Abstract
Spinal Muscular Atrophy is due to the loss of SMN1 gene function. The duplicate gene SMN2 produces some, but not enough, SMN protein because most transcripts lack exon 7. Thus, promoting the inclusion of this exon is a therapeutic option. We show that a somatic gene therapy using the gene for a modified U7 RNA which stimulates this splicing has a profound and persistent therapeutic effect on the phenotype of a severe Spinal Muscular Atrophy mouse model. To this end, the U7 gene and vector and the production of pure, highly concentrated self-complementary (sc) adenovirus-associated virus 9 vector particles were optimized. Introduction of the functional vector into motoneurons of newborn Spinal Muscular Atrophy mice by intracerebroventricular injection led to a highly significant, dose-dependent increase in life span and improvement of muscle functions. Besides the central nervous system, the therapeutic U7 RNA was expressed in the heart and liver which may additionally have contributed to the observed therapeutic efficacy. This approach provides an additional therapeutic option for Spinal Muscular Atrophy and could also be adapted to treat other diseases of the central nervous system with regulatory small RNA genes.
Introduction
The severe, hereditary, neuromuscular disorder Spinal Muscular Atrophy (SMA)1 is due to deletions or mutations of both alleles of the essential survival motor neuron 1 (SMN1) gene.2 A second gene copy located in the same 5q13 region of the human genome (SMN2)3 allows patients to survive embryonic and fetal development, but only partly compensates for the loss of SMN1 in postnatal life. The reason for this reduced functionality is that, in contrast to SMN1, the exon 7 of SMN2 is preferentially skipped, leading to the synthesis of a shortened and unstable protein.4 This increased skipping is due to a C to T transition at position 6 of exon 7 which transforms an exonic splicing enhancer interacting with the splicing factor SRSF1 into an hnRNP A1/A2B1-dependent exonic splicing silencer.5,6,7,8
Because all SMA patients contain at least one SMN2 copy, the splicing of this gene is an attractive therapeutic target. Previous work from our laboratory provided the first proof of principle that a correction of SMN2 splicing can improve the condition of a mouse SMA model. We had developed a modified U7 snRNA gene carrying an antisense sequence complementary to SMN exon 7 and an additional sequence able to bind the positive splicing factor SRSF1 (U7-ESE-B) and had shown that this construct stimulates exon 7 inclusion and SMN protein synthesis in cell culture models, including patient cells.9 We then generated transgenic mice containing the U7-ESE-B transgene and crossed it into the background of severe SMA mice (Smn1−/−, SMN2+/+) that normally survive only for 5–7 days.10 With the additional U7-ESE-B transgene, the mice survived for a median of 150 days, and both the survival and the extent of phenotypic rescue were dependent on the copy number of the transgene.11
Since then, similar results have been obtained with antisense oligonucleotides that bind to various parts of the SMN2 pre-mRNA.12,13 In particular, a phosphorodiamidate morpholino oligonucleotide targeting a splicing silencer in intron 7 (ISS-N1) showed a high efficiency in vitro and significantly prolonged the survival of SMN▵7 mice (Smn1−/−, SMN2+/+, SMN▵7 +/+; normally surviving for ~12 days14).15,16 Additionally, several groups have shown that SMN1 cDNA expression cassettes in self-complementary (sc) adenovirus-associated virus (AAV) vectors can increase survival and lead to a virtually complete rescue in the SMN▵7 mouse SMA model17,18,19,20,21 and in an experimental pig model for SMA.22
Despite these very promising advances, some of which are already being tested in clinical trials,23 the development of other therapeutic approaches for SMA, including our U7 snRNA strategy, should continue. It is still important to explore additional options for treating SMA patients, as it is not yet clear how efficient and safe the currently available options will be in a clinical setting.
Concerning the U7-mediated correction of SMN2 splicing, the major open question has been whether it can be effective in a somatic gene therapy context, as opposed to the previous transgenic approach where SMN2 splicing is corrected from the moment of conception. Here, we demonstrate that this is indeed the case. We have constructed scAAV vectors which contain variable numbers of U7-ESE-B expression cassettes. When scAAV9 vector particles containing four tandem copies of the U7-ESE-B gene are injected into the cerebral ventricle of newborn SMN▵7 mice, this results in a dose-dependent increase in life span and improvement of muscle functions. We also show that the therapeutic U7 RNA is expressed in the heart and liver which may contribute to its therapeutic effect.
Results
Improved efficiency of scAAV compared with ssAAV vectors
The U7 snRNA derivative U7-ESE-B used to correct SMN2 exon 7 splicing contains, from 5′ to 3′ (Figure 1): a repeat of three target sequences for the splicing factor SRSF1, an antisense sequence complementary to nucleotides 31–48 of SMN2 exon 7, an optimized Sm binding site that interacts with the seven Sm proteins and ensures nuclear localization, and a 3′ hairpin that stabilizes the RNA.9,11 To accommodate multiple copies of U7-ESE-B into AAV vectors, we shortened the original gene while retaining all necessary promoter and 3′ elements, i.e., the distal sequence element or octamer sequence, the proximal sequence element, and the 3′ box.24
Figure 1.
Schematic representation of U7-ESE-B snRNA used to improve SMN2 exon 7 splicing. (a) The U7-ESE-B splicing correction cassette9,11 contains an antisense sequence directed to the 3′ part of exon 7 of the human SMN2 gene (orange) and an additional splicing enhancer sequence consisting of repeated binding motifs for splicing factor SRSF1 (yellow). The modified U7 moiety is represented by its 3′-terminal hairpin and its Sm protein core (pictured as a blue ring). Within exon 7, the C–U transition that distinguishes SMN1 and SMN2, the central ESE (SE2) and the UAA stop codon are indicated with pink letters. (b) Structure of the shortened U7-ESE-B cassette (sU7). The distal sequence element or octamer motif (DSE) and the proximal sequence element (PSE) of the U7 promoter as well as the 3′ box determining proper 3′ end formation are indicated by orange boxes. The splicing enhancer sequence (ESE), the antisense sequence and the modified Sm binding site recognizing the seven canonical Sm proteins present in spliceosomal snRNPs (Sm OPT) are shown in similar colors as in a.
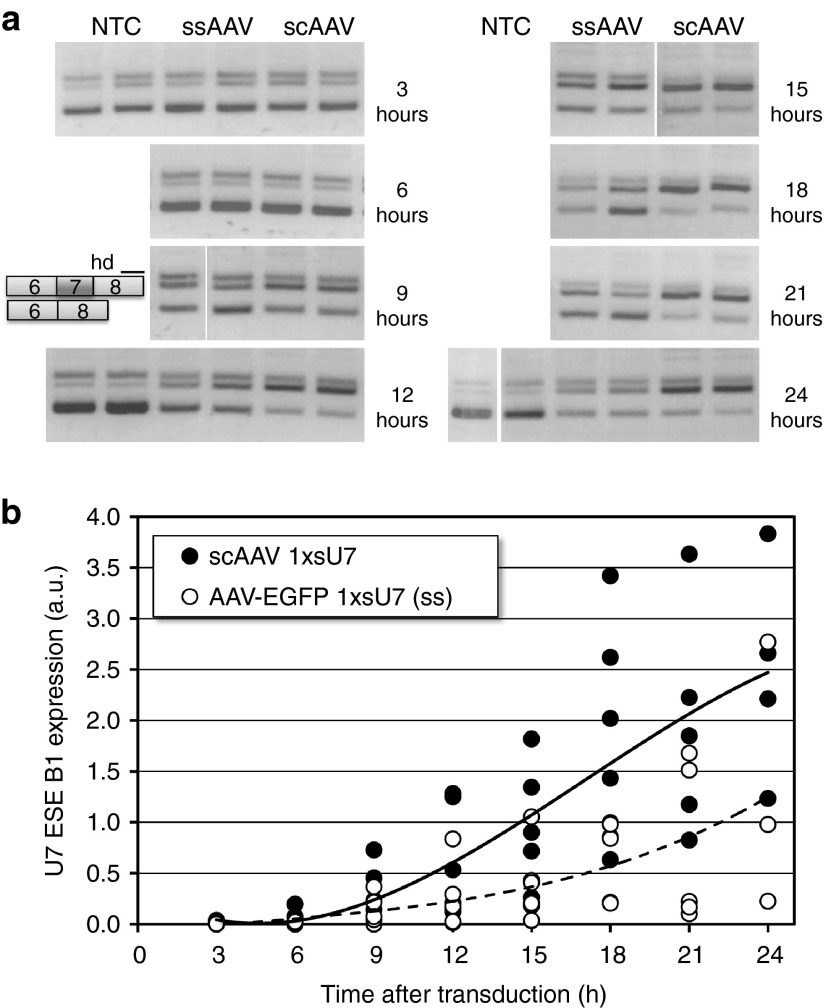
We initially cloned a single copy of this shortened U7-ESE-B gene (1xsU7) into separate vectors allowing the production of either single-stranded (ss) or scAAV particles (see Supplementary Figure S1). Because of an additional enhanced green fluorescence protein (EGFP) cassette present in the ssAAV, the two vectors are of similar size with regard to both the plasmid and the DNA packed into virions. HeLa S2 cells containing a SMN2 minigene reporter9,25 were then transduced with AAV serotype 6 particles. With both ss and scAAV particles, an increase in SMN2 exon 7 inclusion was detectable after 9 hours post-transduction (Figure 2a). However, in the scAAV 1xsU7-transduced cells, the exon 7-containing transcripts prevailed between 12–24 hours post-transduction whereas the cells transduced with AAV-EGFP 1xsU7 (ss) kept considerable levels of RNA lacking exon 7 throughout the experiment. These differences between the two vectors correlated with the measurements of U7-ESE-B RNA by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) (Figure 2b). The ratio of splicing products returned to the uncorrected level after 48 hours, presumably because the AAV genomes were diluted out in these dividing cells (data not shown).
Figure 2.
Comparison of single-stranded (ss) and self-complementary (sc) adenovirus-associated virus (AAV) vectors in cell culture. HeLa S2 cells were transduced with AAV serotype 6 particles of the vectors AAV-EGFP 1xsU7 ss and scAAV 1xsU7 at a concentration of 500 vg/cell and analyzed for the splicing of the SMN2 minigene present in these cells by reverse transcriptase-polymerase chain reaction (RT-PCR) and agarose gel electrophoresis (a) and for the concentration of U7-ESE-B RNA by RT-quantitative PCR (RT-qPCR) (b). The RT-PCR for SMN2 splicing produces amplicons containing and lacking exon 7 as well as a more slowly migrating band corresponding to heterodimers (hd). All samples shown are from a single experiment with duplicate assays for each variable. The samples were processed together but analyzed on four different agarose gels. The RT-qPCR data have been compiled from three biological replicates analyzed with duplicate measurements. The curves were fitted with a third order polynomial function contained in the Excel program. The levels of 7SK snRNA were used for normalization.
Multiple copies of the U7-ESE-B gene result in more efficient splicing correction
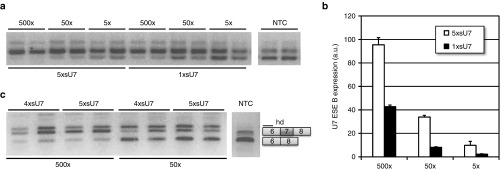
We then tried to improve our scAAV vector by inserting four and five copies of the sU7 cassette (see Supplementary Figure S1). In direct comparison, ~10 times more scAAV 1xsU7 than scAAV 5xsU7 were required to obtain a comparable level of splicing correction in HeLa S2 cells (Figure 3a). Accordingly, scAAV 1xsU7 produced only 20–45% of the U7-ESE-B RNA amount generated by scAAV 5xsU7 of identical titer (Figure 3b). In contrast, scAAV 5xsU7 and scAAV 4xsU7 showed very similar efficiencies of exon 7 splicing correction (Figure 3c). A likely reason for this is that the size of scAAV 5xsU7 exceeds the known packaging capacity of AAV virions.26 Thus, scAAV 5xsU7 genomes may get packaged less efficiently or partly truncated during packaging. This interpretation is supported by the fact that scAAV 4xsU7 generally yielded more viral particles than scAAV 5xsU7 (not shown).
Figure 3.
Comparison of splicing correction by self-complementary adenovirus-associated virus (scAAV) vectors with different numbers of U7-ESE-B cassettes. HeLa S2 cells were transduced with AAV serotype 6 particles of scAAV 5xsU7 and scAAV 1xsU7 (a,b) or scAAV 5xsU7 and scAAV 4xsU7 (c). SMN2 minigene splicing assays a,c and measurements of U7-ESE-B RNA b were performed as in Figure 2. All samples were collected 24 hours post-transduction. The vector concentrations are indicated as vector genome excess over cells. For example, 5× indicates a concentration of 5 vg/cell. Error bars indicate standard deviations from two biological replicates carried out in duplicate.
Dose-dependent improvement of SMA condition in the SMNΔ7 mouse model
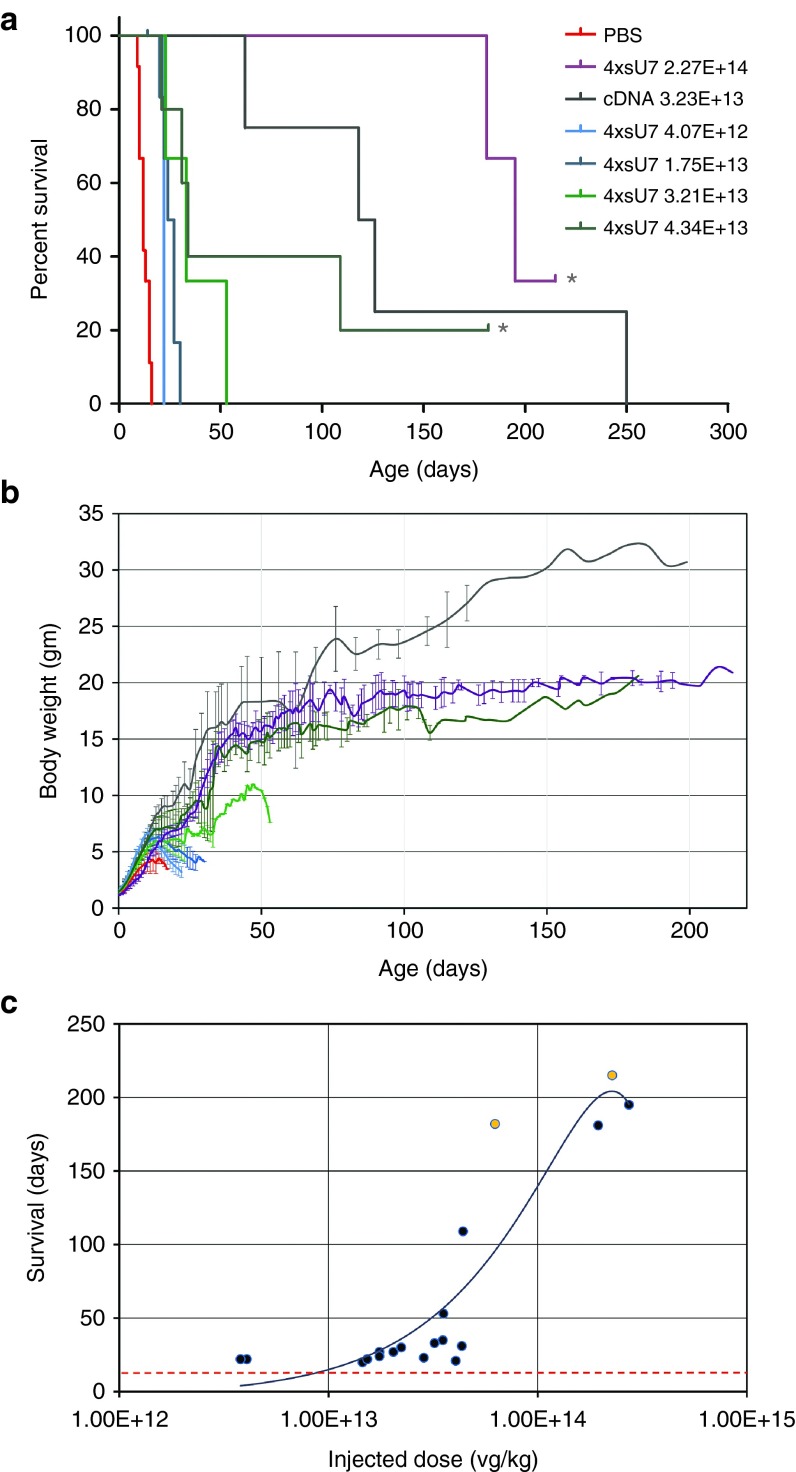
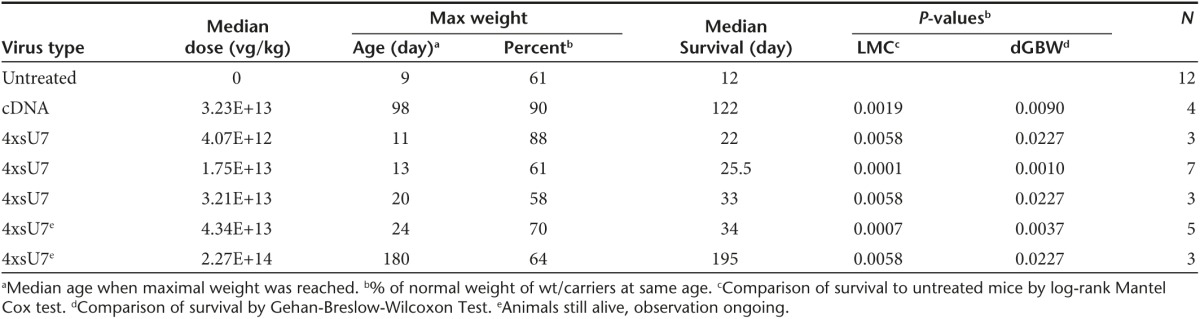
Based on these results, we decided to test scAAV 4xsU7 in somatic gene therapy experiments in the SMNΔ7 mouse model (Smn1 −/−, SMN2 +/+, and SMN▵7 +/+) for a severe form of SMA. Thus, we generated scAAV particles of serotype 9 which had been shown to transduce motoneurons efficiently after intravenous (i.v.) or intracerebroventricular (icv) injection of newborn mice.27,28 We injected 5 µl of purified particles of different concentrations into the cerebral ventricle of mice on the day of birth (P0). For all preparations, except for the least concentrated one, we performed a second injection on the next day (P1). As can be seen from the survival (Figure 4a) and weight development curves (Figure 4b), all these injections significantly increased the survival and growth of the animals, albeit to a different extent, depending on the vector dose injected. With the lowest dose of 4.07 × 1012 vector genomes (vg) per kg body weight, the mice reached a maximum weight of 6.5 g and survived for 22 days, compared with 4.3 g and 12 days, respectively, for the phosphate-buffered saline (PBS)-injected controls (all median values). Two mice treated with the two highest vector doses of 2.27 × 1014 vg/kg and 4.34 × 1013 vg/kg are still alive on days 215 and 182, respectively (at the time of resubmission of this manuscript), and have reached body weights of ~21 g. In comparison, one of four mice treated with 3.23 × 1013 vg/kg body weight of the cDNA vector survived beyond day 200 and progressed to a body weight of 32 g. Thus, a similar therapeutic effect can be reached by the somatic application of the shortened U7-ESE-B cassette as with SMN cDNA, but a higher vector dose is required (see summary in Table 1). Note that we could not observe any effect on growth or survival when wild-type or SMA carrier mice were injected with scAAV9 4xsU7 (data not shown). A plot of the effectively injected doses against the maximal survival of the mice shows that the therapeutic effect is dose-dependent (Figure 4c) and that there seems to be a threshold for prolonged survival of ~4 × 1013 vg/kg.
Figure 4.
Extended survival of SMA Δ7 mice injected with self-complementary adenovirus-associated virus 9 (scAAV9) 4xsU7. (a) Survival curves and (b) weight development. The mice receiving similar amounts of virus (vector genomes per kg body weight) have been grouped and are identically color-coded in a and b. In the groups which received the highest concentrations of scAAV9 4xsU7, mice are still surviving (one of five for 4.34 × 1013 and one of three for 2.27 × 1014; marked by asterisks in a. Deeper dents in the weight curves in b usually indicate when a mouse from the group became severely ill and had to be killed. Error bars in b indicate standard deviations. (c) Dose response curve. The actual virus amount received by each mouse is plotted against the time when it died or had to be killed. Orange points mark mice that are still alive. The red dotted line indicates the median survival of spinal muscular atrophy (SMA) Δ7 mice which either received no injection or were injected with phosphate-buffered saline (PBS) (12 days). The trendline was fitted with a second order polynomial function.
Table 1. Overview of viral injection experiments.
We evaluated the muscle performance of injected mice while they could still be compared with PBS-injected controls, i.e., between days P1–P15. In the tube hanging test, mice injected with ≥5 × 109 vg/kg of scAAV9 4xsU7 surpassed the uninjected or PBS-injected control mice (see Supplementary Figure S2a). This difference was statistically significant from P3–P10 (P = 0.0075, two-sided T-test). For the hindlimb splay test, the difference between the two groups was significant for measurements between P8–P10 (P = 0.028) (see Supplementary Figure S2b). In the righting test, the differences for individual days were only significant on the days P9 and P10 (P < 0.05) (see Supplementary Figure S2c). However, combining all values measured from P1–P15, the mean time to right in mice injected with ≥5 × 109 vg/kg was 30 seconds compared with 43 seconds for the SMA controls (P = 4.3 × 10−5). That this test has a relatively high variability was found in a previously published power analysis.29
Most importantly, the mice treated with the higher doses of scAAV9 4xsU7 survived much longer than control SMNΔ7 mice. They grew to a smaller size compared with wild-type or SMA carrier littermates and often displayed signs of milder SMA models such as partial necrosis of the tail and ear pinnae. Despite these symptoms, they were active and able to feed and drink without assistance. Supplementary Figure S3 shows selected photographs of such mice, and video sequences are available as Supplementary Files S2–S4.
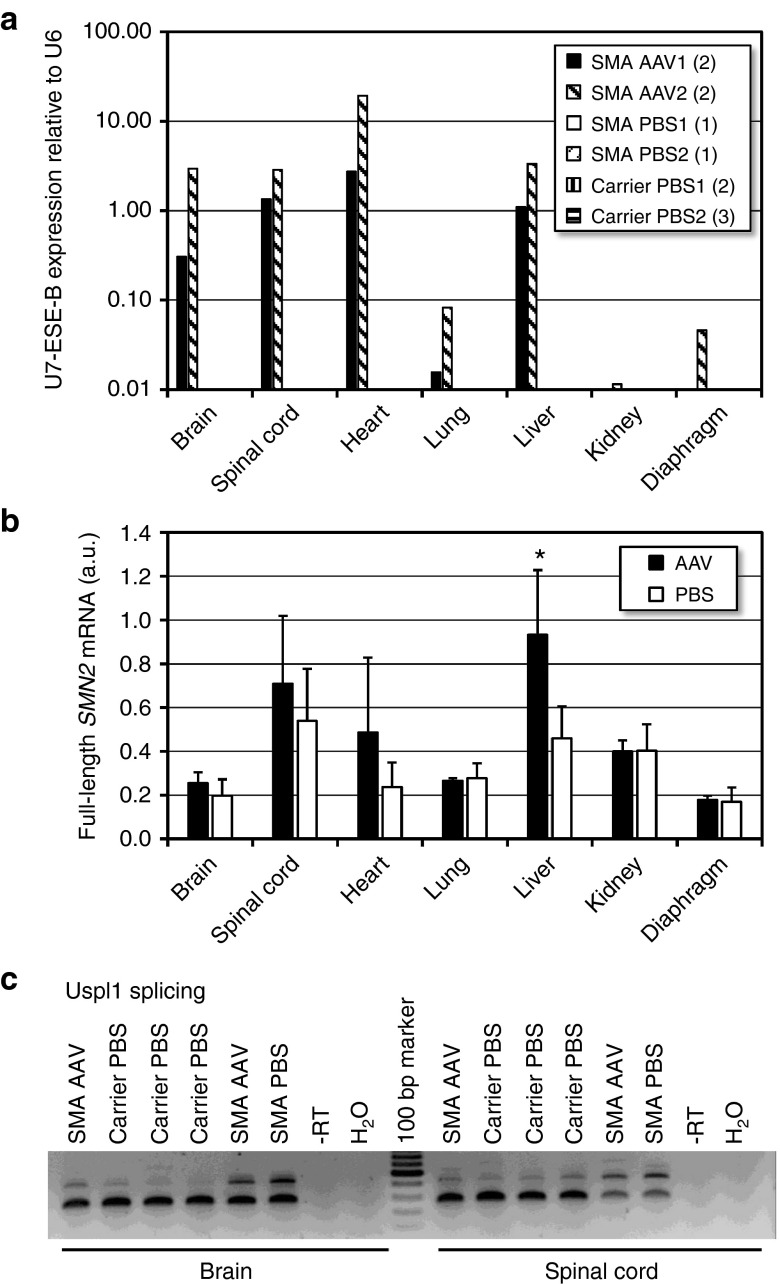
We further measured U7-ESE-B RNA levels by RT-qPCR at P10 in mice injected with 8 × 1013 vg/kg. The therapeutic RNA was expressed in the brain, spinal cord, and liver at a similar level as endogenous U6 snRNA (Figure 5a). Surprisingly, the heart showed the highest expression. In contrast, U7-ESE-B RNA levels were low in the lung and not reproducibly detectable in the diaphragm and kidney. As expected, no tissues from SMA or carrier mice injected with PBS produced positive RT-qPCR signals for U7-ESE-B RNA.
Figure 5.
Functional characterization of Spinal Muscular Atrophy (SMA) Δ7 mice injected with self-complementary adenovirus-associated virus 9 (scAAV9) -4xsU7. (a) U7-ESE-B RNA expression measured by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR). In two separate injection experiments, mice were injected with 8 × 1013 vg/kg of the same preparation of scAAV9 4xsU7 (AAV1 and AAV2) or with phosphate-buffered saline (PBS) as control (PBS1 and PBS2). For each experiment, the data of AAV-injected SMA animals and PBS-injected carriers have been averaged. Numbers in parentheses indicate the numbers of animals. The values of all PBS-injected animals were below the 1% threshold limit of the graph. (b) Expression of exon 7-containing SMN2 mRNA. a.u., arbitrary units, expression levels were normalized with respect to actin mRNA and the numbers multiplied by 1,000. Numbers of AAV-injected (Smn1−/−) animals: two for lung, kidney, diaphragm; four for other tissues. PBS injected controls (Smn1 −/− and +/−): four and seven, respectively. Asterisk: statistical relevance (P = 0.0107, two-sided T-test). Error bars indicate standard deviations. (c) Detection of alternatively spliced Ubiquitin Specific Peptidase Like 1 (Uspl1) transcripts by reverse transcriptase-polymerase chain reaction (RT-PCR). All RNA samples used in this figure were isolated from 10 day old mice.
Additionally, we analyzed whether SMN2 splicing gets corrected at P10 in these mice. A statistically significant increase in the amount of exon 7-containing SMN2 mRNA could be detected in the liver (Figure 5b). For the brain, spinal cord, and heart, there was also a tendency toward higher exon 7 inclusion which was, however, not significant for the limited number of samples analyzed. In contrast, the lung, kidney, and diaphragm that contained very little or no U7-ESE-B RNA also did not show elevated levels of exon 7-containing SMN2 mRNA.
To test whether the U7-ESE-B RNA expression can also reverse molecular events caused by low SMN levels, we analyzed the splicing of ubiquitin specific peptidase like 1 (Uspl1). Transcripts of this gene are alternatively spliced in total brain or spinal cord samples of various SMA models.30,31,32,33 We indeed detected the alternative product in brain and spinal cord samples from 10 day old SMNΔ7 mice, and its relative proportion was reduced in mice which had been treated with scAAV9 4xsU7 (Figure 5c).
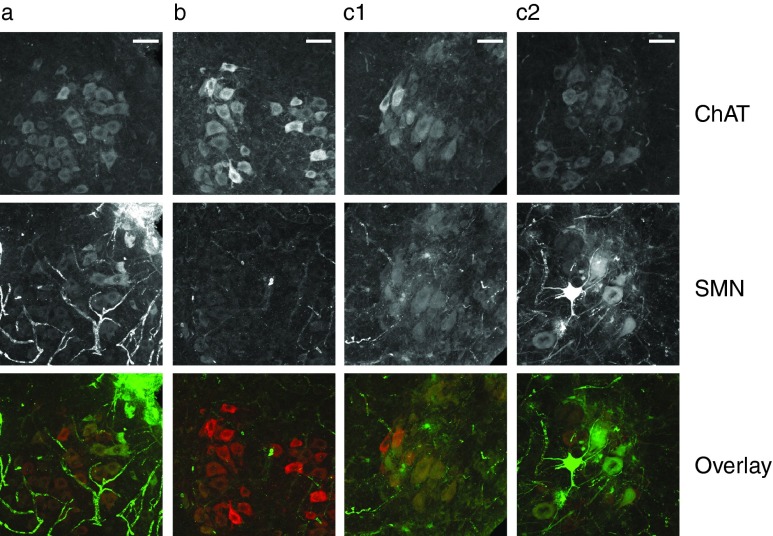
We further tested whether SMN protein is expressed in motoneurons by subjecting spinal cord sections of 10 day old mice to immunohistochemistry. Motoneurons were identified based on their location in the ventral horn, their morphology and their staining with anti-choline acetyl transferase (ChAT) antibody (Figure 6). The staining of motoneurons with anti-SMN antibody was strongly reduced in SMA disease (Figure 6b) compared with carrier mice (Figure 6a). Most importantly, however, in SMA mice injected with scAAV9 4xsU7, most motoneurons showed SMN staining. The intensity, and presumably the transduction by scAAV9 4xsU7, appeared to be variable from cell to cell. While some motoneurons showed only a faint green staining, others had green and red staining of similar intensity, resulting in a yellow overlay (Figure 6, c1). In yet other motoneurons, the staining of SMN even prevailed, reflected by a green appearance of the overlay (Figure 6, c2).
Figure 6.
Detection of SMN protein in spinal cord motoneurons of Spinal Muscular Atrophy (SMA) mice. Spinal cord sections from 10 day old mice were immunolabelled with anti-choline acetyl transferase (ChAT) to identify cholinergic neurons (top) and anti-SMN (middle) to reveal the level and distribution of SMN protein. The lowest row shows an overlay of the ChAT (red) and SMN (green) signals. (a) Smn1 +/− carrier mouse injected with phosphate-buffered saline. (b) Smn1 −/− disease mouse injected with PBS. (c1, c2) Smn1 −/− disease mice injected with scAAV9-4xsU7. Bar size = 50 µm.
To assess the functional morphology of neuromuscular junctions (NMJs), we stained whole mount diaphragms from 10 day old mice with an antibody against the active zone component bassoon. Similar to the situation in 3 day old mice of the severe SMA model,34 the spots of bassoon staining were fewer and covered a smaller fraction of the NMJ area also in these 10 day old mice of the SMNΔ7 model (see Supplementary Figure S4a,b). Importantly, injection of 8 × 1013 vg/kg of scAAV9 4xsU7 conserved these spots to a level that was not significantly different from that of carrier mice (see Supplementary Figure S4c,d). These preliminary findings will have to be corroborated with additional assessments of motor unit functionality, but they nevertheless suggest that the NMJs are much less affected in the animals treated with scAAV9 4xsU7.
Discussion
This study demonstrates that the phenotype of the SMNΔ7 mouse model (Smn1 −/−; SMN2 +/+; and SMNΔ7 +/+) can be considerably alleviated by the postnatal administration of a modified U7 snRNA gene capable of correcting the splicing of the human SMN2 gene. The life span, growth, and muscular functions of the treated mice have all been improved significantly (Figure 4, Supplementary Figure S2, and Table 1). This improvement also extends to the splicing of the SMN2 and Uspl1 mRNAs (Figure 5). Moreover, the immunohistochemical detection of SMN protein indicates that a large proportion of motoneurons have been transduced by the virus (Figure 6) and NMJs of the diaphragm show a near normal staining for the active zone component bassoon whereas this staining is significantly reduced in PBS-injected SMA mice (see Supplementary Figure S4). All this proves that the correction of SMN2 mRNA splicing by the U7-ESE-B gene not only works in a transgenic approach11 but also in a somatic gene therapy setting. Most likely, the remaining symptoms such as the reduced adult body weight and partial necroses could be overcome with yet higher vector doses, as one of the mice treated with the highest titer still does not show any ear or tail necrosis after 215 days. In any case, it is debatable whether these symptoms (which are also observed in milder mouse SMA models) are relevant for human SMA or whether they represent a mouse-specific response to reduced SMN levels.
The success of our approach is largely due to the choice and optimization of the vector. We opted for vectors of the AAV serotype 9 which had been shown to cross the blood-brain barrier and to transduce motoneurons efficiently in newborn mice,22,28 macaques,35 cats,22 and swine.36 Our initial cell culture experiments then indicated that the accumulation of U7-ESE-B snRNA is more rapid and the correction of SMN2 splicing is more efficient with a scAAV vector compared to a ssAAV vector of comparable insert size (Figure 2), in agreement with published information.37 Moreover, the shortening of the U7-ESE-B cassette allowed us to include four tandem copies in a scAAV vector, which further improved the U7-ESE-B snRNA expression and correction of SMN2 exon 7 splicing (Figure 3). Because a construct with five sU7 inserts, i.e., containing more DNA than the theoretical packaging size of scAAV, did not further improve the results and also seemed to yield lower viral titers, we chose the scAAV 4xsU7 construct for in vivo experiments. Additionally, the optimization of viral production and purification greatly contributed to the success of our project. The combination of iodixanol gradient centrifugation, dialysis, and primary concentration on Amicon filters with a final concentration by ultracentrifugation yielded very pure particles of sufficiently high titers to elicit a prolonged therapeutic benefit.
The therapeutic effects obtained in SMNΔ7 mice with the highest vector doses are similar to those reported by others for scAAV9 and scAAV8 vectors containing a SMN1 cDNA expression cassette17,18,19,20,21 or with antisense oligonucleotides correcting SMN2 splicing.13,15,16,38 Concerning the dose of virus necessary to obtain a prolonged survival, our data can best be compared with those of Meyer et al.,27 as both studies used similar vectors as well as the same mouse model and injection technique. For the SMN cDNA vector, Meyer et al. had to inject ~2 × 1013 vg/kg to ensure >50% survival of the mice beyond 100 days.27 This is in line with the reports of other groups who injected scAAV921 or scAAV817 by the icv route. In two direct comparisons, icv injections were shown to be more efficient than i.v. injections.21,27 In our hands, icv injection of ~3 × 1013 vg/kg of the same cDNA vector used by Meyer et al.27 produced a comparable result to their study with almost perfectly superimposable weight gain curves. In contrast, the threshold for survival beyond 100 days with scAAV9 4xsU7 seems to lie at ~4 × 1013 vg/kg (Figure 4c). Thus, a similar therapeutic effect can be achieved, but the U7 approach appears to require a somewhat higher vector dose than the cDNA complementation. It would be interesting to know whether this difference in efficiency is the same in other species and particularly in humans, or if the U7-ESE-B RNA and/or the SMN cDNA are expressed with a different efficiency in other species. In pigs, 1 × 1013 vg/kg to 4 × 1013 vg/kg of an scAAV9 vector carrying human SMN cDNA improved an SMA phenotype provoked by SMN RNA interference.36
The higher vector dose required to obtain a therapeutic benefit with scAAV9 4xsU7 may raise some concern, as it may be associated with a higher rate of genome insertions. Although AAV vectors are considered to remain episomal, integration into the genome has been reported to occur at a frequency of 10–4–10–5 and to be random but without activating nearby genes.39,40,41 To date, one study has described the development of hepatocellular carcinoma about 1 year after i.v. injection of ~7 × 1013 AAV vg/kg into newborn mice.42 However, the risk of AAV vector-mediated insertional mutagenesis remains controversial, as other authors could not induce liver cancer in adult mice after injection of AAV viral vectors at doses up to 1014 vg/kg via the portal vein and 18 months follow-up.43,44 Moreover, AAV vector-mediated cancer development has not been observed so far in nonhuman primates or in human clinical trials.40,41,45 In our case, an additional factor that likely reduces the risk of insertional mutagenesis is the fact that we are using a U snRNA transgene. In contrast to cDNA expression vectors that require a strong promoter/enhancer to drive expression of the transgene, the U7 gene is unlikely to cause enhancer-mediated activation of nearby genes, as U snRNA promoters and transcription complexes are distinct from those of protein-coding genes.46
In any case, neither the scAAV nor the therapeutic U7-ESE-B RNA appears to exert any unwanted side effects. First, control animals injected with the vector did not show any disease symptoms and developed normally. Most importantly, however, we have bred Smn1 +/− carriers of the transgenic SMA/U7 mouse line carrying 5–8 copies of the U7-ESE-B transgene embedded in a lentiviral vector backbone for 17 generations (SMA/U7 mice (For corrected nomenclature, see https://www.jax.org/strain/025102) can be obtained from Jackson Laboratories (Stock# 25102)). These mice never showed any unwanted side effects, had a completely normal behavior, activity and appearance as well as a normal fertility. Moreover, the Smn1 −/− animals of this line showed an improvement and partial reversal of most SMA symptoms (muscle activity, growth and life span, histological and immunohistological features of spinal cord and NMJs, and gene expression profiles).11,30,34,47,48
The question whether SMN expression needs to be restored only in motoneurons, in cells of peripheral organs, or both, has received increased attention following the recent observation of Krainer and coworkers who found that an antisense oligonucleotide correcting SMN2 splicing worked as well when injected subcutaneously, as when the icv route was used.38 Moreover, the beneficial effect of the peripheral application was not abolished by the icv injection of sense oligonucleotides which should have neutralized the antisense oligos in the central nervous system.16 These findings contrast with the observations of various groups that icv injection of scAAV vectors expressing SMN cDNA are more effective than i.v.21,27 or intramuscular49 injections. Moreover, a study using germ-line transgenesis of the severe SMA model with SMN cDNA under the control of different cell type-specific promoters showed that a neuron-specific expression rescued the mice from SMA whereas an expression under a muscle-specific promoter failed to do so, except when it also induced a low level of SMN expression in the central nervous system.50 More recent experiments using cre recombination induced by the ChAT and Nestin promoters indicated that ChAT-cre mediated SMN gene repair (i.e., restricted to cholinergic neurons) is sufficient to restore motor unit function and, in combination with Nestin-cre, which additionally allows the production of functional SMN in glial cells, can extend survival and induce an almost complete phenotypic rescue.51 Even though our data do not directly address the question of peripheral requirements for high SMN levels, it is interesting to note that we detected expression of U7-ESE-B RNA and SMN2 splicing correction in the liver and heart (Figure 5b). High transduction of the liver after icv injection of AAV9 encoding sulfamidase into 2 month old mice has been observed in a study on mucopolysaccharidosis IIIA, but the heart was not transduced with this vector system.52 In contrast, both liver and heart are efficiently transduced after iv injections of AAV9 in mice.53 Our results suggest that scAAV9 injected icv in newborn mice can pass the blood brain barrier and reach peripheral organs that are susceptible to infection by this serotype. This additional expression of U7-ESE-B RNA in the heart and liver may contribute to its therapeutic effect, because cardiac defects have been observed in SMA patients54 and mouse models55,56,57 and a liver dysfunction may play a role, at least in the Taiwanese SMA mouse model.38
In conclusion, our study proves that it is feasible to promote the inclusion of a skipped exon and to obtain a pronounced improvement of a severe SMA phenotype by a somatic gene therapy approach with a modified U7 snRNA derivative. A possible application of this method to human SMA therapy will depend, among other things, on the advance of other therapeutic approaches. Clinical trials are ongonig for AAV-based SMN cDNA vectors and antisense oligonucleotides.23 Additionally, a small molecule drug which corrects SMN2 splicing has shown very promising results in preclinical studies.58,59 However, it is not yet clear which treatment option will be most efficient and have the least side-effects in human SMA patients. A possible argument for gene therapies is that a single treatment can in principle lead to a life-long cure of SMA, whereas an oligonucleotide or small molecule drug will require repeated interventions, even if the compounds are very stable as is the case for some oligonucleotides. On the other hand, a gene therapy is more difficult to apply and may represent an additional burden for the patients, especially if children are treated at a very young age. In any case, there is room for exploring additional treatment options such as our U7-based approach or the recently described method using exon-specific U1 snRNAs which has been demonstrated to alleviate SMA symptoms in a mouse transgenic approach60 but has not yet been tested in a somatic gene therapy setting.
Apart from the relevance of the present findings for SMA, it is conceivable to use an approach similar to ours to introduce other noncoding regulatory RNAs into the central nervous system. This could be used either to modulate specific phenotypes in experimental animals or to improve other kinds of neurological or neuromuscular disorders.
Materials and Methods
Cell lines. HeLa S2 are HeLa cells that have been stably transformed with a human SMN2 minigene25 as described.9 AAV-293 cells containing adenovirus type 5 DNA and expressing the adenovirus E1 gene were obtained from Agilent. All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (BioConcept), 100 U/ml penicillin and 100 U/ml streptomycin (both Lubio Science). This medium is called DMEM+/+; medium without fetal calf serum and antibiotics will be referred to as DMEM−/−. The cells were kept at 37°C in a humid atmosphere with 5% CO2.
AAV production and purification Expression plasmids. To accommodate multiple copies of the U7-ESE-B cassette in AAV vectors, we shortened the original cassette at the 5′ and 3′ ends while retaining all necessary promoter and 3′ elements. The resulting short U7-ESE-B cassettes (sU7) contain 253 and 104 bp of 5′ and 3′ flanking sequences, respectively. As recipient plasmids to generate ss and scAAV, we used pAAV-CMV-eGFP61 and pAAVsc_U7Dtex23,62 respectively. We generated constructs containing one to five copies of sU7. In some vectors, we inserted an EGFP expression cassette in the plasmid backbone to be able to monitor the transfection efficiency during virus production. Maps are shown in Supplementary Figure S1. Sequence files and further information on the cloning are available upon request.
Helper plasmids. For AAV6 production, we used the helper plasmid pDF6.63 For AAV9, the separate helper plasmids Rep2-Cap9 and pHelper were used.28
Detailed procedures for the production and purification of AAV vectors are described in the Supplementary Materials and Methods and Supplementary Table S1. An overview of the method is presented in Supplementary Figure S5.
qPCR based virus titration. Virus suspensions were treated with DNase (10 U DNase I, Roche Diagnostics, Rotkreuz, Switzerland) for 20 minutes at 37°C. Then, the viral capsids were destroyed by an incubation for 20 minutes at 95°C that simultaneously inactivated the DNase. Serial dilutions of the lysed virus preparation and of a reference plasmid were compared by qPCR. Reactions consisted of 10 µl MesaGreen qPCR Mastermix Plus for SYBR Assay (Eurogentec, Liège, Belgium), 0.2 µl of each primer (12 µmol/l; Microsynth AG, Switzerland), 5.6 µl H2O and 4 µl of sample. The qPCR was performed in a Rotor-Gene Q (Qiagen AG, Hombrechtikon, Switzerland) with initial denaturation for 10 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds, 56–60°C (according to the primers used) for 15 seconds, and 10 seconds at 70°C. A melting curve from 65–95°C was then used to assess the purity of the amplicon. The run was evaluated with Rotor Gene Q Series Software (Version 2.2.3, Build 11), Qiagen AG, Hombrechtikon, Switzerland. Primers are listed in Supplementary Table S2.
Plasmid transfections and viral transduction of cells. HeLa S2 cells were seeded at a density of 105 cells per well of a six-well plate. On the next day, the medium was replaced by DMEM −/−. For each well, 1.5 µg of plasmid and 4 µl Fugene HD (Roche) were mixed in 100 µl DMEM -/- and allowed to form complexes at room temperature for 15 minutes. The mixture was then distributed dropwise over the cells. After incubation for 4 hours at 37°C, the medium was replaced by DMEM +/+.
Viral transductions were performed in the mnimal sufficient volume of medium to completely cover the cells. After 6 hours at 37°C, the medium was completed to the standard volume for the corresponding culture flask.
Mouse -breeding and genotyping. SMNΔ7 mice14 were purchased from Jackson Laboratories (FVB.Cg-Tg(SMN2)89Ahmb Smn1tm1MsdTg(SMN2*delta7) 4299AhmB/J; stock number: 005025; Bar Harbor, ME, USA) and kept at the central animal facility, Inselspital Bern under standard IVC conditions according to legal requirements (Swiss federal permit BE14/13). For genotyping, DNA from tail biopsies taken on the day of birth was extracted and analyzed with KAPA Mouse Genotyping Kits (Axonlab, Baden Switzerland) according to the manufacturers' instructions. The primers used (see Supplementary Table S2) allowed us to determine the Smn1 genotype and sex of the animals.
Injections for somatic gene therapy in mice. AAV preparations or PBS (for controls) were injected into the left cerebral ventricle of newborn mice. Prior to injection, the virus was mixed with 0.025% Fast Green (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) to be able to evaluate the success of the injection. For better visibility, the head of the mouse was held onto a small LED torch lamp. Usually, 5 µl of virus were injected at P0 (day of birth) and again at P1 through a glass capillary (microhematocrit tubes non heparinized, ø ext. 1.45 mm, ø int. 0.95 mm, long 75.0 mm, Sovirel France, pulled in a micropipette puller model P97, Sutter Instrument S, Novato, CA) connected via a tube to a 1 ml syringe that was used to control the liquid flow.
Monitoring of mouse development and performance. The overall condition and weight of the mice were initially recorded on a daily basis. As the mice reached a higher age, the intervals were increased to 2–3 days. The motility and strength were determined by the righting reflex (time to right from an inverted position). A second test consisted of measuring how long the mice could hold themselves with their hind legs grabbing the rim of a 50 ml Falcon tube, head downward, before falling into the tube that was padded with paper tissue to soften the fall. Additionally, a score from 0 to 4 was given depending on how far the hind legs were splayed, with 0 indicating that the legs touched each other and 4 representing the maximal possible distance.
RNA extraction and alternative splicing analysis of SMN2, Uspl1, and SMN2 exon 7 specific qPCR. Total RNA from transfected or virus-infected HeLa S2 cells was extracted with homemade Tri-Reagent as described.64 Mouse tissues were homogenized in Tri-Reagent before extraction. Between 0.5–2 µg of RNA was reverse-transcribed with 20 pmoles of random hexamer or oligo d(T) with Affinity-Script Multi-Temp Reverse Transcriptase (Agilent Technologies, Basel, Switzerland) or with the PrimeScript RT Master Mix kit (Takara bIO eUROPE sas, Saint-Germain-en-Laye, France) according to the manufacturers' instructions. PCR for splicing analysis on agarose gels (SMN minigene and Uspl1) was performed with the PCR FastStart 2× PCR Master Mix (Roche) and appropriate primers (see Supplementary Table S2). For the detection of exon 7 containing SMN2 transcripts, the cDNA was analyzed by a SYBR-green based qPCR assay (mix as described for qPCR-based virus titration) using SMN-Ex7-Re(Fw) and SMN-Ex8-Re primers.
Quantitation of U7-ESE-B expression. To analyze U7-ESE-B expression, cDNA was synthesized in a coupled polyadenylation reverse transcription reaction designed for short nonpolyadenylated RNAs.65 This cDNA was then used for a SYBR-green based qPCR analysis (mix as described for qPCR-based virus titration) with qPCR ESE Fw and universal reverse primers.65 As normalizer, U6 snRNA was analyzed with qPCR WT_U6 Fw and the universal reverse primers. The primer sequences are listed in Supplementary Table S2.
Immunofluorescence analysis. Spinal cords were fixed in 4% paraformaldehyde (PFA) followed by incubation in 30% sucrose (w/v) overnight at 4°C each. The following day, the spinal cords were embedded in cryomolds using Tissue-Tek O.C.T. compound (Sakura Finetec, VWR International, Dietikon, Switzerland) and stored at −20°C. Sections of 50 µm thickness were cut with a microtome and transferred into 0.8% sodium azide in PBS. For all further treatments, the sections were incubated freely floating in wells of a 12-well plate, and transfers were done with a thin paint brush. First they were blocked in 3% bovine serum albumin (BSA) in PBS for 3 hours at RT followed by primary antibody incubation in 3% BSA and 0.3% TritonX-100 in PBS overnight at 4°C on a rocker (for antibodies and dilutions see Supplementary Table S3). Following three wash steps in 0.1% TritonX-100 in PBS for 20 minutes each, the sections were incubated with secondary antibody in 3% BSA and 0.3% TritonX-100 in PBS for 5 hours at RT or overnight at 4°C on a rocker in the dark. Subsequently, the sections were washed three times 20 minutes in the dark with 0.1% TritonX-100 in PBS. Finally they were transferred onto glass slides, air-dried, mounted with homemade Moviol and stored at 4°C. The slides were analyzed with an Olympus Fluoview 1000-BX61 microscope and Imaris 7.6 (Bitplane AG, Zürich, Switzerland) software (Olympus Schweiz AG, Volketswil, Switzerland).
Diaphragms were dissected and stored at −20°C in RNAlater (Sigma-Aldrich) for further analysis. During all staining steps, they were incubated freely floating in wells of a 24-well plate. After washing in PBS for 30 minutes, NMJs were detected by incubation with α-Bungarotoxin in 3% BSA and 0.5% TritonX-100 in PBS at RT. Next, the samples were blocked for 3 hours at RT followed by primary antibody incubation in 3% BSA and 0.5% TritonX-100 in PBS for 72 hours at 4°C on a rocker (for antibodies and dilutions see Supplementary Table S3). The incubation with secondary antibodies, all wash steps and the mounting were performed as described above for the spinal cord samples. The samples were analyzed with a Leica DMi8 microscope (Leica Microsystems, Heerbrugg, Switzerland) and Leica application suite X software and further evaluated with FiJi (Image J).
SUPPLEMENTARY MATERIAL Figure S1. AAV vectors used in this work. Figure S2. Muscle performance of SMA Δ7 mice injected with scAAV9 4xsU7 compared to uninjected or PBS-injected mice and wt or carrier littermates. Figure S3. Photographs of SMNΔ7 Smn1−/− mice injected with scAAV9 4xsU7. Figure S4. Conservation of active zones in diaphragm neuromuscular junctions (NMJs) of SMNΔ7 Smn1−/− mice injected with scAAV9 4xsU7. Figure S5. Scheme of AAV production and purification and EM pictures of purified viruses. Supplementary Methods (virus production) and Tables Table S1. Amounts of cells/reagents used for virus production. Table S2. Oligonucleotides used in this work. Table S3. Antibodies and probes used in this work. Description of supplementary videos Supplementary File S2. Male SMA mouse D54-2-1, injected with 2.73 × 1014 vg/kg scAAV9 4xsU7. Video taken on P78. Supplementary File S3. Female SMA mouse D54-2–9, injected with 2.27 × 1014 vg/kg scAAV9 4xsU7, together with slightly larger female carrier littermate D54-2–3 and mother. Video taken on P78. Supplementary File S4. Female SMA mouse D57-1-1, injected with 6.25 × 1013 vg/kg scAAV9 4xsU7, together with adult female. Video taken on P49. Supplementary References
Acknowledgments
This work was supported by the Kanton Bern and the Swiss National Science Foundation through the grants 31003A_135644, 31003A_153199, and through the NCCR RNA and Disease. We thank Pamela Y. Sandoval for cloning help, Mircea-Ioan Iacovache for EM analyses, David Stucki, and Romain Levayer for help with immunofluorescence microscopy and Smita Saxena for critical comments on the manuscript. We also acknowledge AAV vectors and technical advice kindly offered by Bernard L. Schneider and Patrick Aebischer (EPFL Lausanne, Switzerland), Aurélie Goyenvalle and Luis Garcia (Université de Versailles, France) as well as Kathrin Meyer and Brian K. Kaspar (Nationwide Children's Hospital, Columbus, OH, USA).
Supplementary Material
References
- Crawford, TO and Pardo, CA (1996). The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis 3: 97–110. [DOI] [PubMed] [Google Scholar]
- Lefebvre, S, Bürglen, L, Reboullet, S, Clermont, O, Burlet, P, Viollet, L et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 155–165. [DOI] [PubMed] [Google Scholar]
- Lunn, MR and Wang, CH (2008). Spinal muscular atrophy. Lancet 371: 2120–2133. [DOI] [PubMed] [Google Scholar]
- Burnett, BG, Muñoz, E, Tandon, A, Kwon, DY, Sumner, CJ and Fischbeck, KH (2009). Regulation of SMN protein stability. Mol Cell Biol 29: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L and Krainer, AR (2002). Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384. [DOI] [PubMed] [Google Scholar]
- Kashima, T and Manley, JL (2003). A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463. [DOI] [PubMed] [Google Scholar]
- Cartegni, L, Hastings, ML, Calarco, JA, de Stanchina, E and Krainer, AR (2006). Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet 78: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima, T, Rao, N, David, CJ and Manley, JL (2007). hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet 16: 3149–3159. [DOI] [PubMed] [Google Scholar]
- Marquis, J, Meyer, K, Angehrn, L, Kämpfer, SS, Rothen-Rutishauser, B and Schümperli, D (2007). Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol Ther 15: 1479–1486. [DOI] [PubMed] [Google Scholar]
- Monani, UR, Sendtner, M, Coovert, DD, Parsons, DW, Andreassi, C, Le, TT et al. (2000). The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in SMN(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 9: 333–339. [DOI] [PubMed] [Google Scholar]
- Meyer, K, Marquis, J, Trüb, J, Nlend Nlend, R, Verp, S, Ruepp, MD et al. (2009). Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum Mol Genet 18: 546–555. [DOI] [PubMed] [Google Scholar]
- Nlend Nlend, R, Meyer, K and Schümperli, D (2010). Repair of pre-mRNA splicing: prospects for a therapy for spinal muscular atrophy. RNA Biol 7: 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porensky, PN and Burghes, AH (2013). Antisense oligonucleotides for the treatment of spinal muscular atrophy. Hum Gene Ther 24: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, TT, Pham, LT, Butchbach, ME, Zhang, HL, Monani, UR, Coovert, DD et al. (2005). SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 14: 845–857. [DOI] [PubMed] [Google Scholar]
- Hua, Y, Vickers, TA, Okunola, HL, Bennett, CF and Krainer, AR (2008). Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Y, Liu, YH, Sahashi, K, Rigo, F, Bennett, CF and Krainer, AR (2015). Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev 29: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini, MA, Bu, J, Roskelley, EM, Richards, AM, Sardi, SP, O'Riordan, CR et al. (2010). CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest 120: 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust, KD, Wang, X, McGovern, VL, Braun, L, Bevan, AK, Haidet, AM et al. (2010). Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol 28: 271–274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dominguez, E, Marais, T, Chatauret, N, Benkhelifa-Ziyyat, S, Duque, S, Ravassard, P et al. (2011). Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum Mol Genet 20: 681–693. [DOI] [PubMed] [Google Scholar]
- Valori, CF, Ning, K, Wyles, M, Mead, RJ, Grierson, AJ, Shaw, PJ et al. (2010). Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med 2: 35ra42. [DOI] [PubMed] [Google Scholar]
- Glascock, JJ, Shababi, M, Wetz, MJ, Krogman, MM and Lorson, CL (2012). Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem Biophys Res Commun 417: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque, S, Joussemet, B, Riviere, C, Marais, T, Dubreil, L, Douar, AM et al. (2009). Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther 17: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, B, Barkats, M, Martinat, C, Sendtner, M and Gillingwater, TH (2015). Moving towards treatments for spinal muscular atrophy: hopes and limits. Expert Opin Emerg Drugs 20: 353–356. [DOI] [PubMed] [Google Scholar]
- Gruber, A, Soldati, D, Burri, M and Schümperli, D (1991). Isolation of an active gene and of two pseudogenes for mouse U7 small nuclear RNA. Biochim Biophys Acta 1088: 151–154. [DOI] [PubMed] [Google Scholar]
- Zhang, ML, Lorson, CL, Androphy, EJ and Zhou, J (2001). An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther 8: 1532–1538. [DOI] [PubMed] [Google Scholar]
- Wu, Z, Yang, H and Colosi, P (2010). Effect of genome size on AAV vector packaging. Mol Ther 18: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K, Ferraiuolo, L, Schmelzer, L, Braun, L, McGovern, V, Likhite, S et al. (2015). Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol Ther 23: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust, KD, Nurre, E, Montgomery, CL, Hernandez, A, Chan, CM and Kaspar, BK (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 27: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach, ME, Edwards, JD and Burghes, AH (2007). Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis 27: 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, Q, Kayikci, M, Odermatt, P, Meyer, K, Michels, O, Saxena, S et al. (2014). Splicing changes in SMA mouse motoneurons and SMN-depleted neuroblastoma cells: evidence for involvement of splicing regulatory proteins. RNA Biol 11: 1430–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H, Shafey, D, Moores, JN and Kothary, R (2010). Neurodevelopmental consequences of Smn depletion in a mouse model of spinal muscular atrophy. J Neurosci Res 88: 111–122. [DOI] [PubMed] [Google Scholar]
- Bäumer, D, Lee, S, Nicholson, G, Davies, JL, Parkinson, NJ, Murray, LM et al. (2009). Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 5: e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z, Lotti, F, Dittmar, K, Younis, I, Wan, L, Kasim, M et al. (2008). SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve, A, Trüb, J, Saxena, S and Schümperli, D (2016). Central and peripheral defects in motor units of the diaphragm of spinal muscular atrophy mice. Mol Cell Neurosci 70: 30–41. [DOI] [PubMed] [Google Scholar]
- Bevan, AK, Duque, S, Foust, KD, Morales, PR, Braun, L, Schmelzer, L et al. (2011). Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol Ther 19: 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque, SI, Arnold, WD, Odermatt, P, Li, X, Porensky, PN, Schmelzer, L et al. (2015). A large animal model of spinal muscular atrophy and correction of phenotype. Ann Neurol 77: 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, DM (2008). Self-complementary AAV vectors; advances and applications. Mol Ther 16: 1648–1656. [DOI] [PubMed] [Google Scholar]
- Hua, Y, Sahashi, K, Rigo, F, Hung, G, Horev, G, Bennett, CF et al. (2011). Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478: 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, DM, Young, SM Jr and Samulski, RJ (2004). Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet 38: 819–845. [DOI] [PubMed] [Google Scholar]
- Nowrouzi, A, Penaud-Budloo, M, Kaeppel, C, Appelt, U, Le Guiner, C, Moullier, P et al. (2012). Integration frequency and intermolecular recombination of rAAV vectors in non-human primate skeletal muscle and liver. Mol Ther 20: 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppel, C, Beattie, SG, Fronza, R, van Logtenstein, R, Salmon, F, Schmidt, S et al. (2013). A largely random AAV integration profile after LPLD gene therapy. Nat Med 19: 889–891. [DOI] [PubMed] [Google Scholar]
- Donsante, A, Miller, DG, Li, Y, Vogler, C, Brunt, EM, Russell, DW et al. (2007). AAV vector integration sites in mouse hepatocellular carcinoma. Science 317: 477. [DOI] [PubMed] [Google Scholar]
- Bell, P, Wang, L, Lebherz, C, Flieder, DB, Bove, MS, Wu, D et al. (2005). No evidence for tumorigenesis of AAV vectors in a large-scale study in mice. Mol Ther 12: 299–306. [DOI] [PubMed] [Google Scholar]
- Li, H, Malani, N, Hamilton, SR, Schlachterman, A, Bussadori, G, Edmonson, SE et al. (2011). Assessing the potential for AAV vector genotoxicity in a murine model. Blood 117: 3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Farina, I, Fronza, R, Kaeppel, C, Lopez-Franco, E, Ferreira, V, D'Avola, D et al. (2016). Recombinant AAV Integration Is Not Associated With Hepatic Genotoxicity in Nonhuman Primates and Patients. Mol Ther 24: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, S, O'Reilly, D and Murphy, S (2008). Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36(Pt 4): 590–594. [DOI] [PubMed] [Google Scholar]
- Voigt, T, Meyer, K, Baum, O and Schümperli, D (2010). Ultrastructural changes in diaphragm neuromuscular junctions in a severe mouse model for Spinal Muscular Atrophy and their prevention by bifunctional U7 snRNA correcting SMN2 splicing. Neuromuscul Disord 20: 744–752. [DOI] [PubMed] [Google Scholar]
- Voigt, T, Neve, A and Schümperli, D (2014). The craniosacral progression of muscle development influences the emergence of neuromuscular junction alterations in a severe murine model for spinal muscular atrophy. Neuropathol Appl Neurobiol 40: 416–434. [DOI] [PubMed] [Google Scholar]
- Benkhelifa-Ziyyat, S, Besse, A, Roda, M, Duque, S, Astord, S, Carcenac, R et al. (2013). Intramuscular scAAV9-SMN injection mediates widespread gene delivery to the spinal cord and decreases disease severity in SMA mice. Mol Ther 21: 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilina, TO, McGovern, VL, Workman, E, Crawford, TO, Gogliotti, RG, DiDonato, CJ et al. (2008). Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum Mol Genet 17: 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern, VL, Iyer, CC, Arnold, WD, Gombash, SE, Zaworski, PG, Blatnik, AJ III et al. (2015). SMN expression is required in motor neurons to rescue electrophysiological deficits in the SMNΔ7 mouse model of SMA. Hum Mol Genet 24: 5524–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurigot, V, Marcó, S, Ribera, A, Garcia, M, Ruzo, A, Villacampa, P, et al. (2013). Whole body correction of mucopolysaccharidosis IIIA by intracerebrospinal fluid gene therapy. The Journal of Clinical Investigation 123: 3254–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli, C, Soltys, S, Rengo, G and Rabinowitz, JE (2008). Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Rudnik-Schöneborn, S, Heller, R, Berg, C, Betzler, C, Grimm, T, Eggermann, T et al. (2008). Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J Med Genet 45: 635–638. [DOI] [PubMed] [Google Scholar]
- Bevan, AK, Hutchinson, KR, Foust, KD, Braun, L, McGovern, VL, Schmelzer, L et al. (2010). Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet 19: 3895–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier, CR, Satta, R, Lutz, C and DiDonato, CJ (2010). Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet 19: 3906–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi, M, Habibi, J, Yang, HT, Vale, SM, Sewell, WA and Lorson, CL (2010). Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet 19: 4059–4071. [DOI] [PubMed] [Google Scholar]
- Naryshkin, NA, Weetall, M, Dakka, A, Narasimhan, J, Zhao, X, Feng, Z, et al. (2014). SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345: 688–693. [DOI] [PubMed] [Google Scholar]
- Zhao, X, Feng, Z, Ling, KKY, Mollin, A, Sheedy, J, Yeh, S, et al. (2016). Pharmacokinetics, pharmacodynamics and efficacy of a small molecule SMN2 splicing modifier in mouse models of spinal muscular atrophy. Hum Mol Genetdoi: 10.1093/hmg/ddw062. [DOI] [PMC free article] [PubMed]
- Rogalska, ME, Tajnik, M, Licastro, D, Bussani, E, Camparini, L, Mattioli, C et al. (2016). Therapeutic activity of modified U1 core spliceosomal particles. Nat Commun 7: 11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towne, C, Raoul, C, Schneider, BL and Aebischer, P (2008). Systemic AAV6 delivery mediating RNA interference against SOD1: neuromuscular transduction does not alter disease progression in fALS mice. Mol Ther 16: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Goyenvalle, A, Babbs, A, Wright, J, Wilkins, V, Powell, D, Garcia, L et al. (2012). Rescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skipping. Hum Mol Genet 21: 2559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D, Kay, MA and Kleinschmidt, JA (2003). Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther 7: 839–850. [DOI] [PubMed] [Google Scholar]
- Marquis, J, Kämpfer, SS, Angehrn, L and Schümperli, D (2009). Doxycycline-controlled splicing modulation by regulated antisense U7 snRNA expression cassettes. Gene Ther 16: 70–77. [DOI] [PubMed] [Google Scholar]
- Raczynska, KD, Ruepp, MD, Brzek, A, Reber, S, Romeo, V, Rindlisbacher, B et al. (2015). FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res 43: 9711–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.