Abstract
Pyrimidine-rich circular DNA oligonucleotides 1 and 2 display very high binding affinities for complementary DNA and RNA oligomers by forming bimolecular triple helical complexes.
Although circular polynucleotides are abundant in nature, there exist few studies on small synthetic DNA circles.1,2 We report that circular single-stranded oligodeoxynucleotides having two runs of pyrimidines can bind tightly and sequence selectively to single-stranded polypurine DNA and RNA by forming triple helical complexes.
The 34-nucleotide linear precursors to the circles (for 1, 5′-pTTTTTTCACAC-TTTTTTTTTTTTCACACTTTTTT; for 2, 5′-pTTTCTTCACACTTCTTTCTTTTCCACACCTTTTC), along with their DNA templates (3, 5′-AAAAAAAAAAAA; 4, 5′-AAGAAAGAAAAG) were machine synthesized using the β-cyanoethyl phosphoramidite method3 and purified by gel electrophoresis. Circle precursors were phosphorylated on the 5′ end using a commercially available phosphoramidite reagent.4 Also synthesized were DNA oligomers 5′-TTTTTTTTTTTT and 5′-CTTTTCTTTCTT, the Watson–Crick complements of 3 and 4, respectively, to be used as controls for comparison of binding efficiency.
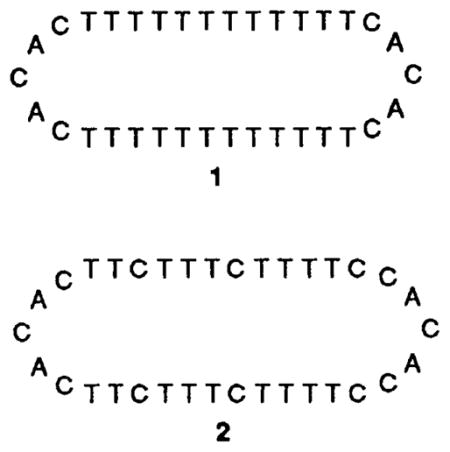
A template-directed chemical ligation was used to condense the 5′-phosphate with the 3′-hydroxy of each precursor oligomer to give the desired macrocycles. For example, the precursor of 1 was hybridized to its complement, 3, bringing the reactive ends adjacent to each other. Cyclization reactions were carried out with the aqueous BrCN–imidazole coupling method, which has been shown to give natural 3′,5′-phosphodiesters for both RNA and DNA using a template to align the reactive ends.5 Isolated yields after gel electrophoresis for the principal new products (1 and 2) of the two reactions were 42 and 58%, respectively.† No product is observed without added template or in the absence of a 5′-phosphate group. The circular structures of products 1 and 2 were confirmed by resistance both to 3′-exonuclease digestion and to 5′ dephosphorylation under conditions in which the linear precursors react completely.
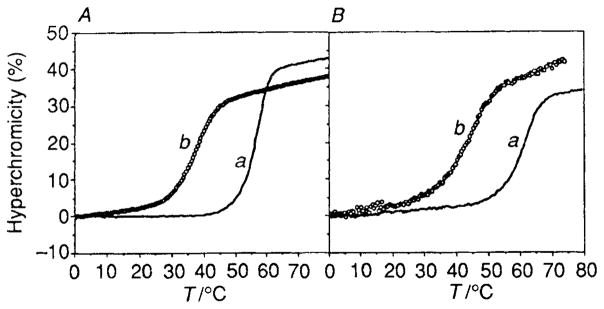
The binding affinities of circles 1 and 2 for their single-stranded templates were measured by comparison of the melting temperatures of the complexes at pH 7 with 100 mmol dm−3 NaCl and 10 mmol dm−3 MgCl2, approximating physiological conditions (Fig. 1 and Table 1). The melting curves at 260 nm show a single transition from bound to unbound species. The free energies (−ΔG°37) of the complexes were derived from the melting data using a two-state model for curve-fitting.6
Fig. 1.
UV absorbance melting curves (260 nm) for the circle complexes and for the corresponding duplexes. Each oligomer strand was present at 3 μmol dm−3 concentration, in a buffer of 100 mmol dm−3 NaCl, 10 mmol dm−3 MgCl2, and 10 mmol dm−3 Tris · HCl (pH 7.0). (A) Complex of circle 1 with d(A)12 (a) compared with duplex d(T)12 · d(A)12 (b). (B) The circle 2 complex with d(AAGAAAGAAAAG) (a) in comparison to the duplex d(CTTTTCTTTCTT) · d(AAGAAAGAAAAG) (b).
Table 1.
Melting transitions (Tm) and free energies of complexation (−ΔG°37) for the circular oligodeoxynucleotides 1 and 2 with their complements and for the analogous duplexes at pH 7.0. Estimated uncertainties in Tm are ±0.5 °C and in free energy, ±10%.
| Complex | Tm/°C | −ΔG°37/kcala |
|---|---|---|
| 5′-AAAAAAAAAAAA 3′-TTTTTTTTTTTT |
36.9 | 8.1 |
|
|
57.4 | 16.7 |
| 5′-AAGAAAGAAAAG 3′-TTCTTTCTTTTC |
44.7 | 9.8 |
|
|
61.8 | 16.5 |
1 cal = 4.184J.
Results show that the circular oligodeoxynucleotides bind to their templates much more strongly than do the natural DNA complementary sequences (Table 1). For example, template 3 with its Watson–Crick complement forms a duplex which melts at 37.1 °C. By comparison, the circular oligomer 1 binds much more strongly, with a Tm of 57.5 °C and a free energy of binding that is 8.6 kcal mol−1 (1 cal = 4.18 J) more favourable than the Watson–Crick duplex. The corresponding association constant at 37 °C is 6 × 1011 dm3 mol−1, which is more than six orders of magnitude greater than that of the duplex. A similar effect is seen for the circle 2 binding to template 4; this complex has a Tm of 62.3 °C, while the analogous duplex melts at 43.8 °C.
The sequence selectivity of binding by the circular oligomers is demonstrated by hybridizing them with the wrong template oligomer. For example, the complex of circle 1 and 4 results in a triple helix with three T–G–T mismatches. The measured Tm is 35.1, or 22.3 °C lower than with the correct template. Similarly, circle 2 hybridized to 3 gives a Tm of only 14.2 °C as a result of three C–A–C mismatches. The ordering of stabilities for these mismatches (T–G–T > C–A–C) is similar to the case for mismatches in duplex DNA, where T–G is more stable than C–A.7
To test binding affinities for RNA, circles 1 and 2 were also hybridized to oligomers having the same sequence as 3 and 4, but with ribose sugars, prepared by the method of Usman.8 Melting studies reveal almost identical affinity of the circles for these RNA oligomers as they show for the DNA oligomers. For example, circle 1 hybridized with r(A)12 has a melting transition of 58.3 °C, and the complex of 2 with r(AAGAAAGAAAAG) has a Tm of 61.9 °C. Both values are within 1 °C of the transitions for the all-DNA complexes.
In addition to the high melting transitions, several other observations support the assumption that the circle · single strand complexes are triple helical. Mixing curves for the two complexes confirm a 1 : 1 stoichiometry,‡ indicating that both binding domains of the circles are involved in the complex; if this were not the case, a 1 : 2 complex would be expected for 1· d(A)12. Additionally, we find that the Tm of circle 2 with 4 is pH dependent: at pH 6 the Tm is 8 °C higher than at neutral pH, while at pH 9 the Tm is lowered by 12 °C. This behaviour is a strong indication of the presence of protonated C + G–C base triads,9 found in pyrimidine · purine · pyrimidine triplexes.10 This pH-dependent binding effect is absent in the analogous duplex (data not shown). The Tm of the complex of 1 and 3 is insensitive to pH over this range, consistent with the model for T–A–T triads, which require no protonation.11 Also supporting a triple helical structure is the observation that the melting transition of the complex of circle 1 could be followed at 284 nm, a wavelength at which triplexes are reported to show transitions but at which Watson–Crick duplexes do not change in absorbance.12
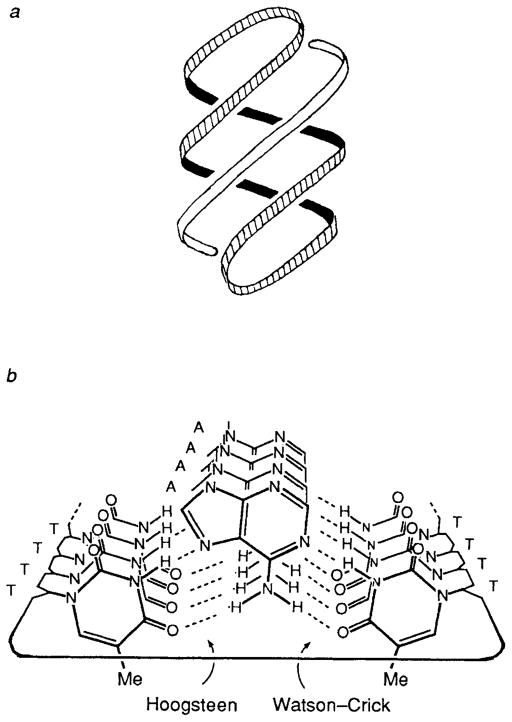
The circular oligodeoxynucleotides evidently achieve this high binding affinity by forming hydrogen bonds on two sides of the central linear polypurine strand (see Fig. 2). Since one side of a circle is complementary in the antiparallel Watson–Crick mode and the other side is complementary in the parallel Hoogsteen mode,13 this allows formation of a 12-base triple helix composed of T–A–T and/or C + G–C triads and capped by two loops of sequence –CACAC–. Although Hoogsteen base pairing in triple helices has been reported to be thermodynamically less favourable than Watson–Crick base pairing,12 we reasoned that the use of both pairing modes in a single molecule should allow for highly stable complexes. This allows, in principle, formation of four hydrogen bonds to adenine by two opposed thymines and five hydrogen bonds to guanine by two cytosines. The cooperative nature of the binding by the Hoogsteen and Watson–Crick domains in the circle complexes is manifested in the high Tm values and single biphasic transitions observed. In effect, the complementary strand is chelated by the outer two strands in the circle.
Fig. 2.
Illustrations of complexes formed between circles and single strands. (a) Helical representation showing triple helical structure. (b) End-on view of complex 1 · d(A)12 showing hydrogen bonds from outer circle bases to the central purine strand. For clarity, the helix is unwound and the phosphate-deoxyribose backbone is simplified.
The binding of polynucleotide single strands with circular DNA oligomers represents a new strategy in nucleotide recognition. Other properties of DNA circles, including binding specificity and the effects of structural variations, are currently under investigation.
Acknowledgments
We thank Professor D. Turner for helpful advice.
Footnotes
Reactions were 50 μmol dm−3 each in circle precursor and template oligonucleotide, with 200 mmol dm−3 imidazole · HCl (pH 7.0), 50 mmol dm−3 BrCN, 100 mmol dm−3 NiCl2, and were allowed to proceed for 24 h at 25 °C and lyophilized. In each case the mixture was then separated by denaturing 20% polyacrylamide gel electrophoresis. The circular products migrated at a rate of 0.89 times the rate for the circle precursors.
Mixing curves (Job’s method) were measured at 260 nm, keeping total oligomer concentrations at 6 μmol dm−3 but varying ratios of circle and complement. Plots of absorbance vs. mole ratio show a break at a ratio of 1:1, confirming this stoichiometry in the complex.
References
- 1.For synthesis of circles 2–10 nt in size, see: Barbato S, DeNapoli L, Picciali G, Santacroce C. Tetrahedron Lett. 1987;289:5727.de Vroom E, Broxterman HJG, Sliedregt LAJM, van der Marel GA, van Boom JH. Nucleic Acids Res. 1988;16:4607. doi: 10.1093/nar/16.10.4607.Capobianco ML, Carcuro A, Tondelli L, Garbesi A, Bonora GM. Nucleic Acids Res. 1990;18:2661. doi: 10.1093/nar/18.9.2661.
- 2.For internally base-paired circles, see: Olivera BM, Scheffler IE, Lehman IR. J Mol Biol. 1968;36:275. doi: 10.1016/0022-2836(68)90381-1.Wemmer DE, Benight AS. Nucleic Acids Res. 1985;13:8611. doi: 10.1093/nar/13.23.8611.Kalisch BW, Krawetz SA, Schoenwaelder KH, van de Sande JH. Gene. 1986;44:263. doi: 10.1016/0378-1119(86)90190-3.Erie DA, Jones RA, Olson WK, Sinha NK, Breslauer KJ. Biochemistry. 1989;28:268. doi: 10.1021/bi00427a037.Ashley GW, Kushlan DM. Biochemistry. 1991;30:2927. doi: 10.1021/bi00225a028.
- 3.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859. [Google Scholar]
- 4.Horn T, Urdea M. Tetrahedron Lett. 1986;27:4705. [Google Scholar]
- 5.Kanaya E, Yanagawa H. Biochemistry. 1986;25:7423. doi: 10.1021/bi00371a026. [DOI] [PubMed] [Google Scholar]; Ferris JP, Huang CH, Hagan WJ. Nucleosides Nucleotides. 1989;8:407. doi: 10.1080/07328318908054184. [DOI] [PubMed] [Google Scholar]; Luebke KJ, Dervan PB. J Am Chem Soc. 1989;111:8733. [Google Scholar]
- 6.Petersheim M, Turner DH. Biochemistry. 1983;22:256. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 7.Aboul-ela F, Koh D, Tinoco I. Nucleic Acids Res. 1985;13:4811. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaringe SA, Francklyn C, Usman N. Nucleic Acids Res. 1990;18:5433. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipsett MN. J Biol Chem. 1964;239:1256. [PubMed] [Google Scholar]
- 10.Arnott S, Seising E. J Mol Biol. 1974;88:509. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- 11.Felsenfeld G, Davies DR, Rich A. J Am Chem Soc. 1957;79:2023. [Google Scholar]
- 12.Pilch DS, Levenson C, Shafer RH. Proc Natl Acad Sci USA. 1990;87:1942. doi: 10.1073/pnas.87.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pilch DS, Brousseau R, Shafer RH. Nucleic Acids Res. 1990;18:5743. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenfeld G, Miles HT. Annu Rev Biochem. 1967;36:407. doi: 10.1146/annurev.bi.36.070167.002203. [DOI] [PubMed] [Google Scholar]; Moser HE, Dervan PB. Science (Washington, DC) 1987:238–645. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]; Rajagopal P, Feigon J. Nature. 1989;339:637. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]