Abstract
The rod cell has an extraordinarily specialized structure that allows it to carry out its unique function of detecting individual photons of light. Both the structural features of the rod and the metabolic processes required for highly amplified light detection seem to have rendered the rod especially sensitive to structural and metabolic defects, so that a large number of gene defects are primarily associated with rod cell death and give rise to blinding retinal dystrophies. The structures of the rod, especially those of the sensory cilium known as the outer segment, have been the subject of structural, biochemical, and genetic analysis for many years, but the molecular bases for rod morphogenesis and for cell death in rod dystrophies are still poorly understood. Recent developments in imaging technology, such as cryo-electron tomography and super-resolution fluorescence microscopy, in gene sequencing technology, and in gene editing technology are rapidly leading to new breakthroughs in our understanding of these questions. A summary is presented of our current understanding of selected aspects of these questions, highlighting areas of uncertainty and contention as well as recent discoveries that provide new insights. Examples of structural data from emerging imaging technologies are presented.
Keywords: photoreceptor, cryo-electron tomography, retinal imaging, retinal degeneration, disease mechanisms, ciliopathies
I. Introduction
Rod photoreceptor cells of the vertebrate retina have a number of unusual features that make them particularly interesting and important objects of study. One is that they have an extraordinarily highly amplified signal transduction cascade that allows them to respond with a reliable physiological signal to the absorption of a single photon of light. The other unique features appear to be related to this fundamental one, and to be necessary for it to be realized. They have an unusual morphology, with a long and, especially in mammals, very thin shape (Fig. 1). Roughly half of their length consists of a modified primary cilium, known as an outer segment, which contains within it a stack of disk-shaped membranes attached to a long axoneme (a 9+0 bundle of microtubules, which transition from doublet microtubules at the base to singlet microtubules in the distal region of the outer segment), a structure not found elsewhere in the body. Likely because of their highly amplified signaling cascade, which places tremendous demands on their energy metabolism, they are highly susceptible to cell death resulting from deprivation of oxygen or nutrients, or from inborn metabolic defects, including mutant proteins. This last feature renders them of great medical and basic scientific interest, because loss of rods can lead to severe visual defects, including total blindness. Despite the great interest of researchers over the years, and much effort expended by them, there are still many features of rods we understand very poorly. Fortunately, in recent years, progress in this area has been accelerating due to the advent of new structural techniques, and to recent enhancements in our ability to analyze genomes and to manipulate those of animal models. This article is intended to review our current understanding of the questions, “what are the important structural features of rods and how do they arise,” and “how do defects in those structures and in the functions of both rod-specific and widely expressed proteins lead to cell death?” It is not intended to be a comprehensive review of these topics, but rather to focus on particular aspects of these questions that have been the subject of ongoing research in our laboratory. Excellent reviews have been published recently on protein trafficking in rods (Anand and Khanna, 2012; Constantine et al., 2012; Hollingsworth and Gross, 2012; Pearring et al., 2013; Schwarz et al., 2012; Wang and Deretic, 2014; Zhang et al., 2012a), which is touched on only briefly here, but is highly relevant for rod structure and diseases of the rod. The same is true for proteins of the intraflagellar-transport (IFT) pathway (Insinna and Besharse, 2008; Malicki, 2012; Nachury et al., 2010; Roepman and Wolfrum, 2007). There have also been a number of useful reviews of retinal degeneration mechanisms and animal models, e.g. (Bales and Gross, 2015; Chan et al., 2016; Daiger et al., 2015; Fletcher et al., 2011; Flisikowska et al., 2014; Gorbatyuk and Gorbatyuk, 2013; Miyadera et al., 2012; Sahel et al., 2010; Siemiatkowska et al., 2014; Veleri et al., 2015; Wright et al., 2010). Recently, he molecular basis for rod cell function has been reviewed (Goldberg et al., 2016).
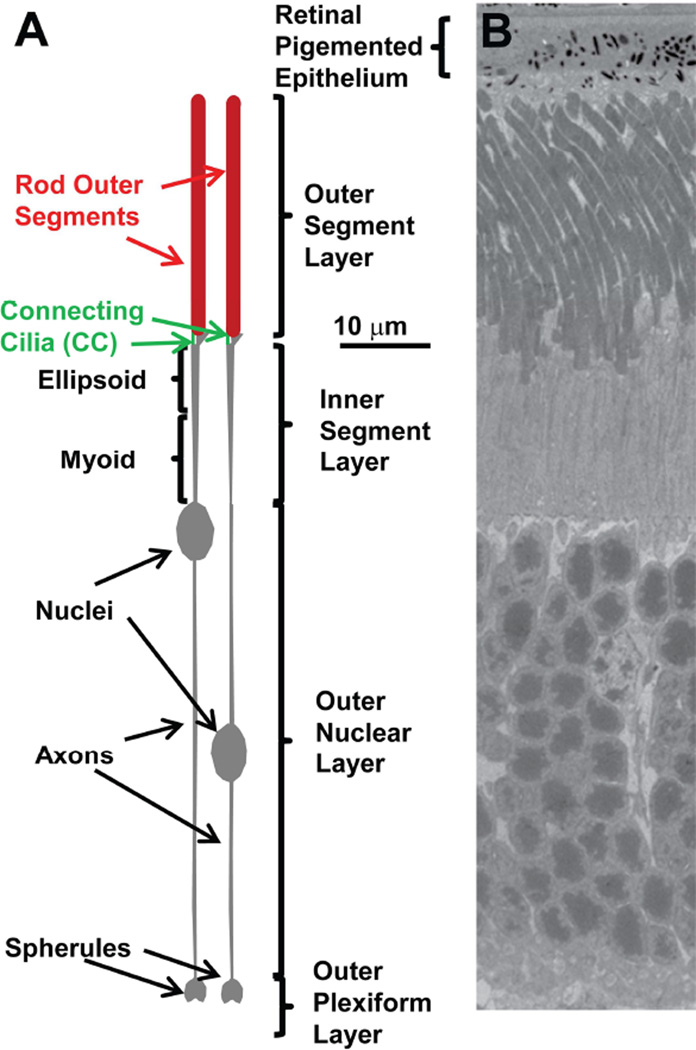
Figure 1.
A. Schematic drawing of mouse rods. Sub-regions of the cell are depicted according to the indicated scale based on dimensions taken from a range of microscopic techniques. This accurate scaling is in contrast to highly distorted scales typically used in cartoons of rods. B. Transmission electron micrograph showing the corresponding retinal layers from a mouse retina. Sample preparation and imaging were as described in (He et al., 2016).
2. Evolution of methods for studying rod cell structure
2.1 Early work
Vertebrate retina was one of the first objects of study when light microscopy developed to the point that cells within tissues could be visualized using selected stains. Ramón y Cajal’s classic 19th century work (Ramón y Cajal, 1892) clearly identified the layers of the retina and noted the essential structural features of the rods. However, many of the most interesting features of the rod, such as the disk membranes, the connecting cilium, the ciliary rootlet, and intracellular vesicles and filaments, are beyond the resolution of conventional light microscopy. The development of electron microscopy in the 1930s and 1940s led to a revolution in the understanding of rod cell ultrastructure in the 1950s and 1960s One of the first studies by Fritjof Sjostrand in 1949 (Sjostrand, 1949) identified many of the familiar features of rod ultrastructure. This was followed by additional classic work from Sjostrand (Sjostrand, 1953a, b), and from DeRobertis (De Robertis, 1956), which called attention to the ciliary structures associated with outer segments and the connecting cilium. Work by Godfrey on chick retina (Godfrey, 1973) noted the substantial differences in apparent dimensions of structures such as disk membranes and inter- and intra-diskal spaces visualized by EM using different stains. Fig. 2 illustrates the appearance of the cilium-associated structures in mouse rods by conventional transmission electron microscopy (TEM) using tissue samples extensively fixed, embedded in resin, sectioned with an ultramicrotome, and stained with heavy metals.
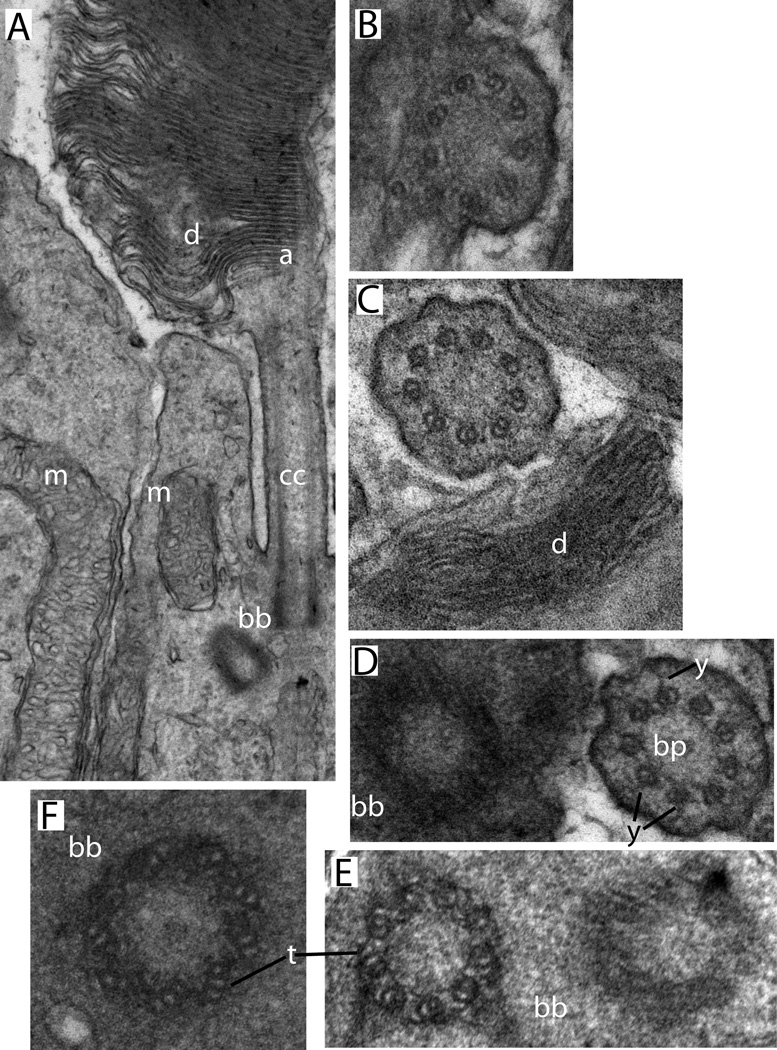
Figure 2.
Conventional transmission electron microscopy of mouse rod sensory cilium. Sample preparation and imaging were as described in (He et al., 2016). A. Longitudinal section showing connecting cilium and adjacent regions of outer and inner segments (Labels: d, disks; cc, connecting cilium; bb, basal body complex; a, axoneme; m, mitochondria). B-F, cross-sectional views through the same regions. B, a region where the CC membrane is continuous with the plasma membrane. C, a connecting cilium sectioned near the base of the outer segment (d, disks). D, a centriole next to a connecting cilium from an adjacent cell (y, y-links; bp, base plate or terminal plate, a structure observed within the lumen of the transition zone immediately above the mother centriole). E, a basal body complex (bb), showing the triplet microtubules (t) of one centriole. F. An isolated centriole with visible microtubule triplets (t).
2.2 Freeze-fracture/deep etch/SEM
In recent years the freeze-fracture technique for preparing membrane samples for scanning electron microscopy has fallen into disuse. However, in past decades it provided invaluable information on the surface properties of rod membranes and inter-bilayer boundaries (Andrews and Cohen, 1981; Clark and Branton, 1968; Corless and Costello, 1982; Leeson, 1970; Roof and Heuser, 1982; Roof et al., 1982; Rosenkranz, 1970, 1973; Rosenkranz and Stieve, 1969). Work by Röhlich (Rohlich, 1975) and Matsusaka (Matsusaka, 1974) revealed key features of the ciliary membrane, including arrays of inter- and external membrane particles known as the “ciliary necklace,” a feature found in the transition zone of many primary cilia. Although the existence of these characteristic structures in cilia has been known for more than 40 years (Flower, 1971; Gilula and Satir, 1972), their molecular composition and function remain unknown.
The deep freeze-etch technique (Kajimura et al., 2000; Roof and Heuser, 1982; Roof et al., 1982) has been particularly valuable in revealing extensive connections between disks and between the plasma membrane and disks. Although these have been confirmed by cryo-ET (Nickell et al., 2007) and biochemistry combined with electron microscopy (Molday and Molday, 1987), they are not routinely observed by conventional TEM of ultrathin sections, presumably because of their narrow width and irregular spacing.
Scanning EM (SEM) has the ability to visualize three-dimensional profiles of surface features of cells or fracture faces, which can be correlated with conventional TEM of ultrathin sections to elucidate fine structure in three dimensions. There have been a number of studies of rods by SEM, e.g., (Molday, 1976; Peters et al., 1983; Wensel, 2012), including in animal models of retinal degeneration, e.g., (Cagianut et al., 1985; Lee et al., 2006; May et al., 2005; Ulshafer and Allen, 1984; Zhang et al., 2009).
2.3 Cryo-electron tomography
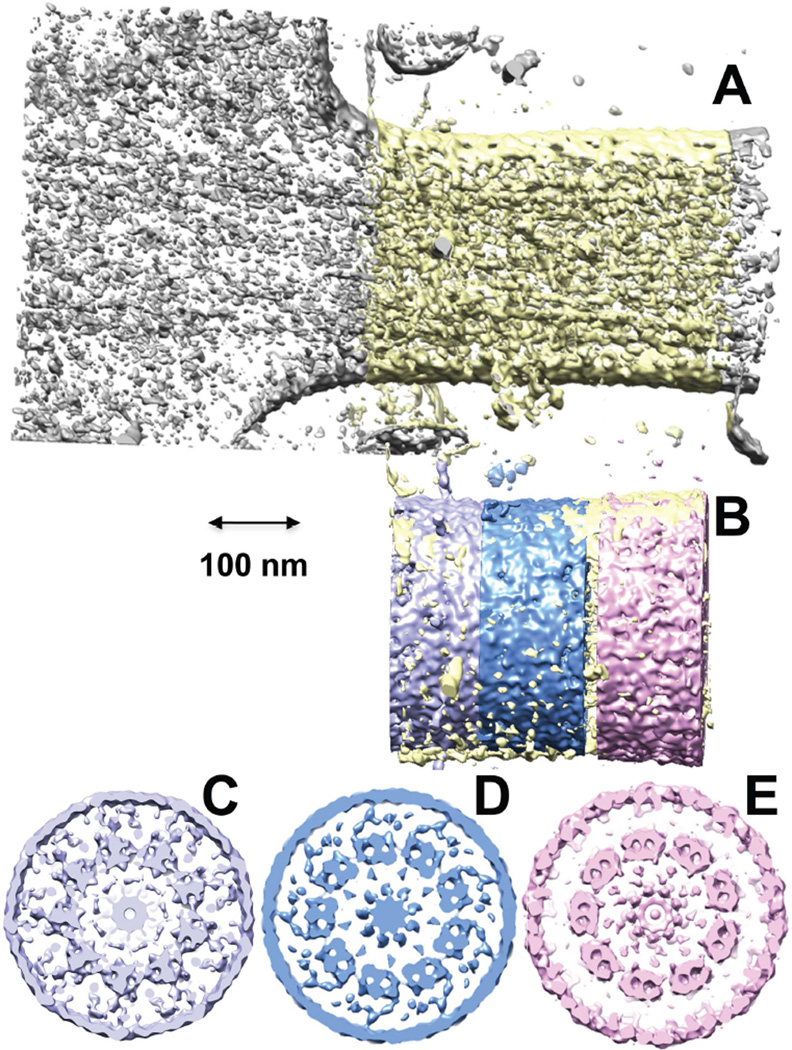
Cryo-electron tomography (cryo-ET) is an emerging technique with the potential to bridge the gap between conventional or super-resolution fluorescence (see below) microscopy and conventional transmission electron microscopy. It also offers the ability to obtain structures of cells that have simply been flash-frozen at speeds that prevent ice crystal formation, and that have not been subjected to the extensive structural distortions that occur when samples are fixed, impregnated with organic solvents, embedded in plastic, sectioned in an ultramicrotome, and stained with heavy metal salts (ref. reviews). To date there have been two cryo-electron tomography studies of rods, one focused on the outer segments and disk structures (Nickell et al., 2007), and the other focused on structures of the distal inner segment, connecting cilium, and proximal outer segment (Gilliam et al., 2012). The latter study included mice with genetic defects causing retinal degeneration with varying degrees of severity, along with wildtype mice. Examples of cryo-ET data from this study are shown in Fig. 3. Both studies revealed clearly a wealth of structural detail on the nanometer scale, and provided unprecedented views of rod structure in three dimensions. For example, sparsely distributed large structures connecting adjacent disks were observed for the first time (Nickell et al., 2007), as were vesicles within the lumen of the connecting cilium (Gilliam et al., 2012). These structures are presumably difficult to observe in ultrathin sections because of the low probability of capturing them in any one section, whereas the availability of three-dimensional density profiles in tomograms enhances the ability to observe them.
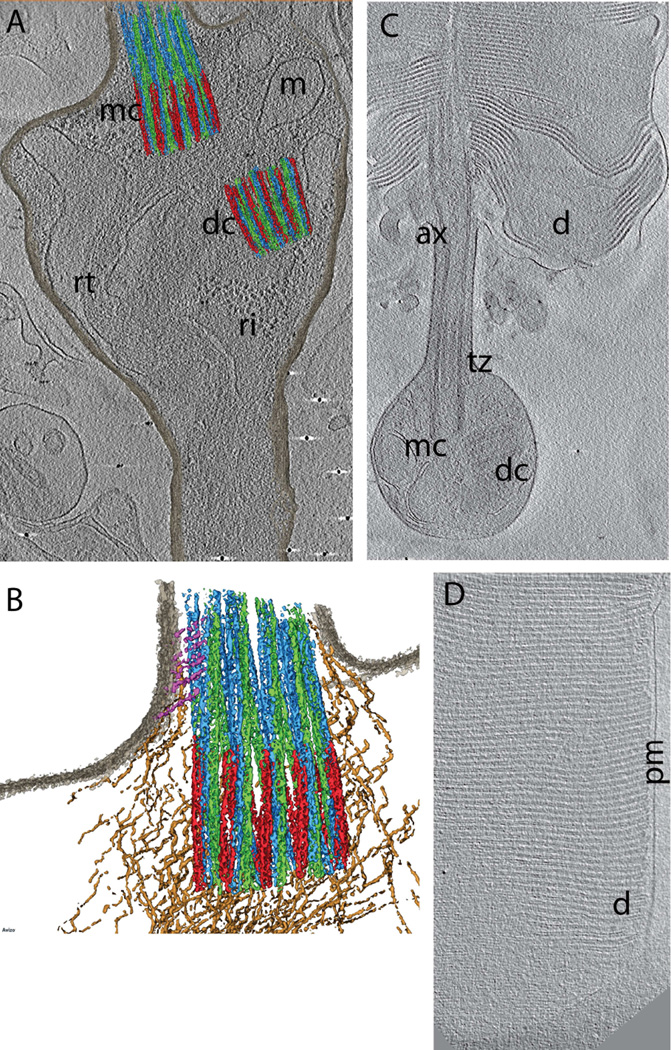
Figure 3.
Cryo-ET data from WT mouse rods (Gilliam et al., 2012). A. Projection image through slice of tomogram (grayscale) with overlaid segmented isosurfaces (color) showing the basal body complex (mother centriole, mc; daughter centriole, dc), the base of the connecting cilium/transition zone, with segmented a, b, and c microtubules (colored red, green and blue, respectively), and adjacent intracellular fibers and organelles, including mitochondria (m), electron-dense ribosomes (ri), and ciliary rootlet (rt). B. Partially segmented image, highlighting the a, b, and c microtubules (colored red, green and blue, respectively), and fibers (tan and magenta) connecting the microtubular structures to one another (tan) and to ciliary and cellular membranes (magenta). C. Slice through a tomogram of a rod cell fragment. The outer segment has been isolated from most of the inner segment, and the inner segment membrane has resealed to contain the mother (mc) and daughter (dc) centrioles as well as numerous membrane vesicles. The outer segment is flattened, revealing the disks (d) emanating from the axoneme (ax) above the transition zone (tz). D. Slice through a tomogram showing the highly ordered stack of disks (d) on the edge of a rod outer segment, and their relationship to the OS plasma membrane (pm).
2.3.1 Sub-tomogram averaging
The technique of sub-tomogram averaging, in which portions of maps containing repeating features within a single specimen, or between similar specimens, are combined to improve signal-to-noise, has been used to reveal a remarkable level of detail in maps of axonemes and basal bodies from motile cilia (Gilliam et al., 2012; Koyfman et al., 2011; Li et al., 2012; Nicastro et al., 2011; Song et al., 2015). Figs. 4 and 5 show some promising early results from our group using data acquired as described (Gilliam et al., 2012; Wensel and Gilliam, 2015) and internally averaged using the nine-fold symmetry of the microtubule bundles.
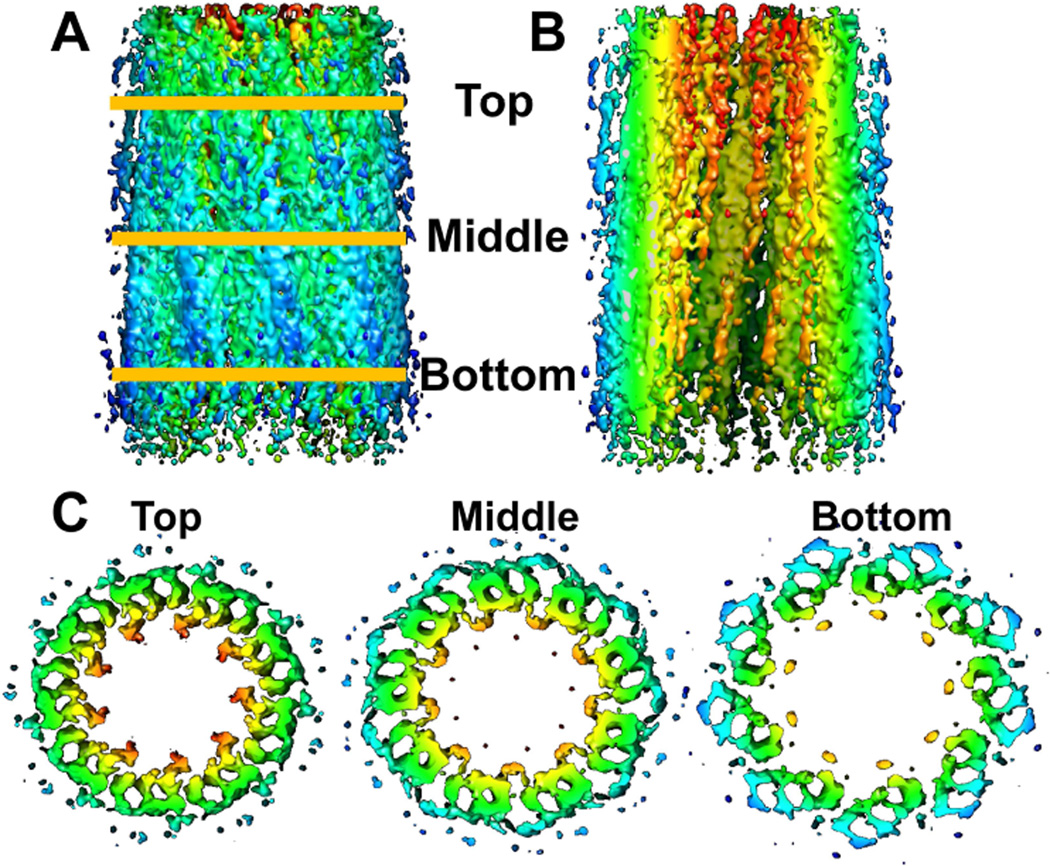
Figure 4.
Enhancement of cryo-ET reconstruction of a daughter centriole by sub-tomogram averaging. A. External view perpendicular to the central axis; B. cut-away view with outer half removed to reveal internal structures; C. Cross-sectional views of slices taken at the indicated axial positions. Colors indicate distance from central axis, orange (closest) to blue (furthest).
Figure 5.
Reconstruction of a transition zone/connecting cilium base from cryo-electron tomography (Cryo-ET) data, with sub-tomogram averaging used to enhance signal-to-noise. A. Tomogram with segmented region highlighted in light yellow. B. segmented version with color-coding of axial displacements; C-E, cross-sectional views of the maps resulting from sub-tomogram averaging of the nine microtubule doublets and membranes, with axial displacement of each section color-coded as in B.
2.3.2. Combining focused ion beam milling of frozen samples with cryo-electron tomography
In focused ion beam milling, a fixed or frozen sample of tissue can be precisely trimmed using a focused ion beam as is available in a number of scanning electron microscopes. The sample can be cut to a thickness (< 500 nm) that makes it amenable to TEM. An initial demonstration of the application of focused-ion-beam milling with more conventional SEM of heavily fixed retina showed great promise for elucidating rod cell structure (Mustafi et al., 2011), but suffered from the defects, e.g., in disk spacing, observed in other samples fixed and stained for conventional TEM. These results suggest that combining ion beam milling of samples subjected to high pressure freezing with cryo-ET (Villa et al., 2013) or with serial scanning electron microscopy using detection of secondary electrons (Villinger et al., 2012) will add substantially to our understanding of rod structure in normal and diseased retinas.
2.4 Subcellular localization with antibodies: immuno-EM and immunofluorescence
The use of staining with antibodies allows combining structural imaging with identification of molecular components of structure. There have been multiple studies applying immuno-electron microscopy to localize antigens in rods, e.g. (Arikawa et al., 1992; Arikawa and Williams, 1991, 1993; Chaitin, 1992; Chaitin and Burnside, 1989; Chaitin et al., 1988; Chaitin and Coelho, 1992; Kachi et al., 1999; Liu et al., 1997; Maerker et al., 2008; Mangini and Pepperberg, 1988; Moritz and Molday, 1996; Nir and Papermaster, 1986; Roof et al., 1991; Sedmak et al., 2009; Sedmak and Wolfrum, 2010; Vigh-Teichmann et al., 1986; Wolfrum, 1995; Wolfrum and Schmitt, 2000). However, this technique is technically challenging and often somewhat limited in information content due to the small number of immuno-labelled particles visualized in a given area of an ultrathin section. Sparsely labeled antigens may have densities comparable to those of background signals. Conventional fluorescence has much poorer resolution, but offers the ability to visualize distributions of samples over large volumes and areas. An early study of dissociated amphibian rods with monoclonal antibodies (Witt et al., 1984) revealed the uniformity of distribution (at the resolution of optical microscopy) of rhodopsin and the phototransduction G protein, transducin. The advent of widely available confocal and multiphoton microscopy greatly enhanced the information content of immunofluorescence images, because of the enhanced resolution and capability of optical sectioning. One of the earlier applications of this technique to rods revealed numerous microtubules radiating away from the basal body and the cilium in frog rods (Hale and Matsumoto, 1993), and a study of effects of a disease-causing mutation on sub-cellular distribution of rhodopsin (Sung et al., 1994) marked the beginning of an era in which application of confocal and multiphoton immunofluorescence has become standard for the characterization of antigen distributions both in both wildtype retina, and in disease models. Around the same time, confocal immunofluorescence was used to localize rab GTPases, important regulators of ciliary trafficking, within the rod (Deretic et al., 1995; Deretic and Papermaster, 1993).
However, these techniques are restricted in resolution by the width of the Airy disk representing the diffraction limit for conventional optical microscopy, so that practically speaking, features less than 300–500 nm apart are not usually resolved. Thus, for example, the determination of sub-cellular localization within the connecting cilium, whose width essentially corresponds to a single pixel in such images, is very challenging with these techniques.
2.5 Super-resolution fluorescence
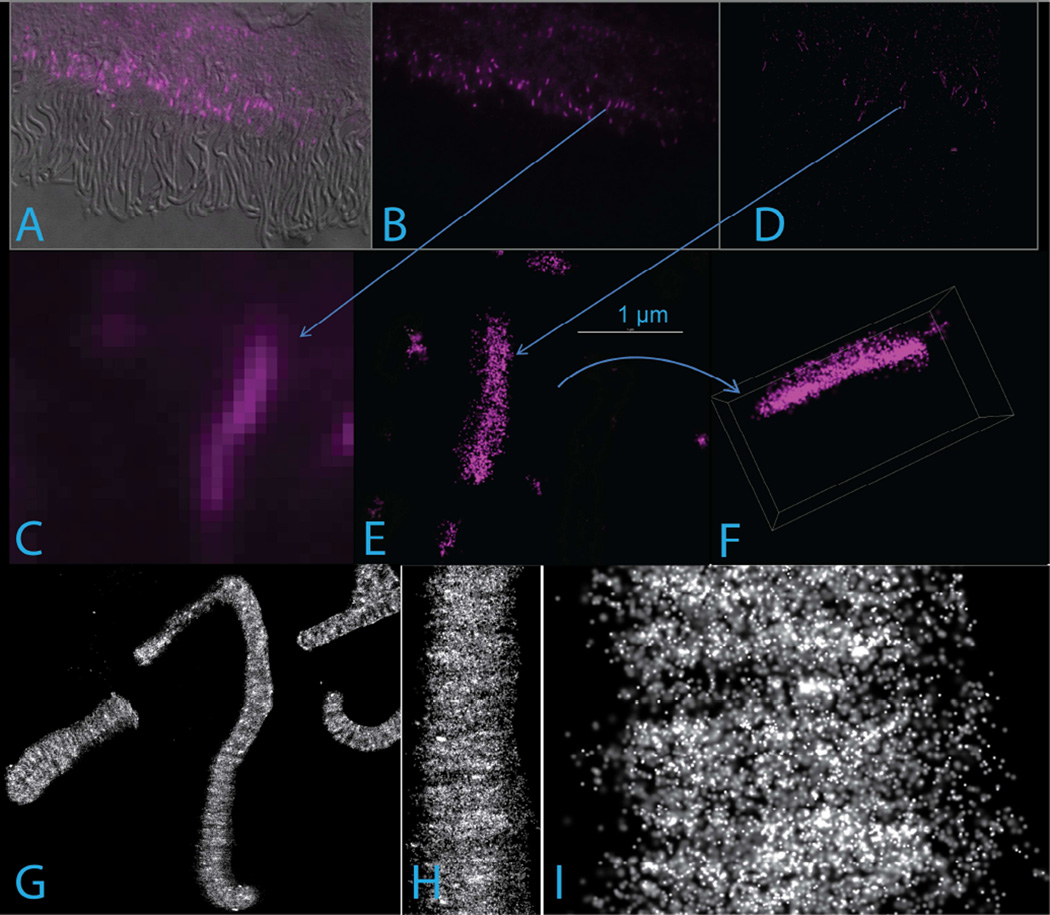
In 2014, the Nobel Prize in Chemistry was awarded to three physicists, Stefan Hell, William Moerner, and Eric Betzig, who had contributed to the development of new forms of optical microscopy that yield resolution well below the diffraction limit which had previously limited even confocal and multiphoton microscopies to resolutions of hundreds of nanometers. This limitation has hindered our ability to locate specific molecules within the small substructures of great importance to photoreceptor biology: post-Golgi vesicles, endosomes, lysosomes, centrioles, the axoneme, the transition zone, etc. Among the most promising of the new imaging technologies are those that fall under the general heading of Single Molecule Localization Microscopies. The most popular versions of these are known by other acronyms: STORM, dSTORM, PALM. All involve using light to switch molecules which are essentially non-fluorescent within a certain wavelength range, into brightly fluorescent states, and then using light to switch them off again. If the excitation intensity and detection optics and camera are sufficient for single molecules to be imaged reliably, then collecting many thousands of images during cycles of sparse activation, imaging, and inactivation makes it possible to build up maps of molecule locations with resolutions reaching 20 nm to 30 nm. This approach has great promise for analyzing rod structure, but such applications have yet to be published. In our laboratory, we have confirmed that labeling of fixed, permeabilized, isolated rods with secondary antibodies suitable for the dSTORM technique can yield images of rod disk membranes (rhodopsin antibodies) and the connecting cilium (centrin 2 or acetylated tubulin antibodies) with resolution approaching 20 nm (Fig. 6). A recent 4-color 3D STORM study of synaptic input fields in retinal interneurons confirmed the suitability of mouse retinal samples for such studies (Sigal et al., 2015).
Figure 6.
Super-resolution STORM images of isolated mouse rods. A-F. Centrin 2 immunofluorescence in mouse retinal sections imaged by wide-field fluorescence (A–C) and differential interference contrast (DIC, A), or 3D STORM (D–F). Arrows show the CC in the low-magnification images (B, D) corresponding to the higher magnification views (C, E). The 3D map whose projection is shown in E has been rotated to produce the projection in F, illustrating the nearly isotropic resolution achieved. Each dot in E and F represents a single localization event, corresponding to individual molecule fluorescence. G-I, STORM images at varying magnifications showing rhodopsin immunofluorescence in isolated mouse rods using monoclonal antibody 1-D4, which recognizes the C-terminus of rhodopsin.
Animals expressing photo-switchable fluorescent protein fusions suitable for PALM have also been described (Gross, 2014; Lodowski and Imanishi, 2015; Lodowski et al., 2013), suggesting another approach likely to be successful. Although working out the proper conditions for single molecule imaging of tissue samples is far from trivial, and almost no super-resolution results for such samples from retina have been reported to date, it is to be expected that there will be a flurry of them in the near future.
2.6 Correlative light and electron microscopy
The great advantage of fluorescence microscopy is that it can be used with antibodies or with genetically encoded fluorescent fusion proteins to determine sub-cellular localization of specific molecules, with greater reliability and sensitivity than immuno-electron microscopy. However, even with super-resolution fluorescence techniques, the resolution obtainable is much poorer than can be obtained by electron microscopy in the form of cryo-ET or conventional TEM. The ideal way to bridge this gap would be to carry out fluorescence and electron microscopy on the same sample. Application of correlative PALM and cryo-ET to bacterial structure has been described (Chang et al., 2014), and it is likely only a matter of time before this approach is applied to rods. New alternative approaches for molecular localization at the EM level include use of miniSOG (mini Singlet Oxygen Generator, a fluorescent flavoprotein engineered from Arabidopsis phototropin 2) fusions, which facilitates visualization both by fluorescence and EM (Shu et al., 2011), and the use of SNAP tag fusions, which can be used to couple to either fluorescent probes or gold particles, as has been described for motile cilia (flagella) of Chlamydomonas reinhardtii (Song et al., 2015).
3. Genetic approaches to understanding the molecular basis of structure and disease
In recent years forward and reverse genetics have become very powerful tools for understanding the molecular basis of rod structure and diseases affecting rod structure and function. In the early days of genetic studies of vision, investigators were limited to studying spontaneously occurring blinding mutations in in-bred animal models, primarily rodents, but also dogs (Aguirre, 1973; Aguirre et al., 1982; Suber et al., 1993; Wrigstad et al., 1994) and chickens (Semple-Rowland et al., 1998). Important early models included the rd1 and rds mice and the RCS rat (LaVail, 1981), but the causative molecular defects in these disease models were not uncovered until gene mapping and DNA sequencing technology had advanced to the point that a mutation in a novel protein, peripherin/rds, could be identified as responsible for disease in rds (Travis et al., 1989) and a mutation in the gene encoding the PDE6 β subunit was found to be responsible for the defect in rd1 (Bowes et al., 1990; Bowes et al., 1993; Pittler and Baehr, 1991). These advances were followed by many others including identification of a Mertk mutation in the RCS rat (D’Cruz et al., 2000). Relevance of these animal models to human disease was underscored by the discovery of disease causing mutations in the human homologues MERTK (Gal et al., 2000) and RDS (Kajiwara et al., 1991). These three models highlight four recurring themes in proteins whose defects cause rod or rod/cone dystrophies: phototransduction proteins (rd1), proteins important for rod cell structure (rds/peripherin, important for structure of disk rims), proteins that cause defects in membrane trafficking or recycling (Mertk, whose principal functional defect seems to be in disk phagocytosis by RPE), and RPE-resident proteins important for maintaining rod health (Mertk). These and additional recurring themes in rod cell defects are discussed in more detail below.
As genetic engineering technology has progressed, a more common approach to animal models for studying disease mechanisms, and for studying the functional roles of specific proteins, has become the use of transgenic, knock-out, or knock-in animals, principally mice (Huang et al., 1993; Kedzierski et al., 1997; Naash et al., 1993; Olsson et al., 1992; Portera-Cailliau et al., 1994), supplemented in a few instances by frogs (Haeri and Knox, 2012; Hollingsworth and Gross, 2013; Lee et al., 2012; Lin-Jones et al., 2003; Moritz et al., 2001; Tam and Moritz, 2006, 2007; Tam et al., 2014), rats (Gorbatyuk et al., 2012; Kroeger et al., 2014; LaVail et al., 1998; Shinde et al., 2012), or pigs (Ross et al., 2012), to create engineered defects. With the dramatic recent increases in throughput and decreases in cost of sequencing human exomes and genomes, human patients are now the most important experimental system for identifying naturally occurring disease-causing mutations. The paradigm has shifted to one in which the mutations initially found in humans are modeled by design in mice. With the advent of a new generation of genome editing tools (Wijshake et al., 2014) such as zinc-finger nucleases (Chou et al., 2012; Jabalameli et al., 2015; Overlack et al., 2012; Pittler et al., 2013; Sandoval, 2012; Sasson and Kelleher, 2014), the CRISPR/Cas9 system (Pelletier et al., 2015; Sander and Joung, 2014), and TALENS (Sommer et al., 2015) (transcriptional activator-like effector nucleases), the generation of such animals is becoming even more efficient. An important use of existing and new animal models is to apply structural methods, as well as biochemical and physiological analyses, to gain insights into structural and functional consequences of mutations, and to understand mechanisms of cell death.
4. Structural Organization of the Rod
The emphasis in this section is on recent results from cryo-electron tomography and from gene ablation studies combined with more conventional imaging techniques. These are considered in light of previous studies using a wide range of techniques.
4.1 Outer Segments
The most characteristic feature of rod cells, which gives them their name, is a long, and, especially in mammals, thin cylindrical rod-like structure, the rod outer segment. This is the principal, if not exclusive, light-sensitive organelle in the rod. It is perhaps best described as a highly modified primary cilium (Horst et al., 1990; Pazour and Bloodgood, 2008; Roepman and Wolfrum, 2007), arising from the distal end of a ciliary transition zone, known as the connecting cilium (see below). An axoneme, consisting of a 9 + 0 bundle of doublet microtubules at its base, runs along its length, but the number of microtubules in the axoneme diminishes toward the distal end as doublets transition into a 9 + 0 arrangement of single microtubules. Attached to the axoneme and enclosed by the plasma membrane is a highly ordered stack of disk membranes.
The length and width of the outer segment, as well as the number of disks in each, vary widely among vertebrates. Mammals have some of the smallest rod outer segments, with a mouse rod outer segment typically measuring 1.4–1.5 µm in diameter and 24–32 µm in length (Carter-Dawson and LaVail, 1979; Price et al., 2012), and the rod of the mudpuppy, Necturus, has outer segments with diameters of 10–13 µm, and a length of 30 µm (Brown et al., 1963). In amphibians, the disk membranes have deep incisures dividing the disks into lobes, whereas mammalian disks typically feature a single incisure. Despite the wide range of volumes, the longitudinal dimensions and spacings of the disks membranes are relatively constant across species, for example, 37 per µm in Necturus (Brown et al., 1963) and 32 per µm in mouse (Gilliam et al., 2012). The mechanism by which this spacing is determined is uncertain, but it is interesting to note that a spacing of 1/32 nm is an integral multiple of the tubulin dimer repeat distance of 1/8 nm. Cryo-ET (Nickell et al., 2007) and freeze-etch/SEM (Rohlich, 1975; Roof and Heuser, 1982; Roof et al., 1982) have revealed multiple connections of unknown composition between adjacent disks, between disk and plasma membrane, and between disks and axoneme.
Functionally, the outer segment is responsible for phototransduction, that is, for the detection of light, and for the conversion of the signal inherent in photon capture to a highly amplified electrical signal, which is converted into an extracellular chemical signal at the synaptic terminus. To carry out this function, the outer segment is packed with high concentrations of the molecules essential for the phototransduction cascade. The most abundant protein, rhodopsin, is present in disk membranes at densities of about 25,000 µm2, and in the plasma membrane at about half that density. Thus rhodopsin accounts for a little less than half of the mass of outer segment membranes, with the rest consisting primarily of phospholipids and cholesterol. It must therefore be considered as having a structural role, as well as serving as the light receptor and activation of the G protein cascade of vision, and this role has important implications for the effects of rhodopsin mutations on rod structure. The biochemistry of the phototransduction cascade, which converts a light signal, photoisomerization of rhodopsin, into an electrical signal, decreased inward current and membrane hyperpolarization, has been reviewed previously (Arshavsky and Burns, 2012; Arshavsky et al., 2002; Burns and Arshavsky, 2005; Gross and Wensel, 2011; Wensel, 2008, 2012).
4.1.1 The structure of basal disks in the rod
The standard model for rod disk formation starts with an evagination of the plasma membrane at the distal end of the connecting cilium, which then folds inward to form one or more basal disks whose lumens are continuous with the extracellular space. As the disks mature, it is thought that they somehow become pinched off so that their lumens are osmotically intact, and the disks are “free-floating”- although it is generally understood that there are many connections between disks and between disks and plasma membrane and microtubules. A number of lines of evidence support this idea, or, more specifically, the idea that one or more basal disks have membranes continuous with the ciliary plasma membrane, and lumens continuous with the extracellular space. First, in cold-blooded species, it has been reported that low molecular weight dyes can penetrate the rod at its base (Laties et al., 1976) Second, numerous electron micrographs collected in many laboratories over the years display basal disks without surrounding plasma membrane, and more distal disks clearly enveloped by an external membrane.
However, there have been reports that challenge this view with respect to mammalian rods, including results from conventional electron microscopy and cryo-electron tomography, which show basal disks predominantly enveloped by an external membrane (Chuang et al., 2007; Gilliam et al., 2012). The lumen of the basal disks was not accessible to ferritin-labeled concanavalin A (Gilliam et al., 2012), but this result could simply reflect the large size of this probe as compared to the width of the intradiskal space. However, an earlier study by conventional EM (Cohen, 1968, 1970) found that lanthanum nitrate, a probe of much smaller size, also did not appear to penetrate the intradiskal space in rods as it did in cones, and in the space between the distal inner segment plasma membrane and the basal outer segment membrane. In studies of monkey, pigeon, rat, and mouse retina, lanthanum infiltration was never observed (Cohen, 1970). However, it was also noted that the region surrounding the axoneme in rods whose plasma membrane had been disrupted allowing lanthanum access to the cytoplasm (but not the disk lumen) was also devoid of lanthanum staining. This result suggests that diffusion barriers other than bilayer membranes, such as gels, could block access to this ion in regions of the cell despite their continuity with accessible regions.
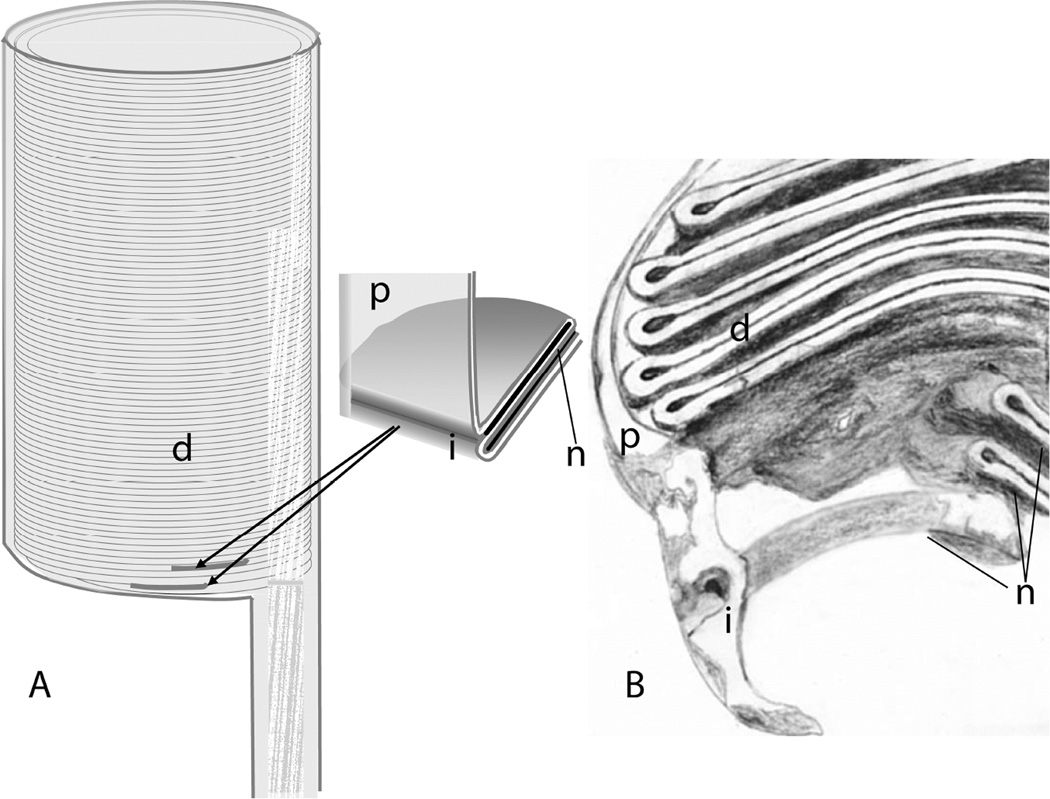
Additional evidence for transient or prolonged continuity between the lumen of basal disks and the extracellular space comes from experiments with fluorescent dyes. Unfortunately, there is some ambiguity with regard to the dye penetration results as well (Laties et al., 1976). Convincing evidence was presented that in rods from the lizard Gekko gekko, dye injected into the vitreous was rapidly incorporated into the lumen of basal disks, and then subsequently became trapped and migrated toward the distal tip as the disks matured. In the same work, only basal staining of mudpuppy rods was presented, which could have reflected penetration of the region between inner and outer segments, and results with mammals did not provide convincing evidence of consistent dye penetration. Gekko gekko may be a special case. The rods of this species appear to have differentiated from cones rather recently, on an evolutionary time scale, and at the molecular level resemble cones more than they do rods (Zhang et al., 2006). Thus, their ultrastructure may not be representative of vertebrate rods in general. In a study with Xenopus laevis rods (Kaplan et al., 1982), intravitreal injection with Lucifer yellow on successive days led to fluorescent bands whose distance from one another varied linearly with the time between injections, suggesting that at some point during disk formation and maturation, the lumen of basal disks is open to the extracellular space, even if only transiently. Both basal disk staining and distal disk staining by Lucifer yellow were observed in Xenopus by Matsumoto and Besharse (Matsumoto and Besharse, 1985). However, some Xenopus rods express cone-like pigment, and it is not clear that all of them have morphologies representative of rods in general, or of those in mammals. Thus, the issue of basal disk structure and mechanism of disk morphogenesis remains an open one requiring additional study. A model consistent with available data but requiring further testing is shown in Fig. 7. The proposal is that during disk morphogenesis the most basal disk (s) is (are) continuous with the plasma membrane and as the disk forms (either through invagination or evagination of the plasma membrane), the lumen of the disk is open to the extracellular space via a small opening, through which diffusion is limited. This opening may be transient, or may persist even after additional disks begin to form so that the membranes of the first few most basal disks may all be continuous with the outer segment plasma membrane. Two recent electron tomography studies using conventionally prepared tissue sections support this model (Burgoyne et al., 2015; Volland et al., 2015). Readers are advised to consult these papers and the accompanying on-line material such as animations to visualize these complex membrane structures.
Figure 7.
A model for the topology of basal disk (s) of mammalian rods. A model consistent with available data proposes that all disks, including the ones nearest to the connecting cilium, are enveloped by the plasma membrane, but that there is continuity with the plasma membrane with the first disk or first few disks, with only a very narrow entry-way into the lumen of the basal disk (s), which would be continuous with the extracellular space. A. Schematic of outside view of basal outer segment, with disks visible through translucent plasma membrane. B. Schematic representation of a section through a tomographic map of an outer segment based, derived from results reported in (Volland et al., 2015). Labeled structures are i, invaginations of plasma membrane; n, nascent disks; d, mature disks, p, plasma membrane
In any such model, a process of separation from the plasma membrane and sealing of the disk rims must take place, and this process must involve protein complexes similar to those involved in other membrane fusion and fission events. Their identities are currently unknown, but it seems likely that some may be rod-specific, as a similar process is unnecessary in cones. Peripherin-2/rds and rom1, tetraspanin proteins localized to disk rims, have been proposed to play a key role in membrane fusion during disk morphogenesis (Boesze-Battaglia et al., 2003; Boesze-Battaglia et al., 1997; Boesze-Battaglia et al., 1998; Boesze-Battaglia et al., 2000), but molecular details of this process remain unclear. A C-terminal amphipathic helix in peripherin-2/rds has been shown to promote tight membrane curvature (Khattree et al., 2013). Prominin1 (CD133) has been localized to membrane protrusions and the bottom of the outer segment, and it has been proposed to play a role in membrane bending (Corbeil et al., 2001; Han et al., 2012).
4.2 Connecting cilium and associated structures
The connecting cilium is a cylindrical structure approximately 300 nm in diameter (Gilliam et al., 2012) and consists of a 9 + 0 bundle of microtubules surrounded by a ~50 nm layer of cytoplasm connecting the inner and outer segments. It has become conventional wisdom among those studying rod structure that the proximal region of this structure is equivalent to the transition zone found in primary cilia from other cells. This structure is immediately adjacent to the basal body from which the axonemal bundle of doublet microtubules emerges (Horst et al., 1987). Within the lumen of the connecting cilium, particles of irregular shape have been observed by cryo-ET (Gilliam et al., 2012), consistent with classic observations by conventional TEM of amorphous material of lower electron density than the microtubules. The molecular composition of these particles is unknown, but they may contain centrin isoforms, which have been reported to stain the lumen of the connecting cilium (Drivas and Bennett, 2014; Drivas et al., 2013; Giessl et al., 2006). This observation has recently been confirmed in our laboratory by super-resolution 3D STORM fluorescence microscopy (M. A. Robichaux and T. G. Wensel, unpublished observations and Fig. 6), which reveals a structure within the connecting cilium less than 200 nm in diameter staining with antibodies specific for centrin 2.
A number of discrete structures within the transition zone have been described for other ciliated cells. The structures in the vertebrate rod appear to be similar but not identical. Transition fibers, structures connecting the microtubules at the junction between the mother centriole and the axoneme to the plasma membrane, have been described as a general feature of cilia. These are generally depicted as triangular weather vane-like structures, with one base of a right triangle aligned along the side of the axoneme or basal body, with the opposite angle attached to the plasma membrane, and are sometimes referred to as “alar sheets” rather than as “fibers”. These have a pinwheel appearance in ultrathin sections. The molecular composition of these structures is unknown Fibers leading from the microtubules of the axoneme to the plasma membrane near the base have been observed in cryo-electron tomograms of mouse connecting cilia (Gilliam et al., 2012). However, triangular weather vane-like structures were not observed. Evidence of these pinwheel vanes in published conventional TEM of rods is rare. However, evidence for connectors or appendages of triangular profile are clearly evident in some studies, e.g. from immature ferret retina (Greiner et al., 1981) and from developing (Sedmak and Wolfrum, 2011) or mature (Li et al., 2015) mouse rods. It is possible that these structures are difficult to visualize in mammalian rods because they are quite small and must be sectioned in a narrow range of angles to be observed, so they may not have been sampled frequently. It is also possible that they do not appear clearly in cryo-tomograms because they have low electron density, but are visible upon staining because they have an affinity for the heavy metal staining material. Their presence and function in rod cells are important, because they have been proposed to limit the size of vesicles that can enter the cilium (Nachury et al., 2010). Indeed, experiments with antibody entry into cultured cells whose plasma membranes, but not ciliary membranes, had been permeabilized by digitonin, revealed that even soluble proteins of diameter > 9 nm are blocked from entry, although diffusion kinetics within the cilium are similar to those in the cytoplasm (Breslow et al., 2013). In any case, their molecular composition is unknown.
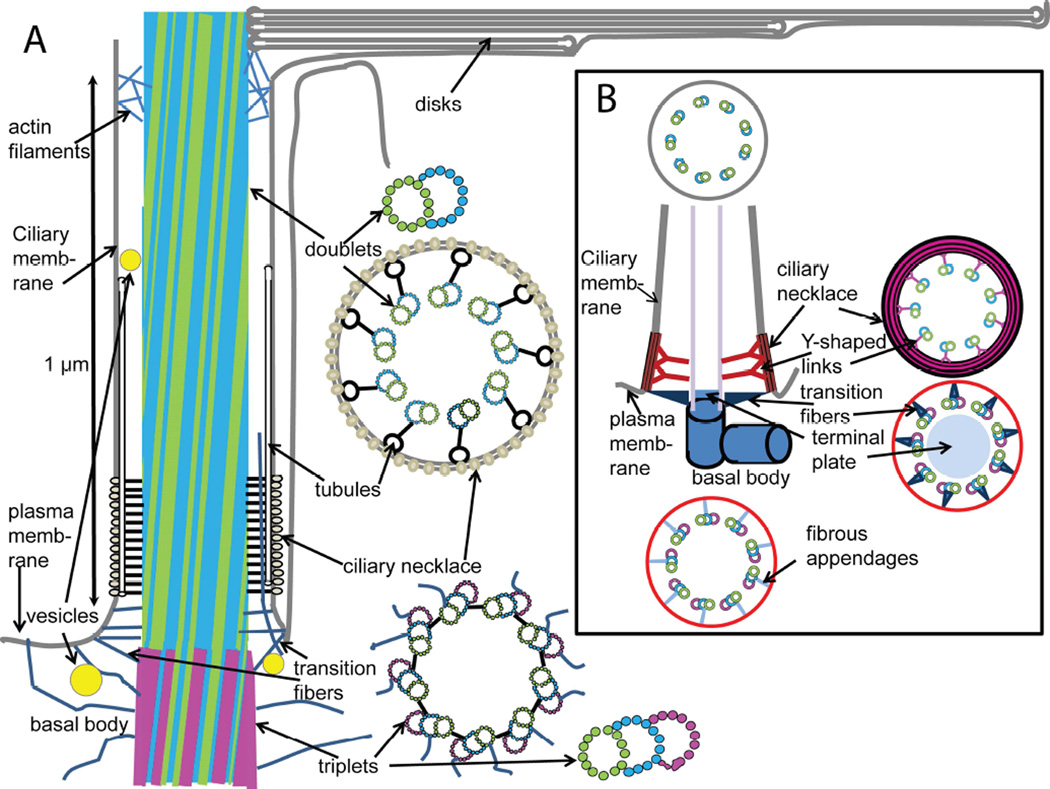
In the transition zone of typical cilia, additional structures connecting the microtubules and plasma membrane are described as “Y-shaped links,” and these have been observed in electron micrographs of cross-sections through the proximal region of the rod connecting cilium (Besharse et al., 1985; Horst et al., 1987) (see also Fig. 2D). In the three-dimensional maps provided by cryo-ET, a somewhat different picture emerges (Gilliam et al., 2012). The tomograms reveal regularly spaced fibers connecting the microtubules to tubular structures adjacent to the plasma membrane and extending along the axis of the axoneme. When the proximal region of the connecting cilium is sectioned perpendicular to the axoneme axis, and through the connecting fibers, the profile appears as the familiar Y-shape. Interestingly, an earlier study of serial sections by conventional TEM of mammalian rods (Tokuyasu and Yamada, 1959) came to a similar set of conclusions: the authors reported fibers leading from the microtubules to tubular structures running the length of the plasma membrane at the base of the connecting cilium, and fibers, but not weather vane-shaped structures connecting the basal body and base of the connecting cilium to the plasma membrane. A model for the structure of the connecting cilium and adjacent structures is depicted in Fig. 8, along with a standard model of a primary cilium (Szymanska and Johnson, 2012).
Figure 8.
Alternative models of basal body and connecting cilium structure based on cryo-electron tomography of isolated mouse rods. Panel A shows a model for the structural organization of the regions of the rod cell adjacent to the connecting cilium, based on available structural data, largely from various EM techniques. Distinctive features include: intersection of fibers emanating from the axoneme with tubules running along the length of the transition zone giving rise to the characteristic “Y-shaped links” in cross section; absence of clearly-defined triangular “alar sheet” transition fibers, and presence of multiple filaments connecting the microtubules to the plasma and ciliary membranes; vesicles adjacent to and within the lumen of the connecting cilium; filaments extending from the cytoplasm into the cilium lumen; thinning of basal body from minus (triplet) to plus (doublet) end; slight tilt of microtubules with respect to ciliary axis; envelopment of basal disks within plasma membrane (not necessarily to exclusion of some regions of continuity; see text). The arrangement of tubulin dimer units (circles) in doublet microtubules is based on the structure determined by cryo-ET and sub-tomogram averaging of axonemes from Chlamydomonas reinhardtii flagella (Nicastro et al., 2011), and that of the triplet microtubules is based on the basal body structure determined using similar approaches to basal bodies from the same organism (Li et al., 2012). The arrangement of the doublet and triplet microtubules is based on cryo-electron tomography and sub-tomogram averaging of mouse rod centrioles (Figs. 3 and 4 and Zhixian Zhang, Michael F. Schmid, Feng He, Theodore Wensel, unpublished observations). At the base of the outer segment, actin filaments are found within the lumen of the axoneme, and extending out between microtubules to contact the disk membranes (Chaitin and Bok, 1986; Chaitin and Burnside, 1989; Chaitin et al., 1984). Not shown is the daughter centriole invariably associated with the depicted structures. Panel B depicts a standard model of primary cilia, based on reference (Szymanska and Johnson, 2012).
It has been proposed that CEP290, also known as NPHP6 or BBS14, forms the Y-shaped links in rod cells (Drivas and Bennett, 2014; Drivas et al., 2013). The evidence for this proposal is far from conclusive, and does not include electron microscopy, although it is supported by electron microscopy of mutant Chlamydomonas reinhardtii (Craige et al., 2010). Conflicting with this proposal are reports that CEP290 localizes to the basal body, or centriolar satellites, and that it co-localizes with the centriolar satellite protein, PCM-1 (Kim et al., 2008). However, consistent with this proposed role in the Y-shaped links are reports of localization to the base of the connecting cilium (Chang et al., 2006; Cideciyan et al., 2011; McEwen et al., 2007; Murga-Zamalloa et al., 2010; Rachel et al., 2012). CEP290 has been reported to interact directly with and co-localize with a number of transition zone/connecting cilium proteins, including RPGR, RGRIP1, NPHP5, AHI1, CCD2a, and RKIP/PEBP1 (Murga-Zamalloa et al., 2011). It has also been reported to interact directly with and co-localize with a number of centriolar/basal body and pericentriolar proteins, including PCM-1, MKK5/BBS6, rab8a, and γ tubulin (reviewed in (Rachel et al., 2012)).
An obstacle to determining the precise structural elements formed by CEP290 is that the structures involved have dimensions and distances from one another on the order of 300 nm or less, which is near to or beyond the practical resolution for conventional fluorescence microscopy. Combining super-resolution immunofluorescence with electron microscopy and cryo-ET could likely help to resolve this question. A recent study of cilia in cultured human retinal pigment epithelial cells (RPE-1) by Stimulated Emission Depletion (STED) fluorescence microscopy, a super-resolution technique, shows promise in this regard (Yang et al., 2015).
4.2.1 The BBSome
One of the most interesting cilium-associated complexes is composed of, or regulated by, the protein products of the BBS genes, mutations in which give rise to Bardet-Biedl syndrome. The structures and roles of the BBSome and its component proteins remain to be clearly determined. There is convincing evidence that seven BBS gene products, (BBS1, 2, 4, 5, 7, 8, and 9) form a complex that binds tightly to Arf-like GTP binding protein, Arl6 (BBS3), in its activated (GTP-bound) state, and can be isolated by affinity chromatography, size exclusion chromatography, cation exchange chromatography, or density gradient ultracentrifugation (Jin et al., 2010; Nachury et al., 2007). There is also evidence that upon addition to synthetic lipid vesicles, this complex, together with Ar1l6 forms a membrane coat (Jin et al., 2010). However, it is not clear what fraction of the cellular pool of each BBSome protein is typically present in the complete complex, and whether there are functions for some of them that do not depend on participation in BBSome function. Although all subunits co-migrated upon size fractionation (Nachury et al., 2007), the complexes analyzed had already been selected for BBSome assembly by affinity purification. It is also not clear where BBSomes are localized, as opposed to localization of individual subunits which are found in multiple locations, some overlapping and some not, and if such membrane coats form in vivo, where they are, what their lipid composition is, and what their function is. The fractionation and co-purification behavior of the BBSome “core complex” subunits suggest that there may be sub-complexes, missing one or more components. How these might differ functionally is unknown, but it is clear from human genetics that absence or malfunction of any of them is detrimental. More precise localization and co-localization experiments, especially those capable of higher spatial resolution, and combining improved antibodies with expression (but preferably not over-expression) of tagged proteins should help to clarify these issues in the near future.
Part of the difficulty in answering these questions lies in experimental shortcomings. One of these is a dearth of antibodies with well-documented specificity not only for immunoblots, but also for immunostaining and immunoprecipitation. This problem exists for many proteins important for ciliary structure or function. This deficiency leads to ambiguity in many studies aimed at localizing endogenous BBSome subunits within photoreceptors. Frequent attempts have been made to overcome this problem by localizing heterologously expressed subunits with tags, but these suffer from ambiguities in interpretation resulting from the artifacts caused by over-expression and from unknown effects of tags. These problems are further compounded by the fact that most such studies are carried out in immortalized cell lines which produce cilia in G1 or G0 phases of the cell cycle, but whose ciliary structures and mechanisms may differ from those in post-mitotic neurons such as rod cells. An instructive example of cell-type specific differences is the finding that mutations in a photoreceptor-specific splice variant of BBS8 leads to non-syndromic retinopathy (Riazuddin et al., 2010).
In addition to forming a complete BBSome, a number of the BBSome subunits are capable of forming subcomplexes with two or more partners. Upon co-expression, binary complexes formed included nearly stoichiometric complex of BBS2 with BBS9, and of BBS9 with BBS8. Additional binary complexes of moderate strength were formed between BBS9 and BBS1, BBS2, BBS4, and BBS5 (Nachury et al., 2007).
Sequence analysis has been used to predict likely structural domains within the BBSome subunits, which have homology to domains found in other membrane coat complexes (Jin, 2010). These include WD40-repeat/β-propeller domains, which typically mediate protein-protein interactions, α-solenoid domains, phosphoinositide pleckstrin homology (PH) domains, helical bundle-forming (α-solenoid) tetratricopeptide repeat domains, and gamma-adaptin “ear” (GAE) domains (Nogi et al., 2002; Robinson, 1990). These domains are found in other vesicle coat complex proteins (McMahon and Mills, 2004; Stagg et al., 2007), such as clathrin, and the components of the COPI and COPII complexes. Analysis of their domain structures, the quaternary structure of the coat complex, and the phenotypes associated with genetic deficiencies should provide insights into their functional roles.
BBS1 is predicted to have a WD40-repeat/beta propeller domain and a GAE domain. A single mutant BBS1 allele, M390R, is involved in >30% of all BBS cases and 80% of all BBS1 disease-associated alleles (Mykytyn, 2007). BBS1 was initially reported not to be involved in complex inheritance patterns, but later studies (Beales et al., 2003) suggested BBS1 is involved in complex inheritance and interactions with other alleles in some families. BBS1 binds directly to Rabin8, the guanine nucleotide exchange factor for the small GTPase, Rab8, and binds to the small G protein, Arl6 (BBS3), in the GTP form through its beta-propeller domain (Jin, 2010). Arl6-GTP is required for formation of a membrane coat by the BBSome, in an interaction likely mediated in part by its N-terminal myristoyl group.
BBS2 has a predicted domain composition which is also found in BBS7 and BBS9, with a β-propeller domain, and a GAE domain with an adjacent α-helical domain, an arrangement that forms a “platform” or “appendage” structure in other coat complexes (McMahon and Mills, 2004). Mice with an engineered deficiency in BBS2 undergo retinal degeneration similar to that observed in other BBS knockouts (Nishimura et al., 2004). At 10 weeks of age, outer segments were short, and beginning to undergo apoptosis; however, rhodopsin was predominantly correctly localized, with only occasional cell body staining, possibly corresponding to apoptotic cells. These results indicate that BBS2 is necessary for cell survival, but not for ciliogenesis or rhodopsin trafficking.
BBS4 has multiple TPR domains and is predicted to have an alpha solenoid structure similar to BBS8. It has been reported that BBS4 is localized to pericentriolar satellites, and binds directly to PCM1 and P150 glued (Kim et al., 2004). However, the physiological relevance of the reported protein-protein interactions is subject to question due to a dearth of controls presented. This paper also concluded, based on siRNA knockdown experiments that BBS4 is essential for cell cycle progression, a conclusion refuted by the viability of BBS4 null mice, illustrating the uncertainty of conclusions relying solely on that approach to understand gene function.
At 2 weeks of age, BBS4 null mice show little evidence of apoptosis or rhodopsin mislocalization, whereas at 4 weeks they have numerous TUNEL-positive puncta, and begin to mislocalize rhodopsin (Kim et al., 2004). Thus, BBS4 is not essential for cilium formation or rhodopsin sorting to the outer segment, but is essential for some other unknown cellular function, likely related to cilium function. Deficiencies in BBS4, which lead to severe compromise of cell health, ultimately cause cell death, which, as it approaches leads to rhodopsin mislocalization perhaps as a secondary, rather than a primary defect. In another study of BBS4-null mice (Abd-El-Barr et al., 2007), the authors proposed rhodopsin mislocalization as a direct consequence of BBS4 inactivation; however, the conclusion was based on comparison of a greatly over-saturated immunofluorescence image of rhodopsin staining in BBS4-null mice. Cryo-electron tomography of BBS4 null mice revealed essentially normal connecting cilium structure, but aberrant disk membrane structure, and an accumulation of small vesicles within the connecting cilium and the immediately adjacent area of the inner segment (Gilliam et al., 2012). Given their similarities in structure, and the fact that both can bind directly to BBS9 and PCM1 (Nachury et al., 2007), it may be that BBS4 and BBS8 can substitute for one another to some extent, albeit with poor efficiency, based on the pathology observed. In addition to localization to the base of the connecting cilium in rods or transition zone in general, BBS4 has been reported to bind directly to the pericentriolar material protein PCM1 and to the centriolar satellite protein AZI1 or CEP131 (Chamling et al., 2014). It was reported that knockdown of AZI1 in zebrafish yields a BBS-like phenotype (Chamling et al., 2014). BBS4 with a fluorescent protein tag has been reported to move within cilia of cultured cells at rates consistent with intraflagellar transport, and to move from a pericentriolar location to the inside of the cilium upon serum starvation (Nachury et al., 2007)
BBS5 contains two pleckstrin homology-like domains, and binds preferentially to phosphatidylinositol-3-phosphate (PI3P) (Nachury et al., 2007), a marker of early endosomes and autophagosomes. Thus, it may cooperate with Arl6, also known as BBS3, in targeting the BBSome to specific vesicles or membrane compartments. A monoclonal antibody directed against BBS5 labels predominantly the axoneme in the outer segment, with barely detectable staining in the connecting cilium and basal body by immuno-EM, and, in contrast, co-localization in the basal body, transition zone and outer segment by immunofluorescence (Smith et al., 2013).
BBS6 is not part of the “core” BBSome, but is essential for BBSome formation. Three BBS genes (BBS6, BBS10, and BBS12) have homology to those encoding type II chaperonins and interact with CCT/TRiC proteins and BBS7 to form a complex termed the BBS-chaperonin complex. This complex is required for BBSome assembly (Zhang et al., 2012b).
BBS7. The domain architecture of BBS7 is similar to that of BBS2 and BBS9, and is essential for BBSome formation (Zhang et al., 2013). Like other BBS knockouts, BBS7 null mice suffer from retinal degeneration and loss of the photoreceptor layer, but appear to be able to form cilia and to carry out some ciliary transport functions (Zhang et al., 2013). It was shown that BBS7 interacts with BBS2 to form core complex of BBS7, BBS2, and BBS9, to which additional BBS subunits are added, with the last being BBS4 (Zhang et al., 2012b).
BBS8 has an α solenoid (TPR) structure similar to BBS4. A knockout of BBS8 yielded, among other BBS-like defects, retinal degeneration, defects in olfactory cilia, and severe defects in planar cell polarity in cochlear development (May-Simera et al., 2010; May-Simera et al., 2015; Tadenev et al., 2011). BBS8 was found to bind to Vangl2, and to co-localize with filamentous actin, as well as with microtubules (May-Simera et al., 2015). BBS8 has been reported to act as an adaptor for cargo undergoing IFT (intraflagellar transport) (Ansley et al., 2003; Tadenev et al., 2011). An interesting recent finding was that there is a splice variant of BBS8 that is unique to the retina (Riazuddin et al., 2010). This variant appears to be the predominant form in rod cells, and mutations that affect only this variant cause non-syndromic retinitis pigmentosa (Riazuddin et al., 2010).
BBS9 has a domain structure similar to that of BBS2 and BBS7, and is the only BBS protein which has been reported to be capable of forming binary complexes with every other BBSome subunit (Nachury et al., 2007). Thus, it likely plays a key role in stabilizing the overall complex, whose stability likely depends on cooperation among multiple weak protein-protein interactions that are not strong enough to sustain binary complexes.
4.3 Primary vs. motile cilia
Much of what we know about the principles of cilium structure, biogenesis, and function derives from studies of motile cilia, especially those of the model organism Chlamydomonas reinhardtii. The experimental tractability of these blue-green algae have made them extremely useful for this purpose, and numerous studies have confirmed the many commonalities of motile cilia in algae and primary cilia in the mammalian retina. However, the differences are also numerous. A recent single cell RNAseq survey of brain cell types (Zeisel et al., 2015) found 69 cell-type restricted transcripts in ependymal cells, which are multi-ciliated cells lining the ventricles. Presumably, these are mRNAs encoding proteins important specifically for motile cilia, and not found at comparable levels in the rest of the brain where primary cilia predominate. In support of this interpretation, most genes implicated in retinal ciliopathies were not included, a notable exception being myosin VIIa. Thus structures and mechanisms identified in motile cilia provide an imperfect model for analogous features of primary cilia. Similar caveats may well apply to potential differences between the highly specialized primary cilia of rods and cones as opposed to those of other cell types, especially those in cells that have not permanently exited the cell cycle, during which cilia disassemble and reassemble, and transformations and migrations occur between centrioles and centrosomes.
4.4 The inner segment
The inner segment can be considered as consisting of two major sub-regions, the ellipsoid, a mitochondria-rich region immediately adjacent to the outer segment, and the myoid, a region of variable length extending to the nucleus, whose depth within the outer nuclear layers is quite variable. This region is enriched in biosynthetic membranes, such as Golgi membranes, rough endoplasmic reticulum, and ribosomes, and typically stains strongly for mRNA species encoding outer segment proteins. However, both mitochondrial and smooth and rough ER membranes are found throughout the inner segment. Dominating electron micrographs of the inner segment is a long striated cytoskeletal element known as the ciliary rootlet, composed largely of the coiled-coil protein, rootletin (Gilliam et al., 2012; Yang et al., 2005; Yang et al., 2002). This extends from near the basal body all the way to the nucleus and beyond.
4.5 Nuclei
Because the nuclei have much larger diameters than the outer or inner segments, they are arranged in rows parallel to the long axes of the rods. Within the nuclei, the chromatin organization depends on whether the species is nocturnal or diurnal. The rods of diurnal retinas have most of their heterochromatin located at the nuclear periphery, euchromatin located more centrally, whereas in nocturnal retinas, the rods have an unusual inverted pattern, with heterochromatin in the center, and euchromatin, nascent transcripts and splicing machinery, distributed around the border of the nucleus (Solovei et al., 2009).
4.6 Axon and nerve terminal
A thin axon extends from the nucleus to the synaptic terminus, known as the rod spherule for its shape. This region contains many synaptic vesicles loaded with the neurotransmitter glutamate, many of them lining the synaptic ribbon, a structural specialization found in only a few neuronal types (Heidelberger et al., 2005; Sterling and Matthews, 2005).
5. Rod Dystrophies
There are a number of retinal dystrophies in which rod cell dysfunction or death is the earliest or primary pathology that is observed. These can be diagnosed as rod dystrophies, rod/cone dystrophies, or the many varieties of retinitis pigmentosa. In these diseases, some of the earliest symptoms are often loss of peripheral vision and night-blindness. Unfortunately, these relatively mild symptoms often progress to a narrower and narrower field of vision, or to a patchy loss of visual field due to geographic atrophy, and eventually to loss of central vision as increasing numbers of cones are lost. There has been tremendous progress over the last 25 years in uncovering the genes whose mutations are involved in these diseases, which now includes close to 200 different genes (Daiger et al., 2013). These are often genes encoding rod-or photoreceptor-specific proteins, or proteins whose function is particularly critical to rod cell survival, such as proteins involved in the visual cycle or disk phagocytosis in the retina pigmented epithelium. Many are genes encoding proteins important for the structure or function of primary cilia and are referred to as either syndromic or non-syndromic ciliopathies, depending on the degree of involvement of pathologies outside the retina. Some of these are discussed below.
5.1 Retinitis pigmentosa and other rod dystrophies
Retinitis pigmentosa (RP) (Daiger et al., 2013; Ferrari et al., 2011; Hartong et al., 2006) is the most common form of retinal dystrophy with a worldwide incidence estimated as about one in 4,000 (Ayuso and Millan, 2010; Boughman et al., 1980; Jay, 1982). This class of diseases is quite heterogeneous, but the common characteristics are a progressive disease primarily affecting rods, at least in early stages, with night-blindness and loss of peripheral vision being among the first detectable symptoms. Loss of rods and rod function can be detected as diminished light responses (especially the scotopic a-wave) in electroretinography (ERG). Rod cell loss leads to exposure of underlying pigment, whose appearance in fundus examinations is responsible for the “pigmentosa” term in the name of this disease. Over time, progressive loss of visual field is observed, leading to increasing cone involvement, tunnel vision, and eventual blindness. Cardinal features apparent in fundus examinations include attenuated retinal vessels, and bone spicule-like pigmentation. There are many genes whose defects lead to RP, including phototransduction protein-encoding genes such as Rhodopsin (RHO), cGMP phosphodiesterase subunit-encoding genes (PDE6A, PDE6B, PDE6G), and cGMP-gated channel-encoding genes (e.g. CNGA1, CNGB1); genes important for disk structure (e.g. ROM1, PRPH2); transcription factors (e.g. NEUROD1, NR2E3, NRL); visual cycle and pigment-epithelium specific genes (e.g. ABCA4, RDH12, LRAT, MERTK, RPE65, BEST1); and numerous genes encoding cilium associated proteins, (e.g. BBS1, BBS2, RPGR, IFT40, IFT172, PROM1, SPATA7, ARL6) among many others.
Additional forms of inherited retinal dystrophies, other than RP, also involve functional defects affecting rods. These include rod-cone dystrophies and a very severe and early onset form of retinal dystrophy known as Leber Congenital Amaurosis. There is considerable overlap among the genes encoding the different retinal dystrophies, and in many cases defects in a given gene can give rise to a range of retinal defects or to syndromic disease, depending on the mutation and genetic background.
5.2 Syndromic vs. non syndromic retinal dystrophies
Roughly one-half (16.7% to 65.2%, based on studies in different countries) of RP cases can be classified as syndromic (Ayuso and Millan, 2010), meaning that they are accompanied by a range of extra-retinal symptoms. The rest are classified as non-syndromic (Estrada-Cuzcano et al., 2012), with retinal degeneration being the only obvious inherited defect. A somewhat surprising observation that has emerged as increasing numbers of defects in different genes have been associated with non-syndromic RP is that while some of them encode rod- or photoreceptor-specific proteins, many encode proteins expressed in many cell types throughout the body, suggesting that rods are particularly sensitive to disruption of certain regulatory pathways.
The inheritance of retinal dystrophies can fall into the categories of autosomal dominant, autosomal recessive, X-linked, mitochondrial, and sporadic- meaning isolated cases for which an inheritance mode (pattern) cannot be established (Daiger et al., 2013; Ferrari et al., 2011; Hartong et al., 2006). In addition, there are examples of digenic or trigenic disease (Beales et al., 2003; Dryja et al., 1997; Eichers et al., 2004; Katsanis et al., 2001; Katsanis et al., 2002). These modes of inheritance serve not only to help assess risk among family members, but also to determine what conclusions can be drawn about gene function from disease phenotype, and what approaches to gene-based therapy should be considered.
5.3 Functional classes of disease genes
Proteins encoded by genes associated with retinal dystrophies fall largely within a few major classes. One such class encompasses phototransduction proteins, such as rhodopsin, PDE6, guanylate cyclase isozymes, guanylate cyclase activating proteins, and cyclic nucleotide-gated channels. Another class includes proteins that play important roles in the visual cycle, the metabolism of retinoids essential for conversion of vitamin A to 11-cis-retinal, the light sensing chromophore of rhodopsin, and for recycling all-trans-retinal back to 11-cis retinal following photoisomerization. Some of these, such as ABCA4 (Molday, 2007; Quazi and Molday, 2014; Shroyer et al., 2001a; Shroyer et al., 2001b), a lipid transport protein of the ATP-binding cassette family, and retinol dehydrogenase isoform 12, rdh12 (Haeseleer et al., 2002; Janecke et al., 2004; Perrault et al., 2004), function in photoreceptors, while others function in the retinal pigmented epithelium (RPE), such as lecithin-retinol acyl transferase (LRAT) (Barry et al., 1989; Saari and Bredberg, 1989) and the isomerohydrolase enzyme RPE65 (Hamel et al., 1993), are localized to the retinal pigmented epithelium (RPE). A third rather large class falls into the category of ciliopathies, either syndromic or non-syndromic. This class includes the BBS proteins, the intraflagellar transport (IFT) proteins, and the Usher syndrome proteins (Adams et al., 2007; Daiger, 2004; Koenig, 2003; Liu et al., 2010). Additional classes include mitochondrial defects, defects in proteins important for transcriptional regulation or mRNA processing, and metabolic enzymes, such as inosine monophosphate dehydrogenase (Daiger et al., 2013).
5.4 Theories of retinal disease
An outstanding problem for the fields of photoreceptor biology and retinal dystrophies is to understand the mechanisms that link gene defects to loss of rods and rod function. Despite years of intensive research using animal models, it would be hard to find a single gene defect for which this question has been definitively answered. It could be argued that autosomal recessive genes encoding proteins essential for phototransduction (rhodopsin, rod transducin, PDE6) or photoreceptor structure (e.g., rhodopsin, rom1, peripherin/rds) or regulation of photoreceptor gene expression (CRX, NRL, NR2E3) would lead to defects in rod structure and function by obvious mechanisms. However, the question remains, why do the cells die in response to these defects? Why do defects in some genes, such as those encoding rod transducin or arrestin or rhodopsin kinase lead to autosomal recessive night blindness without progressive degeneration, whereas mutations in others, such as those encoding rhodopsin, PDE6, or the cGMP-gated cation channel lead to autosomal recessive retinitis pigmentosa?
5.4.1 Equivalent light hypothesis
Based on the observations that relatively modestly increased light exposure can cause retinal degeneration in albino animals, and that certain disease-causing mutations in phototransduction cascade proteins might be expected to produce dysregulation of phototransduction, an “equivalent light” hypothesis has been proposed (Fain and Lisman, 1993; Fain and Lisman, 1999; Lisman and Fain, 1995) suggesting that these mutations may have deleterious effects on cell survival similar to those of excessive light, both through unrestrained operation of the highly amplified phototransduction cascade. This idea was challenged, based on the observation that the G90D mutation in rhodopsin, which causes constitutive signaling, and thereby mimics continual light exposure, leads to photoresponse desensitization and stationary night blindness, but not to cell loss or retinal degeneration (Sieving et al., 2001). Moreover, the G90D mutation actually rescues the retinal degeneration induced by a mutation in the guanylate cyclase activating protein, GCAP-1, which leads to elevated levels of cGMP (Woodruff et al., 2007). Interestingly, a study of effects of phototransduction cascade mutants found evidence that constitutive G-protein activation due to inactivating mutations in rhodopsin kinase or arrestin, but not constitutive membrane hyperpolarization due to inactivation of the cGMP-gated cation channel that is the ultimate downstream target of the phototransduction cascade, led to activation of the Unfolded Protein Response (Wang and Chen, 2014). Thus, constitutive signaling may act on cell survival differently, depending on the step(s) of the cascade affected.
5.4.2 Disruption of Ca2+ homeostasis
Related to the “equivalent light” hypothesis are the hypotheses that either excessively elevated or abnormally reduced levels of intracellular [Ca2+] play a key role in linking rod cell death to abnormalities in phototransduction cascade proteins. Under dark-adapted conditions, resting [Ca2+] has been estimated to be in the range of 250 nM to 670 nM (Korenbrot and Miller, 1989; Sampath et al., 1998; Woodruff et al., 2002), a value that is determined by the balance between leak of Ca2+ into the outer segment through the cGMP-gated cation channel (CNG-channel), and the extrusion of Ca2+ out of the cell by the Na/Ca/K exchanger (Prinsen et al., 2002; Schnetkamp, 1995). Upon illumination, the CNG channel closes as cGMP levels fall, while the extrusion process continues, and [Ca2+] falls to very low levels, perhaps as low as 1 nM or less (Gray-Keller and Detwiler, 1994). Constitutive signaling would be expected to maintain abnormally low intracellular [Ca2+], which would likely be detrimental to the many cellular processes that are regulated by Ca2+ and Ca2+-binding proteins. Conversely, suppression of phototransduction or the basal activity of phototransduction proteins would be expected to lead to an excessively high resting level of [Ca2+], a condition known to be toxic in many cell types. This latter situation may well be involved in the mechanism of cell death in the rd1 mouse, which suffers from an autosomal recessive defect in the cGMP phosphodiesterase PDE6 beta subunit (Bowes et al., 1990; Pittler and Baehr, 1991). A recent study in cultured cells revealed that Ca2+ homeostasis within cilia is markedly different from that in the rest of the cell (Delling et al., 2013). Intra-ciliary [Ca2+] was reported to be regulated by a cilium-specific ion channel, composed of members of the polycystin-1 and polycystin-2 families, PKD1L1 and PKD2L1 (also known as TRPP3), and not to be in equilibrium with [Ca2+] in the cytoplasm (DeCaen et al., 2013; Delling et al., 2013). PKD2L1 is expressed in the retina (Gilliam and Wensel, 2011), and its mRNA levels have been reported to decrease following retinal detachment (Delyfer et al., 2013).
Dysregulation of calcium homeostasis has also been invoked (Sizova et al., 2014) as a contributor to cell death due to misfolded proteins or environmental insults inducing endoplasmic reticulum stress (ER-stress) (Mohlin et al., 2014) and the unfolded protein response (UPR) (Gorbatyuk et al., 2010; Nakanishi et al., 2013; Rana et al., 2014; Zhang et al., 2014a)
5.4.3 Programmed Cell Death or Apoptosis
It has long been held that apoptosis represents a general pathway for cell death in retinal degeneration resulting from rod cell defects (Chang et al., 1993; Gregory and Bird, 1995; Reme et al., 1998). It should be noted that in most published studies, only TUNEL (Terminal deoxynucleotidyl transferase mediated dUTP Nick End) assays were used to test for apoptosis, and correlations, rather than mechanistic cause-and-effect relationships were demonstrated. A universal causative role for apoptosis in rod degeneration has been questioned (Kunchithapautham and Rohrer, 2007a, b; Lohr et al., 2006; Trifunovic et al., 2012) A recent study found that ablating pro-apoptotic genes, CHOP and Ask1, did not suppress photoreceptor cell death in a mouse model of ADRP (Adekeye et al., 2014).
5.4.4 Necrosis/Necroptosis
Programmed necrosis, or necroptosis (Khan et al., 2014), has been proposed as an alternative cell death pathway that may play an important role in rod dystrophies. A recent study concluded, based on effects of knocking down receptor-interacting protein kinase 3 (Rip3) in a cone-rod dystrophy model, that primary photoreceptor cell death occurs by necroptosis, with “bystander” death of nearby rods is associated with caspase activation, and, presumably, apoptosis (Viringipurampeer et al., 2014), while another found that Rip3 knockout in the rd10 rod dystrophy model, necroptosis played an important role in death of cones, but not rods (Murakami et al., 2012).
5.4.5 Autophagy
The role of autophagy, a pathway of self-digestion in response to nutrient deprivation and other stresses, in retinal degeneration is also unclear. It has been proposed that autophagy can play a protective role (Chen et al., 2013; Zhou et al., 2015), but also that it can contribute to photoreceptor cell death (Mohlin et al., 2014; Zhang et al., 2014b), and that the balance between macroautophagy and chaperone-mediated autophagy may shift with age, and thus render aging retinas more susceptible to degeneration (Rodriguez-Muela et al., 2013). Selective knockout of Atg5, a protein essential for normal autophagy, leads to progressive retinal degeneration (Rodriguez-Muela et al., 2013; Zhou et al., 2015) leading to the conclusion that basal autophagy, even in the absence of disease-induced stress, is essential for photoreceptor survival. Selective knockout of Vps34, and Type III phosphatidylinositol 3-kinase essential for synthesis of phosphatidyl inositol 3-phosphate and completion of autophagasome maturation, also leads to rod cell death (He et al., 2016).
5.4.6 Autosomal Dominant Disease Mechanisms: Haploinsufficiency vs. Dominant Negative or Toxic Gain-of-Function Mutations
The problem of understanding mechanisms of cell death is especially daunting in the case of autosomal dominant disorders, such as most of the cases of RP caused by rhodopsin mutations, the most common genetic cause of ADRP. Haploinsufficiency, or pathology due to shortage of wildtype protein, does not explain these diseases, as certain rhodopsin null alleles lead to recessive disease in both humans (E249X mutation (Rosenfeld et al., 1992) and W161X mutation (Kartasasmita et al., 2011) and mice (Lem et al., 1999). Thus, the mutant proteins must act either through a dominant-negative effect, or through a toxic gain-of-function effect (Wilson and Wensel, 2003). These mechanisms can be tested experimentally by over-expressing the wildtype protein in the mutant background. As dominant-negative alleles are defined as producing non-functional proteins that compete with wildtype protein for binding to functional sites or ligands, their phenotype should be at least partly suppressed by increasing levels of wildtype protein, whereas phenotypes in the toxic gain-of-function case should not be affected (Wilson and Wensel, 2003). This test has been applied in one case, that of murine models of retinal degeneration due to the ADRP rhodopsin mutation P23H (Price et al., 2012) using gene dosage experiments. Increasing the levels of rhodopsin mRNA and protein was found to preserve rod cell viability so that as many as four times as many photoreceptor nuclei remained in mice expressing a P23H mutant rhodopsin. This form of rhodopsin is known to fold inefficiently (Liu et al., 1996; Sung et al., 1991), to be unstable (Chen et al., 2014), and to mislocalize to the inner segment (Tam and Moritz, 2006), so it has been assumed to cause cell death through a toxic gain-of-function, such as triggering the unfolded protein response, or toxic stress of the endoplasmic reticulum. The genetic experiments show clearly that the primary mechanism is a dominant negative effect, consistent with the observation that over-expression of wildtype rhodopsin, which has a protective effect on the P23H background, increases, rather than decreases, the percentage of a P23H rhodopsin-EGFP fusion that is transported to the outer segment (Price et al., 2012), and with the observation that P23H rhodopsin has a harmful effect on disk structure in the outer segment (Sakami et al., 2011).
6. Future Directions
Understanding the mechanisms of disease is important for the development of new therapeutic approaches, which are sorely needed for retinal neurodegeneration. New gene delivery and gene editing technologies hold the promise for inducing the expression of beneficial genes or inactivating toxic alleles, but an understanding of mechanism is necessary to determine which genes should be manipulated in which way, and what pathways may be impacted by doing so. This understanding will be facilitated by developing a deeper understanding of the cilium-associated structures of the rod, and the roles of specific proteins within them, which will be facilitated by combining the latest genetic engineering technology with the most advanced structural techniques. The most important gaps to be filled are the molecular identities of the structural features associated with the cilium, and the mechanisms leading from functional defects to rod cell death.
Highlights.
Review of historical and most recent structural studies of vertebrate rod cells
Current state of understanding of basal disk structure and morphogenesis
Cryo-electron tomography and superresolution microscopy of rods
Advances in understanding of cilium-associated structures
BBSome structural and functional insights
Current understanding and uncertainties in mechanisms of rod dystrophies
Acknowledgments
Retinal research in the Wensel laboratory has been supported by the Robert A. Welch Foundation, Q0035, and by grants from the National Eye Institute of the National Institutes of Health, R01-EY007981, R01-EY011900, R01-EY026545, R01-EY011731, F32-EY024815, and P30-EY002520, and by the National Center for Macromolecular Imaging, P41-GM103832, and P41-RR002250. I would like to thank the many colleagues in our laboratory and those of our collaborators for their work which is referenced here.
Abbrevations
- ADRP
autosomal dominant retinitis pigmentosa
- BBS
Bardet-Biedl syndrome, BBSome, membrane coat complex formed by BBS gene products
- Cryo-ET
cryo-electron tomography
- miniSOG
mini singlet oxygen generator, a fluorescent flavoprotein engineered from Arabidopsis phototropin
- PALM
photactivated localization microscopy
- RP
retinitis pigmentosa
- SEM
scanning electron microscopy
- SIM
structured illumination microscopy
- SNAP tag
fusion to a 20 kDa mutant of the DNA repair protein O6-alkylguanine-DNA alkyltransferase that allows covalent labeling with with benzylguanine derivatives
- STED
stimulated emission depletion
- STORM
stochastic optical reconstruction microscopy
- TEM
transmission electron microscopy
- TPR
tetratricopeptide repeats
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd-El-Barr MM, Sykoudis K, Andrabi S, Eichers ER, Pennesi ME, Tan PL, Wilson JH, Katsanis N, Lupski JR, Wu SM. Impaired photoreceptor protein transport and synaptic transmission in a mouse model of Bardet-Biedl syndrome. Vision Res. 2007;47:3394–3407. doi: 10.1016/j.visres.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams NA, Awadein A, Toma HS. The retinal ciliopathies. Ophthalmic Genet. 2007;28:113–125. doi: 10.1080/13816810701537424. [DOI] [PubMed] [Google Scholar]
- Adekeye A, Haeri M, Solessio E, Knox BE. Ablation of the proapoptotic genes CHOP or Ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One. 2014;9:e83871. doi: 10.1371/journal.pone.0083871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre G. Hereditary retinal diseases in small animals. The Veterinary clinics of North America. 1973;3:515–528. doi: 10.1016/s0091-0279(73)50065-0. [DOI] [PubMed] [Google Scholar]
- Aguirre G, Acland G, Chader G. Hereditary retinal degenerations in the dog: specificity of abnormal cyclic nucleotide metabolism to diseases of arrested photoreceptor development. Birth defects original article series. 1982;18:119–133. [PubMed] [Google Scholar]
- Anand M, Khanna H. Ciliary transition zone (TZ) proteins RPGR and CEP290: role in photoreceptor cilia and degenerative diseases. Expert Opin Ther Targets. 2012;16:541–551. doi: 10.1517/14728222.2012.680956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews LD, Cohen AI. Freeze-fracture studies of the structure of rod outer segment membranes: new observations regarding the distribution of particle-free patches and the location of the fracture planes in conventionally prepared retinas. Exp Eye Res. 1981;33:1–10. doi: 10.1016/s0014-4835(81)80076-0. [DOI] [PubMed] [Google Scholar]
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Arikawa K, Molday LL, Molday RS, Williams DS. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J Cell Biol. 1992;116:659–667. doi: 10.1083/jcb.116.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa K, Williams DS. Alpha-actinin and actin in the outer retina: a double immunoelectron microscopic study. Cell Motil Cytoskeleton. 1991;18:15–25. doi: 10.1002/cm.970180103. [DOI] [PubMed] [Google Scholar]
- Arikawa K, Williams DS. Acetylated alpha-tubulin in the connecting cilium of developing rat photoreceptors. Invest Ophthalmol Vis Sci. 1993;34:2145–2149. [PubMed] [Google Scholar]
- Arshavsky VY, Burns ME. Photoreceptor signaling: supporting vision across a wide range of light intensities. J Biol Chem. 2012;287:1620–1626. doi: 10.1074/jbc.R111.305243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- Ayuso C, Millan JM. Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome medicine. 2010;2:34. doi: 10.1186/gm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KL, Gross AK. Aberrant protein trafficking in retinal degenerations: The initial phase of retinal remodeling. Exp Eye Res. 2015 doi: 10.1016/j.exer.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry RJ, Canada FJ, Rando RR. Solubilization and partial purification of retinyl ester synthetase and retinoid isomerase from bovine ocular pigment epithelium. J Biol Chem. 1989;264:9231–9238. [PubMed] [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, Lupski JR, Katsanis N. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse JC, Forestner DM, Defoe DM. Membrane assembly in retinal photoreceptors. III. Distinct membrane domains of the connecting cilium of developing rods. J Neurosci. 1985;5:1035–1048. doi: 10.1523/JNEUROSCI.05-04-01035.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Goldberg AF, Dispoto J, Katragadda M, Cesarone G, Albert AD. A soluble peripherin/Rds C-terminal polypeptide promotes membrane fusion and changes conformation upon membrane association. Exp Eye Res. 2003;77:505–514. doi: 10.1016/s0014-4835(03)00151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Kong F, Lamba OP, Stefano FP, Williams DS. Purification and light-dependent phosphorylation of a candidate fusion protein, the photoreceptor cell peripherin/rds. Biochemistry. 1997;36:6835–6846. doi: 10.1021/bi9627370. [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Lamba OP, Napoli AA, Jr, Sinha S, Guo Y. Fusion between retinal rod outer segment membranes and model membranes: a role for photoreceptor peripherin/rds. Biochemistry. 1998;37:9477–9487. doi: 10.1021/bi980173p. [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Stefano FP, Fenner M, Napoli AA., Jr A peptide analogue to a fusion domain within photoreceptor peripherin/rds promotes membrane adhesion and depolarization. Biochim Biophys Acta. 2000;1463:343–354. doi: 10.1016/s0005-2736(99)00226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JA, Conneally PM, Nance WE. Population genetic studies of retinitis pigmentosa. Am J Hum Genet. 1980;32:223–235. [PMC free article] [PubMed] [Google Scholar]
- Bowes C, Li T, Danciger M, Baxter LC, Applebury ML, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- Bowes C, Li T, Frankel WN, Danciger M, Coffin JM, Applebury ML, Farber DB. Localization of a retroviral element within the rd gene coding for the beta subunit of cGMP phosphodiesterase. Proc Natl Acad Sci U S A. 1993;90:2955–2959. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PK, Gibbons IR, Wald G. The Visual Cells and Visual Pigment of the Mudpuppy, Necturus. J Cell Biol. 1963;19:79–106. doi: 10.1083/jcb.19.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne T, Meschede IP, Burden JJ, Bailly M, Seabra MC, Futter CE. Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1509285113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns ME, Arshavsky VY. Beyond counting photons: trials and trends in vertebrate visual transduction. Neuron. 2005;48:387–401. doi: 10.1016/j.neuron.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Cagianut B, Sandri G, Zilla P, Theiler K. Studies on hereditary retinal degeneration. The rd gene in the mouse. Graefes Arch Clin Exp Ophthalmol. 1985;223:16–22. doi: 10.1007/BF02150568. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Chaitin MH. Double immunogold localization of opsin and actin in the cilium of developing mouse photoreceptors. Exp Eye Res. 1992;54:261–267. doi: 10.1016/s0014-4835(05)80216-7. [DOI] [PubMed] [Google Scholar]
- Chaitin MH, Bok D. Immunoferritin localization of actin in retinal photoreceptors. Invest Ophthalmol Vis Sci. 1986;27:1764–1767. [PubMed] [Google Scholar]
- Chaitin MH, Burnside B. Actin filament polarity at the site of rod outer segment disk morphogenesis. Invest Ophthalmol Vis Sci. 1989;30:2461–2469. [PubMed] [Google Scholar]
- Chaitin MH, Carlsen RB, Samara GJ. Immunogold localization of actin in developing photoreceptor cilia of normal and rds mutant mice. Exp Eye Res. 1988;47:437–446. doi: 10.1016/0014-4835(88)90054-1. [DOI] [PubMed] [Google Scholar]
- Chaitin MH, Coelho N. Immunogold localization of myosin in the photoreceptor cilium. Invest Ophthalmol Vis Sci. 1992;33:3103–3108. [PubMed] [Google Scholar]
- Chaitin MH, Schneider BG, Hall MO, Papermaster DS. Actin in the photoreceptor connecting cilium: immunocytochemical localization to the site of outer segment disk formation. J Cell Biol. 1984;99:239–247. doi: 10.1083/jcb.99.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamling X, Seo S, Searby CC, Kim G, Slusarski DC, Sheffield VC. The centriolar satellite protein AZI1 interacts with BBS4 and regulates ciliary trafficking of the BBSome. PLoS Genet. 2014;10:e1004083. doi: 10.1371/journal.pgen.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Stolz J, Kohl S, Chiang WC, Lin JH. Endoplasmic reticulum stress in human photoreceptor diseases. Brain Res. 2016 doi: 10.1016/j.brainres.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, Sayer JA, Otto EA, Attanasio M, O’Toole JF, Jin G, Shou C, Hildebrandt F, Williams DS, Heckenlively JR, Swaroop A. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GQ, Hao Y, Wong F. Apoptosis: final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- Chang YW, Chen S, Tocheva EI, Treuner-Lange A, Lobach S, Sogaard-Andersen L, Jensen GJ. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Jastrzebska B, Cao P, Zhang J, Wang B, Sun W, Yuan Y, Feng Z, Palczewski K. Inherent instability of the retinitis pigmentosa P23H mutant opsin. J Biol Chem. 2014;289:9288–9303. doi: 10.1074/jbc.M114.551713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sawada O, Kohno H, Le YZ, Subauste C, Maeda T, Maeda A. Autophagy protects the retina from light-induced degeneration. J Biol Chem. 2013;288:7506–7518. doi: 10.1074/jbc.M112.439935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Leng Q, Mixson AJ. Zinc Finger Nucleases: Tailor-made for Gene Therapy. Drugs of the future. 2012;37:183–196. doi: 10.1358/dof.2012.037.03.1779022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JZ, Zhao Y, Sung CH. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007;130:535–547. doi: 10.1016/j.cell.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Rachel RA, Aleman TS, Swider M, Schwartz SB, Sumaroka A, Roman AJ, Stone EM, Jacobson SG, Swaroop A. Cone photoreceptors are the main targets for gene therapy of NPHP5 (IQCB1) or NPHP6 (CEP290) blindness: generation of an all-cone Nphp6 hypomorph mouse that mimics the human retinal ciliopathy. Hum Mol Genet. 2011;20:1411–1423. doi: 10.1093/hmg/ddr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AW, Branton D. Fracture faces in frozen outer segments from the guinea pig retina. Z Zellforsch Mikrosk Anat. 1968;91:586–603. doi: 10.1007/BF00455276. [DOI] [PubMed] [Google Scholar]
- Cohen AI. New evidence supporting the linkage to extracellular space of outer segment saccules of frog cones but not rods. J Cell Biol. 1968;37:424–444. doi: 10.1083/jcb.37.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AI. Further studies on the question of the patency of saccules in outer segments of vertebrate photoreceptors. Vision Res. 1970;10:445–453. doi: 10.1016/0042-6989(70)90001-5. [DOI] [PubMed] [Google Scholar]
- Constantine R, Zhang H, Gerstner CD, Frederick JM, Baehr W. Uncoordinated (UNC)119: coordinating the trafficking of myristoylated proteins. Vision Res. 2012;75:26–32. doi: 10.1016/j.visres.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic. 2001;2:82–91. doi: 10.1034/j.1600-0854.2001.020202.x. [DOI] [PubMed] [Google Scholar]
- Corless JM, Costello MJ. Isolation, rapid freezing, and freeze-fracture methods for frog retinal photoreceptors. Methods Enzymol. 1982;81:585–593. doi: 10.1016/s0076-6879(82)81082-3. [DOI] [PubMed] [Google Scholar]
- Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- Daiger SP. Identifying retinal disease genes: how far have we come, how far do we have to go? Novartis Found Symp. 2004;255:17–27. doi: 10.1002/0470092645.ch3. discussion 27–36, 177–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger SP, Bowne SJ, Sullivan LS. Genes and Mutations Causing Autosomal Dominant Retinitis Pigmentosa. Cold Spring Harbor perspectives in medicine. 2015:5. doi: 10.1101/cshperspect.a017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis E. Electron microscope observations on the submicroscopic organization of the retinal rods. J Biophys Biochem Cytol. 1956;2:319–330. doi: 10.1083/jcb.2.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504:315–318. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delyfer MN, Ait-Ali N, Camara H, Clerin E, Korobelnik JF, Sahel JA, Leveillard T. Transcriptomic analysis of human retinal surgical specimens using jouRNAI. J Vis Exp. 2013 doi: 10.3791/50375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Huber LA, Ransom N, Mancini M, Simons K, Papermaster DS. rab8 in retinal photoreceptors may participate in rhodopsin transport and in rod outer segment disk morphogenesis. J Cell Sci. 1995;108(Pt 1):215–224. doi: 10.1242/jcs.108.1.215. [DOI] [PubMed] [Google Scholar]
- Deretic D, Papermaster DS. Rab6 is associated with a compartment that transports rhodopsin from the trans-Golgi to the site of rod outer segment disk formation in frog retinal photoreceptors. J Cell Sci. 1993;106(Pt 3):803–813. doi: 10.1242/jcs.106.3.803. [DOI] [PubMed] [Google Scholar]
- Drivas TG, Bennett J. CEP290 and the primary cilium. Adv Exp Med Biol. 2014;801:519–525. doi: 10.1007/978-1-4614-3209-8_66. [DOI] [PubMed] [Google Scholar]
- Drivas TG, Holzbaur EL, Bennett J. Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. J Clin Invest. 2013;123:4525–4539. doi: 10.1172/JCI69448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Hahn LB, Kajiwara K, Berson EL. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1997;38:1972–1982. [PubMed] [Google Scholar]
- Eichers ER, Lewis RA, Katsanis N, Lupski JR. Triallelic inheritance: a bridge between Mendelian and multifactorial traits. Ann Med. 2004;36:262–272. doi: 10.1080/07853890410026214. [DOI] [PubMed] [Google Scholar]
- Estrada-Cuzcano A, Roepman R, Cremers FP, den Hollander AI, Mans DA. Non-syndromic retinal ciliopathies: translating gene discovery into therapy. Hum Mol Genet. 2012;21:R111–R124. doi: 10.1093/hmg/dds298. [DOI] [PubMed] [Google Scholar]
- Fain GL, Lisman JE. Photoreceptor degeneration in vitamin A deprivation and retinitis pigmentosa: the equivalent light hypothesis. Exp. Eye Res. 1993;57:335–340. doi: 10.1006/exer.1993.1132. [DOI] [PubMed] [Google Scholar]
- Fain GL, Lisman JE. Light, Ca2+, and photoreceptor death: new evidence for the equivalent-light hypothesis from arrestin knockout mice. Invest Ophthalmol Vis Sci. 1999;40:2770–2772. [PubMed] [Google Scholar]
- Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238–249. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher EL, Jobling AI, Vessey KA, Luu C, Guymer RH, Baird PN. Animal models of retinal disease. Prog Mol Biol Transl Sci. 2011;100:211–286. doi: 10.1016/B978-0-12-384878-9.00006-6. [DOI] [PubMed] [Google Scholar]
- Flisikowska T, Kind A, Schnieke A. Genetically modified pigs to model human diseases. J Appl Genet. 2014;55:53–64. doi: 10.1007/s13353-013-0182-9. [DOI] [PubMed] [Google Scholar]
- Flower NE. Particles within membranes: a freeze-etch view. J Cell Sci. 1971;9:435–441. doi: 10.1242/jcs.9.2.435. [DOI] [PubMed] [Google Scholar]
- Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- Giessl A, Trojan P, Rausch S, Pulvermuller A, Wolfrum U. Centrins, gatekeepers for the light-dependent translocation of transducin through the photoreceptor cell connecting cilium. Vision Res. 2006;46:4502–4509. doi: 10.1016/j.visres.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam JC, Wensel TG. TRP channel gene expression in the mouse retina. Vision Res. 2011;51:2440–2452. doi: 10.1016/j.visres.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Satir P. The ciliary necklace. A ciliary membrane specialization. J Cell Biol. 1972;53:494–509. doi: 10.1083/jcb.53.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AJ. A study of the ultrastructure of visual cell outer segment membranes. J Ultrastruct Res. 1973;43:228–246. doi: 10.1016/s0022-5320(73)80035-8. [DOI] [PubMed] [Google Scholar]
- Goldberg AF, Moritz OL, Williams DS. Molecular basis for photoreceptor outer segment architecture. Prog Retin Eye Res. 2016 doi: 10.1016/j.preteyeres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk M, Gorbatyuk O. Review: retinal degeneration: focus on the unfolded protein response. Mol Vis. 2013;19:1985–1998. [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Gorbatyuk OS, LaVail MM, Lin JH, Hauswirth WW, Lewin AS. Functional rescue of P23H rhodopsin photoreceptors by gene delivery. Adv Exp Med Biol. 2012;723:191–197. doi: 10.1007/978-1-4614-0631-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Knox T, LaVail MM, Gorbatyuk OS, Noorwez SM, Hauswirth WW, Lin JH, Muzyczka N, Lewin AS. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci U S A. 2010;107:5961–5966. doi: 10.1073/pnas.0911991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Keller MP, Detwiler PB. The calcium feedback signal in the phototransduction cascade of vertebrate rods. Neuron. 1994;13:849–861. doi: 10.1016/0896-6273(94)90251-8. [DOI] [PubMed] [Google Scholar]
- Gregory CY, Bird AC. Cell loss in retinal dystrophies by apoptosis--death by informed consent! Br J Ophthalmol. 1995;79:186–190. doi: 10.1136/bjo.79.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner JV, Weidman TA, Bodley HD, Greiner CA. Ciliogenesis in photoreceptor cells of the retina. Exp Eye Res. 1981;33:433–446. doi: 10.1016/s0014-4835(81)80094-2. [DOI] [PubMed] [Google Scholar]
- Gross AK, Parpura V, Sammons JD. Monitoring Kinetic Changes of Proper and Improper Rhodopsin Transport ex vivo. Invest. Ophthal. Vis. Sci. 2014;55 E-Abstract 6010. [Google Scholar]
- Gross AK, Wensel TG. Chapter 18, Biochemical Cascade of Phototransduction. In: Kaufman PA, Alm A, Levin LA, Nilsson SFE, VerHoeve J, Wu SM, editors. Adler’s Physiology of the Eye. 11th. Saunders; 2011. [Google Scholar]
- Haeri M, Knox BE. Rhodopsin mutant P23H destabilizes rod photoreceptor disk membranes. PLoS One. 2012;7:e30101. doi: 10.1371/journal.pone.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Jang GF, Imanishi Y, Driessen CA, Matsumura M, Nelson PS, Palczewski K. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem. 2002;277:45537–45546. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale IL, Matsumoto B. Resolution of subcellular detail in thick tissue sections: immunohistochemical preparation and fluorescence confocal microscopy. Methods Cell Biol. 1993;38:289–324. doi: 10.1016/s0091-679x(08)61008-0. [DOI] [PubMed] [Google Scholar]
- Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J Biol Chem. 1993;268:15751–15757. [PubMed] [Google Scholar]
- Han Z, Anderson DW, Papermaster DS. Prominin-1 localizes to the open rims of outer segment lamellae in Xenopus laevis rod and cone photoreceptors. Invest Ophthalmol Vis Sci. 2012;53:361–373. doi: 10.1167/iovs.11-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- He F, Agosto MA, Anastassov IA, Tse DY, Wu SM, Wensel TG. Phosphatidylinositol-3-phosphate is light-regulated and essential for survival in retinal rods. Sci Rep. 2016;6:26978. doi: 10.1038/srep26978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger R, Thoreson WB, Witkovsky P. Synaptic transmission at retinal ribbon synapses. Prog Retin Eye Res. 2005;24:682–720. doi: 10.1016/j.preteyeres.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth TJ, Gross AK. Defective trafficking of rhodopsin and its role in retinal degenerations. Int Rev Cell Mol Biol. 2012;293:1–44. doi: 10.1016/B978-0-12-394304-0.00006-3. [DOI] [PubMed] [Google Scholar]
- Hollingsworth TJ, Gross AK. The severe autosomal dominant retinitis pigmentosa rhodopsin mutant Ter349Glu mislocalizes and induces rapid rod cell death. J Biol Chem. 2013;288:29047–29055. doi: 10.1074/jbc.M113.495184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst CJ, Forestner DM, Besharse JC. Cytoskeletal-membrane interactions: a stable interaction between cell surface glycoconjugates and doublet microtubules of the photoreceptor connecting cilium. J Cell Biol. 1987;105:2973–2987. doi: 10.1083/jcb.105.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst CJ, Johnson LV, Besharse JC. Transmembrane assemblage of the photoreceptor connecting cilium and motile cilium transition zone contain a common immunologic epitope. Cell Motil Cytoskeleton. 1990;17:329–344. doi: 10.1002/cm.970170408. [DOI] [PubMed] [Google Scholar]
- Huang PC, Gaitan AE, Hao Y, Petters RM, Wong F. Cellular interactions implicated in the mechanism of photoreceptor degeneration in transgenic mice expressing a mutant rhodopsin gene. Proc Natl Acad Sci U S A. 1993;90:8484–8488. doi: 10.1073/pnas.90.18.8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Besharse JC. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabalameli HR, Zahednasab H, Karimi-Moghaddam A, Jabalameli MR. Zinc finger nuclease technology: advances and obstacles in modelling and treating genetic disorders. Gene. 2015;558:1–5. doi: 10.1016/j.gene.2014.12.044. [DOI] [PubMed] [Google Scholar]
- Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, McHenry CL, Nair AR, Ruschendorf F, Heckenlively J, Wissinger B, Nurnberg P, Gal A. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36:850–854. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- Jay M. On the heredity of retinitis pigmentosa. Br J Ophthalmol. 1982;66:405–416. doi: 10.1136/bjo.66.7.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachi S, Nishizawa Y, Olshevskaya E, Yamazaki A, Miyake Y, Wakabayashi T, Dizhoor A, Usukura J. Detailed localization of photoreceptor guanylate cyclase activating protein-1 and −2 in mammalian retinas using light and electron microscopy. Exp Eye Res. 1999;68:465–473. doi: 10.1006/exer.1998.0629. [DOI] [PubMed] [Google Scholar]
- Kajimura N, Harada Y, Usukura J. High-resolution freeze-etching replica images of the disk and the plasma membrane surfaces in purified bovine rod outer segments. J Electron Microsc (Tokyo) 2000;49:691–697. doi: 10.1093/oxfordjournals.jmicro.a023860. [DOI] [PubMed] [Google Scholar]
- Kajiwara K, Hahn LB, Mukai S, Travis GH, Berson EL, Dryja TP. Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature. 1991;354:480–483. doi: 10.1038/354480a0. [DOI] [PubMed] [Google Scholar]
- Kaplan MW, Robinson DA, Larsen LD. Rod outer segment birefringence bands record daily disc membrane synthesis. Vision Res. 1982;22:1119–1121. doi: 10.1016/0042-6989(82)90076-1. [DOI] [PubMed] [Google Scholar]
- Kartasasmita A, Fujiki K, Iskandar E, Sovani I, Fujimaki T, Murakami A. A novel nonsense mutation in rhodopsin gene in two Indonesian families with autosomal recessive retinitis pigmentosa. Ophthalmic Genet. 2011;32:57–63. doi: 10.3109/13816810.2010.535892. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, Scambler PJ, Beales PL, Lupski JR. BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet. 2002;71:22–29. doi: 10.1086/341031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierski W, Lloyd M, Birch DG, Bok D, Travis GH. Generation and analysis of transgenic mice expressing P216L–substituted rds/peripherin in rod photoreceptors. Invest Ophthalmol Vis Sci. 1997;38:498–509. [PubMed] [Google Scholar]
- Khan N, Lawlor KE, Murphy JM, Vince JE. More to life than death: molecular determinants of necroptotic and non-necroptotic RIP3 kinase signaling. Current opinion in immunology. 2014;26:76–89. doi: 10.1016/j.coi.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Khattree N, Ritter LM, Goldberg AF. Membrane curvature generation by a C-terminal amphipathic helix in peripherin-2/rds, a tetraspanin required for photoreceptor sensory cilium morphogenesis. J Cell Sci. 2013;126:4659–4670. doi: 10.1242/jcs.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, Leroux MR, Katsanis N, Beales PL. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Koenig R. Bardet-Biedl syndrome and Usher syndrome. Dev Ophthalmol. 2003;37:126–140. doi: 10.1159/000072043. [DOI] [PubMed] [Google Scholar]
- Korenbrot JI, Miller DL. Cytoplasmic free calcium concentration in dark-adapted retinal rod outer segments. Vision Res. 1989;29:939–948. doi: 10.1016/0042-6989(89)90108-9. [DOI] [PubMed] [Google Scholar]
- Koyfman AY, Schmid MF, Gheiratmand L, Fu CJ, Khant HA, Huang D, He CY, Chiu W. Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc Natl Acad Sci U S A. 2011;108:11105–11108. doi: 10.1073/pnas.1103634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger H, LaVail MM, Lin JH. Endoplasmic reticulum stress in vertebrate mutant rhodopsin models of retinal degeneration. Adv Exp Med Biol. 2014;801:585–592. doi: 10.1007/978-1-4614-3209-8_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunchithapautham K, Rohrer B. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy. 2007a;3:433–441. doi: 10.4161/auto.4294. [DOI] [PubMed] [Google Scholar]
- Kunchithapautham K, Rohrer B. Autophagy is one of the multiple mechanisms active in photoreceptor degeneration. Autophagy. 2007b;3:65–66. doi: 10.4161/auto.3431. [DOI] [PubMed] [Google Scholar]
- Laties AM, Bok D, Liebman P. Procion yellow: a marker dye for outer segment disc patency and for rod renewal. Exp Eye Res. 1976;23:139–148. doi: 10.1016/0014-4835(76)90197-4. [DOI] [PubMed] [Google Scholar]
- LaVail MM. Analysis of neurological mutants with inherited retinal degeneration. Friedenwald lecture. Invest Ophthalmol Vis Sci. 1981;21:638–657. [PubMed] [Google Scholar]
- LaVail MM, Yasumura D, Matthes MT, Lau-Villacorta C, Unoki K, Sung CH, Steinberg RH. Protection of mouse photoreceptors by survival factors in retinal degenerations. Invest Ophthalmol Vis Sci. 1998;39:592–602. [PubMed] [Google Scholar]
- Lee DC, Vazquez-Chona FR, Ferrell WD, Tam BM, Jones BW, Marc RE, Moritz OL. Dysmorphic photoreceptors in a P23H mutant rhodopsin model of retinitis pigmentosa are metabolically active and capable of regenerating to reverse retinal degeneration. J Neurosci. 2012;32:2121–2128. doi: 10.1523/JNEUROSCI.4752-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Burnside B, Flannery JG. Characterization of peripherin/rds and rom-1 transport in rod photoreceptors of transgenic and knockout animals. Invest Ophthalmol Vis Sci. 2006;47:2150–2160. doi: 10.1167/iovs.05-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeson TS. Rat retinal rods: freeze-fracture replication of outer segments. Canadian journal of ophthalmology. 1970;5:91–107. [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Anand M, Rao KN, Khanna H. Cilia in photoreceptors. Methods Cell Biol. 2015;127:75–92. doi: 10.1016/bs.mcb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez JJ, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. Embo J. 2012;31:552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Jones J, Parker E, Wu M, Knox BE, Burnside B. Disruption of kinesin II function using a dominant negative-acting transgene in Xenopus laevis rods results in photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2003;44:3614–3621. doi: 10.1167/iovs.03-0164. [DOI] [PubMed] [Google Scholar]
- Lisman JE, Fain GL. Support for the equivalent light hypothesis for RP. Nat. Med. 1995;1:1254–1255. doi: 10.1038/nm1295-1254. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang Q, Pierce EA. Photoreceptor sensory cilia and inherited retinal degeneration. Adv Exp Med Biol. 2010;664:223–232. doi: 10.1007/978-1-4419-1399-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Garriga P, Khorana HG. Structure and function in rhodopsin: correct folding and misfolding in two point mutants in the intradiscal domain of rhodopsin identified in retinitis pigmentosa. Proc Natl Acad Sci U S A. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Vansant G, Udovichenko IP, Wolfrum U, Williams DS. Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil Cytoskeleton. 1997;37:240–252. doi: 10.1002/(SICI)1097-0169(1997)37:3<240::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Lodowski KH, Imanishi Y. Monitoring of rhodopsin trafficking and mistrafficking in live photoreceptors. Methods Mol Biol. 2015;1271:293–307. doi: 10.1007/978-1-4939-2330-4_19. [DOI] [PubMed] [Google Scholar]
- Lodowski KH, Lee R, Ropelewski P, Nemet I, Tian G, Imanishi Y. Signals governing the trafficking and mistrafficking of a ciliary GPCR, rhodopsin. J Neurosci. 2013;33:13621–13638. doi: 10.1523/JNEUROSCI.1520-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr HR, Kuntchithapautham K, Sharma AK, Rohrer B. Multiple, parallel cellular suicide mechanisms participate in photoreceptor cell death. Exp Eye Res. 2006;83:380–389. doi: 10.1016/j.exer.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Maerker T, van Wijk E, Overlack N, Kersten FF, McGee J, Goldmann T, Sehn E, Roepman R, Walsh EJ, Kremer H, Wolfrum U. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- Malicki J. Who drives the ciliary highway? Bioarchitecture. 2012;2:111–117. doi: 10.4161/bioa.21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangini NJ, Pepperberg DR. Immunolocalization of 48K in rod photoreceptors. Light and ATP increase OS labeling. Invest Ophthalmol Vis Sci. 1988;29:1221–1234. [PubMed] [Google Scholar]
- Matsumoto B, Besharse JC. Light and temperature modulated staining of the rod outer segment distal tips with Lucifer yellow. Invest Ophthalmol Vis Sci. 1985;26:628–635. [PubMed] [Google Scholar]
- Matsusaka T. Membrane particles of the connecting cilium. Journal of ultrastructure research. 1974;48:305–312. doi: 10.1016/s0022-5320(76)80160-8. [DOI] [PubMed] [Google Scholar]
- May-Simera HL, Kai M, Hernandez V, Osborn DP, Tada M, Beales PL. Bbs8, together with the planar cell polarity protein Vangl2, is required to establish left-right asymmetry in zebrafish. Dev Biol. 2010;345:215–225. doi: 10.1016/j.ydbio.2010.07.013. [DOI] [PubMed] [Google Scholar]
- May-Simera HL, Petralia RS, Montcouquiol M, Wang YX, Szarama KB, Liu Y, Lin W, Deans MR, Pazour GJ, Kelley MW. Ciliary proteins Bbs8 and Ift20 promote planar cell polarity in the cochlea. Development. 2015;142:555–566. doi: 10.1242/dev.113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May CA, Lutjen-Drecoll E, Narfstrom K. Morphological changes in the anterior segment of the Abyssinian cat eye with hereditary rod-cone degeneration. Curr Eye Res. 2005;30:855–862. doi: 10.1080/02713680591006219. [DOI] [PubMed] [Google Scholar]
- McEwen DP, Koenekoop RK, Khanna H, Jenkins PM, Lopez I, Swaroop A, Martens JR. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc Natl Acad Sci U S A. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Miyadera K, Acland GM, Aguirre GD. Genetic and phenotypic variations of inherited retinal diseases in dogs: the power of within- and across-breed studies. Mamm Genome. 2012;23:40–61. doi: 10.1007/s00335-011-9361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlin C, Taylor L, Ghosh F, Johansson K. Autophagy and ER-stress contribute to photoreceptor degeneration in cultured adult porcine retina. Brain Res. 2014;1585:167–183. doi: 10.1016/j.brainres.2014.08.055. [DOI] [PubMed] [Google Scholar]
- Molday RS. A scanning electron microscope study of concanavalin A receptors on retinal rod cells labeled with latex microspheres. J Supramol Struct. 1976;4:549–557. doi: 10.1002/jss.400040414. [DOI] [PubMed] [Google Scholar]
- Molday RS. ATP-binding cassette transporter ABCA4: molecular properties and role in vision and macular degeneration. J Bioenerg Biomembr. 2007;39:507–517. doi: 10.1007/s10863-007-9118-6. [DOI] [PubMed] [Google Scholar]
- Molday RS, Molday LL. Differences in the protein composition of bovine retinal rod outer segment disk and plasma membranes isolated by a ricin-gold-dextran density perturbation method. J Cell Biol. 1987;105:2589–2601. doi: 10.1083/jcb.105.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz OL, Molday RS. Molecular cloning, membrane topology, and localization of bovine rom-1 in rod and cone photoreceptor cells. Invest Ophthalmol Vis Sci. 1996;37:352–362. [PubMed] [Google Scholar]
- Moritz OL, Tam BM, Hurd LL, Peranen J, Deretic D, Papermaster DS. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol Biol Cell. 2001;12:2341–2351. doi: 10.1091/mbc.12.8.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsumoto H, Roh M, Suzuki J, Hisatomi T, Ikeda Y, Miller JW, Vavvas DG. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci U S A. 2012;109:14598–14603. doi: 10.1073/pnas.1206937109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Atkins SJ, Peranen J, Swaroop A, Khanna H. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum Mol Genet. 2010;19:3591–3598. doi: 10.1093/hmg/ddq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Ghosh AK, Patil SB, Reed NA, Chan LS, Davuluri S, Peranen J, Hurd TW, Rachel RA, Khanna H. Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. J Biol Chem. 2011;286:28276–28286. doi: 10.1074/jbc.M111.237560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D, Avishai A, Avishai N, Engel A, Heuer A, Palczewski K. Serial sectioning for examination of photoreceptor cell architecture by focused ion beam technology. J Neurosci Methods. 2011;198:70–76. doi: 10.1016/j.jneumeth.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K. Clinical variability in ciliary disorders. Nat Genet. 2007;39:818–819. doi: 10.1038/ng0707-818. [DOI] [PubMed] [Google Scholar]
- Naash MI, Hollyfield JG, al-Ubaidi MR, Baehr W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc Natl Acad Sci U S A. 1993;90:5499–5503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Shimazawa M, Sugitani S, Kudo T, Imai S, Inokuchi Y, Tsuruma K, Hara H. Role of endoplasmic reticulum stress in light-induced photoreceptor degeneration in mice. J Neurochem. 2013;125:111–124. doi: 10.1111/jnc.12116. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci U S A. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Park PS, Baumeister W, Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir I, Papermaster DS. Immunocytochemical localization of opsin in the inner segment and ciliary plasma membrane of photoreceptors in retinas of rds mutant mice. Invest Ophthalmol Vis Sci. 1986;27:836–840. [PubMed] [Google Scholar]
- Nishimura DY, Fath M, Mullins RF, Searby C, Andrews M, Davis R, Andorf JL, Mykytyn K, Swiderski RE, Yang B, Carmi R, Stone EM, Sheffield VC. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci U S A. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi T, Shiba Y, Kawasaki M, Shiba T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Takatsu H, Nakayama K, Wakatsuki S. Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain. Nat Struct Biol. 2002;9:527–531. doi: 10.1038/nsb808. [DOI] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K. Gene repair of an Usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Invest Ophthalmol Vis Sci. 2012;53:4140–4146. doi: 10.1167/iovs.12-9812. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr Top Dev Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retin Eye Res. 2013 doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier S, Gingras S, Green DR. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity. 2015;42:18–27. doi: 10.1016/j.immuni.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Hanein S, Gerber S, Barbet F, Ducroq D, Dollfus H, Hamel C, Dufier JL, Munnich A, Kaplan J, Rozet JM. Retinal dehydrogenase 12 (RDH12) mutations in leber congenital amaurosis. Am J Hum Genet. 2004;75:639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KR, Palade GE, Schneider BG, Papermaster DS. Fine structure of a periciliary ridge complex of frog retinal rod cells revealed by ultrahigh resolution scanning electron microscopy. J Cell Biol. 1983;96:265–276. doi: 10.1083/jcb.96.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci U S A. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittler SJ, Davis D, Johnson LW, Kesterson RA. Generation of GARP2-specific knockout mice using zinc finger nuclease technology. ARVO. 2013 Abstract 2013, Program No. 2023. [Google Scholar]
- Portera-Cailliau C, Sung CH, Nathans J, Adler R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc Natl Acad Sci U S A. 1994;91:974–978. doi: 10.1073/pnas.91.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BA, Sandoval IM, Chan F, Nichols R, Roman-Sanchez R, Wensel TG, Wilson JH. Rhodopsin gene expression determines rod outer segment size and rod cell resistance to a dominant-negative neurodegeneration mutant. PLoS ONE. 2012;7:e49889. doi: 10.1371/journal.pone.0049889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsen CF, Cooper CB, Szerencsei RT, Murthy SK, Demetrick DJ, Schnetkamp PP. The retinal rod and cone Na+/Ca2+-K+ exchangers. Adv Exp Med Biol. 2002;514:237–251. [PubMed] [Google Scholar]
- Quazi F, Molday RS. ATP-binding cassette transporter ABCA4 and chemical isomerization protect photoreceptor cells from the toxic accumulation of excess 11-cis-retinal. Proc Natl Acad Sci U S A. 2014;111:5024–5029. doi: 10.1073/pnas.1400780111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel RA, Li T, Swaroop A. Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia. 2012;1:22. doi: 10.1186/2046-2530-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. La rétine des vertébrés. La Cellule. 1892:9. [Google Scholar]
- Rana T, Shinde VM, Starr CR, Kruglov AA, Boitet ER, Kotla P, Zolotukhin S, Gross AK, Gorbatyuk MS. An activated unfolded protein response promotes retinal degeneration and triggers an inflammatory response in the mouse retina. Cell Death Dis. 2014;5:e1578. doi: 10.1038/cddis.2014.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reme CE, Grimm C, Hafezi F, Marti A, Wenzel A. Apoptotic cell death in retinal degenerations. Prog Retin Eye Res. 1998;17:443–464. doi: 10.1016/s1350-9462(98)00009-3. [DOI] [PubMed] [Google Scholar]
- Riazuddin SA, Iqbal M, Wang Y, Masuda T, Chen Y, Bowne S, Sullivan LS, Waseem NH, Bhattacharya S, Daiger SP, Zhang K, Khan SN, Riazuddin S, Hejtmancik JF, Sieving PA, Zack DJ, Katsanis N. A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am J Hum Genet. 2010;86:805–812. doi: 10.1016/j.ajhg.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cloning and expression of gamma-adaptin, a component of clathrin-coated vesicles associated with the Golgi apparatus. J Cell Biol. 1990;111:2319–2326. doi: 10.1083/jcb.111.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;12:478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman R, Wolfrum U. Protein networks and complexes in photoreceptor cilia. Subcell Biochem. 2007;43:209–235. doi: 10.1007/978-1-4020-5943-8_10. [DOI] [PubMed] [Google Scholar]
- Rohlich P. The sensory cilium of retinal rods is analogous to the transitional zone of motile cilia. Cell Tissue Res. 1975;161:421–430. doi: 10.1007/BF00220009. [DOI] [PubMed] [Google Scholar]
- Roof D, Adamian M, Jacobs D, Hayes A. Cytoskeletal specializations at the rod photoreceptor distal tip. J Comp Neurol. 1991;305:289–303. doi: 10.1002/cne.903050210. [DOI] [PubMed] [Google Scholar]
- Roof DJ, Heuser JE. Surfaces of rod photoreceptor disk membranes: integral membrane components. J Cell Biol. 1982;95:487–500. doi: 10.1083/jcb.95.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DJ, Korenbrot JI, Heuser JE. Surfaces of rod photoreceptor disk membranes: light-activated enzymes. J Cell Biol. 1982;95:501–509. doi: 10.1083/jcb.95.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld PJ, Cowley GS, McGee TL, Sandberg MA, Berson EL, Dryja TP. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- Rosenkranz J. On the fine structure of the frog’s rod outer segments, observed by the freeze-etching technique. Z Zellforsch Mikrosk Anat. 1970;111:228–262. doi: 10.1007/BF00339787. [DOI] [PubMed] [Google Scholar]
- Rosenkranz J. New results on the ultrastructure of frog rod outer segments. Z Zellforsch Mikrosk Anat. 1973;143:45–52. doi: 10.1007/BF00307450. [DOI] [PubMed] [Google Scholar]
- Rosenkranz J, Stieve H. Frog rod outer segments, investigated by the freeze-etching technique. Zeitschrift fur Naturforschung. Teil B: Chemie, Biochemie, Biophysik, Biologie. 1969;24:1356. doi: 10.1515/znb-1969-1038. [DOI] [PubMed] [Google Scholar]
- Ross JW, Fernandez de Castro JP, Zhao J, Samuel M, Walters E, Rios C, Bray-Ward P, Jones BW, Marc RE, Wang W, Zhou L, Noel JM, McCall MA, DeMarco PJ, Prather RS, Kaplan HJ. Generation of an inbred miniature pig model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2012;53:501–507. doi: 10.1167/iovs.11-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari JC, Bredberg DL. Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J Biol Chem. 1989;264:8636–8640. [PubMed] [Google Scholar]
- Sahel J, Bonnel S, Mrejen S, Paques M. Retinitis pigmentosa and other dystrophies. Dev Ophthalmol. 2010;47:160–167. doi: 10.1159/000320079. [DOI] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, Palczewski K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–10567. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath AP, Matthews HR, Cornwall MC, Fain GL. Bleached pigment produces a maintained decrease in outer segment Ca2+ in salamander rods. J Gen Physiol. 1998;111:53–64. doi: 10.1085/jgp.111.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval IM. Characterization of spontaneous and zinc finger-mediated mutagenesis of the rhodopsin gene in photoreceptor cells in mice., Bikochemistry and Molecular Biology. Baylor College of Medicine, Houston, TX. 2012:128. [Google Scholar]
- Sasson SC, Kelleher AD. Site-specific host gene modification by zinc finger nucleases: pointing the way to drug free control of HIV-1? Clinical & translational immunology. 2014;3:e19. doi: 10.1038/cti.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetkamp PP. Calcium homeostasis in vertebrate retinal rod outer segments. Cell Calcium. 1995;18:322–330. doi: 10.1016/0143-4160(95)90028-4. [DOI] [PubMed] [Google Scholar]
- Schwarz N, Hardcastle AJ, Cheetham ME. Arl3 and RP2 mediated assembly and traffic of membrane associated cilia proteins. Vision Res. 2012;75:2–4. doi: 10.1016/j.visres.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Sedmak T, Sehn E, Wolfrum U. Immunoelectron microscopy of vesicle transport to the primary cilium of photoreceptor cells. Methods Cell Biol. 2009;94:259–272. doi: 10.1016/S0091-679X(08)94013-9. [DOI] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol. 2010;189:171–186. doi: 10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U. Intraflagellar transport proteins in ciliogenesis of photoreceptor cells. Biol Cell. 2011;103:449–466. doi: 10.1042/BC20110034. [DOI] [PubMed] [Google Scholar]
- Semple-Rowland SL, Lee NR, Van Hooser JP, Palczewski K, Baehr W. A null mutation in the photoreceptor guanylate cyclase gene causes the retinal degeneration chicken phenotype. Proc Natl Acad Sci U S A. 1998;95:1271–1276. doi: 10.1073/pnas.95.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde VM, Sizova OS, Lin JH, LaVail MM, Gorbatyuk MS. ER stress in retinal degeneration in S334ter Rho rats. PLoS One. 2012;7:e33266. doi: 10.1371/journal.pone.0033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Lupski JR. Null missense ABCR (ABCA4) mutations in a family with stargardt disease and retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2001a;42:2757–2761. [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Wensel TG, Lupski JR. Cosegregation and functional analysis of mutant ABCR (ABCA4) alleles in families that manifest both Stargardt disease and age-related macular degeneration. Hum Mol Genet. 2001b;10:2671–2678. doi: 10.1093/hmg/10.23.2671. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemiatkowska AM, Collin RW, den Hollander AI, Cremers FP. Genomic approaches for the discovery of genes mutated in inherited retinal degeneration. Cold Spring Harbor perspectives in medicine. 2014:4. doi: 10.1101/cshperspect.a017137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieving PA, Fowler ML, Bush RA, Machida S, Calvert PD, Green DG, Makino CL, McHenry CL. Constitutive “light” adaptation in rods from G90D rhodopsin: a mechanism for human congenital nightblindness without rod cell loss. J Neurosci. 2001;21:5449–5460. doi: 10.1523/JNEUROSCI.21-15-05449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal YM, Speer CM, Babcock HP, Zhuang X. Mapping Synaptic Input Fields of Neurons with Super-Resolution Imaging. Cell. 2015;163:493–505. doi: 10.1016/j.cell.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova OS, Shinde VM, Lenox AR, Gorbatyuk MS. Modulation of cellular signaling pathways in P23H rhodopsin photoreceptors. Cell Signal. 2014;26:665–672. doi: 10.1016/j.cellsig.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrand FS. An electron microscope study of the retinal rods of the guinea pig eye. J Cell Physiol. 1949;33:383–403. doi: 10.1002/jcp.1030330309. [DOI] [PubMed] [Google Scholar]
- Sjostrand FS. The ultrastructure of the innersegments of the retinal rods of the guinea pig eye as revealed by electron microscopy. J Cell Physiol. 1953a;42:45–70. doi: 10.1002/jcp.1030420104. [DOI] [PubMed] [Google Scholar]
- Sjostrand FS. The ultrastructure of the outer segments of rods and cones of the eye as revealed by the electron microscope. J Cell Physiol. 1953b;42:15–44. doi: 10.1002/jcp.1030420103. [DOI] [PubMed] [Google Scholar]
- Smith TS, Spitzbarth B, Li J, Dugger DR, Stern-Schneider G, Sehn E, Bolch SN, McDowell JH, Tipton J, Wolfrum U, Smith WC. Light-dependent phosphorylation of Bardet-Biedl syndrome 5 in photoreceptor cells modulates its interaction with arrestin1. Cell Mol Life Sci. 2013;70:4603–4616. doi: 10.1007/s00018-013-1403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Sommer D, Peters AE, Baumgart AK, Beyer M. TALEN-mediated genome engineering to generate targeted mice. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2015;23:43–55. doi: 10.1007/s10577-014-9457-1. [DOI] [PubMed] [Google Scholar]
- Song K, Awata J, Tritschler D, Bower R, Witman GB, Porter ME, Nicastro D. In Situ Localization of N and C Termini of Subunits of the Flagellar Nexin-Dynein Regulatory Complex (N-DRC) Using SNAP Tag and Cryo-electron Tomography. J Biol Chem. 2015;290:5341–5353. doi: 10.1074/jbc.M114.626556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Sterling P, Matthews G. Structure and function of ribbon synapses. Trends Neurosci. 2005;28:20–29. doi: 10.1016/j.tins.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Suber ML, Pittler SJ, Qin N, Wright GC, Holcombe V, Lee RH, Craft CM, Lolley RN, Baehr W, Hurwitz RL. Irish setter dogs affected with rod/cone dysplasia contain a nonsense mutation in the rod cGMP phosphodiesterase beta-subunit gene. Proc Natl Acad Sci U S A. 1993;90:3968–3972. doi: 10.1073/pnas.90.9.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanska K, Johnson CA. The transition zone: an essential functional compartment of cilia. Cilia. 2012;1:10. doi: 10.1186/2046-2530-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadenev AL, Kulaga HM, May-Simera HL, Kelley MW, Katsanis N, Reed RR. Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci U S A. 2011;108:10320–10325. doi: 10.1073/pnas.1016531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. Characterization of rhodopsin P23H–induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:3234–3241. doi: 10.1167/iovs.06-0213. [DOI] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. Dark rearing rescues P23H rhodopsin-induced retinal degeneration in a transgenic Xenopus laevis model of retinitis pigmentosa: a chromophore-dependent mechanism characterized by production of N-terminally truncated mutant rhodopsin. J Neurosci. 2007;27:9043–9053. doi: 10.1523/JNEUROSCI.2245-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Noorwez SM, Kaushal S, Kono M, Moritz OL. Photoactivation-induced instability of rhodopsin mutants T4K and T17M in rod outer segments underlies retinal degeneration in X. laevis transgenic models of retinitis pigmentosa. J Neurosci. 2014;34:13336–13348. doi: 10.1523/JNEUROSCI.1655-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K, Yamada E. The fine structure of the retina studied with the electron microscope. IV. Morphogenesis of outer segments of retinal rods. J Biophys Biochem Cytol. 1959;6:225–230. doi: 10.1083/jcb.6.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis GH, Brennan MB, Danielson PE, Kozak CA, Sutcliffe JG. Identification of a photoreceptor-specific mRNA encoded by the gene responsible for retinal degeneration slow (rds) Nature. 1989;338:70–73. doi: 10.1038/338070a0. [DOI] [PubMed] [Google Scholar]
- Trifunovic D, Sahaboglu A, Kaur J, Mencl S, Zrenner E, Ueffing M, Arango-Gonzalez B, Paquet-Durand F. Neuroprotective strategies for the treatment of inherited photoreceptor degeneration. Curr Mol Med. 2012;12:598–612. doi: 10.2174/156652412800620048. [DOI] [PubMed] [Google Scholar]
- Ulshafer RJ, Allen CB. Scanning electron microscopy of the retina in an animal model of hereditary blindness. Scanning electron microscopy. 1984:841–848. [PubMed] [Google Scholar]
- Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8:109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigh-Teichmann I, Vigh B, Gery I, van Veen T. Different types of pinealocytes as revealed by immunoelectron microscopy of anti-S-antigen and antiopsin binding sites in the pineal organ of toad, frog, hedgehog and bat. Exp Biol. 1986;45:27–43. [PubMed] [Google Scholar]
- Villa E, Schaffer M, Plitzko JM, Baumeister W. Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr Opin Struct Biol. 2013;23:771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Villinger C, Gregorius H, Kranz C, Hohn K, Munzberg C, von Wichert G, Mizaikoff B, Wanner G, Walther P. FIB/SEM tomography with TEM-like resolution for 3D imaging of high-pressure frozen cells. Histochem Cell Biol. 2012;138:549–556. doi: 10.1007/s00418-012-1020-6. [DOI] [PubMed] [Google Scholar]
- Viringipurampeer IA, Shan X, Gregory-Evans K, Zhang JP, Mohammadi Z, Gregory-Evans CY. Rip3 knockdown rescues photoreceptor cell death in blind pde6c zebrafish. Cell Death Differ. 2014;21:665–675. doi: 10.1038/cdd.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland S, Hughes LC, Kong C, Burgess BL, Linberg KA, Luna G, Zhou ZH, Fisher SK, Williams DS. Three-dimensional organization of nascent rod outer segment disk membranes. Proc Natl Acad Sci U S A. 2015;112:14870–14875. doi: 10.1073/pnas.1516309112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Deretic D. Molecular complexes that direct rhodopsin transport to primary cilia. Prog Retin Eye Res. 2014;38:1–19. doi: 10.1016/j.preteyeres.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen J. Induction of the unfolded protein response by constitutive G-protein signaling in rod photoreceptor cells. J Biol Chem. 2014;289:29310–29321. doi: 10.1074/jbc.M114.595207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel TG. Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 2008;48:2052–2061. doi: 10.1016/j.visres.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel TG. The Molecular Biology of Vision. In: Brady S, Albers RW, Price D, Siegel GJ, editors. Siegel’s Basic Neurochemistry. 8th. London: Elsevier; 2012. [Google Scholar]
- Wensel TG, Gilliam JC. Three-dimensional architecture of murine rod cilium revealed by cryo-EM. Methods Mol Biol. 2015;1271:267–292. doi: 10.1007/978-1-4939-2330-4_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijshake T, Baker DJ, van de Sluis B. Endonucleases: new tools to edit the mouse genome. Biochim Biophys Acta. 2014;1842:1942–1950. doi: 10.1016/j.bbadis.2014.04.020. [DOI] [PubMed] [Google Scholar]
- Wilson JH, Wensel TG. The nature of dominant mutations of rhodopsin and implications for gene therapy. Mol Neurobiol. 2003;28:149–158. doi: 10.1385/MN:28:2:149. [DOI] [PubMed] [Google Scholar]
- Witt PL, Hamm HE, Bownds MD. Preparation and characterization of monoclonal antibodies to several frog rod outer segment proteins. J Gen Physiol. 1984;84:251–263. doi: 10.1085/jgp.84.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum U. Centrin in the photoreceptor cells of mammalian retinae. Cell Motil Cytoskeleton. 1995;32:55–64. doi: 10.1002/cm.970320107. [DOI] [PubMed] [Google Scholar]
- Wolfrum U, Schmitt A. Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil Cytoskeleton. 2000;46:95–107. doi: 10.1002/1097-0169(200006)46:2<95::AID-CM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Woodruff ML, Olshevskaya EV, Savchenko AB, Peshenko IV, Barrett R, Bush RA, Sieving PA, Fain GL, Dizhoor AM. Constitutive excitation by Gly90Asp rhodopsin rescues rods from degeneration caused by elevated production of cGMP in the dark. J Neurosci. 2007;27:8805–8815. doi: 10.1523/JNEUROSCI.2751-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff ML, Sampath AP, Matthews HR, Krasnoperova NV, Lem J, Fain GL. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J Physiol. 2002;542:843–854. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- Wrigstad A, Narfstrom K, Nilsson SE. Slowly progressive changes of the retina and retinal pigment epithelium in Briard dogs with hereditary retinal dystrophy. A morphological study. Doc Ophthalmol. 1994;87:337–354. doi: 10.1007/BF01203343. [DOI] [PubMed] [Google Scholar]
- Yang J, Gao J, Adamian M, Wen XH, Pawlyk B, Zhang L, Sanderson MJ, Zuo J, Makino CL, Li T. The ciliary rootlet maintains long-term stability of sensory cilia. Mol Cell Biol. 2005;25:4129–4137. doi: 10.1128/MCB.25.10.4129-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang WJ, Craige B, Witman GB, Tsou MF, Liao JC. Superresolution Pattern Recognition Reveals the Architectural Map of the Ciliary Transition Zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang H, Constantine R, Frederick JM, Baehr W. The prenyl-binding protein PrBP/delta: a chaperone participating in intracellular trafficking. Vision Res. 2012a;75:19–25. doi: 10.1016/j.visres.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Nishimura D, Vogel T, Shao J, Swiderski R, Yin T, Searby C, Carter CS, Kim G, Bugge K, Stone EM, Sheffield VC. BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J Cell Sci. 2013;126:2372–2380. doi: 10.1242/jcs.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem. 2012b;287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Sanders E, Fliesler SJ, Wang JJ. Endoplasmic reticulum stress and the unfolded protein responses in retinal degeneration. Exp Eye Res. 2014a;125:30–40. doi: 10.1016/j.exer.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TZ, Fan B, Chen X, Wang WJ, Jiao YY, Su GF, Li GY. Suppressing autophagy protects photoreceptor cells from light-induced injury. Biochem Biophys Res Commun. 2014b;450:966–972. doi: 10.1016/j.bbrc.2014.06.082. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wensel TG, Yuan C. Tokay gecko photoreceptors achieve rod-like physiology with cone-like proteins. Photochem Photobiol. 2006;82:1452–1460. doi: 10.1562/2006-01-05-RA-767. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Molday LL, Molday RS, Sarfare SS, Woodruff ML, Fain GL, Kraft TW, Pittler SJ. Knockout of GARPs and the beta-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J Cell Sci. 2009;122:1192–1200. doi: 10.1242/jcs.042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Doggett TA, Sene A, Apte RS, Ferguson TA. Autophagy supports survival and phototransduction protein levels in rod photoreceptors. Cell Death Differ. 2015;22:488–498. doi: 10.1038/cdd.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]