Meningococcal disease (meningitis and bloodstream infections) threatens millions of people across the meningitis belt of sub-Saharan Africa. A vaccine introduced in 2010 protects against Africa’s then-most common cause of meningococcal disease, N. meningitidis serogroup A. However, other serogroups continue to cause epidemics in the region—including serogroup W. The rapid identification of strains that have been associated with prior outbreaks can improve the assessment of outbreak risk and enable timely preparation of public health responses, including vaccination. Phylogenetic analysis of newly sequenced serogroup W strains isolated from 1994 to 2012 identified two groups of strains linked to large epidemics in Burkina Faso, one being descended from a strain that caused an outbreak during the Hajj pilgrimage in 2000. We find that applying whole-genome sequencing to meningococcal disease surveillance collections improves the discrimination among strains, even within a single nation-wide epidemic, which can be used to better understand pathogen spread.
KEYWORDS: Africa, Neisseria meningitidis, disease outbreaks, epidemiology, evolution, meningitis, meningococcus
ABSTRACT
Epidemics of invasive meningococcal disease (IMD) caused by meningococcal serogroup A have been eliminated from the sub-Saharan African so-called “meningitis belt” by the meningococcal A conjugate vaccine (MACV), and yet, other serogroups continue to cause epidemics. Neisseria meningitidis serogroup W remains a major cause of disease in the region, with most isolates belonging to clonal complex 11 (CC11). Here, the genetic variation within and between epidemic-associated strains was assessed by sequencing the genomes of 92 N. meningitidis serogroup W isolates collected between 1994 and 2012 from both sporadic and epidemic IMD cases, 85 being from selected meningitis belt countries. The sequenced isolates belonged to either CC175 (n = 9) or CC11 (n = 83). The CC11 N. meningitidis serogroup W isolates belonged to a single lineage comprising four major phylogenetic subclades. Separate CC11 N. meningitidis serogroup W subclades were associated with the 2002 and 2012 Burkina Faso epidemics. The subclade associated with the 2012 epidemic included isolates found in Burkina Faso and Mali during 2011 and 2012, which descended from a strain very similar to the Hajj (Islamic pilgrimage to Mecca)-related Saudi Arabian outbreak strain from 2000. The phylogeny of isolates from 2012 reflected their geographic origin within Burkina Faso, with isolates from the Malian border region being closely related to the isolates from Mali. Evidence of ongoing evolution, international transmission, and strain replacement stresses the importance of maintaining N. meningitidis surveillance in Africa following the MACV implementation.
IMPORTANCE Meningococcal disease (meningitis and bloodstream infections) threatens millions of people across the meningitis belt of sub-Saharan Africa. A vaccine introduced in 2010 protects against Africa’s then-most common cause of meningococcal disease, N. meningitidis serogroup A. However, other serogroups continue to cause epidemics in the region—including serogroup W. The rapid identification of strains that have been associated with prior outbreaks can improve the assessment of outbreak risk and enable timely preparation of public health responses, including vaccination. Phylogenetic analysis of newly sequenced serogroup W strains isolated from 1994 to 2012 identified two groups of strains linked to large epidemics in Burkina Faso, one being descended from a strain that caused an outbreak during the Hajj pilgrimage in 2000. We find that applying whole-genome sequencing to meningococcal disease surveillance collections improves the discrimination among strains, even within a single nation-wide epidemic, which can be used to better understand pathogen spread.
INTRODUCTION
The pathogen Neisseria meningitidis is a common cause of meningitis in the 26 sub-Saharan Africa countries of the so-called “meningitis belt,” where small annual epidemics and periodic large epidemics contribute to the highest incidence of meningococcal meningitis in the world (1). N. meningitidis serogroup A disease has dramatically decreased since the initiation of mass vaccination with the meningococcal A conjugate vaccine (MACV) in 2010 (2, 3), and yet, affordable conjugate vaccines are not available for other serogroups that have caused epidemics in Africa. N. meningitidis serogroup W remains a major cause of meningococcal disease in the region (4).
The first large outbreak of N. meningitidis serogroup W disease occurred during the 2000 Hajj to Saudi Arabia (Islamic pilgrimage to Mecca), with over 300 cases reported worldwide (5, 6). N. meningitidis serogroup W isolates from this outbreak (hereinafter, the Hajj-related outbreak strain) belonged to clonal complex 11 (CC11), a hyperinvasive lineage typically identified with N. meningitidis serogroup C disease (7, 8). All known CC11 N. meningitidis serogroup W isolates originate from a single ancestral strain, with some isolates having been collected as early as 1970 (8, 9). CC11 N. meningitidis serogroup W isolates have become globally distributed and were reported in meningitis belt countries as early as 1993 (9) but were not reported from epidemics until 2001 (10). Although N. meningitidis serogroup W disease caused by both CC11 and CC175 has been reported in meningitis belt countries (11), only N. meningitidis serogroup W CC11 has caused epidemics. A large N. meningitidis serogroup W epidemic occurred in Burkina Faso during 2002, with 12,587 cases reported (12, 13). Since then, additional N. meningitidis serogroup W epidemics have only occurred since 2010 (14, 15), including the CC11 epidemic in Burkina Faso during 2012, with 5,807 cases reported (4, 16, 17).
The occurrence of several CC11 N. meningitidis serogroup W epidemics since 2000 raised concerns that the Hajj-related outbreak strain had become established in the meningitis belt after being carried there by returning pilgrims (12, 14, 18). This was of particular concern because the attack rate among returning pilgrims was reported to be as high as 25 per 100,000 population, with a case fatality rate as high as 37% (19), suggesting that the Hajj-related outbreak strain may have been more virulent than other N. meningitidis serogroup W strains. Isolates that evolved from this strain continue to be identified around the world, and these strains may have become endemic in regions as distant as South Africa, Turkey, and Europe (8, 20). However, CC11 N. meningitidis serogroup W strains that are endemic to the United Kingdom and Chile did not evolve from the Hajj-related outbreak strain (8, 20). While isolates from the 2002 epidemic in Burkina Faso did not evolve from the Hajj-related outbreak strain (12, 20, 21), they are closely related, and it is unclear when these two strains diverged or whether the recent CC11 N. meningitidis serogroup W isolates from meningitis belt countries are descended from the Hajj-related outbreak strain.
Here, we applied whole-genome sequencing to analyze the evolution of N. meningitidis serogroup W populations in the meningitis belt countries from 1994 to 2012. Isolates collected during the 2012 epidemic in Burkina Faso and the preceding years in the belt were analyzed to identify the origin of the 2012 epidemic N. meningitidis serogroup W and the relationship of that epidemic to the 2002 epidemic in Burkina Faso. Isolates collected during the Hajj-related Saudi Arabian outbreak in 2000 and during the following years in Africa were analyzed to evaluate whether the Hajj-related outbreak strain was dispersed to Africa and contributed to later epidemics. We further examined the genetic variation among major lineages to identify loci distinguishing the epidemic-associated isolates from other CC11 N. meningitidis serogroup W strains.
RESULTS
Overview of phylogenetic diversity.
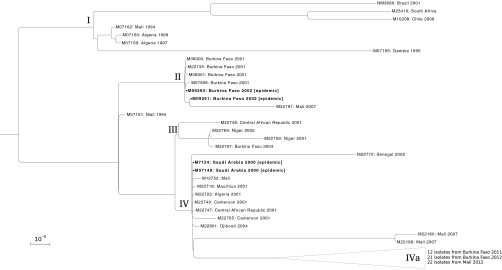
We sequenced the genomes of 92 N. meningitidis serogroup W isolates, 9 from CC175 and 83 from CC11 (Table 1; see also Table S1 in the supplemental material). The CC175 genomes had minimal diversity among them and did not show a clear phylogenetic structure (see Materials and Methods; see also Fig. S1). Within CC11, we identified four major subclades (I through IV) based on their association with large epidemics (Fig. 1; Table 2). Subclades I and III did not contain isolates associated with meningitis epidemics. Subclade I consisted of 7 genomes, with the only 2 meningitis belt isolates being from the 1990s. Subclade III consisted of 4 isolates from the meningitis belt from 2001 to 2004. One strain (M07161) isolated in 1994 from Mali did not fit into a major subclade but was basal to subclades III and IV with high bootstrap support (99%).
TABLE 1 .
Epidemiological context for the isolates analyzed in this study
| Country | ST/CCa | Yr collectedb | No. of isolates |
|---|---|---|---|
| Algeria | ST11/CC11 | 1997 | 1 |
| 1999 | 1 | ||
| 2001 | 1 | ||
| Benin | ST2881/CC175 | 2004 | 1 |
| 2006 | 1 | ||
| 2007 | 1 | ||
| Burkina Faso | ST11/CC11 | 2001 | 4 |
| 2002 | 2 | ||
| 2004 | 1 | ||
| 2011 | 11 | ||
| 2012 | 21 | ||
| ST2961/CC11 | 2011 | 1 | |
| ST2881/CC175 | 2008 | 1 | |
| 2010 | 1 | ||
| Cameroon | ST11/CC11 | 2001 | 2 |
| Central African Republic | ST11/CC11 | 2001 | 2 |
| Djibouti | ST11/CC11 | 2004 | 1 |
| Gambia | ST11/CC11 | 1995 | 1 |
| Mali | ST11/CC11 | 1994 | 2 |
| 2007 | 3 | ||
| 2012 | 21 | ||
| ST11407/CC11 | 2012 | 1 | |
| Mauritius | ST11/CC11 | 2001 | 1 |
| Niger | ST11/CC11 | 2001 | 1 |
| 2002 | 1 | ||
| ST2881/CC175 | 2003 | 1 | |
| 2005 | 1 | ||
| 2006 | 1 | ||
| Saudi Arabia | ST11/CC11 | 2000 | 1 |
| Senegal | ST11/CC11 | 2002 | 1 |
| Togo | ST2881/CC175 | 2007 | 1 |
| Incomplete data | ST11/CC11 | N/A | 2 |
| Previously published | ST11/CC11 | N/A | 4 |
ST/CC, sequence type profile/clonal complex identifier, from MLST.
Boldface indicates years with epidemics and large outbreaks involving CC11.
FIG 1 .
Maximum-likelihood phylogeny of 86 CC11 N. meningitidis serogroup W isolates, rooted on NM3683. Branches with less than 85% bootstrap support are collapsed. Scale bar represents one substitution per 10,000 bases in the whole-genome alignment. Isolates are labeled with country and year of isolation if available. Isolates from the 2002 Burkina Faso epidemic and the 2000 Hajj-related Saudi Arabian outbreak are in boldface. Clades are labeled at their roots and described in the text. Subclade IVa is presented in Fig. 2.
TABLE 2 .
Observations of isolates from clonal complex 11 subcladesa
| Region and country | No. of isolates from indicated subclade inc: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1994 | 1995 | 1997 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2010 | 2011 | 2012 | |
| Hyperendemic meningitis belt | ||||||||||||||||
| Burkina Faso | 4 II | 2 II | 1 III | * | * | 12 IVa | 22 IVa | |||||||||
| Mali | 1 I, 1 NAb | 1 II, 2 IV | 21 IVa | |||||||||||||
| Niger | 1 III | 1 III | * | * | * | |||||||||||
| Nonhyperendemic meningitis belt | ||||||||||||||||
| Benin | * | * | * | |||||||||||||
| Cameroon | 2 IV | |||||||||||||||
| Central African Republic | 1 III, 1 IV | |||||||||||||||
| Gambia | 1 I | |||||||||||||||
| Senegal | 1 IV | |||||||||||||||
| Togo | * | |||||||||||||||
| Not meningitis belt | ||||||||||||||||
| Algeria | 1 I | 1 I | 1 IV | |||||||||||||
| Djibouti | 1 IV | |||||||||||||||
| Mauritius | 1 IV | |||||||||||||||
| Saudi Arabia | 1 IV | |||||||||||||||
Subclades are identified in Fig. 1 and text.
NA, not applicable; isolate M07161 did not fit into a major subclade.
Asterisk indicates single observation of CC175 isolate.
Isolate and sequencing details. Subclades for clonal complex 11 are defined in the text and figures. The number of unique hqSNPs was calculated for the Lyve-set alignment. Antigen subtyping information is based on variable regions. Gene allele identifiers were retrieved from PubMLST. Download Table S1, XLSX file, 0.02 MB (26.9KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination-adjusted phylogeny of 9 single-contig CC175 genomes. ClonalFrameML inferred 95 recombination events, with mean import length (I) of 626 bp, mean divergence (D) of 3.8%, and a relative frequency of recombination to mutation (R) of 0.58. All branches have 100/100 bootstrap support. Download Figure S1, EPS file, 0.03 MB (33.5KB, eps) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence diversity was described further by the number of high-quality single-nucleotide polymorphisms (hqSNPs) between each pair of CC11 isolates in our study (see Table S2 in the supplemental material). After excluding the outgroup, the alignment contained 6,162 variable positions. The diversity within and between each labeled subclade is given in Table 3 (see also Tables S3 and S4).
TABLE 3 .
Core genome similarity between isolates in the major subclades
| Subclade | Range of hqSNP counts or % similarity between indicated subcladesa |
||||
|---|---|---|---|---|---|
| I | II | III | IV | IVa | |
| I | 26–1,410 | 99.97 | 99.96 | 99.97 | 99.95 |
| II | 642–1,488 | 2–238 | 99.98 | 99.98 | 99.97 |
| III | 707–1,467 | 393–697 | 20–310 | 99.99 | 99.98 |
| IV | 662–1,904 | 348–1137 | 132–897 | 0–1,221 | 100 |
| IVa | 938–1,904 | 637–1,137 | 417–897 | 0–1,221 | 0–684 |
Minimum and maximum counts of hqSNPs distinguishing isolates in the subclades are presented on the diagonal and below. Maximum sequence similarity between isolates, based on an alignment of 1,982,813 nucleotides, is presented above the diagonal in boldface. Isolate counts for each subclade are I (n = 7), II (n = 7), III (n = 4), IV (n = 67), and IVa (n = 55).
Number of hqSNPs distinguishing each pair of genomes. Download Table S2, XLSX file, 0.04 MB (45.4KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core genome nucleotide diversity within labeled subclades, minimum hqSNP counts between clades, and discriminatory hqSNP counts for each subclade. Download Table S3, XLSX file, 0.01 MB (12.6KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Locations of SNPs that distinguish major clades from each other, according to the reference genome NM3683. SNPs are clustered if they fall into the same phylogenetic class and less than 500 bp separates them. SNPs are categorized as being either hqSNPs or Mauve SNPs and as being either homoplasic or discriminatory. The SNPs included are discriminatory for subclades II, IV, IVa, III/IV, or II/III/IV. Gene annotations from NM3683 are listed if an SNP is contained in the gene. Annotations are based on PubMLST, when available, and otherwise on the Fam18 reference. Download Table S4, XLSX file, 0.03 MB (29.2KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subclade associated with the epidemic in Burkina Faso during 2002.
Subclade II consists of 2 isolates from the first large N. meningitidis serogroup W epidemic in Burkina Faso (2002), 4 isolates from the previous year, and a later isolate from Mali (2007). The epidemic-associated isolates had 21 hqSNPs separating them, greater than the separation between those two and many isolates from 2001, which is as low as 5 hqSNPs (see Table S5 in the supplemental material). This was in agreement with the phylogenetic analysis that depicted these 6 isolates as a closely related group (Fig. 1) that was clearly distinguishable from the isolate recovered in 2004 (subclade III) and the later isolate from Mali in 2007 (subclade II).
Subclade II was distinguished from the others by 128 hqSNPs found in 16 diverse locations in the genome (see Table S4 in the supplemental material). A large cluster of hqSNPs was present in proximity to the nadC, nicA, and nicB genes (PubMLST identifiers [IDs] NEIS1770 to NEIS1773; 96 hqSNPs).
Core genome similarity of isolates in subclade II, associated with the 2002 Burkina Faso epidemic. Download Table S5, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subclade associated with the outbreak in Saudi Arabia during 2000.
Subclade IV (n = 67) was defined by two isolates from the Hajj-related Saudi Arabian outbreak in 2000 (M07149 and M7124), plus 8 isolates collected from varied locations in Africa between 2001 and 2004. A group of 6 genomes had a maximum of 18 hqSNP differences from each other, each substitution being unique to a single genome (M22722, M22740, M22747, M7124, M07149, and M12752). M07149 had no unique substitutions, indicating that it was very similar to the most recent common ancestor of subclade IV (see Tables S1 and S2 in the supplemental material).
Subclade IV had 24 hqSNPs that distinguished it from other clades (see Table S3 in the supplemental material). These were distributed in 19 genome locations, with small clusters in proximity to the genes greA (NEIS1365; 2 hqSNPs) and lgtA (NEIS1902; 4 hqSNPs). Inclusion of the SNPs from the Mauve alignment also identified pglB (NEIS0399). The clade including subclades III and IV had 91 discriminatory hqSNPs, including one cluster near the argH and galU genes (NEIS0580 and NEIS0581; 46 hqSNPs), and another near the nor and aniA genes (NEIS1548; aniA is not in PubMLST; 26 hqSNPs). There were no polymorphisms shared among the epidemic subclades (II and IV) that distinguished them from the remainder of the collection.
Subclade associated with the epidemic in Burkina Faso during 2012.
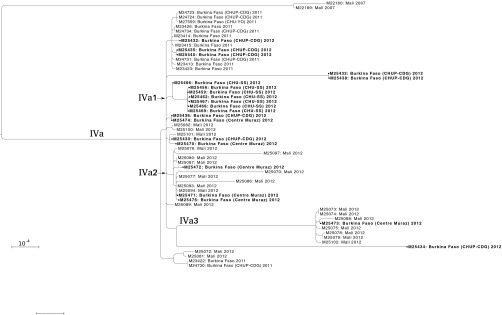
A nested clade within subclade IV, defined as subclade IVa, comprised 55 isolates from Mali and Burkina Faso in 2011 and 2012 (Fig. 2). Burkina Faso experienced a widespread epidemic in 2012, and all 21 isolates from that year were in subclade IVa. These isolates were subdivided according to the National Reference Laboratories that provided the isolates (see Materials and Methods). Phylogeographic structure within Burkina Faso was evident in some of the clades, but there was also evidence of repeated transmission between countries. For instance, the 7 isolates from Centre Hospitalier Universitaire Sanou Souro (CHU-SS) form a monophyletic group (Fig. 2, subclade IVa1). Of the 22 isolates from Mali, 9 were in subclade IVa2 and 7 in subclade IVa3. The only other isolates in those subclades were from Centre Muraz, which covers districts bordering Mali; of the 6 isolates from Centre Muraz, 3 were in IVa2 and 1 in IVa3. In contrast, the isolates from Centre Hospitalier Universitaire Pédiatrique-Charles de Gaulle (CHUP-CDG) in 2012 were widely dispersed (including the most divergent isolates, M25434 and M25433), consistent with this laboratory receiving isolates from varied locations in Burkina Faso.
FIG 2 .
Maximum-likelihood phylogeny of 55 isolates in subclade IVa, rooted on subclade IV. Branches with less than 70% bootstrap support are collapsed. Scale bar represents one substitution per 10,000 bases in the whole-genome alignment. Isolates are labeled with country and year of isolation, with Burkina Faso National Reference Laboratories that provided isolates listed in parentheses. Isolates from Burkina Faso 2012 are in boldface. Subclades mentioned in the text are labeled at their roots.
Isolates from the 2012 Burkina Faso epidemic had a maximum of 684 hqSNPs between any two isolates (see Table S6 in the supplemental material), accounting for the broad diversity of subclade IVa. Some of these isolates were very similar to isolates collected in Burkina Faso during 2011 (6 hqSNPs) and in Mali during 2012 (1 hqSNP), consistent with the phylogeny, where none of these groups were monophyletic. Subclade IVa was distinguished from other subclades by 240 hqSNPs found in 17 locations on the genome (see Table S4).
Core genome SNP diversity of isolates in subclade IVa, associated with the 2012 Burkina Faso epidemic. Download Table S6, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diversity at possible subtyping loci.
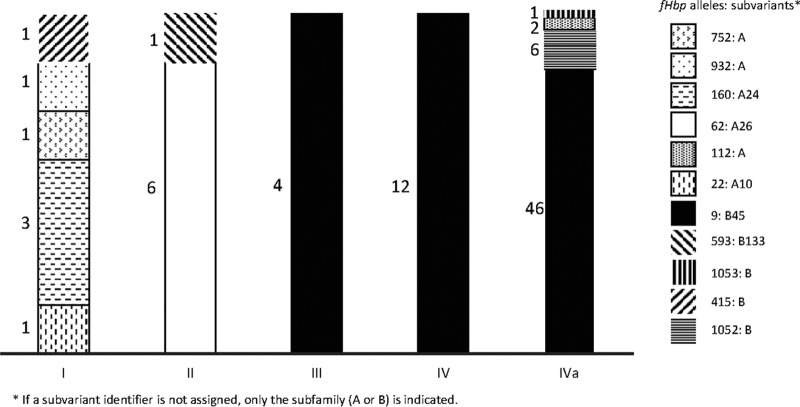
We examined the diversity in several genes that are regularly used to genotype meningococcus (see Table S1 in the supplemental material) (22). The porA subtype was uniform within clonal complexes (P1.5,2 in CC11 and P1.5-1,2-36 in CC175), while the fetA variable region was largely uniform, with a few low-frequency variants. Sequence variation at the fHbp locus has been proposed as a potential marker for the Hajj-related outbreak strain (20); we observed 11 fHbp alleles among the CC11 isolates, encoding six subfamily A proteins and five subfamily B proteins (alternatively known as variant groups 2/3 or group 1, respectively). Alleles associated with both subfamilies were found among isolates of subclades I, II, and IVa (Fig. 3).
FIG 3 .
Frequencies of fHbp alleles in each subclade of CC11. Each bar represents a different subclade, colored by the proportion of isolates from that subclade that carry each allele, with the number of isolates written to the left. The bar for subclade IV does not include isolates from subclade IVa. Each allele encodes a different protein, and protein subvariants are listed in the legend.
To identify additional loci that may distinguish the CC11 subclades from each other, we evaluated the PubMLST allele assignments at several loci where hqSNPs distinguish the subclades. A subset of loci for which alleles were strongly associated with subclades is presented in Table 4 (the full set is in Table S4 in the supplemental material); it includes two loci from the ribosomal multilocus sequence type (MLST) scheme. Reflecting this variation, the subclade IV isolates have ribosomal sequence type (rST) 2332 and subclade IVa isolates have rST 7546 (23).
TABLE 4 .
Select loci with SNPs distinguishing subclades, described by their PubMLST allele assignment
| Locusa | PubMLST IDb | Patternc | Allele in subcladed
|
No. of SNPsf | ||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IVe | IVa | ||||
| mafS7 | NEIS2090 | II | 1 | 18 | 1 | 1g | 1 | 56 |
| nadC | NEIS1770 | II | 1 | 237 | 1 | 1 | 1 | 30 |
| nor | NEIS1548 | III/IVh | 149 | 149 | 1 | 1 | 1 | 59 |
| galU | NEIS0581 | III/IVh | 230 | 230 | 351 | 351 | 351i | 36 |
| nhbA | NEIS2109 | III/IVh | 17 | 17 | 72 | 72 | 72 | 1 |
| opcB | NEIS1877 | II/III/IV | 1 | 2 | 2 | 2j | 2 | 2 |
| folP | NEIS1609 | II/III/IV | 14k | 1 | 1 | 1 | 1 | 67 |
| lgtA | NEIS1902 | IV | 17l | 17 | 17 | 37l | 37 | 2 |
| lgtB | NEIS1901 | IV | 79 | 79 | 79 | 77m | 77 | 4 |
| rpmA | NEIS1848 | IV | 1 | 1 | 1 | 4 | 4 | 1 |
| rpsO | NEIS0552 | IVa | 1 | 1 | 1 | 1 | 16 | 1 |
Locus names are taken from PubMLST annotation if available; otherwise, the mapping of Fam18 annotations to the reference sequence is used.
ID, identifier.
SNP patterns are categorized by which subclades are distinguished from the outgroup.
The predominant allele is reported for each subclade; minor alleles are noted in the footnotes.
Counts for subclade IV do not include isolates from subclade IVa.
The SNP count is the number of polymorphisms separating the two listed PubMLST alleles.
mafS7 is not present in M22765.
M07161 shares alleles with subclades I/II for nor and galU but with III/IV for nhbA.
M25432 has galU allele 2.
lgtA is not found in M22772 and Nm3686.
M22772 has lgtB allele 67.
DISCUSSION
Concern about the epidemic potential of CC11 N. meningitidis serogroup W increased greatly following the multinational outbreaks among Hajj pilgrims returning from Saudi Arabia in 2000 (5, 6, 9, 12, 19). The isolates collected from meningitis belt countries in 2001 and 2002 include representatives of three different CC11 N. meningitidis serogroup W subclades, one of which (subclade IV) may be descended from the strain that caused the Saudi Arabian outbreak (Table 2). Isolates obtained from the Burkina Faso 2002 epidemic are a separate lineage (subclade II), as are some of the isolates from disease cases outside large epidemics (subclade III). Four isolates from subclades II and III were also sequenced for the study of Lucidarme et al. (8) (M22797/30098, M22759/30087, M22769/30089, and M22745/30104), where they were placed into two clusters labeled as the “Burkina Faso/North African strains.”
While subclade IV was not the only cause of CC11 N. meningitidis serogroup W disease in the meningitis belt after 2000, isolates very similar to the Hajj-related outbreak strain were collected in the region. These strains may have been dispersed during the 2000 outbreak, as indicated by the minimal diversity among the subclade IV isolates collected in 7 countries from 2000 to 2004, the absence of phylogenetic structure at the base of subclade IV where 9 branches join (Fig. 1), and the absence of any hqSNPs between the consensus genome sequences of those isolates and the genome of isolate M07149, which was collected during the Saudi Arabian outbreak. Two of these isolates were identified as part of the “Anglo-French Hajj strain” by Lucidarme et al. (8) (M22722/2001076 and M22765/2002029).
The isolates of subclade III were very closely related to the Hajj-related outbreak strain (99.99% sequence identity; minimum of 132 hqSNPs) and share the antigen gene profile of the Hajj-related outbreak strain that was identified by Mustapha et al., specifically the presence of fHbp allele 9 (Fig. 3) (20). However, they are distinguished from the Hajj-related outbreak strain at 19 loci where all subclade IV isolates have derived variants (see Table S4 in the supplemental material), indicating that subclade III isolates are not derived from the Hajj-related outbreak strain. They are further distinguished from the subclade IV strains by their greater sequence diversity and the presence of phylogenetic structure within the clade (Fig. 1), which is evident even when recombination is accounted for (see Fig. S2). Altogether, this indicates that subclade III did not undergo the same population dynamics as the strains that make up subclade IV and is a separate linage.
Recombination-adjusted phylogeny of 41 single-contig CC11 genomes, rooted on NM3683. ClonalFrameML inferred 480 recombination events (279 when the outgroup branch is excluded), with mean import length (I) of 641 bp, mean divergence (D) of 4.4%, and a relative frequency of recombination to mutation (R) of 0.60. Branches with less than 75/100 bootstrap support are collapsed. Download Figure S2, EPS file, 0.1 MB (58.2KB, eps) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Isolates from the Burkina Faso epidemic of 2002 comprised a distinct subclade (II), which clearly diverged from subclades III and IV prior to the 2000 Saudi Arabian outbreak. This was demonstrated by the presence of an isolate from 1994 (M07161) being placed on the lineage leading to subclades III and IV with high confidence (bootstrap value of 99%). In the years after the 2002 epidemic, CC11 N. meningitidis serogroup W was rarely identified among either disease isolates or carriage isolates (11, 24, 25) until isolates from subclade IVa were recovered in 2011 and 2012. This is consistent with the “clonal wave” model of meningococcal strain replacement in meningitis belt countries and communities (26).
Isolates recovered during the 2012 epidemic in Burkina Faso belonged to subclade IVa, a lineage resulting from the clonal expansion and international dispersion of subclade IV that coincided with the Hajj-related Saudi Arabian outbreak in 2000. This subclade contains a different ribosomal MLST profile (7546) than any isolates examined by Lucidarme et al. (8). The geographic location of the subclade IVa ancestral lineage between 2000 and 2011 cannot be inferred from the isolate collection in this analysis, which primarily includes western meningitis belt countries in which a previous study identified low frequencies of CC11 N. meningitidis serogroup W isolates from 2005 to 2010 (11). One possibility is that this lineage was only introduced to the western meningitis belt shortly before 2011; alternatively, a local population may not be represented in this analysis. The phylogeographic structure within subclade IVa indicated that most transmission is geographically restricted during epidemics, but repeated pathogen transmission has still occurred across the border of Mali and Burkina Faso, as indicated by the phylogenetic mixing of isolates from Mali in 2012 with those collected in Burkina Faso in 2011 and 2012 (Fig. 2). Additional isolates from this clade were collected in Niger during 2015 (27), demonstrating that CC11 N. meningitidis serogroup W populations are established in the meningitis belt.
Conclusion.
The N. meningitidis serogroup W isolates associated with the epidemics in Burkina Faso during 2002 and 2012 belong to two distinct subclades, closely related to CC11 isolates recovered from sporadic invasive meningococcal disease cases in meningitis belt countries since 2000. The subclade associated with the Burkina Faso epidemic of 2012 was descended from the Hajj-related outbreak strain, which became globally dispersed near the time of the Hajj-related Saudi Arabian outbreak in 2000. This subclade included all N. meningitidis serogroup W isolates examined that were collected from Mali and Burkina Faso in 2011 and 2012, but it included none of the isolates from the previous decade. These results, along with a large epidemic in Niger during 2015 caused primarily by a N. meningitidis serogroup C strain first identified in 2013 (27), indicate that meningococci can spread rapidly to cause large epidemics, stressing the importance of maintaining N. meningitidis surveillance throughout meningitis belt countries following the MACV implementation. The application of whole-genome sequencing to a greater proportion of representative disease and carriage isolates will allow high-resolution tracking of pathogen dissemination at the scale of both countries and continents, detecting epidemic-associated strains as they become established in new districts and generating hypotheses regarding paths of transmission.
MATERIALS AND METHODS
Strain selection.
A total of 92 isolates from the Centers for Disease Control and Prevention (CDC) culture collection were sequenced for this analysis; 85 originated from the meningitis surveillance systems of 10 meningitis belt countries (Table 1). Another 7 CC11 isolates from other regions were sequenced, and 4 previously published genomes included, to provide a global context for the diversity of meningitis belt populations (Table 1; see also Table S1 in the supplemental material). The WHO Collaborating Centre in Marseille contributed 20 of these isolates. Isolates were selected first to maximize the temporal and geographic diversity of the data set and second to focus on three notable epidemics: Saudi Arabia 2000, Burkina Faso 2002, and Burkina Faso 2012. Isolates from Burkina Faso were identified with the name of the National Reference Laboratory that provided the isolate, when available. These were Centre Hospitalier Universitaire Pédiatrique-Charles de Gaulle (CHUP-CDG), Centre Hospitalier Universitaire Yalgado de Ouagadougou (CHU-YO), Centre Hospitalier Universitaire Sanou Souro (CHU-SS), and Centre Muraz. The serogroup phenotype was confirmed using slide agglutination (28).
Genome sequencing.
Pacific Biosciences (PacBio) RSII sequencing was completed for 48 isolates, using P4-C2 sequencing chemistry. Sequences were assembled using PacBio’s Hierarchical Genome Assembly Process version 3 (29), where 30 Mb of the longest corrected reads was used for the initial assembly (see Table S1 in the supplemental material for details). Contiguous sequence (“contig”) circularity was evaluated by identifying repeats at the ends of the single contig, removing the repeat from one end, transferring the sequence from the 3′ to the 5′ end, and assessing whether the manual join point was supported by remapped reads.
An additional 44 isolates were sequenced on an Illumina HiSeq2500 (or MiSeq) instrument to examine the bacterial diversity during epidemics. Illumina sequencing libraries were prepared from extracted DNA by first shearing it to 600 bp using a Covaris LE220 focused ultrasonicator (Covaris, Inc., Woburn, MA). The sheared DNA was processed with the NEBNext ultra DNA library preparation kit following the manufacturer’s protocol (New England Biolabs, Ipswich, MA), using dual barcoding indices. These libraries were paired end sequenced, using TruSeq rapid SBS (sequencing by synthesis) chemistry, with either 100 bp or 250 bp at each end (see Table S1 in the supplemental material). Base calling and demultiplexing were completed with Casava (version 1.8.2). Reads were filtered to have an expected error rate of <1% (Qual = 20) and assembled by SPAdes (version 3.5) (30), discarding small contigs (<300 bp) or those with low coverage (<10×).
Genome alignment and phylogenetics.
The published genome sequence of NM3683 was used both as a reference for the sequence alignment and as an outgroup to root the phylogeny of CC11. This isolate was collected in Canada in 1970 and previously shown to be an outgroup to the extant CC11 N. meningitidis serogroup W population (20). To identify high-quality single-nucleotide polymorphisms (hqSNPs) for phylogenetic analysis, PacBio assemblies and Illumina read sets were simultaneously aligned to the reference genome using Lyve-Set 1.0 (31). SNPs less than 3 bp apart were excluded, as were any base positions with ambiguous characters. The final alignment covered 90.2% of the reference genome and contained 7,384 variable positions and 1,975,429 invariant positions (see Table S2 in the supplemental material). RAxML 8.1.17 (32) was used to generate a phylogeny, using the GTRGAMMAX model with the Stamatakis ascertainment bias correction and 100 bootstraps. Extended majority rule trees constructed from replicate bootstrap sets differed by <5% weighted Robinson-Foulds distance (33). To identify regions with many mutations, all 41 complete genomes were aligned with progressiveMauve (34), using a hidden Markov model (HMM) identity of 95%, identifying 16,419 SNPs in the core genome alignment (i.e., where all genomes contained a base). A progressiveMauve (34) alignment of the 9 CC175 single-contig genomes identified 3,026 SNPs in the core genome alignment. ClonalFrameML (35) was used to account for recombination in a phylogeny constructed from these alignments of single-contig genomes (see Fig. S1 and S2 in the supplemental material). Custom scripts for evaluation of sequence data and phylogenies used BioPython (36).
MLST alleles were identified by BLAST searches of PubMLST allele lists against the assembled genomes (37). Other genes were identified based on the PubMLST annotation of NM3683 where available and, where it was not available, based on the Fam18 NeMeSys (38) annotation, which was transferred to NM3683 by using Rapid Annotation Transfer Tool (RATT) (39). Discriminatory SNPs are those for which variants correspond to monophyletic groups of isolates, distinguishing the specified clade. Homoplasic SNPs are those for which variants are found in polyphyletic groups of isolates.
Accession number(s).
The genomic data are available in the GenBank database under BioProject accession number PRJNA319252, and individual accession numbers are listed in Table S1 in the supplemental material.
ACKNOWLEDGMENTS
The assembly of this strain collection was enabled by the Bacterial Meningitis Laboratory and Epidemiology team of the CDC’s Meningitis and Vaccine Preventable Diseases Branch. This study made use of the PubMLST website developed by Keith Jolley and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust.
This work was made possible through support from the AMD initiative at the CDC. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Halperin SA, Bettinger JA, Greenwood B, Harrison LH, Jelfs J, Ladhani SN, McIntyre P, Ramsay ME, Sáfadi MA. 2012. The changing and dynamic epidemiology of meningococcal disease. Vaccine 30(Suppl 2):B26–B36. doi: 10.1016/j.vaccine.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 2.Novak RT, Kambou JL, Diomandé FV, Tarbangdo TF, Ouédraogo-Traoré R, Sangaré L, Lingani C, Martin SW, Hatcher C, Mayer LW, Laforce FM, Avokey F, Djingarey MH, Messonnier NE, Tiendrébéogo SR, Clark TA. 2012. Serogroup A meningococcal conjugate vaccination in Burkina Faso: analysis of national surveillance data. Lancet Infect Dis 12:757–764. doi: 10.1016/S1473-3099(12)70168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daugla DM, Gami JP, Gamougam K, Naibei N, Mbainadji L, Narbé M, Toralta J, Kodbesse B, Ngadoua C, Coldiron ME, Fermon F, Page AL, Djingarey MH, Hugonnet S, Harrison OB, Rebbetts LS, Tekletsion Y, Watkins ER, Hill D, Caugant DA, Chandramohan D, Hassan-King M, Manigart O, Nascimento M, Woukeu A, Trotter C, Stuart JM, Maiden MC, Greenwood BM. 2014. Effect of a serogroup A meningococcal conjugate vaccine (PsA-TT) on serogroup A meningococcal meningitis and carriage in Chad: a community study. Lancet 383:40–47. doi: 10.1016/S0140-6736(13)61612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization 2015. Meningococcal disease control in countries of the African meningitis belt, 2014. Wkly Epidemiol Rec 90:123–131. http://www.who.int/wer/2015/wer9013.pdf?ua=1. [PubMed] [Google Scholar]
- 5.Lingappa JR, Al-Rabeah AM, Hajjeh R, Mustafa T, Fatani A, Al-Bassam T, Badukhan A, Turkistani A, Makki S, Al-Hamdan N, Al-Jeffri M, Al Mazrou Y, Perkins BA, Popovic T, Mayer LW, Rosenstein NE. 2003. Serogroup W-135 meningococcal disease during the Hajj, 2000. Emerg Infect Dis 9:665–671. doi: 10.3201/eid0906.020565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera JF, Perrocheau A, Meffre C, Hahné S. 2002. Outbreak of serogroup W135 meningococcal disease after the Hajj pilgrimage, Europe, 2000. Emerg Infect Dis 8:761–767. doi: 10.3201/eid0805.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, Gray S, Kaczmarski E. 2000. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356:2159. doi: 10.1016/S0140-6736(00)03502-9. [DOI] [PubMed] [Google Scholar]
- 8.Lucidarme J, Hill DM, Bratcher HB, Gray SJ, du Plessis M, Tsang RS, Vazquez JA, Taha MK, Ceyhan M, Efron AM, Gorla MC, Findlow J, Jolley KA, Maiden MC, Borrow R. 2015. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect 71:544–552. doi: 10.1016/j.jinf.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer LW, Reeves MW, Al-Hamdan N, Sacchi CT, Taha MK, Ajello GW, Schmink SE, Noble CA, Tondella ML, Whitney AM, Al-Mazrou Y, Al-Jefri M, Mishkhis A, Sabban S, Caugant DA, Lingappa J, Rosenstein NE, Popovic T. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J Infect Dis 185:1596–1605. doi: 10.1086/340414. [DOI] [PubMed] [Google Scholar]
- 10.Taha MK, Parent Du Chatelet I, Schlumberger M, Sanou I, Djibo S, de Chabalier F, Alonso JM. 2002. Neisseria meningitidis serogroups W135 and A were equally prevalent among meningitis cases occurring at the end of the 2001 epidemics in Burkina Faso and Niger. J Clin Microbiol 40:1083–1084. doi: 10.1128/JCM.40.3.1083-1084.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant DA, Kristiansen PA, Wang X, Mayer LW, Taha MK, Ouédraogo R, Kandolo D, Bougoudogo F, Sow S, Bonte L. 2012. Molecular characterization of invasive meningococcal isolates from countries in the African meningitis belt before introduction of a serogroup A conjugate vaccine. PLoS One 7:e46019. doi: 10.1371/journal.pone.0046019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koumaré B, Ouedraogo-Traoré R, Sanou I, Yada AA, Sow I, Lusamba PS, Traoré E, Dabal M, Santamaria M, Hacen MM, Kaboré AB, Caugant DA. 2007. The first large epidemic of meningococcal disease caused by serogroup W135, Burkina Faso, 2002. Vaccine 25(Suppl 1):A37–A41. doi: 10.1016/j.vaccine.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization 2005. Enhanced surveillance of epidemic meningococcal meningitis in Africa: a three-year experience. Wkly Epidemiol Rec 80:313–320. http://www.who.int/wer/2005/wer8037.pdf. [PubMed] [Google Scholar]
- 14.Collard JM, Issaka B, Zaneidou M, Hugonnet S, Nicolas P, Taha MK, Greenwood B, Jusot JF. 2013. Epidemiological changes in meningococcal meningitis in Niger from 2008 to 2011 and the impact of vaccination. BMC Infect Dis 13:576. doi: 10.1186/1471-2334-13-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hossain MJ, Roca A, Mackenzie GA, Jasseh M, Hossain MI, Muhammad S, Ahmed M, Chidiebere OD, Malick N, Bilquees SM, Ikumapayi UN, Jeng B, Njie B, Cham M, Kampmann B, Corrah T, Howie S, D’Alessandro U. 2013. Serogroup W135 meningococcal disease, The Gambia, 2012. Emerg Infect Dis 19:1507–1510. doi: 10.3201/eid1909.130077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacNeil JR, Medah I, Koussoubé D, Novak RT, Cohn AC, Diomandé FV, Yelbeogo D, Kambou JL, Tarbangdo TF, Ouédraogo-Traoré R, Sangaré L, Hatcher C, Vuong J, Mayer LW, Djingarey MH, Clark TA, Messonnier NE. 2014. Neisseria meningitidis serogroup W, Burkina Faso, 2012. Emerg Infect Dis 20:394–399. doi: 10.3201/eid2003.131407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization 2013. Meningococcal disease in countries of the African meningitis belt, 2012—emerging needs and future perspectives. Wkly Epidemiol Rec 88:129–136. http://www.who.int/wer/2013/wer8812.pdf?ua=1. [PubMed] [Google Scholar]
- 18.Forgor AA, Leimkugel J, Hodgson A, Bugri A, Dangy JP, Gagneux S, Smith T, Pluschke G. 2005. Emergence of W135 meningococcal meningitis in Ghana. Trop Med Int Health 10:1229–1234. doi: 10.1111/j.1365-3156.2005.01520.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilder-Smith A, Goh KT, Barkham T, Paton NI. 2003. Hajj-associated outbreak strain of Neisseria meningitidis serogroup W135: estimates of the attack rate in a defined population and the risk of invasive disease developing in carriers. Clin Infect Dis 36:679–683. doi: 10.1086/367858. [DOI] [PubMed] [Google Scholar]
- 20.Mustapha MM, Marsh JW, Krauland MG, Fernandez JO, de Lemos APS, Dunning Hotopp JC, Wang X, Mayer LW, Lawrence JG, Hiller NL, Harrison LH. 2015. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2:1447–1455. doi: 10.1016/j.ebiom.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Njanpop-Lafourcade BM, Parent du Châtelet I, Sanou O, Alonso JM, Taha MK. 2005. The establishment of Neisseria meningitidis serogroup W135 of the clonal complex ET-37/ST-11 as an epidemic clone and the persistence of serogroup A isolates in Burkina Faso. Microbes Infect 7:645–649. doi: 10.1016/j.micinf.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Jolley KA, Brehony C, Maiden MC. 2007. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol Rev 31:89–96. doi: 10.1111/j.1574-6976.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- 23.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, Wimalarathna H, Harrison OB, Sheppard SK, Cody AJ, Maiden MC. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traoré Y, Njanpop-Lafourcade BM, Adjogble KL, Lourd M, Yaro S, Nacro B, Drabo A, Parent du Châtelet I, Mueller JE, Taha MK, Borrow R, Nicolas P, Alonso JM, Gessner BD. 2006. The rise and fall of epidemic Neisseria meningitidis serogroup W135 meningitis in Burkina Faso, 2002–2005. Clin Infect Dis 43:817–822. doi: 10.1086/507339. [DOI] [PubMed] [Google Scholar]
- 25.Kristiansen PA, Diomandé F, Wei SC, Ouédraogo R, Sangaré L, Sanou I, Kandolo D, Kaboré P, Clark TA, Ouédraogo AS, Absatou KB, Ouédraogo CD, Hassan-King M, Thomas JD, Hatcher C, Djingarey M, Messonnier N, Préziosi MP, LaForce M, Caugant DA. 2011. Baseline meningococcal carriage in Burkina Faso before the introduction of a meningococcal serogroup A conjugate vaccine. Clin Vaccine Immunol 18:435–443. doi: 10.1128/CVI.00479-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leimkugel J, Hodgson A, Forgor AA, Pflüger V, Dangy JP, Smith T, Achtman M, Gagneux S, Pluschke G. 2007. Clonal waves of Neisseria colonisation and disease in the African meningitis belt: eight-year longitudinal study in northern Ghana. PLoS Med 4:e101. doi: 10.1371/journal.pmed.0040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kretz CB, Retchless AC, Sidikou F, Issaka B, Ousmane S, Schwartz S, Tate AH, Pana A, Njanpop-Lafourcade B-M, Nzeyimana I, Obama Nse R, Deghmane A-E, Hong E, Brynildsrud OB, Novak RT, Meyer SA, Oukem-Boyer OOM, Ronveaux O, Caugant DA, Taha M-K, Wang X, Niger Response Team . 2016. Whole-genome characterization of the emerging epidemic meningococcal serogroup C and resurgence of serogroup W in Niger, 2015. Emerg Infect Dis 22:1762–1768. doi: 10.3201/eid2210.160468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization 2011. Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 29.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. 2010. How many bootstrap replicates are necessary? J Comput Biol 17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 34.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rusniok C, Vallenet D, Floquet S, Ewles H, Mouzé-Soulama C, Brown D, Lajus A, Buchrieser C, Médigue C, Glaser P, Pelicic V. 2009. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol 10:R110. doi: 10.1186/gb-2009-10-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res 39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolate and sequencing details. Subclades for clonal complex 11 are defined in the text and figures. The number of unique hqSNPs was calculated for the Lyve-set alignment. Antigen subtyping information is based on variable regions. Gene allele identifiers were retrieved from PubMLST. Download Table S1, XLSX file, 0.02 MB (26.9KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination-adjusted phylogeny of 9 single-contig CC175 genomes. ClonalFrameML inferred 95 recombination events, with mean import length (I) of 626 bp, mean divergence (D) of 3.8%, and a relative frequency of recombination to mutation (R) of 0.58. All branches have 100/100 bootstrap support. Download Figure S1, EPS file, 0.03 MB (33.5KB, eps) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of hqSNPs distinguishing each pair of genomes. Download Table S2, XLSX file, 0.04 MB (45.4KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core genome nucleotide diversity within labeled subclades, minimum hqSNP counts between clades, and discriminatory hqSNP counts for each subclade. Download Table S3, XLSX file, 0.01 MB (12.6KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Locations of SNPs that distinguish major clades from each other, according to the reference genome NM3683. SNPs are clustered if they fall into the same phylogenetic class and less than 500 bp separates them. SNPs are categorized as being either hqSNPs or Mauve SNPs and as being either homoplasic or discriminatory. The SNPs included are discriminatory for subclades II, IV, IVa, III/IV, or II/III/IV. Gene annotations from NM3683 are listed if an SNP is contained in the gene. Annotations are based on PubMLST, when available, and otherwise on the Fam18 reference. Download Table S4, XLSX file, 0.03 MB (29.2KB, xlsx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core genome similarity of isolates in subclade II, associated with the 2002 Burkina Faso epidemic. Download Table S5, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core genome SNP diversity of isolates in subclade IVa, associated with the 2012 Burkina Faso epidemic. Download Table S6, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination-adjusted phylogeny of 41 single-contig CC11 genomes, rooted on NM3683. ClonalFrameML inferred 480 recombination events (279 when the outgroup branch is excluded), with mean import length (I) of 641 bp, mean divergence (D) of 4.4%, and a relative frequency of recombination to mutation (R) of 0.60. Branches with less than 75/100 bootstrap support are collapsed. Download Figure S2, EPS file, 0.1 MB (58.2KB, eps) .
Copyright © 2016 Retchless et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.