Abstract
The advent of high throughput next generation sequencing (NGS) has accelerated the pace of discovery of disease-associated genetic variants and genomewide profiling of expressed sequences and epigenetic marks, thereby permitting systems-based analyses of ocular development and disease. Rapid evolution of NGS and associated methodologies presents significant challenges in acquisition, management, and analysis of large data sets and for extracting biologically or clinically relevant information. Here we illustrate the basic design of commonly used NGS-based methods, specifically whole exome sequencing, transcriptome, and epigenome profiling, and provide recommendations for data analyses. We briefly discuss systems biology approaches for integrating multiple data sets to elucidate gene regulatory or disease networks. While we provide examples from the retina, the NGS guidelines reviewed here are applicable to other tissues/cell types as well.
Keywords: ChIP-seq, Chromatin, Epigenetics, Gene Regulatory Network, Genomics, High Throughput Data, Network Analysis, Photoreceptor, Retina, Retinal Disease, RNA-seq, Systems Biology, Transcriptome, Vision, Whole Exome Sequencing, Whole Genome Sequencing, NGS data integration
1. Introduction
Technological advances in genomics and genetics, accelerated exponentially by the Human Genome Project (HGP), have begun to transform most disciplines in biology and medicine. Systems biology and personalized medicine are no longer beyond reach. Whole genome sequencing (WGS) is not limited to a chosen few, and Precision Medicine Initiative has emerged as the approach of the 21st century for prevention and treatment of human disease (www.nih.gov/precision-medicine-initiative-cohort-program). Even evolution and anthropology have embraced the power of genomic technology. The pace of discovery since the middle of the last century is astonishing; it was less than 40 years ago when chain-terminating inhibitors were used for efficient and accurate sequencing of DNA (Sanger et al., 1977b), putting us on the current path in genetics and genomics (Figure 1A). The following decades demonstrated remarkable technological and conceptual progress in human gene mapping and gene discovery, leading eventually to the HGP and the first draft of 3 billion letters of human genome (Lander et al., 2001; Venter et al., 2001). The HGP represented a milestone in biomedicine as it enabled the identification of putative genetic defects by comparing a disease sample with a standard reference genome. Soon thereafter, HapMap (hapmap.ncbi.nlm.nih.gov) and the 1000 Genomes Project (www.1000genomes.org) produced extensive catalogs of human genetic variations (Genomes Project et al., 2015; International HapMap et al., 2010), making it possible to investigate even complex phenotypes and multifactorial diseases using genomewide association studies (GWAS).
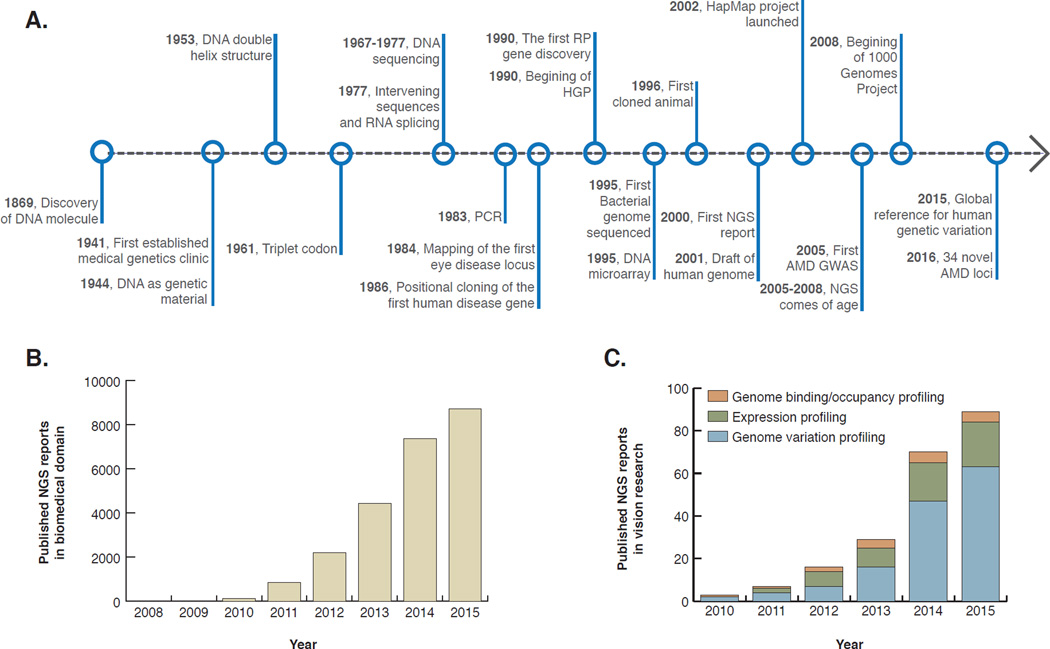
Figure 1. Timeline of human genetics and genomic technologies.
NGS based applications have been utilized widely in vision and other biomedical research. A. From the discovery of DNA molecule until today, substantial scientific and technical advancements in human genetics and eye field are presented in a chronological order. The first NGS report was published a decade after the launch of human genome project. B. Cumulative number of biomedical research papers based on NGS technologies from 2008 to 2015 in PubMed database. We believe the number of scientific reports based on NGS technologies will continue to increase as NGS become more available and affordable. C. Profiling genomic variations is more employed than expression and genome binding profiling in vision research studies. As of December 2015, the total number of NGS based studies doubled in the eye field compared to two years ago. PCR, polymerase chain reaction; RP, retinitis pigmentosa; HGP, human genome project; NGS, next generation sequencing; AMD, age-related macular degeneration; GWAS, genomewide association study.
First massively parallel sequencing approach was reported by Sydney Brenner's group and utilized microbeads for producing gene expression profiles from yeast and a human cell line (Brenner et al., 2000). More widely used next generation sequencing (NGS) methods fueled a revolution in biomedical sciences by addressing the need of generating inexpensive, reproducible, and high throughput nucleic acid sequence data (Bentley et al., 2008; Johnson et al., 2007; Margulies et al., 2005b; Mortazavi et al., 2008; Nagalakshmi et al., 2008; Shendure et al., 2005; Sultan et al., 2008). NGS has opened opportunities and challenges of “big data science” to biologists and clinicians for genomewide evaluation of genetic variations, expression of distinct RNA species, and epigenetic changes associated with development, aging, and disease (Marx, 2013) (Figure 1B,C). "Omics" is now a widely used term for describing high throughput cataloging and/or analysis of cellular molecules. We are moving forward to identify all functional genomic elements (ENCODE Project) (Consortium, 2011; Kellis et al., 2014) and understanding the role of non-coding variants in tissue-specific contexts (GTEx Project) (Consortium, 2015; Gibson, 2015). The massive surge in genomic, transcriptomic, and epigenomic data has led to systems level approaches for quantitative analysis of the dynamic interplay of molecules within a specific cell/tissue. NGS-based approaches have also quickly gained broad applicability in medicine; from genetic diagnosis and disease networks to drug discovery and pharmacogenomics. In Section 2 of this review, we will discuss basic concepts in NGS technology and expand on the Illumina platform that is widely used by genomic biologists.
Retinal degenerative diseases (RDDs) have been early targets of genetic and genomic advances. The X-linked retinitis pigmentosa locus, RP2, was the second gene mapped by polymorphic DNA markers (Bhattacharya et al., 1984), and rhodopsin was the first gene associated with visual dysfunction (Dryja et al., 1990; Farrar et al., 1991) when positional cloning was still in infancy (see Figure 1A). Since then, significant progress has been made in defining the genetic architecture of ocular diseases (Swaroop and Sieving, 2013), specifically in RDDs with 240 genes identified as of January 2016 (sph.uth.edu/retnet). Equally significant was the pioneering discovery of Complement Factor H (CFH) variants that are strongly associated with age-related macular degeneration (AMD) (Klein et al., 2005), a common multifactorial blinding disease, which greatly benefitted from the advances in HGP, genomic technologies, and genomewide association studies (Swaroop et al., 2009). NGS-based methodologies, and in particular whole exome sequencing (WES), are now becoming routine in identifying causal variants associated with Mendelian diseases. The exponential increase in discovery of rare variants by NGS has provided enhanced impetus for causal gene discovery in complex diseases. We discuss these approaches and discoveries in Section 3.
Each of us carries millions of genetic variations that define our unique identity (Genomes Project et al., 2015). While a majority of these variations do not seem to have an obvious pathological impact, some are associated with human traits or clinically identifiable diseases. Despite the remarkable advancements in discovering disease-associated or causal variants (mutations), molecular mechanisms and cellular pathways that underlie the pathophysiology have not been adequately delineated in most cases, in part because of our incomplete understanding of “normal” biological function. Furthermore, signaling and gene regulatory networks that control fundamental biological processes, including organogenesis and aging, are poorly understood in mammals, especially in the context of the nervous system. NGS-based transcriptome analysis (RNA-seq) (discussed in Section 4) allows profiling of global patterns of expression of distinct RNA species (including mRNA, miRNA, lncRNA, and tRNA) that perform unique functions within a given cell/tissue during development or disease pathogenesis. Temporal transcriptome profiling of "normal" and "mutant" retina can help define how genetic changes lead to cellular dysfunction and elucidate gene networks associated with homeostasis and disease. Recent successes in generating expression profiles from single retinal cell types (Kim et al., 2016)(Siegert et al., 2012) or even from single cells (Macosko et al., 2015) are providing novel molecular insights into cell fate determination and disease mechanisms. Epigenetic changes are another major factor in influencing physiology and biological pathways. NGS can be used to evaluate changes in histone modification (by chromatin immunoprecipitation followed by NGS, ChIP-seq) or chromatin structure (DNase I or Tn5 accessibility profiling, DNase-seq or ATAC-seq respectively). Application of NGS in epigenetic profiling is discussed in Section 5. Additionally, ChIP-seq can be utilized to identify targets of DNA-binding proteins or transcriptional regulators, which constitute essential components of gene regulatory networks (GRNs) as demonstrated for Neural Retina Leucine Zipper (NRL) or Cone-Rod Homeobox (CRX) in the context of retinal development and disease (Corbo et al., 2010; Hao et al., 2012; Yang et al., 2015).
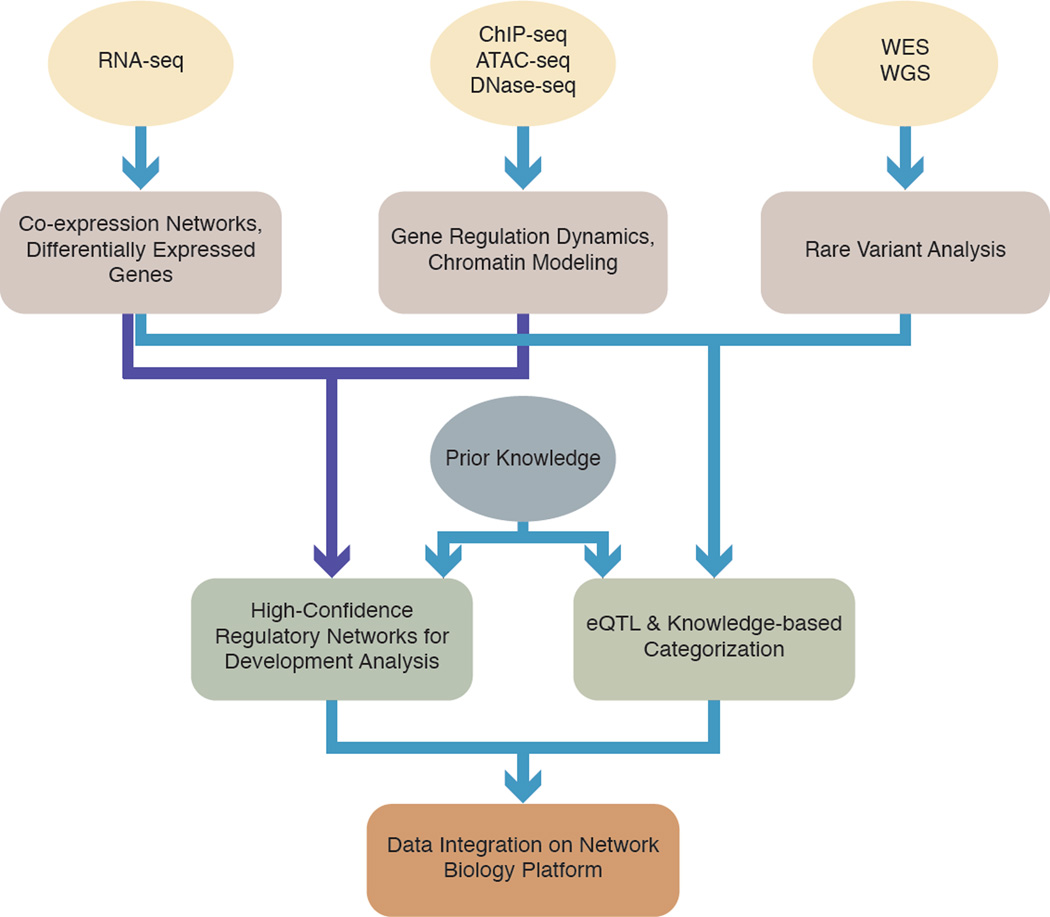
The integration of heterogeneous "omics" data poses major challenges for delineating the complex interplay of genes in pathways and networks. In-depth characterization of genes is valuable in elucidating basic biology and has contributed substantially to our current knowledge. However, cells encompass a complex internal spatial architecture with highly organized compartments, and genes (and their products) do not function in isolation. NGS represents a unique opportunity for investigating genome-scale data sets to build system-level gene networks. The field of vision in general, and retina in particular, has undergone an extensive expansion in gene discovery, which has not been accompanied by detailed cell- or tissue-specific global transcriptome and epigenetic profiling that would permit downstream studies on genotype-phenotype association. In Section 6, we will highlight two system level approaches - network inference and expression quantitative trait locus (eQTL) analysis - to extract meaningful information by integrating large data sets.
This review primarily focuses on general aspects of NGS technology and data analysis. A list of definitions for commonly used terms is provided in Box 1, and description of distinct file types specific to NGS are included in Box 2. We encourage readers to peruse the following recent reviews on NGS and its applications: (Boycott et al., 2013; Bras et al., 2012; Conesa et al., 2016; Davey et al., 2011; Furey, 2012; Koboldt et al., 2013; Ozsolak and Milos, 2011; Yang et al., 2015)
Box 1. Glossary.
Bioinformatics: An interdisciplinary field that encompasses biology and computer science to develop resources and software’s that aid in storage and analysis of omics data.
Systems biology: A discipline that focuses on understanding structure and function of biological systems on multiple stages including molecular, cellular, tissue, and organ levels.
Personalized medicine/Precision medicine: A disease diagnosis and treatment approach that aims in providing targeted therapies to patients based on their individual genetic architecture and disease.
Genome: Set of all genetic information in an organism.
Genomics: Large-scale study of all the genes in an organism.
Transcriptome: Full range of RNA molecules (miRNA, ncRNA, rRNA, tRNA, etc.).
Transcriptomics: Large-scale study of all the Transcripts in a cell.
Epigenetics: Processes affecting gene expression that do not alter DNA sequence.
Genome-wide association studies (GWAS): A method of identifying disease-associated variants across the whole genome.
DNA Sequencing: Process of determining the sequence of bases along a DNA molecule.
Whole Exome Sequencing: Sequencing of protein-coding regions of the genome.
Exome: Protein coding sequences of the genome.
Gene regulatory networks (GRNs): Logic maps that details how genes are regulated.
Expression quantitative trait locus (eQTL): Genomic regions that harbor DNA sequence variants that influence the expression level of one or more genes.
Chromatin: A complex of DNA and proteins that compacts and organizes chromosomes within the nucleus of eukaryotic cells.
Chromatin Immunoprecipitation (ChIP): An assay used in biology to identify protein-DNA interactions, such as transcription factors or histones, in vivo.
Single-end Sequencing: Involves sequencing from only one end of a sequencing library.
Paired-end Sequencing: Involves sequencing from both ends of a sequencing library and provides more accurate alignment and gene/transcript abundance levels.
Multiplex Sequencing: Simultaneous sequencing of multiple sample sequence libraries in the same reaction vessel.
Fragment: A piece of unknown DNA resulting from sequencing library construction originally derived from DNA shearing, enzyme digestion, tagmentation, or reverse transcription of the experimental sample.
Library: Set of all DNA fragments prepared for sequencing for an experiment.
Library size/Sequencing depth/Read depth:
Number of fragments sequenced.
Sequencing Read: Output from sequencing machine of the library fragments.
Coverage: Fraction of the genome mapped by sequenced reads.
Contig: A set of overlapping reads.
Variant Calling: Process of identifying single nucleotide variants from NGS data using various mathematical and computational tools.
Read mapping: Process of aligning sequence reads to a reference genome.
Transcriptome Reconstruction: Process of computational reconstruction of transcripts from sequencing reads
Peak calling: Process of identifying genomic locations where reads align.
Single Cell Sequencing: Sequencing at a single cell level.
Box 2. Frequently used file formats and their descriptions in Illumina sequencing workflows.
BCL (.bcl): Base call (BCL) is a binary file that is generated by Illumina sequencing instrument as an output in each sequencing cycle. bcl2fastq tool merges per-cycle BCL files into FASTQ files that are input of many downstream sequencing analysis tools such as aligners and de novo assemblers.
FASTQ file (.fastq or .fq): Once sequencing is completed, nucleotide sequence and quality score of each read is stored in FASTQ text file format for further analysis steps. An example of the first four lines of a standard FASTQ file is shown below.
Line 1. @HWI-D00541:31:C75NKANXX:4:1101:1333:1963 2:N:0:GCCAAT
Line 2. GCTAGACATTGTTTTATCCAATCTCATCTTGCACTTCTCTAGCATC…
Line 3. +
-
Line 4.
BBBBBFFFFFFFBFFFFFFFFFFFFFFFFFFFFB/BFFB<BFBBFFFFFFFF</7B
The first line always starts with @ character and represents the sequence identifier. The second line is the biological sequence of the read, which is composed of a four-letter nucleotide alphabet (A, T, G and C). The third line is again sequence identifier and always starts with + character. As in this example, the third line may sometimes consist of only a + character. The fourth line describes the quality score of corresponding sequencing read, which is coded with ASCII characters. We recommend readers to look at Cock et al., 2010 article for detailed information regarding FASTQ file variants (Cock et al., 2010).
SAM file (.sam): Sequence Alignment/Map (SAM) is a text file that stores alignment information of reads to reference genome or given sequence (Li et al., 2009). Some aligners such as STAR (Dobin et al., 2013) generate SAM file as an output of alignment process of short reads to reference genome. A SAM file includes a header section starting with @ character and alignment section consisting of multiple lines.
BAM file (.bam): Binary Alignment/Map (BAM) is the binary version of SAM file (Li et al., 2009). As SAM file does, BAM file stores alignment information of reads however BAM file is compressed (has smaller size) and more efficient in many sequencing analysis tools as it is compared to SAM file. A SAM file can be converted to a BAM file (or vice versa) with the help of SAMtools standalone software (Li et al., 2009).
BED file (.bed): A BED file is a tab-delimited text file that might consist of multiple lines each representing a single genomic region or feature such as an exon or gene body. There are three required fields (represented below) in a standard BED file named chrom, chromStart and chromEnd. “chrom” stands for chromosome in which the region is located. “chromStart” represents the starting bp of the region, inclusively, with the first bp in a chromosome designated as 0. “chromEnd” is the end position of the region, exclusively. The other nine fields are optional and provide additional information about the genomic region such as relevant strand information or a context-specific score. Detailed information about BED file and its variants can be found at UCSC Genome Bioinformatics web site: http://genome.ucsc.edu/FAQ/FAQformat
| chr2 | 116848098 | 116877168 |
| chr2 | 118745757 | 118748810 |
| chr2 | 120024806 | 120027453 |
| chr3 | 89394289 | 89398779 |
| chr3 | 94998833 | 95004869 |
Multi-FASTA file (.fa): A multi-FASTA file is a text file that consists of multiple FASTA format sequences. Below is a simple multi-FASTA file in which each sequence. identifier starts with > character and followed by single or multiple lines of biological sequence. We typically use multi-FASTA files of genomes and transcriptomes that can be downloaded multiple sources (e.g. Ensembl or Gencode web sites) while building genome and transcriptome indexes in sequencing data analysis.
>Nucleotide_Sequence1 CGCGCCCGGCCCGTAGCGTCCTCGTCGCCGCCCCCCGCGGACTAGCCCGGGTGGCCTCGTCTCGCAGCC GCACTCCCCGTGAGCCCGCGTGGACGCTCTCGCCTGAGCG >Nucleotide_Sequence 2 CGTAGCGCAGCGATCGGCGCCGGAGATTCGCGACACTGGCGCGCGGGCGAGCGCACGGGCGCTCACCCG ACACTCCGCGCCGCCCGCCGGCCAGGACCCGCGGCGCGACAGTCCGGCAGCGCCGGGGTTAAGCGGCCC AAGTAAATCGCGGCGCCGCGCTACAGCCAGCCT > Nucleotide_Sequence3 GGCCCGCTGAGGCTTGTGCCAGACCACCTCCCCTCCCCCTTTTTGGAAACCTCAGGTACACGACATATC CAGACGCGGGAT
GTF and GFF files (.gtf and .gff respectively): General Transfer Format or Gene Transfer Format (GTF) and General Feature Format (GFF) are text-based annotation files that stores gene structure information of any genome. In both GTF and GFF file, each line represents a single genomic feature (e.g. an exon structure information). A GFF file has nine required field separated by tabs, and the first eight fields of a GTF file are same as GFF file however the ninth field is always start with two mandatory attributes named gene_id value and transcript_id value. GFF and GTF files might be either as an input of aligner software or might be output of a genome assembler tool. Below is example lines from a mouse GTF file downloaded from Ensembl database.
2. Next Generation Sequencing Technologies
The era of genomics was born in 1977 with the sequencing of the bacteriophage phiX174 by Fredrick Sanger (Sanger et al., 1977a). Subsequent improvements made the Sanger method the dominant sequencing approach, which was then employed for the monumentally ambitious HGP (1990–2003) (Collins et al., 2003; Watson, 1990). This endeavor required a large workforce and a 15-year worldwide effort to sequence approximately 3 billion base pairs of the human genome (http://www.genome.gov/11006929). Concomitantly, a privately funded human genome sequencing project employed a strategy of “whole-genome, random shotgun sequencing” in which DNA fragments of known lengths were directly cloned into vectors, sequenced, and assembled computationally (Venter et al., 2001). This approach became the standard for DNA sequencing and evolved into NGS and associated technologies. Notably, a handful of scientists were able to generate a complete human genome sequence in 4 months (Wheeler et al., 2008) by massive parallelization of the biochemical sequencing steps (Margulies et al., 2005a). Though distinct NGS platforms employ different approaches, all techniques make use of massive parallelization of the biochemical and sequencing steps without the need for cloning.
2.1. Applications of NGS Methodology
The plethora of new applications of NGS is truly remarkable as nearly every type of nucleic acid can be assayed by this technology (Lander, 2011). NGS techniques can be broadly classified into applications for investigating genome, transcriptome, and epigenome. Genomic assays include WGS, WES, and targeted resequencing of specific regions to discover variants associated with cell function or disease. NGS-based transcriptome analysis (RNA-seq)(Mortazavi et al., 2008) encompasses quantitative gene expression profiling, discovery of novel transcribed sequences (Trapnell et al., 2010), and non-coding RNA species such as miRNA and lncRNA (Graveley et al., 2011; Guttman et al., 2009). Epigenome methods generally focus on chromatin structure and include DNase-seq (Yaragatti et al., 2008), ATAC-seq (Buenrostro et al.), DNA methylation (Lister et al., 2008), and histone modification ChIP-seq (Barski et al., 2007). We have also included TF ChIP-seq (Johnson et al., 2007; Jothi et al., 2008; Lefrancois et al., 2009; Robertson et al., 2007) in the Epigenome section.
2.2. Current NGS Platforms
In less than a decade, increased throughput of sequencing and dramatic reduction in costs have led to NGS becoming a widely used genomic technology. Since the release of the first commercially available system (GS20 from 454 Life Sciences) with a throughput of 20 megabase pair (Mbp) per run (Margulies et al., 2005b), the NGS technology has improved immensely. The current Illumina HiSeq X system is capable of producing 1.8 terabase pair (Tbp) of sequencing data per run, nearly a 100,000-fold increase within a 10-year period. During this relatively brief span, several NGS systems such as HeliScope from Helicos BioSciences (Thompson and Steinmann, 2010) and 454 GS FLX from Roche have been discontinued, whereas a few others including SOLiD (Shendure et al., 2005) and Ion Torrent (Rothberg et al., 2011) are holding on to their share in the market. The new NGS systems include single molecule sequencers (e.g., RS (Eid et al., 2009) from Pacific Biosciences and minION (Clarke et al., 2009) from Nanopore), which can provide high read lengths and resolution of DNA modifications. Table 1 lists a survey of currently available sequencers and their technical specifications. The juggernaut of the industry are the sequence-by-synthesis (SBS) (Bentley et al., 2008) systems from Illumina that boast a wide range of applications, relative ease of use, multiple levels of throughput, flexibility in configuration, and relatively low sequencing cost. Thus, the following sections will focus on Illumina sequencing technology.
Table 1.
Technical specifications of four major sequencing platforms.
| Sequencing Platform |
Total output (bases per run) |
Total reads (million per run) |
Read Length (bases) |
Run time (days) |
Purpose/Definition | |
|---|---|---|---|---|---|---|
| Illumina | HiSeq X | 1.6 – 1.8 Tb | 6000 | 2×150 | < 3 | Allows sequencing of larger genomes (e.g. mammalian genomes) at population level |
| MiSeq | 300 Mb – 15 Gb |
50 | 2×300 | 0.2 – 2.7 | Designed for particularly small genomes (e.g. bacterial genomes) and amplicon sequencing |
|
| Life Technologies | Solid 5500 Systems |
80 Gb – 320 Gb |
1200 M – 2400 M |
50 – 2×50 bp |
7 | Offers application-per- lane sequencing that allows transcriptome, exome and genome sequencing concurrently in a single run. Additionally, pay- per-lane sequencing feature makes Solid 5500 Systems cost- effective because reagent usage does not require for unused lanes. |
| Ion Torrent 520 Chip |
600 Mb – 2 Gb | 3 | 200 – 400 bp |
0.1 | Ion S5 System allows generation of diverse sequencing data ranging from targeted re-sequencing to genome sequencing with as little as 10 ng sample. |
|
| Ion Torrent 540 Chip |
10 – 15 Gb | 60 | 200 – 400 bp |
0.1 | ||
| PacBio | Sequel System |
500 Mb – 16 Gb |
55 – 880 M | up to 60 kb | < 0.1 – 0.3 | Useful in the studies of de novo assembly of large genomes. Sequel System can be utilized for generating variation, expression and/or regulation related sequencing data. |
| PacBio RS II | 500 Mb – 16 Gb |
55 – 880 M | up to 60 kb | < 0.1 – 0.3 | Much more suitable for sequencing small genomes although animal and plant genomic studies is also possible. |
|
| Nanopore | PromethION | up to 12 Tb1 | 1250 M1 | 230 – 300 kb1 |
2 | Ideal for large sample numbers. PromethION can sequence up to 48 samples in a single run |
| MinION | up to 42 Gb1 | up to 4.4 M1 | 230 – 300 kb1 |
2 | Portable sequencing instrument. MinION can be run with a desktop or laptop computer and data can be performed in real time. |
|
Measured at “fast” mode in which 500 bases pass through the pore per second.
2.3. Illumina Sequencing Methodology
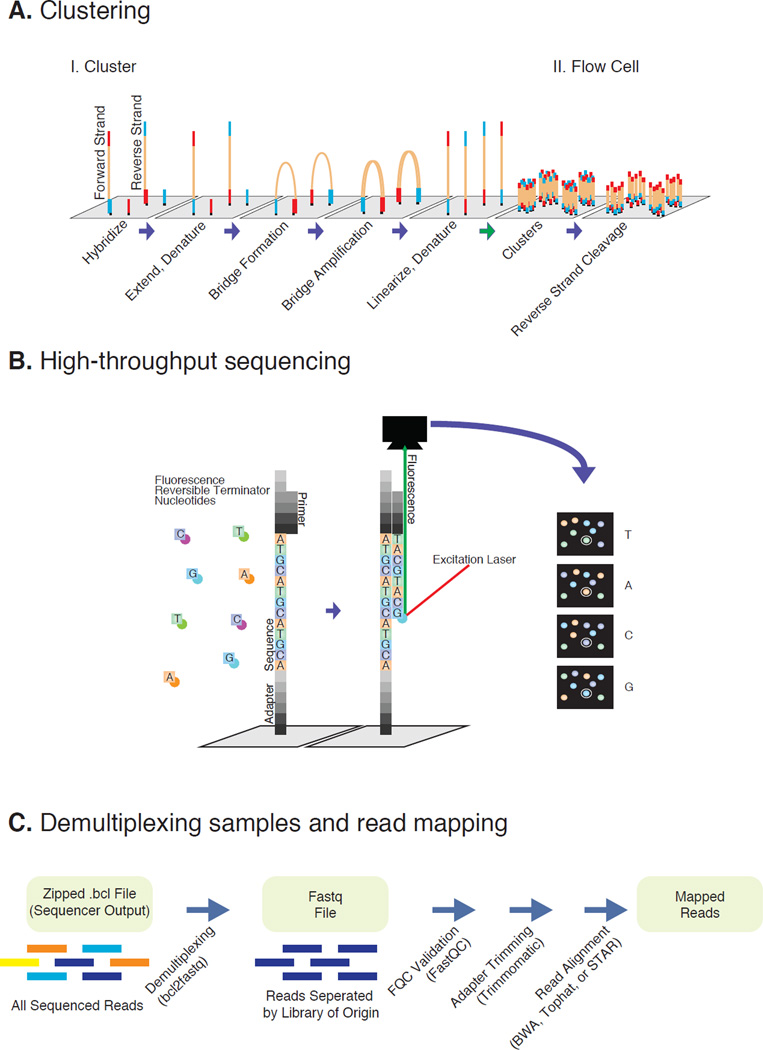
The NGS technologies using the Illumina platform employ a massively parallel SBS methodology which involves sequencing the ends of millions, or even billions, of DNA fragments in parallel and performing read assembly for analysis. The routine sequencing protocol includes three steps: sample library construction, cluster generation, and SBS. All sample libraries are composed of double-stranded DNA inserts flanked by known adapter sequences capable of hybridizing to the oligonucleotides on the Sequencer’s flow cell surface. The flow cell is the heart of the technology, consisting of a thick, glass fluidic device, reminiscent of a microscope slide, with single or multiple channels (lanes) coated with a lawn of two designed oligonucleotides (Fedurco et al., 2006; Turcatti et al., 2008). Cluster generation proceeds when denatured DNA libraries are allowed to randomly hybridize to the oligonucleotide lawn in the channels by their adapter ends (Figure 2A). A covalently attached DNA fragment is created by extension of the flow cell oligonucleotides using the hybridized library fragment as a template. The original library strands are then denatured and washed away, leaving only the newly synthesized strand. A complementary copy of the covalently bound strand is then generated through bridge amplification, a process by which the strand bends to hybridize to an adjacent and complementary oligonucleotide, thereby allowing the polymerase to extend the complementary strand. Denaturation then results in two covalently bound complementary copies of the original DNA fragment. Bridge amplification is repeated 24 times to produce clusters of DNA clones in which half of the molecules represent the forward orientation and the other half the reverse. The reverse orientation strands are then cleaved and washed away leaving only clusters of identical forward strands (ready for SBS). A sequencing primer flanking the unknown insert region is hybridized just prior to the SBS. Fluorescently labeled and reversibly terminated nucleotides are then flowed across the lawn of clusters allowing only the first nucleotide base to be incorporated (Figure 2B). The clusters on the flow-cell surface are then imaged by laser excitation, revealing a single color corresponding to the incorporated nucleotide. The fluorophore is cleaved off after imaging, and the terminator is reversed allowing for the incorporation of the next base. This process is repeated until the predetermined sequence (read) length is reached. The Illumina technology allows for single-end (SE) or paired-end (PE) sequencing and single or dual indexing of libraries for multiplex (multiple) loading of libraries. In PE sequencing both ends of the DNA insert are sequenced, generating two reads per library fragment. To generate a second read, another round of bridge amplification is then performed, followed by cleavage of the forward strand prior to performing the second round of SBS. Both reads are processed together computationally. To take full advantage of the massive numbers of clusters generated in each experimental run and reduce costs, and depending on the depth of sequencing required, many samples can be multiplexed and sequenced simultaneously. The adapters of each library may contain distinct index sequences that are used to identify the library from which each read originates. Up to 24 libraries may be pooled together in each lane of the flow cell using single indexing, and as many as 96 libraries combined when dual indexing is used.
Figure 2. Illumina sequencing and data processing workflow.
A. Denaturated NGS library fragments are flowed across a flow cell and hybridize on a lawn of complementary Illumina adapter oligos. Complementary fragments are extended, amplified via bridge amplification PCR, and denaturated, resulting in clusters of identical single-stranded library fragments. B. Fragments are primed and sequenced utilizing reversible terminator nucleotides. Base pairs are identified after laser excitation and fluorescence detection. C. Raw data is demultiplexed into individual libraries and assessed for quality. Removing adapter reads reduces technical noise. Finally reads are aligned onto assembly of interest.
2.4. NGS Experimental Design
Careful planning can maximize the success of NGS experiments, yielding useful data for extracting biologically relevant information. In addition to the basic precepts, such as the number of samples, replication and controls, the experimental design should consider the depth at which each sample is to be sequenced, the optimal length of sequence reads, whether PE or SE sequencing is more appropriate, and which samples are combined for multiplexing. PE sequencing is usually preferred to SE sequencing because of gain in coverage and enhanced accuracy of alignment, especially for mapping and quantification of RNA-seq data. Long sequencing reads (over 50 bp) are not needed for miRNA-seq and epigenomic assays, and SE sequencing may be sufficient in many experiments. In addition to a reduction in costs, multiplexing of samples can minimize lane or NGS-run bias and should be employed based on depth considerations. For example, it is better to pool 24 libraries and sequence these on three lanes of a flow cell, as compared to preparing three pools of 8 libraries and sequencing each pool in a single lane.
2.5. Basic Raw Data Pre-processing and Quality Assessment
Sequencing reads from the Illumina platform are generated in a binary base call (BCL) file format that is incompatible with most open source analysis software. Therefore, the first pre-processing step involves the conversion of BCL files to the universally accepted FASTQ format (Figure 2C; Box 1). Only high quality reads that successfully pass Illumina's "Chastity Filtering" are kept for further sequence analysis, which can be performed by a multitude of available software. The next step in data pre-processing is to eliminate the Illumina adapters, and poly(A) or poly(T) sequences (added during cDNA and library preparation) that may be present at the end of the reads. We recommend Trimmomatic software (Bolger et al., 2014) though others such as Cutadapt and FASTX-Toolkit can be used. Quality control (QC) software (such as FASTX-Toolkit, FastQC, NGS QC Toolkit, and PRINSEQ (Schmieder and Edwards, 2011) can then be applied to provide crucial information about the quality of sequencing reads including quality score distribution along the reads, GC content, read length, and level of sequence duplication. Many of these tools can perform both sequence trimming and QC analysis. Once the FASTQ files have been validated, sequence alignment and bioinformatic analysis is performed based on the goals of the experiment, as discussed in Sections 3–5.
3. Genome
WGS, WES, and targeted re-sequencing are powerful and relatively unbiased methods for discovering disease-associated genetic variations and genes. WGS provides a unique window to investigate genetic or somatic variations, leading to new avenues for exploration of normal and disease phenotypes. However, the massive quantity of data and the requirement of significant computational resources make WGS cost prohibitive for routine genetic and biological studies at this stage (as of March 2016). In contrast, WES focuses on capturing and sequencing protein-coding regions (exomes), limiting the data to a more functionally informative part of the genome. WES has become a popular choice for genetic studies, primarily for disease gene identification and clinical diagnosis, yielding coding and splice-site variants from a large number of samples within a relatively short time-span (e.g., WES of 96 samples can be completed within a week). This method is best suited for identifying highly penetrant variants associated with Mendelian diseases. Targeted re-sequencing involves the capture and sequencing of selected genomic regions and complements WES and WGS. In this review, we focus on WES, describing various capture methods, computational tools and applications.
3.1. Whole Exome Sequencing
Whole exome sequencing (WES) involves capture and sequencing of the coding regions (the exomes) of genome (Ng et al., 2009; Priya et al., 2012). WES has become the method of choice for mutation identification since a majority of disease-causing variants for monogenic disease are detected in the protein-coding regions that comprise less than 2% of the genome (Bamshad et al., 2011). Currently, four major exome capture/enrichment kits are available: Nextera Rapid Capture Exome and TruSeq exome enrichment (both from Illumina), SureSelect XT Human All Exon (Agilent), and SeqCap EZ Human Exome Library (Roche/NimbleGen). They differ in the target selection regions, bait (capture probe) length, bait density used for capture of exonic sequences, and the total number of targeted nucleotides (see Table 2 for distinguishing features). For instance, NimbleGen, and Agilent targets cover 64.1 Mb and 51.1 Mb, respectively, whereas the two Illumina platforms target 62.08 Mb of the human genome. The four kits only share 26.2 Mb of the total targeted nucleotides. We have used Nextera Rapid Capture Exome platform extensively. Briefly, this protocol uses transposase-mediated fragmentation and adapter ligation using 50 ng of DNA, followed by two rounds of exome capture with biotinylated DNA baits complementary to the target exomes (Figure 3). A performance comparison of the four platforms demonstrated high target enrichment for the consensus coding sequence (CCDS), with the Nextera exhibiting a sharp increase in read depth for GC-rich sequences compared to other technologies (Chilamakuri et al., 2014).
Table 2.
Capture technologies for WES
| Platform | Target Capture Region Length |
Bait Length |
Bait Density |
Notes |
|---|---|---|---|---|
| NimbleGen | 64.1 Mb | Not Available |
2.8 Mb | Requires 1–2 µg of DNA, utilizes overlapping biotinylated DNA probe design. Adapter addition is through ligation. |
| Agilent | 51.1 Mb | 66.48 Mb | 1.63 – 3.5 Mb |
Requires 2–3 µg of DNA, utilizes biotinylated cRNA as bait from non- overlapping probes that are directly adjacent to each other. Adapter addition is through ligation. |
| Nextera | 62.08 Mb | 33.01 Mb | 1.44 Mb | Requires 50 ng of DNA. The gapped capture probes rely on paired-end reads to extend outside the bait sequences and to fill the gaps. Utilizes transposons for fragmentation and adapter ligation without the need of mechanical shearing. |
| TrueSeq | 62.08 Mb | 33.01 Mb | - | This capture platform from Illumina is similar to Nextera except that the input requirement is higher (1 µg) and ultrasonication is used for DNA fragmentation. |
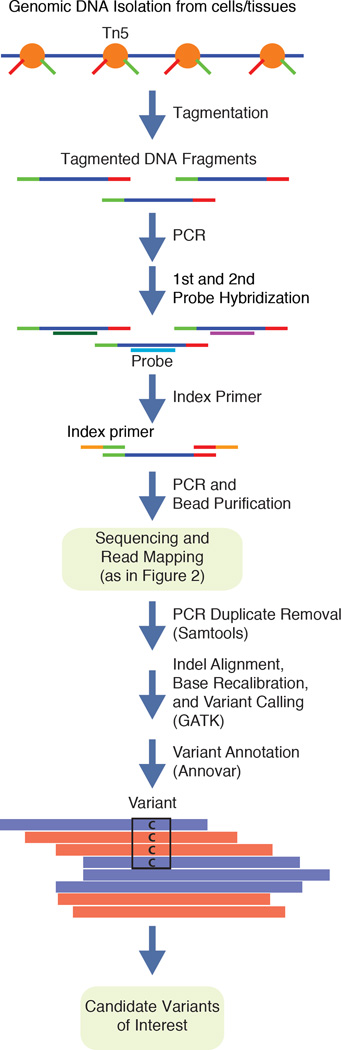
Figure 3. WES workflow and analysis.
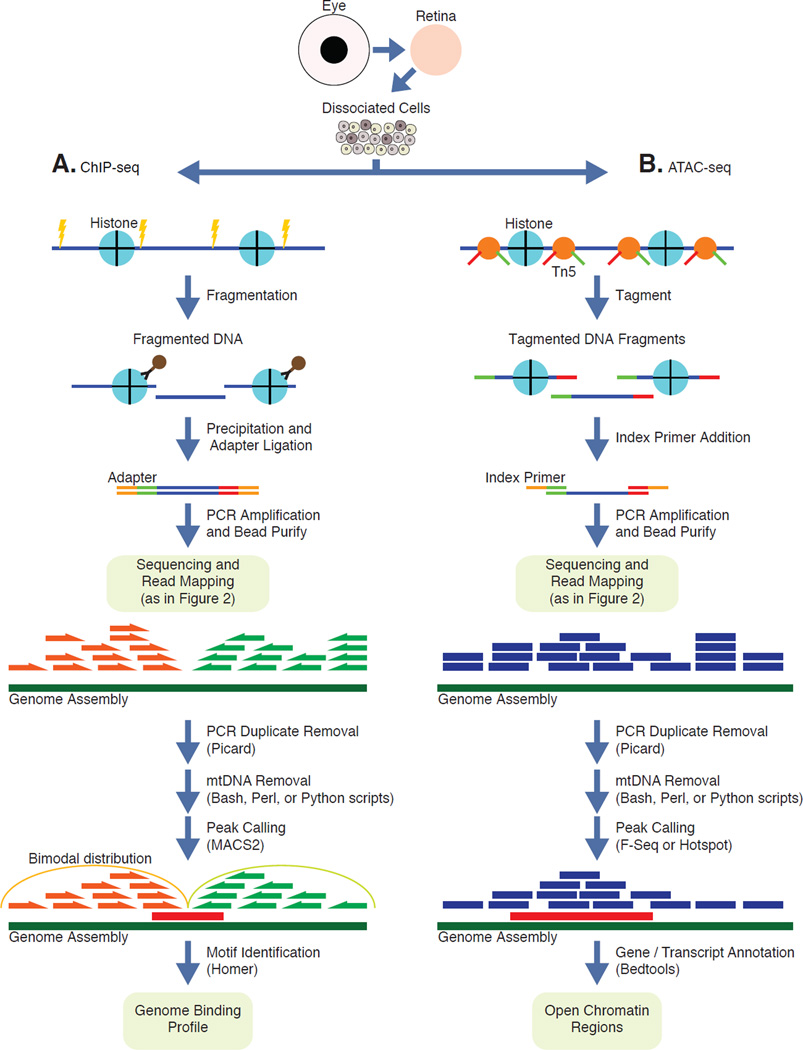
Genomic DNA from cells or tissue is tagmented using hyperactive Tn5 transposase coupled with Illumina sequencing adapters as described in (http://www.illumina.com/products/nextera-rapid-capture-custom-enrichment-kit.html). After PCR amplification, DNA probes specific to exonic sequences are used to isolate coding sequences using two-step hybridization. Library amplification with index primers allow for multiplexing a variety of libraries in the same sequencing flow cell. After sequencing and read mapping steps, PCR duplicates are removed using available computational tools. Realignment around indels, base recalibration, variant calling and annotations are all WES-specific computational processes to extract variant information.
WES is relatively comprehensive, inexpensive, and rapid for identifying coding and splice site mutations compared with other variant detection methods and adopted extensively in clinical diagnostics. However, several limitations exist. None of the WES capture probe sets seem to target all of the exons listed in the Consensus Coding Sequence project (CCDS) (Farrell et al., 2014), RefSeq (O'Leary et al., 2016), or Ensembl (Cunningham et al., 2015) databases. The capture step is not uniform and tends to have bias against high GC rich regions. In addition, identification of variants is restricted by capture design, which would generally skip unannotated or as yet unidentified exons and variants residing in the non-coding and/or regulatory region of the genome. Furthermore, only 92–95% of exons are captured in WES and mutations may be missed even if the region is included in the capture probe design. Capture kits are also not very efficient in identifying structural variants such as translocations and inversions.
3.2. Primary Analysis of WES data
After quality control analysis of NGS data (described in Section 2.5), we have four additional steps in our WES pipeline to identify genomic variants with high accuracy (Figure 3).
3.2.1. Sequence Alignment
The short reads generated in the NGS are first aligned to a reference genome with mapping tools (Table 3), such as BWA and STAR, producing sequence alignment/map (SAM) files. Accurate and efficient mapping of WES data is essential for variant identification.
Table 3.
Commonly Used Tools for RNA-Seq Data Analysis Workflow.
| Process | Tool | Description | |
|---|---|---|---|
|
Short read alignment |
Splice- unaware |
BWA | Burrows–Wheeler Transform algorithm based tool that accurately maps reads (up to 1Mbp) to a given reference genome. |
| Bowtie2 | Memory-efficient aligner for mapping very short reads (ranging from 50 to 100bp) to large genomes. |
||
| Splice- aware |
TopHat2 | Short read aligner for the discovery of novel splice sites at exon-exon junctions. |
|
| STAR | Spliced read aligner for de novo identification of novel splice junctions. STAR is significantly faster at read mapping compared with other sequence aligners. |
||
|
Transcript assembly |
De novo | Trinity | Full-length transcript assembler for the identification of novel transcripts from Illumina RNA-seq data. Trinity uses a de Brujin graph-based method for the construction of transcript structures and provides accurate transcript assemblies when a reference genome is unavailable. |
| Oases | Short read assembler for RNA-seq samples with missing reference genomes. Oases enables the discovery of novel exon structures in previously undescribed transcripts from Illumina, SOLID and 454 data. |
||
| Ab initio | Cufflinks | Transcriptome analysis suite for RNA-seq assembly as well as quantification and differential expression analysis. Cufflinks assembles short reads and predicts novel exon structures on the reference genome. |
|
| Scripture | A tool developed for stringently describing abundantly expressed novel transcripts. It is more appropriate to use Cufflinks instead of Scripture to identify non-coding RNA that are generally expressed at low levels (Cabili, M. N., et al. (2011). |
||
| Quantification | eXpress | A transcript quantification tool based on the Expectation Maximization (EM) algorithm for estimation of transcript-level abundances. Additionally, it provides analysis diverse options for allele-specific expression, ChIP-seq and metagenomic data. |
|
| RSEM | Another commonly used EM algorithm-based tool for accurate quantification of transcripts. RSEM is also capable of estimating gene-level expression levels. |
||
| Kallisto | An ultra-fast and alignment-free transcript quantification tool. Kallisto can approximate expression levels of transcripts in minutes on a standard desktop computer using a pseudoalignment approach, and it does not require large memory for quantification relative to other quantification tools. |
||
| Cuffquant | Developed as a part of Cufflinks. Cuffquant calculates a gene expression table at the transcript level. The output of Cuffquant may be used as an input to Cuffdiff 2 once properly normalized using Cuffnorm. |
||
|
Differential expression |
Cuffdiff 2 | Differential expression analysis tool, part of the Cufflinks transcriptome analysis suite, for group comparison at transcript resolution. Cufflinks 2 supports robust alternate splicing and differential promoter usage analyses. |
|
| edgeR | An R/Bioconductor package for performing count- based differential expression analysis. edgeR provides diverse statistical test options, including generalized linear models and negative binomial distribution based modeling. It is also suitable for differential analysis of other data types i.e. ChIP-seq and proteome peptide count data. |
||
| DESeq | Another R/Bioconductor package for statistical analysis of replicated high-throughput count data. DESeq is built on a negative binomial distribution model and can also be utilized for differential expression at exon level resolution to resolve splice event differences between samples. |
||
| limma | A linear modeling-based tool for testing differential gene expression for both microarray and RNA-seq data. limma is an R/Bioconductor package that provides diverse functionalities to users, including data-preprocessing, transformation and normalization. |
||
|
Alternate splicing analysis |
MATS | Detects altered exon usage. MATS uses a Bayesian statistical model for testing differential splicing events between groups and offers statistical significance testing individually for each possible event using a Markov chain Monte Carlo method. |
|
| Diffsplice | A computational tool for the identification of splicing patterns without transcript annotation data or pre- defined splicing motifs. Diffsplice utilizes a non- parametric approach to summarize significance level of differential splicing events. |
||
| SplicingC ompass |
An R package that identifies differential alternative splicing events at the gene level rather than exon level. SplicingCompass creates a read count vector for each gene and calculates geometric angles between the vectors for discovering potential spliced genes. |
||
| DSGseq | A negative binomial model-based tool that uses exon read counts for two-group comparison of alternative splicing signatures. DSGseq supports the identification of novel splicing events as well as differential splicing. |
||
3.2.2. Post-Processing
The post-processing steps are required to minimize the number of false positive variants arising in the WES data and include the removal of duplicate reads generated by PCR amplification, realignment around insertion-deletion variants (indels), and base quality recalibrations. Local realignment of the BAM files is essential to minimize the mismatching bases, thus eliminating false positive single-nucleotide variants near indels. This extended post-processing realignment step is not required for transcriptome and epigenome data analysis. Finally, base quality recalibration adjusts the quality score of each base using the known variants in a database and helps improve the accuracy of variant calling.
3.2.3. Variant Analysis
A number of open source tools are available for variant calling (Table 4). Application of multiple sample variant calling software is recommended to reduce false positive variants. We can also improve variant calling in regions with fewer reads by utilizing reads from multiple samples concurrently.
Table 4.
Commonly Used Tools for Variant Analysis.
| Process | Tool | Description |
|---|---|---|
|
Variant Calling |
CRISP | Compares allele count distribution across multiple pools or evaluates the probability of identifying multiple non-reference base calls occuring due to sequencing errors to identify variants. |
| GATK | Built to process data originating from Illumina sequencing technology, but can be adapted to other sequencing technologies. Implements MapReduce functionality to achieve parallelism for faster data processing. |
|
| SAMTOOLS | Can identify variants from single or multiple samples. BCFtools utility in SAMTools suite is used to identify SNPs and short INDELs from a single alignment file. |
|
| SNVer | Implements binomial–binomial model for significance testing of inferred allele frequency against sequencing error. |
|
| VarScan 2 | A heuristic and statistical algorithm that detects and classifies variants based on somatic status. |
|
| SomaticSniper | Implements Bayesian Statistics to compare liklihoods of genotypes in cases and controls obtained from existing germline genotyping algorithm. |
3.3. Secondary Analysis
The primary analysis of WES data provides a large number of genomic variants. Additional steps are needed to understand the role of these variations in the context of the disease trait under investigation. These steps include variant annotation, estimation of variant incidence in the population (frequency), and customized filtering steps to identify candidate disease-causing variants (variant prioritization).
3.3.1. Variant Annotation
A single exome analysis can reveal 20,000–30,000 variants. Thus, assigning functional information (annotation) to the variants is important. The first step in gene annotation focuses on determining whether a single nucleotide variant reflects synonymous, non-synonymous, non-sense codon, or consensus splice site changes. In addition, a variant can be an indel that may impact transcript structure. The next step involves estimating the incidence (minor allele frequency, MAF) of the variant in the general population. Large-scale genomic studies such as the 1000 Genomes Project, ESP6500, dbSNP, ExAC (Table 5) have catalogued sequence variants from thousands of exomes and genomes, which serve as a valuable resource for allele frequency estimations. Another aspect of annotation includes base conservation and functional predictions, which can be accessed using LJB23 database (Liu et al., 2011), Combined Annotation Dependent Depletion (CADD) database, and Genomic Evolutionary Rate Profiling (GERP) score (Table 5). Three major tools are used to classify variants: ANNOVAR, SnpEff and Variant Effect Predictor (Table 5). The choice of software and reference transcript annotation can have a large impact on variant interpretation (McCarthy et al., 2014). ANNOVAR is a popular software for variant analysis because of its capability to integrate most of the functions discussed here (Yang and Wang, 2015).
Table 5.
Web links (in alphabetical order) of resources & tools.
3.3.2. Variant Prioritization
Identification of a disease causing or associated variant in exome sequencing experiments requires a customized filtering process depending on the question being pursued, as discussed below. A number of reviews provide general recommendations for identifying disease variants (Bamshad et al., 2011; Ratnapriya and Swaroop, 2013).
3.3.3. Filtering WES Data in Mendelian/Monogenic Diseases
Traditional positional cloning methods for identifying Mendelian disease genes involve collecting large pedigrees, performing linkage analysis to map the disease locus, and screening candidate genes for a segregating rare variant. This process is time consuming and requires a minimum number of individuals to reach statistical significance in linkage analysis. WES and targeted re-sequencing have dramatically altered the analysis landscape, and we can now identify mutations in small families or even single affected individuals.
The search of causal variants includes applying a cut-off for MAF and focusing on variants with a major effect on gene function (non-synonymous, truncation, and splice variants). Inheritance pattern is another filter that can be applied; for example, a recessively inherited disease variant is likely homozygous whereas a dominant disease variant is heterozygous. However, there can be exceptions to these rules. For instance, recessive disease variants can be compound heterozygous. Segregation with affection status is another critical filter that can be applied to family-based studies. In a large cohort, the search for either identical variants or additional rare variants in the same gene can further strengthen the evidence for causality. WES was first employed in vision research for the analysis of an Ashkenazi Jewish family with three affected siblings and resulted in the identification of a mutation in a novel gene, DHDDS, as a cause of retinitis pigmentosa (Zuchner et al., 2011). At this stage, 62 retinal disease genes have been identified using WES, three by targeted capture, and another two by taking advantage of WGS (RetNet; see Table 5).
A rapid pace of evolution in variant detection methods has made it possible to obtain more accurate diagnosis and prognosis in clinical practice and yielded opportunities for precision medicine initiatives (Amendola et al., 2016). However, previously unknown and dynamic aspects of the genome in health and disease are presenting great challenge for interpreting the effect of a specific variant in causing the phenotype. Each individual carries thousands of unique variants in the genome (Genomes Project et al., 2015). WES of an individual can identify as many as 100 loss of function variants that may not have any dramatic effect on the phenotype (MacArthur et al., 2012; Sulem et al., 2015), yet many loss of function mutations lead to lethality or disease. A large number of variants (dozens to hundreds, depending on the study design) can pass the filtering methods described above, and determination of causal variant(s) needs careful examination. Finding a rare variant even in a known disease gene is not sufficient to suggest causality. If a pedigree is available, one must look into the mode of inheritance and perform linkage or homozygosity mapping to narrow down genomic regions to focus the search. In the absence of the family data, one has to depend on the overall burden of rare variants in a disease population compared to healthy controls. Distinct complementary approaches can therefore help in identifying few candidate variants and genes that must be evaluated further (using in vivo and in vitro model systems) to elucidate their functional impact in causing the disease.
3.3.4. Filtering in Complex Diseases
The analysis and filtering of NGS data for a complex disease requires a different strategy to identify candidate causal variants in biologically relevant genes and pathways. GWAS has been a popular method for identifying genetic risk variants in complex diseases by comparing a large number of common and/or rare variants between individuals with a phenotype of interest (cases) and a set of unrelated (matched) controls (http://www.ebi.ac.uk/gwas/). Such studies have yielded a catalog of common SNPs associated with complex diseases affecting vision (Bailey et al., 2016; Fritsche et al., 2016; Grassi et al., 2011; Kirin et al., 2013). However, associated alleles are not causal, and majority of association signals are located in the non-coding region of the genome with ill-defined function (Chakravarti et al., 2013).
The concept of causal variants in complex disease is still evolving. AMD is one of the best-studied complex diseases, where 52 common and rare variants at 34 genetic loci have been identified so far using GWAS and Exome-Chip approaches (Fritsche et al., 2013; Fritsche et al., 2016). However, the pathological role of the candidate genes or variants is not completely clear at a majority of the AMD loci and underlying disease mechanisms are largely unknown. Targeted re-sequencing of some of these loci has led to the discovery of high-risk rare, coding variants, providing crucial functional clues about causal genes. For example, a rare penetrant mutation, R1210C, was identified at the CFH locus (Raychaudhuri et al., 2011). Rare variants in other complement genes have also been identified in advanced AMD patients by targeted and WGS (Fritsche et al., 2016; Zhan et al., 2013). Rare variants exhibit very high odds ratio and are likely to be causal, as these are observed at very low frequency in the general population. However, these events are also rare in the disease population with few disease individuals carrying the disease-causing rare coding mutation at an associated locus. Even a genomewide survey of exome variants with low to moderate frequency (Exome Chip) did not lead to novel associations in a large AMD study (Fritsche et al., 2016). Several explanations can be put forth. It is possible that the contribution of rare variants is small and common non-coding variants with regulatory functions are indeed disease-causing. Alternatively, the rare variants likely arise independently in genomes and a sequencing based approach (such as WGS) focusing on all rare events, rather than Exome Chip, might more successful. Indeed, one would predict that the effect of a single variant/gene is not large and thus we must focus on biological pathways relevant to the disease biology or an integrated approach combining transcriptome and epigenome analysis with GWAS (discussed in Section 6).
We note that several statistical methods have been developed to evaluate the impact of multiple independent rare variants that cause functional damage in a combinatorial manner; these methods can be broadly classified as burden and non-burden tests. Burden tests collapse rare variants in a genetic region into a single burden variable, and then model the phenotype using the burden variables to test for the cumulative effects of rare variants in the region. These models include collapsing methods such as CAST, CMC, RareCover, and aSum and aggregation methods such as WSS, KBAC, and RBT (Lee et al., 2014). Non-burden tests such as VT, C-alpha, EREC, and SKAT (Lee et al., 2014) aggregate individual variant score test statistics with weights when SNP effects are modeled.
3.4. Interpretation of Genetic Variations
Elucidating the functional impact of thousands of variants identified in WES or other NGS studies poses major challenges in for genetic diagnosis and personalized medicine. Rules of genetics are now being redefined. Even healthy individuals have been identified to harbor mutations in at least 8 severe Mendelian conditions but with no phenotype (Chen et al., 2016), suggesting incomplete penetrance and/or existence of alleles that might be protective. The roles of synonymous (Plotkin and Kudla, 2011) and non-coding variants (Sakabe et al., 2012) in disease causation are also becoming evident. Contributions of more than one mutations/variants in Mendelian disease are being recognized as modifiers (Genin et al., 2008; Slavotinek and Biesecker, 2003), compensatory mutations (Jordan et al., 2015) or triallelic inheritance (Eichers et al., 2004). Modifier alleles might also explain vast clinical/phenotypic heterogeneity that is commonly observed in RDDs (Ebermann et al., 2010; Khanna et al., 2009; Priya et al., 2014).
NGS presents immense opportunity to decipher exciting attributes of human history, biology and disease than merely cataloging primary genetic defects. Nonetheless, guidelines for systematically investigating the causality of the variants in human disease through functional assays are highly desirable (MacArthur et al., 2014). One needs to take account of biological context such as tissue types and species when designing such approaches. The lack of high throughput functional assays has been a major bottleneck in the field. For years, scientists have used mice and other model organisms for elucidating how genetic defects might cause retinal disease (Veleri et al., 2015). More recently, the use of human pluripotent stem cells (hPSCs), especially induced pluripotent stem cells (iPSCs), has significantly expanded the focus on investigating human disease (Merkle and Eggan, 2013). hPSCs are becoming routine for developmental studies and screening small molecules to rescue disease-associated phenotypes (Kaewkhaw et al., 2015; Kaewkhaw et al., 2016), offering immense opportunities in combination with NGS to make precision medicine a reality in the near future.
4. Transcriptome
The pattern of gene expression in a cell/tissue can broadly reflect its functional state. NGS-based expression profiling by RNA-seq (Marioni et al., 2008; Mortazavi et al., 2008) allows comprehensive qualitative and quantitative mapping of all transcripts (Garber et al., 2011). Prior to NGS, transcriptome profiling techniques had limited scope and accuracy and were not quantitatively precise. Northern blotting and qRT-PCR analysis could not be employed at genomewide scale. Expressed sequence tag (EST) analysis (Adams et al., 1991; Gieser and Swaroop, 1992) and serial analysis of gene expression (SAGE) (Blackshaw et al., 2001; Blackshaw et al., 2003) were instrumental in profiling novel and known transcripts but were labor-intensive and had limited breadth and quantitative capability. Gene expression microarrays (Brown and Botstein, 1999) have been the mainstay of genomewide profiling during the last decade, yet several issues inherent to hybridization-based methods were not easily overcome; these included varying background noise, requirements for high RNA amounts, dependence of annotated probe sets included on the array, and lack of precise quantification. The massively parallel capabilities (as discussed in Section 2) of NGS has expanded the scope of transcriptional landscape dramatically with miniscule quantities of total RNA, low background noise, and quantification accuracy rivaling qRT-PCR, which has been the "gold-standard" for quantitative studies.
Massive datasets produced by RNA-seq create unique computational challenges for analysis. For convenience, we have divided the analysis in two parts – primary and secondary. The primary analysis includes read mapping, transcriptome reconstruction, expression quantification (Garber et al., 2011), and differential expression (DE) analysis. Read mapping refers to the alignment of short reads to the reference transcriptome and/or genome. Sequencing reads can also be used to generate contigs for de novo assembly and novel transcript identification. Transcriptome reconstruction focuses on identifying different transcript isoforms. Expression quantification refers to evaluation of transcript abundance at the gene or isoform level. Higher-level secondary analyses are generally required to extract biologically relevant information after the primary analysis is completed. The secondary data analysis can include DE analysis, de novo assembly, expression cluster analysis, co-expression networks construction, and differential alternative splicing (DAS). DE analysis aims to identify dissimilarly expressed genes in different experimental conditions. De novo transcript assembly permits the discovery of novel unannotated transcribed sequences. Cluster analysis focuses on grouping the genes based on a specific characteristic, such as co-expression or shared biological function. Co-expression network construction refers to elucidation of gene regulatory networks from expression data. DAS examines differential isoform expression across the biological samples or conditions. Thus, RNA-seq provides us the necessary data for a comprehensive evaluation of broader transcriptional landscape.
4.1 Library Construction, Data Generation, and Primary Analysis
The basic steps in performing RNA-seq include library construction and generation of sequence data followed by primary analysis. Depending on the goals of the experiment (e.g., RNA species being investigated), a meticulous experimental design is essential for extracting biologically relevant information.
4.1.1. Library Construction and Data Generation
RNA-seq library construction protocols include similar basic steps, which require elimination of ribosomal RNA (rRNA), reverse transcription of the desired RNA species, fragmentation, adapter ligation, and enrichment (Figure 4). A number of issues must be considered to obtain high quality global expression profiles. First and foremost, the RNA species being investigated (e.g., mRNA or miRNA) should be enriched in the library since rRNA represents the dominant transcript species within any given cell/tissue; the commonly used protocols use oligo-dT beads for enrichment of mRNA. To minimize inherent 3' bias in this protocol (due to RNA degradation), high quality total RNA is necessary for library generation. A widely applied method to evaluate RNA quality is by using Agilent 2100 Bioanalyzer, which calculates RNA integrity number (RIN) on a scale of 1–10. We recommend RNA with RIN >= 7.0 for RNA-seq analysis. An alternative approach is to eliminate rRNA using beads containing complementary rRNA sequences; this approach permits an unbiased examination of the transcriptional landscape but requires a significantly higher sequencing depth per sample (Li et al., 2014a). The rRNA removal protocol is also essential when investigating non-polyA transcripts or if the quality of RNA is poor (RIN < 7.0). Nonetheless, this approach allows successful analysis of even substantially degraded RNA (Li et al., 2014a). We should mention that genomic DNA elimination is required for this protocol prior to reverse transcription.
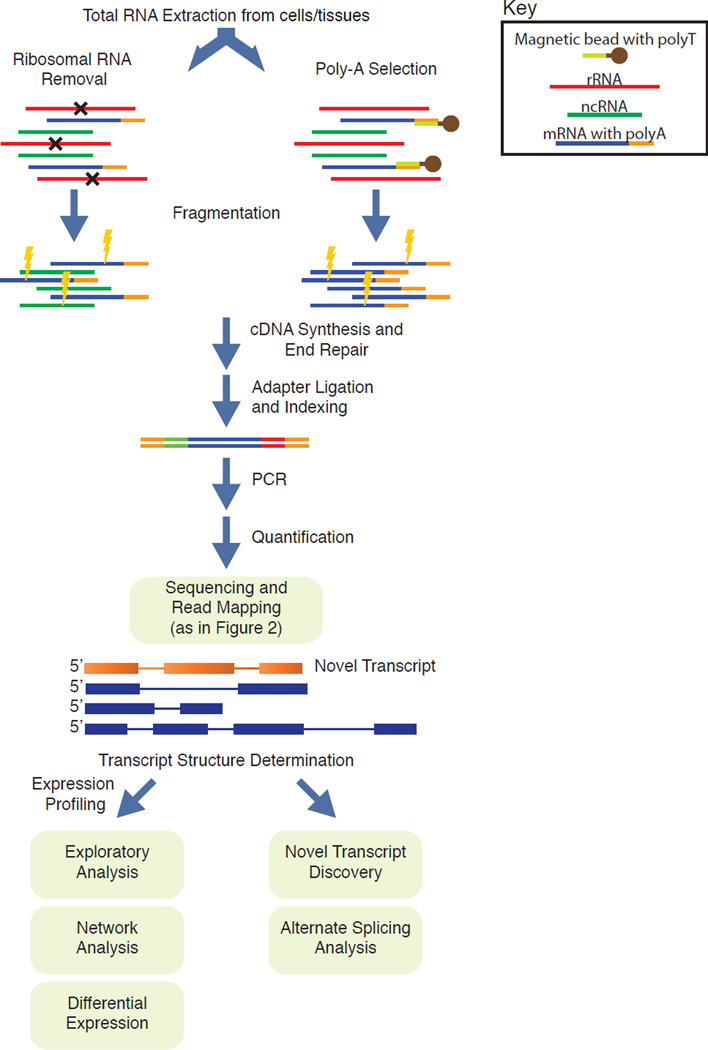
Figure 4. RNA sequencing workflow and analysis.
Total RNA is extracted and ribosomal RNA is either removed to enrich for other RNA species, or polyA-tailed RNA are isolated using poly(T) oligomer magnetic beads as described in (http://www.illumina.com/products/truseq_stranded_total_rna_library_prep_kit.html). RNA is then fragmented using sonication, followed by cDNA synthesis, end repair, adapter ligation, and indexing. After PCR amplification and library quantification, RNA reads are mapped to known transcripts and the whole genome to facilitate transcript identification and quantification. Multiple secondary analyses exist to understand the expression profile of cells and whole tissues.
The library preparation protocol should keep the fidelity of the genomic strand from which it is transcribed for correct alignment and assignment of sequencing reads to specific transcripts since many genes have overlapping transcribed regions on the same or opposite strand of the genomic DNA; therefore, a directional library protocol is necessary (Brooks et al., 2012). The desired length and depth of the sequencing depends on the RNA species being investigated. For example, 10–50 million sequence reads are sufficient for quantification of almost all of the known protein coding transcripts. Over 100 million sequence reads per sample might be necessary to evaluate low to moderately expressing novel transcripts or for non-polyA RNA species. The sequencing depth for mature miRNA libraries can be as little as 1–10 million reads. Except for miRNA, longer sequence lengths are desirable for sequencing reads to overlap neighboring exon boundaries (i.e., to cross over the intron) thereby leading to more accurate transcript assignment. In addition, PE sequencing can lead to more accurate read alignments for mRNA profiling.
The amount of starting RNA is an important consideration before initiating a specific library preparation protocol. We generally use 20–100 ng of total RNA for standard oligo-dT enrichment protocol and perform paired end sequencing to produce at least 30 million directional sequence reads. Although many library kits are available, we use the following kits: TruSeq Stranded mRNA Library Prep Kit (Illumina) for mRNA, TruSeq Stranded Total RNA Library Prep Kit (Illumina) for total RNA, TruSeq Small RNA Library Prep Kit for miRNA, and SMARTer Ultra Low Input RNA Kit (Clontech) and Nextera XT DNA Library Preparation Kit (Illumina) for single cell or low amounts of starting RNA.
4.1.2. Sequencing Read Alignment
Alignment of RNA-seq reads is the crucial first step in transcriptome profiling. Over 90% of human transcripts are derived from more than one exon (Harrow et al., 2012); therefore, at least some of the reads must cross over introns and include exon-exon junctions for determining proper transcript structure. Early alignment algorithms used a priori knowledge of known splice junction sites for aligning RNA-seq reads to the genome but lacked the ability to discover novel or rare splice events. More recent approaches include the alignment of sequence reads to a reference transcriptome prior to the reference genome, which can overcome multiple mapping issues associated with pseudogenes. We have effectively performed spliced read alignment using TopHat2 (Trapnell et al., 2009), and STAR (Kim et al., 2013). TopHat was one of the first algorithms that performed spliced-read alignments and also aligned reads to novel transcript isoforms and genes. More recently, STAR has become popular due to its successful alignment of spliced-reads, accuracy, and speed (Engstrom et al., 2013). In addition, STAR can now provide concurrent gene level quantification. Whole transcriptome level sequence alignments can be completed within minutes with appropriate computer configurations and memory (e.g., using 4 cores and 72 Gb RAM on Biowulf computing cluster of NIH - https://hpc.nih.gov).
4.1.3 Gene and Transcript Quantification
Successful secondary transcriptome analysis is facilitated by accurate quantification of RNA-seq reads, which can be performed at the gene or transcript level depending upon study objectives. Gene-level quantification summarizes read counts assigned to coordinates of genomic features such as genes and exons and is appropriate in cases where considerable 3' bias exists in the data, short and/or single-end sequencing reads (e.g., <50 nucleotides) with low read-depth (e.g., <30 million) are obtained, or the reference transcriptome is poorly annotated (such as for non-model species). The algorithms for gene-level analysis include HTSeq (Anders et al., 2015), featureCounts function in SubRead software (Liao et al., 2014), RSEM (Li and Dewey, 2011), and STAR (see Table 5 for web sites). In contrast to gene-level analysis, transcript-level algorithms additionally assign reads probabilistically to putative transcripts of a given gene and can provide a more accurate representation of the transcriptome state, specifically when applied to high quality, PE sequencing reads. Software applications for transcript level quantification include RSEM, Cufflinks (Trapnell et al., 2010), and eXpress (Roberts and Pachter, 2013).
Regardless of the quantification method, expression values from the algorithms are obtained as feature counts, RPKM/FPKM (Reads/Fragments Per Kilobase of the exon model per Million reads) (Mortazavi et al., 2008; Trapnell et al., 2010), or TPM (Transcripts Per Million) (Li and Dewey, 2011). Feature counts are the most basic unit of measure and do not account for the length of a transcript or the depth at which the sample RNA was sequenced. Thus, feature counts are used primarily for normalization, DE analysis, and quantification of expression. RPKM and FPKM are used for SE or PE sequencing, respectively, to account for the transcript length and sequencing depth, thereby allowing comparison across samples. Normalization (discussed later) of the counts is essential prior to calculating RPKM/FPKM values to reduce the disproportionate impact of highly transcribed genes. TPM calculates the relative abundance of a transcript by normalizing for sequence depths of specific transcripts rather than of the whole transcriptome dataset; it is thus a preferred metric for quantifying gene/transcript abundance (Alamancos et al., 2015; Burns et al., 2015; Shalek et al., 2013).
4.2. Secondary Data Analysis
Primary analysis of RNA-seq yields genome-aligned reads (BAM files) and quantified expression data that provides basic information on transcribed sequences. However, additional bioinformatic analysis is required for deciphering molecular insights into cellular functions. Here, we have focused on comparative analysis of transcriptomes, identification of novel transcripts and isoforms, differential alternative splicing (DAS), and generation of co-expression networks.
4.2.1 DE Analysis
DE analysis is generally used to compare transcriptomes of two or more groups of samples. From simple two-group comparison (sample A versus sample B) to more complex multivariate analyses, one needs to be aware of various considerations that make DE of RNA-seq different from microarray or qRT-PCR. Before performing DE analysis of RNA-seq data, it is critical to remove low-expressing transcripts or genes (e.g., those expressed at <1 FPKM), normalize for differences in sequencing depth, model overdispersion, and experimental factors. The data is then subjected to DE analyses using various tools that provide fold change in gene expression and statistical significance.
Typically, a large number of genes or transcripts are not expressed at high enough levels to be considered above background noise and are eliminated to avoid an adverse impact on normalization algorithms. This filter can be set to an arbitrary number, such as 1 count per million or 1 FPKM in 10% of the samples or even all replicates in any group. In retina RNA-seq data, this filter can remove as much as 40–50% of all annotated transcripts (Brooks et al., 2012). Normalization is then required to account for differences in sequencing depth. It uses various algorithms, such as median, upper quartile, full quantile (Bullard et al., 2010), relative log expression (RLE) (Anders and Huber, 2010), or trimmed median of the mean (TMM) (Robinson and Oshlack, 2010). Alternatively, RNA spike-in or housekeeping gene matrices may be used for normalization (Jiang et al., 2011; Risso et al., 2014). We find TMM to be widely applicable for our DE analyses (Conesa et al., 2016) (Brooks et al., manuscript in preparation). After normalization, FPKM values are exported for secondary analyses including clustering and inference of co-expression network(s).
The count data generated from digital gene expression experimentations, such as SAGE and RNA-seq, demonstrate more variance than what is expected from a Poisson distribution model (overdispersion) (Robinson and Smyth, 2007), leading to an increase in type-I error (false positives) in DE analysis. This observed overdispersion should be compensated prior to DE analysis since traditional DE algorithms, such as student’s t-test and ANOVA, assume a normal distribution of data. Several software packages can perform this task utilizing different methodologies; these include DESeq (Anders and Huber, 2010), edgeR (Robinson et al., 2010), Cuffdiff 2 (Trapnell et al., 2013), and limma (Law et al., 2014). After evaluating various methods, we have settled on limma for our DE analysis. The flexibility in the limma package allows us to model many different experimental factor configurations, minimize type-I errors, and permit the correction of experimental batch factors. The final steps in DE analysis are to filter the data for fold change and determine statistical significance. We initially set thresholds to have two-fold or greater change and a false discovery rate (FDR) of less than 5%.
4.2.1 Co-expression Network Inference
Co-expression networks can be developed based on expression patterns in transcriptome data sets; e.g., if two genes exhibit a strong correlation in their pattern of expression, these genes are predicted to demonstrate similar transcriptional regulatory mechanisms (Figure 5). A number of co-expression network inference algorithms have been developed, including Boolean Networks, Probabilistic Boolean Networks, Bayesian Networks, Dynamic Bayesian Networks, Differential Equations Methods, Linear Methods, Neural Network Methods and Information Theoretic Methods (see (Hecker et al., 2009) for a review).
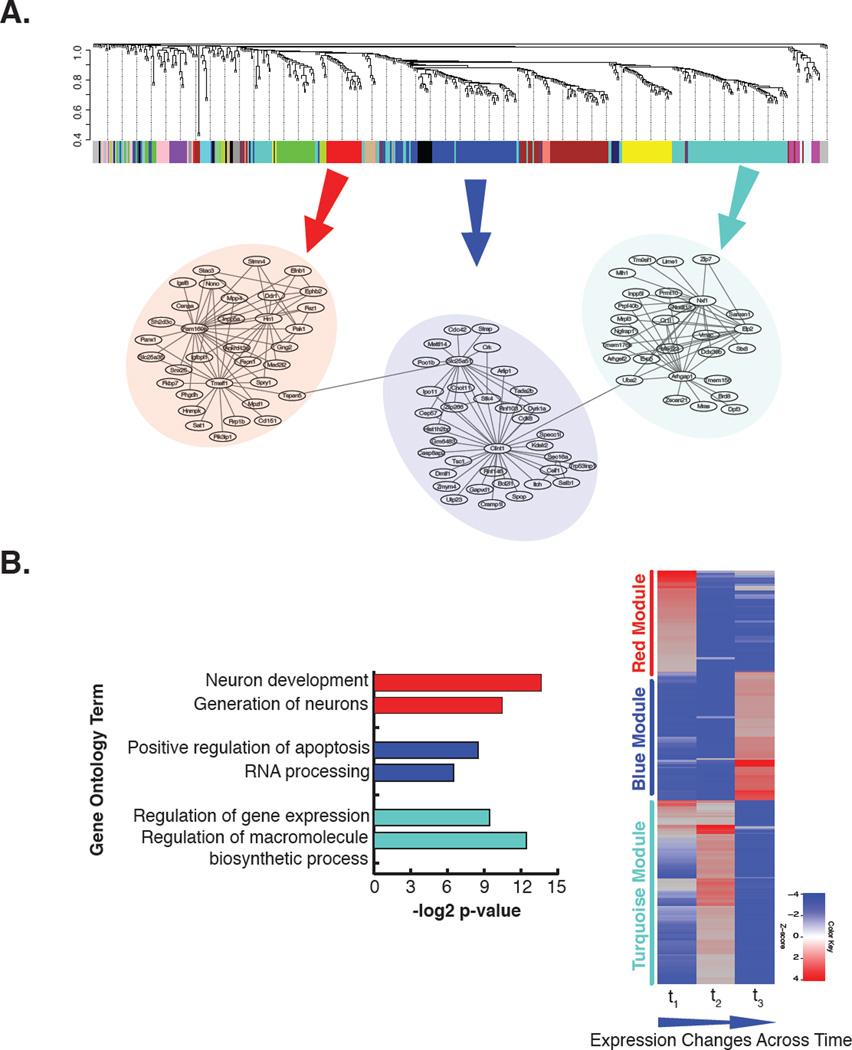
Figure 5.
Construction of co-expression networks and functional enrichment of network modules A. Co-expressed genes (also called linked genes) can be identified and grouped into network modules (represented with different colors). Here we demonstrate example network structures of three network modules (red, blue and turquoise), built using Weighted Gene Co-expression Network Analysis tool (WGCNA)(Langfelder and Horvath, 2008). In each network module, genes are represented as nodes and co-expressed genes are linked. In co-expression networks, it is believed that highly connected genes (also called hub genes) represent biologically significant genes since their dysregulation may affect many other linked genes. B. Genes with similar expression patterns tend to group in the same network module, and are more likely to be in the similar biological processes/pathways. Biological relevance of network modules can be elucidated with diverse online functional enrichment analysis tools. Here we show the first two most significant Gene Ontology (GO) terms related to each module after functional GO enrichment analysis using DAVID online tool (Huang da et al., 2009). Additionally, heatmap shows expression patterns of genes in each network module across time points.
A systematic performance analysis of network inference approaches concluded that no single method can be used on distinct data sets to obtain high confidence networks and that multiple inference methods may yield complementary information which can be combined for constructing a biologically relevant co-expression network (Marbach et al., 2012a). After evaluating different approaches, the average rank method was implemented for integrating networks generated by distinct algorithms (Marbach et al., 2012a). The quality of network(s) inferred by the average rank method depends on the quality of the data sets and efficiency of prediction algorithms.
4.2.2. Analysis of Alternatively Spliced Transcript Isoforms
Alternative splicing (AS) generates extensive transcript diversity in eukaryotes by producing multiple RNA molecules from a single gene. As many as 95% of human and other mammalian genes with multiple exons undergo AS (Pan et al., 2008; Wang et al., 2008), contributing to cellular and phenotypic complexity during development and disease (Revil et al., 2010; Singh and Cooper, 2012). While exon-microarrays first introduced genomewide profiling of distinct transcripts, NGS-based RNA-seq technology has dramatically accelerated the identification of novel transcript isoforms generated by DAS events. A number of computational tools have been developed for DAS analysis to obtain valuable information on skipped exons, alternative 5’ and 3’ splice site usage, mutually exclusive exons, and intron retention in transcribed sequences.
DAS analysis tools can be classified as count-based or multi-reads, depending on the methods for quantification of isoform levels (Pachter, 2011). In the count-based models, the total number of reads uniquely mapped to each genomic feature (exons in DAS analysis) is counted individually before testing for the statistical significance of count difference between the control and experimental groups. On the contrary, the reads can be mapped to multiple isoforms in the multi-reads model, which then test the difference in relative transcript abundance statistically across distinct conditions. DAS analysis requires higher sequencing depth compared with differential expression analysis, as much as 100 million reads of 101 nucleotides PE sequencing (Liu et al., 2013).
A number of DAS analysis tools (see Table 3) permit the use of biological replicates across different groups. MATS, DSGseq, and SplicingCompass are count-based models, whereas Diffsplice represent multi-reads models (see Table 5 for web sites). All methods can utilize genome-aligned reads (as SAM and/or BAM file formats) as input, except for MATS, which can also use unaligned reads (FASTQ file format) if the reference genome sequence is provided. Evaluation of different DAS analysis tools with simulated and real RNA-seq data sets indicated that no single algorithm can satisfactorily elucidate all possible splice events and that the choice of DAS method is based on sample size, sequencing depth and quality, and availability of reference transcript annotation (Liu et al., 2014).
Regulation and functional consequences of AS can be inferred by integrating distinct RNA-seq data sets using network based methods (Li et al., 2014b). For example, a tensor-based pattern mining method has been used to correlate exon splicing in different genes and across diverse conditions for identifying exon clusters that are regulated by a specific splicing factor (Dai et al., 2012). Such co-splicing clusters indicate regulation of exon usage in different contexts and may reveal insights into post-transcriptional control of gene expression. Along these lines, a label propagation algorithm can be utilized for systematic functional evaluation of distinct transcript isoforms (Li et al., 2014c). In this approach, gene-isoform relations are modeled by aggregating multiple co-expression networks of transcript isoforms into a single one and then assigning specific roles based on their interaction with genes of known function.
4.2.3. Novel Transcript Identification using Transcriptome Assembly Techniques
Genome databases, such as RefSeq and Ensembl, include hundreds of thousands of transcribed sequences from numerous organisms and cell lines. However, transcript catalogs of unique neuronal cell types (e.g., distinct retinal cells) are still far from complete. Transcriptome profiling by RNA-seq provides both qualitative and quantitative depth, permitting the detection of low-expressed and novel transcripts, unannotated or fusion transcripts, and isoforms generated by alternate splicing or alternate promoter usage (Guttman et al., 2010; Kim and Salzberg, 2011; Trapnell et al., 2010). Transcriptome assembly (also called transcriptome reconstruction) of the sequence reads can be performed by genome-guided or genome independent assembly tools (Garber et al., 2011; Martin and Wang, 2011).
Genome-guided (also called reference-based or ab initio) transcriptome assembly is utilized when high-quality reference genome is available for the target transcriptome, and sequence reads are first aligned to the reference genome through a splice-aware aligner tool, such as TopHat or STAR. The genome-aligned reads are then assembled into transcripts using a graph-based algorithm (Guttman et al., 2010; Trapnell et al., 2010). In this assembly process, the first step is clustering of overlapping reads in each genomic region, followed by construction of all possible isoforms (transcripts) using one of the genome-guided assembly tools, such as Scripture (Guttman et al., 2010) or Cufflinks (Trapnell et al., 2010).
In the genome independent (also called de novo) transcriptome assembly approach, overlapping sequence reads are first organized by De Brujin graph method and then assembled into transcript structures (Martin and Wang, 2011). An accurate transcriptome assembly in de novo approach relies upon the length of reads and sequencing errors since no reference genome is available. Thus, relatively longer sequence reads are preferred for a complete and high quality de novo transcriptome assembly (Martin and Wang, 2011). Oases (Schulz et al., 2012) and Trinity (Grabherr et al., 2011) are among the common genome independent assembly tools, which vary in terms of their performance and sensitivity. De novo assembly generally requires extensive computational resources and long processing times depending on the size of the transcriptome data set (Simpson and Durbin, 2012). Trinity can perform transcriptome assembly with a reference genome in de novo mode, which is useful when the reference genome of the organism in question does not exist or is incomplete but the reference genome of a closely related species is available (Grabherr et al., 2011).
Alignment and assembly steps are critical for the success of novel transcript discovery. A large fraction of the novel assembled transcripts do not seem to be biologically relevant and likely represent transcriptional noise. Hence, different combinations of aligner-assembler pairs may be utilized to obtain novel high confidence transcripts, which can then be validated by independent methods including in silico replication by another assembly algorithm, q-RTPCR and/or in situ hybridization. Annotations associated with identical reference genome assembly from databases such as Ensembl (Cunningham et al., 2015), UCSC (Karolchik et al., 2003), and RefSeq (Pruitt et al., 2014) should be employed for accurate identification of novel transcripts.
Transcriptome reconstruction methods have begun to unravel complex dynamics of transcriptomes, yet numerous challenges remain (Guttman et al., 2010; Trapnell et al., 2010): (i) We may not be able to detect the transcripts expressed at very low levels (e.g., lncRNAs) because of technical limitations, (ii) Construction of accurate transcript structures is difficult due to short sequence reads from unprocessed mRNAs containing intronic regions, and (iii) Sequence reads may be too short or not of sufficient quality to accurately map to transcriptome or genome (Martin and Wang, 2011; Trapnell et al., 2010).
5. Epigenome
Epigenome encompasses a broad array of non-genomic regulatory factors that do not alter DNA sequence yet play a significant role in modulating biological processes including development and disease pathogenesis (Sadakierska-Chudy and Filip, 2015; Sadakierska-Chudy et al., 2015). The analyses of epigenetic changes associated with disease states have provided new avenues for possible therapies of RP and AMD (Berner and Kleinman, 2016; Zentner et al., 2015). For example, a histone deacetylase inhibitor, valproic acid, seems to affect the severity of photoreceptor loss in two mouse models of retinal degeneration, Rd1 and Rd10 (Mitton et al., 2014). More recently, P300 (a histone acetyltransferase that catalyzes acetylation of H3K27) and LSD1 (a histone demethylase with enhancer repression functions) have been employed with nuclease-deficient Cas9 system, opening a new window of opportunity to direct epigenome editing for designing treatment paradigms (Hilton et al., 2015; Kearns et al., 2015).
NGS technology has greatly facilitated global profiling of epigenetic modifications. In this section, we will primarily focus on chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) to identify DNA binding sequences for transcription factors and to localize histone modifications, and on DNase I digestion and transposase-mediated methods for determining chromatin accessibility (Barski et al., 2007; Buenrostro et al., 2015; Song and Crawford, 2010). We also highlight the guidelines and working standards for ChIP-seq and DNase-seq experiments provided by ENCODE and modENCODE consortia and describe relevant details for computational analysis of epigenetic data.
5.1. ChIP-seq
Identification of genomic regions occupied by DNA-binding proteins and elucidation of chromatin state by chromatin immunoprecipitation followed by NGS (ChIP-seq) (Figure 6) provide better understanding of regulatory factors associated with patterns of gene expression. The ChIP-seq assay involves sonication of formaldehyde-fixed (or unfixed for histone modification (HM) ChIP-seq) chromatin from cells/tissues, followed by precipitation of the protein or protein modification of interest using a target-specific antibody conjugated to magnetic beads. Double-stranded DNA is then isolated from the precipitated protein-DNA complexes and used to generate libraries for NGS (Barski et al., 2007; Johnson et al., 2007; Sheaffer and Schug, 2016).
Figure 6. ChIP and ATAC sequencing workflow and analysis.
Retina is dissected from the eye, and cells are dissociated for follow-up studies.
A. (ChIP-seq): Isolated chromatin is fixed (or unfixed) and fragmented as described in (Sheaffer and Schug, 2016). Magnetic beads conjugated to antibody specific to the target protein are used to precipitate fragments bound to the protein of interest. Adapter ligation and indexing is followed by sequencing and quality control assessment. Reads are mapped onto the genome assembly and PCR duplicates are removed to minimize experimental artifacts. Mitochondrial DNA may also be removed to improve data set comparisons. Peak calling using one of the many available tools and DNA-binding sequencing motif analysis may facilitate the elucidation of the genome binding profile for the precipitated protein.
B. (ATAC-seq): Unfixed chromatin is obtained and tagmented as described in (Buenrostro et al., 2013). PCR amplification with index primers facilitates multiplexing strategies. Sequencing and mapping are followed by PCR duplicate and mitochondrial DNA removal using available bioinformatics tools or custom scripts. Open chromatin peak calling allows for the identification of active regulatory regions and may be combined with other NGS data sets to improve systems integration analyses.
Identification of antibodies specific to the protein or histone modification of interest constitutes the initial and critical step in conducting transcription factor (TF) ChIP-seq or HM ChIP-seq. The quality of the data obtained relies significantly on the specificity of the antibody used for immunoprecipitation of the target protein domain from fragmented chromatin. Antibodies may show poor reactivity to their target or bind to other DNA-binding proteins nonspecifically, leading to false inferences of protein binding localization (Landt et al., 2012). The ENCODE consortium recommends evaluation of antibody specificity using primary and secondary tests (Landt et al., 2012). A large number of ChIP-tested antibodies are commercially available for examining various histone modifications; however, many TF antibodies are not suitable for ChIP analysis as they either lack specificity or affinity to the target protein domain. The number of replicates for each experiment and library sequencing depth are critical considerations as well. At least two independent biological replicates should be performed to enhance the confidence in findings from ChIP-seq datasets. The depth of sequencing depends on the size of the genome occupied by the protein under examination (Jung et al., 2014; Landt et al., 2012). According to the ENCODE and modENCODE consortia, point-source factors (e.g., NRL and CRX) localize to specific points within the chromatin structure, whereas broad-source factors (such as H3K4me and H3K36me3) have broad chromatin footprints (Corbo et al., 2010; Feng et al., 2012; Hao et al., 2012; Roadmap Epigenomics et al., 2015). For point-source factors, each biological replicate is recommended to have at least 10 million uniquely mapped reads when using mammalian cells. Twenty million reads are preferred for broad-source factors to obtain equivalent resolution. A total of 40–50 million reads is recommended for HM ChIP-seq experiments. About 500,000 to a few million cells are required for each ChIP-seq library, including controls (Sheaffer and Schug, 2016).
TF ChIP-seq has been used to profile genomewide binding sites of NRL, a transcription factor necessary for determining rod cell fate in the mammalian retina (Mears et al., 2001), to better understand transcriptional regulatory networks associated with rod photoreceptor development. The transcriptional targets of NRL include genes involved in phototransduction, gene regulation, morphogenesis, and even epigenetic modulation (such as Kdm5b, a histone demethylase involved in gene silencing) (Hao et al., 2012). CRX is another key photoreceptor specific transcription factor that controls photoreceptor differentiation (Chen et al., 1997; Furukawa et al., 1997). The addition of CRX ChIP-seq data (Corbo et al., 2010) to the NRL targetome has revealed an overlap of over 50% of targets, and all known rod-specific genes appear to have binding sites for both CRX and NRL (Hao et al., 2012).
Histone modification ChIP-seq has been performed for whole retina, and H3K4me2 histone marks have been observed to be associated with active rod genes, which lack repressor methylation H3K27me3 (Popova et al., 2014). However, whole tissue histone modification data might miss valuable epigenetic information on low expressed genes and those expressed in minor cell types, and such studies should be performed with purified cell types (Kim et al., 2016). New methods have been developed to perform single cell ChIP-seq experiments (Rotem et al., 2015).
We recommend high library diversity (i.e., using a variety of indices) to take full advantage of sample multiplexing as Illumina sequencing technology relies on the identification of individual DNA strand clusters via fluorescence (See section 2.3 and Figure 2) (Krueger et al., 2011). Illumina software must locate and define each library fragment cluster within the flow cell; thus, insufficient sequence diversity within each lane of a flow cell may lead to misidentification of closely spaced clusters with similar sequences that may result in poor quality of data. Therefore, many distinct samples are needed in a single flow cell lane for the best read quality in ChIP-seq experiments.
5.2. DNase-seq
DNase I digestion followed by sequencing (DNase-seq) is used to identify genomic regions having regulatory elements such as promoters or enhancers (Thurman et al., 2012; Wilken et al., 2015). DNase I cleaves the DNA preferentially in open chromatin regions (Cockerill, 2011; Suck, 1994) that are often considered active or primed for activity due to their accessibility to TF binding and their proximity to active histone marks, such as H3K27ac or H3K4me (Ziemann et al., 2013). Thus, the ends of fragments produced by DNase I digestion of chromatin correspond to accessible genomic regions.
After digestion of chromatin with DNase I, adaptors are ligated to DNA fragments to produce libraries for NGS (Song and Crawford, 2010). Mapping of fragment sequences onto the genome can provide cut site frequencies at each base by counting the number of fragments with endpoints at a specific site (Wilken et al., 2015). Genomic regions with a high frequency of cut sites are referred as DNase I hypersensitivity sites (DHS) that can be correlated to active gene expression and to regulatory regions of interest. Resolution of DNase-seq experiments may be high enough to identify genomic regions that are protected from DNase I digestion by bound transcription factors, thereby yielding possible cis-regulatory domains associated with nearby genes. It is also possible to predict binding of transcription regulatory proteins by searching for known sequence motifs within TF footprints present in open chromatin (Madrigal and Krajewski, 2012). DNase-seq has been used extensively resulting in a broad range of tools specific to TF footprinting and DHS identification (Table 6). We also have a better understanding of the sequence bias of DNase I enzyme and tools have recently been developed that take it into consideration (He et al., 2014; Madrigal, 2015; Yardimci et al., 2014).
Table 6.
Tools for analyzing NGS epigenetic data.
| Process | Tool | Description |
|---|---|---|
| Quality Validation | fastQC/ChIPQC | Ensure high data quality throughout the analysis pipeline. Check for overrepresented sequences and Kmers. |
| Adapter Trimming | Trimmomatic | Remove adapter sequences present in sequenced short fragments (smaller than sequenced read length). |
| Read Alignment | Bowtie2 | Align reads to genome in order to understand read distribution. |
|
PCR Duplicate Removal |
Picard | Remove reads sharing exact mapping position in order to reduce PCR duplicate bias. |
| mtDNA Removal | Samtools | Remove reads mapping to mitochondrial DNA in order to focus on epigenetics of autosomal DNA. |
| Peak Calling | MACS2, F-Seq, Hotspot, HPeak, SPP, PeakSeq, and Peakzilla |
Identify genomic regions with a significant overrepresentation of mapped reads. |
|
Peak/Genome Annotation |
Bedtools Biomart/ BEDOPS |
Assign peaks to genes/transcripts, or cluster genomic regions by peak profiles. |
| Motif Discovery | MEME, DREME, FCmotif |
Identify transcription factor binding DNA motifs within peaks. |
|
Differential Peak Analysis |
ODIN/DiffBind | Compare peaks present in different samples to identify significant differences. |
Genomewide profiling of open chromatin by DNase-seq has demonstrated complex regional diversity in cis-regulatory domains within mouse neural retina, cerebellum, cerebral cortex, and whole brain (Ueki et al., 2015). Significant overlap of DHS was observed between brain and retina. Interestingly, DHS could be identified even for some of the genes that are uniquely, yet highly, expressed in small cell populations, such as S-opsin (Opn1sw) in S-cones or Pou4f2 in retinal ganglion cells of the mouse retina; however, prior understanding of gene regulation for cell-type specific genes is required to extract such information. Given that neuronal tissues contain numerous morphologically and functionally distinct cell types, whole tissue profiling provides data of limited value with respect to cis-regulatory mechanisms. We therefore recommend DNase-seq analysis be performed with purified cell populations or even single cells when possible.
5.3. ATAC-seq
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) may be used to identify regions of active regulatory regions within the genome (Figure 6) (Buenrostro et al., 2015). ATAC-seq is a three-step protocol that requires only 500 to 50,000 cells, compared to around 50 million required for DNase-seq, making it a faster and more efficient alternative to DNase-seq (Buenrostro et al., 2013; John et al., 2013; Kulakovskiy et al., 2009). ATAC-seq uses a hyperactive form of the Tn5 transposase to fragment chromatin and add Illumina sequencing adapters in a single step called tagmentation (Adey et al., 2010). These adapter-tagged fragments are then PCR-amplified with unique DNA index sequences. Prior to sequencing, and after PCR cleanup using Agencourt AMPure XP beads (available from Beckman Coulter), the libraries are quantified using Agilent 2100 Bioanalyzer or PCR-based methods. The former would provide library size distribution, which may help in troubleshooting library preparation methods. For high quality data, DNA quantity peaks are clearly noticeable for fragments at 200 bp intervals corresponding to nucleosome binding regions (Buenrostro et al., 2013). Final library concentrations may vary depending on the number of PCR cycles and starting cell types; however, the DNA library amount should be greater than 2nM for sequencing on Illuima HiSeq 2500.
Paired-end sequencing enhances accuracy of alignment and helps in identifying PCR duplicates. Read lengths for ATAC-seq experiments can be limited to 34–50 bp for accurate mapping to the genome, although larger read length may be desired for accurate mapping onto highly repetitive genomes (Buenrostro et al., 2013; Buenrostro et al., 2015). Sequencing depth > 50 million reads is recommended for the analysis of open chromatin and 200 million reads for TF footprinting.
ATAC-seq has many benefits compared with DNase-seq and produces comparable data. We note that ATAC-seq has been successfully performed for single cell open chromatin profiling (Buenrostro et al., 2015). However, computational tools needed for Tn5 sequence bias correction (Green et al., 2012) are still under development.
5.4. Primary Data Analysis
The primary steps in computational analysis of epigenomic data include evaluation of sequence read quality, adapter trimming, read alignment, and identification of genomic regions of interest through peak calling (Figures 2C and 6). First, fastQC tool (Table 6) may be used to perform data quality analysis, producing a read report that includes per base quality, sequence duplication levels, and adapter content. Second, processing tools such as Trimmomatic are utilized to remove adapter sequences that are added during library preparation (Bolger et al., 2014). Paired-end sequencing improves the accuracy of trimming since both forward and reverse reads from a library fragment should contain the same amount of adapter contamination. Third, we routinely apply the bowtie2 read alignment tool to map reads to genome assemblies and have observed >75% alignments for successful TF ChIP-seq experiments in the retina. Bowtie2 utilizes paired-end read data to improve alignment accuracy by attempting to align the two reads from a DNA fragment within a pre-specified distance from one another. This maximum fragment length variable (defined by the maxins parameter in Bowtie2) should be larger than most library fragment lengths (e.g., 2 kb). Finally, we observed high percentage of duplicated reads in epigenetic data sets compared to whole genome sequencing due to relatively small starting DNA concentrations and high number of PCR cycles necessary in ChIP-seq and ATAC-seq experiments. These duplicate reads confound the identification of read-enriched regions necessary peak identification (see section 5.4.1) (Dozmorov et al., 2015). Therefore, PCR over-amplification is examined by qPCR using an aliquot of DNA prior to library amplification (Buenrostro et al., 2015). Both Picard MarkDuplicates and SAMtools rmdup (Li et al., 2009) functions can remove PCR duplicates from the aligned reads (Table 6). Picard has the option of identifying duplicates generated during the sequencing process and evaluating paired-end data with paired-reads that do not align to the same chromosome. Some of the peak callers, such as MACS, can also find identical reads generated during PCR (Feng et al., 2012).
Mitochondrial DNA should also be removed using any programming language capable of processing text efficiently (e.g., Perl, Python, or Bash) if the goal is to analyze the nuclear epigenome. In our experience, mitochondrial DNA reads may vary greatly depending on a number of factors, including cell type, cell condition (e.g., fresh or frozen), tissue condition, and age. Changes in cell lysis conditions and cell storage may help in reducing mitochondrial DNA. For comparative analysis, the data sets are normalized by total read count to avoid any differences related to read depth.
For ATAC-seq data sets, read alignment data needs to be compensated for 9 bp duplications that are introduced during Tn5 tagmentation step (Adey et al., 2010) (see Figure 6B). Thus, precise cut sites for Tn5 transposase should be determined by shifting all positive-strand reads 4 bp downstream and all negative-strand reads 5 bp upstream in the genome (Buenrostro et al., 2013). Peak callers are then used to identify significantly accessible genomic regions (Boyle et al., 2008).
5.4.1 Peak Identification
Peak-calling algorithms are often used to recognize protein-binding sites or chromatin state in ChIP-seq and significantly accessible genomic regions in DNase-seq and ATAC-seq data (Table 6). Many peak callers specialize in identifying specific patterns in the data through the use of different statistical models. MACS is a common ChIP-seq peak caller for identifying TF binding sites and both sharp and broad histone modification sites by examining positive and negative strand reads that flank protein-binding regions (Feng et al., 2012; Liu, 2014). This pattern is used to predict fragment size using a sliding window algorithm. Significantly enriched regions after peak modeling and background correction are called peaks. MACS may also consider a control data set to better model the background noise. Other commonly used ChIP-seq peak callers are listed in Table 6.
F-Seq appears to be a suitable peak caller for the identification of open chromatin in both DNase-seq and ATAC-seq data using a kernel density estimation model to describe the data (Boyle et al., 2008; Buenrostro et al., 2013; Koohy et al., 2014). This model determines regions with significant levels of read overlap to define peaks, and a bandwidth parameter is used to define the width of peaks. Other common peak callers for defining significant sites in open chromatin data are ZINBA and Hotspot (John et al., 2011; Rashid et al., 2011). Peak detection methods are under continuous development, and a variety of tools must be tested for obtaining the best results.
5.4.2 Differential Peak Analyses
Differential regions (DR) are regions that have been found to be significantly different between data sets. The algorithms used to detect these DR may vary significantly, and may work on sharp, broad, or both types of peak signals (e.g., MACS), and some may consider biological replicates (e.g., DiffBind) (Steinhauser et al., 2016). The number and size of DRs identified also varies significantly depending on the detection method (Table 6). Therefore, it is important to select tools that are more likely to be optimized for the data set under scrutiny, and to validate significant results experimentally, if possible.
5.4.3 Identification of Enriched TF Motif Sequences
Identifying protein-DNA interactions is important for understanding gene regulatory mechanisms that drive cellular and organism level changes. DNA-binding proteins, such as TFs, often bind with a bias for certain sequence motifs. DNA-binding motifs are frequently represented via a position weight matrix (PWM) that contains the log-likelihood of each base appearing at each position of the motif as compared to a random background model (Stormo et al., 1982). A wide range of TF binding motif identification tools exists (Table 6); these tools vary in their underlying mathematical and computational methods (Tran and Huang, 2014). The use of multiple algorithms in parallel to obtain recurring motifs using ensemble methods has been recommended (Lihu and Holban, 2015). The identification of sequence bias inherent to DNase I and Tn5 for DNase-seq and ATAC-seq respectively, has also led to development of novel algorithms which attempt to compensate for these technical effects (Madrigal, 2015). Multiple well-annotated databases of validated TF-binding motifs exist (Bryne et al., 2008; Newburger and Bulyk, 2009; Wingender et al., 1996); some of these databases also provide web-based platforms for motif mining. These web tools search for candidate TF binding sites in data produced from open chromatin assays, where each tool provides customized parameters that can be chosen by the user.
5.5 Secondary Analysis
In this section, we discuss additional computational approaches for elucidating transcriptional regulatory mechanisms.
5.5.1 Elucidating TF-Mediated Gene Regulation
Genomewide TF binding sites (TFBS), as identified by peak detection algorithms, can aid in decoding complexities associated with gene regulation. Location of TFBS may uncover regulatory targets of TFs; e.g., if a peak is identified in close proximity of a gene’s transcription start site (TSS), it is inferred as a putative target gene; however, further experimental validation is required to delineate biologically relevant interactions. TF ChIP-seq experiments are reasonably accurate but target inference is restricted to a single TF. In eukaryotes, multiple TFs cooperatively (synergistically or antagonistically) control gene expression (Chen et al., 2015); hence, an array of TF ChIP-seq experiments is required to construct combinatorial and higher order regulatory networks. Significant efforts have been made by ENCODE and high throughput ChIP-seq projects to identify principles and dynamics of gene regulation (Garber et al., 2012; Gerstein et al., 2012). Genomewide maps of DNase I footprints and open chromatin regions identified by DNase-seq or ATAC-seq experiments complement the ChIP-seq data by providing active regulatory regions that can be interrogated for locations of TF binding motif sequences to uncover novel TF interactions (Neph et al., 2012; Vierstra et al., 2014).
6. Systems Integration
Cells in an organism contain numerous distinct small and large molecules that work together for maintaining physiology and survival. NGS methods yield high quality data that measures the levels or activity of some of these molecules at genome scale albeit independently. Systems biology aims to elucidate normal and abnormal functioning of the whole cell, tissue and the organism by integrating multiple data (such as genetic, epigenetic, proteomic, functional) on molecules into pathways and networks (Barabasi et al., 2011; Chuang et al., 2010; Gustafsson et al., 2014; Kitano, 2002; Vidal et al., 2011). In this section, we highlight two specific systems frameworks that integrate NGS data sets to uncover gene regulatory networks and to identify variants that influence expression levels of gene(s) (called expression quantitative trait loci or eQTL). Table 7 lists available open source tools for regulatory network inference and eQTL analysis.
Table 7.
Systems Integration Tools
| Method | Tool | Availability |
|---|---|---|
|
Network Inference |
Integrative Inference Of Regulatory Networks |
http://compbio.mit.edu/flynet/ |
| Binding And Expression Target Analysis (BETA) |
http://cistrome.org/BETA/ | |
| Integrated Regulatory Network Framework |
Available upon request to authors. | |
| Physical Module Networks |
http://www.compbio.cs.huji.ac.il/PMN/Welcome.html | |
| eQTL | Matrix Eqtl | https://cran.r-project.org/web/packages/MatrixEQTL/ |
| Eqtl | https://cran.r-project.org/web/packages/eqtl/ | |
| Genetictools | https://cran.r-project.org/web/packages/GeneticTools/ |
Systems biology methods assist in nullifying issues caused by false positives and false negatives obtained by analyzing single “omics” data set (Ge et al., 2003). For example, not all DE genes identified by the loss of NRL in photoreceptors are direct transcriptional targets and integration of this data with NRL ChIP-seq can reveal biologically relevant targets with high confidence. Just as methods to analyze NGS data sets are not completely optimized and one can use different software, no benchmarking tools exist in systems biology. We note that systems biology paradigms are flexible and that multiple data sets can be combined using different parameters to answer a specific query (Figure 7).
Figure 7. Integrating NGS data for systems-level understanding.
Knowledge integration resulting from analyzing sequencing data from genomics, epigenomics, transcriptomics, and literature can help decode networks and variants underlying development and disease. A systems level study can yield better identification of disease genes and drug targets.
6.1. Regulatory Network Inference
Precise control of gene regulation is central to biological processes and is modulated by complex yet highly organized and integrated networks that may span multiple levels (Thompson et al., 2015). Binding of TFs to DNA elements and chromatin re-organization factors including those associated with DNA methylation and histone modifications can stabilize transcription initiation complex at the promoter region and are largely responsible for controlling gene expression patterns. NGS-based transcriptome profiling provides quantified gene expression levels, which can be integrated with other molecular profiles (such as DNA methylome or histone methylation and acetylation) to initiate the construction of genome scale gene regulatory networks (GRNs) underlying development and disease (Yang et al., 2015). Inference of GRN using a single genomewide dataset would likely yield physical association networks in which many regulatory interactions may not be functionally relevant (Marbach et al., 2012b). Such networks especially those inferred from the expression data would likely contain large number of false positives (Ge et al., 2003). Integration of different NGS datasets is thus desired to infer a functional regulatory network, which is still a major challenge especially for mammalian systems. Computational and mathematical methods are begun to emerge lately to systematically integrate high-throughput data sets for elucidating GRNs. For example, BETA (Binding and expression target analysis) combines ChIP-seq and RNA-seq data to elucidate gene regulation (Wang et al., 2013). BETA also conducts network motif analysis to identify putative collaborating factors that contribute to gene expression.
NGS data can be integrated for systems studies at the transcriptional and post-transcriptional levels to infer genetic interactions, such as those between genes/transcripts and miRNAs (Cheng et al., 2011). A comprehensive systems integration approach to reverse engineer networks has been formulated in a machine-learning framework that uses both supervised and unsupervised methods to predict regulatory edges by integrating RNA-seq, TF ChIP-seq and histone modification datasets along with prior knowledge of conserved motifs of TF as input features (Marbach et al., 2012b). A rank combined framework that assimilates data from NGS and other high throughput technologies (such as microarray or ChIP-chip) has been successfully implemented to discover novel central regulators that participate in modulating cellular plasticity in Th17 cells (Ciofani et al., 2012). This method ranks regulatory interactions based on multiple metrics including correlation and TF binding scores obtained from RNA-seq and microarray experiments, and DE scores computed from wild type and knockout experiments.
6.2. eQTL
Every human being carries millions of genetic variants in the genome (Genomes Project et al., 2015; Sudmant et al., 2015). While a majority of these variants may be benign, many can potentially impact cell/tissue specific gene expression patterns (Consortium, 2015) that would in turn affect downstream traits ranging from morphological and functional diversity to disease and response to treatments (Albert and Kruglyak, 2015; Cookson et al., 2009). eQTL refers to a genomic region, which harbors sequence/structural variants that influence gene expression levels; these regulatory variants can be identified by combining global expression profiles from cells/tissues and under different conditions with genomewide genetic variations (Gilad et al., 2008). NGS methods can permit more precise quantification of eQTL variations across cell types or tissues and discern their regulatory impact of gene expression (Bahcall, 2015; Majewski and Pastinen, 2011). Regulatory variations that maps in close proximity to the target gene(s) in the genome are classified as cis-eQTLs and those mapping far away (even on a different chromosome) are called trans-eQTLs.
The concept of eQTLs is extremely attractive because it allows us to correlate genetic diversity to phenotypic manifestations. In many inherited retinal diseases, the presence of a genetic variation is not always associated with identical clinical phenotypes and other genomic variants are predicted to modify the impact of causal mutation; e.g., the presence of a common RPGRIP1L variant (A229T) can lead to highly penetrant retinal degeneration phenotype in ciliopathies (Khanna et al., 2009). However, in case of common multifactorial diseases the causality is difficult to establish, and GWAS of complex traits have discovered a vast majority of genetic susceptibility variants that have relatively small impact on the disease risk. Most GWAS-identified disease associated variants are localized in gene deserts or non-coding genome regions (Chakravarti et al., 2013) and likely manifest their influence through modulation of gene expression (i.e., eQTL). Functional implications of a disease-associated eQTL should however be evaluated in the context of cells/tissues that are influenced by the disease. For example, genetic studies have discovered 45 independent common variants associated with AMD (Fritsche et al., 2016), and it is prudent to look at their impact on gene expression in the retina, retinal pigment epithelium or choroid. GTEx project was initiated with a goal to create the resource for systematic study of genetic variations in multiple reference human tissues (Consortium, 2015). GTEx consortium, however, does not include ocular cells/tissues. Our laboratory has therefore undertaken this task by generating RNA-seq data and genotypes for over 400 human retina samples.
7. Concluding Remarks
Precision medicine is designed to provide targeted therapies to patients based on their individual genetic architecture and disease. Though the current use of precision medicine in clinical settings is limited, substantial efforts, such as the Precision Medicine Initiative (www.nih.gov/precisionmedicine-initiative-cohort-program), are underway to facilitate a more rapid progression of the field. NGS methodologies have allowed the collection of valuable genomic information from individuals, thereby accelerating patient-oriented decision-making including assessment of disease risk and/or prediction of drug response. Systems biology approaches, such as network modeling of genetic variants, are expected to uncover and expedite the better understanding of disease causality and enable the design of customized treatments based on an individual's genetic and epigenetic profile.
In this review, we have primarily focused on genome, transcriptome and epigenome analysis with a goal to provide a comprehensive perspective of NGS methods and tools to scientists in the vision field. WES and targeted sequencing are becoming standard methodology for identifying genetic variants/mutations in retinal research and clinical diagnosis. NGS-based methods are increasingly being used to evaluate drug targets and small molecules and their broader impact on retinal cell/tissue function (Chen and Palczewski, 2015; Kaewkhaw, 2016). Over 240 genes have so far been associated with retinal diseases; however, underlying pathways leading to disease pathology are poorly understood. A vast majority of functional studies have focused on one or few genes, which provide useful though limited insight into molecular and cellular pathways. Genes and their products (such as proteins, miRNA) are part of combinatorial functional networks, and any genetic (sequence variation/mutation) or epigenetic change can potentially have an impact on multiple physiological processes leading to cellular/organismic adaptation or disease (Yang et al., 2015). It is therefore imperative that we elucidate global profiles of transcriptome and epigenome, together with those of lncRNA, miRNA and other noncoding transcribed sequences, under normal (control) and abnormal (disease) conditions focusing on different cell types in the retina/eye and generate gene interaction networks. While a majority of reported NGS data in retina/eye originates from small number of samples and whole tissues (Farkas et al., 2013; Tian et al., 2015), several groups have initiated NGS studies on specific cell types (Macosko et al., 2015; Mo et al., 2016; Siegert et al., 2012)(Kim et al., 2016). An array of NGS data thus provides unprecedented opportunities to address fundamental questions related to gene/genome organization, function and regulation, which in turn would dramatically influence our understanding of cell physiology and pathogenic mechanisms. A higher-order integrative analysis of different datasets would permit extraction of useful pathway and network information at systems level. We have briefly touched upon two such aspects and discussed strategies for elucidating regulatory networks and delineating causality of variants in complex disease.
The rapidly changing and ever evolving NGS field is now leading to applications where one can examine genomic, transcriptomic, and epigenomic landscape even at the level of an individual cell; such studies are expected to have a broad impact on biology and medicine (Wang and Navin, 2015). Single-cell RNA-seq has accelerated molecular profiling of rare cell populations in complex tissues. Drop-seq, a massively parallel transcriptome analysis approach, has been successfully applied to the mouse retina, leading to the identification of 39 distinct cell populations representing specific subtypes of one of the seven major retinal cells (Macosko et al., 2015). Additionally, NGS-based epigenome technologies have adapted to solve specific questions on the translation of genotype information into phenotype; these include ChIP-exo (Rhee and Pugh, 2012) that can detect TF binding sites at the single nucleotide resolution, and FAIRE-seq (Bianco et al., 2015; Giresi and Lieb, 2009) that identifies open chromatin regions in fixed nuclei. Both ATAC-seq and ChIPmentation (Schmidl et al., 2015) take advantage of the flexibility and efficiency provided by the use of the Tn5 transposase in library preparation in order to assay open chromatin and protein binding, respectively. NGS assays must continue to adapt to the increasing demand for rapid, accurate, and cost-effective generation of epigenomewide analysis.
NGS based approaches are rapidly gaining broad applicability in biology and medicine. However, current computational methods are not able to harness the full potential of large genomic and epigenomic data sets being generated by innovations in NGS technology. Thus, a greater focus is needed on developing novel tools for computational and systems level analysis. Machine learning including neural networks and support vector machines and other mathematical models are being built to integrate knowledge from NGS data generated at diverse molecular levels, but systems integration models are yet to be standardized. The pace of data production has added a new dimension of challenges to computational and systems biologists; thus, additional innovations are needed for extracting useful information to have a desired impact of NGS in solving challenges of biology and medicine.
Acknowledgments
We thank Jung-Woong Kim, Hyun-Jin Yang, Tiziana Cogliati and Lina Zelinger for discussions and comments on the manuscript. Our work is supported by the Intramural Research Program of the National Eye Institute, National Institutes of Health. Some of the NGS analysis required the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
List of Abbreviations
- AS
Alternative Splicing
- BETA
Binding And Expression Target Analysis
- bp
Base Pair
- CCDS
Consensus Coding Sequence
- ChIP
Chromatin Immunoprecipitation
- DAS
Differential Alternative Splicing
- DR
Differential Regions
- emPCR
Emulsion PCR
- eQTL
Expression Quantitative Trait Loci
- ENCODE
Encyclopedia of DNA Elements
- EST
Expressed Sequence Tag
- FDR
False Discovery Rate
- GO
Gene Ontology
- GRN
Gene Regulatory Network
- GTEx
Gene Tissue Expression
- GWAS
Genomewide Association Study
- HGP
Human Genome Project
- HM
Histone Modification
- kb
Kilobase
- lncRNA
Long non-coding RNA
- LM
Linear Method
- NGS
Next Generation Sequencing
- PCA
Principal Component Analysis
- PE
Paired-End
- PWM
Position Weight Matrix
- RDD
Retinal Degenerative Disease
- RIN
RNA Integrity Number
- RLE
Relative Log Expression
- SBS
Sequence-By-Synthesis
- SCS
Single Cell Sequencing
- SE
Single-End
- TF
Transcription Factor
- TFBS
Transcription Factor Binding Site
- TMM
Trimmed Median Of The Mean
- TSS
Transcription Start Site
- WES
Whole Exome Sequencing
- WGCNA
Weighted Gene Co-expression Networks Analysis
- WGS
Whole Genome Sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamancos GP, Pages A, Trincado JL, Bellora N, Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21:1521–1531. doi: 10.1261/rna.051557.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, Cooper GM, Dorschner MO, Dulik MC, Ghazani AA, Ghosh R, Green RC, Hart R, Horton C, Johnston JJ, Lebo MS, Milosavljevic A, Ou J, Pak CM, Patel RY, Punj S, Richards CS, Salama J, Strande NT, Yang Y, Plon SE, Biesecker LG, Rehm HL. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahcall OG. Human genetics: GTEx pilot quantifies eQTL variation across tissues and individuals. Nat Rev Genet. 2015;16:375. doi: 10.1038/nrg3969. [DOI] [PubMed] [Google Scholar]
- Bailey JN, Loomis SJ, Kang JH, Allingham RR, Gharahkhani P, Khor CC, Burdon KP, Aschard H, Chasman DI, Igo RP, Jr, Hysi PG, Glastonbury CA, Ashley-Koch A, Brilliant M, Brown AA, Budenz DL, Buil A, Cheng CY, Choi H, Christen WG, Curhan G, De Vivo I, Fingert JH, Foster PJ, Fuchs C, Gaasterland D, Gaasterland T, Hewitt AW, Hu F, Hunter DJ, Khawaja AP, Lee RK, Li Z, Lichter PR, Mackey DA, McGuffin P, Mitchell P, Moroi SE, Perera SA, Pepper KW, Qi Q, Realini T, Richards JE, Ridker PM, Rimm E, Ritch R, Ritchie M, Schuman JS, Scott WK, Singh K, Sit AJ, Song YE, Tamimi RM, Topouzis F, Viswanathan AC, Verma SS, Vollrath D, Wang JJ, Weisschuh N, Wissinger B, Wollstein G, Wong TY, Yaspan BL, Zack DJ, Zhang K, Study EE, Consortium A, Weinreb RN, Pericak-Vance MA, Small K, Hammond CJ, Aung T, Liu Y, Vithana EN, MacGregor S, Craig JE, Kraft P, Howell G, Hauser MA, Pasquale LR, Haines JL, Wiggs JL. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016 doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner AK, Kleinman ME. Therapeutic Approaches to Histone Reprogramming in Retinal Degeneration. Adv Exp Med Biol. 2016;854:39–44. doi: 10.1007/978-3-319-17121-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SS, Wright AF, Clayton JF, Price WH, Phillips CI, McKeown CM, Jay M, Bird AC, Pearson PL, Southern EM, et al. Close genetic linkage between X-linked retinitis pigmentosa and a restriction fragment length polymorphism identified by recombinant DNA probe L1.28. Nature. 1984;309:253–255. doi: 10.1038/309253a0. [DOI] [PubMed] [Google Scholar]
- Bianco S, Rodrigue S, Murphy BD, Gevry N. Global Mapping of Open Chromatin Regulatory Elements by Formaldehyde-Assisted Isolation of Regulatory Elements Followed by Sequencing (FAIRE-seq) Methods Mol Biol. 2015;1334:261–272. doi: 10.1007/978-1-4939-2877-4_17. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Kuo WP, Park PJ, Tsujikawa M, Gunnersen JM, Scott HS, Boon WM, Tan SS, Cepko CL. MicroSAGE is highly representative and reproducible but reveals major differences in gene expression among samples obtained from similar tissues. Genome Biol. 2003;4:R17. doi: 10.1186/gb-2003-4-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci. 2012;13:453–464. doi: 10.1038/nrn3271. [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brooks MJ, Rajasimha HK, Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol Biol. 2012;884:319–334. doi: 10.1007/978-1-61779-848-1_23. [DOI] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21–29. doi: 10.1002/0471142727.mb2129s109. 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Kelly MC, Hoa M, Morell RJ, Kelley MW. Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun. 2015;6:8557. doi: 10.1038/ncomms9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell. 2013;155:21–26. doi: 10.1016/j.cell.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li H, Liu F, Zheng X, Wang S, Bo X, Shu W. An integrative analysis of TFBS-clustered regions reveals new transcriptional regulation models on the accessible chromatin landscape. Sci Rep. 2015;5:8465. doi: 10.1038/srep08465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, Sleiman P, Cheng WY, Chen W, Shah H, Shen Y, Fromer M, Omberg L, Deardorff MA, Zackai E, Bobe JR, Levin E, Hudson TJ, Groop L, Wang J, Hakonarson H, Wojcicki A, Diaz GA, Edelmann L, Schadt EE, Friend SH. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Palczewski K. Systems Pharmacology Links GPCRs with Retinal Degenerative Disorders. Annu Rev Pharmacol Toxicol. 2015 doi: 10.1146/annurev-pharmtox-010715-103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yan KK, Hwang W, Qian J, Bhardwaj N, Rozowsky J, Lu ZJ, Niu W, Alves P, Kato M, Snyder M, Gerstein M. Construction and analysis of an integrated regulatory network derived from high-throughput sequencing data. PLoS Comput Biol. 2011;7:e1002190. doi: 10.1371/journal.pcbi.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilamakuri CS, Lorenz S, Madoui MA, Vodak D, Sun J, Hovig E, Myklebost O, Meza-Zepeda LA. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics. 2014;15:449. doi: 10.1186/1471-2164-15-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J. 2011;278:2182–2210. doi: 10.1111/j.1742-4658.2011.08128.x. [DOI] [PubMed] [Google Scholar]
- Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, Weber BH, Langmann T. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Li W, Liu J, Zhou XJ. Integrating many co-splicing networks to reconstruct splicing regulatory modules. BMC Syst Biol. 2012;6(Suppl 1):S17. doi: 10.1186/1752-0509-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov MG, Adrianto I, Giles CB, Glass E, Glenn SB, Montgomery C, Sivils KL, Olson LE, Iwayama T, Freeman WM, Lessard CJ, Wren JD. Detrimental effects of duplicate reads and low complexity regions on RNA- and ChIP-seq data. BMC Bioinformatics. 2015;16(Suppl 13):S10. doi: 10.1186/1471-2105-16-S13-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Ebermann I, Phillips JB, Liebau MC, Koenekoop RK, Schermer B, Lopez I, Schafer E, Roux AF, Dafinger C, Bernd A, Zrenner E, Claustres M, Blanco B, Nurnberg G, Nurnberg P, Ruland R, Westerfield M, Benzing T, Bolz HJ. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Invest. 2010;120:1812–1823. doi: 10.1172/JCI39715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichers ER, Lewis RA, Katsanis N, Lupski JR. Triallelic inheritance: a bridge between Mendelian and multifactorial traits. Annals Med. 2004;36:262–272. doi: 10.1080/07853890410026214. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Steijger T, Sipos B, Grant GR, Kahles A, Ratsch G, Goldman N, Hubbard TJ, Harrow J, Guigo R, Bertone P, Consortium R. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Kenna P, Redmond R, Shiels D, McWilliam P, Humphries MM, Sharp EM, Jordan S, Kumar-Singh R, Humphries P. Autosomal dominant retinitis pigmentosa: a mutation in codon 178 of the rhodopsin gene in two families of Celtic origin. Genomics. 1991;11:1170–1171. [PubMed] [Google Scholar]
- Farrell CM, O'Leary NA, Harte RA, Loveland JE, Wilming LG, Wallin C, Diekhans M, Barrell D, Searle SM, Aken B, Hiatt SM, Frankish A, Suner MM, Rajput B, Steward CA, Brown GR, Bennett R, Murphy M, Wu W, Kay MP, Hart J, Rajan J, Weber J, Snow C, Riddick LD, Hunt T, Webb D, Thomas M, Tamez P, Rangwala SH, McGarvey KM, Pujar S, Shkeda A, Mudge JM, Gonzalez JM, Gilbert JG, Trevanion SJ, Baertsch R, Harrow JL, Hubbard T, Ostell JM, Haussler D, Pruitt KD. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014;42:D865–D872. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:e22. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Jr, Buitendijk GH, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht AM, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan CC, Cheng CY, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdottir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JP, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis N, C NK, Khan JC, Kim IK, Kiyohara Y, Klein BE, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Said S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJ, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, Sahel JA, Schaumberg DA, Scholl HP, Schwartz SG, Scott WK, Shahid H, Sigurdsson H, Silvestri G, Sivakumaran TA, Smith RT, Sobrin L, Souied EH, Stambolian DE, Stefansson H, Sturgill-Short GM, Takahashi A, Tosakulwong N, Truitt BJ, Tsironi EE, Uitterlinden AG, van Duijn CM, Vijaya L, Vingerling JR, Vithana EN, Webster AR, Wichmann HE, Winkler TW, Wong TY, Wright AF, Zelenika D, Zhang M, Zhao L, Zhang K, Klein ML, Hageman GS, Lathrop GM, Stefansson K, Allikmets R, Baird PN, Gorin MB, Wang JJ, Klaver CC, Seddon JM, Pericak-Vance MA, Iyengar SK, Yates JR, Swaroop A, Weber BH, Kubo M, Deangelis MM, Leveillard T, Thorsteinsdottir U, Haines JL, Farrer LA, Heid IM, Abecasis GR Consortium, A.M.D.G. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–439. 439e431–439e432. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Said S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanche H, Deleuze JF, Igo RP, Jr, Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA, Jr, Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Leveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Walhout AJ, Vidal M. Integrating 'omic' information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124:357–368. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project, C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Human genetics. GTEx detects genetic effects. Science. 2015;348:640–641. doi: 10.1126/science.aab3002. [DOI] [PubMed] [Google Scholar]
- Gieser L, Swaroop A. Expressed sequence tags and chromosomal localization of cDNA clones from a subtracted retinal pigment epithelium library. Genomics. 1992;13:873–876. doi: 10.1016/0888-7543(92)90173-p. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011;20:2472–2481. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B, Bouchier C, Fairhead C, Craig NL, Cormack BP. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob DNA. 2012;3:3. doi: 10.1186/1759-8753-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M, Nestor CE, Zhang H, Barabasi AL, Baranzini S, Brunak S, Chung KF, Federoff HJ, Gavin AC, Meehan RR, Picotti P, Pujana MA, Rajewsky N, Smith KG, Sterk PJ, Villoslada P, Benson M. Modules, networks and systems medicine for understanding disease and aiding diagnosis. Genome Med. 2014;6:82. doi: 10.1186/s13073-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Kim DS, Klocke B, Johnson KR, Cui K, Gotoh N, Zang C, Gregorski J, Gieser L, Peng W, Fann Y, Seifert M, Zhao K, Swaroop A. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Lambeck S, Toepfer S, van Someren E, Guthke R. Gene regulatory network inference: data integration in dynamic models-a review. Biosystems. 2009;96:86–103. doi: 10.1016/j.biosystems.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- International HapMap, C. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, Gingeras TR, Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Canfield TK, Lee K, Vong S, Weaver M, Wang H, Vierstra J, Reynolds AP, Thurman RE, Stamatoyannopoulos JA. Genome-scale mapping of DNase I hypersensitivity. Curr Protoc Mol Biol. 2013;Chapter 27(Unit 21):27. doi: 10.1002/0471142727.mb2127s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jordan DM, Frangakis SG, Golzio C, Cassa CA, Kurtzberg J, Task Force for Neonatal, G. Davis EE, Sunyaev SR, Katsanis N. Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–229. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YL, Luquette LJ, Ho JW, Ferrari F, Tolstorukov M, Minoda A, Issner R, Epstein CB, Karpen GH, Kuroda MI, Park PJ. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res. 2014;42:e74. doi: 10.1093/nar/gku178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem cells. 2015;33:3504–3518. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Swaroop M, Homma K, Nakamura J, Brooks M, Kaya KD, Chaitankar V, Michael S, Tawa G, Zou J, Rao M, Zheng W, Cogliati T, Swaroop A. Treatment Paradigms for Retinal and Macular Diseases Using 3-D Retina Cultures Derived From Human Reporter Pluripotent Stem Cell Lines. Invest Ophthal Vis Sci. 2016;57:ORSFl1–ORSFl11. doi: 10.1167/iovs.15-17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ University of California Santa, C. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, Maubaret C, Diaz-Font A, MacDonald I, Muzny DM, Wheeler DA, Morgan M, Lewis LR, Logan CV, Tan PL, Beer MA, Inglehearn CF, Lewis RA, Jacobson SG, Bergmann C, Beales PL, Attie-Bitach T, Johnson CA, Otto EA, Bhattacharya SS, Hildebrandt F, Gibbs RA, Koenekoop RK, Swaroop A, Katsanis N. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-W, Yang H-J, Oel AP, Brooks MJ, Jia L, Plachetzki DC, Li W, Allison WT, Swaroop A. Recruitment of rod photoreceptors from short wavelength sensitive cones during the evolution of nocturnal vision in mammals. Dev Cell. 2016;37 doi: 10.1016/j.devcel.2016.05.023. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirin M, Chandra A, Charteris DG, Hayward C, Campbell S, Celap I, Bencic G, Vatavuk Z, Kirac I, Richards AJ, Tenesa A, Snead MP, Fleck BW, Singh J, Harsum S, Maclaren RE, den Hollander AI, Dunlop MG, Hoyng CB, Wright AF, Campbell H, Vitart V, Mitry D. Genome-wide association study identifies genetic risk underlying primary rhegmatogenous retinal detachment. Hum Mol Genet. 2013;22:3174–3185. doi: 10.1093/hmg/ddt169. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koohy H, Down TA, Spivakov M, Hubbard T. A comparison of peak callers used for DNase-Seq data. PLoS One. 2014;9:e96303. doi: 10.1371/journal.pone.0096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR, Osborne CS. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One. 2011;6:e16607. doi: 10.1371/journal.pone.0016607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Favorov AV, Makeev VJ. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics. 2009;25:2318–2325. doi: 10.1093/bioinformatics/btp434. [DOI] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J International Human Genome Sequencing, C. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tighe SW, Nicolet CM, Grove D, Levy S, Farmerie W, Viale A, Wright C, Schweitzer PA, Gao Y, Kim D, Boland J, Hicks B, Kim R, Chhangawala S, Jafari N, Raghavachari N, Gandara J, Garcia-Reyero N, Hendrickson C, Roberson D, Rosenfeld J, Smith T, Underwood JG, Wang M, Zumbo P, Baldwin DA, Grills GS, Mason CE. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol. 2014a;32:915–925. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Dai C, Kang S, Zhou XJ. Integrative analysis of many RNA-seq datasets to study alternative splicing. Methods. 2014b;67:313–324. doi: 10.1016/j.ymeth.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kang S, Liu CC, Zhang S, Shi Y, Liu Y, Zhou XJ. High-resolution functional annotation of human transcriptome: predicting isoform functions by a novel multiple instance-based label propagation method. Nucleic Acids Res. 2014c;42:e39. doi: 10.1093/nar/gkt1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lihu A, Holban S. A review of ensemble methods for de novo motif discovery in ChIP-Seq data. Brief Bioinform. 2015;16:964–973. doi: 10.1093/bib/bbv022. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Loraine AE, Dickerson JA. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics. 2014;15:364. doi: 10.1186/s12859-014-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol. 2014;1150:81–95. doi: 10.1007/978-1-4939-0512-6_4. [DOI] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ferguson JF, Xue C, Silverman IM, Gregory B, Reilly MP, Li M. Evaluating the impact of sequencing depth on transcriptome profiling in human adipose. PLoS One. 2013;8:e66883. doi: 10.1371/journal.pone.0066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Genomes Project C, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal P. On Accounting for Sequence-Specific Bias in Genome-Wide Chromatin Accessibility Experiments: Recent Advances and Contradictions. Front Bioeng Biotechnol. 2015;3:144. doi: 10.3389/fbioe.2015.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal P, Krajewski P. Current bioinformatic approaches to identify DNase I hypersensitive sites and genomic footprints from DNase-seq data. Front Genet. 2012;3:230. doi: 10.3389/fgene.2012.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Consortium D, Kellis M, Collins JJ, Stolovitzky G. Wisdom of crowds for robust gene network inference. Nat Methods. 2012a;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 2012b;22:1334–1349. doi: 10.1101/gr.127191.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Vinson JP, Program NCS, Miller W, Jaffe DB, Lindblad-Toh K, Chang JL, Green ED, Lander ES, Mullikin JC, Clamp M. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc Natl Acad Sci U S A. 2005a;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005b;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Marx V. Next-generation sequencing: The genome jigsaw. Nature. 2013;501:263–268. doi: 10.1038/501261a. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Humburg P, Kanapin A, Rivas MA, Gaulton K, Cazier JB, Donnelly P. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Guzman AE, Deshpande M, Byrd D, DeLooff C, Mkoyan K, Zlojutro P, Wallace A, Metcalf B, Laux K, Sotzen J, Tran T. Different effects of valproic acid on photoreceptor loss in Rd1 and Rd10 retinal degeneration mice. Mol Vis. 2014;20:1527–1544. [PMC free article] [PubMed] [Google Scholar]
- Mo A, Luo C, Davis FP, Mukamel EA, Henry GL, Nery JR, Urich MA, Picard S, Lister R, Eddy SR, Beer MA, Ecker JR, Nathans J. Epigenomic landscapes of retinal rods and cones. Elife. 2016;5 doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter L. Models for transcript quantification from RNA-Seq. 2011 arXiv preprint arXiv:1104.3889. [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Barnstable CJ, Zhang SS. Cell type-specific epigenetic signatures accompany late stages of mouse retina development. Adv Exp Med Biol. 2014;801:3–8. doi: 10.1007/978-1-4614-3209-8_1. [DOI] [PubMed] [Google Scholar]
- Priya R, Zipprer D, Zhan X, Friedman JS, Schwartz SB, Sharon D, Banin E, Abecasis GR, Jacobson SG, Swaroop A. Genomewide search for genetic modifiers in patients with Leber congenital amaurosis using whole exome sequencing. Invest Ophthal Vis Sci. 2014;55:3282. [Google Scholar]
- Priya RR, Rajasimha HK, Brooks MJ, Swaroop A. Exome sequencing: capture and sequencing of all human coding regions for disease gene discovery. Methods Mol Biol. 2012;884:335–351. doi: 10.1007/978-1-61779-848-1_24. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnapriya R, Swaroop A. Genetic architecture of retinal and macular degenerative diseases: the promise and challenges of next-generation sequencing. Genome Med. 2013;5:84. doi: 10.1186/gm488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, Ripke S, Gowrisankar S, Vemuri S, Montgomery K, Yu Y, Reynolds R, Zack DJ, Campochiaro B, Campochiaro P, Katsanis N, Daly MJ, Seddon JM. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics. 2010;11:399. doi: 10.1186/1471-2164-11-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol Chapter. 2012;21(Unit 21):24. doi: 10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Filip M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox Res. 2015;27:172–197. doi: 10.1007/s12640-014-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Kostrzewa RM, Filip M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox Res. 2015;27:84–97. doi: 10.1007/s12640-014-9497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977a;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977b;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Schug J. ChIP-Seq: Library Preparation and Sequencing. Methods Mol Biol. 2016;1402:101–117. doi: 10.1007/978-1-4939-3378-5_9. [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, Roska B. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15:487–495. S481–S482. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A, Biesecker LG. Genetic modifiers in human development and malformation syndromes, including chaperone proteins. Hum Mol Genet. 2003;12(1):R45–R50. doi: 10.1093/hmg/ddg099. [DOI] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser S, Kurzawa N, Eils R, Herrmann C. A comprehensive comparison of tools for differential ChIP-seq analysis. Brief Bioinform. 2016 doi: 10.1093/bib/bbv110. 10.1093/bib/bbv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold L, Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D. DNA recognition by DNase I. J Mol Recognit. 1994;7:65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, Konkel MK, Malhotra A, Stutz AM, Shi X, Paolo Casale F, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJ, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HY, Jasmine Mu X, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Genomes Project C, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, Hjartarson E, Sigurdsson GT, Jonasdottir A, Jonasdottir A, Sigurdsson A, Magnusson OT, Kong A, Helgason A, Holm H, Thorsteinsdottir U, Masson G, Gudbjartsson DF, Stefansson K. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O'Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Sieving PA. The golden era of ocular disease gene discovery: race to the finish. Clin Genet. 2013;84:99–101. doi: 10.1111/cge.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Regev A, Roy S. Comparative Analysis of Gene Regulatory Networks: From Network Reconstruction to Evolution. Annu Rev Cell Dev Biol. 2015;31:399–428. doi: 10.1146/annurev-cellbio-100913-012908. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Steinmann KE. Single molecule sequencing with a HeliScope genetic analysis system. Curr Protoc Mol Biol Chapter. 2010;7(Unit7):10. doi: 10.1002/0471142727.mb0710s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Kazmierkiewicz KL, Bowman AS, Li M, Curcio CA, Stambolian DE. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics. 2015;105:253–264. doi: 10.1016/j.ygeno.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NT, Huang CH. A survey of motif finding Web tools for detecting binding site motifs in ChIP-Seq data. Biol Direct. 2014;9:4. doi: 10.1186/1745-6150-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcatti G, Romieu A, Fedurco M, Tairi AP. A new class of cleavable fluorescent nucleotides: synthesis and optimization as reversible terminators for DNA sequencing by synthesis. Nucleic Acids Res. 2008;36:e25. doi: 10.1093/nar/gkn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Wilken MS, Cox KE, Chipman LB, Bermingham-McDonogh O, Reh TA. A transient wave of BMP signaling in the retina is necessary for Muller glial differentiation. Development. 2015;142:533–543. doi: 10.1242/dev.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8:109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, Thurman RE, Johnson AK, Vong S, Lee K, Bates D, Neri F, Diegel M, Giste E, Haugen E, Dunn D, Wilken MS, Josefowicz S, Samstein R, Chang KH, Eichler EE, De Bruijn M, Reh TA, Skoultchi A, Rudensky A, Orkin SH, Papayannopoulou T, Treuting PM, Selleri L, Kaul R, Groudine M, Bender MA, Stamatoyannopoulos JA. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. The human genome project: past, present, and future. Science. 1990;248:44–49. doi: 10.1126/science.2181665. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Wilken MS, Brzezinski JA, La Torre A, Siebenthall K, Thurman R, Sabo P, Sandstrom RS, Vierstra J, Canfield TK, Hansen RS, Bender MA, Stamatoyannopoulos J, Reh TA. DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin. 2015;8:8. doi: 10.1186/1756-8935-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Ratnapriya R, Cogliati T, Kim JW, Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog Retin Eye Res. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaragatti M, Basilico C, Dailey L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 2008;18:930–938. doi: 10.1101/gr.073460.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci GG, Frank CL, Crawford GE, Ohler U. Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res. 2014;42:11865–11878. doi: 10.1093/nar/gku810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Kasinathan S, Xin B, Rohs R, Henikoff S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat Commun. 2015;6:8733. doi: 10.1038/ncomms9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Larson DE, Wang C, Koboldt DC, Sergeev YV, Fulton RS, Fulton LL, Fronick CC, Branham KE, Bragg-Gresham J, Jun G, Hu Y, Kang HM, Liu D, Othman M, Brooks M, Ratnapriya R, Boleda A, Grassmann F, von Strachwitz C, Olson LM, Buitendijk GH, Hofman A, van Duijn CM, Cipriani V, Moore AT, Shahid H, Jiang Y, Conley YP, Morgan DJ, Kim IK, Johnson MP, Cantsilieris S, Richardson AJ, Guymer RH, Luo H, Ouyang H, Licht C, Pluthero FG, Zhang MM, Zhang K, Baird PN, Blangero J, Klein ML, Farrer LA, DeAngelis MM, Weeks DE, Gorin MB, Yates JR, Klaver CC, Pericak-Vance MA, Haines JL, Weber BH, Wilson RK, Heckenlively JR, Chew EY, Stambolian D, Mardis ER, Swaroop A, Abecasis GR. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet. 2013;45:1375–1379. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann M, Kaspi A, Lazarus R, El-Osta A. Motif analysis in DNAse hypersensitivity regions uncovers distal cis elements associated with gene expression. Bioinformation. 2013;9:212–215. doi: 10.6026/97320630009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Dallman J, Wen R, Beecham G, Naj A, Farooq A, Kohli MA, Whitehead PL, Hulme W, Konidari I, Edwards YJ, Cai G, Peter I, Seo D, Buxbaum JD, Haines JL, Blanton S, Young J, Alfonso E, Vance JM, Lam BL, Pericak-Vance MA. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am J Hum Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]