Abstract
The large-scale conformation of DNA molecules play a critical role in many basic elements of cellular functionality and viability. By targeting the structural properties of DNA, many cancer drugs, such as anthracyclines, effectively inhibit tumor growth, but can also produce dangerous side effects. To enhance the development of innovative medications, rapid screening of structural changes to DNA can provide important insight into their mechanism of interaction. In this study, we report changes to circular DNA conformation using All-Atom Molecular Dynamics simulations and characterized experimentally by translocation through a silicon nitride solid-state nanopore from intercalation with ethidium bromide. Our measurements reveal three distinct current blockade levels and a six-fold increase in translocation times for ethidium bromide-treated circular DNA as compared to untreated circular DNA. We attribute these increases to changes in the supercoiled configuration hypothesized to be branched or looped structures formed in the circular DNA molecule. Further evidence of the conformational changes is demonstrated by qualitative atomic force microscopy analysis. These results expand the current methodology for predicting and characterizing DNA tertiary structure, and advances nanopore technology as a platform for deciphering structural changes of other important biomolecules.
Keywords: DNA conformation, nanopore, DNA translocation, ethidium bromide, intercalation, molecular dynamics
Graphical abstract
Over the course of its life cycle, a cell's DNA undergoes many carefully orchestrated conformational changes that facilitate vital cellular processes such as replication and transcription.1,2 Consequently, unresolved conformational defects in the structure can interfere with critical interactions, resulting in genetic anomalies that culminate in cell death.3,4 As a result, numerous cancer treatments target DNA, seeking to induce structural changes to inhibit tumor growth.5,6 Medications such as the widely used anthracycline class of anti-tumor drugs interfere with cellular functionality through interrupting protein-DNA interactions.7 The specific molecular mechanisms are not completely understood, but the increased torsional stress on the DNA from the intercalation is thought to be a significant contributor.8 Although these important chemotherapy drugs have proven to be effective treatments for many forms of human cancers, their associated cardiotoxicity has pushed for the advancement of novel analogs and formulations.9-11 The development of innovative cancer drugs can be greatly enhanced by high-throughput, low-cost methods to characterize their effects on DNA integrity and configuration.12 However, conventional investigations of conformational changes of DNA frequently require extensive preparation, labeling, or substrate adhesion, which may induce unexpected artificial conformational changes.13-16 Here we show that the structural changes induced by the intercalation of ethidium bromide (EtBr) in circular DNA can be identified using a solid-state nanopore. Our experiments are supported by all-atom molecular dynamics simulations that provide fundamental insight into the unwinding of the supercoiled DNA molecule, resulting in the formation of branched (looped) structures and unique signatures in the translocation events.
Although routinely considered as a next-generation platform to achieve rapid and inexpensive DNA sequencing,17-19 the most promising emerging application of solid-state nanopores is to provide a label-free molecular probe to investigate individual biomolecular structures in solution. This approach has been previously used to characterize linear DNA structures, protein binding to linear DNA, and other modifications to biomolecules.20-24 However, the evaluation of circular DNA has been limited to comparisons with linear conformations.25,26 Such comparisons are not adequate for predicting structural changes in DNA conformation in vivo, which requires a DNA molecule with a constrained configuration in the absence of cellular infrastructure. Circular covalently closed DNA has been shown to be a good model for representing the structure of DNA in vivo,1,27,28 since it contains the constraints of closed domains found in DNA that give rise to its large-scale conformational properties. Being constrained, intercalation into circular DNA causes localized unwinding of the molecule, which subsequently causes a structural transformation according to the Calugareanu-Fuller-White model.29 EtBr is a very popular prototypical intercalating molecule that has been used for several decades to fluorescently label DNA and alter its mobility during electrophoresis.30 EtBr's interaction with DNA has been well characterized experimentally and serves as a good model for anthracyclines such as daunorubicin and doxorubicin.31 Previous investigations have shown that upon intercalation, EtBr reduces the negative charge of DNA32,33 while increasing the effective molecular weight.30 The intercalation also initiates a negative unwinding of the helix by reducing the helix angle by 26° per intercalated base pair while increasing the distance between adjacent base pairs, consequently lengthening the DNA molecule.27,34-38 However, while the intercalation of EtBr has been extensively studied, modeling the changes to conformation has been largely overlooked and no direct measurements of the resultant structural changes in a fluid environment have been reported. By investigating the induced conformational changes with a solid-state nanopore, the findings reported here will enhance the current methodology for investigating DNA tertiary structure to advance the understanding of molecular interactions between DNA and intercalating cancer medications.
Results and Discussion
To further our understanding of the changes to DNA conformation that occur due to the intercalation of ethidium between base pairs, all-atom molecular dynamics simulations were conducted to predict and identify unwinding of the DNA in the presence of EtBr. We began with a negatively supercoiled DNA ring of 180 base pairs and linking number (Lk) of 14 in 1M KCl, as depicted in the schematic representation of the simulation system in Figure 1a. Three separate simulations were carried out on the DNA ring. We first simulated the DNA ring for 200 ns and calculated its writhe based on the change in the position of the center of mass of the base pairs. In the other two simulations, 6 and 12 ethidium ions, respectively, were intercalated between the base pairs at approximately equidistant positions (Table S1, Supporting Information), and each system simulated and allowed to evolve for 200 ns. A sample configuration of an ethidium ion located between two base pairs is shown in Figure 1b, and the time evolution of the writhe for the three simulations is shown in Figure 1c. In the first run, i.e., the free DNA ring, the writhe becomes stable after about 20 ns of simulation and fluctuates around -0.65 ± 0.03 for the remainder of the 100 ns run. In the other two systems containing 6 and 12 ethidium ions, the calculated writhe increases during the first 50 ns of the simulation, reaching an average writhe of -0.45 ± 0.09 and -0.16 ± 0.04 over the final 100 ns of the simulation, respectively. The initial and final snapshots of these two systems are shown in Figures 1d and 1e. The figures show that the intercalated ethidium ions unwind the DNA ring, and the extent of unwinding correlates with the number of ethidiums in the system. Comparing the average writhe of the DNA ring with that of the DNA-ethidium complexes, it is possible to estimate the amount of unwinding per intercalated ethidium ion. The second simulation containing 6 ethidium ions yielded an unwinding of 12.0° per intercalated ethidium while the 12 ethidium simulation produced a 14.7° unwinding per ethidium, smaller than the reported 26° per ethidium.36,37 Experimentally, DNA intercalation has been demonstrated to require several seconds to reach a maximum unwinding,39 making it reasonable that a 200 ns-long simulation is not sufficient to capture the complete conformational changes that correspond to a 26° increase in writhe per ethidium. The degree of unwinding has also been reported to correlate with the extent of intercalation,39 so simulations of 6 and 12 ethidium ions per 180 base pairs may not be sufficient to achieve an average unwinding of 26° per intercalated base pair.
Figure 1.
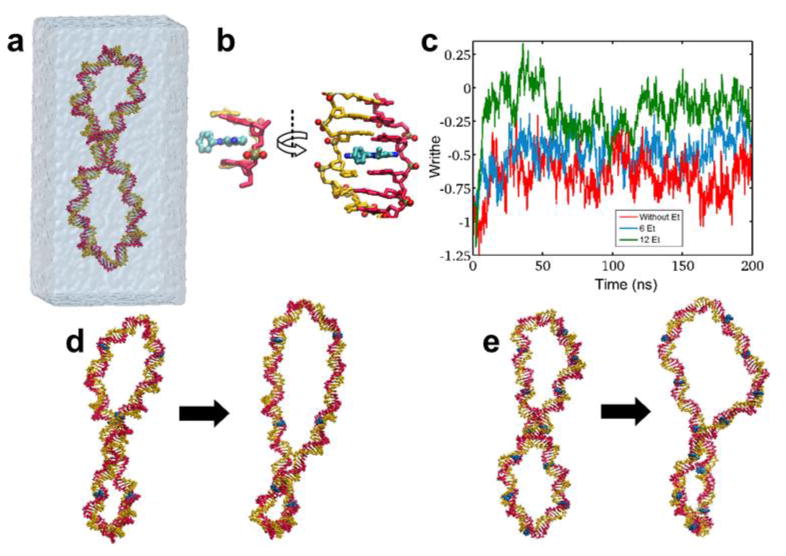
Molecular Dynamics Simulations of Intercalated DNA. (a) Schematic representation of the simulation system containing the DNA ring in explicit solvent. (b) Top and side view of an ethidium ion (blue) intercalated between two DNA base pairs (yellow and red) in a simulation of the DNA ring with 12 intercalated ethidiums. (c) Writhe versus time for three simulations: DNA ring without ethidium (red), DNA ring with 6 (blue), and DNA ring with 12 ethidiums (green). (d and e) Schematic representation of conformational changes after 200 ns of a DNA ring treated by 6 and 12 ethidiums, respectively.
Although the calculated unwinding is not as large as previous experimental reports, the simulations still reveal significant changes in the writhe over the 200 ns evolution period. Because of the intercalant-induced unwinding of the constrained molecule, the changes in writhe are expected to yield changes in the twist which we anticipate to manifest as branching observable with nanopore translocation. The predicted structural changes were assessed by supercoiled DNA translocation experiments through a solid-state nanopore sensor, which offers fast label-free detection and characterization of biomolecules in solution as depicted in Figure 2a. The translocation of a molecule through a nanopore (Fig. 2b) produces a characteristic drop in ionic current that is used to analyze the structural configuration. To characterize the effects caused by the intercalation of ethidium ions (Fig. 2c), we first predicted changes to the translocation of DNA with further molecular dynamics simulations. The translocation of a 50 bp linear DNA molecule through a 3.8 nm diameter, 10.5 nm thick nanopore shown in Figure S1 (Supporting Information) reveals that the intercalation of ethidium ions does not have a significant impact on the current drop for the nanopore used in the simulation. However, with the reported increases in DNA mass and length as well as the decreased charge,27,30,33-35 an increase in the translocation time would be expected. From the simulations, it is anticipated that changes in the current drop will arise only from branching and not occur due to an increase in the intercalated DNA diameter.
Figure 2.
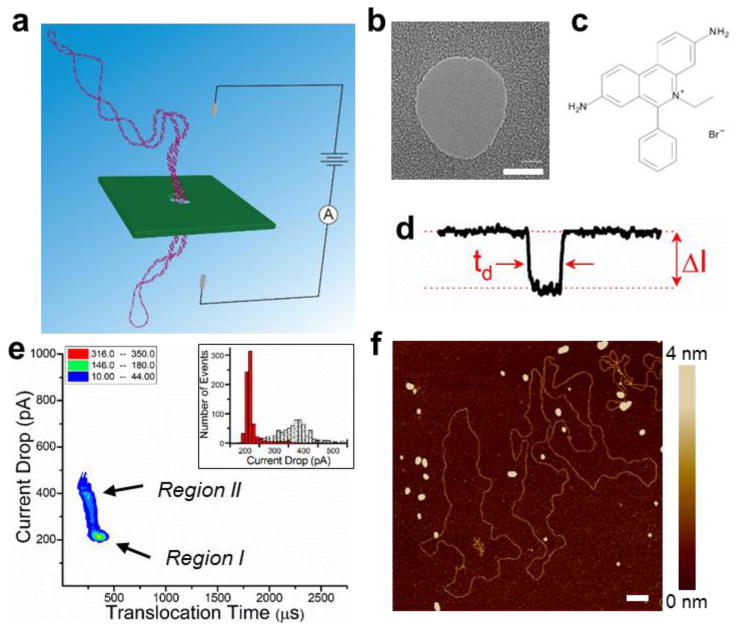
Experimental setup and analysis of circular DNA translocation through a solid-state nanopore. (a) Schematic of a nanopore experiment. (b) TEM image of a typical nanopore. Scale bar is 10 nm. (c) The molecular structure of ethidium bromide. (d) Translocation events are analyzed for the current drop, ΔI, and the translocation time, td. (e) The density plot of the circular DNA translocations events suggests two discrete DNA conformations without EtBr intercalation. (f) AFM height images of circular DNA on mica reveal a generally uniform structure without EtBr intercalation. Horizontal scale bar is 100 nm. A large area AFM image is shown in Figure S2a (Supporting Information).
Briefly, the nanopore experiments utilize a single ∼20 nm pore drilled in a 20 nm thick Si3N4 membrane (Fig. 2b) sandwiched between two flow cells containing electrolyte, thus forming the only electrical path between the reservoirs. An applied voltage across the nanopore generates an ionic current that is interrupted by the translocation of DNA through the pore. The change in current, ΔI, and the translocation time, td, (Fig. 2d) of the translocation events are characteristic for the biomolecules. A detailed discussion of the experimental parameters is provided in the Methods section.
The density plot and the accompanying histograms of the translocation data from circular DNA (Fig. 2e) revealed two distinct regions of current drop, which suggests two populations of different DNA molecular conformations. The majority of the translocations occur in Region I, corresponding to longer translocation time and smaller current drop. The small temporal distribution of the events indicates a relatively uniform population of DNA, and a current drop of ∼200 pA suggests the simultaneous translocation of two double strands (N = 2) consistent with DNA in the circular covalently closed conformation. We further investigated this by atomic force microscopy (AFM), and found a generally uniform circular conformation as demonstrated by the representative AFM height image in Figure 2f and Figure S2a (Supporting Information). The inclusion of circular nicked DNA in the population would also exhibit a similar uniform structural configuration.40,41 However, its presence would be clearly observable in the translocation plots.25,26 The second region of events occurred with a shorter translocation time, but with twice the current drop (∼400 pA). This suggests that four double strands (N = 4) are occupying the nanopore, which consequently decreases the effective length of the DNA and leads to a decreased translocation time. Linear DNA translocation is known to exhibit a similar behavior of decreased translocation time for a doubling of the current drop, which is attributed to a folded DNA conformation.25,26,42,43 Although we cannot entirely exclude the possibility of folded conformations in Region II, folded events seem unlikely with supercoiled DNA due to the stiffness of the molecule. Previous work with nicked circular DNA, which is a more relaxed conformation than supercoiled, did not reveal the occurrence of folded events.25,26
To identify the configuration of the DNA during translocation, we analyzed the translocation events that comprise the two regions on the density plot. Region I in Figure 2e was thought to be a circular closed configuration. The translocation events shown in Figure 3a confirm a uniform conformation, indicating a structure represented by the illustration in Figure 3b. The events comprising Region II indicate different structural features that give rise to the multilevel current drops shown in Figure 3c. These typical events feature two discrete current drops, each of magnitude 200 pA. The second current drop occurring in the middle of the translocation event suggests the presence of a branched (looped) structure similar to the conformation illustrated in Figure 3d and demonstrated in the representative AFM image in Figure 2f. These data also support the hypothesis that the secondary current drops are not caused by folded events. We hypothesized that the addition of an intercalant to unwind the circular DNA would initiate further branching of the circular DNA due to the unwinding predicted by the simulations and previous experimental studies.36-38
Figure 3.
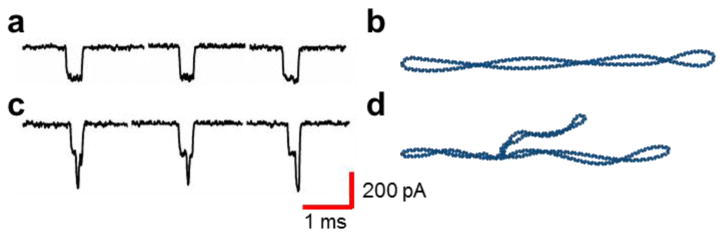
Analysis of the translocation events reveals different topological conformations. (a) Uniform translocation events suggest a circular covalently closed structure. (b) An illustration of circular covalently closed DNA. (c) Translocation events with a peak current drop of ∼400 pA composed of two discrete current drops of ∼200 pA. The second current drop within the event suggests a branched structural conformation. (d) An illustration of a branched circular DNA structure.
Under identical translocation conditions, we added EtBr to the DNA to initiate structural changes. As shown by the density plot in Figure 4a, the translocation time of the events increased by at least 85% and there were 3 regions of current drops as shown by the inset histogram. Comparing the density plot in Figure 2e with Figure 4a, we observed that the majority of events have shifted from having a current drop of 200 pA to events with a 400 pA current drop. To determine if the changes in translocation could be due to changes in conformation, we used AFM to identify relative changes in the DNA structure after intercalation. The AFM height image in Figure 4b and Figure S2b (Supporting Information) shows a significant increase in branched structures relative to those seen in Figure 2f, which we attribute to the interaction with EtBr.
Figure 4.
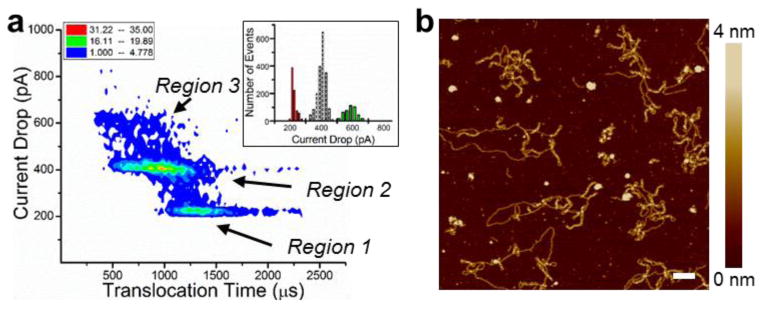
Intercalation of ethidium bromide induces significant changes in the translocation and DNA structure. (a) The density plot of the intercalated (0.5 μM EtBr) circular DNA translocations reveals four discrete DNA conformations. The histogram shows only three populations. (b) AFM height images of circular DNA after treatment with ethidium bromide (initial EtBr concentration ∼0.1 μM) exhibit significant branching structures. Horizontal scale bar is 100 nm. A large area AFM image and height profiles are shown in Figure S2b and Figure S3 (Supporting Information) respectively.
Again, we analyzed the translocation events to determine the structural conformation of the DNA. For uniform supercoiled DNA intercalated with ethidium ions, the simulations suggest the translocation events will remain at 200 pA, but with a longer translocation time. The events in Region 1, shown in Figure 5a, were uniform with a current drop of 200 pA, which suggests that the DNA remains in the circular closed configuration. However, the translocation time increased by ∼265% which we attribute to the increased length, increased mass, and decreased charge arising from ethidium ion intercalation. An illustration of the increased length is shown in Figure 5b and is consistent with the results of the simulation.
Figure 5.
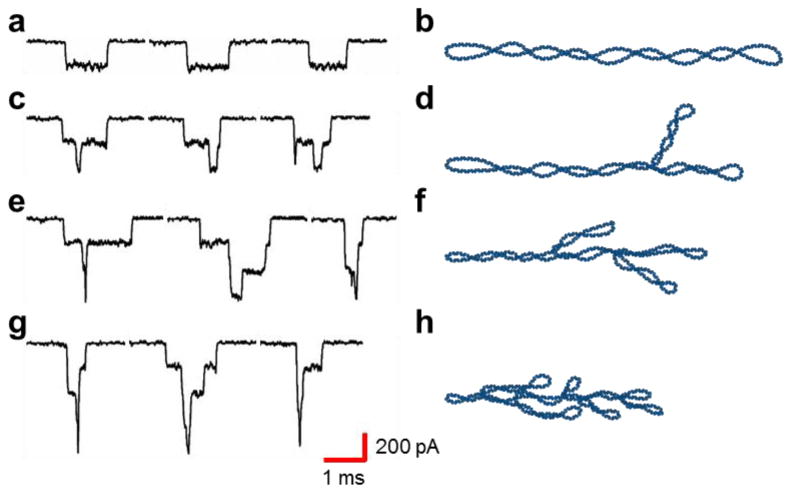
The translocation events of the intercalated circular DNA reveal changes in the structural conformations. (a) Uniform translocation events have a longer translocation time attributed to the lengthening of the molecule, increased mass, and decreased charge. (b) An illustration of the longer intercalated DNA. (c, e, g) Translocation events with multiple discrete current drops suggesting increasing amounts of branching. (d, f, h) Illustrations of multiple branched structures.
As indicated by Figure 4a, upon addition of EtBr the majority of events shifted to Region 2, indicating that intercalation caused branching in the DNA conformation. Although the current blockage remained 400 pA, the translocation time of branched DNA increased by ∼286%, which is consistent with changes to molecular length, mass, and net charge. The branching structures can be identified by the discrete current drops in Figure 5c, suggesting a conformation like that illustrated in Figure 5d.
The translocation events comprising Region 3 have multiple discrete current drops, shown in Figure 5e, with a maximum current drop of 600 pA. Again the current drops occur in multiples of 200 pA, consistent with the translocation of multiply branched structures as illustrated in Figure 5f. Further analysis of the translocation events reveals that approximately 5% of the total events correspond to a current drop of over 700 pA, shown in Figure 5g, that were not represented in the histogram. Additional primary branches or secondary branches (branched branches), as illustrated in Figure 5h, would be expected to produce an 800 pA current, which is observed for about one-half of the events. We attribute the discrepancy in current drop to signal filtering since the translocation of these individual features is so fast.
The analysis of the event morphology suggests branched structures translocating the nanopore, implying that multiple pairs of helices occupy the nanopore simultaneously. Although the hydrodynamic radius of this particular plasmid used has not been reported, previous work using smaller nanopores in similar high salt concentrations (1 M KCl) demonstrated the simultaneous translocation of multiple helices. Circular DNA translocation using 10 nm diameter pores report branched conformation events (N = 4),44 and up to 5 helices of linear DNA have been shown to concurrently translocate using a ∼10 nm pore.26 The nanopores used in our investigation had approximately 4 times the cross sectional area, and up to 8 times greater volume than the nanopores used in the previous reports, so it is reasonable that multiple pairs of helices (N = 4, N = 6) can translocate the nanopore simultaneously. Furthermore, comparing the translocation of the closed circular conformation with and without intercalated EtBr, we found that translocation of intercalated DNA does not change the current drop (within the experimental error). EtBr is known to also bind external to the DNA helix,45-49 and high concentrations have been reported to increase the hydrodynamic radius;33 however, our results suggest that the concentration of EtBr used was low enough not to significantly change the hydrodynamic radius of the circular DNA.
In summary, we predict and evaluate changes to DNA conformation due to the intercalation of EtBr into circular DNA. All-atom molecular dynamics simulations predict that the intercalation will induce unwinding of the helix. Although not observed on the time-scales used in simulation (200 ns), we hypothesize that the unwinding gives rise to branches in the DNA molecule that we experimentally characterized using solid-state nanopore translocation and further identified with AFM. This work represents a significant advancement in simulating changes to circular DNA conformation and is supported by experimental results. Our findings provide new fundamental insight into characterizing DNA structural conformations, which is expected to have important implications for anti-cancer drug treatment and design.
Methods
Molecular dynamics simulations
Molecular dynamics simulations were carried out using the program NAMD.50 The AMBER forcefield parmbsc051 was used to describe interaction of the atoms. A circular covalently closed DNA molecule composed of 180 base pairs with four separate regions of sequences has been used. The sequence of the simulated DNA ring is d(GpC)45d(GpG)45d(GpC)45d(GpG)45, which consists of four regions to make it easier to get a “figure 8” shape for the ring.52 We avoided A-T base-pairs in the sequence to reduce the probability of bubble formation and local denaturation. The initial conformation of the DNA ring is generated by the 3DNA software,53 assuming a perfect circular geometry for the ring with base pair step parameters corresponding to a linking number of 14 as an input. Such a DNA ring is an underwound conformation compared to the equilibrium values of the intrinsic twist of the base pairs.54-56 To equilibrate this circular DNA, we applied generalized Born implicit solvent implemented in NAMD.57 We start with 10,000 steps of minimization, then carry out 20 ns simulations at a constant temperature of 298 K and an ion concentration of 1 M. After this stage, the DNA configuration changes to a “figure 8” shape and can be used as input for an explicit all-atom MD simulation. We used the final configuration and solvated it with TIP3P water molecules in a periodic box of 120 × 150 × 290 Å3 and added K+ and Cl- ions to achieve a salt concentration of 1 M. Such a system contains approximately 520,000 atoms. For simulations including ethidium bromide molecules, we increased the size of the water box to 140 × 160 × 350 Å3 to be able to capture conformational changes of the DNA ring while keeping enough distance between the DNA and its periodic images. In this case, the simulation box consists of about 730,000 atoms. The Particle-Mesh Ewald (PME) summation method58 has been employed for electrostatic interactions and the timestep chosen to be 1 fs. The system has been minimized for 10,000 steps and then simulated for 5 ns at a constant temperature of 298 K and constant pressure of 1 atm while harmonic constraints were imposed on all DNA atoms. The constraints were then released and the system was equilibrated for 25 ns at constant temperature. At this stage we take the final equilibrated configuration and start three separate simulations. The first simulation system consists of the DNA ring in the water box. In the second and third systems, we manually insert six and twelve ethidium ions between the DNA base pairs, uniformly distributed along the length of the DNA ring. The interatomic parameters and partial charges of the ethidium ions were taken from a previous investigation59 and the initial positions of the ethidiums were manually adjusted to make it as close as possible to the available crystallographic information.60 Accordingly, six and twelve Cl- ions were randomly selected and removed from the two simulation systems, to maintain charge neutrality of the system. Each system was minimized for 20,000 steps and equilibrated at constant temperature for 2 ns with harmonic constraints applied to both DNA and ethidiums. The constraints were released and the system minimized again for 10,000 steps. After this initial minimization and equilibration, each simulation is continued at constant temperature for 200 ns to observe the effect of the ethidiums on conformational changes of the DNA ring. As a control simulation, the DNA ring has also been simulated in the absence of ethidium bromide for 100 ns. The simulation trajectories are sampled every 1 ps for further analysis. Writhe is calculated numerically by discretization of the Gauss double integral along a polygon of 180 segments.61
DNA molecules
The circular plasmid pTYB21 (7514 bp) in 10 mM Tris and 1 mM EDTA was purchased from New England Biolabs and used for translocation and AFM investigations without further purification.
AFM imaging
Sample Preparation
For non-EtBr (ethidium bromide) treated samples, 20 μL of 1× TAE buffer (40 mM Tris base, 20 mM acetic acid, and 1 mM EDTA, pH 8.0) containing 20 mM MgCl2 and 5 μL of 4 nM circular DNA (pTYb21) in 1× TAE buffer were incubated together on a freshly cleaved mica surface for 5 minutes. The divalent magnesium cations served to screen the negatively charged DNA from the negatively charged mica surface, thereby enhancing adsorption of DNA on the mica. The samples were then rinsed with 1 mL aliquot of ultrapure (18.2 MΩ·cm) water to remove non-adsorbed DNA before adding 20-40 μL of 12 mM NiCl2 in 1× TAE buffer.
For EtBr treated samples, 20 μL of 1× TAE buffer containing 20 mM MgCl2 and 5 μL of 1× TAE buffer containing 4 nM circular DNA (pTYb21) were incubated together on a freshly cleaved mica surface for 5 minutes. The samples were then rinsed with a 1 mL aliquot of ultrapure (18.2 MΩ·cm) water before adding 20 μL of 15 mM MgCl2 in 1× TAE buffer. 2 μL of 1.26 μM EtBr were sequentially added to the sample, mixed using a pipette and allowed to incubate for 5-10 minutes. 20 μL of solution was removed from the sample and immediately replaced by 20 μL of 12mM NiCl2 in TAE buffer before imaging with the AFM.
Imaging Conditions
All images were acquired in fluid (15 mM MgCl2 in 1× TAE or 12 mM NiCl2 in 1× TAE) using a Bruker MultiMode 8 AFM operated in PeakForce Tapping mode (20-40 mV PeakForce setpoint, corresponding to ≤ 1 nN) with a Bruker ScanAsyst Fluid+ probe (k : 0.7 N/m, f0 : 120-180 kHz). Additional imaging buffer was added as necessary during imaging to counteract evaporation.
Image Editing Procedure
All image processing was carried out using NanoScope Analysis Version 1.50 or WSxM 5.0.62 All images were cropped then first-order plane fit to remove sample tilt. If necessary, a first order line-by-line flatten was subsequently applied to account for slight line-to-line fast axis piezo offsets.
Nanopore translocation
Silicon nitride nanopores were fabricated in 20 nm thick suspended membrane TEM grids (Norcada) using a JEOL 2010F field emission gun transmission electron microscope. All investigations were conducted using nanopores with diameters of a least ∼20 nm, to ensure adequate capacity for simultaneous translocation of multiple strands. Each nanopore was fixed between two fluid reservoirs (∼100 μl each) filled with a buffered ionic solution (1 M KCl, 10 mM Tris, 1 mM EDTA). Two Ag/AgCl electrodes were immersed in the ionic solution and connected to the headstage of an Axopatch 200B amplifier (Molecular Devices), biased at 100 mV. The amplifier was linked to a Digidata 1440 (Molecular Devices) data acquisition system for recording and analysis. Circular covalently closed DNA was added to the cis reservoir and the recorded signal was filtered with the 10 kHz low pass hardware filter from the Axopatch amplifier. Intercalation experiments were performed in identical conditions with the addition of 50 μM EtBr added to the cis chamber (0.5 μM final concentration). Attempts to increase the EtBr concentration resulted in longer translocation events with decreased current drop resembling linear DNA translocation (data not shown) that we hypothesize to be intercalant-induced DNA breaking.8,63 Further characterization of these events is beyond the scope the work presented here. Data analysis was performed with Clampfit (Molecular Devices) and Origin8.5 (OriginLab) software packages and custom routines developed with MATLAB (MathWorks).
Supplementary Material
Acknowledgments
The authors would like to acknowledge the use of the High Performance Computing Cluster as well as startup funding and a CURA award at the University of Illinois at Chicago. We also would like to thank F. Remacle at University of Liège of for providing the ethidium forcefield. Further financial support was provided by the National Science Foundation Grant No. 0856815, Idaho STEP (Science Talent Expansion Program), bridge funding from the Division of Research and Economic Development and the generous startup funding provided by the College of Arts and Science and the College of Engineering at Boise State University.
Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Awards Number P20GM109095 and P20GM103408. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available. Table of ethidium positions for MD simulations. MD simulation of linear DNA translocation. Large area AFM height images of DNA on mica and AFM height profiles. The Supporting Information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli N. Topological Challenges to DNA Replication: Conformations at the Fork. Proc Natl Acad Sci USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zakharova SS, Jessie W, Backendorf C, Egelhaaf SU, Lapp A, van der Maarel JRC. Dimensions of Plectonemically Supercoiled DNA. Biophys J. 2002;83:1106–1118. doi: 10.1016/S0006-3495(02)75234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J. The DNA-Damage Response in Human Biology and Disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roos WP, Kaina B. DNA Damage-Induced Cell Death by Apoptosis. Trends in Molecular Medicine. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Martinez R, Chacon-Garcia L. The Search of DNA-Intercalators as Antitumoral Drugs: What It Worked and What Did Not Work. Current Medicinal Chemistry. 2005;12:127–151. doi: 10.2174/0929867053363414. [DOI] [PubMed] [Google Scholar]
- 6.Gurova K. New Hopes from Old Drugs: Revisiting DNA-Binding Small Molecules as Anticancer Agents. Future oncology. 2009;5:1685. doi: 10.2217/fon.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hortobagyi GN. Anthracyclines in the Treatment of Cancer. Drugs. 1997;54:1–7. doi: 10.2165/00003495-199700544-00003. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA Torsion, and Chromatin Dynamics. Biochim Biophys Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thigpen JT. Innovations in Anthracycline Therapy: Overview. Commun Oncol. 2005;2:3–7. [Google Scholar]
- 10.Rabbani A, Finn RM, Ausio J. The Anthracycline Antibiotics: Antitumor Drugs That Alter Chromatin Structure. BioEssays. 2004;27:50–56. doi: 10.1002/bies.20160. [DOI] [PubMed] [Google Scholar]
- 11.Greene J, Hennessy B. The Role of Anthracyclines in the Treatment of Early Breast Cancer. J Oncol Pharm Practice. 2015;21:201–212. doi: 10.1177/1078155214531513. [DOI] [PubMed] [Google Scholar]
- 12.Palchaudhuri R, Hergenrother PJ. DNA as a Target for Anticancer Compounds: Methods to Determine the Mode of Binding and the Mechanism of Action. Curr Opin Biotechnol. 2007;18:497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Pastre D, Pietrement O, Zozime A, Le Cam E. Study of the DNA/Ethidium Bromide Interactions on Mica Surface by Atomic Force Microscope: Influence of the Surface Friction. Biopolymers. 2005;77:53–62. doi: 10.1002/bip.20185. [DOI] [PubMed] [Google Scholar]
- 14.Nagami F, Zuccheri G, Samorı B, Kuroda R. Time-Lapse Imaging of Conformational Changes in Supercoiled DNA by Scanning Force Microscopy. Anal Biochem. 2002;300:170–176. doi: 10.1006/abio.2001.5435. [DOI] [PubMed] [Google Scholar]
- 15.Schvartzman JB, Martinez-Robles ML, Hernandez P, Krimer DB. Plasmid DNA Topology Assayed by Two-Dimensional Agarose Gel Electrophoresis. In: Makovets S, editor. DNA Electrophoresis: Methods and Protocols. Springer; New York, New York, USA: 2013. [DOI] [PubMed] [Google Scholar]
- 16.Schmatko T, Muller P, Maaloum M. Surface Charge Effects on the 2d Conformation of Supercoiled DNA. Soft Matter. 2014;10:2520–2529. doi: 10.1039/c3sm53071j. [DOI] [PubMed] [Google Scholar]
- 17.Dekker C. Solid-State Nanopores. Nature Nanotech. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 18.Healy K, Schiedt B, Morrison AP. Solid-State Nanopore Technologies for Nanopore-Based DNA Analysis. Nanomedicine. 2007;2:875–897. doi: 10.2217/17435889.2.6.875. [DOI] [PubMed] [Google Scholar]
- 19.Venkatesan BM, Bashir R. Nanopore Sensors for Nucleic Acid Analysis. Nature Nanotech. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 20.Carlsen AT, Zahid OK, Ruzicka JA, Taylor EW, Hall AR. Selective Detection and Quantification of Modified DNA with Solid-State Nanopores. Nano Lett. 2014;14:5488–5492. doi: 10.1021/nl501340d. [DOI] [PubMed] [Google Scholar]
- 21.Shim J, Humphreys GI, Venkatesan BM, Munz JM, Zou X, Sathe C, Schulten K, Kosari F, Nardulli AM, Vasmatzis G, Bashir R. Detection and Quantification of Methylation in DNA Using Solid-State Nanopores. Scientific Reports. 2013;3:1389. doi: 10.1038/srep01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of Methylcytosine from Hydroxymethylcytosine in DNA Molecules. J Am Chem Soc. 2011;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae MH, Aluru NR, Pop E, Bashir R. Stacked Graphene-Al2o3 Nanopore Sensors for Sensitive Detection of DNA and DNA-Protein Complexes. ACS Nano. 2012;6:441–50. doi: 10.1021/nn203769e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim J, Kim Y, Humphreys GI, Nardulli AM, Kosari F, Vasmatzis G, Taylor WR, Ahlquist DA, Myong S, Bashir R. Nanopore-Based Assay for Detection of Methylation in Double-Stranded DNA Fragments. ACS Nano. 2015;9:290–300. doi: 10.1021/nn5045596. [DOI] [PubMed] [Google Scholar]
- 25.Fologea D, Brandin E, Uplinger J, Branton D, Li J. DNA Conformation and Base Number Simultaneously Determined in a Nanopore. Electrophoresis. 2007;28:3186–3192. doi: 10.1002/elps.200700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of Double-Strand DNA through a Silicon Oxide Nanopore. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 27.Keller W. Determination of the Number of Superhelical Turns in Simian Virus 40 DNA by Gel Electrophoresis. Proc Natl Acad Sci USA. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levene SD. Topology in Biology: From DNA Mechanics to Enzymology. In: Monastyrsky MI, editor. Topology in Molecular Biology. Springer; New York, New York, USA: 2007. [Google Scholar]
- 29.Ganbibov A, Yakubovskaya E, Lukin M, Favorov P, Reshetnyak A, Montastyrsky M. Dynamics of DNA Supercoiling. In: Monastyrsky MI, editor. Topology in Molecular Biology. Springer; New York, New York, USA: 2007. [Google Scholar]
- 30.Sigmon J, Larcom LL. The Effect of Ethidium Bromide on Mobility of DNA Fragments in Agarose Gel Electrophoresis. Electrophoresis. 1996;17:1524–1527. doi: 10.1002/elps.1150171003. [DOI] [PubMed] [Google Scholar]
- 31.Viglasky V, Valle F, Adamcik J, Joab I, Podhradsky D, Dietler G. Anthracycline-Dependent Heat-Induced Transition from Positive to Negative Supercoiled DNA. Electrophoresis. 2003;24:1703–1711. doi: 10.1002/elps.200305388. [DOI] [PubMed] [Google Scholar]
- 32.Waring MJ. Complex Formation between Ethidium Bromide and Nucleic Acids. J Mol Biol. 1965;13:269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- 33.Liang D, Zhang J, Chu B. Study of Ethidium Bromide Effect on Dsdna Separation by Capillary Zone Electrophoresis and Laser Light Scattering. Electrophoresis. 2003;24:3348–3355. doi: 10.1002/elps.200305604. [DOI] [PubMed] [Google Scholar]
- 34.Reese HR. Effects of DNA Charge and Length on the Electrophoretic Mobility of Intercalated DNA. Biopolymers. 1994;34:1349–1358. doi: 10.1002/bip.360341007. [DOI] [PubMed] [Google Scholar]
- 35.Nielson PE, Zhen W, Henriksen U, Buchardt O. Sequence-Influenced Interactions of Oligoacridines with DNA Detected by Retarded Gel Electrophoretic Migrations. Biochemistry. 1988;27:67–73. doi: 10.1021/bi00401a012. [DOI] [PubMed] [Google Scholar]
- 36.Liu LF, Wang JC. On the Degree of Unwinding of the DNA Helix by Ethidium: Ii. Studies by Electron Microscopy. Biochimica et Biophysica Acta - Nucleic Acids and Protein Synthesis. 1975;395:405–412. [PubMed] [Google Scholar]
- 37.Wang JG. The Degree of Unwinding of the DNA Helix by Ethidium: I. Titration of Twisted Pm2 DNA Molecules in Alkaline Cesium Chloride Density Gradients. J Mol Biol. 1974;89:783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- 38.Tsai CC, Jain SC, Sobell HM. X-Ray Crystallographic Visualization of Drug-Nucleic Acid Intercalative Binding: Structure of an Ethidium-Dinucleoside Monophosphate Crystalline Complex, Ethidium: 5-Iodouridylyl(3′-5′)Adenosine. Proc Natl Acad Sci USA. 1975;72:628–632. doi: 10.1073/pnas.72.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi M, Harada Y. Direct Observation of the Reversible Unwinding of a Single DNA Molecule Caused by the Intercalation of Ethidium Bromide. Nucleic Acids Res. 2007;35:e125. doi: 10.1093/nar/gkm529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Y, Marszalek PE. Detecting DNA Damage at a Single-Molecule Level by Atomic Force Microscopy. In: Mendez-Vilas A, Diaz J, editors. Microscopy: Science, Technology, Applications and Education. Formatex; Badajoz, Spain: 2010. [Google Scholar]
- 41.Umemura K, Nagami F, Okada T, Kuroda R. Afm Characterization of Single Strand-Specific Endonuclease Activity on Linear DNA. Nucleic Acids Res. 2000;29:e39. doi: 10.1093/nar/28.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fologea D, Gershow M, Ledden B, McNabb DS, Golovchenko JA, Li J. Detecting Single Stranded DNA with a Solid State Nanopore. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. DNA Translocation through Graphene Nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 44.Uplinger J, Thomas B, Rollings R, Fologea D, McNabb DS, Li J. K+, Na+ and Mg2+ on DNA Translocation in Silicon Nitride Nanopores. Electrophoresis. 2012;33:3448–3457. doi: 10.1002/elps.201200165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baranovskii SF, Chernyshev DN, Buchel'nikov AS, Evstigneev MP. Thermodynamic Analysis of Complex Formation of Ethidium Bromide with DNA in Water Solutions. Biophysics. 2011;56:214–219. [PubMed] [Google Scholar]
- 46.McMurray CT, Small EW, Van Holde KE. Binding of Ethidium to the Nucleosome Core Particle. 2. Internal and External Binding Modes. Biochemistry. 1991;30:5644–5652. doi: 10.1021/bi00237a002. [DOI] [PubMed] [Google Scholar]
- 47.Nafisi S, Saboury AA, Keramat N, Neault JF, Tajmir-Riahi HA. Stability and Structural Features of DNA Intercalation with Ethidium Bromide, Acridine Orange and Methylene Blue. J Mol Struct. 2007;827:35–43. [Google Scholar]
- 48.Nordmeier E. Absorption Spectroscopy and Dynamic and Static Light-Scattering Studies of Ethidium Bromide Binding to Calf Thymus DNA: Implications for Outside Binding and Intercalation. J Phys Chem. 1992;96:6045–6055. [Google Scholar]
- 49.Olmsted J, III, Kearns DR. Mechanism of Ethidium Bromide Fluorescsnce Enhancement on Binding to Nucleic Acids. Biochemistry. 1977;16:3647–3654. doi: 10.1021/bi00635a022. [DOI] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with Namd. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez A, et al. Refinement of the Amber Force Field for Nucleic Acids: Improving the Description of A/Γ Conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell JS, Harris SA. Thermodynamics of Writhe in DNA Minicircles from Molecular Dynamics Simulations. Phys Rev Lett. 2013;110:148105. doi: 10.1103/PhysRevLett.110.148105. [DOI] [PubMed] [Google Scholar]
- 53.Lu XJ, Olson WK. 3dna: A Software Package for the Analysis, Rebuilding and Visualization of Three-Dimensional Nucleic Acid Structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris SA, Laughton CA, Liverpool TB. Mapping the Phase Diagram of the Writhe of DNA Nanocircles Using Atomistic Molecular Dynamics Simulations. Nucleic Acids Res. 2008;36:21–29. doi: 10.1093/nar/gkm891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell JS, Laughton CA, Harris SA. Atomistic Simulations Reveal Bubbles, Kinks and Wrinkles in Supercoiled DNA. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez A, et al. Towards a Molecular Dynamics Consensus View of B-DNA Flexibility. Nucleic Acids Res. 2008;36:2379–2394. doi: 10.1093/nar/gkn082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanner DE, et al. Parallel Generalized Born Implicit Solvent Calculations with Namd. Journal of Chemical Theory and Computation. 2011;7:3635–3642. doi: 10.1021/ct200563j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darden T, York D, Pedersen L. Particle Mesh Ewald: An N⋅Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 59.Fresch B, Remacle F. Atomistic Account of Structural and Dynamical Changes Induced by Small Binders in the Double Helix of a Short DNA. PCCP. 2014;16:14070–14082. doi: 10.1039/c4cp01561d. [DOI] [PubMed] [Google Scholar]
- 60.Sobell HM, Tsai CC, Jain SC, Gilbert SG. Visualization of Drug-Nucleic Acid Interactions at Atomic Resolution: Iii. Unifying Structural Concepts in Understanding Drug-DNA Interactions and Their Broader Implications in Understanding Protein-DNA Interactions. J Mol Biol. 1977;114:333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- 61.Klenin K, Langowski J. Computation of Writhe in Modeling of Supercoiled DNA. Biopolymers. 2000;54:307–317. doi: 10.1002/1097-0282(20001015)54:5<307::AID-BIP20>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 62.Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. Wsxm: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev Sci Instrum. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 63.Ghosh D, Hossain M, Saha C, Dey SK, Kumar GS. Intercalation and Induction of Strand Breaks by Adriamycin and Daunomycin: A Study with Human Genomic DNA. DNA and Cell Biology. 2012;31:378–387. doi: 10.1089/dna.2011.1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


