Abstract
Laboratory mice (Mus musculus) have become the major model species for inner ear research. The major uses of mice include gene discovery, characterization, and confirmation. Every application of mice is founded on assumptions about what mice represent and how the information gained may be generalized. A host of successes support the continued use of mice to understand hearing and balance. Depending on the research question, however, some mouse models and research designs will be more appropriate than others. Here, we recount some of the history and successes of the use of mice in hearing and vestibular studies and offer guidelines to those considering how to apply mouse models.
Electronic supplementary material
The online version of this article (doi:10.1007/s10162-016-0589-1) contains supplementary material, which is available to authorized users.
Keywords: mice, inbred, outbred, recombinant inbred, knockout, cochlea, deafness, presbycusis, age-related hearing loss, noise-induced hearing loss, genetic hearing loss, hair cells, auditory neurons, lateral wall, stria vascularis, vestibular, vestibular testing, saccule, utricle, maculae, semicircular canal, ampullae
Introduction
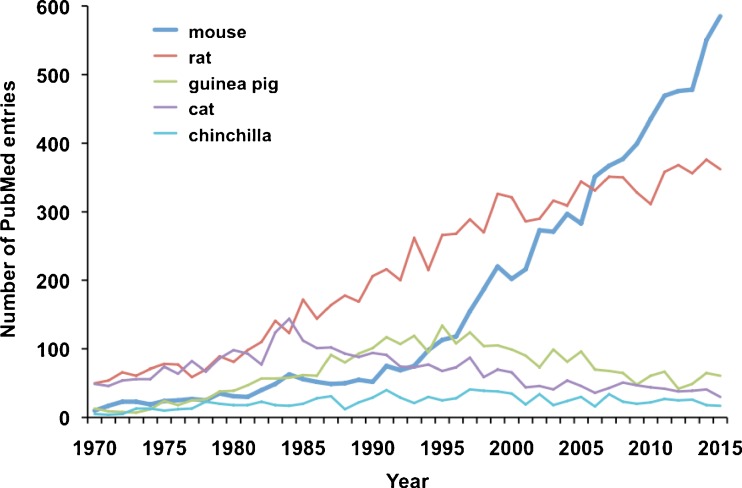
Thirty years ago, most of what was known about hearing and balance in mammals was inferred from studies of cats, rats, and guinea pigs (Fig. 1). By about 2008, the scope of research by using the laboratory mouse (or house mouse, Mus musculus) surpassed all other models and by 2015 exceeded all other models combined. Mice have come a long way from a niche—and even derided—model to predominance. Their rise is founded on the innovation that is the inbred strain and the near elimination of uncontrolled genetic variance. Phenotypic differences among inbred strains of mice first were studied primarily as a way of establishing the genetic foundations of hearing and balance. But, developments of the 1990s in manipulation of DNA exhibited a steep trajectory, carrying mice along as the mammalian inner ear model. The molecular wave has since made every mechanistic question an inherently “genetic” question and swept mice into a majority of laboratories. Nevertheless, mice occupy an ecological niche different from ours and have been shaped by evolutionary pressures unlike our own. They eat a different type of diet and have a different life span and metabolic rate. Any inbred mouse strain is like one highly consanguineous population—or one person—for comparative purposes. Consequently, any commercial mouse model we may apply represents discarding of genetic diversity. To be sure, mice have been a singularly successful model in our understanding of hearing and balance and will continue to play a vital role. However, all applications of mice are based on assumptions that we may be unaware of or take for granted. The goal of this review is not to generate an exhaustive list of potentially useful mouse models. Rather, we state some assumptions relevant to particular research contexts and offer guidelines for effective use of mice. In the interest of brevity, there appear statements about what “mice” do or do not do, while one of our major points is that this depends dramatically on genetic background. In the application of mice to inner ear disease, the question is not whether mice effectively model human pathology, but rather which mice may model the pathology of which humans?
FIG. 1.
The striking increase in the use of the mouse in auditory and vestibular research. Plot compares mouse, rat, guinea pig, cat, and chinchilla studies retrieved from PubMed with the query “species” AND (hearing OR deafness OR auditory OR vestibular) by year.
Brief History of Mice in Biomedical Research
Mice were well situated to exploit human agriculture from its first appearance in central Asia about 10,000 years ago (Silver 1995). As soon as humans began to cultivate and store grain, mice recognized an easily raided, endless food supply. The word “mus” is Sanskrit for “thief,” reflecting early human opinions of mice. Indeed, domestic cats owe their present cushy status to their initial role as mousers. The development of “fancy mice” as pets in Asia and Japan in the 1800s broadened our relationship with mice and gave rise to selective breeding practices. Fancy mice were bred for unusual coat colors and patterns. These sometimes coincided with abnormal hearing and balance that imparted characteristic behaviors (Yerkes 1904). Like dog breeds, fancy mice bearing particular coat patterns were only partially inbred and represented random mixes of the four major recognized wild subspecies of M. musculus distributed across Asia and Europe (musculus, domesticus, castaneus, and bactrianus) (see Figs. 2.2 and 2.3 in Silver 1995). Breeding mice as a hobby spread from Asia to Europe and the USA. A pivotal step in the promotion of mice from pet shop to research tool in the USA occurred around 1900 when Abbie Lathrop, a mouse hobbyist and amateur scientist, set up a shop near the Bussey Institute, which was run by Harvard geneticist William Castle. Castle’s initial research stocks, and many later inbred strains, were derived from mice bred by Lathrop. It was through Castle’s influence, along with student Clarence Little, that the need for genetically homogeneous inbred strains was first recognized and put into practice. Little established the first inbred strain (DBA) starting in 1909 and co-founded The Jackson Laboratory (JAX) in 1929.
Application to Genetics
The rediscovery of Mendel’s work, also around 1900, underscored the inheritance of discrete phenotypes according to surprisingly simple principles. The genius of Mendel’s experiments was that he used highly inbred true-breeding lines of pea plants, which minimized confounds posed by uncontrolled genetic heterogeneity. However, critics seized upon the seemingly more “blended” inheritance of traits in animals as proof that the underlying principles were different. The unrecognized problem, of course, was that these traits were typically polygenic and polyallelic (determined by multiple versions of the gene). What was needed was the animal equivalent of Mendel’s true-breeding peas. Mice, in which inbreeding had been taken to a level unmatched in any other mammal, were ideal for that purpose. Commercial research models such as guinea pigs, chinchillas, and gerbils are outbred, deliberately bred so that any two animals will differ genetically. By virtue of at least 20 generations of brother-sister mating, any two mice of the same inbred strain will be >99 % genetically identical and homozygous at essentially all autosomal loci. The first clear extension of Mendel’s principles to mammals (circa 1905) was based on the inheritance of distinct coat colors and patterns in inbred mice (Silver 1995). Now, more than 20 Nobel prizes have been awarded for medical discoveries that could not have been made without inbred mouse strains, including the genetics of graft rejection, immune tolerance, and acquired immunity (Festing 2010).
Application to Hearing and Vestibular Function
Due to the random fixation of recessive alleles, some fancy mice and inbred strains derived from them exhibited behavioral signs of balance disorders (circling, head tilting and tossing), and some of these were associated with hearing deficits. Before evoked response methods became common in the 1960s, hearing assessment was restricted to startle responses. The genetics of hearing loss gained momentum from post-World War II interest in the effects of ionizing radiation on genetic abnormalities. Inbred mice were frequently used models for radiation experiments, which yielded a host of useful phenotypes. Since identification of the underlying genes would have to await technological advances, the initial task was simply to determine by complementation how many separate genes were likely involved. Among the first to distinguish models and to characterize their anatomic and physiologic defects were M.S. Deol and H. Grüneberg (see references in Steel et al. 1983; Steel 1995; Johnson et al. 2001). Nevertheless, mouse work remained a small niche in hearing research. Prior to 1994, mice were the objects of just a handful of research papers in hearing per year, while basic physiological principles of hearing were derived by using larger and more easily manipulated animal models. The 1970s featured rapid growth of work in audiogenic seizures in mice (e.g., Saunders et al. 1972; Chen et al. 1973; Saunders 1974; Chen et al. 1976; Chen 1978), although this work has gone surprisingly undeveloped. By the early 1980s, other complex traits like age-associated hearing loss or noise vulnerability emerged as a theme of mouse research in work by Ehret, Henry, Willott, and others (Ehret 1974; Henry 1982a; Shnerson and Pujol 1982; Henry 1983a; Henry 1984; Hunter and Willott 1987). The primary assertion from such studies was that mouse model differences could be inferred to be genetic, although the identity of the underlying genes was still decades away. Differences between C57BL/6 (B6) and CBA-related strains were first noted and put to work very effectively. For example, because B6 mice show early progressive cochlear degeneration, while CBAs do not, one could study the effects of peripheral hearing loss on central auditory anatomy and function even without knowing the specific genes involved (Willott 1986; Hunter and Willott 1987; Willott and Bross 1990). Tools for hearing assessment in mice were refined during this time, principally by K.R. Henry (Henry and Haythorn 1978; Henry and Lepkowski 1978; Henry 1979; Henry and Chole 1979; Henry 1985; Henry et al. 1985; Henry 1989) in the auditory periphery and by J.F. Willott (Willott and Shnerson 1978; Shnerson and Willott 1980; Willott et al. 1982; Kulig and Williot 1984; Willott 1984; Willott et al. 1988) in the auditory CNS. A particularly significant “trilogy” of books originating with Willott bears mention. The first (Auditory Psychobiology of the Mouse, 1983), edited and co-authored by Willott, established mice as a resource for hearing research and assembled much essential information under one cover. The second (Aging and the Auditory System: Anatomy, Physiology, and Psychophysics, 1991), authored by Willott, focused on the value of mouse strain comparisons for extracting principles about hearing in aging. The third book (Handbook of Mouse Auditory Research: From Behavior to Molecular Biology, 2001) appeared after the mouse revolution was in full gear, expanding and updating many of the themes and methods from the first book.
Behavioral studies in “dancing mice” were published in the late 1800s and early 1900s (see Yerkes 1904). Imbalance behaviors may be due to deficits in visual, vestibular, or somatic sensory systems or at any number of levels along the neuraxis. The morphological studies by Yerkes were inconclusive as to whether inner ear abnormalities, brain defects, or both accounted for the observed behaviors in dancing mice. Later studies in selected mouse mutants (e.g., Deol and Lane 1966; Anniko et al. 1980) would provide supporting evidence that inner ear variation could account for observed imbalance behaviors. Although studies continue to be published describing behavior or morphology as the hallmark of inner ear vestibular dysfunction, neither feature provides definitive evidence regarding the functional status of the semicircular canals or gravity receptor organs. To that end, tools for assessing inner ear vestibular function in mice have been validated. Eye movement recordings of the vestibulo-ocular reflex (VOR) infer canal function (van Stahl et al. 2000; Iwashita et al. 2001; van Alphen et al. 2001), and vestibular evoked potentials (VsEPs) elicited by linear acceleration stimuli directly test gravity receptor function (Jones et al. 1999; Jones and Jones 1999; Jones et al. 2002). More recently, vestibular evoked myogenic potentials (VEMPs) have been claimed to infer gravity receptor function in mice, although this measure has yet to be widely used (Sheykholeslami et al. 2009). Just as hearing assessment tools have contributed to an ever-expanding understanding of genetic hearing loss, development and application of functional vestibular assessments will be critical for elucidating genetic vestibular impairment.
Arrival of Molecular Methods
Around 1994, papers dealing with mouse hearing and balance showed a discernible up-tick, reflecting the rapid arrival of molecular techniques (Fig. 1). 1n 1992, Friedman and Ryan had published an influential review on transgenic mice (Friedman and Ryan 1992), and the first characterizations began to appear (Rauch 1992). It was around this time that the first mouse orthologs of human deafness genes were identified (Birkenmeier et al. 1989; Steel and Smith 1992; Hughes et al. 1994; Tassabehji et al. 1994; Battinelli et al. 1996) and mouse deafness genes were first tied to proteins critical for hearing (Avraham et al. 1995; Gibson et al. 1995). Prior to 1999, most mouse studies applied “forward genetics,” whereby useful defects are identified in existing mouse stocks. These were initially characterized anatomically and physiologically, followed by mapping and identification of candidate genes. Parallel mapping endeavors in mice and humans facilitated discovery of deafness genes in a leap-frog manner.
Around 1999, “reverse genetics” notably accelerated. In reverse genetics, one makes an educated guess about a gene and selectively modifies it, then determines the effect on form and function. Two basic manipulations became available: insertion of up to 1000 extra copies of a gene at a random location in the genome (the technical meaning of “transgenic”) and targeted deletion of a gene (knockout (KO)) (Crawley 2000; Jackson and Abbott 2000). The initial goal was stable, constitutive (every cell in the body), assimilation of exogenous DNA, which required transfection of embryonic stem (ES) cells or fertilized oocytes. Stem cell transfection worked best in 129 related strains, while oocyte injection worked well in B6 mice. Both B6 and many 129 substrains show progressive hearing loss and are problematic for hearing research. The 129 strains also came in a confusing variety of substrains, some of which were determined to be genetically contaminated (Simpson et al. 1997). Transfected 129 ES cells were typically microinjected into mouse blastulas from strains with a different coat color, so that chimeric coat-colored mice then represented the mice with the transgene. Germline positive animals were then determined by a further cross, often to B6 or CD-1 mice, which also feature progressive hearing loss. The result has been that, up to the present, most transgenic or KO models of interest have forced investigators to deal with potentially confounding polygenic background hearing loss. To minimize this problem, one could test young mice and hope for a robust phenotype that stood out against the background hearing loss. Alternatively, one could move the engineered allele to a “good hearing” background through at least 10 serial backcrosses (that is, serially mating progeny back to the desired strain), selecting carriers of the transgene as breeders in each generation. This cumbersome process of serial backcrosses to a desired strain removes the alleles of the undesired strain at an average rate of 50 % per generation. The goal is to obtain mice that are “congenic” to the desired strain—identical in all respects except for the addition of the transgene. This process might require 2 years but can be shortened to <5 generations through “speed congenic” methods, in which mice carrying the fewest alleles from the undesired background are selected as breeders. At the end of either process, however, the inserted DNA will often still include “tag-along” genes from the original ES cell strain that may affect the phenotype. Randomly inserted transgenes are also prone to hidden KO effects, wherein the insertion interrupts the function of a gene or nearby transcriptional control elements.
Because of funding pressures, it is tempting for investigators to use results from inactivation KO models to infer causal links to human disease. However, constitutive KOs rarely address whether elimination of a gene produces a particular human disease phenotype. Only a subset of human genetic disease will reflect completely inactivated alleles. More often, the inactivation will be partial, will yield less of the gene product, or yield a subtle change in protein function. Often, the disease-causing allele will not be present in homozygous form. Thus, whenever possible, heterozygous mice should be examined for potential abnormal phenotypes.
Constitutive KO models may not only fail to accurately model a disease condition but may also promote cause confounding morbidity or lethality. A key refinement was to render expression of the KO allele conditional by age or cell type by using Cre-LoxP recombination (Crawley 2000; Jackson and Abbott 2000). Now, we have entered into the era of CRISPR/Cas9-based gene editing, which is based on means evolved by bacteria for excising foreign DNA from their genome (Gaj et al. 2013). By greatly extending the types of cells and conditions under which cells can be transfected and boosting success rates, CRISPR/Cas9 is revolutionizing both the production of KO and transgenic mice and possibilities for gene therapy. Two variations on CRISPR/Cas9 permit gene deletion through non-homologous end-joining (NHEJ) or gene editing through homology-directed repair (HDR). These operations may be rendered conditional, are orders of magnitude more efficient than the prior methods, and allow direct transfection of fertilized embryos, thus eliminating the need for ES cells (Wang et al. 2013; Yang et al. 2013). That said, the success rate for HDR is presently only ∼3 %, so that hundreds of cells must be injected, and local in vivo application (say, perilymph) will likely transfect few target cells. By contrast, gene deletion through NHEJ is highly efficient (>80 %), so that silencing of dominant hearing loss mutations by direct transfection of inner ear cells holds promise as a clinical tool (Zou et al. 2015). CRISPR/Cas9 methods applied to mice permit the simultaneous editing or deletion of multiple loci and are not limited to particular strains. Also, while it is not always desirable, CRISPR/Cas9 can alter both alleles of the target gene. In a single generation, one could therefore produce homozygous carriers for multiple engineered alleles. To date, the number of applications of CRISPR/Cas9 to hearing research has been relatively few (Zou et al. 2015; Mianné et al. 2016), and it is still common to find that the knockout or transgenic model one needs is still available only on a B6 or mixed B6/129/CD-1 background. Notably, large-scale efforts to generate ES cell lines carrying engineered mutations of coding genes, such as The KnockOut Mouse Project (KOMP) and European Conditional Mouse Mutagenesis (EUCOMM) program, are using the C57BL/6N strain as their standard. These roughly 10-year-old endeavors preceded the discovery of CRISPR/Cas9 and remain a valuable resource for generating constitutive and conditional KOs. It seems just a matter time, however, before CRISPR/Cas9 renders ES cell technology obsolete for targeted mutagenesis (Skarnes 2015).
Rationale for Using Mice to Elucidate Human Biology
Mice offer a wide range of advantages as models of human health and disease (Schughart et al. 2013; Bowl and Dawson 2015). Among these, they are sexually mature by 6 weeks of age, and most commercial mouse strains reproduce well, providing large numbers in a short period of time. The mouse life span is relatively short, 2 to 3 years, enabling collection of a lifetime of data over a reasonable length of time. Housing large numbers of mice can be accommodated economically and in relatively small space compared to most other model species. Behavioral, physiological, anatomical, and molecular studies can be completed on the same set of mice, thereby reducing the number of animals needed for study.
The logic underpinning all hearing and vestibular experiments in any mammal is that their inner ears and brains work much the same way as in humans. Even instances where they may not still illuminate our understanding of our own species. Curing disease was not really the goal of animal research for decades. We first had to understand how natural systems work. Why did evolution solve the same problems in different ways? What are the universal features? Myopic notions of “translatability” now constrict the scope of research and research models and risk choking off the flow of basic discoveries (Brenowitz and Zakon 2015). In any event, the foundation of most mouse work in hearing and balance is the extensive overlap of human and mouse genes and proteins critical for these functions. Presently, more than 140 loci and 100 identified genes are known to cause deafness in humans (Girotto et al. 2014). According to Steel (2014), as many as 1000 genes may ultimately be involved, either by promoting deafness in a direct (Mendelian) fashion or by magnifying the effects of environmental or personal risk factors. About 99 % of mouse genes have human orthologs (Bowl and Dawson 2015). In most cases, mutations of mouse genes exhibit deafness phenotypes similar to those associated with comparable human mutations. However, there are exceptions (such as Gjb3, Crym, Dfna5, and Coch mutations). The incomplete overlap of known deafness genes suggests that inner ear function in mice versus humans can involve functionally related genes and/or different genetic modifiers. In such cases, it is worth understanding exactly why “substitute” genes and modifiers in mice are able to correct for mutations that cause deafness in humans. About two thirds of ∼340 mouse genes known to cause inner ear dysfunction (Table S2) have not yet been linked to deafness in humans, but this most likely simply reflects how far mouse work has outpaced the human gene search (Steel 2014). Mendelian genetic hearing loss often reflects loss of function of highly conserved “bottleneck” genes, such as those that encode hair cell-specific structures like stereocilia. These are amenable to proof-of-concept testing with knockout alleles that need not reproduce specific human mutations. Even negative results in transgenic and knockout models often provide insights to the deeper biology: Are the human and mouse orthologs different functionally, or just differentially expressed? What does it tell us about a process—or prospects for therapy—if the two species fill a need using different genes?
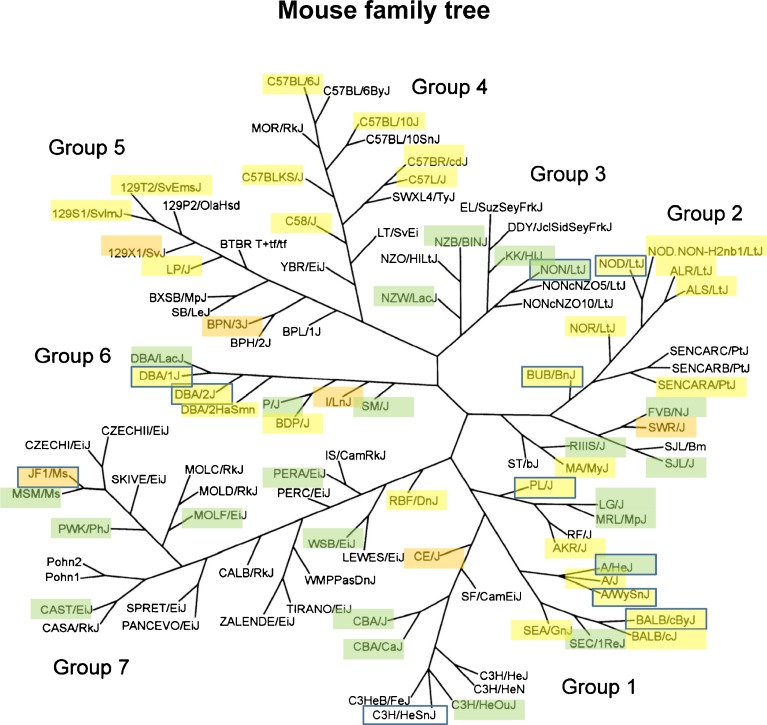
Before concluding that the result of a knockout experiment is universally negative, the next step should always be to repeat the experiment on genetic backgrounds quite different from the one initially tested. Just as no single person can be taken to exemplify the human genome or proteome, no inbred mouse strain represents all mice nor is really asserted to model all humans in any respect. Conversely, a clear phenotype observed on a specific inbred background indicates only what is possible and might occur in humans if orthologous genes and modifiers act similarly. There is no “neutral” strain for the way a trait manifests, and any result should be repeated on other genetic backgrounds. Funding and resource constraints often force investigators to skip this step, causing results to be missed or over-generalized. As a guide to strain selection, the schematic evolutionary tree of mouse inbred strains in Figure 2 shows that commercial inbred strains can be assigned to seven genetically divergent groups. Strains have been highlighted to indicate those presently known to carry the Cdh23 753A (ahl) allele, which accelerates age-related hearing loss (yellow, also see Table S1), other strains with identified progressive hearing loss (orange), and those suggested to possess stable hearing to at least 12 weeks of age (green). To explore background effects on one’s chosen mutation or treatment, one might thus select “good-hearing” strains from widely separated groups in Figure 2. Of course, whether a strain is considered good hearing or not depends upon when it is assessed and using what maximum test frequency. To ascertain the background effects on a particular mutation, one might outcross the original strain to selected recipient strains, intercross the F1 hybrids, and then phenotype F2 mice from each cross that are homozygous for the mutation. Alternatively, one could invest the effort of producing a congenic line on each strain or use CRISPR/Cas9 to generate the mutation independently in each strain. It is worth noting that, with the exception of groups 3 and 7, strains with the ahl allele or progressive hearing loss of unknown origin are widely distributed. In addition to the Cdh23 753A allele, Table S4 shows several additional known genes and quantitative trait loci (QTLs, locations that carry an unknown gene that impacts phenotype) that exert similar effects. Some of these overlap, so that some strains carry multiple alleles (e.g., B6, NOD/LtJ, DBA/2J).
FIG. 2.
Mouse family tree. 102 strains are organized into seven groups: Bagg albino derivatives (Group 1), Swiss mice (Group 2), Japanese and New Zealand inbred strains (Group 3), C57/58 strains (Group 4), Castle’s mice (Group 5), C.C. Little’s DBA and related strains (Group 6), and wild-derived strains (Group 7). The length and angle of the branches do not reflect the actual evolutionary distances between strains. Strains known to carry the Cdh23 753A allele (yellow, based on Table S1), show other progressive hearing loss (orange, from Fig. 3, Table S4, or Myint et al. 2016) or vestibular dysfunction (boxes, from Fig. 4), are indicated. Strains indicated as “good hearing” at 12 weeks of age (Myint et al. 2016) are indicated in green. (Modified with permission from Petkov et al. 2004).
The very idea of a standardized inbred mouse strain flies in the face of cumulative random mutational events, and it should seem strange that any life form may be treated as immutable and come with such guarantees of standardization. It is the nature of DNA to mutate, and for isolated populations to exhibit genetic drift, that is, to accumulate random mutations that are unique to each population. Yet, we can in fact expect a commercial inbred strain that has carried the same name for decades to be invariant for most research purposes. Slowing the rate of genetic drift is accomplished by proven breeding protocols and by periodic (every five generations at JAX) re-introduction of cryopreserved embryos (Stevens et al. 2007). It follows that, once an investigator has purchased commercial breeders and established a colony, the potential for genetic drift again arises. By the 10th generation, the probability of new mutations approaches unity (Silver 1995). At that point, the investigator is no longer breeding the intended commercial strain, but a unique substrain. Most likely, the newly accumulated mutations will not impact the phenotype of interest. But, there is no guarantee, and the arrival of a confounding mutation is likely to be missed.
Limitations
Important differences between mice and humans should be acknowledged. Mice are quadrupeds, so the relative contributions of, or reliance on, vestibular, visual, and somatosensory inputs for balance and locomotor output may differ from that of bipedal species including humans. By contrast with humans’ typical “predator” forward-facing eyes, mice have typical “prey” side-facing eyes with a more restricted range of motion. This may have implications for the neural circuitry governing binocular fusion and oculo-motor behavior. The mouse retina is not foveated and possesses a different complement of photoreceptors from ours (Jeon et al. 1998). If they were human, they would qualify as legally blind for their poor visual acuity (Baker 2013). Mice rely far less on vision than on olfaction and hearing (Brenowitz and Zakon 2015). In fact, the mouse rd1 (Pde6b rd1) mutation, which often imparts complete blindness, is common among wild mice (Farber et al. 1994). Despite the different reliance on vision in mice and humans, there is extensive overlap between many of the genes known to cause human retinitis pigmentosa and retinal degeneration in mice, including phoshodiesterase 6b (Pde6b), peripherin 2 (Prph 2), and myosin 7a (Myo7a) (Chang 2016).
Compared to larger mammals, the mouse brain features different functional weightings of subcortical auditory and vestibular nuclei and different interconnectivity of hearing and balance centers (e.g., Aitkin et al. 1984; Cryan and Holmes 2005; Schreiner and Winer 2007; Hofman 2012). The lissencephalic cerebral cortex in mice comprises far less of total brain mass and is less than half as thick as that in primates (Gilman et al. 2016). Moreover, the shape and function of cortical pyramidal cells are sufficiently different in mice and primates that they may form a qualitatively different type of columnar circuit (Gilman et al. 2016). Such differences, however, have not undermined the use of mouse models to extract putatively fundamental properties of cortical function, particularly with the advent of transgenic tools for selective activation of cells and circuits (e.g., Li et al. 2013; Moore and Wehr 2013; Hamilton et al. 2013). The main organizational feature that distinguishes mice and other rodents from primates appears to be the degree of “encephalization” of coding in the latter. That is, in rodents, particular stimulus characteristics may be extracted at lower levels in the CNS (e.g., Piscopo et al. 2013).
There are also limitations to overall genetic similarity of mice and humans. The most stark differences derive not so much from changes in protein-coding genes but rather from much more rapid divergence in regulatory sequences that lie near or within a given gene (cis-regulatory sequences) (Vierstra et al. 2014). These have led to tissue-by-tissue differences in gene expression, even though overall regulatory programs (gene sets activated by particular transcription factors) have been conserved. Probably as a result, humans and mice have developed different gene redundancies by tissue, so that we may solve a particular functional problem with a different protein isoform or an entirely different protein. Even Mendelian human genetic diseases, such as cystic fibrosis or Duchene’s muscular dystrophy, yield very different phenotypes when the same genes are mutated in humans and mice (Uhl and Warner 2015). Notably, a recent comparison of cochlear gene expression patterns in marmosets and C57BL/6J mice concluded that discrepancies in deafness phenotype for 20 major deafness genes are best accounted for by species differences in gene expression, not coding differences (Hosoya et al. 2016a). The same was suggested to apply for discrepant human/marmoset versus mouse phenotypes related to DFNB4 and Pendred syndrome (Hosoya et al. 2016b). Among cochlear cell types, outer sulcus cells in particular may express different complements of ion exchangers in humans and mice, suggesting that these cells serve roles that do not completely overlap (Hosoya et al. 2016b).
Aligning Life Stages
Humans and mice fall along an orderly relation in mammals between metabolic rate and life span and are each allotted roughly one billion heartbeats per lifetime (Dobson 2003), prompting attempts to align ages for experimental purposes. From the perspective of hearing and balance, early developmental stages are difficult to align since mice are altricial while humans are precocial. The other stages we might wish to align include adolescence, middle age, menopause/estropause, and old age. Pre-adolescence is important because of mouse data suggesting that the cochlea is more vulnerable to ototoxins during roughly the first month of life (Henry et al. 1981; Prieve and Yanz 1984). Adolescence is important for a large body of animal data suggesting that the cochlea is more vulnerable to noise, beginning at the onset of adult-like sensitivity, peaking around the time of sexual maturity (∼6 weeks in mice), and ending some time in early adulthood (∼4 months in mice) (Henry 1982a, b, 1983b). Exactly how and whether distinct early vulnerability windows for ototoxicity and noise apply to humans is unclear (Henry and McGinn 1992; Pujol 1992; Henley and Rybak 1995), yet the potential implications for human risk are considerable.
Aligning human/mouse middle age and old age is of interest for relating hearing loss to age-associated risk factors in humans, such as cardiovascular disease, diabetes, obesity, hypertension, and hypercholesterolemia (Agrawal et al. 2009; Lin et al. 2011). Menopause in females offers a definitive event. Women generally reach menopause at 48–55 years, while female mice undergo estropause (the mouse analog) at 11–16 months (Syed et al. 2010). Unlike humans, wherein females tend to outlive males (Iachine et al. 2006), a survey of 31 inbred mouse stains showed a high degree of strain dependence (Yuan et al. 2009). As a recent paper makes clear (Geifman and Rubin 2013), there is no single age normalization that applies across organ systems or health issues. This may complicate studies of interactions between age-associated co-morbidities and sensory loss. Mice are not particularly prone to complex human age-associated diseases such as Alzheimer’s or cardiovascular disease unless they are “humanized” with predisposing alleles or fed special diets (Vanhooren and Libert 2013). Even then, they may only manifest specific useful characteristics, such as the formation of β-amyloid plaques, without the behavioral consequences (Kokjohn and Roher 2009). Numerous mouse genetic models of Parkinson’s disease likewise produce some aspects of the human disease, yet none reproduce the definitive feature, loss of dopaminergic neurons (Blesa and Przedborski 2016). Huntington’ disease, another delayed neurodegenerative condition, is more completely modeled in transgenic mice, perhaps because of its narrower genetic etiology (CAG repeats in the HTT gene) (Pouladi et al. 2013). Unlike humans, mice can synthesize their own vitamin C, a difference with wide-ranging implications for aging processes. Like other rodents, mice have comparatively long chromosomal telomeres and high levels of telomerase activity (Vanhooren and Libert 2013). This may protect organs and tissues that self-renew through mitosis.
Drug Metabolism
The rapid metabolic rate of mice and other differences in drug metabolism have meant that mice must be given much larger systemic doses of any ototoxic or therapeutic drug than might be given to larger animals. With aminoglycosides and platinum-based agents, this has meant applying them only to mice less than 1 month old (Henry et al. 1981; Wu et al. 2001), boosting dosing by four to eight times (Poirrier et al. 2010), or combining them with a potentiator such as furosemide (Hirose and Sato 2011; Xia et al. 2014). For therapeutics, systemic doses in mice must be larger by a factor of up to 10× versus other animals or humans (Dowdell et al. 2009; Davis et al. 2010). Other complicating factors include species differences in cytochrome P450 enzymes, which metabolize about 75 % of drugs, as well as differences in gastrointestinal microbiome (Uhl and Warner 2015). Overall, rats and mice accurately predict human drug toxicity in about 43 % of cases. This is not a rodent weakness but rather an animal model limitation: When all animal models are considered, the success increases only to 71 % (Uhl and Warner 2015). Since the effect of any drug will likely be influenced by genetic background, it is recommended that any study of drug toxicity or therapeutic value be undertaken in more than one inbred mouse strain.
Cochlear Anatomy, Function, and Gene Expression
Mouse hearing extends to 100 kHz, well beyond the upper frequency limit of human hearing. This could be associated with significant anatomic and physiologic differences not yet appreciated. The microarchitecture of the organ of Corti and lateral wall are difficult to define and quantify and prone to fixation artifacts, so that fine anatomic differences would be difficult to evaluate. A recent quantitative analysis of spatial gradients in the conformation of the organ of Corti (Soons et al. 2015) determined the mouse to be typically mammalian. However, another recent paper determined that the cellular distributions of key components of EP generation are different in mice and humans (Liu et al. 2016). These differences include expression of Kir4.1 (KCNJ10) in both basal and intermediate cells of human stria vascularis, but only in intermediate cells of mice. Also, the Na+/K+/Cl− co-exchanger (SLC12A2) is expressed in strial intermediate cells of humans, but not mice. This re-distribution of critical elements may alter the respective roles of basal and intermediate cells in EP generation and its pathophysiology. Mice and humans may also differ with regard to the elaboration of root cells (a subset of outer sulcus cells extending root processes into the spiral ligament) (Santi et al. 2016). Mice (at least B6 mice) have more of these cells, which are thought to help regulate endolymph composition.
Cochlear single unit studies in mice are few (e.g., Taberner and Liberman 2005) but indicate prevailing similarities to other mammals. The most striking differences pertain to input/output and spike timing characteristics that are thought to be influenced by phase locking in mammals with good low-frequency hearing (<4 kHz). Cats, guinea pigs, and especially gerbils tend toward apical-basal divergence of single fiber characteristics that suggests an emphasis on spike timing in the cochlear apex and spike rate in the base (Ohlemiller and Siegel 1994; Huet et al. 2016). Mice hear poorly at the frequencies where phase locking is prominent and may have no cochlear neurons tuned below ∼2 kHz (Taberner and Liberman 2005). From the standpoint of single unit characteristics, the entire mouse cochlea may mirror the cochlear base of other mammals.
Mice as Models of Hearing in Aging
At the level of cellular pathology, mouse models do a pretty good job of recreating specific aspects of human presbycusis (Kikkawa et al. 2012; Bowl and Dawson 2015). All the specific human cochlear cell pathologies noted by Schuknecht and others (Schuknecht 1964; Schuknecht and Gacek 1993) can be found in mice, as well pathology that corresponds to all Schuknecht’s suggested types of presbycusis (Ohlemiller 2006). Like most humans, most inbred strains do not model these in isolation, and in neither case does the shape of the audiogram offer a definitive diagnostic (Landegger et al. 2016). As in humans, most mouse age-associated cochlear pathology appears to fall under the rubric of “sensory” presbycusis, wherein hair cell loss becomes limiting for hearing. A variety of natural and engineered mutations worsen this phenotype (Bowl and Dawson 2015). While little is yet known about human genetic predisposition to “neural” presbycusis, both engineered mutations and insults can magnify this type of lesion in mice (e.g., Lang et al. 2006; Kujawa and Liberman 2009). Human “strial” (metabolic) presbycusis actually remains theoretical, beyond example temporal bones showing such extensive strial pathology that the EP was almost certainly reduced (Schuknecht et al. 1974). Strial presbycusis may disproportionately appear in women (Gates et al. 1999), particularly after menopause (Hederstierna et al. 2010). Notably, the mouse analog precedes a demonstrated disproportionate EP reduction in female CBA/CaJ mice (Ohlemiller et al. 2010), perhaps through mechanisms that may operate in post-menopausal women (Guimaraes et al. 2004; Price et al. 2009). Moreover, strial marginal cell pathology constitutes both the primary initial age-associated pathology in human stria (Schuknecht et al. 1974) and the pathology most predictive of EP decline in aging mice (Ohlemiller 2009; Ohlemiller et al. 2010; Ohlemiller et al. 2016). The detail with which mice can be studied has revealed some age-associated pathologies (e.g., root cell loss) (Ohlemiller et al. 2010) whose effects on hearing remain unclear, and we do not yet know if humans show the same changes.
Mice remain a barely tapped resource for the study of human presbycusis. Table S3 lists a large number of known genes that impact either age-associated or noise-associated hearing loss in mice, some of which may play a similar role in humans. Age and noise susceptibility have been listed together because of the common notion that much apparent aging pathology is injury, and because in humans these are difficult to distinguish. Few of the models in Table S3 or S4 could be considered well-characterized, and a staggering amount of work remains to be done to see what clues they hold for human presbycusis. This also holds for the many strains yet unstudied, some of which appear in Figure 2. As Harper has emphasized (Harper 2008), the process of creating easily bred and handled inbred mice has led to a jettisoning of traits (and their underlying alleles) that are associated with longevity, such as slow maturation, small body size, and peroxidation-resistant cell membranes. Some useful phenotypic and allelic variation might be recovered in studies of wild-derived inbred strains (Fig. 2, group 7) (Miller et al. 2002). These strains are fully inbred and should not be confused with “wild-caught” mice, yet they carry alleles lost from the more popular research strains.
General Considerations in Choosing a Mouse Model
Mice may not represent the best model for some types of inner ear studies. Some manipulations require a larger bulla, more than 2.5 cochlear turns (the approximate number in B6 and CBA/J mice) (Saunders and Garfinkle 1983; Muller et al. 2005) or a cochlea that affords access to all three scalae in multiple cochlear turns. At no location does the mouse cochlea offer non-traumatic access to scala vestibuli, and only in the basal turn is scala tympani accessible. If mice are chosen as the best animal model for a particular study, the first point is always to use complete standardized nomenclature (http://www.informatics.jax.org/mgihome/nomen/) for the strain that is used. Seemingly small differences in names may represent substantial genetic divergence and may mean the difference between success and failure in replicating a study. Reviewers and journals should not accept incomplete terminology or vagueness regarding where mice were obtained. Access to major research strains is facilitated by agreements among major suppliers (JAX, Taconic, Harlan Sprague Dawley) to sell the others’ mice, replete with the exact substrain designation. Hence, for example, one may purchase C57BL/6J mice around the world from major suppliers with the confidence that they are the same mice and will produce the same experimental results. The goal of one’s research may require the identification of mice with a particular phenotype. Substantial online resources exist for this purpose. Among these are the Mouse Phenome Database (http://phenome.jax.org/), International Mouse Phenotyping Consortium (IMPC) (http://www.mousephenotype.org/), and German Mouse Clinic (http://www.mouseclinic.de/) (see Peters et al. 2007).
Not all applications of mice ask explicitly genetic questions at the outset. One may simply wish to test a drug or manipulation while keeping genetic variability to a minimum. Using an inbred mouse strain or an F1 hybrid is a sensible approach. F1 hybrids are the first-generation progeny from a cross of two inbred strains. Every F1 mouse carries one allele from each strain at every locus (except for X and Y chromosomes), yet all F1s are genetically identical. If the two parent strains are very different genetically, F1s gain robustness (hybrid vigor) from having more kinds of alleles. The experimental design benefits from the use of healthier mice and increased generality of results, since more gene variants are incorporated. Note that the generation produced by crossing F1s (yielding F2s) will feature random recombination and will not be standardized, so that one must again cross the parent strains and generate new F1s. As part of the experimental design, one should plan to test multiple, distantly related inbred strains or F1 hybrids formed from these. Otherwise, there is a risk of writing off a drug that just happened not to produce an effect on the single strain examined, or conversely, falsely assuming that the drug is widely effective regardless of genetic background.
Avoiding Problematic Strain Backgrounds
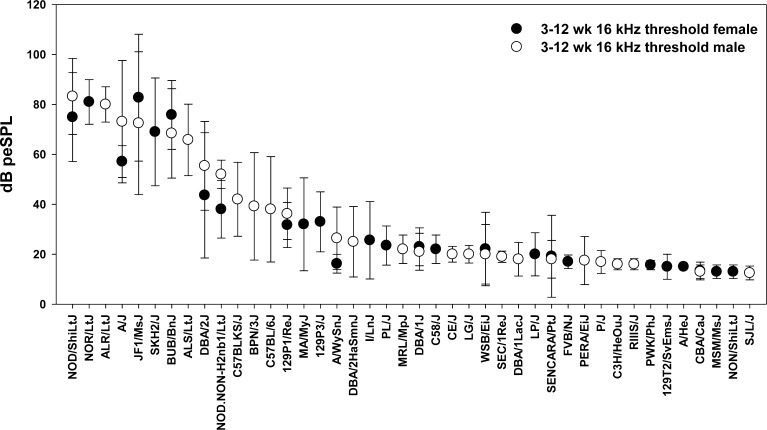
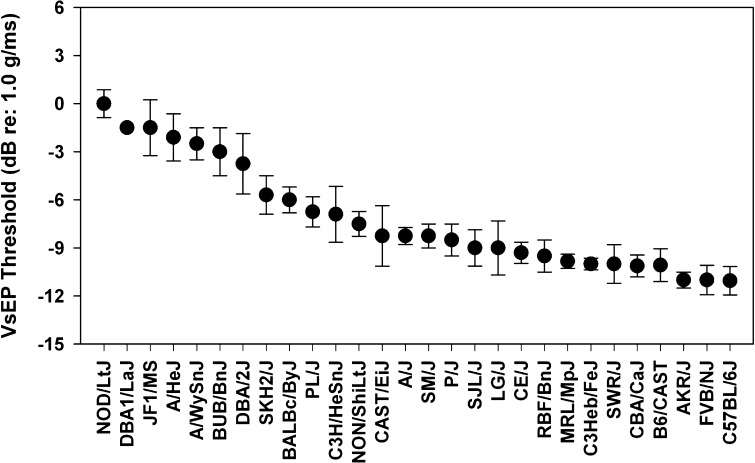
Strains with known hearing or vestibular loss should be avoided, except in the case where the effect of a drug is specifically to be tested against known deficits, like the progressive hearing loss exhibited by C57BL/6 mice. Generally, it makes little sense to use mice with known hearing defects for noise or ototoxic studies or for normative studies of development, gene expression, proteomics, or protein localization. Pure tone 16 kHz thresholds for 42 inbred strains aged up to 12 weeks are shown in Figure 3, so that one might pick good-hearing young mouse models from the right end of this graph. Figure 4 presents VsEP thresholds for many of the same inbred strains shown in Figure 3 (organized with “good” gravity receptor function to the right). Comparing the two figures, one can quickly see that the strains are ordered differently along the x-axis, so that abnormal hearing and vestibular function need not coincide (also compare boxed and highlighted strains in Fig. 2). Strains with progressive hearing loss may gradually lose threshold sensitivity on both low and high ends of the audibility range. One might decide to simply work within the uncompromised part of the cochlea, but that may not be accurately assessed by auditory brainstem response (ABR) testing. Moreover, the progressive loss may reflect the influence of “pro-injury” alleles that are relevant to any injury paradigm.
FIG. 3.
ABR thresholds at 16 kHz for 42 inbred mouse strains sorted by level. All mice were tested at 3 to 12 weeks of age. (Modified from http://phenome.jax.org/db/qp?rtn=views/measplot&brieflook=1404&projhint=Johnson1).
FIG. 4.
VsEP thresholds for 28 inbred mouse strains sorted by threshold level.
Caveats also apply to the use of genetically heterogeneous mice such as outbred stocks (see below). When the question to be asked pertains to a normal process (e.g., development), the effects of any manipulation (noise, ototoxins), or the efficacy of a therapeutic agent, heterogeneous mice will simply add statistical genetic variance that may obscure the result. By contrast, if the experimental goal is specifically to identify genetic factors that contribute to the variability in response to a treatment, then heterogeneous populations created just for this purpose, such as diversity outbred mice (described below), may be appropriate.
The appropriateness of a mouse strain may depend upon what is being studied. Many inner ear studies are aimed at genes and proteins that are highly conserved. These will often have a profound and relatively unmodifiable effect on peripheral function. Blindness, such as occurs in CBA/J, C3H/HeJ, and FVB/NJ mice, probably poses no confound for peripherally based hearing studies, although one might wonder if top-down influences like efferent function are altered. But what about studies of central auditory function in blind mice? Most cortical sensory fields appear multi-modal (e.g., Bizley et al. 2007; Campi et al. 2010), not to mention possible redistribution of auditory tasks to encompass visual cortex (e.g., Laemle et al. 2006) in normal-hearing blind mice. It is probably best if normative studies of central auditory function are performed in models with no known sensory or CNS deficits (e.g., CBA/CaJ, SJL/J), and some attention paid to the characteristics of the sensory environment. Many basic features one might study may exhibit little plasticity, but how clear is it where the line can be drawn?
Some examples may illustrate principles outlined here for selecting a mouse model at the outset of a study, along with unknowns we quickly encounter. If we wish to test a drug that may reduce noise or ototoxic injury, we might set criteria of (1) a normal inner ear at time of lesion and (2) enhanced susceptibility to insult. Three to four genetically divergent good-hearing strains might be selected by using Figures 2, 3, and 4. Yet, in designing the experiment, we quickly run into unknowns. Which strains are more vulnerable to noise or ototoxins? Beyond a few heavily studied strains (e.g., Wu et al. 2001; Ohlemiller et al. 2011a), simply too little is known, particularly with regard to ototoxicity. We might pick young but mature mice (say, 1 month of age) when both noise vulnerability and ototoxicity appear heightened (see below). But, we still must plan a series of dose-response studies and are left with the question of why younger mice are more vulnerable, lest our mechanistic results ultimately fail to generalize to older mice. Often, even arbitrary choices of strains will reveal surprising and useful interstrain differences in susceptibility. These can enhance the value of our drug study and lead to important new studies not anticipated. Surprising results may come in the realm of strain differences in the efficacy of our drug, the extent of noise or ototoxic lesions, or even in the form of injury: Within the inner ear, strain A may lose more hair cells than strain B for a given functional deficit or show more strial injury. Beyond the inner ear, strain A may show fewer central consequences of a peripheral lesion. In molecular studies, good-hearing inbred strains may differ greatly—albeit benignly—in inner ear cell-by-cell gene or protein expression. This inconvenience is also a strength, in that differences that do not produce pathology are less likely to be interesting. But, investigators may find themselves awash in genes to be considered and left to wonder if different treatments or functional metrics would have revealed a phenotype. For forward genetics studies aimed at identifying human-like pathology (and the underlying genes), Figures 2, 3, and 4 present a wealth of poorly characterized strains with known hearing and vestibular deficits. Whatever pathology is found is likely to be polygenic, so that crosses to two to three other genetically divergent good-hearing strains, followed by backcrosses to the initial strain may dissect the phenotype into categories representing the summed and distinct effects of different genes. These could be partially mapped in the same backcross (N2) or F2 intercross mice.
Lack of Melanin Versus Lack of Melanocytes
Some comments on the use of albino versus “amelanocytic” models may be helpful. The former possess no melanin, while the latter lack the cells that produce melanin. Production of melanin in tissues other than skin, hair, and iris was presumably maintained evolutionarily because it conferred advantages, and albinism is an abnormality that results from loss-of-function mutations in melanin synthesis. Known or suspected protective cellular functions of melanin include actions as a chelator of metals, binder of toxins, and an antioxidant (del Marmol and Beermann 1996; Riley 1997; Schraermeyer and Heimann 1999). In the inner ear, melanin is produced by intermediate cells of the stria vascularis (Wright and Lee 1989) but may also be widely distributed (Wolff 1931). In humans, lack of melanin, as indicated by skin or iris color, has been associated with greater hearing loss with age (Lin et al. 2012) and more robust noise-induced temporary threshold shift (TTS) (Barrenäs and Lindgren 1991; Da Costa et al. 2008). Animal data, including mouse data (Bartels et al. 2001), on the benefits of melanin are largely negative or equivocal. Positive findings in guinea pigs relating to noise, aging, and ototoxicity have been undermined by the lack of genetically matched controls (Conlee et al. 1986, 1988; Conlee et al. 1989). However, elimination of melanin in C57BL/6 congenic mice was found to promote strial marginal cell loss and EP reduction (Ohlemiller et al. 2009). Absence of melanin in the inner ear is not comparable to its effects on the visual system, where it plays a role in normal development (Jeffery 1997). For this reason, albino mice are not suitable for normative visual studies (Creel 1980). Nevertheless, since not all functions of melanin in the inner ear are well understood nor exactly which sites of melanin production are critical, it may be advisable to avoid albino mouse models for normative studies of inner ear function. Amelanocytic models involve a different process wherein melanocytes fail to migrate to their proper fields during development. Steel and colleagues characterized several mouse models of this type (Steel et al. 1987; Steel and Barkway 1989; Steel 1991; Cable et al. 1993; Steel 1995). These mice lack strial intermediate cells, which are required for EP generation. They therefore have little or no EP and, for reasons that are not entirely clear, tend toward wholesale degeneration of both stria and organ of Corti. They are genetically related to some forms of human deafness.
Lesioning the Mouse Inner Ear
Often, we wish to know whether a particular gene or therapeutic impacts the vulnerability of the inner ear to ototoxins or noise. Therapeutics aimed at regeneration are preferably tested in models with near complete loss of hair cells, so that the optimal type/level of noise or dose of ototoxicant is of interest. Creation of cochlear lesions comes with requirement to decide what kind of lesion we wish to obtain. Mutations, high noise levels, impulse noise, or ototoxic protocols can potentially damage the stria, wipe out all OHCs, produce holes in the reticular lamina, or kill varying numbers of IHCs. In some proof-of-concept experiments, the goal may be to inflict as much damage as possible. Yet, it may be worth asking if we are creating a lesion that exists in nature or represents a type of injury evolution has ever encountered. Accordingly, apparent failure of a therapeutic to correct an unnatural lesion may be misleading, and some thought should be given to how to “titrate” the lesion to its clinical counterpart. In applying mouse models, this may mean optimizing the mouse strain, age, and method of lesioning.
The type and dose of any ototoxicant or therapeutic agent required can differ for hearing versus vestibular organs. Although more strain comparison data are sorely needed, it appears difficult to achieve a cochlear hair cell wipeout in mice with noise above the basal turn (e.g., Wang et al. 2002). The relation between outer hair cell loss and permanent threshold shift (PTS) appears to differ for mice versus chinchillas, cats, rats, guinea pigs, and possibly humans (Bredberg 1968; Liberman and Kiang 1978; Hamernik et al. 1989; Altschuler et al. 1992; Chen and Fechter 2003). In general, hair cell loss may less reliably account for the extent of PTS in mice. Instead, permanent hair cell damage and changes in the shape of the organ of Corti may be more predictive (Ou et al. 2000a; Ou et al. 2000b; Wang et al. 2002). For a given insult, the extent of hair cell loss will nevertheless be strain dependent. Extant parametric studies cover only a few strains and noise conditions, so that one should plan pilot studies to evaluate the extent of lesions. Beyond the first month of life, mice are relatively impervious to aminoglycosides or cisplatin (Henry et al. 1981; Prieve and Yanz 1984; Wu et al. 2001). The reasons for this steep gradient of age dependence are not clear. Near-complete outer hair cell wipeouts have been achieved in adult mice through systemic application of kanamycin combined with the loop diuretic furosemide in a single dose (Oesterle et al. 2008) or subchronic dosing regimen (Hirose and Sato 2011). A potentially cleaner total hair cell wipeout may be achieved by using transgenic models in which hair cells can be selectively killed by using an agent such as diphtheria toxin (e.g., Kaur et al. 2015).
For noise lesions, in selecting a noise exposure level or duration, it should be kept in mind that any single choice tests one point on a typically unknown noise energy-versus-noise-induced PTS (NIPTS) relation. Such dose-response data are almost entirely missing from the literature. Although the relation is likely to be sigmoidal, it will often be unknown at the outset whether any strain or age differences in noise susceptibility will manifest as a difference in the shape of the curve or simply a shift on the x-axis. Likewise, we cannot know a priori whether our particular mutation or therapy shifts the curve on the x-axis or alters its shape. If too much noise is applied, ceiling effects may hamper the interpretation of results. A given NIPTS may be achieved by using a wide variety of noise exposure levels and durations. Up to some level that depends on noise type (kurtotic versus Gaussian), noise level and duration can be traded to similar effect, as long as total energy is constant (Qiu et al. 2006). However, for any type of noise, there exists a level where “microinjury” and metabolic fatigue will give way to overt tearing of the reticular lamina. These are different modes of injury, with different prospects for remediation. For broadband or octave band noise up to several hours in duration, the threshold noise level for lamina breach in CBA/CaJ and B6 mice at least 4 months old is 113–116 dB SPL (Ohlemiller in prep. Wang et al. 2002). Finally, in relating the noise exposure band to changes in the audiogram, it should be considered that the mouse ear canal shows a sharp resonance of nearly 20 dB (eardrum versus tragus, based on young C57BL/6 mice) at 16–30 kHz (Saunders and Garfinkle 1983). This may artifactually steepen the high-frequency roll-off of thresholds after noise exposure, particularly if thresholds above ∼32 kHz are not examined. The best hearing strains of mice can hear at frequencies up to 80–100 kHz. Attempts to use audio-range equipment (≤20 kHz) or any assessment that tests no higher than 30 kHz will leave the likely most fragile portion of the cochlea untested. However, it is worth noting that behavioral and some electrophysiological data (Ehret 1983; Ou et al. 2000a) suggest a dip in the audiogram near 50–70 kHz, a point where one might otherwise assume thresholds are rolling off monotonically. This frequency range corresponds to the peak energy of some communication sounds (Haack et al. 1983; Whitney and Nyby 1983). Thresholds in this range may conceivably be maintained by selective pressure and impacted by factors such as behavioral state or gender.
Gender Considerations
Gender should be considered in mouse experimental design, and NIH now requires that both genders be included in experimental plans. Based on human and animal studies, the effects of aging and possibly noise exposure are modulated by sex (McFadden et al. 1999; Henry 2004; Hederstierna et al. 2010; Ohlemiller et al. 2010). Features of cochlear anatomy such as total number of hair cells and basilar membrane length can be sexually dimorphic (Al-Mana et al. 2008). The cochlea and the entire auditory system are rich with receptors for sex hormones (Caras 2013). Potential roles include adjustment of sensitivity or responsiveness to communication sounds, courtship song or mating behavior, or detection and responsiveness to pup isolation cries. Quite possibly, sensitivity of the cochlea itself is subject to hormonal regulation.
The Meaning and Significance of TTS
Recent work has also emphasized the significance of TTS as a predictive metric for long-term hearing outcomes (see below). TTS is solely a threshold shift that statistically resolves to zero. It can be measured at any time up to when it disappears (typically hours to days, depending on severity). TTS is not the acute portion of NIPTS. That has a different name, the compound threshold shift (CTS) (Mills 1973). Regardless of when it is measured, TTS does not reflect the same physical processes as CTS, and the term TTS should never be used in the context of any exposure that yields NIPTS. Confusion around this point has muddied the discussion over the significance of TTS.
Current applications of TTS include its potential utility as a predictor of cochlear damage risk. Individuals who are more prone to TTS might also be more prone to NIPTS (Feuerstein et al. 2014; Moshammer et al. 2015), and remediation of TTS by a therapeutic might also indicate promise against NIPTS (Le Prell et al. 2012; Le Prell and Lobarinas 2015). In addition, the recent surge of interest in hidden hearing loss and “synaptopathy” has heightened interest in TTS as a risk factor for primary neural loss. This phenomenon appears robust in CBA/CaJ (Kujawa and Liberman 2006, 2009) and FVB/nJ mice (Paquette et al. 2016) but potentially less so in B6 (Shi et al. 2015). What may distinguish mouse strains and species in the severity of synaptopathy from a single exposure is the extent of synaptic repair (Shi et al. 2016; Song et al. 2016). If TTS proves to be a risk factor for primary neural loss, how much TTS imparts what degree of risk, and how broad is the exposure range that separates “risk-free” TTS, dangerous TTS, and NIPTS? Recent work in CBA/CaJ mice indicates that the entire span of these states may be <6 dB (Jensen et al. 2015). That is, in CBA/CaJ, these critical distinctions lie on a knife’s edge. Attempts to extend the science of synaptopathy to other mouse and animal models should begin with similar ranging experiments to ensure that analogous states are compared. How the narrow dynamic range of distinct cochlear injury states in mice extrapolates to human risk remains to be seen.
Pre-Existing Hearing Loss
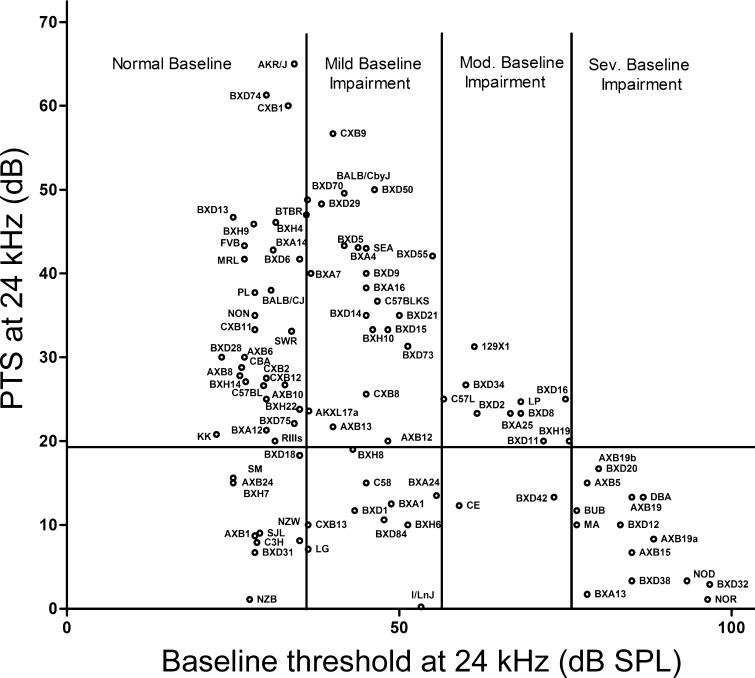
Animals with existing cochlear injury and hearing loss are “protected” from further injury, partly because the same cells cannot be lost or rendered dysfunctional twice and partly because a pre-existing threshold shift reduces the “working dynamic range” of threshold shifts we may observe after insults. Figure 5 compares initial thresholds with NIPTS at 24 kHz for a single type of noise exposure in a large set of mouse strains (for complete data set, see Myint et al. 2016). Strains with higher initial thresholds exhibit less NIPTS for far less interesting reasons than strains that appear in the lower left corner of the plot. Genetic backgrounds associated with progressive hearing loss (e.g., C57BL/6, BALB/c, DBA/2J) or progressive vestibular dysfunction are not suitable for most noise or ototoxic protocols after a few months of age. By 4–6 months of age, cochlear hair cell loss in some strains may envelop the basal ∼2 mm of the cochlea (Spongr et al. 1997; Willott et al. 1998; Ding et al. 2001; Hequembourg and Liberman 2001), rendering much of the cochlea uninformative for other manipulations. More generally, groups of mice with very different physiological hearing thresholds, whether due to hair cell loss, middle ear differences, or different developmental stages, cannot be compared after a single type of noise exposure: They will effectively receive different exposures. The confounding effect of prior injury provides a cogent rationale for reporting the animals’ initial thresholds in any publication. Editors and reviewers should require this information, certainly in papers that apply mice with known progressive hearing loss or strains that are not well characterized in the literature. Inclusion of baseline thresholds also enhances the readers’ confidence that threshold assessment was properly performed.
FIG. 5.
Baseline ABR thresholds at 24 kHz versus noise-induced threshold shifts (14 days after 108 dB octave band noise centered at 10 kHz) in 100 inbred strains. Thresholds greater than ∼50 dB SPL appear associated with less PTS. This can reflect real physiological differences of interest or an artifact of a reduced dynamic testing range. (Reproduced with permission from Myint et al. 2016).
Unilateral Noise Exposure and Anesthesia
Noise exposures are generally carried out in awake animals moving freely in a homogeneous sound field. Ideally, even the time of exposure should be standardized in light of evidence that cochlear noise vulnerability appears subject to a diurnal cycle (Meltser et al. 2014). In some studies, the goal may be unilateral NIPTS, so that an anesthetized animal may be positioned to only expose one ear (e.g., Longenecker and Galazyuk 2011; Turner et al. 2012; Hickox and Liberman 2014). If the goal is explicitly to study unilateral NIPTS, this may be reasonable. If, however, the goal is to accommodate testing paradigms that require normal hearing in one ear (such as gap detection), the effects of unilateral hearing loss may not be straightforward (Lobarinas et al. 2013). In addition, one may wonder whether unilateral exposure establishes a generalizable type of central auditory pathology (e.g., tinnitus) (Galazyuk and Hébert 2015). Anesthesia may introduce a number of effects that should be addressed by the authors in the publications. First, some anesthetics may exert protective effects on sensory elements (Rubinstein and Pluznik 1976; Hildesheimer et al. 1991; Giraudet et al. 2002; Kim et al. 2005). Second, anesthesia may alter descending modulatory influences (middle ear muscles, cochlear efferent reflexes). Third, anesthesia cools the animal, which may be protective against NIPTS (Henry 2003), and steps should be taken to maintain proper body temperature. Finally, we know that noise lesions not only the cochlea but also stations several synapses centrally (e.g., Basta et al. 2005). The net effect of anesthesia on the complex central sequelae of noise will be impossible to predict and would properly require an entire set of experiments on its own. In sum, it seems clear that experimental designs should, if possible, avoid noise exposure of anesthetized animals. Unfortunately, there is trend among institutional animal studies committees to force investigators to anesthetize their animals, based on well-intentioned notions of reducing the animals’ stress. This view, typically imposed by those with little knowledge of noise research or the noise literature, will have the opposite effect to that intended: Many studies involving noise exposure in anesthetized animals will ultimately have to be repeated to remove confounds, increasing the total number of animals needed.
Local Drug Application to the Mouse Inner Ear
An additional point on unilateral manipulations in mice pertains to the local application of drugs to one ear, whether to the middle ear or inner ear fluids. Due to their small head size, mice may be particularly prone to the Schreiner effect whereby drugs that reach the perilymph may be communicated to cerebrospinal fluid (CSF) via the cochlear aqueduct (Schreiner 1999; Barkdull et al. 2007; Ciuman 2009). From there, they may reach the opposite inner ear, so that it is not clear whether the opposite ear can be considered an appropriate untreated control. Once an applied drug reaches the CSF, moreover, unintended central nervous effects may also occur. The small size of the mouse bulla can also cause them to swallow or aspirate a portion of compounds that are injected trans-tympanically. These can reach the pharyngeal cavity through the Eustachian tube in the anterior bulla, posing a choking or asphyxia hazard to the animal and effectively become systemically applied. Also, given that the major routes of inner ear entry of drugs injected into the middle ear are likely through both round and oval windows (see AN Salt resources at http://oto2.wustl.edu/cochlea/), the basal-apical dispersion patterns will vary hugely with cochlear length and thus with animal model. Mice, which among research mammals feature the highest ratio of round window/oval window size to cochlear length, are excellent for basic testing of locally applied therapeutic agents but may yield non-general results for basal-apical dispersion of drugs. A related consideration is that some drugs may pass directly into the cochlea through channels in the cochlear capsule, especially in the cochlear apex (Salt and Plontke 2009). The cochlear capsule in mice is particularly thin, as is the stapes and the bone that covers the cochlear nerve just distal to the internal auditory meatus. These features also may render the inner ear distribution of drugs applied to the middle ear of mice somewhat atypical.
Mouse Resources for Gene Discovery and Identification
Over 400 strains of inbred mice are bred commercially around the world. Each of these is like a factory, churning out new mutations. Naturally occurring mutations on inbred backgrounds come with the added benefit of being congenic to the strain of origin. If we wish, we can accelerate the rate of mutation with mutagens. Of course, to take advantage of these new mutations, comprehensive screening programs must be in place. Mice have been applied with great success to identify and characterize genes that underlie human Mendelian genetic inner ear disorders. Their promise with regard to complex genetic inner ear disorders is still unfolding. New mouse lines are appearing that greatly facilitate this process with added convenience, increased mapping resolution, and increased allelic variance.
Gene Discovery Is Not Over
It bears emphasis that gene discovery—mining of natural mutations in mouse repositories—is not “over.” The inner ear is a highly complex organ with intricate structure and extraordinary sensitivity, so it is no wonder that many genes are involved in its development and function. Human genetic deafness reflects nature’s blind modification experiments with genes that turned out to be too limiting and critical to withstand changes. These natural experiments continue, both in human populations and in huge commercial populations of laboratory mice. When pathology naturally arises in humans or mice, it means that exacting conditions have been met: Just the right subtle or overt change has occurred in some critical protein. It may be a protein we did not know existed or had no clue what it did. Perhaps the critical change was in where the protein is produced or in what amount. For pathology to manifest, alleles for all interacting proteins had to be of the right type. Nature stumbled into just the right conditions, conditions we could never have guessed, would never have known to hypothesize. Each disease-causing mutation has presented an opportunity to understand the relation between molecules and diseases. Indeed, much of our entire understanding of complex systems like the inner ear arose from the study of natural mutations. Naturally arising mutations in more than 340 genes have been reported to cause inner ear malformation or dysfunction in mice (Table S2). These only partially overlap with over 200 known genes and loci in humans. Spontaneous mutations continually appearing in mouse stocks will often affect known genes, yet some will be novel and ultimately prove to be homologs of human deafness genes. The rate of new gene identification by the forward mutation approach is decelerating, so that it makes sense to expedite this process through the analysis of chemically induced mutation, as is being performed by Harwell, Oak Ridge National Laboratory, and several other centers (see Table 1 in Peters et al. 2007). Many known mutations have yet to be analyzed for their potential effects on auditory or vestibular function (see the IMPC https://www.mousephenotype.org/). Phenotypes of most mutations are analyzed at relatively young ages. Evaluations at later time points could uncover new mutations with late onset, age-related effects. New hypomorphic (partly functional) mutations may be discovered that encode partially functioning proteins that cause subtle phenotypes or modify phenotypes of other genes. In some cases, a mutation in a single gene may not cause hearing loss by itself but will if combined with a mutation of a different gene with redundant or predisposing function. Such interactions cannot be predicted and cannot be built into reverse genetic experiments. In addition to coding sequence mutations, new mutations in regulatory elements are certain to be discovered. The conditional expression of known mutations in selected inner ear cells, or at different stages of development, may uncover new hearing-related genes that could not be detected by constitutive knockout mutations.
Mendelian Traits
Early onset genetic deafness is typically inherited in a Mendelian manner. Mendelian traits are those that show good correspondence between genotype and phenotype. They involve typically one large effect gene and are little affected by environment. A host of single genes that impair hearing and balance have been identified by leap-frog work in mice and humans (Steel 1995; Steel and Kimberling 1996; Dror and Avraham 2010; Steel 2014; Müller and Barr-Gillespie 2015). Candidate genes identified in human pedigree mapping have been confirmed in mice by using reverse genetics, and natural gene variants that cause hearing and balance problems in mice have led to identification of genes for human disorders (e.g., Girotto et al. 2014). In mice, candidate genes can be further tested by fine mapping, targeted inactivation, RNA silencing (Rudnicki and Avraham 2012), immunolocalization of gene products, and gene expression studies. While over 100 deafness genes have been identified in humans, the majority of clinical cases involve fewer than about 10 genes. Thus, in any case where deafness appears hereditary, direct sequencing of known deafness genes by using platforms like OtoSCOPE (Shearer and Smith 2012) is becoming a cost-effective approach. A recent application of massively parallel sequencing and OtoSCOPE in 1119 patients (Sloan-Heggen et al. 2016) led to isolation of the causative gene in 440 (39 %) of cases. While this success is impressive and highly indicative of the rapid pace of progress, the remaining 61 % testifies that the majority of genes remain to be discovered and added to testing platforms. An alternative approach that might reveal novel deafness genes might be non-targeted whole-exome sequencing, but this is not yet practical because of the high degree of genetic heterogeneity, let alone the daunting prospect of by using whole genomic sequencing to encompass the vast majority of highly variable DNA that is not exonic. We are left with a gap in accounting for Mendelian genetic inner ear disease. Mice remain critical, both as a fountain of new mutations and for reverse genetic approaches to test candidate human genes.
Complex Traits
Prospects for new gene discovery include those that promote complex disease traits. Complex traits are those that are influenced by many genes and by gene-environment interactions. They include cardiovascular disease, diabetes, cancer risk, dementia, and susceptibilities to vision loss, hearing loss, and vestibular loss—all conditions for which risk rises with age. The gene variants sought do not “cause” these conditions, but rather confer some amount of risk estimated by statistical association. To find genes that influence the probability of a particular complex disease trait, one begins not by targeting a sample or pedigree with affected individuals but instead by measuring a related biometric in a large population and correlating this with particular genetic markers. Any identified loci will be candidates for influencing the trait and could modulate disease risk. To date, few reproducible genes for complex disease traits have emerged from human genome-wide association studies (GWAS) (Bennett et al. 2010). The problem is the high degree of uncontrolled biological noise (both genetic and environmental variation) in human populations. A nearly universal finding in the analysis of complex traits is one of a residual mismatch between the risk that can be attributed to all significant SNPs and the percent risk that appears to be genetic. This mismatch is known as the “missing heritability” problem (Parker and Palmer 2011). A key question has been whether the missing genetic factors correspond to rare mutations (<5 % of the population), each conferring a relatively large amount of risk, or common mutations, each conferring just a few percent of risk. Rare mutations might be variants of critical genes that overlap with those underlying Mendelian traits, while common variants might represent other types of genes, e.g., homeostatic and protective genes. Rare alleles might be difficult to detect in a population precisely because they are rare, while common alleles are difficult to detect due to their weak effects. The hope driving new human GWAS is that if enough sufficiently large samples are examined by whole genome sequencing, both types of mutations will be revealed.
Complex trait analysis intersects hearing mostly around quantitative traits related to aminoglycoside exposure, presbycusis, and PTS caused by continual noise exposure. These conditions possess a genetic component. A common perception of presbycusis, like other diseases of aging, is that it often represents accumulated injury (Ohlemiller 2012, 2013; Ohlemiller 2015). Thus, some predisposing genes may not encode components critical for hearing, but rather modulatory, homeostatic, or protective factors. They may, however, include hypomorphic variants of a wide variety of genes. Unfortunately, few of the reported human GWAS results for complex hearing loss have been reproduced (Fransen et al. 2003; Carlsson et al. 2005; Van Laer et al. 2006; Van Eyken et al. 2007). A recent study (Fransen et al. 2015) questions whether any of the genes that promote presbycusis (or intermixed PTS) exert effects that are large enough to be pulled out of the biological noise. It is not questioned that such genes exist. Over 20 % of risk appears heritable (Fransen et al. 2015). Rather, all the underlying genes may be limited to 1–2 % of that risk and may show non-linear interactions (epistasis). This human complexity makes the extension of GWAS to mice all the more critical. Mouse models facilitate control of genetic and environmental variance and should help reveal genes that merit close examination in human complex hearing and vestibular loss. Across a number of human GWAS, there has been encouraging agreement between the genes—or types of genes—that contribute to complex forms of hearing loss in humans and those implicated in mouse studies. This adds confidence that we may hope to tease out subtle genetic influences that give rise to complex human-hearing phenotypes by using mice. Factors limiting mouse GWAS have included limited allelic variation and low mapping resolution. Until recently, GWAS of inbred mouse strains were not practical because of linkage disequilibrium caused by their related ancestry. Time and generations have “broken up” the human genome into segments (haplotypes) spanning generally less than 100 kb (Parker and Palmer 2011). Assuming that large enough samples can be amassed, this means that SNP-based human GWAS can theoretically achieve resolution spanning just one or a few genes. Traditional approaches to gene mapping in mice (i.e., intercross, backcross) have required analyzing thousands of mice to achieve megabase (that is, 10–100× lower) resolution. Another issue is the limited number of alleles in play in mouse studies if one begins with just two or a few inbred strains. Yet, though we might gain more alleles for test by adding more strains, we risk decreasing the frequency of each allele in the test population, thus creating detection problems. New mouse resources for mapping, described below, go a long way in resolving these historic issues.
Recombinant Inbred Strains and the Collaborative Cross
Recombinant inbred (RI) strains are not new but are being expanded in important ways. RI strains provide the ability to map genes without having to genotype mice. To generate an RI series, mice of two genetically distinct (polymorphic) inbred strains are interbred, and the resulting hybrid progeny are then used as founders to establish individual lines of mice that are inbred for at least 20 generations (Silver 1995). Mice within each RI strain are genetically identical, although the individual RI strains differ from one another in terms of their inherited patterns of strain-specific alleles. Moreover, each individual RI strain has been genotyped for thousands of genetic markers randomly inherited from the parental strains, so that it is known which parental strain contributed each segment of the genome. To map a trait, the experimenter first obtains phenotype data for as many strains of an RI series as possible and then submits the data to an online resource that aligns phenotype and marker patterns across the RI series. The mapping resolution of an RI series depends on the genetic differences between progenitor strains, the degree of variance of the trait of interest across the RI series, and the number of RI strains developed. The largest and most frequently used series is comprised of more than 100 individual BXD RI strains, which were derived from C57BL/6J and DBA/2J. The BXD notation incorporates a single letter for each parent strain (B for C57BL/6J and D for DBA/2J) with an “X” to indicate that these were crossed, and the particular strain number is appended (e.g., BXD35 to indicate the 35th strain of the RI series). Another large RI series is comprised of 36 individual AXB and BXA RI strains, which were derived from a reciprocal cross between A/J (A) and C57BL/6J (B). The reciprocal nature of the cross (as indicated by the different order of the A and B) stipulates which parental strain provided the male and female of every breeding pair used to produce a given RI strain. This is kept the same for every generation and is done because a particular gene may be epigenetically silenced in either sperm or egg in a highly stereotyped way. The reciprocal cross ensures that a particular allele is not systematically silenced (thus not tested) in every RI strain of a series. RI strains have been used to map both Mendelian and complex hearing traits in mice (Hitzemann et al. 2001; Noben-Trauth et al. 2010; Nagtegaal et al. 2012; Ohlemiller et al. 2016). GeneNetwork (http://www.genenetwork.org/home.html) is a useful web-based resource for the analysis of mouse RI strains.
The primary limitation posed by RI strains is that they are limited in genetic diversity: At each locus, there can be no more than two alleles, which does not model natural genetic diversity of large populations very well, and decreases the likelihood of detecting associations. The Collaborative Cross (CC) is a large series of RI strains that addresses this limitation. The series is derived from an eight-way cross of genetically distinct founder strains: A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ (Threadgill and Churchill 2012). These were selected because they capture nearly 90 % of the genetic variance of laboratory mice. Genetic associations have been mapped in CC strains with submegabase resolution, that is, within a few genes, and approaching human GWAS. Hundreds of CC strains are in development. All are characterized by dense maps for marker distribution from the founder stains. Because the CC strains are inbred, just as for RI strains, all mice within each line are identical and no genotyping is required. All the experimenter need do is measure the biometric of interest in as many strains as possible, then align markers and strain distribution pattern with an online utility (such as http://churchill.jax.org/research/cgd.shtml) to generate a preliminary map. For each strain, it may be necessary to phenotype several mice to decrease experimental variation. This may be cumbersome over a large number of strains, especially if the measure is something involved like the response to noise exposure. If the resulting QTLs lack significance, or the map requires more resolution, the experimenter can confine additional tests to CC strains with known recombinations within the chromosomal regions of interest.
Hybrid Mouse Diversity Panel
The Hybrid Mouse Diversity Panel (HMDP) is a collection of 100 inbred and recombinant inbred strains designed for mapping of complex trait QTLs at high resolution (Bennett et al. 2010). The panel presently includes 29 classic inbred strains and 71 strains selected from the BXD, AXB/BXA, and BXH RI series. Mapping is accomplished by comparing the strain distribution pattern for the trait of interest with dense marker sets established for each strain, similar to the strategy for RI strains. Because the test strains are all inbred, genotyping of mice is not needed. Maps constructed by using the HMDP can detect associations accounting for as little as 10 % of phenotypic variability with submegabase resolution, although the HMDP captures less genetic diversity than the Collaborative Cross. Derived maps can be refined by expanding the sample with strains having known recombinations in regions of interest. While much effort may still be expended in phenotyping, all in all, the mapping resolution that can be achieved per animal by using the HDMP is phenomenal. A recent HDMP study of frequency-specific effects on ABR thresholds yielded nine significant QTL regions, each with relatively few candidate loci (Crow et al. 2015). To achieve this required phenotyping of ∼500 mice, yet this compares quite favorably with larger samples often needed to detect fewer, less manageable, QTL regions by using backcrosses, intercrosses, and congenic lines. Notably, all but one of the genes (Otogl, otogelin, a human deafness gene) implicated were novel. It may seem odd that such a comprehensive study would turn up so few genes identified from other human and mouse studies. This may jointly reflect (1) that mice were examined at 5 weeks of age (thus too young to identify Cdh23 753A or other gene variants with a delayed impact on hearing), (2) the large amount of genetic variance captured by the HDMP (thus high potential to detect new small-effect loci), and (3) low minor allele frequencies across the 100 strains (that is, previously reported loci may have appeared in too few of the strains to be detected). The finding of so many novel loci supports that contention that many deafness loci remain to be detected. It is worth considering that such a study could have been done in many ways (different ages, addition of various injury paradigms) that would likely have turned up whole other sets of QTLs. Another recent HDMP study identified Nox3 as a gene that promotes noise-induced hearing loss in mice (Lavinsky et al. 2015).
Outbred Strains and Diversity Outbreds: Uses and Pitfalls
Commercial outbred strains such as NMRI, ICR, SABRA, and CD-1 are intentionally bred to foster genetic heterogeneity, both in terms of homozygosity/heterozygosity and multiple allelic combinations. In theory, no two mice of an outbred strain are clones. However, they are often derived from a small number of strains, so that their phenotype may be dominated by alleles common to the source strains or that were randomly fixed. Outbreds are bred according to protocols that may differ among commercial breeders, who make few claims about the degree of standardization one may expect. Relaxed standardization and heightened fecundity allow breeders to produce and sell these mice relatively cheaply. Breeders tend to be selected for large litter size, which effectively co-selects for size, since larger mice have larger litters (Land 1970). To cut costs and avoid limitations on “generalizability” of results from any single inbred strain, it has been tempting, for example, in pharmacology testing, to apply outbred mice as an approximation to the genetic variability of human populations. This approach has been highly criticized for false economy, since more animals are needed to counter the increased data variance (Festing 2010). Problems with outbreds potentially apply to hearing research. For example, CD-1 mice show rapidly progressive hearing loss whose etiology is of some interest (Shone et al. 1991; Le Calvez et al. 1998; Wu and Marcus 2003; Riva et al. 2007; Mahendrasingam et al. 2011). However, it is illogical to claim that these mice possess a single hearing phenotype if they are truly genetically heterogeneous. The appearance of a single phenotype must reflect chance fixation or mutations common to the source strains and will likely differ across vendors. Moreover, it is likely that modifier genes will impart differences among mice, even from the same vendor. CD-1s were derived from Lynch’s Swiss mice, from which Swiss Webster and Black Swiss outbreds were also derived (Chia et al. 2005). While it is not clear which progressive hearing loss alleles CD-1 mice harbor, Black Swiss mice carry the ahl5 and ahl6 alleles (Drayton and Noben-Trauth 2006). A much more appropriate use of commercial outbreds is in complex trait mapping (Chia et al. 2005). Compared to inbred mice, commercial outbreds incorporate greater genetic diversity, although less than can be achieved by using the Collaborative Cross. The breeding strategies employed to maintain these strains have fostered high rates of recombination in at least some outbred strains that theoretically permit mapping with submegabase resolution (Yalcin and Flint 2012). The disadvantages of commercial outbred strains are largely corrected in diversity outbred (DO) mice. These mice are derived from the same eight genetically distinct founder strains as were used to form the Collaborative Cross. DO mice, which display a broad range of phenotypes, are maintained at JAX as a randomized breeding colony with a population size of 175 pairs and can be purchased on the JAX website (Svenson et al. 2012). DO mice can be used for genome-wide association mapping of complex trait loci at submegabase resolution; however, large sample sizes (200–800 mice) are recommended, and mice must be individually genotyped for a large array of markers (Gatti et al. 2014). Compared to other commercial outbreds, DO mice also provide a more genetically diverse population for generalizing conclusions from toxicology and other drug screens.
Advantages and Quirks of Widely Used Inbred Strains
Inbred mice that are raised in highly controlled environments offer maximum subject uniformity and minimal variance for almost any type of data. This is highly appealing from the standpoint of budgeting and study design. Inbred mice have no counterpart in nature and represent breeding endeavors that selected only against a lethal dose of homozygous deleterious alleles. Nearly all present day inbred strains contain non-lethal deleterious alleles and represent disease models of some kind. Most commercial inbred strains are derived from a mix of the major natural M. musculus subspecies (musculus, domesticus, castaneus, and bactrianus), although the major contributor is domesticus (Silver 1995). The mitochondrial genes of all major inbred strains derive wholly from domesticus, while the Y sex chromosome (which is little altered by recombination) comes principally from musculus. Although they are mostly used in gene mapping because of their genetic diversity, one can purchase inbred strains that are recently derived from wild populations of each of these subspecies (Fig. 2, group 7).
The complete name of any inbred mouse strain tells the story—if cryptically—of its origin and derivation. Examination of the inbred lineages in Figure 2 reveals that each of the seven major groups includes strains with generally similar names, indicating some degree of relatedness. The names feature the major strain name (e.g., “C57BL”) followed by a “/” and a string of letters that refer to the individual or commercial breeders that received the strain and then further bred the mice in isolation. Each new breeder serially created a new substrain that diverged genetically from the strain they received. These substrains are not interchangeable. They cannot be combined in the same study or serve as controls for mice of other substrains. Any two studies that use different substrains, even if applying the identical experimental protocol, are different studies, and the same results cannot be assumed.
Below, we consider in detail four inbred strains that are disproportionately represented in mouse hearing publications. In addition to these, many other inbred mouse strains have been used in auditory and vestibular research, including A/J, C3H, CAST, FVB, NOD, and 129 related strains. Origins and general characteristics of inbred mouse strains can be found in Festing’s index of major mouse strains (http://www.informatics.jax.org/external/festing/mouse/STRAINS.shtml) as well as in mouse strain datasheets available online from distributors such as The Jackson Laboratory (https://www.jax.org/mouse-search?p=205:1:4621098353963360727) and Charles River (http://www.criver.com/find-a-model).
C57BL/6
This strain owes its name to the breeding of female #57 to male #52 of Lathrop’s stock by Little in 1921 (Silver 1995). Substrain number “6” was just one of several isolated by further breeding. The mice arrived at JAX for development and distribution in 1948. Some of the earliest characterizations of hearing in inbred mice focused on the progressive, age-related hearing loss of C57BL/6 mice (Mikaelian et al. 1974; Mikaelian 1979). These mice begin life with normal hearing but soon show high-frequency hearing loss that gradually extends to lower frequencies. They also show a less commonly recognized low-frequency hearing loss that may be causally distinct (Ohlemiller 2006). While more than one locus has turned out to drive the high-frequency hearing loss (Johnson et al. 1997; Nemoto et al. 2004), a significant influence is the hypomorphic Cdh23 753A allele for which these mice are homozygous (Noben-Trauth et al. 2003) and which has turned out to be present in many other inbred strains (Table S1). The encoded protein forms part of the hair cell stereociliary tip-link apparatus (Siemens et al. 2004). The mutation contributes not only to a generally accelerated loss of OHCs but intriguingly also to NIPTS susceptibility (Davis et al. 2001). The CDH23 protein interacts with Ca++ (e.g., de Brouwer et al. 2003), so that the Cdh23 753A allele magnifies the influence of other deafness genes that likewise encode proteins that bind or transport Ca++ (e.g., Schultz et al. 2005), as well as other tip-link and hair bundle components (e.g., Geng et al. 2013). Finally, CDH23 is a known deafness gene, underlying both DFNB12 and USH1D (e.g., Pennings et al. 2004). B6 mice have repeatedly been proffered as an “accelerated aging” model to test therapeutics against age-related hearing loss. They have also been used to speed up aging studies to fit into NIH funding cycles. This approach will continue to have supporters and detractors among reviewers, so that one should be prepared to make a strong case for using these mice. Despite accelerated hearing loss in B6 mice, there is little to no loss of canal or gravity receptor function in this strain (Shiga et al. 2005; Mock et al. 2016). Although a few hypotheses have been proposed (Schwander et al. 2009; Manji et al. 2011; Mock et al. 2016), the mechanism for preserved vestibular function in B6 mice remains to be determined.
While age and noise-induced hearing loss studies suggest that B6 mice have relatively fragile OHCs, they appear relatively resistant to aminoglycosides (Wu et al. 2001). Their cochlear lateral wall and endocochlear potential also resist injury from noise exposures that impose much greater injury in CBA-related and BALB/c mice (Ohlemiller and Gagnon 2007; Ohlemiller et al. 2011c). B6 mice also appear to be outliers in some studies of pre-conditioning, wherein a non-injurious stressor is protective against a later injurious stressor. Pre-conditioning protection has been demonstrated in the mouse inner ear for a number of mild stressors, including moderate noise (Yoshida and Liberman 2000), mild hypoxia (Gagnon et al. 2007), hyperthermia (Yoshida et al. 1999), and low-dose kanamycin (Fernandez et al. 2010). While not all these pre-conditioning paradigms have been tested in B6 mice, head-to-head comparisons with CBA/J indicated a lack of protection only in B6 mice for hypoxia and kanamycin (Gagnon et al. 2007; Ohlemiller et al. 2011b). B6 mice carrying null alleles for catalase also showed paradoxical protection of hearing with age, and after noise (Rellinger et al. 2012). Purely protective paradigms like antioxidant treatment have been reported to work in B6 mice by some investigators, but not others (Davis et al. 2010; Heman-Ackah et al. 2010). What may differentiate these results is whether the experimental manipulation is stressful or protective. A recent essay (Kraev 2014) summarizes concerns over atypical stress responses in C57BL/6J mice potentially resulting from a null genotype for the Nnt gene, which encodes the enzyme nicotinamide nucleotide transhydrogenase. This mitochondrial enzyme increases the production of reactive oxygen species (ROS) under stressful conditions, so that its elimination appears protective (Nickel et al. 2015). It was suggested that experiments related to stress responses be repeated in closely related C57BL/6N mice or other substrains that carry functioning Nnt alleles. These mice appear to have an aging profile similar to C57BL/6J (Mianné et al. 2016), but other stress manipulations have not been tested. In summary, it is worth considering that C57BL/6J mice may not yield generalizable results in some types of experiments.
Amid the current heightened interest in synaptopathy and resulting primary neuronal loss (Kujawa and Liberman 2006, 2009), one feature reported only in C57BL/6J mice merits mention: They may recapitulate a feature of the human spiral ganglion whereby the somata of radial afferent neurons form unmyelinated “aggregates” that may be connected electrically (Cohen et al. 1990; Hequembourg and Liberman 2001; Jyothi et al. 2010). In humans, where most spiral ganglion cell bodies are unmyelinated (Tylstedt et al. 1997; Tylstedt and Rask-Andersen 2001; Glueckert et al. 2005), this feature may promote survival and sound responsiveness in neurons that have become disconnected from their hair cell targets. In B6 mice, aggregates are observed principally in the apical half of the cochlea and appear to multiply with age. In older B6 mice where most neurons have been lost as a secondary effect of the Cdh23 753A mutation, the surviving neurons tend to be those within aggregates, so that formation of aggregates may likewise represent a means of preserving neurons in these mice. While it is not clear exactly why this feature appears in B6 mice, it is magnified in B6 congenics carrying a particular variant of the Ly5 (Ptprc or CD45) gene (Jyothi et al. 2010).
B6 mice are fairly docile, breed well, respond well to anesthetics, and are resistant to middle ear infections. They are, however, prone to atherosclerotic lesions (Bult et al. 2016). Their general characteristics make them easy to work with, although often preferably in the absence of the Cdh23 753A mutation. Two congenic lines are commercially available. One carries the wild-type Cdh23 allele from CAST/Ei mice (Johnson et al. 1997) and the other from CBA/CaJ mice (Kane et al. 2012). Both of these lines retain normal mid-frequency and high-frequency hearing beyond 1 year, although they still show rapidly progressive low-frequency hearing loss. In our hands, the CAST-derived congenics breed poorly. B6 mice with a genetically engineered knock-in of the Cdh23 753G protective allele have been produced through CRISPR/Cas gene editing (Mianné et al. 2016) and also by the traditional ES cell approach (KRJ personal communication). Single nucleotide knock-in mice eliminate the possibility of deleterious effects from co-integrated linked genes in the congenic strains.
BALB
All BALB mice originate from an albino line acquired and developed by H.J. Bagg (hence the name) in 1913 (Silver 1995; Bult et al. 2016). The “c” substrain (the c added by G.D. Snell in 1932) arrived at JAX for distribution in 1947, while the “cBy” substrain was developed in 1975, having been bred in isolation by DW Bailey (hence “By”). BALB/cJ and BALB/cByJ were early popular hearing models for the effects of albinism and for their B6-like progressive hearing loss (Willott et al. 1998). BALB/cJ and BALB/cByJ appear to possess a similar hearing phenotype, although this has not been examined in detail. It is now known that BALB/cJ and BALB/cByJ carry the Cdh23 753A allele (Johnson et al. 2000; Table S1) as well as other unknown alleles (Erway et al. 1993) that contribute to a more rapidly progressive hearing loss than in B6 mice (Ohlemiller 2002). The divergence may reflect EP reduction that B6 mice do not exhibit (Ohlemiller et al. 2006). The EP reduction correlates with marginal cell loss, and a QTL for marginal cell density on chromosome 12 (Maced) was recently reported based on work in BALB (Ohlemiller et al. 2016). BALBs appear exquisitely vulnerable to noise, both during the early vulnerable period and later (Ohlemiller et al. 2000; Ohlemiller et al. 2011a), and show acute or permanent EP reduction by intense noise (Ohlemiller et al. 2011c). Gravity receptor function in BALB mice shows slightly elevated VsEP thresholds with significantly reduced amplitudes suggesting a loss of gravity receptor sensitivity (Jones et al. 2006). Male BALB/cJ mice are very aggressive and should not be housed together.
CBA
CBA-related strains date to a 1920 cross by L.C. Strong of a Bagg albino female to a DBA male (Silver 1995). The “J” substrain arrived at JAX for development and distribution in 1948, while the “Ca” substrain (for T.C. Carter) arrived in 1966. CBA/J and CBA/CaJ provided good-hearing controls in the early B6 studies (e.g., Henry and Chole 1980; Henry 1982a; Li 1992). Perhaps due to the early overlap in their uses, some papers have simply stated “CBA” as the type of mice used, which would be fine if these strains were identical, which they are not. Having diverged ∼80 years ago, they are separated by >2000 polymorphisms (Bult et al. 2016). Notable among these are the Pdeb rd1 retinal degeneration mutation, which renders CBA/J mice completely blind, plus other unknown mutations that impart to them different hearing thresholds after 1 year and different sensitivities to noise only during the early vulnerable period to noise (Ohlemiller et al. 2011a). The age-related divergence can largely be accounted for by effects of aging on the EP and cochlear stria vascularis in CBA/CaJ, so that these mice appear to model Schuknecht’s strial presbycusis (Ohlemiller et al. 2010). CBA/J mice are relatively poor breeders and are prone to middle ear and respiratory infections (McGinn et al. 1992). With regard to manipulations such as aging, noise, and ototoxins, CBA/CaJ mice are the single best characterized good-hearing strain for hearing research. This simplifies decision making about what age or gender to test and how much noise or ototoxin to apply.
CBA/CaJ mice show normal gravity receptor function at younger ages but undergo more age-related decline than B6. Tests of canal function across age have not been reported. If the research goal is genetics of the gravity sensing organs, B6 might be the better choice than CBA/CaJ since the Cdh23 753A mutation does not impact vestibular function, vestibular sensitivity is better in B6 than in CBA, and gravity receptor sensitivity in B6 mice is well maintained into advanced age (Mock et al. 2016). If one is interested in studying both hearing and vestibular sensory systems, CBA/CaJ would be a reasonable choice for background strain.
DBA
DBA represents the oldest inbred mouse strain, having been founded by Little in 1909 (Silver 1995). They were named for a combination of three coat color genes (dilute, brown, non-agouti) that were the subject of the initial experiments. What would become DBA/1J and DBA/2J arrived at JAX for development and distribution in 1948, having passed through different breeders. Both substrains are particularly prone to audiogenic seizures around 1 month of age. The DBA/2J strain exhibits a very early onset hearing loss caused primarily by a combination of the Cdh23 753A variant and a missense mutation of the fascin-2 gene (ahl8, later renamed Fscn2 R109H) (Shin et al. 2010), both of which affect hair cell stereociliary bundle function and stability. Recent mapping efforts have further identified a QTL on chr. 5 (M5ahl8) that modifies the effects of the Fscn2 R109H mutation (Johnson et al. 2015). Mice of the DBA/1J strain and other DBA/2 substrains such as DBA/2NCrl and DBA/2De do not carry the Fscn2 R109H variant and exhibit a slower progression of hearing loss than DBA/2J mice. Gravity receptor function is also severely impaired in DBA/2J mice at a young age (Jones et al. 2005; Jones et al. 2006).
Overview of Mouse Mutant Inner Ear Models
A detailed treatment of known genetic mouse models of hearing and balance dysfunction is beyond the scope of this review. Table S2 provides an exhaustive list. Some of these models recapitulate human deafness both genetically and phenotypically. Others offer useful phenotypes where the causative genes remain unknown. Human genetic hearing loss can originate with pathology (or faulty development) of hair cells, stria vascularis, spiral ligament fibrocytes, or extracellular matrix. Mutant mice have been found that model all these. Genes that govern hair cell function and development are notably over-represented in Table S2. Hair cells constitute a probable evolutionary bottleneck of specialized structures encoded by unique genes. In mammals, they are not replaced and are non-redundant. At least in terms of sound detection, mammals appear to possess excess neurons and excess ion pumping capacity (strial/lateral wall/Reissners’ membrane). Although the organ of Corti hosts over a dozen cell types, only hair cells are routinely quantified. Some roles of Deiters and pillar cells are clear, although there are presently no known primary genetic pathologies of these cells. Of the other cell types, we have little sense of their unique purpose or redundancy of function. This is precisely why detailed examination of more inbred strains, and induced and spontaneous mutations in these strains, is still needed. Such studies will remain blind and non-conducive to specific hypotheses at the outset. And while molecular tools such as single-cell expression profiling and transgenic fluorescent labeling can be quite usefully applied, there can be no replacement for ABR/DPOAE testing and traditional histopathology. Sick cells will have altered gene and protein expression. High-quality histopathology remains the essential way to detect sick cells.
Age-associated hearing loss in mice, a complex trait that may be influenced by noise and ototoxins, involves a panoply of genes that tend to be related to oxidative stress, mitochondrial dysfunction, ion homeostasis, DNA repair, structural integrity, and protective processes (Table S3). Several human genes show specific overlap with known mouse genes or affect similar processes. Several mouse loci imparting age-associated hearing loss remain to be identified (Table S4).
Genetics of Vestibular Impairment Lag Genetics of Hearing Impairment
Considering the large number of genes and mouse models identified for human hearing loss (Table S2), it is striking that no genetic causes for non-syndromic peripheral vestibulopathies have been identified in humans (Eppsteiner and Smith 2011; Gazquez and Lopez-Escamez 2011; Jen 2011). A few central disturbances in balance or motor control such as episodic ataxia or spinocerebellar ataxia have been linked to neuronal genes KCNA1 and CACNA1 (Jen 2008; Gazquez and Lopez-Escamez 2011; Baloh 2012). Studies on peripheral vestibular disorders such as Menière’s disease or bilateral vestibular hypofunction have reported no definitive genetic linkage (Birgerson et al. 1987; Frykholm et al. 2006; Klockars and Kentala 2007)). Recently, one study of familial benign paroxysmal positional vertigo (BPPV) identified linkage on chromosome 15 (Gizzi et al. 2015), and a recently reported QTL on syntenic chromosome 9 in A/J mice was associated with a VsEP functional deficit coinciding with an otoconial abnormality (Vijayakumar et al. 2015). Whether these independent findings in humans and mice will coalesce around the same gene remains to be determined.
There are several reasons for the paucity of data on hereditary vestibular impairment. First, the inner ear vestibular system contains five organs: three ampullae and two gravity sensing organs. There is no single test that evaluates function of all five organs. Therefore, investigators must use a battery of assessments to fully evaluate the vestibular periphery or select those that are best suited to evaluate the organ(s) of interest. Limiting an assessment to a single end organ or subset of organs may miss deficits impacting untested organs. Second, the vast majority of tests for vestibular function evaluate the final motor output of control systems that maintain eye position or postural balance during movement. Such measures encompass the input, integration, and output components of nervous system function, making it more difficult to ascertain the functional status of the vestibular periphery (Löscher 2010; Jones and Jones 2014) and then correlate that function with other data such as genetic data. Third, relatively few investigators are systematically looking for vestibular dysfunction in human or animal genetic studies. Canal or gravity receptor functional assessment is often not part of routine phenotyping in suspected cases of hereditary abnormalities or broad-scale studies of genetic mouse mutants. Fourth, if peripheral vestibular impairment is present at birth or deficits appear gradually, the central nervous system likely compensates for the altered vestibular sensory input rendering any deficit undetectable via gross behavioral observations, VOR, VEMP, or other motor responses. In cases where motor delays, imbalance, or abnormal postural behaviors are apparent, canal or gravity receptor function is often not tested as a possible etiology so the functional status of the vestibular periphery remains unknown.
Relationship of Gene Expression with Auditory and Vestibular Function
Many gene transcripts and proteins have been shown to be expressed throughout the inner ear or prevalent in both auditory and vestibular sensory epithelia. One often assumes that any gene with demonstrated expression in the inner ear must play a crucial role in function and that more abundant expression suggests greater functional significance. Furthermore, it is often assumed that genes expressed in both auditory and vestibular sensory epithelia serve equally critical roles in both sensory modalities. In reality, the functional role for genes is more complex than can be predicted by expression levels or patterns. One example is cochlin (encoded by the gene COCH), which is the most abundant protein in the inner ear (Robertson et al. 2006). Based on that observation alone, one might hypothesize that mutations in COCH would result in a profound functional deficit. While COCH mutations do cause progressive sensorineural hearing loss (i.e., DFNA9) and progressive vestibular dysfunction, the severity of dysfunction at onset is relatively mild compared to the profound deficits that result from mutations in genes with less abundant or more restricted expression.
Summary
The genes, or gene types, that promote Mendelian and complex inner ear phenotypes show remarkable overlap between humans and mice, and both forward and reverse genetic studies of mouse genes and inner ear pathology remain invaluable. We believe that gene discovery in mice is not over, and the discovery potential for large-scale screening of commercial inbreds and mice with induced mutations is undiminished. Surprisingly few inbred and congenic lines with hearing and/or vestibular deficits are well characterized. This is particularly true with regard to the effects of noise and aging, so that our impressions of how mice are affected by these are presently based on a too-small sample tested under too few conditions. The value of reverse genetic experiments, including characterization of knockout, knock-in and transgenic mice, is enhanced if these are conducted on multiple inbred strain backgrounds. There is no neutral background and no universal phenotype caused by any allele. Outbred stocks should be avoided in studies of normal processes such as development, and in therapeutic trials, as genetic heterogeneity across animals and suppliers will obscure results and hamper reproducibility. Authors should be exacting and complete in their terminology for mouse strain and substrain. They should also provide complete information regarding mouse age and gender, as either can affect results.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
(XLSX 24 kb)
(DOCX 455 kb)
(DOCX 94 kb)
(DOC 84 kb)
Compliance with Ethical Standards
Funding Agencies
KKO: Washington University Medical School Department of Otolaryngology
SAJ: Nebraska Tobacco Settlement Biomedical Research Foundation
KRJ: NIH NIDCD RO1 DC004301, R01 DC005827
References
- Agrawal Y, Platz EA, Niparko JK. Risk factors for hearing loss in US adults: data from the National Health and Nutrition Examination Survey, 1999 to 2002. Otol Neurotol. 2009;30:139–145. doi: 10.1097/MAO.0b013e318192483c. [DOI] [PubMed] [Google Scholar]
- Aitkin LM, Irvine DRF, Webster W.R (1984) Central neural mechanisms of hearing. Comprehensive Physiology
- Al-Mana D, Ceranic B, Djahanbakhch O, Luxon LM. Hormones and the auditory system: a review of physiology and pathophysiology. Neurosci. 2008;153:881–900. doi: 10.1016/j.neuroscience.2008.02.077. [DOI] [PubMed] [Google Scholar]
- Altschuler RA, Raphael Y, Prosen C, Dolan DF, Moody DB. Acoustic stimulation and overstimulatoni in the cochlea: a comparison between basal and apical turns of the cochlea. In: Dancer AL, Henderson D, Salvi RJ, Hamernik RP, editors. Noise-Induced Hearing Loss. St. Louis: Mosby Year Book; 1992. pp. 60–73. [Google Scholar]
- Anniko M, Sobin A, Wersäll J. Vestibular hair cell pathology in the Shaker-2 mouse. Arch Otorhinolaryngol. 1980;226:45–50. doi: 10.1007/BF00455401. [DOI] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet. 1995;11:369–375. doi: 10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- Baker M. Through the eyes of a mouse. Nature. 2013;502:156–158. doi: 10.1038/502156a. [DOI] [PubMed] [Google Scholar]
- Baloh RW. Episodic ataxias 1 and 2. Handb Clin Neurol. 2012;103:595–602. doi: 10.1016/B978-0-444-51892-7.00042-5. [DOI] [PubMed] [Google Scholar]
- Barkdull GC, Hondarrague Y, Meyer T, Harris JP, Keithley EM. AM-111 reduces hearing loss in a guinea pig model of acute Labyrinthitis. Laryngoscope. 2007;117:2174–2182. doi: 10.1097/MLG.0b013e3181461f92. [DOI] [PubMed] [Google Scholar]
- Barrenäs M-L, Lindgren F. The influence of eye color on susceptibility to TTS in humans. Br J Audiol. 1991;25:303–307. doi: 10.3109/03005369109076602. [DOI] [PubMed] [Google Scholar]
- Bartels S, Ito S, Trune DR, Nuttall AL. Noise-induced hearing loss: the effect of melanin in the stria vascularis. Hear Res. 2001;154:116–123. doi: 10.1016/s0378-5955(01)00213-1. [DOI] [PubMed] [Google Scholar]
- Basta D, Tzschentke B, Ernst A. Noise-induced cell death in the mouse medial geniculate body and primary auditory cortex. Neurosci Lett. 2005;381:199–204. doi: 10.1016/j.neulet.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Battinelli EM, Boyd Y, Craig IW, Breakefield XO, Chen ZY. Characterization and mapping of the mouse NDP (Norrie disease) locus (Ndp) Mamm Genome. 1996;7:93–97. doi: 10.1007/s003359900026. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang W-P, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castallani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgerson L, Gustavson K, Stahle J. Familial Menière’s disease: a genetic investigation. Am J Otol. 1987;8:323–326. [PubMed] [Google Scholar]
- Birkenmeier EH, Davisson MT, Beamer WG, Ganschow RE, Vogler CA, Gwynn B, Lyford KA, Maltais LM, Wawrzyniak CJ. Murine mucopolysaccharidosis type VII: characterization of a mouse with a beta-glucuronidase deficiency. J Clin Invest. 1989;83:1258–1266. doi: 10.1172/JCI114010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizley JK, Nodal FR, Bajo VM, Nelken I, King AJ. Physiological and anatomical evidence for multisensory interactions in auditory cortex. Cereb Cortex. 2007;17:2172–2189. doi: 10.1093/cercor/bhl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa J, Przedborski, S (2016) Parkinson’s disease: animal models and dopaminergic cell vulnerability. Parkinson’s Disease: Cell Vulnerability and Disease Progression, p. 9 [DOI] [PMC free article] [PubMed]
- Bowl MR, Dawson SJ. The mouse as a model for age-related hearing loss-a mini-review. Gerontology. 2015;61:149–157. doi: 10.1159/000368399. [DOI] [PubMed] [Google Scholar]
- Bredberg G. Cellular pattern and nerve supply of the human organ of Corti. Acta Otolaryngol. 1968;236(Suppl):1–135. [PubMed] [Google Scholar]
- Brenowitz EA, Zakon HH. Emerging from the bottleneck: benefits of the comparative approach to modern neuroscience. TINS. 2015;38:273–278. doi: 10.1016/j.tins.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE, Mouse Genome Database Group Mouse genome database 2016. Nucleic Acids Res. 2016;44(D1):D840–D847. doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable J, Jackson IJ, Steel KP. Light (Blt), a mutation that causes melanocyte death, affects stria vascularis function in the mouse inner ear. Pigment Cell Res. 1993;6:215–225. doi: 10.1111/j.1600-0749.1993.tb00605.x. [DOI] [PubMed] [Google Scholar]
- Campi KL, Bales KL, Grunewald R, Krubitzer L. Connections of auditory and visual cortex in the prairie vole (Microtus ochrogaster): evidence for multisensory processing in primary sensory areas. Cereb Cortex. 2010;20:89–108. doi: 10.1093/cercor/bhp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caras ML. Estrogenic modulation of auditory processing: a vertebrate comparison. Front Neuroendocrinol. 2013;34:285–299. doi: 10.1016/j.yfrne.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson P-I, Van Laer L, Borg E, Bondeson M-L, Thys M, Fransen E, Van Camp G. The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear Res. 2005;202:87–96. doi: 10.1016/j.heares.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Chang B (2016) Animal Models of Retinitis Pigmentosa (RP). Animal Models of Ophthalmic Diseases. Springer International Publishing, pp. 101–116
- Chen CS. Acoustic trauma-induced developmental change in the acoustic startle response and audiogenic seizures in mice. Exp Neurol. 1978;60:400–403. doi: 10.1016/0014-4886(78)90094-8. [DOI] [PubMed] [Google Scholar]
- Chen G-D, Fechter LD. The relationship between noise-induced hearing loss and hair cell loss in rats. Hear Res. 2003;177:81–90. doi: 10.1016/s0378-5955(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Chen CS, Gates GR, Bock GR. Effect of priming and tympanic membrane destruction on development of audiogenic seizure susceptibility in BALBc mice. Exp Neurol. 1973;39:277–284. doi: 10.1016/0014-4886(73)90230-6. [DOI] [PubMed] [Google Scholar]
- Chen CS, Gates GR, Reynoldson JA. Effect of Morphine and Naloxone on Priming-Induced Audiogenic Seizures in BALB/c Mice. Br J Pharmacol. 1976;58:517–520. doi: 10.1111/j.1476-5381.1976.tb08618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat Genet. 2005;37:1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- Ciuman RR. Communication routes between intracranial spaces and inner ear: function, pathophysiologic importance and relations with inner ear diseases. Am J Otolaryngol. 2009;30:193–202. doi: 10.1016/j.amjoto.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Cohen GM, Park JC, Grasso JS. Comparison of demyelination and neural degeneration in spiral and Scarpa’s ganglion of C57BL/6 mice. J Elect Microsc Tech. 1990;15:165–172. doi: 10.1002/jemt.1060150208. [DOI] [PubMed] [Google Scholar]
- Conlee JW, Abdul-Baqi KJ, McCandless GA, Creel DJ. Differential susceptibility to noise-induced permanent threshold shift between albino and pigmented guinea pigs. Hear Res. 1986;23:81–91. doi: 10.1016/0378-5955(86)90177-2. [DOI] [PubMed] [Google Scholar]
- Conlee JW, Abdul-Baqi KJ, McCandless GA, Creel DJ. Effects of aging on normal hearing loss and noise-induced threshold shift in albino and pigmented guinea pigs. Acta Otolaryngol. 1988;106:64–70. doi: 10.3109/00016488809107372. [DOI] [PubMed] [Google Scholar]
- Conlee JW, Gill SS, McCandless PT, Creel DJ. Differential susceptibility to gentamicin ototoxicity between albino and pigmented guinea pigs. Hear Res. 1989;41:43–52. doi: 10.1016/0378-5955(89)90177-9. [DOI] [PubMed] [Google Scholar]
- Crawley JN. What’s Wrong with my Mouse? New York: Wiley and Sons; 2000. [Google Scholar]
- Creel D. Inappropriate use of albino animals as models in research. Pharmacol Biochem Behav. 1980;12:969–977. doi: 10.1016/0091-3057(80)90461-x. [DOI] [PubMed] [Google Scholar]
- Crow A.L, Ohmen J, Wang J, Lavinsky J, Hartiala J, Li Q, Li X, Salehide P, Eskin E, Pan C, Lusis AJ (2015) The genetic architecture of hearing impairment in mice: evidence for frequency specific genetic determinants. G3: Genes, Genomes, Genetics, g3-115 [DOI] [PMC free article] [PubMed]
- Cryan JF, Holmes A. The ascent of mouse: advances in modelling human depression and anxiety. Nat Rev Drug Discov. 2005;4:775–790. doi: 10.1038/nrd1825. [DOI] [PubMed] [Google Scholar]
- Da Costa DA, Castro JC, Macedo MEG. Iris pigmentation and susceptibility to noise-induced hearing loss. Int J Audiol. 2008;47:115–118. doi: 10.1080/14992020701704776. [DOI] [PubMed] [Google Scholar]
- Davis RR, Newlander JK, Ling X-B, Cortopassi GA, Kreig EF, Erway LC. Genetic basis for susceptibility to noise-induced hearing loss in mice. Hear Res. 2001;155:82–90. doi: 10.1016/s0378-5955(01)00250-7. [DOI] [PubMed] [Google Scholar]
- Davis RR, Custer DA, Krieg E, Alagramam K. N-Acetyl L-Cysteine does not protect mouse ears from the effects of noise. J Occ Med Toxicol. 2010;5:11. doi: 10.1186/1745-6673-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brouwer AP, Pennings RJ, Roeters M, Van Hauwe P, Astuto LM, Hoefsloot LH, Huygen PL, van den Helm B, Deutman AF, Bork JM, Kimberling WJ. Mutations in the calcium-binding motifs of CDH23 and the 35delG mutation in GJB2 cause hearing loss in one family. Hum Genet. 2003;112:156–163. doi: 10.1007/s00439-002-0833-0. [DOI] [PubMed] [Google Scholar]
- del Marmol V, Beermann F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 1996;381:165–168. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- Deol MS, Lane PW. A new gene affecting the morphogenesis of the vestibular part of the inner ear in the mouse. J Embryol Exp Morphol. 1966;16:543–548. [PubMed] [Google Scholar]
- Ding D-L, McFadden SL, Salvi RJ. Cochlear hair cell densities and inner ear staining techniques. In: Willott JF, editor. Handbook of mouse auditory research: from behavior to molecular biology. New York, NY: CRC Press; 2001. pp. 189–204. [Google Scholar]
- Dobson GP. On being the right size: heart design, mitochondrial efficiency and lifespan potential. Clin Exp Pharmacol Physiol. 2003;30:590–597. doi: 10.1046/j.1440-1681.2003.03876.x. [DOI] [PubMed] [Google Scholar]
- Dowdell KC, Pesnicak L, Hoffmann V, Steadman K, Remaley AT, Cohen JI, Straus SE, Rao VK. Valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, diminishes lymphoproliferation in the Fas-deficient MRL/lpr−/− murine model of autoimmune lymphoproliferative syndrome (ALPS) Exp Hematol. 2009;37:487–494. doi: 10.1016/j.exphem.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayton M, Noben-Trauth K. Mapping quantitative trait loci for hearing loss in Black Swiss mice. Hear Res. 2006;212:128–139. doi: 10.1016/j.heares.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68:293–308. doi: 10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Ehret G. Age-dependent hearing loss in normal hearing mice. Naturwissenschaften. 1974;61:506–507. doi: 10.1007/BF00622976. [DOI] [PubMed] [Google Scholar]
- Ehret G. Psychoacoustics. In: Willott JF, editor. The auditory psychobiology of the mouse. Spring field, Illinois: Charles C Thomas; 1983. pp. 13–56. [Google Scholar]
- Eppsteiner RW, Smith RJ. Genetic disorders of the vestibular system. Curr Opin Otolaryngol Head Neck Surg. 2011;19:397–402. doi: 10.1097/MOO.0b013e32834a9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erway LC, Willott JF, Archer JR, Harrison DE. Genetics of age-related hearing loss in mice: I. Inbred and F1 hybrid strains. Hear Res. 1993;65:125–132. doi: 10.1016/0378-5955(93)90207-h. [DOI] [PubMed] [Google Scholar]
- Farber DB, Flannery JG, Bowes-Rickman C. The rd mouse story: seventy years of research on an animal model of inherited retinal degeneration. Prog Retin Eye Res. 1994;13:31–64. [Google Scholar]
- Fernandez EA, Ohlemiller KK, Gagnon PM, Clark WW. Protection against noise-induced hearing loss in young CBA/J mice by low-dose kanamycin. J Assoc Res Otolaryngol. 2010;11:235–244. doi: 10.1007/s10162-009-0204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing MF. Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicol Pathol. 2010;38:681–690. doi: 10.1177/0192623310373776. [DOI] [PubMed] [Google Scholar]
- Feuerstein A, Herbst A, Wallner P. Another biomarker of susceptibility to noise induced hearing loss. Biomonitoring. 2014;1:1. [Google Scholar]
- Fransen E, Lemkens N, Van Laer L, Van Camp G. Age-related hearing impairment (ARHI): environmental risk factors and genetic prospects. Exp Gerontol. 2003;38:353–359. doi: 10.1016/s0531-5565(03)00032-9. [DOI] [PubMed] [Google Scholar]
- Fransen E, Bonneux S, Corneveaux JJ, Schrauwen I, Di Berardino F, White CH, Ohmen JD, Van de Heyning P, Ambrosetti U, Huentelman MJ, Van Camp G. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur J Hum Genet. 2015;23:110–115. doi: 10.1038/ejhg.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RA, Ryan AF. Transgenic mice. Current applications to the study of the auditory and vestibular systems. Otolaryngol Clin North Am. 1992;25:1017–1026. [PubMed] [Google Scholar]
- Frykholm C, Larsen H, Dahl N, Klar J, Rask-Andersen H, Friberg U. Familial Ménière’s disease in five generations. Otol Neurotol. 2006;27:681–686. doi: 10.1097/01.mao.0000226315.27811.c8. [DOI] [PubMed] [Google Scholar]
- Gagnon PM, Simmons DD, Bao J, Lei D, Ortmann AJ, Ohlemiller KK. Temporal and genetic influences on protection against noise-induced hearing loss by hypoxic preconditioning in mice. Hear Res. 2007;226:79–91. doi: 10.1016/j.heares.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazyuk A, Hébert S. Gap-prepulse inhibition of the acoustic startle reflex (GPIAS) for tinnitus assessment: current status and future directions. Front Neurol. 2015;6:88. doi: 10.3389/fneur.2015.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates GA, Couropmitree NN, Myers RH. Genetic associations in age-related hearing thresholds. Arch Otolaryngol Head Neck Surg. 1999;125:654–659. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- Gatti DM, Svenson KL, Shabalin A, Wu LY, Valdar W, Simecek P, Goodwin N, Cheng R, Pomp D, Palmer A, Chesler EJ. Quantitative trait locus mapping methods for diversity outbred mice. G3. 2014;4(9):1623–1633. doi: 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazquez I, Lopez-Escamez JA. Genetics of recurrent vertigo and vestibular disorders. Curr Genomics. 2011;12:443–450. doi: 10.2174/138920211797248600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geifman N, Rubin E. The mouse age phenome knowledgebase and disease-specific inter-species age mapping. PLoS One. 2013;8:e81114. doi: 10.1371/journal.pone.0081114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Sotomayor M, Kinder KJ, Gopal SR, Gerka-Stuyt J, Chen DHC, Hardisty-Hughes RE, Ball G, Parker A, Gaudet R, Furness D. Noddy, a mouse harboring a missense mutation in protocadherin-15, reveals the impact of disrupting a critical interaction site between tip-link cadherins in inner ear hair cells. J Neurosci. 2013;33:4395–4404. doi: 10.1523/JNEUROSCI.4514-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- Gilman JP, Medalla M, Luebke JI (2016) Area-Specific Features of Pyramidal Neurons-a Comparative Study in Mouse and Rhesus Monkey. Cereb. Cortex, p. bhw062 [DOI] [PMC free article] [PubMed]
- Giraudet F, Horner KC, Cazals Y. Similar half-octave TTS protection of the cochlea by xylazine/ketamine or sympathectomy. Hear Res. 2002;174:239–248. doi: 10.1016/s0378-5955(02)00698-6. [DOI] [PubMed] [Google Scholar]
- Girotto G, Vuckovic D, Buniello A, Lorente-Canovas B, Lewis M, Gasparini P, Steel KP. Expression and replication studies to identify new candidate genes involved in normal hearing function. PLoS One. 2014;9:e85352. doi: 10.1371/journal.pone.0085352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi MS, Peddareddygari LR, Grewal RP. A familial form of benign paroxysmal positional vertigo maps to chromosome 15. Int J Neurosci. 2015;125:593–596. doi: 10.3109/00207454.2014.953157. [DOI] [PubMed] [Google Scholar]
- Glueckert R, Pfaller K, Kinnefors A, Rask-Andersen H, Schrott-Fischer A. The human spiral ganglion: New insights into ultrastructure, survival rate, and implications for cochlear implants. Audiol Neurootol. 2005;10:258–273. doi: 10.1159/000086000. [DOI] [PubMed] [Google Scholar]
- Guimaraes P, Zhu X, Cannon T, Kim S, Frisina RD. Sex differences in distortion product otoacoustic emissions as a function of age in CBA mice. Hear Res. 2004;192:83–89. doi: 10.1016/j.heares.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Haack B, Markl H, Ehret G. Sound communication between parents and offspring. In: Willott JF, editor. The auditory psychobiology of the mouse. Spring field, Illinois: Charles C. Thomas; 1983. pp. 57–97. [Google Scholar]
- Hamernik RP, Patterson JH, Turrentine GA, Ahroon WA. The quantitative relation between sensory cell loss and hearing thresholds. Hear Res. 1989;38:199–212. doi: 10.1016/0378-5955(89)90065-8. [DOI] [PubMed] [Google Scholar]
- Hamilton LS, Sohl-Dickstein J, Huth AG, Carels VM, Deisseroth K, Bao S. Optogenetic activation of an inhibitory network enhances feedforward functional connectivity in auditory cortex. Neuron. 2013;80(4):1066–1076. doi: 10.1016/j.neuron.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM. Wild-derived mouse stocks: an underappreciated tool for aging research. Age. 2008;30(2–3):135–145. doi: 10.1007/s11357-008-9057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstierna C, Hultcrantz M, Collins A, Rosenhall U. The menopause triggers hearing decline in healthy women. Hear Res. 2010;259:31–35. doi: 10.1016/j.heares.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Heman-Ackah SE, Juhn SK, Huang TC, Wiedmann TS. A combination antioxidant therapy prevents age-related hearing loss in C57BL/6 mice. Otolaryngol Head Neck Surg. 2010;143:429–434. doi: 10.1016/j.otohns.2010.04.266. [DOI] [PubMed] [Google Scholar]
- Henley CM, Rybak LP. Ototoxicity in developing animals. Brain Res Rev. 1995;20:68–90. doi: 10.1016/0165-0173(94)00006-b. [DOI] [PubMed] [Google Scholar]
- Henry KR. Differential changes of auditory nerve and brain stem short latency evoked potentials in the laboratory mouse. Electroencephalogr Clin Neurophysiol. 1979;46:452–459. doi: 10.1016/0013-4694(79)90146-9. [DOI] [PubMed] [Google Scholar]
- Henry KR. Influence of genotype and age on noise-induced auditory losses. Behav Genet. 1982;12:563–573. doi: 10.1007/BF01070410. [DOI] [PubMed] [Google Scholar]
- Henry KR. Age-related changes in sensitivity of the postpubertal ear to acoustic trauma. Hear Res. 1982;8:285–294. doi: 10.1016/0378-5955(82)90020-x. [DOI] [PubMed] [Google Scholar]
- Henry KR. Ageing and audition. In: Willott JF, editor. The auditory psychobiology of the mouse. Springfield, Illinois: Charles C. Thomas; 1983. pp. 470–494. [Google Scholar]
- Henry KR. Lifelong susceptibility to acoustic trauma: changing patterns of cochlear damage over the life span of the mouse. Audiology. 1983;22:372–383. doi: 10.3109/00206098309072797. [DOI] [PubMed] [Google Scholar]
- Henry KR. Noise and the young mouse: genotype modifies the sensitive period for effects on cochlear physiology and audiogenic seizures. Behav Neurosci. 1984;98:1073–1082. doi: 10.1037//0735-7044.98.6.1073. [DOI] [PubMed] [Google Scholar]
- Henry KR. Tuning of the auditory brainstem OFF responses is complementary to tuning of the auditory brainstem ON response. Hear Res. 1985;19:115–125. doi: 10.1016/0378-5955(85)90115-7. [DOI] [PubMed] [Google Scholar]
- Henry KR. Detuning of cochlear action potential tuning curves at high sound pressure levels: influence of temporal, spectral and intensity variables. Audiology. 1989;28:19–36. doi: 10.3109/00206098909081608. [DOI] [PubMed] [Google Scholar]
- Henry KR. Hyperthermia exacerbates and hypothermia protects from noise-induced threshold elevation of the cochlear nerve envelope response in the C57BL/6J mouse. Hear Res. 2003;179:88–96. doi: 10.1016/s0378-5955(03)00097-2. [DOI] [PubMed] [Google Scholar]
- Henry KR. Males lose hearing earlier in mouse models of late-onset age-related hearing loss; Females lose hearing earlier in mouse models of early-onset hearing loss. Hear Res. 2004;190:141–148. doi: 10.1016/S0378-5955(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Henry KR, Chole RA. Cochlear electrical activity in the C57BL/6 laboratory mouse: volume-conducted vertex and round window response. Acta Otolaryngol. 1979;87:61–68. doi: 10.3109/00016487909126387. [DOI] [PubMed] [Google Scholar]
- Henry KR, Chole RA. Genotypic differences in behavioral, physiological and anatomical expressions of age-related hearing loss in the laboratory mouse. Audiology. 1980;19:369–383. doi: 10.3109/00206098009070071. [DOI] [PubMed] [Google Scholar]
- Henry KR, Haythorn MM. Effects of age and stimulus intensity of the far field auditory brain stem potentials in the laboratory mouse. Dev Psychobiol. 1978;11:161–168. doi: 10.1002/dev.420110208. [DOI] [PubMed] [Google Scholar]
- Henry KR, Lepkowski CM. Evoked potential correlates of genetic progressive hearing loss: age-related changes from the ear to the inferior colliculus of C57BL/6 and CBA/J mice. Acta Otolaryngol. 1978;86:366–374. doi: 10.3109/00016487809107515. [DOI] [PubMed] [Google Scholar]
- Henry KR, McGinn MD. The mouse as a model for human audition. A review of literature. Audiology. 1992;31:181–189. doi: 10.3109/00206099209081653. [DOI] [PubMed] [Google Scholar]
- Henry KR, Chole RA, McGinn MD, Frush DP. Increased ototoxicity in both young and old mice. Arch Otolaryngol. 1981;107:92–95. doi: 10.1001/archotol.1981.00790380022006. [DOI] [PubMed] [Google Scholar]
- Henry KR, Fast GA, Nguyen HH, Paolinelli MC, Ayars NM. Extra high-frequency auditory thresholds: fine structure, reliability, temporal integration and relation to ear canal resonance. Audiology. 1985;24:92–103. doi: 10.3109/00206098509081543. [DOI] [PubMed] [Google Scholar]
- Hequembourg S, Liberman MC. Spiral ligament pathology: a major aspect of age-related cochlear degeneration in C57BL/6 mice. J Assoc Res Otolaryngol. 2001;2:118–129. doi: 10.1007/s101620010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickox AE, Liberman MC. Is noise-induced cochlear neuropathy key to the generation of hyperacusis or tinnitus? J Neurophysiol. 2014;111:552–564. doi: 10.1152/jn.00184.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildesheimer M, Henkin Y, Muchnik C, Anafi R, Sahartov E, Rubinstein M. Sedation effect on temporary threshold shift induced by acoustic overstimulation. Hear Res. 1991;51:161–166. doi: 10.1016/0378-5955(91)90014-z. [DOI] [PubMed] [Google Scholar]
- Hirose K, Sato E. Comparative analysis of combination kanamycin-furosemide versus kanamycin alone in the mouse cochlea. Hear Res. 2011;272:108–116. doi: 10.1016/j.heares.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Bell J, Rasmussen E, McCaughran J. Chapter 21. Mapping the genes for the acoustic startle response (ASR) and prepulse inhibition of the ASR in the BxD recombinant inbred series: effect of high-frequency hearing loss and cochilear pathology. In: Willott JF, editor. Handbook of mouse auditory research: from behavior to molecular biology. Boca Raton: CRC Press; 2001. pp. 441–455. [Google Scholar]
- Hofman MA. Design principles of the human brain: an evolutionary perspective. Evolution of the primate brain: from neuron to behavior. London: Elsevier; 2012. pp. 373–390. [DOI] [PubMed] [Google Scholar]
- Hosoya M, Fujioka M, Ogawa K, Okano H (2016a). Distinct Expression Patterns of Causative Genes Responsible for Hereditary Progressive Hearing Loss in Non-Human Primate Cochlea. Sci. Rep. 6 [DOI] [PMC free article] [PubMed]
- Hosoya M, Fujioka M, Kobayashi R, Okano H, Ogawa K (2016b) Overlapping expression of anion exchangers in the cochlea of a non-human primate suggests functional compensation. Neurosci. Res [DOI] [PubMed]
- Huet A, Batrel C, Tang Y, Desmadryl G, Wang J, Puel JL, Bourien J (2016) Sound coding in the auditory nerve of gerbils. Hearing Res [DOI] [PubMed]
- Hughes AE, Newton VE, Liu XZ, Read AP. A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat Genet. 1994;7:509–512. doi: 10.1038/ng0894-509. [DOI] [PubMed] [Google Scholar]
- Hunter KP, Willott JF. Aging and the auditory brainstem response in mice with severe or minimal presbycusis. Hear Res. 1987;30:207–218. doi: 10.1016/0378-5955(87)90137-7. [DOI] [PubMed] [Google Scholar]
- Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Iwashita M, Kanai R, Funabiki K, Matsuda K, Hirano T. Dynamic properties, interactions and adaptive modifications of vestibulo-ocular reflex and optokinetic response in mice. Neurosci Res. 2001;39:299. doi: 10.1016/s0168-0102(00)00228-5. [DOI] [PubMed] [Google Scholar]
- Jackson IJ, Abbott CM, editors. Mouse genetics and transgenics: a practical approach. New York: Oxford University Press; 2000. [Google Scholar]
- Jeffery G. The albino retina: an abnormality that provides insight into normal retinal development. Trends Neurosci. 1997;20:165–169. doi: 10.1016/s0166-2236(96)10080-1. [DOI] [PubMed] [Google Scholar]
- Jen JC. Recent advances in the genetics of recurrent vertigo and vestibulopathy. Curr Opin Neurol. 2008;21:3–7. doi: 10.1097/WCO.0b013e3282f41ca0. [DOI] [PubMed] [Google Scholar]
- Jen JC. Genetics of vestibulopathies. Adv Otorhinolaryngol. 2011;70:130–134. doi: 10.1159/000322900. [DOI] [PubMed] [Google Scholar]
- Jensen JB, Lysaght AC, Liberman MC, Qvortrup K, Stankovic KM. Immediate and delayed cochlear neuropathy after noise exposure in pubescent mice. PLoS One. 2015;10:e0125160. doi: 10.1371/journal.pone.0125160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Erway LC, Cook SA, Willott JF, Zheng QY. A major gene affecting age-related hearing loss in C57BL/6J mice. Hear Res. 1997;114:83–92. doi: 10.1016/s0378-5955(97)00155-x. [DOI] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Erway LC. A major gene affecting age-related hearing loss is common to at least 10 inbred strains of mice. Genomics. 2000;70:171–180. doi: 10.1006/geno.2000.6377. [DOI] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Letts VA. Chapter 27. Genetic analysis of non-transgenic mouse mutations affecting ear morphology or function. In: Willott JF, editor. Handbook of mouse auditory research: from behavior to molecular biology. New York: CRC Press; 2001. pp. 401–428. [Google Scholar]
- Johnson KR, Longo-Guess CM, Gagnon LH. A QTL on Chr 5 modifies hearing loss associated with the fascin-2 variant of DBA/2J mice. Mamm Genome. 2015;26:338–347. doi: 10.1007/s00335-015-9574-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Jones SM. Short latency compound action potentials from mammalian gravity receptor organs. Hear Res. 1999;136:75–85. doi: 10.1016/s0378-5955(99)00110-0. [DOI] [PubMed] [Google Scholar]
- Jones SM, Jones TA. Genetics of peripheral vestibular dysfunction: lessons from mutant mouse strains. J Am Acad Audiol. 2014;25:289–301. doi: 10.3766/jaaa.25.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Erway LC, Bergstrom RA, Schimenti JC, Jones TA. Vestibular responses to linear acceleration are absent in otoconia-deficient C57BL/6JEi-het mice. Hear Res. 1999;135:56–60. doi: 10.1016/s0378-5955(99)00090-8. [DOI] [PubMed] [Google Scholar]
- Jones SM, Subramanian G, Avniel W, Guo Y, Burkard RF, Jones TA. Stimulus and recording variables and their effects on mammalian vestibular evoked potentials. J Neurosci Methods. 2002;118:23–31. doi: 10.1016/s0165-0270(02)00125-5. [DOI] [PubMed] [Google Scholar]
- Jones SM, Johnson KR, Yu H, Erway LC, Alagramam KN, Pollak N, Jones TA. A quantitative survey of gravity receptor function in mutant mouse strains. J Assoc Res Otolaryngol. 2005;6:297–310. doi: 10.1007/s10162-005-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Jones TA, Johnson KR, Yu H, Erway LC, Zheng QY. A comparison of vestibular and auditory phenotypes in inbred mouse strains. Brain Res. 2006;1091:40–46. doi: 10.1016/j.brainres.2006.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyothi V, Li M, Kilpatrick LA, Smythe NM, LaRue AC, Zhou D, Schulte BA, Schmiedt RA, Lang H. Unmyelinated auditory type I spiral ganglion neurons in congenic Ly5.1 mice. J Comp Neurol. 2010;518:3254–3271. doi: 10.1002/cne.22398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane KL, Longo-Guess CM, Gagnon LH, Ding D, Salvi RJ, Johnson KR. Genetic background effects on age-related hearing loss associated with Cdh23 variants in mice. Hear Res. 2012;283:80–88. doi: 10.1016/j.heares.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Zamani D, Tong L, Rubel EW, Ohlemiller KK, Hirose K, Warchol ME. Fractalkine signaling regulates macrophage recruitment into the cochlea and promotes the survival of spiral ganglion neurons after selective hair cell lesion. J Neurosci. 2015;35:15050–15061. doi: 10.1523/JNEUROSCI.2325-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa Y, Seki Y, Okumura K, Ohshiba Y, Miyasaka Y, Suzuki S, Ozaki M, Matsuoka K, Noguchi Y, Yonekawa H. Advantages of a mouse model for human hearing impairment. Exp Anim. 2012;61:85–98. doi: 10.1538/expanim.61.85. [DOI] [PubMed] [Google Scholar]
- Kim JU, Lee HJ, Kang HH, Shin JW, Ku SW, Ahn JH, Kim YJ, Chung JW. Protective effect of isoflurane anesthesia on noise‐induced hearing loss in mice. Laryngoscope. 2005;115:1996–1999. doi: 10.1097/01.mlg.0000180173.81034.4d. [DOI] [PubMed] [Google Scholar]
- Klockars T, Kentala E. Inheritance of Meniere’s disease in the Finnish population. Arch Otolaryngol Head Neck Surg. 2007;133:73–77. doi: 10.1001/archotol.133.1.73. [DOI] [PubMed] [Google Scholar]
- Kokjohn TA, Roher AE. Amyloid precursor protein transgenic mouse models and Alzheimer’s disease: understanding the paradigms, limitations, and contributions. Alzheimers Dement. 2009;5:340–347. doi: 10.1016/j.jalz.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraev A. Parallel universes of Black Six biology. Biol Direct. 2014;9:1–10. doi: 10.1186/1745-6150-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC. Acceleration of age-related hearing loss by early noise: evidence of a misspent youth. J Neurosci. 2006;26:2115–2123. doi: 10.1523/JNEUROSCI.4985-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC. Adding insult to injury: Cochlear nerve degeneration after ‘temporary’ noise-induced hearing loss. J Neurosci. 2009;29:14077–14085. doi: 10.1523/JNEUROSCI.2845-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulig J, Williot JF. Frequency difference limens of C57BL/6 and DBA/2mice: relationship to auditory neuronal response properties and hearing impairment. Hear Res. 1984;16:169–174. doi: 10.1016/0378-5955(84)90006-6. [DOI] [PubMed] [Google Scholar]
- Laemle LK, Strominger NL, Carpenter DO. Cross-modal innervation of primary visual cortex by auditory fibers in congenitally anophthalmic mice. Neurosci Lett. 2006;396:108–112. doi: 10.1016/j.neulet.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Land RB. Genetic and phenotypic relationships between ovulation rate and body weight in the mouse. Genet Res. 1970;15:171–182. doi: 10.1017/s0016672300001506. [DOI] [PubMed] [Google Scholar]
- Landegger LD, Psaltis D, Stankovic KM. Human audiometric thresholds do not predict specific cellular damage in the inner ear. Hear Res. 2016;335:83–93. doi: 10.1016/j.heares.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang H, Schulte BA, Zhou D, Smythe NM, Spicer SS, Schmiedt RA. Nuclear factor kB deficiency is associated with auditory nerve degeneration and increased noise-induced hearing loss. J Neurosci. 2006;26:3541–3550. doi: 10.1523/JNEUROSCI.2488-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Crow AL, Pan C, Wang J, Aaron KA, Ho MK, Li Q, Salehide P, Myint A, Monges-Hernadez M, Eskin E. Genome-wide association study identifies Nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS Genet. 2015;11:e1005094. doi: 10.1371/journal.pgen.1005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Calvez S, Avan P, Gilain L, Romand R. CD1 hearing-impaired mice. I: distortion product otoacoustic emission levels, cochlear function and morphology. Hear Res. 1998;120:37–50. doi: 10.1016/s0378-5955(98)00050-1. [DOI] [PubMed] [Google Scholar]
- Le Prell CG, Lobarinas E (2015) Strategies for Evaluating Antioxidant Efficacy in Clinical Trials Assessing Prevention of Noise-Induced Hearing Loss. In: Miller, J., Le Prell, C.G., (Eds.), Free Radicals in ENT Pathology. Springer International New York, pp. 163–192
- Le Prell CG, Dell S, Hemsley B, Hall JW, Campbell KCM, Antonelli PJ, Green GE, Miller JM, Guire K. Digital music exposure reliably induces temporary threshold shift in normal-hearing human subjects. Ear Hear. 2012;33:e44–e358. doi: 10.1097/AUD.0b013e31825f9d89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS. Influence of genotype and age on acute acoustic trauma and recovery in CBA/Ca and C57BL/6J mice. Acta Otolaryngol. 1992;112:956–967. doi: 10.3109/00016489209137496. [DOI] [PubMed] [Google Scholar]
- Li LY, Li YT, Zhou M, Tao HW, Zhang LI. Intracortical multiplication of thalamocortical signals in mouse auditory cortex. Nat Neurosci. 2013;16(9):1179–1181. doi: 10.1038/nn.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Kiang N-YS. Acoustic trauma in cats: cochlear pathology and auditory nerve activity. Acta Otolaryngol. 1978;358:1–63. [PubMed] [Google Scholar]
- Lin FR, Thorpe R, Gordon-Salant S, Ferrucci L. Hearing loss prevalence and risk factors among older adults in the United States. J Gerontol A Biol Sci Med Sci. 2011;66:582–590. doi: 10.1093/gerona/glr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Maas P, Chien W, Carey JP, Ferrucci L, Thorpe R. Association of skin color, race/ethnicity, and hearing loss among adults in the USA. J Assoc Res Otolaryngol. 2012;13:109–117. doi: 10.1007/s10162-011-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Edin F, Blom H, Magnusson P, Schrott-Fischer A, Glueckert R, Santi PA, Li H, Laurell, G, Rask-Andersen H (2016) Super-resolution structured illumination fluorescence microscopy of the lateral wall of the cochlea: the Connexin26/30 proteins are separately expressed in man. Cell Tiss. Res. 1–15 [DOI] [PubMed]
- Lobarinas E, Hayes SH, Allman BL. The gap-startle paradigm for tinnitus screening in animal models: limitations and optimization. Hear Res. 2013;295:150–160. doi: 10.1016/j.heares.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longenecker RJ, Galazyuk AV. Development of tinnitus in CBA/CaJ mice following sound exposure. J Assoc Res Otolaryngol. 2011;12:647–658. doi: 10.1007/s10162-011-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher W. Abnormal circling behavior in rat mutants and its relevance to model specific brain dysfunctions. Neurosci Biobehav Rev. 2010;34:31–49. doi: 10.1016/j.neubiorev.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Mahendrasingam S, MacDonald JA, Furness DN. Relative time course of degeneration of different cochlear structures in the CD/1 mouse model of accelerated aging. J Assoc Res Otolaryngol. 2011;12:437–453. doi: 10.1007/s10162-011-0263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji SSM, Miller KA, Williams LH, Andreasen L, Siboe M, Rose E, Bahlo M, Kuiper M, Dahl H-HM. Molecular pathogenesis of genetic and inherited diseases an ENU-induced mutation of Cdh23 causes congenital hearing loss, but no vestibular dysfunction, in mice. Am J Pathol. 2011;179:903–914. doi: 10.1016/j.ajpath.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden SL, Henselman LW, Zheng X-Y. Sex differences in auditory sensitivity of chinchillas before and after exposure to impulse noise. Ear Hear. 1999;20:164–174. doi: 10.1097/00003446-199904000-00007. [DOI] [PubMed] [Google Scholar]
- McGinn MD, Bean-Knudsen D, Ermel RW. Incidence of otitis media in CBA/J and CBA/CaJ mice. Hear Res. 1992;59:1–6. doi: 10.1016/0378-5955(92)90094-4. [DOI] [PubMed] [Google Scholar]
- Meltser I, Cederroth CR, Basinou V, Savelyev S, Lundkvist GS, Canlon B. TrkB-mediated protection against circadian sensitivity to noise trauma in the murine cochlea. Curr Biol. 2014;24:658–663. doi: 10.1016/j.cub.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mianné J, Chessum L, Kumar S, Aguilar C, Codner G, Hutchison M, Parker A, Mallon AM, Wells S, Simon MM, Teboul L. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med. 2016;8:1. doi: 10.1186/s13073-016-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian DO. The development and degeneration of hearing in the C57/bl6 mouse: relation of the electrophysiologic responses from the round window to cochlear anatomy and behavioral responses. Laryngoscope. 1979;89:1–15. doi: 10.1288/00005537-197901000-00001. [DOI] [PubMed] [Google Scholar]
- Mikaelian DO, Warfield D, Norris O. Genetic progressive hearing loss in the C57b16 mouse. Acta Otolaryngol. 1974;77:327–334. doi: 10.3109/00016487409124632. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harper JM, Dysko RC, Durkee SJ, Austad SN. Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med. 2002;227(7):500–508. doi: 10.1177/153537020222700715. [DOI] [PubMed] [Google Scholar]
- Mills JH. Threshold shifts produced by exposure to noise in chinchillas with noise-induced hearing loss. J Speech Hear Res. 1973;16:700–708. doi: 10.1044/jshr.1604.700. [DOI] [PubMed] [Google Scholar]
- Mock BE, Vijayakuma S, Pierce J, Jones TA, Jones SM (2016) Differential effects of Cdh23753A on auditory and vestibular functional aging in C57BL/6J mice. Neurobiol Aging 43:13–22 [DOI] [PMC free article] [PubMed]
- Moore AK, Wehr M. Parvalbumin-expressing inhibitory interneurons in auditory cortex are well-tuned for frequency. J Neurosci. 2013;33(34):13713–13723. doi: 10.1523/JNEUROSCI.0663-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshammer H, Kundi M, Wallner P, Herbst A, Feuerstein A, Hutter HP. Early prognosis of noise-induced hearing loss. Occup Environ Med. 2015;72:85–89. doi: 10.1136/oemed-2014-102200. [DOI] [PubMed] [Google Scholar]
- Müller U, Barr-Gillespie PG. New treatment options for hearing loss. Nat Rev Drug Discov. 2015;14:346–365. doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- Muller M, von Hunerbein K, Hoidis S, Smolders JWT. A physiological place-frequency map of the cochlea in the CBA/J mouse. Hear Res. 2005;202:63–73. doi: 10.1016/j.heares.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Myint A, White CH, Ohmen JD, Li X, Wang J, Lavinsky J, Salehi P, Crow AL, Ohyama T, Friedman RA. Large-scale phenotyping of noise-induced hearing loss in 100 strains of mice. Hear Res. 2016;332:113–120. doi: 10.1016/j.heares.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagtegaal AP, Spijker S, Crins TTH, Borst JGG. A novel QTL underlying early-onset, low‐frequency hearing loss in BXD recombinant inbred strains. Genes Brain Behav. 2012;11:911–920. doi: 10.1111/j.1601-183X.2012.00845.x. [DOI] [PubMed] [Google Scholar]
- Nemoto M, Morita Y, Mishima Y, Takahashi S, Nomura T, Ushiki T, Shiroishi T, Kikkawa Y, Yonekawa H, Kominami R. Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochem Biophys Res Commun. 2004;324:1283–1288. doi: 10.1016/j.bbrc.2004.09.186. [DOI] [PubMed] [Google Scholar]
- Nickel AG, von Hardenberg A, Hohl M, Löffler JR, Kohlhaas M, Becker J, Reil JC, Kazakov A, Bonnekoh J, Stadelmaier M, Puhl SL. Reversal of mitochondrial transhydrogenase causes oxidative stress in heart failure. Cell Metab. 2015;22:472–484. doi: 10.1016/j.cmet.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Latoche JR, Neely HR, Bennett B. Phenotype and genetics of progressive sensorineural hearing loss (Snhl1) in the LXS set of recombinant inbred strains of mice. PLoS One. 2010;5:e11459. doi: 10.1371/journal.pone.0011459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol. 2008;9:65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. Reduction in sharpness of frequency tuning but not endocochlear potential in aging and noise-exposed BALB/cJ mice. J Assoc Res Otolaryngol. 2002;3:444–456. doi: 10.1007/s10162-002-2041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. Contributions of mouse models to understanding of age- and noise-related hearing loss. Brain Res. 2006;1091:89–102. doi: 10.1016/j.brainres.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK. Mechanisms and genes in human strial presbycusis from animal models. Brain Res. 2009;1277:70–83. doi: 10.1016/j.brainres.2009.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. Chapter 1: current issues in noise exposure. In: Tremblay KL, Burkard RF, editors. Translational Perspectives in Hearing Science, Special Topics. San Diego: Plural Publishing; 2012. pp. 1–34. [Google Scholar]
- Ohlemiller KK. Gene/environment interactions in acquired hearing loss. In: Toriello HSS, editor. Hereditary hearing loss and its syndromes. New York: Oxford Press; 2013. pp. 58–84. [Google Scholar]
- Ohlemiller KK. Chapter 3: a question of balance: free radicals in inner ear homeostasis. In: Miller J, Le Prell CG, editors. Free radicals in ENT medicine. New York: Springer; 2015. pp. 21–55. [Google Scholar]
- Ohlemiller KK, Gagnon PM. Genetic dependence of cochlear cells and structures injured by noise. Hear Res. 2007;224:34–50. doi: 10.1016/j.heares.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Siegel JH. Cochlear basal and apical differences reflected in the effects of cooling on responses of single auditory nerve fibers. Hear Res. 1994;80:174–190. doi: 10.1016/0378-5955(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Wright JS, Heidbreder AF. Vulnerability to noise-induced hearing loss in ‘middle-aged’ and young adult mice: a dose-response approach in CBA, C57BL, and BALB inbred strains. Hear Res. 2000;149:239–247. doi: 10.1016/s0378-5955(00)00191-x. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Lett JM, Gagnon PM. Cellular correlates of age-related endocochlear potential reduction in a mouse model. Hear Res. 2006;220:10–26. doi: 10.1016/j.heares.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Rice MR, Lett JM, Gagnon PM. Absence of strial melanin coincides with age associated marginal cell loss and endocochlear potential decline. Hear Res. 2009;249:1–14. doi: 10.1016/j.heares.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Dahl AR, Gagnon PM. Divergent aging characteristics in CBA/J and CBA/CaJ mouse cochleae. J Assoc Res Otolaryngol. 2010;11:605–623. doi: 10.1007/s10162-010-0228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Rybak Rice ME, Rellinger EA, Ortmann AJ. Divergence of noise vulnerability in cochleae of young CBA/J and CBA/CaJ mice. Hear Res. 2011;272:13–20. doi: 10.1016/j.heares.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Rybak Rice ME, Rosen AD, Montgomery SC, Gagnon PM. Protection by low-dose Kanamycin against noise-induced hearing loss in mice: dependence on dosing regimen and genetic background. Hear Res. 2011;280:141–147. doi: 10.1016/j.heares.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Rosen AR, Rellinger EA, Montgomery SC, Gagnon PM. Different cellular and genetic basis of noise-related endocochlear potential reduction in CBA/J and BALB/cJ mice. J Assoc Res Otolaryngol. 2011;12:45–58. doi: 10.1007/s10162-010-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Kiener AL, Gagnon PM. QTL Mapping of Endocochlear Potential Differences between C57BL/6J and BALB/cJ mice. J Assoc Res Otolaryngol. 2016;17:173–194. doi: 10.1007/s10162-016-0558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HC, Bohne BA, Harding GW. Noise damage in the C57BL/CBA mouse cochlea. Hear Res. 2000;145:111–122. doi: 10.1016/s0378-5955(00)00081-2. [DOI] [PubMed] [Google Scholar]
- Ou HC, Harding GW, Bohne BA. An anatomically based frequency-place map for the mouse cochlea. Hear Res. 2000;145:123–129. doi: 10.1016/s0378-5955(00)00082-4. [DOI] [PubMed] [Google Scholar]
- Paquette ST, Gilels F, White PM. Noise exposure modulates cochlear inner hair cell ribbon volumes, correlating with changes in auditory measures in the FVB/nJ mouse. Sci Rep. 2016;6:25056. doi: 10.1038/srep25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Palmer AA. Dark matter: are mice the solution to missing heritability? Front Genet. 2011;2:32. doi: 10.3389/fgene.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings RJ, Topsakal V, Astuto L, de Brouwer AP, Wagenaar M, Huygen PL, Kimberling WJ, Deutman AF, Kremer H, Cremers CW. Variable clinical features in patients with CDH23 mutations (USH1D-DFNB12) Otol Neurotol. 2004;25:699–706. doi: 10.1097/00129492-200409000-00009. [DOI] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo DM, El-Danaf RN, Huberman AD, Niell CM. Diverse visual features encoded in mouse lateral geniculate nucleus. J Neurosci. 2013;33:4642–4656. doi: 10.1523/JNEUROSCI.5187-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirrier AL, Van den Ackerveken P, Kim T-S, Vandenbosch R, Nguyen L, Lefebvre PP, Malgrange B. Ototoxic drugs: difference in sensitivity between mice and guinea pigs. Toxicol Lett. 2010;193:41–49. doi: 10.1016/j.toxlet.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat Rev Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- Price K, Zhu X, Guimaraes PF, Vasilyeva ON, Frisina RD. Hormone replacement therapy diminishes hearing in peri-menopausal mice. Hear Res. 2009;252:29–36. doi: 10.1016/j.heares.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieve BA, Yanz JL. Age-dependent changes in susceptibility to otoxic hearing loss. Acta Otolaryngol. 1984;98:428–438. doi: 10.3109/00016488409107584. [DOI] [PubMed] [Google Scholar]
- Pujol R. Sensitive developmental period and acoustic trauma: facts and hypotheses. In: Dancer AL, editor. Noise-induced hearing loss. St. Louis: Mosby; 1992. pp. 196–203. [Google Scholar]
- Qiu W, Hamernik RP, Davis B. The kurtosis metric as an adjunct to energy in the prediction of trauma from continuous, nonGaussian noise exposure. J Acoust Soc Am. 2006;120:3901–3906. doi: 10.1121/1.2372455. [DOI] [PubMed] [Google Scholar]
- Rauch SD. Malformation and degeneration in the inner ear of mos transgenic mice. Ann Otol Rhinol Laryngol. 1992;101:430–436. doi: 10.1177/000348949210100510. [DOI] [PubMed] [Google Scholar]
- Rellinger EA, Gagnon PM, Ohlemiller KK (2012) Eliminating catalase paradoxically reduces age- and noise-associated threshold elevation in C57BL/6 mice. Abstr., Assn. Res. Otolaryngol. 35, 330
- Riley PA. Molecules in focus: melanin. Int J Biochem Cell Biol. 1997;11:1235–1239. doi: 10.1016/s1357-2725(97)00013-7. [DOI] [PubMed] [Google Scholar]
- Riva C, Donadieu E, Magnan J, Lavieille JP. Age-related hearing loss in CD/1 mice is associated to ROS formation and HIF target proteins up-regulation in the cochlea. Exp Gerontol. 2007;42:327–336. doi: 10.1016/j.exger.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Rubinstein M, Pluznik N. Effect of anesthesia on susceptibility to acoustic trauma. Ann Otol Rhinol Laryngol. 1976;85:276–280. doi: 10.1177/000348947608500213. [DOI] [PubMed] [Google Scholar]
- Rudnicki A, Avraham KB. microRNAs: the art of silencing in the ear. EMBO Mol Med. 2012;4:849–859. doi: 10.1002/emmm.201100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt AN, Plontke SK. Principles of local drug delivery to the inner ear. Audiol Neurootol. 2009;14:350–360. doi: 10.1159/000241892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi PA, Aldaya R, Brown A, Johnson S, Stromback T, Cureoglu S, Rask-Andersen H (2016) Scanning Electron Microscopic Examination of the Extracellular Matrix in the Decellularized Mouse and Human Cochlea. J. Assoc. Res. Otolaryngol [DOI] [PMC free article] [PubMed]
- Saunders JC. The physiological effects of priming for audiogenic seizures in mice. Laryngoscope. 1974;84:750–756. doi: 10.1288/00005537-197405000-00006. [DOI] [PubMed] [Google Scholar]
- Saunders JC, Garfinkle TJ. Peripheral anatomy and physiology I. In: Willott JF, editor. The auditory psychobiology of the mouse. Springfield, Illinois: Charles C. Thomas; 1983. pp. 131–168. [Google Scholar]
- Saunders JC, Bock GR, Chen CS, Gates GR. The effects of priming for audiogenic seizures on cochlear and behavioral responses in BALB/c mice. Exp Neurol. 1972;36:426–436. doi: 10.1016/0014-4886(72)90003-9. [DOI] [PubMed] [Google Scholar]
- Schraermeyer U, Heimann K. Current understanding on the role of retinal pigment epithelium and its pigmentation. Pigment Cell Res. 1999;12:219–236. doi: 10.1111/j.1600-0749.1999.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Schreiner L. Recent experimental and clinical findings retarding an interlabyrinthine connection. Laryngo-Rhino-Otologie. 1999;78:387–393. doi: 10.1055/s-2007-996893. [DOI] [PubMed] [Google Scholar]
- Schreiner CE, Winer JA. Auditory cortex mapmaking: principles, projections, and plasticity. Neuron. 2007;56:356–365. doi: 10.1016/j.neuron.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schughart K, Libert C, Kas MJ. Controlling complexity: the clinical relevance of mouse complex genetics. Eur J Hum Genet. 2013;21:1191–1196. doi: 10.1038/ejhg.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuknecht HF. Further observations on the pathology of presbycusis. Arch Otolaryngol. 1964;80:369–382. doi: 10.1001/archotol.1964.00750040381003. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF, Gacek MR. Cochlear pathology in presbycusis. Ann Otol Rhinol Laryngol. 1993;102:1–16. doi: 10.1177/00034894931020S101. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF, Watanuki K, Takahashi T, Belal AA, Kimura RS, Jones DD. Atrophy of the stria vascularis, a common cause for hearing loss. Laryngoscope. 1974;84:1777–1821. doi: 10.1288/00005537-197410000-00012. [DOI] [PubMed] [Google Scholar]
- Schultz JM, Yang Y, Caride AJ, Filoteo AG, Penheiter AR, Lagziel A, Morell RJ, Mohiddin SA, Fananapazir L, Madeo AC, Penniston JT. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. NEJM. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- Schwander M, Xiong W, Tokita J, Lelli A, Elledge HM, Kazmierczak P, Sczaniecka A, Kolatkar A, Wiltshire T, Kuhn P, Holt JR, Kachar B, Tarantino L, Müller U. A mouse model for nonsyndromic deafness (DFNB2) links hearing loss to defects in tip links of mechanosensory hair cells. Proc Natl Acad Sci U S A. 2009;106:5252–5257. doi: 10.1073/pnas.0900691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Smith RJ. Genetics: advances in genetic testing for deafness. Curr Opin Pediatr. 2012;24:679. doi: 10.1097/MOP.0b013e3283588f5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheykholeslami K, Megerian CA, Zheng QY. Vestibular evoked myogenic potentials in normal mice and Phex mice with spontaneous endolymphatic hydrops. Otol Neurotol. 2009;30:535–544. doi: 10.1097/MAO.0b013e31819bda13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Liu K, Wang H, Zhang Y, Hong Z, Wang M, Wang X, Jiang X, Yang S. Noise induced reversible changes of cochlear ribbon synapses contribute to temporary hearing loss in mice. Acta Otolaryngol. 2015;135:1093–1102. doi: 10.3109/00016489.2015.1061699. [DOI] [PubMed] [Google Scholar]
- Shi L, Chang Y, Li X, Aiken SJ, Liu L, Wang J. Coding deficits in noise-induced hidden hearing loss may stem from incomplete repair of ribbon synapses in the cochlea. Front Neurosci. 2016;10:231. doi: 10.3389/fnins.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga A, Nakagawa T, Nakayama M, Endo T, Iguchi F, Kim TS, Naito Y, Ito J. Aging effects on vestibulo-ocular responses in C57BL/6 mice: comparison with alteration in auditory function. Audiol Neurootol. 2005;10:97–104. doi: 10.1159/000083365. [DOI] [PubMed] [Google Scholar]
- Shin JB, Longo-Guess CM, Gagnon LH, Saylor KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL, Gillespie PG, Johnson KR. The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J Neurosci. 2010;30:9683–9694. doi: 10.1523/JNEUROSCI.1541-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnerson A, Pujol R. Age-related changes in the C57BL/6J mouse cochlea. I. Physiological findings. Dev Brain Res. 1982;2:65–75. doi: 10.1016/0165-3806(81)90059-6. [DOI] [PubMed] [Google Scholar]
- Shnerson A, Willott JF. Ontogeny of the acoustic startle response in C57BL/6J mouse pups. J Comp Physiol Psychol. 1980;94:36–40. doi: 10.1037/h0077648. [DOI] [PubMed] [Google Scholar]
- Shone G, Raphael Y, Miller JM. Hereditary deafness occurring in cd/1 mice. Hear Res. 1991;57:153–156. doi: 10.1016/0378-5955(91)90084-m. [DOI] [PubMed] [Google Scholar]
- Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, Muller U. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- Silver LM. Mouse genetics. Oxford: Oxford Press; 1995. [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Skarnes WC. Is mouse embryonic stem cell technology obsolete? Genome Biol. 2015;16:1. doi: 10.1186/s13059-015-0673-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, Ephraim SS, Shibata SB, Booth KT, Campbell CA, Ranum PT. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Shen P, Li X, Shi L, Liu L, Wang J, Yu Z, Stephen K, Aiken S, Yin S, Wang J. Coding deficits in hidden hearing loss induced by noise: the nature and impacts. Sci Rep. 2016;6:25200. doi: 10.1038/srep25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soons JA, Ricci AJ, Steele CR, Puria S. Cytoarchitecture of the mouse organ of Corti from base to apex, determined using in situ two-photon imaging. J Assoc Res Otolaryngol. 2015;16:47–66. doi: 10.1007/s10162-014-0497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spongr VP, Flood DG, Frisina RD, Salvi RJ. Quantitative measures of hair cell loss in CBA and C57BL/6 mice throughout their life span. J Acoust Soc Am. 1997;101:3546–3553. doi: 10.1121/1.418315. [DOI] [PubMed] [Google Scholar]
- Steel KP. Similarities between mice and humans with hereditary deafness. Ann N Y Acad Sci. 1991;630:69–79. doi: 10.1111/j.1749-6632.1991.tb19576.x. [DOI] [PubMed] [Google Scholar]
- Steel KP. Inherited hearing defects in mice. Annu Rev Genet. 1995;29:675–701. doi: 10.1146/annurev.ge.29.120195.003331. [DOI] [PubMed] [Google Scholar]
- Steel KP. What’s the use of genetics? Perspectives on auditory research. New York: Springer; 2014. pp. 569–584. [Google Scholar]
- Steel KP, Barkway C. Another role for melanocytes: their importance for normal stria vascularis development in the mammalian inner ear. Development. 1989;107:453–463. doi: 10.1242/dev.107.3.453. [DOI] [PubMed] [Google Scholar]
- Steel KP, Kimberling W. Approaches to understanding the molecular genetics of hearing and deafness. In: Van De Water T, Popper AN, Fay RR, editors. Clinical aspects of hearing. New York: Springer; 1996. pp. 10–40. [Google Scholar]
- Steel KP, Smith RJ. Normal hearing in Splotch (Sp/+), the mouse homologue of Waardenburg syndrome type 1. Nat Genet. 1992;2:75–79. doi: 10.1038/ng0992-75. [DOI] [PubMed] [Google Scholar]
- Steel K, Niaussat MM, Bock GR. The genetics of hearing. In: Willott JF, editor. The auditory psychobiology of the mouse. Springfield, Illinois: Charles C. Thomas; 1983. pp. 341–394. [Google Scholar]
- Steel KP, Barkway C, Bock GR. Strial dysfunction in mice with cochleo-saccular abnormalities. Hear Res. 1987;27:11–26. doi: 10.1016/0378-5955(87)90022-0. [DOI] [PubMed] [Google Scholar]
- Stevens JC, Banks GT, Festing MF, Fisher EM. Quiet mutations in inbred strains of mice. Trends Mol Med. 2007;13:512–519. doi: 10.1016/j.molmed.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed FA, Mödder UI, Roforth M, Hensen I, Fraser DG, Peterson JM, Oursler MJ, Khosla S. Effects of chronic estrogen treatment on modulating age‐related bone loss in female mice. J Bone Miner Res. 2010;25:2438–2446. doi: 10.1002/jbmr.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberner AM, Liberman MC. Response properties of single auditory nerve fibers in the mouse. J Neurophysiol. 2005;93:557–569. doi: 10.1152/jn.00574.2004. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Leverton K, Turnbull K, Seemanova E, Kunze J, Sperling K, Strachan T, Read AP. PAX3 gene structure and mutations: close analogies between Waardenburg syndrome and the Splotch mouse. Hum Mol Genet. 1994;3:1069–1074. doi: 10.1093/hmg/3.7.1069. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the collaborative cross. G3. 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Larsen D, Hughes L, Moechars D, Shore S. Time course of tinnitus development following noise exposure in mice. J Neurosci Res. 2012;90:1480–1488. doi: 10.1002/jnr.22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylstedt S, Rask-Andersen H. A 3-D model of membrane specializations between human auditory spiral ganglion cells. J Neurocytol. 2001;30:465–473. doi: 10.1023/a:1015628831641. [DOI] [PubMed] [Google Scholar]
- Tylstedt S, Kinnefors A, Rask-Andersen H. Neural interaction in the human spiral ganglion: a TEM study. Acta Otolaryngol. 1997;117:505–512. doi: 10.3109/00016489709113429. [DOI] [PubMed] [Google Scholar]
- Uhl EW, Warner NJ. Mouse models as predictors of human responses: evolutionary medicine. Curr Pathobiol Rep. 2015;3:219–223. doi: 10.1007/s40139-015-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen AM, Stahl JS, De Zeeuw CI. The dynamic characteristics of the mouse horizontal vestibulo-ocular and optokinetic response. Brain Res. 2001;890:296–305. doi: 10.1016/s0006-8993(00)03180-2. [DOI] [PubMed] [Google Scholar]
- Van Eyken E, Van Camp G, Van Laer L. The complexity of age-related hearing impairment: contributing environmental and genetic factors. Audiol Neurootol. 2007;12:345–358. doi: 10.1159/000106478. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Carlsson PI, Ottschytsch N, Bondeson M-L, Konings A, Vandevelde A, Dieltjens N, Fransen E, Snyders D, Borg E, Raes A, Van Camp G. The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Hum Mutat. 2006;27:786–795. doi: 10.1002/humu.20360. [DOI] [PubMed] [Google Scholar]
- van Stahl JS, Alphen AM, De Zeeuw CI. A comparison of video and magnetic search coil recordings of mouse eye movements. J Neurosci Methods. 2000;99:101–110. doi: 10.1016/s0165-0270(00)00218-1. [DOI] [PubMed] [Google Scholar]
- Vanhooren V, Libert C. The mouse as a model organism in aging research: usefulness, pitfalls and possibilities. Ageing Res Rev. 2013;12:8–21. doi: 10.1016/j.arr.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, Thurman RE. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar S, Lever TE, Pierce J, Zhao X, Bergstrom D, Lundberg YW, Jones TA, Jones SM. Vestibular dysfunction, altered macular structure, and trait localization in A/J inbred mice. Mamm Genome. 2015;26:154–172. doi: 10.1007/s00335-015-9556-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hirose K, Liberman MC. Dynamics of noise-induced cellular injury and repair in the mouse cochlea. J Assoc Res Otolaryngol. 2002;3:248–268. doi: 10.1007/s101620020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney G, Nyby J. Sound communication among adults. In: Willott JF, editor. The auditory psychobiology of the mouse. Springfield, Illinois: Charles C. Thomas; 1983. pp. 98–130. [Google Scholar]
- Willott JF. Changes in frequency representation in the auditory system of mice with age-related hearing impairment. Brain Res. 1984;309:159–162. doi: 10.1016/0006-8993(84)91022-9. [DOI] [PubMed] [Google Scholar]
- Willott JF. Effects of aging, hearing loss, and anatomical location on thresholds of inferior colliculus neurons in C57BL/6 and CBA mice. J Neurophysiol. 1986;56:391–408. doi: 10.1152/jn.1986.56.2.391. [DOI] [PubMed] [Google Scholar]
- Willott JF, Bross LS. Morphology of the octopus cell area of the cochlear nucleus in young and aging C57BL/6J and CBA/J mice. J Comp Neurol. 1990;300:61–81. doi: 10.1002/cne.903000106. [DOI] [PubMed] [Google Scholar]
- Willott JF, Shnerson A. Rapid development of tuning characteristics of inferior colliculus neurons of mouse pups. Brain Res. 1978;148:230–333. doi: 10.1016/0006-8993(78)90395-5. [DOI] [PubMed] [Google Scholar]
- Willott JF, Demuth RM, Lu SM, Van Bergem P. Abnormal tonotopic organization in the ventral cochlear nucleus of the hearing-impaired DBA/2 mouse. Neurosci Lett. 1982;34:13–17. doi: 10.1016/0304-3940(82)90085-4. [DOI] [PubMed] [Google Scholar]
- Willott JF, Hunter KP, Coleman JR. Aging and presbycusis: effects on 2-deoxy-D-glucose uptake in the mouse auditory brain stem in quiet. Exp Neurol. 1988;99:615–621. doi: 10.1016/0014-4886(88)90178-1. [DOI] [PubMed] [Google Scholar]
- Willott JF, Turner JG, Carlson S, Ding D, Bross LS, Falls WA. The BALB/c mouse as an animal model for progressive sensorineural hearing loss. Hear Res. 1998;115:162–174. doi: 10.1016/s0378-5955(97)00189-5. [DOI] [PubMed] [Google Scholar]
- Wolff D. Melanin in the inner ear. Arch Otolaryngol. 1931;14:195–211. [Google Scholar]
- Wright CG, Lee DH. Pigmented cells of the stria vascularis. Acta Otolaryngol. 1989;108:190–200. doi: 10.3109/00016488909125518. [DOI] [PubMed] [Google Scholar]
- Wu T, Marcus DC. Age-related changes in cochlear endolymphatic potassium and potential in CD-1 and CBA/CaJ mice. J Assoc Res Otolaryngol. 2003;4:353–362. doi: 10.1007/s10162-002-3026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W-J, Sha S, McLaren JD, Kawamoto K, Raphael Y, Schacht J. Aminoglycoside ototoxicity in adult CBA, C57BL, and BALB mice and the Sprague-Dawley rat. Hear Res. 2001;158:165–178. doi: 10.1016/s0378-5955(01)00303-3. [DOI] [PubMed] [Google Scholar]
- Xia L, Chen Z, Su K, Yin S, Wang J. Comparison of cochlear cell death caused by cisplatin, alone and in combination with furosemide. Toxicol Pathol. 2014;42:376–385. doi: 10.1177/0192623313483213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Flint J. Association studies in outbred mice in a new era of full-genome sequencing. Mamm Genome. 2012;23:719–726. doi: 10.1007/s00335-012-9409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerkes RM. The dancing mouse: a study in animal behavior. New York: MacMillan; 1904. [Google Scholar]
- Yoshida N, Liberman MC. Sound conditioning reduces noise-induced permanent threshold shift in mice. Hear Res. 2000;148:213–219. doi: 10.1016/s0378-5955(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Kristiansen A, Liberman MC. Heat stress and protection from permanent acoustic injury in mice. J Neurosci. 1999;19:10116–10124. doi: 10.1523/JNEUROSCI.19-22-10116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R, Tsaih SW, Petkova SB, Evsikova D, Marin C, Xing S, Marion MA, Bogue MA, Mills KD, Peters LL, Bult CJ. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8:277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B, Mittal R, Grati MH, Lu Z, Shu Y, Tao Y, Feng Y, Xie D, Kong W, Yang S, Chen ZY. The application of genome editing in studying hearing loss. Hear Res. 2015;327:102–108. doi: 10.1016/j.heares.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 24 kb)
(DOCX 455 kb)
(DOCX 94 kb)
(DOC 84 kb)