Abstract
This study is expected to investigate the association of ATP/GTP binding protein-like 4 (AGBL4), LDL receptor related protein 8 (LRP8) and proprotein convertase subtilisin/kexin type 9 (PCSK9) gene single nucleotide variants (SNVs) with lipid metabolism in 2,552 individuals (Jing, 1,272 and Han, 1,280). We identified 12 mutations in this motif. The genotype and allele frequencies of these variants were different between the two populations. Multiple-locus linkage disequilibrium (LD) elucidated the detected sites are not statistically independent. Possible integrative haplotypes and gene-by-gene (G × G) interactions, comprising mutations of the AGBL4, LRP8 and PCSK9 associated with total cholesterol (TC, AGBL4 G-G-A, PCSK9 C-G-A-A and G-G-A-A-C-A-T-T-T-G-G-A), triglyceride (TG, AGBL4 G-G-A, LRP8 G-A-G-C-C, PCSK9 C-A-A-G, A-A-G-G-A-G-C-C-C-A-A-G and A-A-G-G-A-G-C-C-C-G-A-A), HDL cholesterol (HDL-C, AGBL4 A-A-G and A-A-G-A-A-G-T-C-C-A-A-G) and the apolipoprotein(Apo)A1/ApoB ratio (A1/B, PCSK9 C-A-A-G) in Jing minority. However, in the Hans, with TG (AGBL4 G-G-A, LRP8 G-A-G-C-C, PCSK9 C-A-A-G, A-A-G-G-A-G-C-C-C-A-A-G and A-A-G-G-A-G-C-C-C-G-A-A), HDL-C (LRP8 A-A-G-T-C), LDL-C (LRP8 A-A-G-T-C and A-A-G-A-A-G-T-C-C-A-A-G) and A1/B (LRP8 A-C-A-T-T and PCSK9 C-A-A-G). Association analysis based on haplotype clusters and G × G interactions probably increased power over single-locus tests especially for TG.
Cardiovascular disease (CVD) ranks as the leading cause of morbidity and mortality globally1, and increases extraordinarily in the developing country2. Cardiometabolic risk3,4 especially lipid metabolism dysfunction5 represents a key event in atherosclerosis, a pathogenesis of CVD. High total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C) and apolipoprotein (Apo) B concentrations, as well as low high-density lipoprotein cholesterol (HDL-C), ApoA1 levels and the ApoA1/ApoB ratio (A1/B) are considered as complex traits to which both genetic and environmental factors contribute6,7.
Despite hundreds of genome-wide hits from genome-wide association studies (GWAS), a large portion of variations in lipid metabolism attributable to heritability remains unexplained8. Because of stringent statistical cutoffs necessary in the GWAS methodology, it is argued many common variants with an appreciable effect on phenotypic variations are reported as false negatives and dismissed9. To correct away the hidden heritability, fine mapping follow from high-density replicated GWAS data need only use the tag single nucleotide variants (SNVs) and regions of linkage disequilibrium (LD) independent of annotation or relationship to nearby genes10.
Recently, the compelling genes for modifying lipid metabolism emerged from very large replicated GWAS: the ATP/GTP binding protein-like 4 gene (AGBL4 [MIM 616476]), the LDL receptor related protein 8 gene (LRP8 [MIM 602600]) and the proprotein convertase subtilisin/kexin type 9 gene (PCSK9 [MIM 607786])11,12,13,14,15,16. The objective of this study was to perform association analysis to identify integrative mutations, haplotypes and gene-by-gene (G × G) interactions of the AGBL4 (rs320017 A > G, rs320018 A > G and rs320019 G > A), LRP8 (rs6694764 G > A, rs1288519 A > C, rs872315 G > A, rs1288520 C > T and rs1288521 C > T) and PCSK9 (rs533375 C > T, rs584626 A > G, rs585131 A > G and rs540796 G > A) associated with lipid phenotypic variations in the Jing and Han populations. Furthermore, we wanted to test if the association analysis of these loci based on haplotype clusters and G × G interactions increase power over single-locus tests.
Results
Study participants
Demographic, epidemiological and clinical characteristics of the 2, 552 analyzed study subjects are summarized in Table 1. The values of body mass index (BMI), waist circumference (WC) and the percentage of individuals whom consumed alcohol were higher, as well as the level of systolic blood pressure (SBP) was lower in Jing than Han (P < 0.05–0.001). For plasma lipid phenotypic variations, there were higher plasma TC and TG levels, as well as lower A1/B in Jing (P < 0.001, for each). However, no difference was noted in fasting plasma glucose, HDL-C and LDL-C levels between the two ethnic groups (P > 0.05 for all).
Table 1. Demographic, epidemiological and clinical characteristics.
| Characteristics | Jing | Han | test-statistic | P-value |
|---|---|---|---|---|
| Number (n) | 1272 | 1280 | ||
| Gender (Male/Female) | 624/648 | 636/644 | 0.102 | 0.750 |
| Age (years)1 | 57.27 ± 12.85 | 56.85 ± 13.32 | 0.818 | 0.414 |
| Height (cm) | 158.51 ± 7.93 | 157.63 ± 8.02 | 2.796 | 0.005 |
| Weight (kg) | 58.88 ± 10.03 | 56.78 ± 9.36 | 5.489 | 4.453E-08 |
| Body mass index (kg/m2) | 23.37 ± 3.17 | 22.82 ± 3.16 | 4.405 | 1.101E-05 |
| Underweight(BMI < 18.5) | 52(4.1) | 90(7.0) | ||
| Normal weight(18.5 ≤ BMI < 24) | 729(57.3) | 761(59.5) | ||
| Overweight(24 ≤ BMI < 28) | 387(30.4) | 356(27.8) | ||
| Obesity(28 ≤ BMI) | 104(8.2) | 73(5.7) | 17.554 | 0.001 |
| Waist circumference (cm) | 80.24 ± 9.26 | 77.93 ± 8.72 | 6.484 | 1.073E-10 |
| Male(Waist circumference ≤85) | 398(63.8) | 520(81.8) | ||
| Male(Waist circumference >85) | 226(36.2) | 116(18.2) | 51.484 | 7.218E-13 |
| Female(Waist circumference ≤80) | 376(58.0) | 412(64.3) | ||
| Female(Waist circumference >80) | 272(42.0) | 229(35.7) | 5.297 | 0.021 |
| Systolic blood pressure (mmHg) | 131.37 ± 20.92 | 134.84 ± 29.18 | −1.968 | 0.049 |
| Diastolic blood pressure (mmHg) | 80.81 ± 10.55 | 81.05 ± 10.29 | −0.561 | 0.575 |
| Pulse pressure (mmHg) | 50.56 ± 16.84 | 53.79 ± 27.63 | −1.920 | 0.055 |
| Cigarette smoking [n (%)] | ||||
| Nonsmoker | 1008(79.25) | 989(77.26) | ||
| ≤20 Cigarette smoking/day | 63(4.95) | 59(4.61) | ||
| >20 Cigarette smoking/day | 201(15.80) | 232(18.13) | 2.506 | 0.286 |
| Alcohol consumption [n (%)] | ||||
| Nondrinker | 971(76.34) | 870(67.97) | ||
| ≤25 g/day | 157(12.34) | 90(7.03) | ||
| >25 g/day | 144(11.32) | 320(25.00) | 90.450 | 2.286E-20 |
| Blood glucose level (mmol/L) | 6.70 ± 1.71 | 6.63 ± 1.11 | 1.353 | 0.176 |
| Total cholesterol (mmol/L) | 5.15 ± 0.91 | 4.88 ± 0.85 | 7.877 | 4.935E-15 |
| Triglyceride (mmol/L)2 | 1.43(1.12) | 1.32(1.07) | −4.439 | 9.018E-06 |
| High-density lipoprotein cholesterol (mmol/L) | 1.78 ± 0.53 | 1.81 ± 0.46 | −1.544 | 0.123 |
| Low-density lipoprotein cholesterol (mmol/L) | 2.86 ± 0.43 | 2.83 ± 0.43 | 1.477 | 0.140 |
| Apolipoprotein (Apo) A1 (g/L) | 1.31 ± 0.24 | 1.33 ± 0.20 | −2.305 | 0.021 |
| ApoB (g/L) | 1.06 ± 0.25 | 1.04 ± 0.24 | 2.693 | 0.007 |
| ApoA1/ApoB | 1.30 ± 0.38 | 1.35 ± 0.38 | −3.528 | 4.256E-04 |
1Mean ± SD determined by t-test.
2Median (interquartile range) tested by the Wilcoxon-Mann-Whitney test.
Single-mutation association
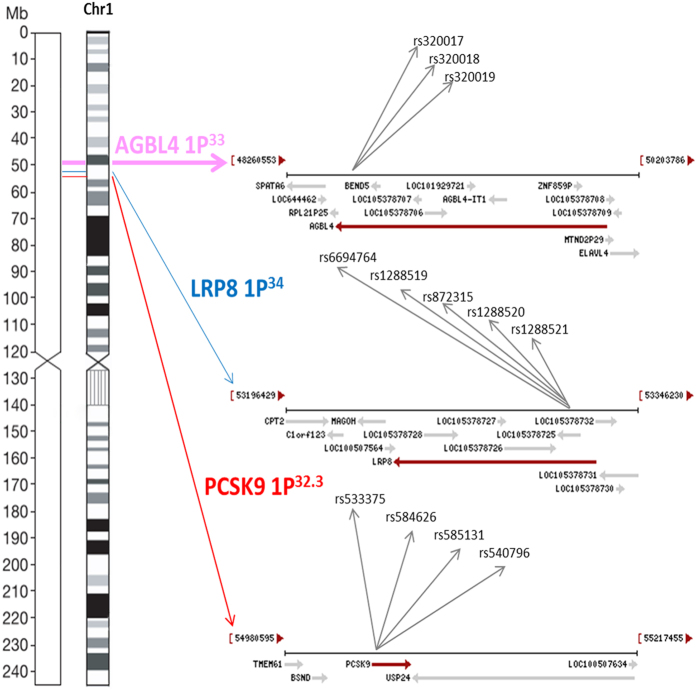
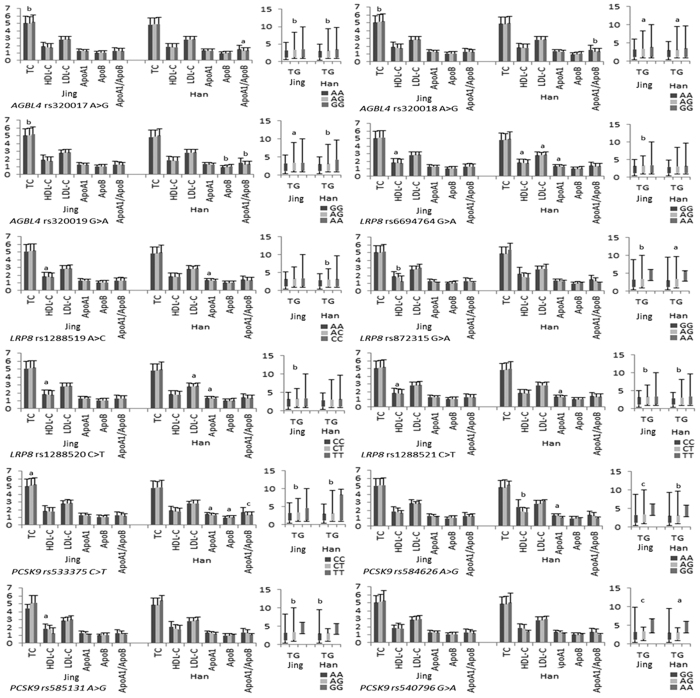
The detected 12 mutations in this motif are located in a closely genomic region of chromosome 1 (Fig. 1). As shown in Tables 2 and 3, the genotype and allele frequencies of these variants were different between the two populations (P < 0.05–0.001). All mutations exhibit the Hardy-Weinberg equilibrium (HWE, P > 0.05 for all). We tested each mutation individually for association with plasma lipid levels separately in each population. We discovered the association of the AGBL4, LRP8 and PCSK9 mutations with TC (rs320017, rs320018, rs320019 and rs533375), TG (rs320017, rs320018, rs320019, rs6694764, rs872315, rs1288520, rs1288521, rs533375, rs584626, rs585131 and rs540796) and HDL-C (rs6694764, rs1288519, rs872315, rs1288520, rs1288521 and rs585131) in Jing minority. However, in the Hans, with TG (rs320017, rs320018, rs320019, rs1288519, rs872315, rs1288521, rs533375, rs584626, rs585313 and rs540796), HDL-C (rs6694764 and rs584626), LDL-C (rs6694764 and rs1288520), ApoA1 (rs6694764, rs1288519, rs1288520, rs1288521, rs533375 and rs584626), ApoB (rs320019 and rs5333375) and A1/B (rs320017, rs320018, rs320019 and rs533375). (P < 0.05–0.001; Fig. 2).
Figure 1. The positions of the AGBL4, LRP8 and PCSK9 mutations.
Table 2. Prevalence of genotype frequencies in the Jing and Han populations [n (%)].
| Mutation | Genotype | Jing (n = 1272) | Han (n = 1280) | X2 | P-value |
|---|---|---|---|---|---|
| AGBL4 rs320017 A > G | AA | 768(60.38) | 836(65.31) | 6.704 | 0.035 |
| AG | 438(34.43) | 388(30.31) | |||
| GG | 66(5.19) | 56(4.38) | |||
| PHWE | 0.730 | 0.202 | |||
| AGBL4 rs320018 A > G | AA | 762(59.90) | 827(64.61) | 6.137 | 0.046 |
| AG | 443(34.83) | 397(31.01) | |||
| GG | 67(5.27) | 56(4.82) | |||
| PHWE | 0.802 | 0.344 | |||
| AGBL4 rs320019 G > A | GG | 769(60.46) | 834(65.16) | 6.054 | 0.048 |
| AG | 438(34.43) | 387(30.23) | |||
| AA | 65(5.11) | 59(4.61) | |||
| PHWE | 0.797 | 0.105 | |||
| LRP8 rs6694764 G > A | GG | 405(31.84) | 465(36.33) | 6.657 | 0.036 |
| AG | 632(49.69) | 611(47.73) | |||
| AA | 235(18.47) | 204(15.94) | |||
| PHWE | 0.674 | 0.889 | |||
| LRP8 rs1288519 A > C | AA | 425(33.41) | 484(37.81) | 6.270 | 0.043 |
| AC | 618(48.59) | 597(46.64) | |||
| CC | 229(18.00) | 199(15.55) | |||
| PHWE | 0.868 | 0.507 | |||
| LRP8 rs872315 G > A | GG | 1180(92.77) | 1224(95.63) | 9.588 | 0.008 |
| AG | 88(6.92) | 54(4.21) | |||
| AA | 4(0.31) | 2(0.16) | |||
| PHWE | 0.091 | 0.090 | |||
| LRP8 rs1288520 C > T | CC | 431(33.88) | 502(39.22) | 10.453 | 0.005 |
| CT | 590(46.38) | 574(44.84) | |||
| TT | 251(19.74) | 204(15.94) | |||
| PHWE | 0.057 | 0.064 | |||
| LRP8 rs1288521 C > T | CC | 490(38.52) | 554(43.28) | 6.169 | 0.046 |
| CT | 587(46.15) | 552(43.13) | |||
| TT | 195(15.33) | 174(13.59) | |||
| PHWE | 0.356 | 0.053 | |||
| PCSK9 rs533375 C > T | CC | 886(69.65) | 963(75.23) | 10.534 | 0.005 |
| CT | 340(26.73) | 285(22.27) | |||
| TT | 46(3.62) | 32(2.50) | |||
| PHWE | 0.064 | 0.051 | |||
| PCSK9 rs584626 A > G | AA | 1116(87.74) | 1162(90.78) | 6.649 | 0.036 |
| AG | 148(11.63) | 114(8.91) | |||
| GG | 8(0.63) | 4(0.31) | |||
| PHWE | 0.207 | 0.501 | |||
| PCSK9 rs585131 A > G | AA | 1118(87.89) | 1172(91.56) | 9.332 | 0.009 |
| AG | 150(11.79) | 105(8.20) | |||
| GG | 4(0.32) | 3(0.23) | |||
| PHWE | 0.663 | 0.689 | |||
| PCSK9 rs540796 G > A | GG | 1092(85.85) | 1159(90.55) | 15.222 | 4.949E-04 |
| AG | 172(13.52) | 119(9.30) | |||
| AA | 8(0.63) | 2(0.15) | |||
| PHWE | 0.666 | 0.560 |
AGBL4, the ATP/GTP binding protein-like 4 gene; LRP8, the LDL receptor related protein 8 gene; PCSK9, the Proprotein convertase subtilisin/kexin type 9 gene; HWE, Hardy-Weinberg equilibrium.
Table 3. Prevalence of allele frequencies in the Jing and Han populations [n(%)].
| Mutation | Allele | Jing (n = 1272) | Han (n = 1280) | X2 | P-value |
|---|---|---|---|---|---|
| AGBL4 rs320017 | A/G | 1974(77.59)/570(22.41) | 2060(80.47)/500(19.53) | 6.363 | 0.012 |
| AGBL4 rs320018 | A/G | 1967(77.32)/577(22.68) | 2051(80.12)/509(19.88) | 5.964 | 0.015 |
| AGBL4 rs320019 | G/A | 1976(77.67)/568(22.33) | 2055(80.27)/505(19.73) | 5.197 | 0.023 |
| LRP8 rs6694764 | G/A | 1442(56.68)/1102(43.32) | 1541(60.20)/1019(39.80) | 6.484 | 0.011 |
| LRP8 rs1288519 | A/C | 1468(57.70)/1076(42.30) | 1565(61.13)/995(38.87) | 6.220 | 0.013 |
| LRP8 rs872315 | G/A | 2448(96.23)/96(3.77) | 2502(97.73)/58(2.27) | 9.480 | 0.002 |
| LRP8 rs1288520 | C/T | 1452(57.08)/1092(42.92) | 1578(61.64)/982(38.36) | 11.024 | 0.001 |
| LRP8 rs1288521 | C/T | 1567(61.60)/977(38.40) | 1660(64.84)/900(35.16) | 5.789 | 0.016 |
| PCSK9 rs533375 | C/T | 2112(83.02)/432(16.98) | 2211(86.37)/349(13.63) | 11.038 | 0.001 |
| PCSK9 rs584626 | A/G | 2380(93.55)/164(6.45) | 2438(95.23)/122(4.77) | 6.816 | 0.009 |
| PCSK9 rs585131 | A/G | 2386(93.79)/158(6.21) | 2449(95.66)/111(4.34) | 8.983 | 0.003 |
| PCSK9 rs540796 | G/A | 2356(92.61)/188(7.39) | 2437(95.20)/123(4.80) | 14.904 | 1.131E-04 |
AGBL4, the ATP/GTP binding protein-like 4 gene; LRP8, the LDL receptor related protein 8 gene; PCSK9, the Proprotein convertase subtilisin/kexin type 9 gene.
Figure 2. Single-mutation association with lipid phenotypic variations.
Haplotype-based association
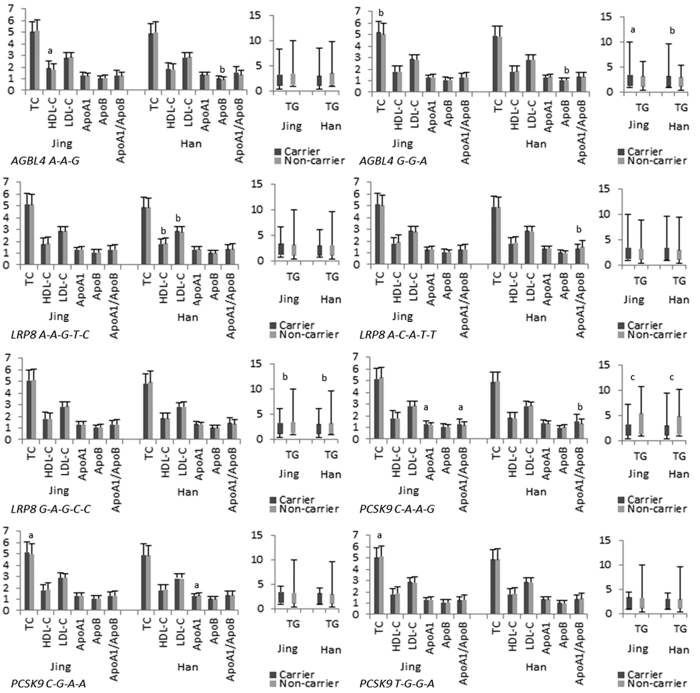
Multiple-locus linkage disequilibrium (LD) elucidated the detected sites were not statistically independent separately in each population. Figures 3 and 4 show the LD blocks and the haplotypes for blocks separately in the Jing and Han ethnic groups. As shown in Table 4, the commonest haplotypes were AGBL4 A-A-G, LRP8 G-A-G-C-C and PCSK9 C-A-A-G (>50% of the samples). The frequencies of the AGBL4 A-A-G, AGBL4 G-G-A, LRP8 A-A-G-T-C, LRP8 A-C-A-T-T, LRP8 G-A-G-C-C, PCSK9 C-A-A-G, PCSK9 C-G-A-A and PCSK9 T-G-G-A haplotypes were quantitative significantly different between the Jing and Han populations (P < 0.05–0.001). We confirmed that the AGBL4, LRP8 and PCSK9 haplotypes were associated with TC (AGBL4 G-G-A and PCSK9 C-G-A-A), TG (AGBL4 G-G-A, LRP8 G-A-G-C-C and PCSK9 C-A-A-G), HDL-C (AGBL4 A-A-G), ApoA1 (PCSK9 C-A-A-G), and A1/B (PCSK9 C-A-A-G) in Jing minority. However, they were associated with TG (AGBL4 G-G-A, LRP8 G-A-G-C-C and PCSK9 C-A-A-G), HDL-C (LRP8 A-A-G-T-C), LDL-C (LRP8 A-A-G-T-C), ApoA1 (PCSK9 C-G-A-A), ApoB (AGBL4 G-G-A) and A1/B (LRP8 A-C-A-T-T and PCSK9 C-A-A-G) in Han Chinese. (P < 0.05–0.001; Fig. 5).
Figure 3. The LD plot represents pair-wise r2 and haplotypes frequency in the Jing population.
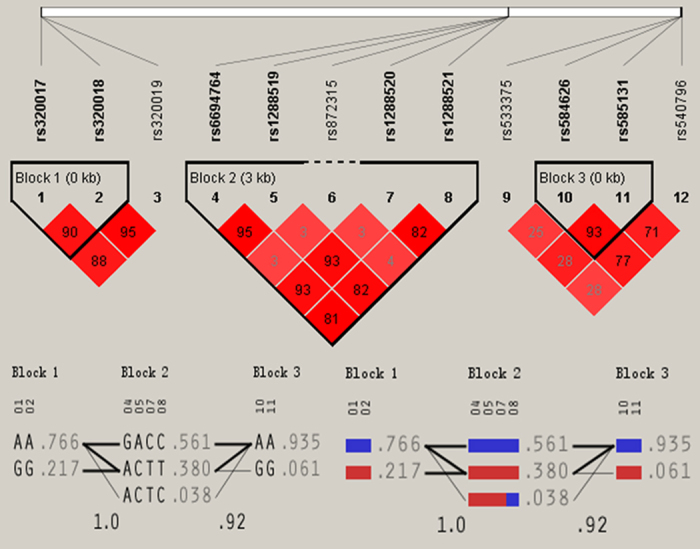
Figure 4. The LD plot represents pair-wise r2 and haplotypes frequency in the Han population.
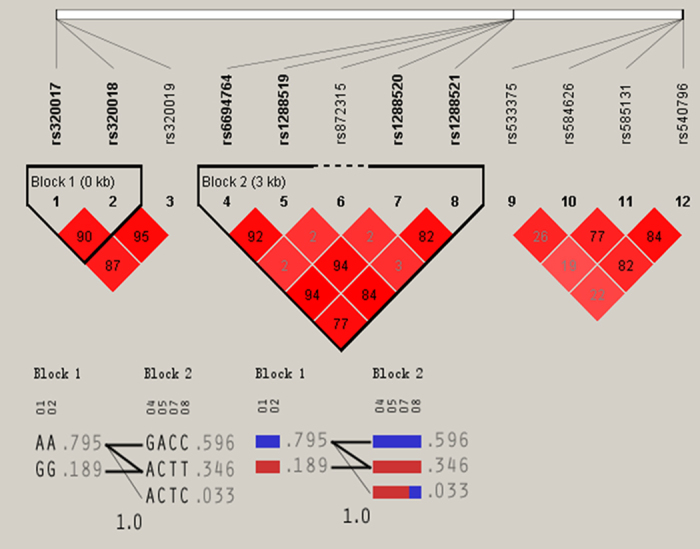
Table 4. Prevalence of haplotype frequencies in the Jing and Han populations [n (frequency)].
| Haplotype | Jing | Han | X2 | P-value | Odds Ratio [95%CI] |
|---|---|---|---|---|---|
| AGBL4 A-A-A | 0.00(0.000) | 4.95(0.002) | 4.927 | 0.026456 | — |
| AGBL4 A-A-G | 1948.87(0.766) | 2030.84(0.793) | 5.509 | 0.018941 | 0.853 [0.747~0.974] |
| AGBL4 A-G-A | 24.62(0.010) | 24.21(0.009) | 0.007 | 0.935631 | 1.023 [0.582~1.798] |
| AGBL4 G-A-A | 5.12(0.002) | 3.15(0.001) | 0.481 | 0.487924 | 1.636 [0.402~6.663] |
| AGBL4 G-A-G | 13.01(0.005) | 12.06(0.005) | 0.043 | 0.836051 | 1.086 [0.495~2.383] |
| AGBL4 G-G-A | 538.26(0.212) | 472.69(0.185) | 5.830 | 0.015777 | 1.185 [1.032~1.360] |
| AGBL4 G-G-G | 13.60(0.005) | 12.10(0.005) | 0.098 | 0.754777 | 1.132 [0.521~2.460] |
| AGBL4 A-G-G | 0.51(0.000) | 0.00(0.000) | 0.517 | 0.472063 | — |
| LRP8 A-A-G-C-C | 13.00(0.005) | 24.19(0.009) | 3.322 | 0.068348 | 0.538 [0.274~1.059] |
| LRP8 A-A-G-T-C | 3.03(0.001) | 12.06(0.005) | 5.361 | 0.020611 | 0.252 [0.071~0.889] |
| LRP8 A-C-A-T-T | 88.00(0.035) | 54.00(0.021) | 8.594 | 0.003384 | 1.663 [1.180~2.344] |
| LRP8 A-C-G-C-C | 12.08(0.005) | 12.01(0.005) | 0.001 | 0.975872 | 1.012 [0.455~2.254] |
| LRP8 A-C-G-C-T | 0.00(0.000) | 0.80(0.000) | 0.791 | 0.373701 | — |
| LRP8 A-C-G-T-C | 96.89(0.038) | 84.86(0.033) | 0.906 | 0.341247 | 1.155 [0.858~1.554] |
| LRP8 A-C-G-T-T | 879.03(0.346) | 831.08(0.325) | 2.499 | 0.113912 | 1.098 [0.978~1.234] |
| LRP8 G-A-A-C-C | 8.00(0.003) | 4.00(0.002) | 1.361 | 0.243302 | 2.015 [0.606~6.700] |
| LRP8 G-A-G-C-C | 1417.91(0.557) | 1522.66(0.595) | 7.322 | 0.006827 | 0.858 [0.768~0.959] |
| LRP8 G-A-G-C-T | 0.00(0.000) | 2.09(0.001) | 2.074 | 0.149794 | — |
| LRP8 G-C-G-C-C | 0.00(0.000) | 0.22(0.000) | 0.219 | 0.639983 | — |
| LRP8 G-C-G-C-T | 0.00(0.000) | 12.03(0.005) | 11.985 | 0.000540 | — |
| LRP8 A-A-G-C-T | 1.01(0.000) | 0.00(0.000) | 1.014 | 0.313942 | — |
| LRP8 A-A-G-T-T | 8.96(0.004) | 0.00(0.000) | 9.032 | 0.002663 | — |
| LRP8 G-A-G-T-C | 16.09(0.006) | 0.00(0.000) | 16.243 | 5.63e-005 | — |
| PCSK9 C-A-A-A | 9.15(0.004) | 3.01(0.001) | 3.150 | 0.075953 | 3.069 [0.833~11.303] |
| PCSK9 C-A-A-G | 2086.39(0.820) | 2191.61(0.856) | 12.173 | 0.000488 | 0.766 [0.660~0.890] |
| PCSK9 C-A-G-A | 0.00(0.000) | 8.02(0.003) | 7.978 | 0.004749 | — |
| PCSK9 C-G-A-A | 8.01(0.003) | 0.51(0.000) | 5.953 | 0.014713 | 15.703 [2.003~123.131] |
| PCSK9 C-G-A-G | 0.00(0.000) | 0.54(0.000) | 0.539 | 0.462699 | — |
| PCSK9 C-G-G-A | 4.42(0.002) | 3.86(0.002) | 0.042 | 0.836807 | 1.154 [0.294~4.528] |
| PCSK9 C-G-G-G | 4.03(0.002) | 3.46(0.001) | 0.048 | 0.827146 | 1.173 [0.279~4.941] |
| PCSK9 T-A-A-G | 259.47(0.102) | 235.37(0.092) | 1.472 | 0.224998 | 1.122 [0.932~1.351] |
| PCSK9 T-G-A-A | 0.00(0.000) | 11.93(0.005) | 11.888 | 0.000569 | — |
| PCSK9 T-G-A-G | 0.00(0.000) | 6.02(0.002) | 5.993 | 0.014383 | — |
| PCSK9 T-G-G-A | 143.43(0.056) | 95.67(0.037) | 10.327 | 0.001317 | 1.539 [1.181~2.006] |
| PCSK9 T-A-A-A | 22.99(0.009) | 0.00(0.000) | 23.237 | 1.46e-006 | — |
| PCSK9 T-A-G-G | 2.00(0.001) | 0.00(0.000) | 2.018 | 0.155421 | — |
| PCSK9 T-G-G-G | 4.10(0.002) | 0.00(0.000) | 4.133 | 0.042083 | — |
AGBL4, the ATP/GTP binding protein-like 4 gene; LRP8, the LDL receptor related protein 8 gene; PCSK9, the Proprotein convertase subtilisin/kexin type 9 gene.
Figure 5. Haplotype-based association with lipid-related traits.
G × G interaction-based association
As shown in Table 5, the commonest G × G interaction was A-A-G-G-A-G-C-C-C-A-A-G (>50% of the samples). The frequencies of the A-A-G-A-A-G-T-C-C-A-A-G, A-A-G-G-A-G-C-C-C-A-A-G, A-A-G-G-A-G-C-C-C-G-A-A, G-G-A-A-C-A-T-T-T-G-G-A and G-G-A-A-C-G-T-T-T-G-G-A G × G interactions were significantly different between Jing and Han populations (P < 0.05–0.001). We identified that the G × G interactions among the detected mutations of AGBL4, LRP8 and PCSK9 were related with TC (G-G-A-A-C-A-T-T-T-G-G-A), TG (A-A-G-G-A-G-C-C-C-A-A-G and A-A-G-G-A-G-C-C-C-G-A-A), HDL-C (A-A-G-A-A-G-T-C-C-A-A-G) and ApoB (A-A-G-A-A-G-T-C-C-A-A-G) in Jing minority. However, in the Hans, with TG (A-A-G-G-A-G-C-C-C-A-A-G and A-A-G-G-A-G-C-C-C-G-A-A) and LDL-C (A-A-G-A-A-G-T-C-C-A-A-G). (P < 0.05–0.001; Fig. 6).
Table 5. Prevalence of G × G interaction frequencies in the Jing and Han populations [n (frequency)].
| G × G interactions | Jing | Han | X2 | P-value | Odds Ratio[95%CI] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | |||||
| A | A | G | A | A | G | C | C | C | A | A | G | 13.00(0.005) | 24.20(0.009) | 3.325 | 0.068240 | 1.858 [0.945~3.653] |
| A | A | G | A | A | G | C | T | C | A | A | G | 1.01(0.000) | 0.00(0.000) | 1.014 | 0.313942 | — |
| A | A | G | A | A | G | T | C | C | A | A | G | 4.00(0.002) | 12.07(0.005) | 4.015 | 0.045102 | 3.009 [0.970~9.340] |
| A | A | G | A | A | G | T | T | C | A | A | G | 7.99(0.003) | 0.00(0.000) | 8.054 | 0.004555 | — |
| A | A | G | A | G | G | C | C | C | A | A | G | 12.09(0.005) | 12.00(0.005) | 0.001 | 0.973787 | 0.987 [0.443~2.197] |
| A | A | G | A | C | G | T | C | C | A | A | G | 95.90(0.038) | 84.09(0.033) | 0.882 | 0.347570 | 0.867 [0.643~1.168] |
| A | A | G | A | C | G | T | T | C | A | A | G | 369.92(0.145) | 356.86(0.139) | 0.377 | 0.539053 | 0.952 [0.814~1.114] |
| A | A | G | A | C | G | T | T | T | A | A | G | 3.08(0.001) | 0.00(0.000) | 3.097 | 0.078447 | — |
| A | A | G | G | A | A | C | C | C | G | G | A | 4.00(0.002) | 2.00(0.001) | 0.680 | 0.409551 | 0.496 [0.091~2.713] |
| A | A | G | G | A | A | C | C | C | G | G | G | 4.00(0.002) | 2.00(0.001) | 0.680 | 0.409551 | 0.496 [0.091~2.713] |
| A | A | G | G | A | G | C | C | C | A | A | A | 8.00(0.003) | 3.00(0.001) | 2.309 | 0.128579 | 0.372 [0.099~1.403] |
| A | A | G | G | A | G | C | C | C | A | A | G | 1401.90(0.551) | 1507.63(0.589) | 7.460 | 0.006325 | 1.167 [1.045~1.304] |
| A | A | G | G | A | G | C | C | C | G | A | A | 8.00(0.003) | 0.50(0.000) | 5.982 | 0.014474 | 0.062 [0.008~0.492] |
| A | A | G | G | A | G | T | C | C | A | A | G | 16.10(0.006) | 0.00(0.000) | 16.248 | 5.61e-005 | — |
| A | G | A | A | C | A | T | T | T | G | G | A | 12.01(0.005) | 12.00(0.005) | 0.000 | 0.985825 | 0.993 [0.445~2.214] |
| A | G | A | A | C | G | T | T | T | A | A | G | 12.00(0.005) | 12.10(0.005) | 0.000 | 0.996678 | 1.002 [0.450~2.230] |
| A | G | G | A | C | G | T | T | T | A | A | G | 1.00(0.000) | 0.00(0.000) | 1.006 | 0.315782 | — |
| G | A | A | A | C | G | T | T | C | A | A | G | 5.00(0.002) | 3.10(0.001) | 0.459 | 0.498128 | 0.616 [0.149~2.541] |
| G | A | G | A | C | A | T | T | T | G | G | A | 11.29(0.004) | 0.00(0.000) | 11.389 | 0.000743 | — |
| G | A | G | A | C | G | T | T | C | A | A | G | 1.72(0.001) | 0.00(0.000) | 1.734 | 0.187919 | — |
| G | G | A | A | C | A | T | T | T | G | G | A | 60.53(0.024) | 36.00(0.014) | 6.513 | 0.010727 | 0.585 [0.386~0.887] |
| G | G | A | A | C | A | T | T | T | G | G | G | 4.16(0.002) | 0.00(0.000) | 4.194 | 0.040582 | — |
| G | G | A | A | C | G | T | T | C | A | A | G | 147.37(0.058) | 160.01(0.063) | 0.472 | 0.492153 | 1.084 [0.861~1.366] |
| G | G | A | A | C | G | T | T | T | A | A | A | 24.16(0.009) | 0.00(0.000) | 24.431 | 7.84e-007 | — |
| G | G | A | A | C | G | T | T | T | A | A | G | 241.74(0.095) | 222.87(0.087) | 0.979 | 0.322457 | 0.908 [0.750~1.099] |
| G | G | A | A | C | G | T | T | T | A | G | G | 1.00(0.000) | 0.00(0.000) | 1.006 | 0.315782 | — |
| G | G | A | A | C | G | T | T | T | G | G | A | 60.00(0.024) | 36.00(0.014) | 6.270 | 0.012298 | 0.590 [0.389~0.896] |
| G | G | G | A | C | G | T | T | C | A | A | G | 12.00(0.005) | 12.00(0.005) | 0.000 | 0.987907 | 0.994 [0.446~2.216] |
| G | G | G | A | C | G | T | T | T | A | G | G | 1.00(0.000) | 0.00(0.000) | 1.006 | 0.315782 | — |
| A | A | A | A | C | G | T | T | C | A | A | G | 0.00(0.000) | 4.90(0.002) | 4.874 | 0.027288 | — |
| A | A | G | A | C | G | C | T | C | A | A | G | 0.00(0.000) | 0.80(0.000) | 0.799 | 0.371406 | — |
| A | A | G | G | A | G | C | C | C | A | G | A | 0.00(0.000) | 8.00(0.003) | 7.962 | 0.004789 | — |
| A | A | G | G | A | G | C | C | C | G | A | G | 0.00(0.000) | 0.50(0.000) | 0.496 | 0.481221 | — |
| A | A | G | G | A | G | C | C | C | G | G | A | 0.00(0.000) | 1.50(0.001) | 1.491 | 0.222007 | — |
| A | A | G | G | A | G | C | C | C | G | G | G | 0.00(0.000) | 1.50(0.001) | 1.491 | 0.222007 | — |
| A | A | G | G | A | G | C | T | C | A | A | G | 0.00(0.000) | 2.10(0.001) | 2.089 | 0.148303 | — |
| A | A | G | G | C | G | C | C | C | A | A | G | 0.00(0.000) | 1.01(0.000) | 1.008 | 0.315460 | — |
| A | A | G | G | C | G | C | T | C | A | A | G | 0.00(0.000) | 11.23(0.004) | 11.180 | 0.000831 | — |
| G | A | G | A | C | G | T | T | T | G | G | A | 0.00(0.000) | 12.00(0.005) | 11.952- | 0.000549 | — |
| G | G | A | A | C | A | T | T | T | G | A | G | 0.00(0.000) | 6.00(0.002) | 5.970 | 0.014571 | — |
| G | G | A | A | C | G | T | T | T | G | A | A | 0.00(0.000) | 12.00(0.005) | 11.952 | 0.000549 | — |
| G | G | A | G | C | G | C | T | T | A | A | G | 0.00(0.000) | 0.03(0.000) | 0.028 | 0.867129 | — |
A, AGBL4 rs320017 A > G; B, AGBL4 rs320018 A > G; C, AGBL4 rs320019 G > A; D, LRP8 rs6694764 G > A; E, LRP8 rs1288519 A > C; F, LRP8 rs872315 G > A; G, LRP8 rs1288520 C > T; H, LRP8 rs1288521 C > T; I, PCSK9 rs533375 C > T; J, PCSK9 rs584626 A > G;K, PCSK9 rs585131 A > G;L, PCSK9 rs540796 G > A; AGBL4, the ATP/GTP binding protein-like 4 gene; LRP8, the LDL receptor related protein 8 gene; PCSK9, the Proprotein convertase subtilisin/kexin type 9 gene.
Figure 6. G × G interaction-based association with plasma lipid levels.
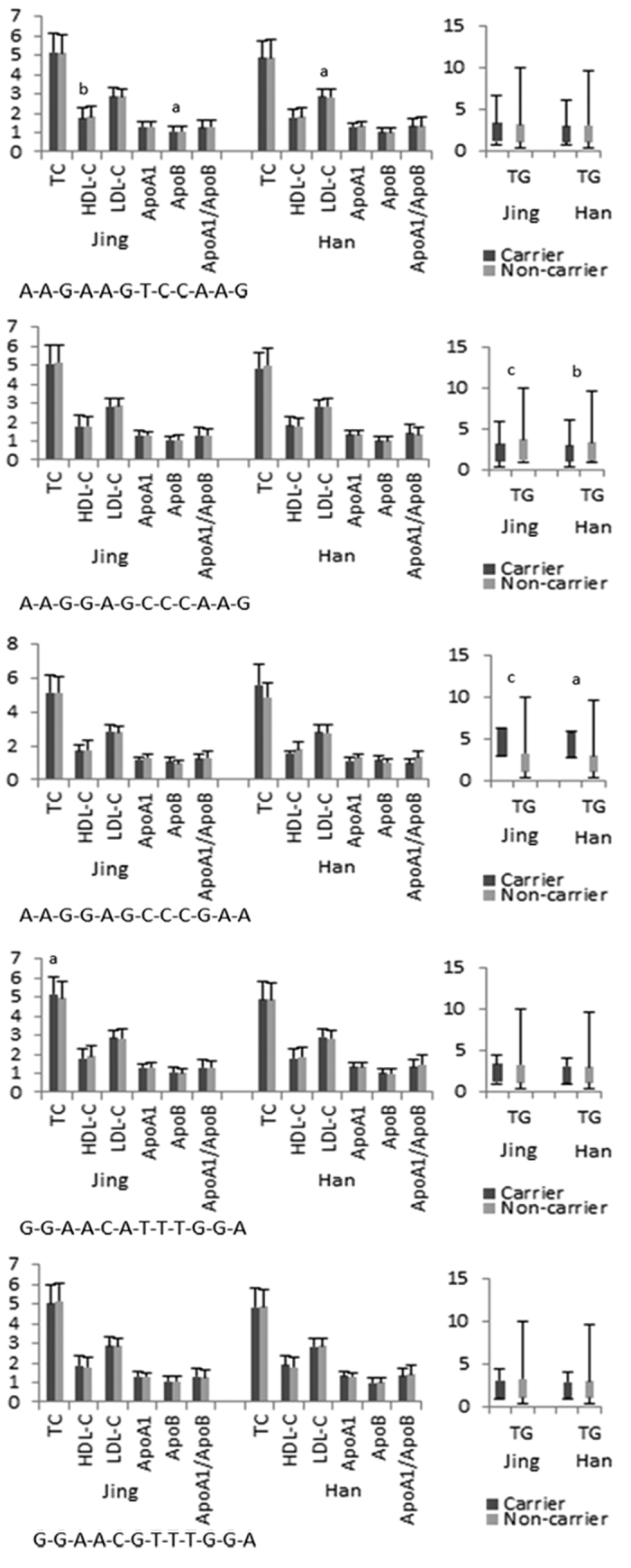
Integrative association analysis of mutation, haplotype and G × G interaction
Table 6 depicts the integrative association analysis of mutation, haplotype and G × G interaction of AGBL4, LRP8 and PCSK9 with lipid phenotypic variations separately in the two ethnic groups. Generalized linear models adjusted for age, gender, BMI, WC, SBP, DBP, pulse pressure, cigarette smoking, alcohol consumption and fasting plasma glucose level demonstrated mutations, haplotypes and G × G interactions of AGBL4, LRP8 and PCSK9 quantitative significantly correlated with lipid-related traits. (P < 0.05–0.001). Furthermore, the association analysis based on haplotype clusters and G × G interactions probably increased power over single-locus tests especially for TG.
Table 6. Association of integrative AGBL4, LRP8 and PCSK9 mutations, haplotypes and G × G interactions with lipid-related traits in the Jing and Han populations.
| Lipid | Mutation/Hapolype/G × G interaction | Affected phenotype/Other phenotype | Unstandardized Coefficients |
Standardized Coefficients | t | P-value | |
|---|---|---|---|---|---|---|---|
| B | Std.error | Beta | |||||
| Jing | |||||||
| TC | PCSK9 rs533375 | CC/CT/TT | 0.194 | 0.090 | 0.117 | 2.160 | 0.031 |
| PCSK9 rs585131 | A/G | 0.823 | 0.363 | 0.296 | 2.270 | 0.023 | |
| AGBL4 G-G-A | Carriers/Non-carriers | 0.167 | 0.050 | 0.089 | 3.368 | 0.001 | |
| PCSK9 C-G-A-A | Carriers/Non-carriers | 0.178 | 0.085 | 0.056 | 2.100 | 0.036 | |
| PCSK9 T-G-G-A | Carriers/Non-carriers | 0.151 | 0.076 | 0.053 | 1.992 | 0.047 | |
| G-G-A-A-C-A-T-T-T-G-G-A | Carriers/Non-carriers | 0.234 | 0.104 | 0.059 | 2.236 | 0.026 | |
| TG | AGBL4 rs320017 | AA/AG/GG | 0.272 | 0.099 | 0.174 | 2.752 | 0.006 |
| AGBL4 rs320017 | A/G | 0.894 | 0.205 | 0.473 | 4.369 | 1.352E-05 | |
| AGBL4 rs320018 | A/G | −1.287 | 0.244 | −0.683 | −5.274 | 1.573E-07 | |
| AGBL4 rs320019 | G/A | 0.533 | 0.208 | 0.282 | 2.556 | 0.011 | |
| LRP8 rs6694764 | GG/AG/AA | 0.980 | 0.165 | 0.739 | 5.933 | 3.845E-09 | |
| LRP8 rs6694764 | G/A | 0.623 | 0.190 | 0.314 | 3.283 | 0.001 | |
| LRP8 rs1288519 | AA/AC/CC | −1.588 | 0.171 | −1.203 | −9.278 | 7.367E-20 | |
| LRP8 rs1288519 | A/C | −1.048 | 0.224 | −0.535 | −4.675 | 3.265E-06 | |
| LRP8 rs1288520 | CC/CT/TT | 0.537 | 0.141 | 0.417 | 3.819 | 1.405E-04 | |
| LRP8 rs1288520 | C/T | 0.437 | 0.219 | 0.224 | 1.991 | 0.047 | |
| LRP8 rs1288521 | CC/CT/TT | 0.226 | 0.088 | 0.171 | 2.576 | 0.010 | |
| PCSK9 rs533375 | CC/CT/TT | 0.348 | 0.081 | 0.205 | 4.301 | 1.831E-05 | |
| PCSK9 rs584626 | AA/AG/GG | 0.724 | 0.182 | 0.277 | 3.986 | 7.113E-05 | |
| PCSK9 rs540796 | GG/AG/AA | −0.621 | 0.118 | −0.250 | −5.276 | 1.557E-07 | |
| PCSK9 rs540796 | G/A | −0.754 | 0.146 | −0.284 | −5.163 | 2.829E-07 | |
| AGBL4 A-A-G | Carriers/Non-carriers | 0.357 | 0.096 | 0.093 | 3.723 | 2.052E-04 | |
| AGBL4 G-G-A | Carriers/Non-carriers | −0.234 | 0.047 | −0.123 | −4.979 | 0.000 | |
| LRP8 G-A-G-C-C | Carriers/Non-carriers | 0.349 | 0.057 | 0.151 | 6.110 | 1.329E-09 | |
| PCSK9 C-A-A-G | Carriers/Non-carriers | 1.110 | 0.105 | 0.255 | 10.577 | 4.166E-25 | |
| A-A-G-G-A-G-C-C-C-A-A-G | Carriers/Non-carriers | 0.400 | 0.056 | 0.175 | 7.149 | 1.482E-12 | |
| A-A-G-G-A-G-C-C-C-G-A-A | Carriers/Non-carriers | −1.302 | 0.335 | −0.097 | −3.888 | 1.065E-04 | |
| HDL-C | LRP8 rs6694764 | GG/AG/AA | 0.221 | 0.112 | 0.291 | 1.977 | 0.048 |
| LRP8 rs1288519 | AA/AC/CC | −0.236 | 0.116 | −0.313 | −2.040 | 0.042 | |
| AGBL4 A-A-G | Carriers/Non-carriers | 0.123 | 0.061 | 0.056 | 2.016 | 0.044 | |
| LRP8 A-A-G-T-C | Carriers/Non-carriers | 0.071 | 0.029 | 0.067 | 2.414 | 0.016 | |
| A-A-G-A-A-G-T-C-C-A-A-G | Carriers/Non-carriers | 0.082 | 0.029 | 0.078 | 2.792 | 0.005 | |
| LDL-C | LRP8 rs1288521 | CC/CT/TT | 0.108 | 0.049 | 0.177 | 2.226 | 0.026 |
| ApoA1 | PCSK9 C-A-A-G | Carriers/Non-carriers | −0.075 | 0.031 | −0.066 | −2.417 | 0.016 |
| ApoB | LRP8 rs1288520 | CC/CT/TT | 0.091 | 0.044 | 0.262 | 2.060 | 0.040 |
| LRP8 rs1288521 | CC/CT/TT | 0.061 | 0.028 | 0.171 | 2.220 | 0.027 | |
| LRP8 rs1288521 | C/T | 0.090 | 0.034 | 0.176 | 2.618 | 0.009 | |
| A-A-G-A-A-G-T-C-C-A-A-G | Carriers/Non-carriers | −0.028 | 0.014 | −0.056 | −2.050 | 0.041 | |
| ApoA1/ApoB | LRP8 rs1288521 | C/T | −0.121 | 0.052 | −0.154 | −2.311 | 0.021 |
| PCSK9 C-A-A-G | Carriers/Non-carriers | −0.098 | 0.049 | −0.054 | −2.004 | 0.045 | |
| Han | |||||||
| TC | AGBL4 rs320018 | AA/AG/GG | −0.551 | 0.209 | −0.370 | −2.632 | 0.009 |
| AGBL4 rs320019 | GG/AG/AA | 0.505 | 0.187 | 0.341 | 2.698 | 0.007 | |
| TG | AGBL4 rs320017 | AA/AG/GG | 0.543 | 0.117 | 0.311 | 4.635 | 3.942E-06 |
| AGBL4 rs320017 | A/G | 1.059 | 0.253 | 0.507 | 4.178 | 3.147E-05 | |
| AGBL4 rs320018 | AA/AG/GG | −1.793 | 0.213 | −1.030 | −8.416 | 1.049E-16 | |
| AGBL4 rs320018 | A/G | −1.447 | 0.300 | -.695 | −4.826 | 1.562E-06 | |
| AGBL4 rs320019 | GG/AG/AA | 1.262 | 0.190 | 0.730 | 6.628 | 5.035E-11 | |
| LRP8 rs6694764 | GG/AG/AA | −0.446 | 0.153 | −0.311 | −2.906 | 0.004 | |
| LRP8 rs1288519 | AA/AC/CC | 1.173 | 0.160 | 0.820 | 7.334 | 4.005E-13 | |
| LRP8 rs1288519 | A/C | 0.540 | 0.249 | 0.263 | 2.173 | 0.030 | |
| LRP8 rs872315 | GG/AG/AA | 0.489 | 0.176 | 0.106 | 2.779 | 0.006 | |
| LRP8 rs872315 | G/A | 0.616 | 0.181 | 0.127 | 3.412 | 0.001 | |
| LRP8 rs1288520 | CC/CT/TT | −1.049 | 0.176 | −0.743 | −5.974 | 3.020E-09 | |
| LRP8 rs1288520 | C/T | −0.569 | 0.230 | −0.279 | −2.473 | 0.014 | |
| LRP8 rs1288521 | CC/CT/TT | 0.327 | 0.096 | 0.227 | 3.409 | 0.001 | |
| LRP8 rs1288521 | C/T | 0.273 | 0.118 | 0.136 | 2.312 | 0.021 | |
| PCSK9 rs533375 | CC/CT/TT | 0.550 | 0.089 | 0.276 | 6.174 | 8.956E-10 | |
| PCSK9 rs533375 | C/T | 0.430 | 0.099 | 0.187 | 4.358 | 1.419E-05 | |
| PCSK9 rs584626 | AA/AG/GG | 0.421 | 0.199 | 0.129 | 2.116 | 0.035 | |
| PCSK9 rs584626 | A/G | 0.849 | 0.268 | 0.247 | 3.166 | 0.002 | |
| PCSK9 rs540796 | GG/AG/AA | −0.603 | 0.221 | −0.182 | −2.730 | 0.006 | |
| PCSK9 rs540796 | G/A | −1.051 | 0.341 | −0.309 | −3.083 | 0.002 | |
| AGBL4 A-A-G | Carriers/Non-carriers | 0.890 | 0.109 | 0.205 | 8.133 | 9.903E-16 | |
| AGBL4 G-G-A | Carriers/Non-carriers | −0.319 | 0.054 | −0.151 | −5.942 | 3.632E-09 | |
| LRP8 A-A-G-T-C | Carriers/Non-carriers | 0.129 | 0.051 | 0.065 | 2.509 | 0.012 | |
| LRP8 A-C-A-T-T | Carriers/Non-carriers | −0.522 | 0.127 | −0.106 | −4.099 | 4.409E-05 | |
| LRP8 G-A-G-C-C | Carriers/Non-carriers | 0.511 | 0.068 | 0.191 | 7.550 | 8.322E-14 | |
| PCSK9 C-A-A-G | Carriers/Non-carriers | 1.299 | 0.146 | 0.225 | 8.917 | 1.638E-18 | |
| A-A-G-A-A-G-T-C-C-A-A-G | Carriers/Non-carriers | 0.119 | 0.051 | 0.060 | 2.323 | 0.020 | |
| A-A-G-G-A-G-C-C-C-A-A-G | Carriers/Non-carriers | 0.538 | 0.067 | 0.203 | 8.049 | 1.919E-15 | |
| HDL-C | PCSK9 rs533375 | CC/CT/TT | −0.100 | 0.050 | −0.109 | −2.005 | 0.045 |
| LRP8 rs6694764 | G/A | 0.185 | 0.091 | 0.194 | 2.029 | 0.043 | |
| LRP8 rs1288521 | C/T | −0.130 | 0.062 | −0.140 | −2.097 | 0.036 | |
| PCSK9 rs533375 | C/T | −0.133 | 0.052 | −0.126 | −2.586 | 0.010 | |
| LRP8 A-A-G-T-C | Carriers/Non-carriers | −0.059 | 0.026 | −0.064 | −2.276 | 0.023 | |
| LDL-C | LRP8 rs1288520 | C/T | 0.224 | 0.112 | 0.255 | 1.999 | 0.046 |
| LRP8 rs1288521 | C/T | −0.129 | 0.057 | −0.150 | −2.248 | 0.025 | |
| LRP8 A-A-G-T-C | Carriers/Non-carriers | −0.059 | 0.024 | −0.069 | −2.464 | 0.014 | |
| A-A-G-A-A-G-T-C-C-A-A-G | Carriers/Non-carriers | −0.057 | 0.024 | −0.066 | −2.375 | 0.018 | |
| ApoA1 | AGBL4 rs320017 | A/G | −0.147 | 0.055 | −0.347 | −2.665 | 0.008 |
| AGBL4 rs320018 | A/G | 0.146 | 0.065 | 0.346 | 2.236 | 0.026 | |
| PCSK9 rs540796 | G/A | −0.153 | 0.074 | −0.223 | −2.064 | 0.039 | |
| PCSK9 C-G-A-A | Carriers/Non-carriers | 0.049 | 0.021 | 0.061 | 2.282 | 0.023 | |
| ApoB | AGBL4 rs320018 | A/G | −0.158 | 0.079 | −0.316 | −1.992 | 0.047 |
| AGBL4 rs320019 | G/A | 0.149 | 0.059 | 0.295 | 2.506 | 0.012 | |
| PCSK9 rs540796 | G/A | 0.240 | 0.090 | 0.294 | 2.660 | 0.008 | |
| AGBL4 A-A-G | Carriers/Non-carriers | −0.084 | 0.028 | −0.080 | −2.945 | 0.003 | |
| ApoA1/ApoB | PCSK9 rs533375 | CC/CT/TT | 0.082 | 0.039 | 0.106 | 2.092 | 0.037 |
| AGBL4 rs320018 | A/G | 0.365 | 0.123 | 0.454 | 2.966 | 0.003 | |
| AGBL4 rs320019 | G/A | −0.205 | 0.092 | −0.254 | −2.231 | 0.026 | |
| PCSK9 rs584626 | A/G | 0.244 | 0.110 | 0.184 | 2.214 | 0.027 | |
| PCSK9 rs540796 | G/A | −0.430 | 0.140 | −0.328 | −3.074 | 0.002 | |
| AGBL4 A-A-G | Carriers/Non-carriers | 0.165 | 0.044 | 0.098 | 3.727 | 2.026E-04 | |
| LRP8 A-C-A-T-T | Carriers/Non-carriers | −0.137 | 0.051 | −0.072 | −2.704 | 0.007 | |
| PCSK9 C-A-A-G | Carriers/Non-carriers | 0.185 | 0.059 | 0.083 | 3.126 | 0.002 | |
HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; Apo, apolipoprotein; AGBL4, the ATP/GTP binding protein-like 4 gene; LRP8, the LDL receptor related protein 8 gene; PCSK9, the Proprotein convertase subtilisin/kexin type 9 gene.
Discussion
The main finding of the present study encompass (i) it elucidated the frequencies of mutation, haplotype and the G × G inter-locus interaction among AGBL4, LRP8 and PCSK9 genes in the Jing ethnic minority and Han population, which may be proposed as an potential supplement to the 1000 Genomes database (ii) it gave integrative mutation, haplotype and G × G interaction evidence to prove there are possible interaction between the AGBL4, LRP8 and PCSK9 genes and serum lipid concentrations; and (iii) it demonstrated association analysis based on haplotype clusters and G × G interactions probably increased power over single-locus tests especially for TG.
Aspects of primary prevention differ in some respects in ethnic minority groups when compared with general population17. Jing, as a group of migrants from Vietnam to south of China, maintains the higher cardiometabolic risk especially higher TC and TG, and lower A1/B ratio than local Han population living in the same natural and social environments. It is important to recognize that definitions of cardiometabolic risk especially dyslipidemia derived in local Han population perhaps inappropriate for ethnic minority groups. Resulting disease risks may remain difference in second and third generation migrants, even though blood pressure, fasting plasma glucose level and cigarette smoking lifestyle are converging towards those of the general Han population. Our present study pronounces differences in genetics values. The challenge now is to ensure that prevention and treatment services are ready to respond to these demographic and ethnic structure. Epidemiological survey has revealed that the Jing ethnic minority maintains genetic homogeneity. In the present study, all of the mutations satisfied with HWE separately in each population. It has been proved that the Jing and Han populations have different genetic ancestry from a statistical point of view. Our results showed that there was quantitative significantly different distributions of the detected 12 mutations of AGBL4, LRP8 and PCSK9 genes, their haplotypes and their G × G inter-locus interactions between the Jing and Han populations. These genetic heterogeneity may be correlated with the heterogeneousness of cardiometabolic risk especially dyslipidemia between the Jing and Han populations.
Environmental exposures cannot be ignored. We summarized the values of weight, BMI and WC were significantly different between the two populations. Maybe they are related with the custom of fish intake. Jing is an oceanic ethnic minority like Kinh populations in North Vietnam, survival relying on fishing18. Maybe there are differences in saturated fatty acid (SFA), polyunsaturated tatty acid (PUFA; n-3 PUFA and n-6 PUFA), and monounsaturated fatty acid (MUFA)19 intake to compare with the local Han population in their diet structure. Unfortunately it is only a hypothesis, because lack of dietary intake data. Consensus exists pertaining to the scientific evidence regarding effects of various those bad dietary fatty acids rich in fish on cardiometabolic risk including lipid phenotypic variations reported in a previous study20. What’s more, the cardiometabolic risk is known to be lower in light-to-moderate alcohol drinkers than in abstainers21. The effects of alcohol on lipid metabolism, especially the HDL cholesterol-elevating effects, are thought to greatly contribute to the cardio-protective action of alcohol22. On the other hand, excessive alcohol consumption has been shown to cause hypertriglyceridemia23,24, which is a prevalent risk factor for CVD. With regard to mechanisms underlying the effects of alcohol on lipid metabolism25,26,27, alcohol consumption has been shown to increase the activity of lipoprotein lipase and decrease the activity of cholesteryl ester transfer protein, resulting in elevation of HDL cholesterol28. Hypertriglyceridemia induced by excessive alcohol drinking may be mainly due to an increase in the synthesis of large very low-density lipoprotein (VLDL) particles in the liver. Consistently, the % of participants who consumed alcohol was different between the two groups. Wine culture plays a pivotal role in the history of China Han ethnic group. Many Han populations are good at alcohol consumption, especially in festivals.
Our data come from nuclear family and pedigree data, unfortunately, pedigree information were not documented. Heritability is a measure of familial resemblance29. Estimating the heritability of a trait represents one of the first steps in the gene mapping process. Or we can estimate heritability for quantitative traits from nuclear and pedigree data using the ASSOC program in the Statistical Analysis for Genetic Epidemiology (S.A.G.E.) software package. Estimating heritability rests on the assumption that the total phenotypic variance of a quantitative trait can be partitioned into independent genetic and environmental components30.
A number of clinical studies have demonstrated that inhibition of PCSK9 alone and in addition to statins potently reduces lipid phenotypic variation concentrations31,32. Plasma lipid phenotypic variation especially plasma TG level is heritable and modifiable33. Several groups have successfully to identify signals for TG and other lipid traits, including HDL-C, LDL-C, and TC34. However, the lead GWAS signals may not themselves be functional rather in LD with the actual underlying susceptibility mutations. The limitation in GWAS derives from the fact that the human genome is superficially screened using single independently tag SNVs. It is acknowledged that complex disease is not caused by or associated with one single variant. The functional mutation often acts through regional gene mutations, including haplotypes and G × G interactions. Therefore, GWAS, epigenome-wide association studies (EWAS) and transcriptome-wide association studies (TWAS) are only a starting and require subsequent fine mapping and functional validation to identify the actual susceptibility variants and gene interactions. AGBL4, LRP8 and PCSK9 genes are neighbors. Integrative mutations, haplotypes and G × G interactions evidence connects AGBL4, LRP8 and PCSK9 gene to lipid phenotypic variations perhaps can further elaborate the clinical application of PCSK9 inhibitors.
There are several limitations in our study. Firstly, the number of participants available for minor allele frequency (MAF) of some mutations was not high enough to calculate a strong power as compared with many previous GWAS and replication studies. Secondly, as an association analysis and observation study, inherent methodologic limitations that generate bias and confounding mean that causal inferences cannot reliably be drawn. Thirdly, take into consideration the randomized clinical trials (RCTs) provide the best opportunity to control for confounding and avoid certain biases. Consequently, well-designed, high-quality further therapeutic intervention study, including prophylactic agent, treatment, surgical approach, or diagnostic test is needed. Moreover, there are still many unmeasured environmental and genetic factors including TFA, SFA, PUFA (including n-3 PUFA and n-6 PUFA) and MUFA that needed to be considered. In addition, the relevance of this finding has to be defined in further high caliber of studies including incorporating the genetic information of AGBL4, LRP8 and PCSK9 gene mutations, haplotypes and G × G interactions in vivo and vitro functional studies to confirm the impact of a variant on a molecular level including transcription and expression. The last but not the least, discussion of race and ethnicity in medicine must rigorously avoid polarization and the further perpetuation of disparate health care.
In summary, there are potential interaction between the AGBL4, LRP8 and PCSK9 genes and serum lipid concentrations. And the association analysis based on haplotype clusters and G × G interactions probably increased power over single-locus tests especially for TG. These genetic heterogeneity may be correlated with the heterogeneousness of cardiometabolic risk between the Jing and Han populations. Differences in lipid phenotypic variations between the two populations might partially attribute to AGBL4, LRP8 and PCSK9 gene mutations, haplotypes and G × G interactions.
Materials and Methods
Ethical approval
The study were carried out following the rules of the Declaration of Helsinki of 1975 (http://www.wma.net/en/30publications/10policies/b3/), revised in 2008. All participants from contributing populations gave written informed consent to participate in epidemiologic investigation and genetic analysis. All study protocols in this motif have approval from the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University (No: Lunshen-2011-KY-Guoji-001; Mar. 7, 2011).
Subjects
Two groups of study population including 1272 unrelated participants of Jing (624 males, 49.06% and 648 females, 50.94%) and 1280 unrelated subjects of Han (636 males, 49.69% and 644 females, 50.31%) were randomly selected from our previous stratified randomized samples35. All participants were rural fishery (Jing) and/or agricultural (Han) workers from the three islands of Wanwei, Wutou and Shanxin in the county of Fangchenggang in the province of Guangxi, China, near the Sino-Vietnamese border. The participants’ age ranged from 18 to 80 years with a mean age of 57.27 ± 12.85 years in Jing and 56.85 ± 13.32 years in Han; respectively. The gender ratio and age distribution were matched between the two groups. All participants were essentially healthy with no history of coronary artery disease, stroke, diabetes, hyper- or hypo-thyroids, and chronic renal disease. They were free from medications known to affect lipid profiles.
Epidemiological survey
The epidemiological survey was carried out using internationally standardized method, following a common protocol36. Information on demographics, socioeconomic status, and lifestyle factors were collected with standardized questionnaires. Cigarette smoking status was categorized into groups of cigarettes per day: ≤20 and >2037. Alcohol consumption was categorized into groups of grams of alcohol per day: ≤25 and >2538. Several parameters such as blood pressure, height, weight and WC were measured, while BMI (kg/m2) was calculated. BMI was categorized into four groups: underweight (BMI < 18.5), normal weight (18.5 ≤ BMI < 24), overweight (24 ≤ BMI < 28) and Obesity (28 ≤ BMI)39. Likewise, WC was categorized into groups including normal group (WC ≤ 85 for male and WC ≤ 80 for female) and abdominal obesity (WC > 85 for male and WC > 80 for female)40.
Biochemical measurements
A fasting venous blood sample of 5 ml was drawn from the participants. The levels of fasting plasma TC, TG, HDL-C and LDL-C in the samples were determined by enzymatic methods with commercially available kits. Fasting plasma ApoA1 and ApoB levels were assessed by the immuneturbidimetric immunoassay.
Diagnostic criteria
The normal values of fasting plasma TC, TG, HDL-C, LDL-C, ApoA1 and ApoB levels, as well as the A1/B ratio in our Clinical Science Experiment Center were 3.10–5.17, 0.56–1.70, 1.16–1.42, 2.70–3.10 mmol/L, 1.20–1.60, 0.80–1.05 g/L, and 1.00–2.50; respectively41.
Mutation selection
We selected 12 mutations in the AGBL4, LRP8 and PCSK9 with the following assumption: (i) tag SNVs, which were established by Haploview (Broad Instituteof MIT and Harvard, Cambridge, MA, USA, version 4.2); (ii) functional mutations (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) in functional areas of the gene fragment from NCBI dbSNP Build 132 (http://www-ncbi-nlm-nih-gov.ezp-prod1.hul.harvard.edu/SNP/); (iii) a known minor allele frequency (MAF) higher than 1% in European ancestry from the Human Genome Project Database; and (iiii) mutations might be associated with the lipid-related traits or cardiometabolic risk in the latest studies.
Genotyping
Genomic DNA was extracted from leucocytes of venous blood using the phenol-chloroform method. Genotyping of 12 mutations was performed by PCR and Sanger sequencing. The characteristics of each mutation and the details of each primer pair, annealing temperature, length of the PCR products are summarized in Supplemental Tables 1 and 2. The PCR products of the samples were sequenced with a sequencer ABI Prism 3100 Genetic Analyzer (Applied Biosystems, International Equipment Trading Ltd., Vernon Hills, IL, USA) in Shanghai Sangon Biological Engineering Technology & Services Co. Ltd., Shanghai China.
Statistical Analyses
The statistical analysis were performed with the statistical software SPSS 21.0 (SPSS Inc., Chicago, IL, USA). Quantitative variables were presented as the mean ±SD for those, that are normally distributed, whereas the medians and interquartile ranges for TG, which is not normally distributed. General characteristics between the two groups were compared by the ANCOVA. The distributions of the genotype, allele, haplotype and G × G interaction between the two groups were analyzed by the chi-squared test; The HWE, Pair-wise LD, frequencies of haplotype and G × G interaction comprising the mutations were calculated using Haploview (version 4.2; Broad Institute of MIT and Harvard). The association of the genotypes, haplotypes and G × G interactions with lipid phenotypic variations was tested by the Univariant. Any variants associated with the lipid phenotypic variations at a value of P < 0.05 were considered statistically significant. Generalized linear models were used to assess the association of the genotypes (common homozygote genotype = 1, heterozygote genotype = 2, rare homozygote genotype = 3), alleles (the minor allele non-carrier = 1, the minor allele carrier = 2), haplotypes (the haplotype non-carrier = 1, the haplotype carrier = 2) and G × G interactions (the G × G interaction non-carrier = 1, the G × G interaction carrier = 2) with lipid phenotypic variations. The model of age, gender, BMI, WC, SBP, DBP, pulse pressure, cigarette smoking, alcohol consumption and fasting plasma glucose level were adjusted for the statistical analysis. The pattern of pair-wise LD between the selected mutations was measured by D′ and r2 using the Haploview software.
Additional Information
How to cite this article: Guo, T. et al. Integrative mutation, haplotype and G × G interaction evidence connects ABGL4, LRP8 and PCSK9 genes to cardiometabolic risk. Sci. Rep. 6, 37375; doi: 10.1038/srep37375 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors acknowledge the essential role of the funding of the National Natural Science Foundation of China (No: 81160111) and the Innovation Project of Guangxi Graduate Education in this motif.
Footnotes
Author Contributions The project concept and design: T.G. and R.-X.Y. Collection and characterization of the demographic and epidemiological data: T.G., R.-X.Y., L.-M.Y., F.H., L.P., W.-X.L., D.-Z.Y and S.-L.P. Performed the experiments: T.G., W.-X.L. and D.-Z.Y. Performed statistical analysis: T.G., R.-X.Y. and S.-L.P. Wrote the draft of the manuscript: T.G. Critical revision of the manuscript for important intellectual content: R.-X.Y. All authors read, reviewed and approved the final manuscript.
References
- Catapano A. L. et al. ESC/EAS Guidelines for the Management of Dyslipidaemias: The Task Force for the Management of Dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS)Developed with the special contribution of the European Assocciation for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J, ehw272 (2016). [DOI] [PubMed] [Google Scholar]
- Sacco R. L. et al. The Heart of 25 by 25: Achieving the Goal of Reducing Global and Regional Premature Deaths From Cardiovascular Diseases and Stroke: A Modeling Study From the American Heart Association and World Heart Federation. Circulation 133, e674–e690 (2016). [DOI] [PubMed] [Google Scholar]
- Santulli G. et al. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J Clin Invest 125, 1968–1978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G. & Marks A. R. Essential Roles of Intracellular Calcium Release Channels in Muscle, Brain, Metabolism, and Aging. Curr Mol Pharmacol 8, 206–222 (2015). [DOI] [PubMed] [Google Scholar]
- Sperling L. S. et al. The CardioMetabolic Health Alliance: Working Toward a New Care Model for the Metabolic Syndrome. J Am Coll Cardiol 66, 1050–1067 (2015). [DOI] [PubMed] [Google Scholar]
- Stone N. J. et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 63, 2889–2934 (2014). [DOI] [PubMed] [Google Scholar]
- Nordestgaard B. G. & Varbo A. Triglycerides and cardiovascular disease. Lancet 384, 626–635 (2014). [DOI] [PubMed] [Google Scholar]
- Global Lipids Genetics Consortium, Willer C. J. et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- On beyond GWAS. Nat Genet 42, 551 (2010). [DOI] [PubMed] [Google Scholar]
- Williams S. M. & Haines J. L. Correcting away the hidden heritability. Ann Hum Genet 75, 348–350 (2011). [DOI] [PubMed] [Google Scholar]
- Petros Z. et al. Genome-wide association and replication study of anti-tuberculosis drugs-induced liver toxicity. BMC Genomics 17, 755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. C. et al. Clinical Relevance of Plasma DNA Methylation in Colorectal Cancer Patients Identified by Using a Genome-Wide High-Resolution Array. Ann Surg Oncol 22, S1419–S1427 (2015). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Premature myocardial infarction novel susceptibility locus on chromosome 1P34–36 identified by genomewide linkage analysis. Am J Hum Genet 74, 262–271 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G. Q. et al. An LRP8 variant is associated with familial and premature coronary artery disease and myocardial infarction. Am J Hum Genet 81, 780–791 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. et al. Association between the DOCK7, PCSK9 and GALNT2 Gene Polymorphisms and Serum Lipid levels. Sci Rep 6, 19079 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron N., Bergeron N., Phan B. A., Ding Y., Fong A. & Krauss R. M. Proprotein convertase subtilisin/kexin type 9 inhibition: a new therapeutic mechanism for reducing cardiovascular disease risk. Circulation 132, 1648–1666 (2005). [DOI] [PubMed] [Google Scholar]
- Chaturvedi N. Ethnic differences in cardiovascular disease. Heart 89, 681–686 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieu N. T. et al. Serum fatty acids, lipoprotein (a) and apolipoprotein composition of rural, suburban and urban populations in North Vietnam. Asia Pac J Clin Nutr 9, 66–72 (2000). [PubMed] [Google Scholar]
- Kris-Etherton P. M. & Innis S. Ammerican Dietetic Assocition & Dietitians of Canada. Position of the American Dietetic Association and Dietitians of Canada: dietary fatty acids. J Am Diet Assoc 107, 1599–1611(2007). [PubMed] [Google Scholar]
- Hammad S., Pu S. & Jones P. J. Current Evidence Supporting the Link Between Dietary Fatty Acids and Cardiovascular Disease. Lipids 51, 507–517 (2016). [DOI] [PubMed] [Google Scholar]
- Corrao G., Rubbiati L., Bagnardim V., Zambon A. & Poikolainen K. Alcohol and coronary heart disease: a meta-analysis. Addiction 95, 1505–1523 (2000). [DOI] [PubMed] [Google Scholar]
- Ellison R. C. et al. Lifestyle determinants of high-density lipoprotein cholesterol: the National Heart, Lung, and Blood Institute Family Heart Study. Am Heart J 14, 529–535 (2004). [DOI] [PubMed] [Google Scholar]
- Castelli W. P. et al. Alcohol and blood lipids. The cooperative lipoprotein phenotyping study. Lancet 2, 153–155 (1977). [DOI] [PubMed] [Google Scholar]
- Van de Wiel A. The effect of alcohol on postprandial and fasting triglycerides. Int J Vasc Med 2012, 862504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson J. E. & Austin M. A. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J Cardiovasc Risk 3, 213–219 (1996). [PubMed] [Google Scholar]
- Durrington P. N. Triglycerides are more important in atherosclerosis than epidemiology has suggested. Atherosclerosis 141, S57–S62 (1998). [DOI] [PubMed] [Google Scholar]
- Labreuche J., Touboul P. J. & Amarenco P. Plasma triglyceride levels and risk of stroke and carotid atherosclerosis: a systematic review of the epidemiological studies. Atherosclerosis 203, 331–345 (2009). [DOI] [PubMed] [Google Scholar]
- Hannuksela M. L., Rämet M. E., Nissinen A. E. T., Liisanantti M. K. & Savolainen M. J. Effects of ethanol on lipids and atherosclerosis. Pathophysiology 10, 93–103 (2004). [DOI] [PubMed] [Google Scholar]
- Bochud M. Estimating heritability from nuclear family and pedigree data. Methods Mol Biol 850, 171–186 (2012). [DOI] [PubMed] [Google Scholar]
- van Dongen J. et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun 7, 11115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban D., Pöss J., Böhm M. & Laufs U. Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis. J Am Coll Cardiol 62, 1401–1408 (2013). [DOI] [PubMed] [Google Scholar]
- Laufs U., Custodis F. & Werner C. PCSK9 inhibitors. Herz, doi: 10.1007/s00059-016-4429-1 (2016). [DOI] [PubMed] [Google Scholar]
- Weissglas-Volkov D. et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet 50, 298–308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancáková A. et al. Hyperglycemia and a common variant of GCKR are9 associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes 61, 1895–1902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. et al. Association of the variants and haplotypes in the DOCK7, PCSK9 and GALNT2 genes and the risk of hyperlipidaemia. J Cell Mol Med 20, 243–265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. et al. Association of the SPT2 chromatin protein domain containing 1 gene rs17579600 polymorphism and serum lipid traits. Int J Clin Exp Pathol 8, 12995–13010 (2015). [PMC free article] [PubMed] [Google Scholar]
- Okuyemi K. S., Ahluwalia J. S., Richter K. P., Mayo M. S. & Resnicow K. Differences among African American light, moderate, and heavy smokers. Nicotine Tob Res 3, 45–50 (2001). [DOI] [PubMed] [Google Scholar]
- Kerr W. C., Mulia N. & Zemore S. E. U.S. trends in light, moderate, and heavy drinking episodes from 2000 to 2010. Alcohol Clin Exp Res 38, 2496–2501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Obesity: preventing and managing the global epidemic. Report of a WHO Consultation. World Health Organ Tech Rep Ser 894, 1–253 (2000). [PubMed] [Google Scholar]
- Han T. S., van Leer E. M., Seidell J. C. & Lean M. E. Waist circumference action levels in the identification of cardiovascular risk factors: prevalence study in a random sample. BMJ 311, 1401–1405 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrington P. Dyslipidaemia. Lancet 362, 717–731 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.