Abstract
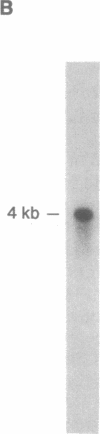
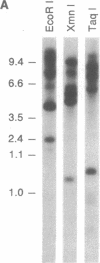
The adherence of Entamoeba histolytica to colonic mucins and to host cells appears to be predominantly mediated by a 170-kDa surface lectin of the amoebae. By using an antiserum to the purified lectin, the corresponding cDNA was isolated from an expression library of the pathogenic E. histolytica isolate HM-1:IMSS. Northern blot analysis indicated a transcript of approximately 4 kilobases, and Southern blot analyses suggested that multiple genes may encode the lectin or closely related proteins in HM-1:IMSS trophozoites. The cDNA-deduced amino acid sequence revealed an N-terminal signal peptide and a mature protein of 1270 amino acids corresponding to a molecular mass of 143 kDa, which comprises a short C-terminal cytoplasmic domain with potential phosphorylation sites, a transmembrane region, and a large extracellular portion with nine potential asparagine-linked glycosylation sites. The extracellular portion may be separated into a cysteine-poor domain and a cysteine-rich domain, the latter of which shows in part repetitive structural elements with a low degree of sequence homology to wheat germ agglutinin and to pDd63, a developmentally expressed protein of Dictyostelium discoideum.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chadee K., Petri W. A., Jr, Innes D. J., Ravdin J. I. Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J Clin Invest. 1987 Nov;80(5):1245–1254. doi: 10.1172/JCI113199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond L. S., Harlow D. R., Cunnick C. C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72(4):431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Petri W. A., Jr, Chapman M. D., Snodgrass T., Mann B. J., Broman J., Ravdin J. I. Subunit structure of the galactose and N-acetyl-D-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. J Biol Chem. 1989 Feb 15;264(5):3007–3012. [PubMed] [Google Scholar]
- Petri W. A., Jr, Smith R. D., Schlesinger P. H., Murphy C. F., Ravdin J. I. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J Clin Invest. 1987 Nov;80(5):1238–1244. doi: 10.1172/JCI113198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri W. A., Jr, Snodgrass T. L., Jackson T. F., Gathiram V., Simjee A. E., Chadee K., Chapman M. D. Monoclonal antibodies directed against the galactose-binding lectin of Entamoeba histolytica enhance adherence. J Immunol. 1990 Jun 15;144(12):4803–4809. [PubMed] [Google Scholar]
- Ravdin J. I., Murphy C. F., Salata R. A., Guerrant R. L., Hewlett E. L. N-Acetyl-D-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. I. Partial purification and relation to amoebic virulence in vitro. J Infect Dis. 1985 May;151(5):804–815. doi: 10.1093/infdis/151.5.804. [DOI] [PubMed] [Google Scholar]
- Tannich E., Horstmann R. D., Knobloch J., Arnold H. H. Genomic DNA differences between pathogenic and nonpathogenic Entamoeba histolytica. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5118–5122. doi: 10.1073/pnas.86.13.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S., Olafsdottir S. Structural differences in the two major wheat germ agglutinin isolectins. J Biol Chem. 1986 Jun 5;261(16):7191–7195. [PubMed] [Google Scholar]