Abstract
A new multimodal confocal microscope has been developed, which includes a parallel Partial Wave Spectroscopic (PWS) microscopy path. This combination of modalities allows molecular-specific sensing of nanoscale intracellular structure using fluorescent labels. Combining molecular specificity and sensitivity to nanoscale structure allows localization of nanostructural intracellular changes, which is critical for understanding the mechanisms of diseases such as cancer. To demonstrate the capabilities of this multimodal instrument, we imaged HeLa cells treated with valinomycin, a potassium ionophore that uncouples oxidative phosphorylation. Colocalization of fluorescence images of the nuclei (Hoechst 33342) and mitochondria (anti-mitochondria conjugated to Alexa Fluor 488) with PWS measurements allowed us to detect a significant decrease in nuclear nanoscale heterogeneity (Σ), while no significant change in Σ was observed at mitochondrial sites. In addition, application of the new multimodal imaging approach was demonstrated on human buccal samples prepared using a cancer screening protocol. These images demonstrate that nanoscale intracellular structure can be studied in healthy and diseased cells at molecular-specific sites.
Keywords: nanocytology, confocal microscopy, hyperspectral microscopy, chromatin, mitochondria
1. Introduction
Information about nanoscale structure has been difficult to obtain with far-field microscopy techniques due to its diffraction limited resolution. Fluorescence super-resolution modalities have overcome this resolution limit; however, these techniques can only examine a finite number of labeled structures at a time and cannot work in non-fluorescent samples. Partial Wave Spectroscopic (PWS) Microscopy is a novel, far-field, spectroscopic microscopy technique that quantitatively senses the nanoscale distribution of mass density within a biological sample. PWS does not require exogenous labels and thus works even in unstained samples. In addition, because PWS does not rely on fluorescence it is not specific to a single labeled molecule. The nanoscale sensitivity of the PWS microscope has been achieved through a spectral analysis technique that characterizes the interference from light scattered in the visible spectrum (500–700 nm) within each diffraction limited voxel of a widefield, reflected-light microscope image [1]. Scattered light collected by the microscope arises from variations in the refractive index of the sample, which directly correspond to changes in the local macromolecular density arising from the distribution of intracellular solids including proteins, lipids, DNA, and RNA. Due to this relationship between local density and refractive index, the spectrum of the scattered light contains nanoscale length information about the spatial variations in density. To quantify the heterogeneity of this nanoscale macro-molecular density arrangement, limited only by the signal-to-noise ratio, the standard deviation of the spectrum of the scattered light (Σ) captured by the PWS microscope is calculated. Utilizing this approach, it has been shown that with a 5% noise floor, PWS microscopy is sensitive to a range of length scales from ~22 nm to at least 200 nm depending on sample structure and thickness [1–4]. Techniques have been proposed with similar instrumentation and methods that have achieved comparable nanoscale sensitivity [5].
Due to this nanoscale sensitivity, PWS microscopy has proven to be an effective tool for detecting changes in intracellular nanostructure associated with disease. In particular, PWS microscopes have been extensively used for minimally-invasive detection of malignant transformation by capturing nano-architectural changes that occur distal to the primary tumor site. This detection largely relies on the field effect of carcinogenesis, a phenomena inherent in tumor formation, which is manifested by the transformation of structure and function of cells throughout an affected organ [6]. Previous clinical investigations with PWS microscopy have detected significantly higher intracellular Σ from histologically normal sites in cancer patients compared with controls in multiple organ systems [3, 7–9]. In particular, the increased heterogeneity of the spatial density arrangement of nuclear chromatin appears to be the most significant change in early and field carcinogenesis that is observed by PWS [10]. However, the question remains as to which other underlying cellular nano-structures are transformed during field carcinogenesis that contribute to the observed differences in the heterogeneity of macromolecular density between healthy and malignant tissues. Previous studies of field carcinogenesis in colon cancer demonstrated that nuclear transformation as measured by PWS is in part linked to epigenetic transformation of chromatin acetylation [11]. Finally, investigation into early-stage tumorigenic alterations using transmission electron microscopy showed changes in the nanoscopic organization of chromatin, a marker that can be detected with PWS [12].
While these previous studies have suggested chromatin condensation and cytoskeleton organization as potential mechanisms for tumorigenic transformation and the origin of cellular changes observed by PWS microscopy, the majority of the biological assays used in these experiments were performed in parallel with PWS. Although multimodality population measurements can be done in parallel to PWS with existing instruments, it is impossible with these approaches to directly determine at an individual cell level in which specific organelles or molecular structures changes in the nanoscale mass distribution are occurring, or the causative molecular events. Direct colocalization comparisons and spatially matched quantitative measures cannot be done on a cell-by-cell basis using these parallel techniques. Therefore, to directly correlate biological processes and molecules with the nanoscale organization of macromolecular structure, a new multimodal PWS instrument integrated with a commercial white light confocal microscope was constructed. This new microscope combines the nano-scale sensitivity of PWS with the molecular specificity of confocal microscopy, allowing for consecutive measurements between PWS and processes targeted by immunofluorescence, including DNA/RNA (through fluorescence in situ hybridization, FISH), protein markers, and organelle structures. The multi-modal instrumentation described here, combining molecular specificity with PWS, is a critical research tool to enable studies that co-localize PWS nanoscale structural changes with specific molecular compartments and structures on a cell-by-cell basis.
With the experiments contained in this work we illustrate how this new multimodal instrumentation provides a means to study the biological interplay of functional processes and structural organization, highlighting the study of carcinogenesis as one potential application. The organization of organelles (e.g. chromatin, endoplasmic reticulum, golgi, and mitochondria) and their subsequent structural and functional transformation during carcinogenesis have implications in the development of diagnostic and therapeutic tools. As examples, the nucleus and mitochondria are examined here due to their observed structural and functional changes during tumor formation. Due to the critical role mitochondria serve in cellular metabolism and signaling, alterations in structure and function is implicated in many diseases [13]. Likewise, nuclear dysregulation, in particular epigenetic and genetic transformation, has been shown as a universal requirement for tumorigenesis. Nuclear structure is largely determined by the organization of chromatin, which consists of DNA, histones, and numerous other proteins directly attached to DNA. Many studies have demonstrated that the hierarchical organization of chromatin plays a critical role in the regulation of transcription, replication, DNA repair, and chromosomal stability [14]. Finally, mitochondrial dysregulation has been shown to regulate growth and arrest of the cell through secondary messengers and chromatin remodeling enzymes. As such, there is a critical need for tools, such as the multimodal PWS instrument described herein, to study multiple organelles in parallel.
One main convergence point between mitochondria and chromatin structure is their dependence on proper intracellular ionic conditions. Studies have shown that altering potassium homeostasis influences both the structure and function of mitochondria and chromatin [15]. Using valinomycin, a potassium ionophore that uncouples oxidative phosphorylation and causes potassium depletion, previous work has found that the drug induced slight changes in chromatin structure along with mitochondrial swelling, particularly at longer incubation times (3 and 6 hrs) [15]. In this experiment, we used this model of valinomycin treatment to examine nano-scale differences in organelle structures using the new multimodal PWS system. The methods and experimental demonstration described here may serve as a platform for a diverse range of applications, including multiple labeling of key cellular structures in car cinogenesis (e.g. mitochondria), as well as performing cancer cytogenetics and PWS simultaneously on clinical samples.
2. Materials and methods
2.1 Instrument description
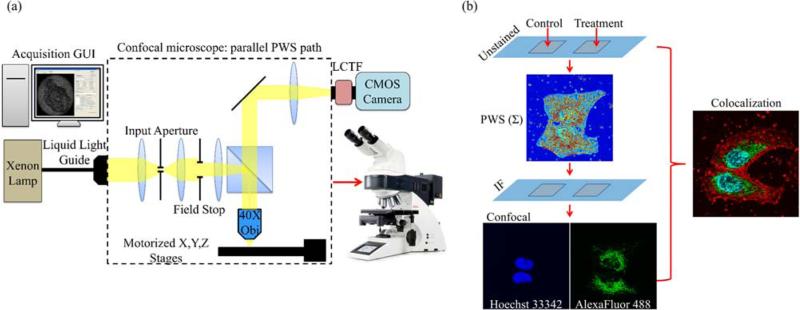
We have integrated a PWS instrument into a Leica TCS SP5 (Leica, Wetzlar, Germany) confocal microscope to simultaneously examine fluorescent molecular markers and nanoscale structure as measured by Σ in individual cells. This particular confocal system provides several unique capabilities that make it well suited for co-localization analysis with PWS data. The system includes a white light supercontinuum laser that allows the selection of any excitation wavelength between 470 nm and 670 nm for confocal experiments. It also utilizes an acousto-optic beam splitter and a prism in the collection path to achieve tunable, spectrally-filtered excitation and collection. This unique combination of hardware enables the use of any combination of fluorophores for colocalization analysis with PWS, including those with similar emission spectra.
PWS instrumentation was built into the Epi-illumination path parallel to the confocal path in an up-right version of the SP5 confocal. Through this path, broadband light from a Xenon lamp was used to illuminate the sample through a 40X, 0.6 numerical aperture (NA) objective lens (HCX FLUOTAR 40X/0.6 CORR; Leica, Wetzlar, Germany). A custom illumination aperture was added through a filter slot in the illumination path to reduce the illumination numerical aperture to approximately 0.15. Raw PWS data was collected by acquiring a spectrum of the backscattered light from 500–700 nm in 1 nm steps using a liquid crystal tunable filter (Varispec VIS, bandwidth 7 nm, PerkinElmer, Waltham, MA) and a CMOS camera (Hamamatsu Orca Flash 2.8, Hamamatsu City, Japan) connected to the top camera port of the microscope. A custom software interface was developed in MATLAB (MathWorks, Natick, MA) to automate collection of the PWS data. PWS measurements took approximately 30 seconds per cell. The microscope could be switched between the two operating modes, PWS and confocal, for back-to-back measurements as needed. Figure 1 schematically illustrates the addition of the PWS hardware to the Leica SP5 confocal and shows the workflow to acquire PWS and confocal fluorescence images from the same cells. Statistical analysis of the spectra at each pixel in the PWS raw data cube (x, y, λ) yields an image of the spatial distribution of Σ, which indicates the local heterogeneity of nanoscale mass density at each diffraction limited voxel in the sample.
Figure 1.
(a) Schematic of the PWS addition to the confocal microscope. A custom software acquisition GUI controls data acquisition through the parallel Epi-illumination port in the confocal microscope. Light from a Xenon lamp is incident on the sample through a custom input aperture to reduce the illumination numerical aperture. Backscattered light is collected with a 40X objective and spectrally filtered by the LCTF before the image is acquired at the camera. Typically images are acquired in the visible spectrum, 500–700 nm with 1nm steps. (b) Workflow for combined usage of the confocal and PWS acquisition paths. A multi-chamber slide was used for control and treatment cell groups. PWS data was acquired on the unstained sample and analysis of the spectra generated images showing the distribution of nanoscale structure in the sample. After PWS measurements, the slide is removed and the fluorescent labels are added. Confocal images are the acquired using stored coordinates from the PWS experiment. Colocalization of the PWS data with confocal fluorescence allows molecular-specific study of the cellular nanostructure.
2.2 Preparation of cancer cell lines
The human cervical cancer HeLa cells were grown in RPMI-1640 medium (ATCC, Manassas, VA), supplemented with 10% fetal bovine serum + L-glutamine in a 5% CO2 environment at 37 °C. For the experiments, ~25,000 cells were plated in each chamber of the sterile glass chamber slides (LabTek, Waltham, MA) and incubated overnight under standard conditions. Two of the four chambers of HeLa cells were treated with 1 μM valinomycin for 2 hrs. To fix the cells, the chamber slides were washed with 1X phosphate buffered saline (PBS) and then fixed with 95% ethanol. Chamber slides, thus prepared, were stored at 4 °C keeping cells submerged in 70% ethanol. PWS measurements were acquired prior to immunofluorescence staining. Following PWS measurements, slides were rinsed twice in PBS and then cells were permeabilized using 0.1% Triton X-100 in PBS for 20 min at room temperature. Blocking was carried out using 1% bovine serum albumin in PBS for 20 min. Cells were incubated with anti-mitochondria antibody (AbCam, Cambridge, UK) overnight at 4 °C. The next day, slides were incubated with goat anti-mouse conjugated to Alexa-Fluor488 for 2 hrs at room temperature. Chromatin counterstaining was carried out using Hoechst 33342 (Molecular Probes, Waltham, MA) for 15 min. Finally, the slides were mounted in ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA) and a glass coverslip was placed over the sample.
2.3 Preparation of buccal cells
The buccal cell samples were acquired via a cheek brushing using a Cytobrush (Cooper Surgical, Trumbull, CT). Human samples were collected in accordance with the Institutional Review Board at Northwestern University. Collected cells were deposited onto a glass slide and fixed in 95% ethanol for 15 min. PWS measurements were then acquired before immunofluorescence staining. The immunofluorescence staining protocol was the same as was used for the HeLa cells (Section 2.2), except that a single co-incubation step of phalloidin conjugated to AlexaFluor488 (Life Technologies, Carlsbad, California) and Hoechst 33342 was performed for 20 min at room temperature.
2.4 PWS and confocal image collection
For PWS acquisition, 15–25 non-overlapping cells were randomly selected from each chamber (two of untreated controls and two of valinomycin-treated). Coordinates of each cell were recorded and a reference location on the slide was used as the coordinate origin to aid in relocation of the same cells. At each cell position, an image cube was acquired from with spectra ranging from 500–700 nm in 1 nm steps. Following PWS measurements, cells were processed for immunofluorescence staining. Specimens were then imaged using the confocal arm (Leica SP5 with Leica Application Suite) of the multimodal PWS instrument. The same cells that had been measured using PWS were relocated using the reference point and coordinates. Individual cells were zoomed in on using a 63X oil immersion objective (HCX PL APO 63X/1.4-0.60; Leica, Wetzlar, Germany) and a multi-channel confocal fluorescence image was acquired at the focal plane. After both PWS and confocal measurements, a total of 26 controls and 25 valinomycin treated cells had been acquired from both modalities for colocalization analysis. While in this instance a high magnification, high NA oil immersion objective was chosen to optimize confocal image resolution, it would be equally valid to perform experiments using the same objective for both techniques to simplify the method and perform high-throughput colocalization experiments on many samples for quantitative data analysis. In addition, PWS measurements can be conducted on samples that are already fixed and stained as typically absorption and emission spectra associated with fluorescence are weak enough to not alter the back-scattered spectrum collected during PWS measurements – although this should be tested with any new dye. For this experiment, the PWS measurements were performed prior to immunofluorescence staining to minimize any possible effects due to the sample processing associated with the staining protocols on nanoscale structure.
2.5 Image processing and colocalization
To achieve colocalization of the confocal fluorescence images and the PWS data, a custom software interface was developed in MATLAB. This software was used to account for differences in the imaging hardware between the two parallel microscope paths; the confocal fluorescence images were re-scaled, rotated, and translated to match the images from the PWS path. Corrected versions of each confocal channel were saved as tiffs and used for all co-localization analysis. Multi-channel fluorescence images were combined using ImageJ [16]. The nanoscale-sensitive PWS images (Σ) were calculated as the standard deviation of the backscattered spectrum at each pixel normalized by a glass reflection measurement using code developed in MATLAB. Overlay images of the organelle fluorescence with contrast provided by Σ were generated in Mathematica (Wolfram, Champaign, IL). For statistical analysis, each fluorescence channel was evaluated independently for staining quality based on specificity and presence of stain in the desired cellular structure. Cells that were not specifically stained for the structure of interest were excluded from statistical analysis.
3. Results and discussion
3.1 Demonstration of multimodal imaging and colocalization
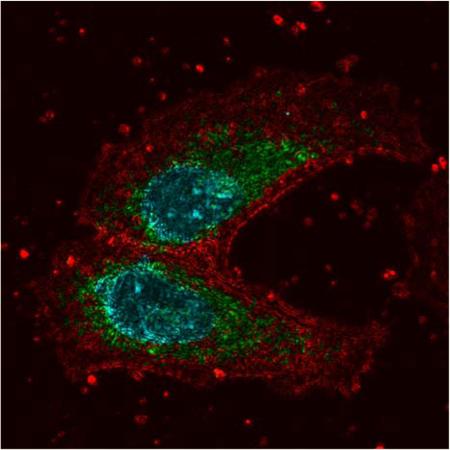
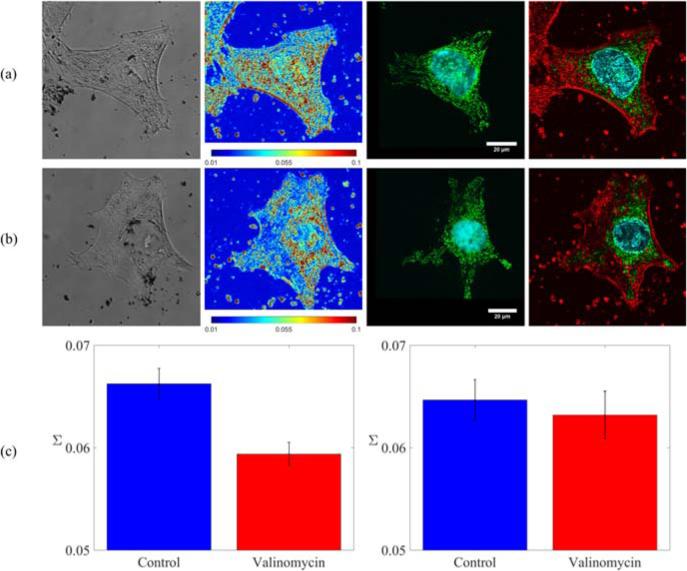
The multimodal capacity of the microscope was demonstrated by imaging the distribution of intracellular nanostructure with PWS and overlaying those results with molecular-specific images from the confocal path. Colocalization of these images allowed experiments to be conducted that observed changes in the nanoscale structural distribution at molecular-specific sites in the cell. Two organelles of particular interest to correlate with PWS are the nucleus and mitochondria, given their observed structural and functional changes during tumor formation. Thus, these structures were examined following valinomycin treatment to examine structural changes in response to changes in the intracellular ionic environment. Figure 2a–b shows representative images from each of the parallel arms of the multimodal microscope for control and valinomycin-treated HeLa cells. Label-free PWS data, as well as confocal images of Hoechst 33342 labeled DNA and labeled mitochondria (anti-mitochondria antibody, Alexa-Fluor488) are shown. Overlays of the Σ image with the confocal channels, Figure 2a–b column 4, allow quantitative investigation of the nanoscale distribution of structure at organelle-specific sites, in this case chromatin (Hoechst 33342) and mitochondria (anti-mitochondria antibody, AlexaFluor488). In these overlay images, intensity corresponds to the heterogeneity of macromolecular structure (Σ) with green regions marking the mitochondrial sites, cyan the chromatin, and red the regions that corresponded to Σ only.
Figure 2.
(a) Representative PWS and confocal images of a control HeLa cell. Left to right: (1) grayscale reflection image, (2) PWS image of the distribution of the heterogeneity of nanoscale macromolecular density (Σ, scaled from 0.01 to 0.1), (3) confocal fluorescence where DNA was labeled with Hoechst 33342 and mitochondria was labeled with anti-mitochondria antibody conjugated to AlexaFluor488 (4) overlay of the confocal fluorescence and PWS images. Intensity was set by the PWS (Σ) image with regions corresponding to mitochondria and DNA colored green and cyan respectively. (b) Representative PWS and confocal images for a valinomycin treated cell of the same types as in (a). (c) Left to right: (1) heterogeneity of nuclear nanostructure (Σ) for control and valinomycin treated cells and (2) heterogeneity of nanoscale structure (Σ) from mitochondrial regions for control and valinomycin treated cells. A significant decrease was observed in mean nuclear Σ for valinomycin treated cells, p < 0.001. No significant difference was observed between control and valinomycin treated cells in mean Σ at mitochondrial sites.
3.2 Nanoscale structural changes observed at organelle-specific sites
To demonstrate the ability to quantitatively sense nanoscale structural changes at organelle-specific sites, we imaged HeLa cells that had undergone treatment with 1 μM valinomycin for 2 hrs. Valinomycin treatment has previously been shown to induce slight changes in chromatin structure along with mitochondrial swelling, particularly at longer incubation times (3 and 6 hrs) [15]. In this experiment, no visible change in the microstructure was observable in the brightfield reflection images. However, from the confocal images, there was an observed qualitative change in the size and shape of the mitochondria following valinomycin treatment, as they became rounder and less elongated. At the nanoscale, we measured a significant decrease, Figure 2c, in the heterogeneity of nuclear macromolecular density (mean nuclear Σ) for cells treated with valinomycin compared to controls (p < 0.001). This decrease in Σ can be visually observed in the nucleus of the representative cell images in columns 2 and 4 of Figure 2a–b. In contrast, no significant difference in nanostructural heterogeneity (Σ) was observed in mitochondrial regions segmented from the fluorescence images, despite the visible microstructural changes observed via fluorescence. Previous experiments using fluorescence microscopy and electron microscopy have found that a 16-hour treatment with valinomycin only causes outer membrane fusion and no changes to the inner membrane [17]. As a result, it is possible that after only a 2-hour treatment with valinomycin there is a change in mitochondrial morphology visible with confocal due to outer membrane fusion, but that nanoscale structure as measured by PWS remains relatively unchanged due to a lack of inner membrane fusion.
3.3 Application to clinical data (buccal cells)
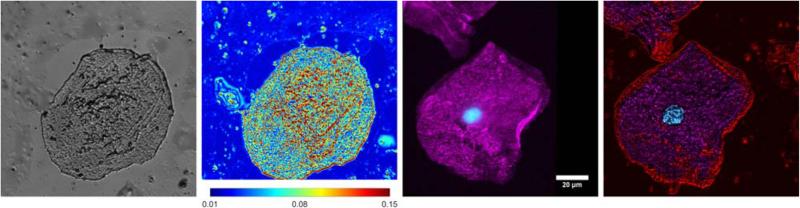
As a secondary demonstration of multimodal molecular imaging and nanoscale sensing, we next imaged human buccal cells with both PWS and confocal microscopy. PWS has previously shown clinical utility as a screening tool for cancer, where an increase in the heterogeneity of nanoscale mass density (Σ) has been observed in the field effect of carcinogenesis in multiple organ sites [7–9, 18, 19]. In particular, previous experiments have shown that lung cancer can be detected through this increase in Σ from buccal swabs using PWS instrumentation [19]. The biological mechanisms and origins of the changes in nanoscale intracellular structure that occur during carcinogenesis resulting in a measurable increase in Σ are not well characterized. In Figure 3, we demonstrate that through colocalization of confocal images with PWS measurements of Σ, the molecular origins of the nanoscale structural changes detected by PWS can be studied in samples used in clinical cancer screening protocols. In this case, buccal cells were prepared using the clinical sample protocol and PWS data was acquired. The chromatin was then labeled with Hoechst 33342 and actin was labeled with phalloidin conjugated to Alexa-Fluor488. These structural markers were chosen for these cells due to the low viability of buccal cells and the fact that structural changes such as DNA damage have been previously observed as a consequence of carcinogenesis [20, 21]. In Figure 3 we show a representative human buccal cell imaged with both PWS and confocal modalities.
Figure 3.
Representative PWS and confocal images of a buccal cell. Left to right: (1) grayscale reflection image, (2) PWS image of the distribution of the heterogeneity of nanoscale macromolecular density (Σ, scaled from 0.01 to 0.15), (3) confocal fluorescence where DNA was labeled with Hoescht 33342 and actin was labeled with phalloidin conjugated to Alexa-Fluor488, (4) overlay of the confocal fluorescence and PWS images. Intensity was set by the PWS (Σ) image with regions corresponding to actin and DNA colored cyan and magenta respectively. The red color corresponds to Σ only.
3.4 Experimental implications
As the results presented here suggest, this multimodal instrumentation combining molecular-specific confocal microscopy and nanoscale-sensitive PWS microscopy is capable of identifying the molecular origins of changes in nanoscale biological structure. As compared to previous experiments using PWS microscopy, it would not have been possible to localize changes in the nanoscale structure to particular organelles and molecules. Instead, it was only possible to detect nanoscale changes in structure across an entire cell population without molecular specificity. However, here we have shown that just by targeting two organelles, nucleus and mitochondria, the nanoscale structural changes can be studied within individual cellular compartments. Further studies can be performed on any cellular organelle, molecular fingerprint, or structure of interest provided it can be fluorescently labeled. The implications of this method are significant, as it allows for changes in cellular nanostructure as a result of disease within particular cellular compartments to be studied. In addition, this method can be used to develop therapies and to determine their effectiveness at the cellular level by identifying if disease state changes in the cellular nanostructure can be repaired or prevented in response to therapy. Future work with this method can encompass fluorescence in situ hybridization (FISH) of key genes in clinical cancer samples. Additional applications include extending the method to live cell imaging or super-resolution techniques such as stochastic optical reconstruction microscopy, (STORM), structured illumination microscopy (SIM), or stimulated emission depletion microscopy (STED), to significantly improve the lateral resolution of fluorescence images, extending the visual co-localization from the microscale to the nanoscale.
4. Conclusions
In this work, we developed protocols and analysis tools to directly correlate molecular-specific biological markers with the nanoscale heterogeneity of macromolecular density. With this multimodal approach comprised of PWS microscopy and confocal fluorescence, we have shown that the molecular origins of nanoscale structural changes sensed by PWS can be localized to particular organelles or molecular components of the cell. We have shown that PWS detects nanoscale structural changes that occur in the nucleus of HeLa as a result of valinomycin treatment, while similar quantitative differences were not detected in the mitochondria, despite visible changes in the mitochondrial microstructure. Furthermore, colocalization of nanostructural measurements and molecular-specific fluorescence was established in samples prepared using a clinical protocol. This demonstrates that the techniques and analysis described here can be used to study the origins of the increase in the heterogeneity of macromolecular density associated with early carcinogenic events in clinical samples. These results exhibit the innovative ability of correlating biologically meaningful markers with the nanoscale density distribution as measured by PWS.
Supplementary Material
Acknowledgements
The results presented here are based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant DGE-0824162, the National Institutes of Health under Grants: U54CA193419, R01CA155284, R01CA200064, and R01EB016983. Drs. Subramanian and Backman are co-founders and/or shareholders in Nanocytomics LLC.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's website.
Author biographies Please see Supporting Information online.
References
- 1.Cherkezyan L, Capoglu I, Subramanian H, Rogers JD, Damania D, Taflove A, Backman V. Phys Rev Lett. 2013;111:033903. doi: 10.1103/PhysRevLett.111.033903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherkezyan L, Subramanian H, Backman V. Opt Lett. 2014;39:4290–4293. doi: 10.1364/OL.39.004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramanian H, Pradhan P, Liu Y, Capoglu IR, Li X, Rogers JD, Heifetz A, Kunte D, Roy HK, Taflove A, Backman V. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20118–20123. doi: 10.1073/pnas.0804723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian H, Pradhan P, Liu Y, Capoglu IR, Rogers JD, Roy HK, Brand RE, Backman V. Opt Lett. 2009;34:518–520. doi: 10.1364/ol.34.000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Bista RK, Khalbuss WE, Qiu W, Uttam S, Staton K, Zhang L, Brentnall TA, Brand RE, Liu Y. J Biomed Opt. 2010;15:066028. doi: 10.1117/1.3523618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slaughter DP, Southwick HW, Smejkal W. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Damania D, Roy HK, Kunte D, Hurteau JA, Subramanian H, Cherkezyan L, Krosnjar N, Shah M, Backman V. Int. J. Cancer. 2013;133:1143–1152. doi: 10.1002/ijc.28122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy HK, Brendler CB, Subramanian H, Zhang D, Maneval C, Chandler J, Bowen L, Kaul KL, Helfand BT, Wang CH, Quinn M, Petke wicz J, Paterakos M, Backman V. PLoS ONE. 2015;10:e0115999. doi: 10.1371/journal.pone.0115999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backman V, Roy HK. J. Cancer. 2013;4:251–261. doi: 10.7150/jca.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damania D, Subramanian H, Tiwari AK, Stypula Y, Kunte D, Pradhan P, Roy HK, Backman V. Biophys J. 2010;99:989–996. doi: 10.1016/j.bpj.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stypula-Cyrus Y, Damania D, Kunte DP, Dela Cruz M, Subramanian H, Roy HK, Backman V. PLoS ONE. 2013;8:e64600. doi: 10.1371/journal.pone.0064600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherkezyan L, Stypula-Cyrus Y, Subramanian H, White C, Dela Cruz M, Wali RK, Goldberg MJ, Bianchi LK, Roy HK, Backman V. BMC Cancer. 2014;14:189. doi: 10.1186/1471-2407-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunnari J, Suomalainen A. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, Dechassa ML, Tremethick DJ. Nature reviews Molecular cell biology. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein B, Wörndl K, Lütz-Meindl U, Kerschbaum HH. Apoptosis. 2011;16:1101–1117. doi: 10.1007/s10495-011-0642-9. [DOI] [PubMed] [Google Scholar]
- 16.Schneider CA, Rasband WS, Eliceiri KW. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombes A, Rojo M. Embo Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian H, Roy HK, Pradhan P, Goldberg MJ, Muldoon J, Brand RE, Sturgis C, Hensing T, Ray D, Bogojevic A, Mohammed J, Chang JS, Backman V. Cancer Research. 2009;69:5357–5363. doi: 10.1158/0008-5472.CAN-08-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy HK, Subramanian H, Damania D, Hensing TA, Rom WN, Pass HI, Ray D, Rogers JD, Bogojevic A, Shah M, Kuzniar T, Pradhan P, Backman V. Cancer Research. 2010;70:7748–7754. doi: 10.1158/0008-5472.CAN-10-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endler G, Greinix H, Winkler K, Mitterbauer G, Mannhalter C. Bone Marrow Transpl. 1999;24:95–98. doi: 10.1038/sj.bmt.1701815. [DOI] [PubMed] [Google Scholar]
- 21.Osswald K, Mittas A, Glei M, Pool-Zobel BL. Mutat Res-Rev Mutat. 2003;544:321–329. doi: 10.1016/j.mrrev.2003.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.