SUMMARY
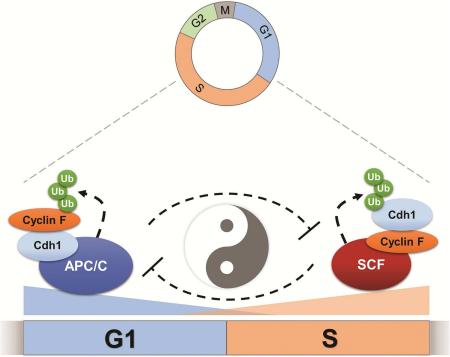
The Anaphase Promoting Complex/Cyclosome (APC/C) is an ubiquitin ligase and core component of the cell cycle oscillator. During G1-phase APC/C binds to its substrate receptor Cdh1 and APC/CCdh1 plays an important role in restricting S-phase entry and maintaining genome integrity. We describe a reciprocal feedback circuit between APC/C and a second ubiquitin ligase, the SCF (Skp1-Cul1-F box). We show that Cyclin F, a cell cycle regulated substrate receptor (F-box protein) for the SCF, is targeted for degradation by APC/C. Furthermore, we establish that Cdh1 is itself a substrate of SCFCyclin F. Cyclin F loss impairs Cdh1 degradation and delays S-phase entry, and this delay is reversed by simultaneous removal of Cdh1. These data indicate that the coordinated, temporal ordering of Cyclin F and Cdh1 degradation, organized in a double-negative feedback loop, represents a fundamental aspect of cell cycle control. This mutual antagonism could be a feature of other oscillating systems.
Graphical Abstract
INTRODUCTION
During cell cycle progression, the ubiquitin proteasome system controls the destruction of numerous proteins. The best studied of these are Cyclins, the activating subunits of Cyclin Dependent Kinases (CDKs). Degradation of the mitotic Cyclins is catalyzed by the Anaphase Promoting Complex/Cyclosome (APC/C), a multi-subunit E3 ubiquitin ligase and core component of the cell cycle oscillator (reviewed in Sivakumar and Gorbsky, 2015). APC/C was discovered for its role in promoting Cyclin B destruction (King et al., 1995) and is now known to regulate dozens of proteins involved in cell cycle progression (Zhang et al., 2014). The APC/C is required for orderly and unidirectional progress through the cell cycle, and genome maintenance (Sivakumar and Gorbsky, 2015). APC/C utilizes two related substrate receptor proteins. In mid-mitosis, APC/C binds Cdc20, leading to the degradation of Cyclin B and Securin, and promoting anaphase onset. In late mitosis and throughout G1, APC/C binds to the Cdc20 related protein Cdh1/Fzr1 (hereafter referred to as Cdh1), targeting a myriad of proteins, including the Aurora A kinase and FoxM1 transcription factor (Zhang et al., 2014). Cdc20 and Cdh1 both bind substrates through canonical degron motifs termed D and KEN boxes, although others have been described more recently (Barford, 2011). While many APC/C substrates have been identified, estimates suggest there are more still to be discovered.
APC/CCdh1 must be turned off at the end of G1 to allow for S-phase entry. Cdh1 overexpression in G1 cells prevents S-phase entry, whereas Cdh1 depletion or genetic inactivation accelerates G1 progression (Sigl et al., 2009; Sørensen et al., 2001; Yuan et al., 2014). The role of APC/C in restricting S-phase entry is conserved in budding and fission yeast, flies, worms, chickens, mice, and humans (Fay et al., 2002; García-Higuera et al., 2008; Kitamura et al., 1998; Sigrist and Lehner, 1997; Sudo et al., 2001; Wirth et al., 2004). Consistently, Cdh1 is a haploinsufficient tumor suppressor in mice (García-Higuera et al., 2008).
Several reported mechanisms account for APC/C inactivation during cell cycle progression. APC/C activity is held in check by the spindle assembly checkpoint during mitosis (Musacchio, 2015). APC/C is also kept inactive during S and G2 phases, prior to spindle checkpoint activation. Emi1 (early mitotic inhibitor 1, FBXO5, Rca1 in flies) was discovered as a key inhibitor of APC/C (Reimann et al., 2001). When Emi1 is inactivated in flies and mammals it results in DNA re-replication during late S and G2 phases due to unscheduled APC/C activation (DiFiore and Pines, 2007; Grosskortenhaus and Sprenger, 2002; Machida and Dutta, 2007). Cdh1 itself is controlled by both phosphorylation and ubiquitylation. Cdh1 phosphorylation by Cyclin A/CDK2 occludes binding to the APC/C core complex, preventing ligase activation (Kramer et al., 2000; Lukas et al., 1999). In addition, Cdh1 degradation can occur through the SCFβTRCP E3 ubiquitin ligase, and this degradation is also dependent on Cyclin A/Cdk2 (Fukushima et al., 2013). Furthermore, it has been reported that APC/C can auto-ubiquitylate Cdh1, as well as its own E2 enzymes, Ubch10 and Ube2S (Listovsky et al., 2004; Rape and Kirschner, 2004; Williamson et al., 2009a). However, despite these myriad mechanisms implicated in APC/C inactivation, it remains controversial how APC/CCdh1 is turned off prior to S-phase entry (Pines, 2011).
The Cullin RING Ligases (CRL) represent the largest E3 ligase family in humans and together control the destruction of hundreds of proteins involved in virtually all aspects of cellular physiology (Emanuele et al., 2011; Petroski and Deshaies, 2005). SCF/CRL1 was the first described Cullin Ligase, and remains the best understood. SCF ligases recognize their targets through a family of 69 interchangeable substrate receptors termed F-box proteins. Cyclin F (CCNF, FBXO1) is the founding member of the F-box family and was identified in a yeast gain-of-function screen searching for suppressors of temperature sensitivity in cdc4-1 cells (Bai et al., 1994). Cyclin F expression oscillates during the cell cycle and the Cyclin F amino acid sequence is most similar to Cyclin A. However, Cyclin F is an atypical Cyclin in that it does not bind to CDKs (D'Angiolella et al., 2013). Cyclin F heterozygous mice (Cyclin F+/−) develop normally and homozygous loss of Cyclin F (Cyclin F−/−) is lethal, with null embryos dying around day E10.5 (Tetzlaff et al., 2004).
Cyclin F has few known substrates, and all that have been described thus far are linked to either microtubule organization or DNA metabolism and repair. Among the described Cyclin F substrate are the centrosome protein CP110, and the nucleolar and spindle associated protein Nusap1 (D'Angiolella et al., 2010; Emanuele et al., 2011). The RRM2 subunit of ribonucleotide reductase is an important Cyclin F substrate that regulates genome stability (D'Angiolella et al., 2012). The exonuclease and DNA repair factor Exo1 was reported to be a Cyclin F substrate, as was the replication licensing factor Cdc6 (Elia et al., 2015; Walter et al., 2016). Cyclin F is unique among F-box proteins in that its transcript levels oscillate throughout the cell cycle. The Cyclin F mRNA was identified as cell cycle regulated in five independent mRNA profiling studies that examined cell cycle transcriptional dynamics, making it the only F-box protein common among all studies (Bar-Joseph et al., 2008; Grant et al., 2013; Peña-Diaz et al., 2013; Sadasivam et al., 2012; Whitfield et al., 2002). The importance of Cyclin F in cell cycle progression and animal development, combined with its dynamic expression pattern throughout the cell cycle, prompted us to examine the mechanisms regulating Cyclin F degradation and to identify additional substrates that it controls.
RESULTS
Cyclin F is an APC/C substrate
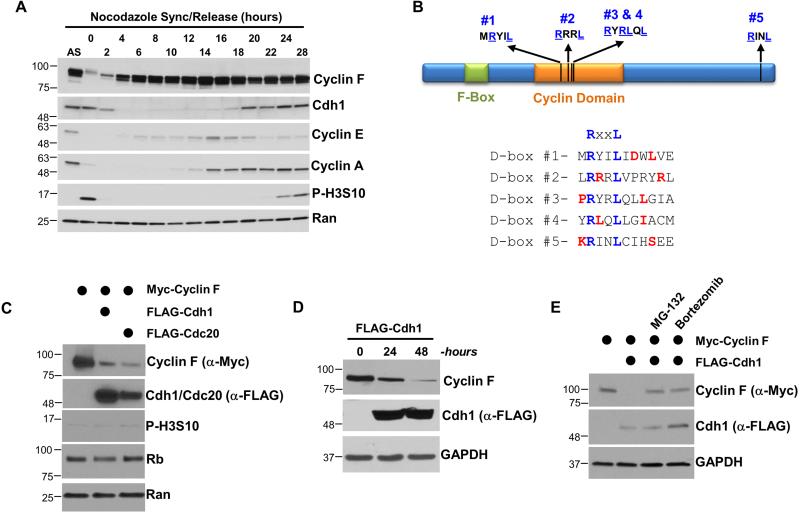
Several lines of evidence suggested that Cyclin F might be an APC/C substrate. The Cyclin F amino acid sequence is most similar to Cyclin A, a bona fide APC/C substrate (den Elzen and Pines, 2001). Cyclin F protein levels peak in G2/M phase and are subsequently diminished in G1, consistent with APC/C mediated degradation (Bai et al., 1994; D'Angiolella et al., 2012). This can be observed in U2OS and HeLa cells synchronized in mitotic prometaphase with nocodazole, isolated by “shake-off”, and followed by immunoblot after release into the cell cycle (Figures 1A and S1A). Cyclin F levels are lowest in early G1 phase and then accumulate in late G1 and throughout the rest of cell cycle. Cyclin A accumulation begins after Cyclin F, consistent with a previous result (Bai et al., 1994). Finally, Cdh1 levels decrease as cells enter S-phase, and then begin to accumulate later in the cell cycle, also consistent with prior reports (Benmaamar and Pagano, 2005; Fukushima et al., 2013; Kramer et al., 2000). We observed similar oscillations in protein abundance in HeLa cells synchronized at G1/S by a double thymidine block and release (Figures S2A, S2B).
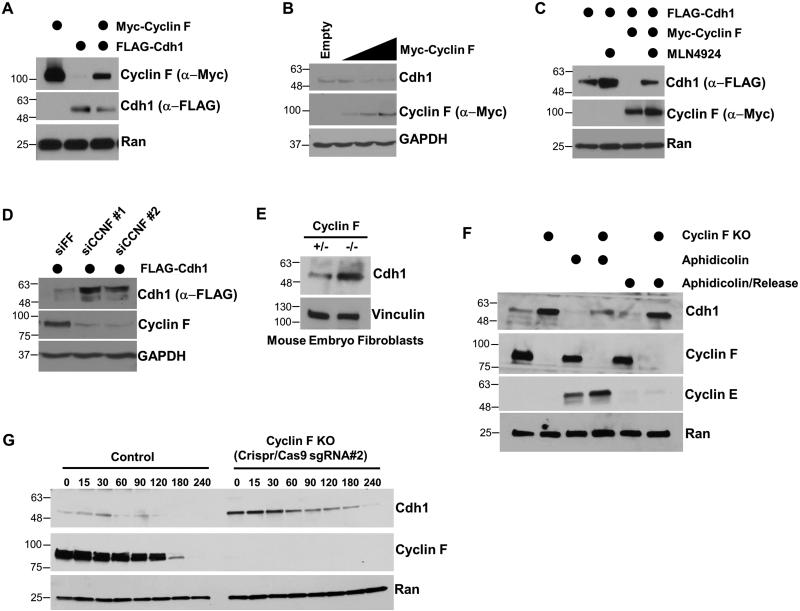
Figure 1. Cdh1 regulates the abundance and stability of Cyclin F.
(A) U2OS cells were synchronized in mitosis with nocodazole, isolated by “shake-off”, and analyzed by immunoblot after release into the cell cycle. (B) Domain structure of human Cyclin F showing the position of its Cyclin homology domain, F-box domain, and five putative APC/C degron motifs. Preferred residues around the putative D-box motifs are shown in red. (C) FLAG-Cdc20 or FLAG-Cdh1 were ectopically expressed in 293T cells in combination with Myc-Cyclin F. Cells were harvested after 24 hours and analyzed by immunoblot. (D) FLAG-Cdh1 was ectopically expressed in 293T cells and reduced the level of endogenous Cyclin F at 24 and 48 hours after transfection. (E) Myc-Cyclin F and FLAG-Cdh1 were ectopically expressed in 293T cells for 24 hours. Cells were treated with the proteasome inhibitors MG-132 (10 μM) or bortezomib (100 nM) four hours prior to harvesting.
Prolonged mitotic arrest in nocodazole led to Cyclin F and Cyclin A degradation, consistent with prior reports (Figure 1A) (D'Angiolella et al., 2012; den Elzen and Pines, 2001; Geley et al., 2001). Consistently, the time at which Cyclin F decreases around mitosis is indistinguishable from that of Cyclin A, an early mitotic APC/C target (Figure S2A). Cyclin F contains five potential APC/C D-box degron motifs, which are conserved evolutionarily (Figures 1B and S1B; amino acid sequence RxxL) (Pfleger and Kirschner, 2000). Preferred residues surrounding the core D-box consensus were identified by the Barford Lab (He et al., 2013) and all five potential D-boxes in Cyclin F are surrounded by preferred residues (Figure 1B).
To determine if Cyclin F is an APC/C substrate we first applied a simple, in vivo assay, taking advantage of the ability to activate APC/C in mitotic cells by pharmacologically inhibiting CDK1. Cyclin F levels decreased significantly following CDK inactivation with roscovitine in nocodazole arrested HeLa cells (Figure S1C). Next, we examined Cyclin F abundance following overexpression of the APC/C substrate receptors Cdc20 and Cdh1. The expression of Cdc20 and Cdh1 significantly reduced exogenous Myc-Cyclin F protein levels in 293T cells analyzed 24 hours after transfection (Figure 1C). Importantly, the Cdc20 and Cdh1 overexpression conditions did not alter the cell cycle, as assayed by flow cytometric analysis of DNA content (Figure S2C). The mRNA levels of Cyclin F, and the well-characterized cell cycle genes Cdc20, Plk1 and FoxM1 were also unchanged (Figures S2D). Transfection of FLAG-Cdh1 also reduced endogenous Cyclin F protein levels (Figure 1D). Moreover, the decline in Cyclin F caused by Cdh1 overexpression was fully reversed by proteasome inhibition using either MG-132 or Bortezimib (Figure 1E), demonstrating that Cdh1 causes the proteasome-mediated degradation of Cyclin F. These data suggest that APC/CCdh1 regulates Cyclin F degradation.
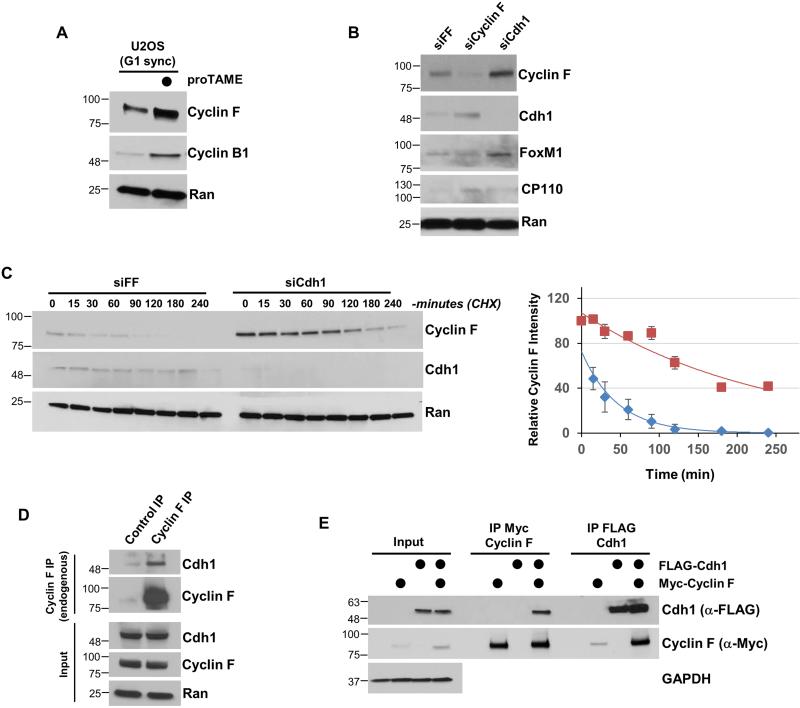
We next examined the impact of pharmacological APC/C inactivation on Cyclin F. We treated cells with the small molecule inhibitor proTAME after synchronization in G1, the primary time during the cell cycle when APC/CCdh1 is active (Cappell et al., 2016; Zeng et al., 2010). U2OS cells were synchronized using a nocodazole block-and-release and then acutely treated with proTAME. APC/C inhibition lead to an increase in the abundance of Cyclin F, and the APC/C substrate Cyclin B (Figure 2A). Cyclin F levels were also increased in asynchronously dividing 293T, HeLa, and U2OS cells treated with proTAME for just 2.5 hours (Figure S1D).
Figure 2. Cyclin F degradation is regulated by the APC/C.
(A) U2OS cells synchronized in G1 phase by mitotic block and release and treated with the APC/C inhibitor proTAME for 90 minutes. (B) Cyclin F or Cdh1 were depleted from T47D cells using siRNA for 48 hours. Negative control siRNA (siFF) targets firefly luciferase. (C) Depletion of Cdh1 using siRNA extended the half-life of Cyclin F. Cells were depleted of Cdh1 by siRNA (same as B), synchronized in G1 by nocodazole block and release, and then treated with cycloheximide (CHX) to analyze Cyclin F stability. On the right is a semi-quantitative analysis of the Cyclin F signal. Negative control (blue diamonds), Cdh1 depletion (maroon squares). Cyclin F signal relative to the Ran loading control is shown on the Y axis (error bars indicate standard deviation of mean). (D) Endogenous Cyclin F was precipitated from 293T whole cell extracts. Cells were treated with MG-132 prior to lysis. (E) Myc-Cyclin F and FLAG-Cdh1 were transfected into 293T cells, and each was separately recovered and analyzed by immunoblot. Cells were treated with MG-132 for four hours prior to lysis.
Cdh1 depletion by siRNA resulted in an increase in Cyclin F abundance compared to cells treated with control siRNAs targeting firefly luciferase (FF; Figure 2B). The APC/CCdh1 substrate FoxM1 is also increased by Cdh1 depletion. Likewise, Cdc20 depletion augments Cyclin F levels to a similar degree as Cyclin A, a known target of APC/CCdc20 (den Elzen and Pines, 2001; Geley et al., 2001). Treatment with proteasome inhibitors blunted the effect of Cdc20 depletion on both proteins, indicating that Cyclin F, like Cyclin A, is targeted to the proteasome by APC/C in mitosis (Figure S3A). We next examined changes in Cyclin F stability because pharmacological APC/C inactivation, and gain- and loss-of-function genetic approaches suggested that APC/C regulates Cyclin F post-transcriptionally. We monitored Cyclin F half-life after inhibiting protein synthesis with cycloheximide (CHX). Cells were depleted for 48 hours with siRNA targeting Cdh1 or FF, synchronized into G1 using nocodazole block and release, and then treated with CHX. Cdh1 depletion significantly elevated Cyclin F levels in G1 cells and extended its half-life to 173.2 minutes from 31.5 minutes (Figure 2C). We conclude that the abundance and stability of Cyclin F are governed by APC/CCdh1 during G1 phase.
The above data strongly suggest that Cyclin F is an APC/C substrate. Consistently, endogenous Cyclin F co-precipitated endogenous Cdh1 from 293T cell extracts (Figure 2D). In addition, exogenously expressed Cyclin F co-precipitated endogenous Cdh1 and Cdc20 (Figure S3B). We ectopically expressed Myc-Cyclin F and FLAG-Cdh1 in 293T cells to establish a system to test the interaction between Cyclin F and Cdh1. Cells were treated with the proteasome inhibitor MG-132 prior to lysis to normalize protein levels across conditions and increase the abundance of low abundance proteins, improving the ability to detect E3-substrate interactions by co-immunoprecipitation (coIP). Under these conditions, we could readily coIP Myc-Cyclin F and FLAG-Cdh1, regardless of which protein was immuno-purified (Figure 2E).
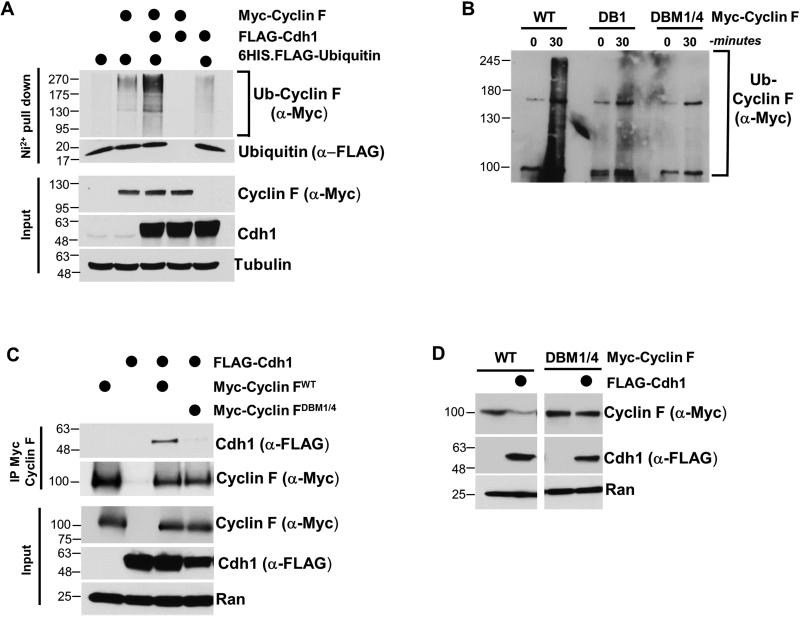
To determine if Cdh1 could enhance the ubiquitylation of Cyclin F in cells, Cyclin F and Cdh1 were co-expressed in the presence of a hexa-histidine tagged (6HIS) ubiquitin. Since ubiquitin causes the degradation of substrates, cells were treated with MG-132 prior to lysis and ubiquitin pulldown. Lysis was performed under strong denaturing conditions and ubiquitin was recovered on nickel agarose. We immunoblotted for Cyclin F to evaluate its relative level of ubiquitylation. Cyclin F ubiquitylation was significantly enhanced by the ectopic expression of Cdh1, suggesting that APC/CCdh1 enhances Cyclin F ubiquitylation in vivo (Figure 3A). Next, we analyzed Cyclin F ubiquitylation by APC/CCdh1 in vitro. APC/C was isolated from G1-synchronized HeLa cell extracts using an anti-Cdc27 antibody. Isolated APC/C was used in an ubiquitylation reaction with added ubiquitin, E1, E2, Cdh1 and ATP. APC/C efficiently ubiquitylated wild-type Cyclin F in vitro, providing strong evidence that Cyclin F is a direct APC/C substrate (Figure 3B).
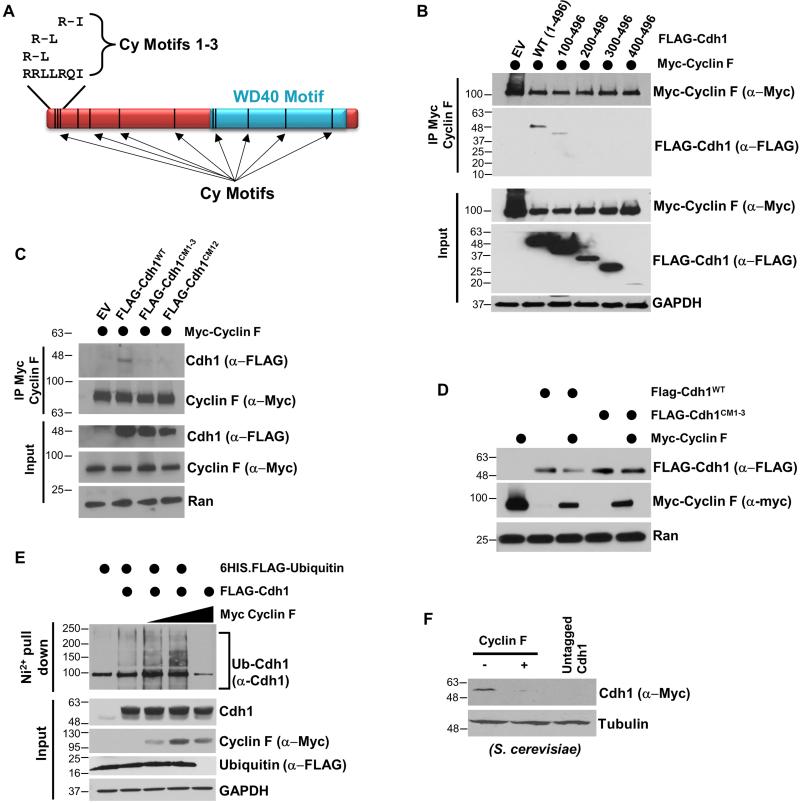
Figure 3. Cdh1 regulates Cyclin F ubiquitylation through canonical D-box degron motifs.
(A) Myc-Cyclin F, FLAG-Cdh1 and 6HIS-ubiquitin were expressed in 293T cells, and ubiquitin conjugates were recovered on nickel agarose under denaturing conditions (6M Guanidine). Ubiquitylation of Cyclin F was analyzed by immunoblot. Cells were treated with MG-132 prior to harvesting and lysis. (B) APC/C ubiquitylates Cyclin F in vitro. APC/C was immuno-purified from G1 HeLa cell extracts and mixed with in vitro translated Cyclin F (WT or mutants), E1, E2, ATP, ubiquitin and Cdh1. (C) Myc-Cyclin FWT or Myc-Cyclin F harboring mutations in D-boxes 1 and 4 (DBM1/4) were co-transfected with FLAG-Cdh1, and analyzed using Myc IP after treatment with MG132 for 4 hours. (D) FLAG-Cdh1 was ectopically expressed in 293T cells in combination with Myc-Cyclin FWT or Myc-Cyclin FDBM1/4. Cdh1 catalyzed the degradation of Myc-Cyclin FWT but not Myc-Cyclin FDBM1/4 .
Cyclin F contains five potential D-box motifs (Figure 1B). We predicted that altering the sequence of one or more of the putative D-boxes in Cyclin F would interfere with Cdh1 binding. We disrupted the D-boxes in Cyclin F by changing the essential arginine and leucine residues to alanine in each motif individually (RxxL to AxxA). We measured binding between Myc-Cyclin F and FLAG-Cdh1 by coIP in 293T cells (treated with MG-132 prior to lysis). Several of the D-boxes appeared to contribute to Cdh1-Cyclin F binding (Figure S4A). Further analysis revealed that mutating D-boxes one and four together (Cyclin FDBM1/4) abrogated the ability of Cyclin F to interact with Cdh1 by coIP (Figure 3C). We therefore tested the ability of this non-interacting Cyclin F mutant to be ubiquitylated in vitro. We did not detect any appreciable ubiquitylation when using Cyclin FDBM1/4 as a substrate in vitro (Figure 3B). The conserved arginine and leucine residues in D-boxes 1 and 4 are predicted to be on the surface of Cyclin F based on homology modeling (Figure S3C-S3E). In addition, mutation of only the first D-box in Cyclin F (Cyclin FDBM1), which partially impaired binding, gave an intermediate defect on ubiquitylation in vitro. These data strongly argue that Cyclin F is targeted for ubiquitylation and degradation by APC/CCdh1. Accordingly, whereas Cdh1 catalyzes the degradation of Cyclin FWT when the two proteins are ectopically expressed together in cells, Cyclin FDBM1/4 is resistant to Cdh1 mediated degradation in this assay (Figure 3D). We conclude that Cyclin F is an APC/C substrate.
Cdh1 is a Cyclin F substrate
Cdh1 levels are decreased in S-phase cells (Figure 1A and S2A) (Benmaamar and Pagano, 2005; Kramer et al., 2000). Pagano and colleagues first noted the dependence of this S-phase degradation on Cul1, although they did not establish the relevant substrate receptor (Benmaamar and Pagano, 2005). Wei and colleagues reported that Cdh1 could be targeted by SCFβTRCP, in a Cyclin A/CDK2 and Plk1 dependent mechanism (Fukushima et al., 2013). However, since both kinases are activated later in the cell cycle it is unclear if they account for Cdh1 destruction in late G1 and early S-phase.
We noted that the reappearance of Cyclin F was coincident with the decrease in Cdh1 (Figure 1A). Additionally, Cdh1 and Cyclin F levels both decreased when the proteins were expressed together, compared to expressing either alone (Figure 4A). Together, these data suggest the possibility that Cdh1 might be also a substrate of SCFCyclin F. Titrated overexpression of Cyclin F reduced endogenous Cdh1 levels (Figure 4B). Cyclin F expression also reduced the abundance of exogenously expressed Cdh1, suggesting that the reduction in Cdh1 is caused by post-translational mechanisms (Figures 4A and 4C). Importantly, the Cyclin F overexpression conditions did not alter the cell cycle or the expression of Cdh1, Cdc20, Plk1, and FoxM1 (Figures S2C and S2D). To confirm that the reduction in Cdh1 following ectopic Cyclin F expression is dependent on the activity of Cullin Ring Ligases, we inhibited their activity using a small-molecule inhibitor of the neddylation cascade, MLN4924 (Soucy et al., 2009). Myc-Cyclin F and FLAG-Cdh1 were co-expressed in 293T cells for 18 hours, and then treated with MLN4924 for 6 hours prior to harvesting for immunoblot. The degradation of Cdh1 caused by Cyclin F expression could be reversed by MLN4924 (Figure 4C). Accordingly, depletion of Cyclin F using siRNA increased endogenous Cdh1 abundance (Figure 2B). Furthermore, siRNA depletion of Cyclin F using either of two independent oligonucleotides increased the abundance of exogenously expressed FLAG-Cdh1 (Figure 4D). To confirm the importance of Cyclin F in regulating Cdh1 we examined Cyclin F null mouse embryo fibroblasts (MEFs) (Tetzlaff et al., 2004). Cyclin F was more abundant in Cdh1 knockout MEFs relative to WT controls (Figure 4E). This was true in both asynchronous MEFs, and in those synchronized in G0/G1 by serum deprivation (Figure S4C). These data demonstrate that Cdh1 abundance is regulated by Cyclin F in mouse and human cells, and suggests that Cdh1 is itself a substrate of SCFCyclin F.
Figure 4. Cdh1 abundance and stability are regulated by Cyclin F.
(A) Ectopic expression of Myc-Cyclin F and FLAG-Cdh1 in 293T cells reduces the level of both proteins. (B) Myc-Cyclin F reduced the abundance of endogenous Cdh1 in a dose dependent manner when expressed in 293T cells. (C) Myc-Cyclin F overexpression reduces the abundance of FLAG-Cdh1 when expressed in 293T cells. The degradation of Cdh1 was rescued by the neddylation inhibitor MLN4924. (D) Depletion of Cyclin F using two independent siRNA reagents increases exogenously expressed FLAG-Cdh1 in 293T cells. (E) Cdh1 levels were analyzed in Cyclin F null (−/−) MEFs and in a corresponding control cell line (+/−). (F) Control and Cyclin F knockout HeLa cells were analyzed either in asynchronous populations (lanes 1 and 2), synchronized in S-phase with aphidicolin (lanes 3 and 4), or 6 hours after aphidicolin release (lanes 5 and 6). During aphidicolin block serum was reduced to mitigate DNA damage. (G) Control and Cyclin F knock-out HeLa cells were treated with cycloheximide and Cdh1 half-life was analyzed.
We generated Cyclin F knockout cells in a HeLa background using Crispr/Cas9 technology to further assess the consequence of Cyclin F loss in human cells. We generated two independent single guide RNAs (sgRNA#1 and sgRNA#2) that target the 5’ end of the Cyclin F locus, and used them to generate two independent Cyclin F knockout cell lines. Cyclin F knockout HeLa cells show an increase in steady-state Cdh1 levels (Figures 4F, 4G, S4D and S4E). Notably, one of the Cyclin F knockout cell lines (generated using guide #2) showed a more significant increase in Cdh1 levels (Figure S4D, explained below). To test the ability of Cyclin F to control Cdh1 in S-phase, when Cdh1 is targeted for degradation, we examined cells synchronized using aphidicolin. In this experiment, we temporarily reduced serum in the media (for all conditions) to mitigate aphidicolin induced DNA damage. Cdh1 abundance was significantly higher in Cyclin F knock-out cells compared to controls (Figure 4F). This was also true in S-phase synchronized cells, suggesting that Cyclin F regulates Cdh1 during S-phase (Figure 4F). Finally, after release from arrest (and following the re-introduction of serum), Cyclin F knockout cells still had significantly increased levels of Cdh1, suggesting that these results were not related to serum withdrawal. Similar results were found in an experiment in which serum was not removed during aphidicolin treatment (data not shown).
We further utilized these HeLa Cyclin F knockout cell lines to examine Cdh1 stability after CHX treatment. We performed these assays in both Cyclin F knockout cell lines (made using sgRNA#1 and sgRNA#2). Cdh1 half-life was significantly longer in Cyclin F knockout cells compared to controls (Figures 4G, S4E and S4F). Interestingly, longer immunoblot exposures revealed very low, but detectable Cyclin F expression in the sgRNA#1 knockout cell line. This fortuitously provided us with, in essence, an allelic series for Cyclin F. Accordingly, Cdh1 showed an intermediate half-life in the sgRNA#1 cell line (t1/2=60.0 min), in which the knockout is incomplete (Figure S4E), exhibiting a stability between that the control (t1/2=43.3 min) and the sgRNA#2 line (t1/2=90.0 min), in which Cyclin F levels are undetectable (Figure S4F). A significantly extended Cdh1 stability was also observed after Cyclin F depletion with siRNA (Figure S4B). We conclude that Cyclin F regulates the stability of Cdh1.
Since Cyclin F is not present in early G1, and Cdh1 is degraded in S-phase, we predicted that SCFCyclin F targets Cdh1 for degradation during S-phase. We synchronized control and Cyclin F knockout cells at various points in the cell cycle after release from a double thymidine block and examined Cdh1 stability by CHX chase. Cdh1 half-life was extended in S and G2 phases by Cyclin F knockout, measured two and eight hours after release, respectively (Figure S5). In addition, Cdh1 stability was indistinguishable between Cyclin F knockout and control cells in G1, measured 13 hours after release. We therefore conclude that Cyclin F regulates Cdh1 stability during S and G2 phases.
Canonical Cyclins that bind CDKs recognize Cy motifs when engaging substrates for phosphorylation (Schulman et al., 1998). Cyclin F also binds to Cy motifs in its substrates to control their ubiquitylation (D'Angiolella et al., 2013). Intriguingly, the Cdh1 amino acid sequence contains twelve potential Cy motifs (Figure 5A). We tested the possibility that Cyclin F directly catalyzes Cdh1 ubiquitylation and degradation by searching for Cy motifs in Cdh1 that regulate binding to and degradation by Cyclin F. We narrowed down the interacting region using truncation mutagenesis. An amino-terminal truncation deleting the first 100 amino acids in Cdh1 impaired its ability to bind to Cyclin F by coIP (Figure 5B). We therefore examined possible Cy motifs in this region. Within the first 13 amino acids of Cdh1 there are three partially overlapping Cy motifs (Figure 5A). One of these motifs has the sequence RRL, which is identical to the Cyclin F interacting Cy motif in the known substrate CP110 (D'Angiolella et al., 2010). We tested if these three Cy motifs are important for Cyclin F binding by substituting the conserved residues for alanine (a.a. 7-14 changed from RRLLRQIV to AAAAAQAV). This Cdh1 Cy motif 1-3 mutant (Cdh1CM1-3) is impaired in its ability to bind to Cyclin F by coIP (Figure 5C, S6). Notably, this mutation retains some binding to Cyclin F, as we still observe residual Cyclin F co-purifying with both Cdh1CM1-3 and the truncated version lacking the first 100 a.a, particularly on longer immunoblot exposures (Figure 5B, 5C, S6). Cdh1CM1-3 still binds to Cyclin A, indicating it is generally folded properly and functional (Figure S6). Interestingly, Cy motif 12 (CM12; amino acids 445-447, sequence RVL) which mediates binding to Cyclin A (Sørensen et al., 2001), also contributes to Cyclin F binding (Figure 5C). We conclude that the overlapping Cy motifs in the amino-terminal 13 residues of Cdh1 specifically control binding to Cyclin F.
Figure 5. Cyclin F regulates Cdh1 degradation through binding to canonical Cy motif sequences.
(A) Schematic of Cdh1 showing the position of the WD40 repeat domain and 12 putative Cy-motifs. (B) Truncated versions of Cdh1 were tested for binding to Cyclin F by coIP in transfected 293T cells treated with MG132 prior to IP. (C) Myc-Cyclin F and FLAG-Cdh1 (WT and mutants) were expressed in 293T cells, and binding was analyzed by IP following treatment with MG-132. (D) Myc-Cyclin F, Flag-Cdh1WT and FLAG-Cdh1CM1-3 were expressed together in 293T cells and Cdh1 degradation was examined. The non-binding Cdh1CM1-3 mutant is resistant to Cyclin F mediated degradation. (E) FLAG-Cdh1, Myc-Cyclin F and 6HIS-ubiquitin were expressed together in 293T cells. 6HIS-ubiquitin was isolated under denaturing conditions and endogenous Cdh1 was analyzed by immunoblot. Cells were treated with MG132 prior to harvesting. (F) Human Cyclin F was expressed in a yeast strain where endogenous yeast Cdh1 was tagged with Myc.
Identifying Cy motifs in Cdh1 that control binding to Cyclin F suggests that SCFCyclin F directly catalyzes Cdh1 ubiquitylation. Consistently, Cyclin F expression catalyzes the degradation of Cdh1WT (Figures 4A, 4B, 4C, and 5D), but not Cdh1CM1-3, when co-expressed in 293T cells (Figure 5D). We next tested the ability of Cyclin F to enhance Cdh1 ubiquitylation in cells using the 6HIS-ubiquitin pulldown assay (treated with MG-132 and pulled-down under denaturing conditions). In this assay Cyclin F enhanced the ubiquitylation of Cdh1 (Figure 5E). Taken together, these data show that Cdh1 is targeted for ubiquitylation and degradation by Cyclin F. Thus, SCFCyclin F and APC/CCdh1 biochemically antagonize one-another, forming a reciprocal feedback circuit.
Cyclin F was identified as a suppressor of temperature sensitive cdc4-1 yeast cells (Bai et al., 1994). Cdc4 is a yeast F-box protein, responsible for targeting the CDKi Sic1 for degradation. Consequently, cdc4-1 cells grown at the restrictive temperature have elevated Sic1 levels and arrest in G1 because they are unable to activate CDK (Bai et al., 1996; Schwob et al., 1994). The lethality of cdc4-1 cells (grown at the restrictive temperature) can be suppressed by expressing Skp1, which activates the impaired SCFCdc4 ligase. Alternatively, cdc4-1 cells can be rescued by expression of the yeast B-type cyclin Clb4, which activates CDKs despite high Sic1 levels (Bai et al., 1996).
Unlike Skp1, expression of Cyclin F does not diminish Sic1 levels, excluding the possibility that Cyclin F restores viability by reactivating SCFCdc4. Moreover, Cyclin F cannot bind CDKs, excluding the possibility that it restores viability in a manner analogous to Clb4 (Bai et al., 1996). We reasoned that overproduction of human Cyclin F might target yeast Cdh1 for degradation. Cdh1 destruction might allow for the accumulation of APC/C substrates like Clb4 and the other B-type cyclins. Therefore, the original plasmid that identified Cyclin F as a suppressor of cdc4-1 lethality (YEP PGAL - Cyclin F) was introduced into S. cerevisiae that have endogenous Cdh1 tagged with 13xMyc. Cdh1-Myc levels were virtually undetectable following Cyclin F overexpression in yeast (Figure 5F). We conclude that Cyclin F can regulate the abundance of Cdh1 in yeast, mouse, and human cells.
Cyclin F and Cdh1 are antagonistic in regulating G1 progression and S-phase entry
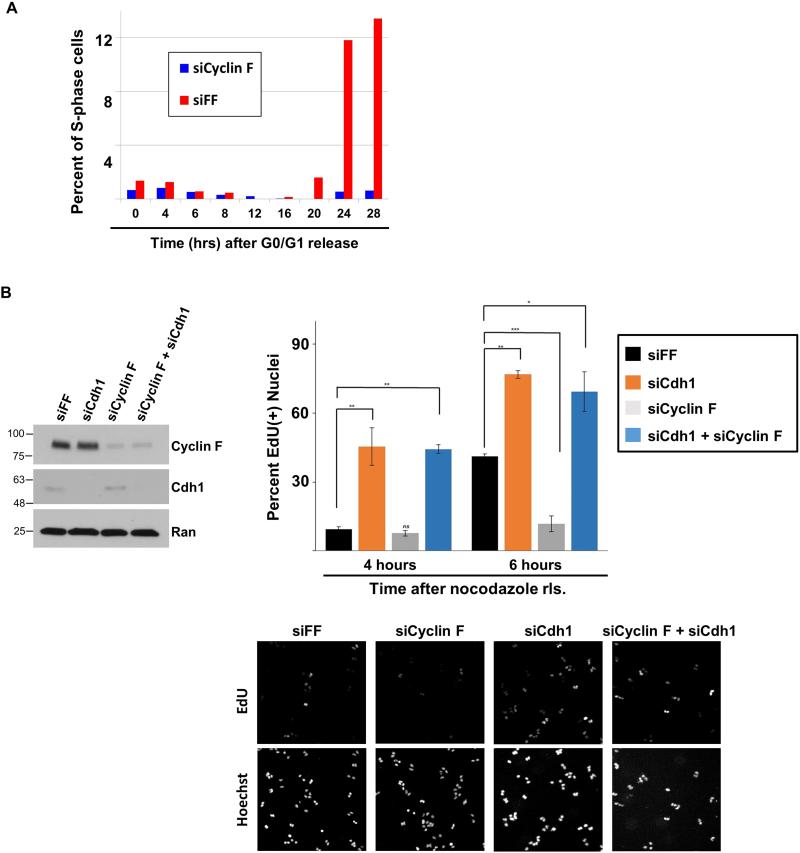
APC/C is inactivated at the G1-S boundary and we found that Cyclin F protein begins to accumulate prior to Cyclin A in late G1 (Figure 1A). Since Cdh1 plays an evolutionarily conserved role in restricting S-phase entry, we reasoned that Cyclin F would be important for the timely initiation of DNA replication. Cyclin F was depleted using siRNA from non-transformed RPE1 cells. Serum was then withdrawn to synchronize the population in G0/G1. We monitored S-phase entry over time using cell cycle flow cytometric analysis of DNA content after re-feeding with media containing serum. Cyclin F depletion substantially blunted S-phase entry kinetics in this assay (Figure 6A). While 13.4% of control depleted cells had entered S-phase by 28 hours after re-feeding, the Cyclin F depleted cell population had no increase in the percentage of S-phase cells by that time (Figure 6A). These data indicate an important role for Cyclin F in mediating cell cycle entry in non-transformed cells.
Figure 6. Cyclin F regulation of G1 progression is dependent on Cdh1.
(A) Non-transformed RPE1 cells were treated with siRNA targeting Cyclin F or FF. Cells were synchronized in G0/G1 by serum withdrawal. After refeeding, entry in S-phase was monitored at the designated time points by flow cytometry. (B) U2OS cells were synchronized in mitosis with nocodazole following depletion with siRNA targeting FF, Cdh1, Cyclin F, or Cyclin F and Cdh1 together. After release from nocodazole, cells were pulsed with EdU for 30 minutes, fixed, and analyzed for EdU incorporation. The percent of nuclei that are EdU positive is shown (performed in triplicate, * p≤0.01; ** p≤0.004; *** p≤ 0.0005 ; p values were calculated using un-paired t-test). Immunoblot on left shows knockdown at zero time point (in mitosis). Representative images of EdU positive cells (top) and DNA content (bottom) are shown below.
The experiment above monitors exit from G0 and transit through G1. To isolate the role of Cyclin F in G1 progression we monitored cells progressing through G1 after synchronization in mitosis. U2OS cells were transfected with siRNA for 24 hours, followed by the addition of nocodazole. Mitotic cells were isolated by “shake-off”, washed extensively and then re-plated in fresh media. The BrdU analog EdU was added 30 minutes prior to fixing cells on plates. This allowed us to identify cells that had progressed into S-phase using cell imaging. In this assay we compared cells depleted of Cyclin F, Cdh1, or both proteins simultaneously. We checked cell lysates at the zero time point to ensure that Cyclin F and Cdh1 had been depleted (Figure 6B-left). Depletion of either protein had no detectable effect on the abundance of the other, suggesting post-translational regulation of their ability to control each other during mitosis. Cdh1 depletion accelerated S-phase entry, in agreement with previous reports, (Figure 6B) (García-Higuera et al., 2008; Sigl et al., 2009; Yuan et al., 2014). Forty-one percent of control cells were incorporating EdU at six hours after nocodazole release, whereas seventy-seven percent of Cdh1 depleted cells had entered S-phase at the same time point. Cyclin F depletion blunted the accumulation of EdU positive cells, consistent with a role for Cyclin F in mediating S-phase entry. Only 12% of Cyclin F-depleted cells had incorporated EdU, compared to 41% of control cells and 71% of Cdh1-depleted cells, 6 hours after release (Figure 6B). Importantly, the decreased rate of S-phase entry in Cyclin F depleted cells was completely reversed by co-depletion of Cdh1 (Figure 6B). Similar results were obtained in a separate experiment when cells were analyzed for EdU incorporation by flow cytometry (Figure S7A). In addition, we observed a similar effect on S-phase entry after Cyclin F and Cdh1 depletion using multiple siRNA oligonucleotides in both HeLa and U2OS cells (Figure S7B and S7C). Together, these results demonstrate an important role for Cyclin F in the timing of S-phase entry and that the S-phase entry delay in Cyclin F depleted cells is dependent on Cdh1. This suggests antagonism between Cdh1 and Cyclin F in regulating the initiation of DNA replication.
DISCUSSION
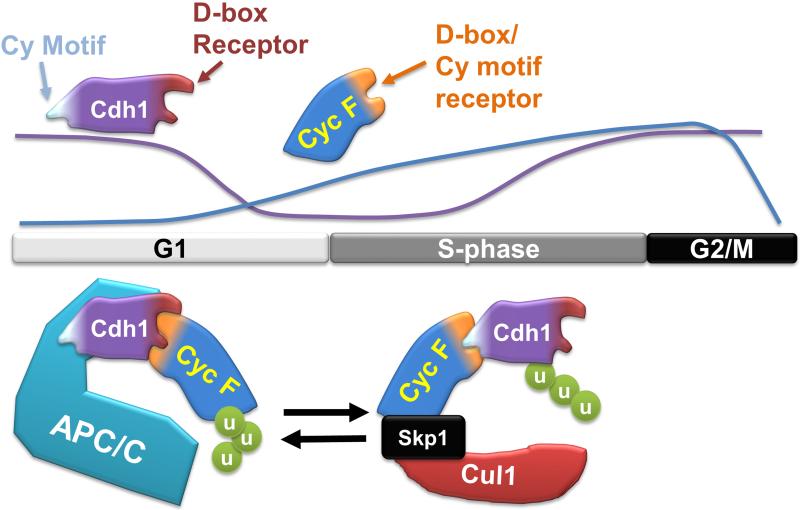
Our data indicate that the coordinated, temporal ordering of Cyclin F and Cdh1 degradation is a key aspect of cell cycle control (summarized in Figure 7). We show that Cyclin F and Cdh1 antagonize each other biochemically, since Cyclin F is a substrate of the APC/C, and Cdh1 is a substrate of SCFCyclin F. Further, Cyclin F depletion slows G1 progression, and this is fully rescued by co-depletion of Cdh1. We therefore conclude that Cdh1 and Cyclin F exist in a mutually antagonistic feedback circuit. The regulation of APC/C and SCFCyclin F by one another represents a unique mechanism for temporally controlling E3 ligases. This direct, ubiquitin dependent, feedback could represent a common feature among other oscillating systems.
Figure 7. A model depicting the interplay between APC/CCdh1 and SCFCyclin F during cell cycle progression.
Cyclin F levels are diminished in mitosis and early G1, and then re-appear late in G1 coincident with the loss of Cdh1. The location of relative motifs in Cdh1 and Cyclin F that mediate their interaction are shown. The proposed configurations of the Cyclin F-Cdh1 interaction when bound to either APC/C in G1 or the SCF in S-phase is depicted graphically.
Recently, Cyclin F was reported to control the stability of the replication licensing protein Cdc6. We recovered Cdc6 in a proteomic screen for Cyclin F interactors and have confirmed their binding by coIP (unpublished data) (Walter et al., 2016). The regulation of Cdc6 by Cyclin F prevents DNA re-replication. We have shown here that depletion of Cyclin F increases the abundance Cdh1, whose overexpression also induces re-replication (Sørensen et al., 2000). Together, these results suggest that Cyclin F might restrain re-replication by repressing Cdc6 and Cdh1 levels. Moreover, since depletion of the APC/C inhibitor Emi1 also causes to re-replication (Machida and Dutta, 2007), we speculate that Cyclin F and Emi1 redundantly regulate APC/C inactivation in S/G2 phase of the cell cycle.
Mechanistic insights into the APC/C – SCFCyclin F circuit
The APC/C is activated in mitosis and remains on throughout G1 phase. During this time Cyclin F stability and abundance are regulated by APC/C. Our data suggest that Cyclin F degradation begins in mitosis and that Cyclin F is destroyed in a manner analogous to its most closely related Cyclin, Cyclin A. This suggests that Cyclin F turnover likely begins with APC/CCdc20, and then it is maintained at low levels throughout G1 phase by APC/CCdh1. In support of this notion, Cyclin F is largely degraded in nocodazole treated cells, and its abundance and stability are maintained at low levels in G1 phase by APC/CCdh1 (D'Angiolella et al., 2012). Cyclin F then accumulates in late G1 and into S-phase, and during this time Cyclin F targets Cdh1 for degradation. Accordingly, the levels of Cdh1 and Cyclin F are anti-correlated at the G1/S boundary following mitotic synchronization, and there is antagonism between Cyclin F and Cdh1 in regulating S-phase entry. Moreover, Cyclin F regulates Cdh1 in S and G2 phases, but not in G1 cells (when Cyclin F levels are lowest).
Based on these observations, we predict that the cell exists in at least two different biochemical states with respect to the Cdh1-Cyclin F circuit. The first state is one that is permissive for APC/C targeting of Cyclin F and occurs in mitosis and early G1 phase. Alternatively, the cellular milieu can be permissive for SCFCyclin F targeting of Cdh1 in S and G2 phases, contributing to APC/C inactivation. Cdh1 and Cyclin F eventually co-accumulate, both reaching their highest levels around G2/M. Thus, there is likely a third state that permits the increasing levels of both proteins and which suppresses their antagonism. The identification of upstream elements and the mechanisms by which they dictate these different states is an important area of future study.
An interesting feature of the Cyclin F-Cdh1 interaction pertains to the sites that mediate binding. A compound Cy motif in amino terminus of Cdh1 is important for its regulation by Cyclin F. The site presumably contacts the hydrophobic patch in the cyclin homology domain of Cyclin F that serves as a Cy motif receptor. We predict that when this amino-terminal Cy motif in Cdh1 binds to the cyclin homology domain in Cyclin F, their interaction is permissive for Cdh1 ubiquitylation (Figure 7). Notably, the hydrophobic patch on Cyclin F is also the region of the protein that contains the D-boxes responsible for APC/C targeting. In fact, the two critical D-boxes that we mapped form part of the core of the hydrophobic patch. One of these D-boxes is the well-established MRYIL motif, analogous to the MRAIL motif in Cyclins A and B (D'Angiolella et al., 2010; Schulman et al., 1998). As depicted in Figure 7, we predict that these D-box motifs tether Cyclin F to the APC/C, enabling Cyclin F ubiquitylation in mitosis and early G1. A version of Cyclin F harboring mutations in these D-box consensus motifs is resistant to Cdh1 degradation in vivo and cannot be ubiquitylated by APC/C in vitro.
Thus, we hypothesize that Cyclin F and Cdh1 engage each other in at least two possible conformational states, depending on which protein is being targeted for degradation. One occurs in early G1, in which the D-box receptor on Cdh1 is bound to the hydrophobic patch/D-box in Cyclin F. The second exists in S-phase when the Cy motif in Cdh1 is bound to the hydrophobic patch/D-box in Cyclin F. Consistently, changing amino acids in the D-box/Cy receptor on Cyclin F completely blocked binding to Cdh1 by coIP, since these changes abolish the formation of both binding states. Furthermore, we observed some residual binding when the Cy motif in Cdh1 was altered, since the proteins could still bind in the alterative conformation. We speculate that post-translational modifications surrounding these sites may mediate the ability of Cyclin F to target Cdh1 for ubiquitylation and vice versa.
APC/C regulation throughout the cell cycle
The APC/C is the most complex E3 ligase that has been described with a mass of >1.2 mDa and 14 individual protein components, some present in duplicate copies (Sivakumar and Gorbsky, 2015). The role of APC/CCdh1 in restraining cell cycle entry is conserved from yeast to humans. Its importance in controlling cell cycle events and genome stability is highlighted by the multiple mechanisms that have evolved to control its activity at different points in the cell cycle. APC/C is kept inactive by the spindle checkpoint in early mitosis (Musacchio, 2015). In addition, Emi1 potently inhibits APC/C activity during interphase. In both humans and flies, Emi1 depletion results in DNA re-replication due to the unscheduled re-activation of APC/C in late S and G2 phases (DiFiore and Pines, 2007; Machida and Dutta, 2007). These data are most consistent with a primary role for Emi1 in controlling APC/C later in the cell cycle. Consistently, reports in flies and animal cells have shown that Emi1 is dispensable for G1-S progression (DiFiore and Pines, 2007; Grosskortenhaus and Sprenger, 2002). Recent results have also shown an alternative role for Emi1 in the irreversibility of APC/C inactivation in S-phase (Cappell et al., 2016).
APC/CCdh1 is also regulated by phosphorylation and ubiquitylation of Cdh1. Cyclin A-Cdk2 sits at the nexus of both of these pathways. Cyclin A/CDK2 phosphorylation of Cdh1 impairs its binding to the APC/C core complex (Kramer et al., 2000; Lukas et al., 1999). In addition, sequential phosphorylation of Cdh1 first by Cyclin A/CDK2, and then by Plk1, triggers Cdh1 degradation through the SCFβTRCP ubiquitin ligase, via recognition of a non-canonical phospho-degron motif (Fukushima et al., 2013). However, careful kinetic analysis by Zetterberg and colleagues demonstrated that the accumulation of Cyclin A begins after the start of DNA replication, making it unlikely that Cyclin A/CDK2 inhibits APC/CCdh1 prior to the start of S-phase (Erlandsson et al., 2000).
Other researches have suggested that APC/C-dependent degradation of its own E2s contributes to its inactivation, arguing that the APC/C is an autonomous oscillator that self-inactivates at the end of G1 (Rape and Kirschner, 2004; Williamson et al., 2009a). However, the notion that the APC/C is inactivated through E2 auto-ubiquitylation has not been universally accepted (Pines, 2011). Others have found that the levels of the APC/C E2s are insufficiently reduced to impair APC/C activity, and that UbcH10 accumulates as cells approach S-phase (Walker et al., 2008).
Finally, APC/CCdh1 was reported to auto-ubiquitylate Cdh1 (Listovsky et al., 2004). This interpretation was based on the identification of two D-box consensus motifs in Cdh1 itself. These potential D-boxes were required for Cdh1 degradation in S-phase (Listovsky et al., 2004). Interestingly, both of these potential D-boxes are also Cy-motifs (amino acids 7-10:RRLL and amino acids 28-31:RRTL). Notably, the first of these D-box/Cy motif hybrids (RRLL at amino acids 7-10) is part of the compound, N-terminal Cy motif that mediates Cyclin F binding and which is required for Cyclin F mediated Cdh1 degradation. Thus, we propose that SCFCyclin F dependent degradation explains the requirement for D-box/Cy motif hybrid sequences in Cdh1 that control its S-phase destabilization.
We propose that Cyclin F is a key mediator of Cdh1 degradation and APC/C inactivation at G1/S. Importantly, Cyclin F has the ability to act prior to Cyclin A, since it accumulates earlier during the cell cycle (Figure 1A, S2; Bai et al., 1994). In addition, since Cyclin F catalyzes Cdh1 ubiquitylation, it provides a robust and irreversible mechanism of APC/C inactivation. This is not unlike the inactivation of CDK1 in mitosis, triggered by the APC/C dependent degradation of Cyclin B. Since Cyclin F and Cdh1 are tightly intertwined biochemically, we predict that they possess the necessary affinities for one another to mediate their rapid ubiquitylation and degradation. These results raise the possibility that the sole reason APC/C catalyzes Cyclin F degradation is to prevent its own inactivation in early G1-phase, thereby preventing premature S-phase entry. However, since Cyclin F is non-essential in HeLa and MEF cells, Cdh1 inactivation can be compensated through other mechanisms. Functionally shifting the responsibility of APC/C inactivation might be analogous to the ability of cells to survive without canonical Cyclins and CDKs (Sherr and Roberts, 2004). Delineating the contributions of these various components to APC/C inactivation and S-phase entry in a single experimental system that directly measures the start of DNA replication is an important area of future study.
EXPERIMENTAL PROCEDURES
Mammalian cell culture and immunoblotting
HEK 293T and HeLa cells were obtained from ATCC, and grown in DMEM complete medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals). T47D cells were also obtained from ATCC and grown in RMPI-1640 medium with 10% FBS. RPE1-hTRET (FRT) cells, a kind gift from Peter Jackson, were grown in DMEM complete medium (Gibco) supplemented with 10% FBS. The APC/C inhibitor proTame was purchased from Boston Biochem.
All transfection experiments were performed in HEK 293T. Transfections were performed using Lipofectamine 2000 (Life Technologies). Cells that had been transfected with protein expression vectors were cultured for 24-48h prior to analysis. Samples for protein analysis by immunoblot were lysed in NETN buffer [20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 0.5 mM EDTA and 0.5 % (v/v) Nonidet P-40 (NP-40)] supplemented with 2µg/ml pepstatin, 2µg/ml apoprotinin, 10µg/ml leupeptin, 1mM AEBSF [4-(2 Aminoethyl) benzenesulfonyl fluoride], 1mM Na3VO4, and 1mM NaF. Lysis was performed on ice for ~20 minutes. Protein concentration was determined using Bradford (Bio-Rad). Standard immunoblotting procedures were followed. Membranes were blocked in 5% nonfat dried milk [diluted in Phosphate buffered saline 0.05% tween-20 (PBST)].
RNA interference and Crispr/Cas9 knockout reagents, including oligonucleotide sequences, are described in the supplemental materials and methods.
Immuno-precipitation (IP)
FLAG-tagged Cdh1 and Myc-tagged Cyclin F (or mutants) were expressed in HEK 293T cells for 24 h. All cells were treated with MG-132 (10μM for 4 hours) prior to lysis, dislodged by trypsinization, washed with PBS, lysed in NETN, and clarified by centrifugation at 15,000 rpm for 15 min. Anti-FLAG M2, or anti-Myc beads (20 μl per IP, Sigma, Cat No F2426, E6779 and E6654 respectively) were used to precipitate specific proteins (4h to overnight at 4°C). The beads were washed with NETN three times and eluted with Laemmli sample buffer at 70°C for 10 minutes. For endogenous Cyclin F IP, 293T cells were treated with 10μM MG-132 for 6h. Lysates were precleared with Protein A/G beads and mixed with Cyclin F polyclonal Rabbit antibody (sc-952) or normal Rabbit IgG (sc-2027) and 75μl of equilibrated protein A/G beads.
In vitro and in vivo ubiquitination assay
Mutant or WT Cyclin F proteins were translated in vitro in rabbit reticulocyte lysate (Promega). G1 HeLa extract was prepared as described (Williamson et al., 2009b). APC/C was captured on Protein A/G beads with an anti-Cdc27 antibody (Santa Cruz Biotech), by rotating the bead slurry with G1 extract on a rotary mixer for ~4h. In vitro ubiquitin assays were performed by incubating 2.5 μl of in vitro translated substrate in 20-25μl reactions containing UBAB buffer (2.5mM Tris-HCL pH 7.5, 5mM NaCl and 1mM MgCl2), energy mix (15mM creatine phosphate, 2mM ATP and 2mM MgCl2 pH 8.0), 4mM DTT, 1.5 μg purified ubiquitin, 100nM E1 (UBE1), 100nM of E2 (UbcH5c or UBE2D3) and 5μl of the APC/C-bead slurry. Ubiquitin, E1 and E2 enzymes were purchased from Life Sensors. MG-132 was added to prevent deubiquitylation. The reactions were terminated after 60 minutes by boiling in sample buffer.
A FLAG-HIS ubiquitin construct (6-His-Flag-Ub) was a gift from Dr. Philippe Soubeyran. For in vivo ubiquitylation assays, 80% of the cell suspension was lysed in buffer 1 [6 M Guanidine-HCl, 0.1 M Na2HPO4/ NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 15 mM imidazole and 10 mM β-mercaptoethanol (βME)] and used for pulldown of HIS-ubiquitin conjugated proteins. The remaining 20% was used to analyze inputs by immunoblot. The lysates used for pulldown were sonicated to reduce viscosity, and loaded onto 50 μl of Ni2+-NTA resin pre-washed with buffer 1. Binding was performed at room temperature for 4 h. The beads were successively washed with 750 μl of each of the following buffers: buffer 1; buffer 2 (8 M Urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 10 mM βME); buffer 3 (8 M Urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 6.3, 10mM βME) plus 0.2% Triton X-100; buffer 3 and then buffer 3 plus 0.1% Triton X-100. After the last wash, beads were eluted by incubating in 50 μl of buffer 4 (200 mM imidazole, 0.15 M Tris/HCl pH 6.7, 30% glycerol, 0.72 M βME, 5% SDS) for 20 min. Eluates were analyzed by immunoblot.
Cell cycle analysis
Hela S3 cells were synchronized in S phase with double treatments with 2.5 mM thymidine. For flow cytometry analysis on RPE1, cells were treated with siRNA targeting FF (control) of Cyclin F for 24 hours. Serum was withdrawn for 24 hours, and after refeeding, entry in S-phase was monitored by propidium iodide staining using standard protocols. U2OS cells were synchronized in mitosis with nocodazole after siRNA. After release from nocodazole block, cells were pulsed for 30 minutes with 10 μM EdU (Sigma # T511285), and then trypsinized into a single cell suspension, fixed in 3.7% formaldehyde and washed in PBS prior to EdU labeling by azide-alkyne cycloaddition click chemistry. Alternatively, cells were fixed on plates in 3.7% formaldehyde. Click reactions are a total of 500 μl in PBS, and include: Alexa Fluor 488 Azide (final concentration of 1 μM), CuSO4 (final concentration of 1mM), ascorbic acid (final concentration of 100mM, made fresh). The labeled cells were counter stained for DNA with 7-AAD or Hoechst and either analyzed directly by flow cytometry or imaging.
Statistical analysis
Statistical significance was tested by t-test or paired t-test using the Prism statistical analysis software (Graph Pad).
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks to Jean Cook for critical reading of the manuscript. We thank the following individuals for providing reagents, with specific details in the methods section: Stephen J. Elledge (Harvard Medical School, HHMI), Christopher M. Yellman (University of Texas at Austin), Peter Jackson (Stanford University), and Philippe Soubeyran (Institut Paoli-Calmettes, Aix-Marseille Université). We thank the UNC Flow Cytometry Core Facility (supported in part by P30 CA016086 Cancer Center Core Support Grant to the Lineberger Cancer Center). This work was supported by start-up funds from the University Cancer Research Fund, and grants from the Susan G. Komen Foundation (CCR14298820), Jimmy-V Foundation, and US Public Health Service (R01GM120309) (MJE, RC, TB, AA, JLK, and CAM); North Carolina State University (DL and DJB); grants from US Public Health Service (R01CA63113, R01CA173023; TBB, JAD) and an F31 training grant (CA189328; TBB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
MJE, RC, DJB, and JAD conceptualized the project and designed experiments. RC carried out the majority of experiments. MJE and RC assembled the figures and wrote the paper. DJB and JAD edited the paper. TB, AA, DL, CAM, JLK, and TBB also carried out experiments.
REFERENCES
- Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barford D. Structural insights into anaphase-promoting complex function and mechanism. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2011;366:3605–3624. doi: 10.1098/rstb.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmaamar R, Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T. Irreversible APCCdh1 Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell. 2016;166:167–180. doi: 10.1016/j.cell.2016.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF Cyclin F controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Forrester FM, Jeong Y, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-Mediated Degradation of Ribonucleotide Reductase M2 Controls Genome Integrity and DNA Repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Esencay M, Pagano M. A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis. Trends Cell Biol. 2013;23:135–140. doi: 10.1016/j.tcb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AEH, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell. 2015:1–15. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AEH, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen Y-N, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson F, Linnman C, Ekholm S, Bengtsson E, Zetterberg a. A detailed analysis of cyclin A accumulation at the G(1)/S border in normal and transformed cells. Exp. Cell Res. 2000;259:86–95. doi: 10.1006/excr.2000.4889. [DOI] [PubMed] [Google Scholar]
- Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–517. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Ogura K, Wan L, Lu Y, Li V, Gao D, Liu P, Lau AW, Wu T, Kirschner MW, et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell Rep. 2013;4:803–816. doi: 10.1016/j.celrep.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Higuera I, Manchado E, Dubus P, Cañamero M, Méndez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Brooks L, Zhang X, Mahoney JM, Martyanov V, Wood T. a, Sherlock G, Cheng C, Whitfield ML. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell. 2013;24:3634–3650. doi: 10.1091/mbc.E13-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Sprenger F. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell. 2002;2:29–40. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- He J, Chao WCH, Zhang Z, Yang J, Cronin N, Barford D. Insights into Degron Recognition by APC/C Coactivators from the Structure of an Acm1-Cdh1 Complex. Mol. Cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Maekawa H, Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol. Biol. Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov a V, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, Brandeis M. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Sørensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Peña-Diaz J, Hegre S. a, Anderssen E, Aas P. a, Mjelle R, Gilfillan GD, Lyle R, Drabløs F, Krokan HE, Sætrom P. Transcription profiling during the cell cycle shows that a subset of Polycomb-targeted genes is upregulated during DNA replication. Nucleic Acids Res. 2013;41:2846–2856. doi: 10.1093/nar/gks1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Reimann JDR, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio J. a. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman B. a, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Sigl R, Wandke C, Rauch V, Kirk J, Hunt T, Geley S. Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J. Cell Sci. 2009;122:4208–4217. doi: 10.1242/jcs.054197. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CS, Lukas C, Kramer ER, Peters J, Bartek J, Lukas J. Nonperiodic Activity of the Human Ubiquitin Ligase Results in Continuous DNA Synthesis Uncoupled from Mitosis Nonperiodic Activity of the Human Anaphase-Promoting Complex –Cdh1 Ubiquitin Ligase Results in Continuous DNA Synthesis Uncoupled from Mitosis. Mol. Cell. Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CS, Lukas C, Kramer ER, Peters J, Bartek J, Lukas J. A Conserved Cyclin-Binding Domain Determines Functional Interplay between Anaphase-Promoting Complex – Cdh1 and Cyclin A-Cdk2 during Cell Cycle Progression. Mol. Cell. Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy T. a, Smith PG, Milhollen M. a, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Sudo T, Ota Y, Kotani S, Nakao M, Takami Y, Takeda S, Saya H. Activation of Cdh1-dependent APC is required for G 1 cell cycle arrest and DNA damage-induced G 2 checkpoint in vertebrate cells. EMBO J. 2001;20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Bai C, Finegold M, Harper JW, Mahon KA, Stephen J, Wilson J, Elledge SJ. Cyclin F Disruption Compromises Placental Development and Affects Normal Cell Cycle Execution. Mol. Cell. Biol. 2004;24:2487–2498. doi: 10.1128/MCB.24.6.2487-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A, Acquaviva C, Matsusaka T, Koop L, Pines J. UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. J. Cell Sci. 2008;121:2319–2326. doi: 10.1242/jcs.031591. [DOI] [PubMed] [Google Scholar]
- Walter D, Hoffmann S, Komseli E-S, Rappsilber J, Gorgoulis V, Sørensen CS. SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication. Nat. Commun. 2016;7:10530. doi: 10.1038/ncomms10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of Genes Periodically Expressed in the Human Cell Cycle and Their Expression in Tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U. S. A. 2009a;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Jin L, Rape M. Preparation of Synchrnoized Human Cell Extracts to Study Ubiquitination and Degradation. Methods Mol. Biol. 2009b;545:301–312. doi: 10.1007/978-1-60327-993-2_19. [DOI] [PubMed] [Google Scholar]
- Wirth KG, Ricci R, Giménez-Abián JF, Taghybeeglu S, Kudo NR, Jochum W, Vasseur-Cognet M, Nasmyth K. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 2004;18:88–98. doi: 10.1101/gad.285404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Srividhya J, De Luca T, Lee J-HE, Pomerening JR. Uncovering the role of APC-Cdh1 in generating the dynamics of S-phase onset. Mol. Biol. Cell. 2014;25:441–456. doi: 10.1091/mbc.E13-08-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh D-C, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wan L, Dai X, Sun Y, Wei W. Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. Biochim. Biophys. Acta. 2014;1845:277–293. doi: 10.1016/j.bbcan.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.