. J Clin Invest 124: 3489–3500, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA 106: 4453–4458, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev 29: 317–333, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smushkin G, Vella A. Genetics of type 2 diabetes. Curr Opin Clin Nutr Metab Care 13: 471–477, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolove PG, Bushell WN. The chi square periodogram: its utility for analysis of circadian rhythms. J Theor Biol 72: 131–160, 1978. [DOI] [PubMed] [Google Scholar]
- 44.Szabat M, Page MM, Panzhinskiy E, Skovso S, Mojibian M, Fernandez-Tajes J, Bruin JE, Bround MJ, Lee JT, Xu EE, Taghizadeh F, O'Dwyer S, van de Bunt M, Moon KM, Sinha S, Han J, Fan Y, Lynn FC, Trucco M, Borchers CH, Foster LJ, Nislow C, Kieffer TJ, Johnson JD. Reduced insulin production relieves endoplasmic reticulum stress and induces beta cell proliferation. Cell Metab 23: 179–193, 2016. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9: 764–775, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150: 1223–1234, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA 106: 21453–21458, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]