Abstract
Caveolin-1 (CAV1) is an oncogenic membrane protein associated with endocytosis, extracellular matrix organisation, cholesterol distribution, cell migration and signaling. Recent studies reveal that CAV1 is involved in metabolic alterations – a critical strategy adopted by cancer cells to their survival advantage. Consequently, research findings suggest that CAV1, which is altered in several cancer types, influences tumour development or progression by controlling metabolism. Understanding the molecular interplay between CAV1 and metabolism could help uncover druggable metabolic targets or pathways of clinical relevance in cancer therapy. Here we review from a cancer perspective, the findings that CAV1 modulates cell metabolism with a focus on glycolysis, mitochondrial bioenergetics, glutaminolysis, fatty acid metabolism, and autophagy.
Keywords: CAV1, Glycolysis, Glutaminolysis, Mitochondrion, Fatty acid metabolism, Autophagy, Metabolic targets
Background
Cancer cells have anomalous metabolic features, which enable them to adapt to constraints in the microenvironment [1, 2]. Otto Warburg proposed that cancer cells have impaired mitochondrial function and as such rely on high turnover of a rather inefficient aerobic glycolysis instead of oxidative phosphorylation (OXPHOS) [3–5]. While aerobic glycolysis (often called Warburg effect) largely holds true in several tumours, it is now known that tumours also retain normal mitochondrial function, using glutaminolysis to support proliferation via tricarboxylic acid (TCA) cycle [5, 6]. New evidences further highlight the importance of lipid metabolism, the serine pathway, autophagic alanine secretion, and macropinocytosis-mediated use of extracellular proteins for sustaining cancer nutrition and growth activities [5, 7–11]. Metabolic alterations in tumours are coordinated by several genes, prominent among which are TP53, MYC oncogene and hypoxia inducible factor 1α (HIF1A) [8, 9, 12–16]. In addition, metabolic enzymes such as pyruvate kinase muscle isoform (PKM2), lactate dehydrogenase A (LDHA), phosphofructose kinase A (PFKA) isoform, isocitrate dehydrogenase (IDH) and phosphoglycerate kinase (PGK1) confer oncogenic phenotypes when altered [17]. Furthermore, metabolites notably called “oncometabolites” (e.g. hydroxyglutarate, fumarate, lactate and ketones) can promote tumourigenesis in diverse ways, including by increasing cancer stemness, upregulating oncogene expression and reactive oxygen species amplification [18–22]. A detailed insight on molecular regulators of metabolic alterations could enhance the prospects of identifying cancer drug targets. In this article, we review evidences that CAV1 – a protein known to regulate cholesterol distribution, signal transduction, cell migration, and endocytic vesicular trafficking [23–25] – represents one such regulator of cell metabolism, especially in cancer cells.
Relevance of Caveolin-1 in cancer
Caveolin-1 is a 22 kDa protein encoded by CAV1 gene, and occupies flask-shaped plasma membrane invaginations called caveolae. It is one of three known caveolins (CAV1, 2 and 3) and is ubiquitously expressed in all cell types as is CAV2; CAV3 is mostly found in skeletal muscles [23, 25]. Besides earlier studies that implicated CAV1 in endocytosis, signaling and lipid disorders, research activities in the last two decades also focused on clarifying its relevance in cancer [23–33]. Consequently, CAV1 was found to be overexpressed in cancers of liver, colon, breast, kidney, lung, among others [29], and acts as a tumour promoter or suppressor depending on tumour type and stage [23, 33]. Regarding its tumour promoting function, it has been reported that high expression of CAV1 drives tumourigenesis by inhibiting apoptosis, facilitating anchorage-independent growth, drug resistance as well as metastasis [30, 33–39]. For instance, CAV1 expression in liver cancer patients was found to positively correlate with differentiation status, increased portal vein invasion, intrahepatic metastasis, and to predict overall survival outcome [37]. Accordingly, in vitro mechanistic study showed that CAV1 overexpression induced known mediators of migration and invasion, namely matrix metalloproteinases 2 and 9, and vascular epidermal growth factor [37]. Indeed, CAV1 and caveolae, which mediate molecular trafficking and contain signaling molecules such as non-receptor tyrosine kinases and endothelial nitric oxide synthase (eNOS), have long been proposed as potential therapeutic targets for disrupting tumour angiogenesis, progression and metastasis [40].
On the other hand, CAV1 acts as a tumour suppressor in some settings in that its low expression favours tumour progression [41–43]. For instance, in NIH3T3 cells oncogenically transformed by H-Ras induction, high CAV1 expression in the mitochondria reduced cell proliferation [43]. Codeficiency of CAV1 and the tumour suppressor, adenomatous polyposis coli, enhanced colorectal tumourigenesis in mice [44]. Furthermore, loss of stromal CAV1 in human breast cancer is associated with increased tumour recurrence, metastasis and poor clinical outcome [41]. Consistently, and contrary to its tumour promoting function highlighted above [37], high CAV1 expression improved overall survival in liver cancer patients, ostensibly by countering eNOS activity [42]. Altogether, accumulating evidences consistently support that CAV1 plays an important role in cancer progression – the specific nature of which seems to depend on several factors, including cancer type and stage, lesions on CAV1 or its associated genes, its protein expression level and subcellular localisation. The fact that CAV1 could serve as a clinical biomarker [45, 46] further emphasizes its importance in cancer. However, despite knowledge of its expression pattern and roles in different cancers, it is still unclear whether CAV1 expression is a property that accompanies or directly drives altered metabolism, or if changes in energy balance modulate CAV1 level towards or against cancer progression.
CAV1 in glycolysis
The preference of cancer cells for aerobic glycolysis is an evasive pro-survival strategy. This makes glycolysis an attractive therapeutic target in cancer, especially if its molecular regulators are identified and well characterized. Several studies reveal that CAV1 is involved in the modulation of glycolytic activities (Figs. 1 and 2). For instance, CAV1 expressing colon cancer cells undergo increased glycolysis upon exposure to inhalation anaesthesia (isoflurane), and are thus protected from tumour necrosis factor associated apoptosis [47]. High CAV1 expression in advanced colon cancer increased glucose uptake and ATP production by stimulating transcription of glucose transporter 3 (GLUT3, encoded by SLC2A3) [48]. Further, knockdown of CAV1 reduced cellular glucose uptake and lactate output (indicative of suppressed Warburg effect), reduced intracellular ATP, and triggered autophagy through activation of AMPK-p53 signaling [48]. In prostate cancer, immunoprecipitation and signaling studies showed that CAV1 interacts with insulin- and IGF-1 receptors (IR/IGF-1R), and can stimulate IR kinase activities by also interacting with low-density lipoprotein receptor-related protein 6 (LRP6). Accordingly, both LRP6 and CAV1 stimulate IR/IGF-1R signaling and, through AKT signaling activation, enhance glucose uptake, lactate output and cell proliferation – these effects correlated positively with hexokinase 2 (HK2) and GLUT3 expression [49]. Noteworthy, however, the authors reported that the expression of some glycolytic enzymes like enolase 1 (ENO1), PKM2 and LDHA were unchanged upon CAV1 knockdown.
Fig. 1.
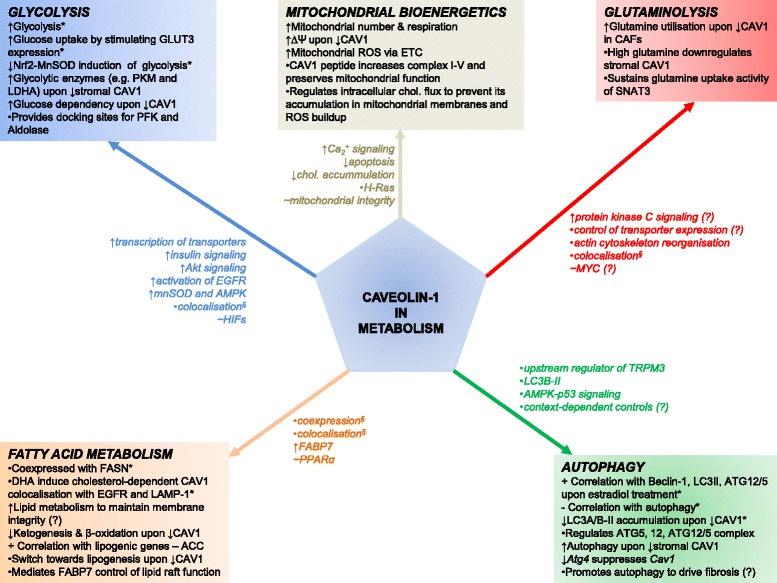
CAV1 influences metabolic processes in normal and cancer cells
Fig. 2.
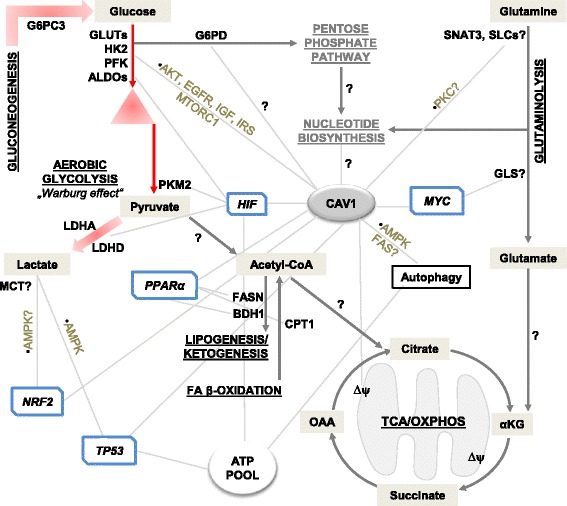
Schematic representation of metabolic processes and targets associated with CAV1 based on research findings
Furthermore, CAV1 enhanced sensitivity to anti-diabetic drug (metformin) in a study comparing two non-small-cell lung cancer cell lines, Calu-1 and Calu-6, which express high and low CAV1 levels, respectively [50]. Specifically, metformin reduced phosphorylation of IGF-1 receptor substrates [AKT and Forkhead transcription factor 3a (FOXO3A)], suppressed IGF1-dependent cell proliferation, and increased AMPK phosphorylation as well as AMP/ATP ratio in Calu-1. In line, ectopic overexpression of CAV1 in Calu-6 enhanced sensitivity to metformin, as observed by increased AMP/ATP ratio and AMPK phosphorylation [50]. This CAV1-mediated metformin sensitivity may extend beyond glycolysis since the drug also acts on complex I of the electron transport chain in cancer cells; hence it offers further clue for investigating CAV1 expression and response to anti-metabolic drugs.
CAV1 provides a docking site for glycolytic enzymes. For instance, in caveolae of vascular smooth muscle cells and lymphocytes, the rate-limiting glycolytic enzyme phosphofructose kinase (PFK) as well as aldolase co-localised with CAV1 by binding to its scaffolding domain [51]. Likewise, the intracellular metabolite profile of endothelial cells revealed a decrease in glycolytic intermediates (e.g. 3-phosphoglycerate, fructose-6-phosphate and glucose-6-phosphate) upon CAV1 knockdown, indicating perturbed glycolysis, even though the effect on glucose consumption was not reported [52]. Thus, it highlights an interesting possibility that CAV1 could influence the localisation of relevant glycolytic mediators in cancer as depicted in Fig. 3b.
Fig. 3.
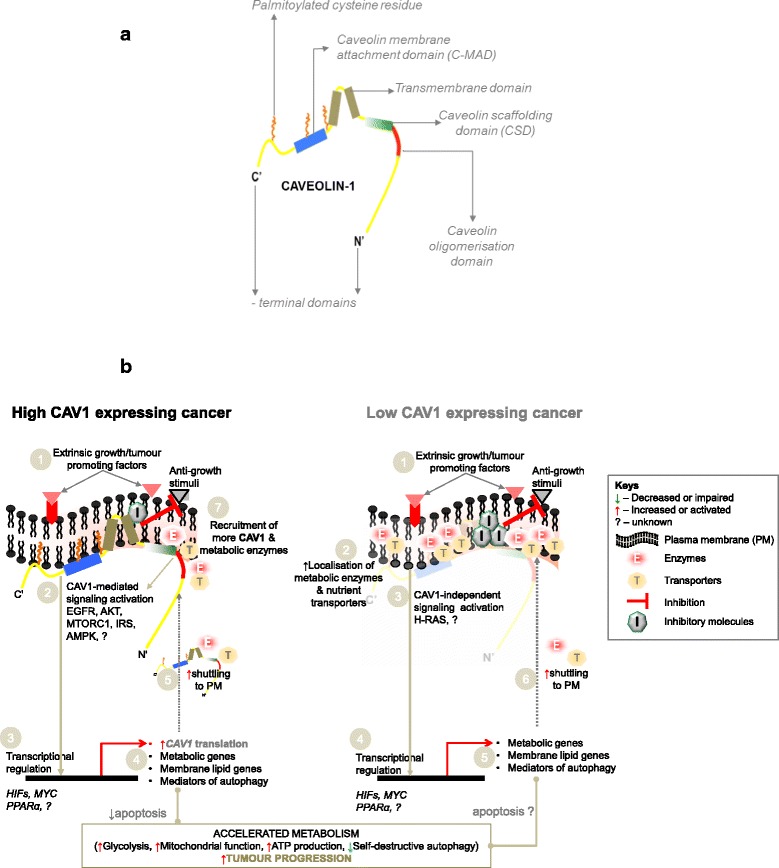
CAV1 and a hypothetical model of its role in cancer cell metabolism. a. A simplified diagram of CAV1. b. Model depicting CAV1 and its potential influence on metabolism in cancer cells expressing it in high or low levels
There are contexts in which CAV1 level is inversely related to glycolytic phenotypes. This has been reported in its crosstalk with HIFs, which are crucial transcriptional regulators of genes encoding glycolytic enzymes, glucose transporters, and also mediators of angiogenesis and tumour metastasis [53]. For instance, CAV1 is a direct target of HIF1 and HIF2, and is upregulated under hypoxia in clear cell renal cell carcinoma [54]. However, overexpression of activated HIF1α reduced CAV1 level, caused upregulation of glycolytic enzymes towards enhanced aerobic glycolysis, and increased lactate output in immortalized fibroblasts (hTERT-BJ1 cells). The fibroblasts expressing activated HIF1α also promoted xenograft tumour growth upon co-injection with breast cancer cells (MDA-MB-231) in nude mice [55]. This implies that downregulation of CAV1 in the stromal fibroblast compartment – in parallel with increased aerobic glycolysis – is a mechanism by which HIF1α promotes cancer progression. Furthermore, upregulation of several glycolytic enzymes, including aldolase A, enolase 1, PGK1, PKM2 and LDHA was observed in bone marrow derived stromal cells from Cav1 knockout (KO) mice [41]. In invasive ductal carcinoma, CAV1 is reduced at the early stage of progression and predicts poor survival outcome. Mechanistically, reduced CAV1 expression enabled the induction of transcription factor NRF2 (NF-E2-related factor 2), which activates anti-oxidant manganese superoxide dismutase (MnSOD) that triggers AMPK-dependent glycolysis [56]. As a proof, ectopic expression of CAV1 in invasive ductal carcinoma cells (MCF7) suppressed NRF2 expression, the induction of MnSOD, and decreased aerobic glycolytic phenotype as measured by extracellular acidification and lactate output [56]. Further evidences have suggested that low CAV1 expression correlate with high reliance on glucose metabolism. For example, CAV1 deficiency increased carbohydrate metabolism in mice as determined by high respiratory exchange ratio, and prevented a metabolic switch to free fatty acids including during fasting. Specifically, hepatic gluconeogenesis emerged as a possible pathway to sustain glucose supply in Cav1-KO mice as revealed by upregulation of glucose-6-phosphatase (G6PC3) and LDHD [57]. Moreover, in a study on three different CAV1-deficient mouse strains, loss of CAV1 delayed liver regeneration and decreased survival after partial hepatectomy [58]. In the strain where regeneration still occurred, the loss of CAV1 was compensated by increased dependence on hepatic carbohydrate metabolism, as blocking glycolysis with glucose analogue (2-deoxy-glucose) further reduced survival and suppressed liver regeneration [58]. In a cancer context, low CAV1 and high glycolytic phenotypes would suggest a tumour suppressor function, but contradicts its role as a tumour promoter as discussed earlier. Taken together, the underlying mechanisms for the apparently context-dependent links between CAV1 and glycolysis remain largely unclear. More studies are required to elucidate the influence of CAV1 on glucose utilisation and glycolytic enzymes, especially in CAV1 overexpressing tumours, and to determine whether glycolysis modulates the expression and function of CAV1 in normal and cancer cells.
CAV1 in mitochondrial bioenergetics
Beyond aerobic glycolysis, cancer cells still retain intact mitochondrial functions and can use alternative metabolites to support energy production. CAV1 has been associated with mitochondrial number and bioenergetic function in various cell types. For instance, mitochondria of hepatic cells in hypercholesterolemic rabbits were found to have high localisation of CAV1 [59]. Intravenous injection of mice with antennapedia-CAV1 (AP-CAV1) peptide–to increase CAV1 translocation across cell membrane–led to a darker electron-dense mitochondrial matrix, lower superoxide dismutase and catalase activity as compared to controls, suggesting that CAV1 is required for maintaining mitochondrial architecture and function. AP-CAV1 treatment also restored respiratory chain subunit proteins (complexes I–V), preserved mitochondrial function and suppressed apoptotic cell death [59]. Consistently, increased CAV1 expression was observed in an in vitro model of active microglia along with increased mitochondrial number, respiration, and glycolysis [60]. Colon cancer cells (HCT116) that overexpress CAV1 also had its abundant localisation in the mitochondria and, in the low expressing cells (HT29), overexpression of CAV1 led to its enrichment in the mitochondria and reduced apoptosis [61]. These findings provide clues that CAV1 could enhance mitochondrial functions to support tumour progression, details of which are yet to be fully understood. On the other hand, high CAV1 in the mitochondria suppresses proliferation in H-Ras driven tumour cell model [43]. Specifically, neoplastic transformation triggered by oncogenic H-Ras12v in NIH3T3 cells suppresses basal intracellular calcium (Ca2+) level, Ca2+ influx and also CAV1 expression. Further, reintroduction of CAV1 enhanced Ca2+ uptake into mitochondria, suppressed cell growth, colony formation, and induced apoptosis, implying that high CAV1 level may suppress mitochondrial function as a mechanism to mediate a suppressor activity in H-Ras-driven tumours [43].
CAV1 may modulate mitochondrial function by regulating cholesterol flux. In line with this notion, loss of CAV1 in mouse embryonic fibroblasts (MEFs) led to cholesterol accumulation in mitochondrial membrane, and increased reactive oxygen species (ROS) and cell death upon OXPHOS activation [62]. Increased circulating ROS was also observed in Cav1-KO mice, in which gonadal white adipose tissue presented with mitochondrial dysfunction. The increase in ROS was attributed to high mitochondrial membrane potential as further validated in vitro with CAV1-deficient MEFs [57]. However, the authors suggested that impaired mitochondrial function is a tissue-specific consequence of CAV1 loss, as comparison of oxygen consumption rates in liver and lung tissues revealed divergent results [57]. In cultured bovine aortic endothelial cells, knockdown of CAV1 caused an increase in mitochondrial ROS production, intracellular H2O2, and reduced the intracellular redox balance indicator [the ratio of reduced to oxidized glutathione (GSH/GSSG)] [52]. In the same study, Cav1-KO mice had increased oxidative stress as determined by high level of 8-isoprostane (a biomarker of lipid peroxidation and oxidative stress). Noteworthy, although membrane potential was increased, suppressed CAV1 expression did not significantly alter mitochondria abundance [52]. Altogether, these observations strongly link CAV1 expression to mitochondrial function, and could be relevant in understanding how CAV1 affects mitochondrial bioenergetics in cancer.
Glutaminolysis – CAV1 in the alternative tumour energy pathway?
Glutaminolysis is a metabolic process in which glutamine is converted to α-ketoglutarate for onward utilisation in TCA cycle. Through this process, glutamine serves as alternative energy-rich substrate for cancer cells to meet the excessive nutritional demands that result from rapid proliferation [6, 63]. Currently, there is a limited insight on the association between CAV1 and glutaminolysis (Figs. 1 and 2). However, it has been proposed that glutamine availability inversely correlates with CAV1 expression in tumour stromal compartment, and that fibroblasts lacking CAV1 secrete more glutamine [64]. Accordingly, cancer associated fibroblasts cultured in high glutamine media had suppressed CAV1 expression and increased autophagy [64]. It is unclear how the observed loss of CAV1 and high glutamine availability work to drive tumour progression; however, the authors suggested that autophagy in the fibroblasts may serve as a key source of energy-rich glutamine to fuel mitochondrial activity in adjacent cancer cells [64]. This proposition aligns with inverse association of CAV1 with autophagy induction as reported by some studies (discussed later), but does not establish a direct link between enhanced glutaminolysis and high CAV1 expression as also seen in cancer.
Enhanced glutaminolysis via glutaminase induction is a known transcriptional function of MYC in cancer [14, 65]. Its encoded protein, c-MYC, regulates CAV1 and both promote tumour progression in prostate and colon cancer [49, 66–69]. Hence, although not yet proven, CAV1 could play a role in c-MYC-driven glutamine metabolism in tumours. It is also probable that CAV1 regulates the expression and localisation of amino acid transporters in plasma membrane. Consistent with the latter, the glutamine uptake activity of sodium neutral amino acid transporter 3 (SNAT3) was abolished following its coexpression with dominant-negative CAV1 mutant in Xenopus laevis oocytes [70]. The CAV1 mutant, when expressed 2 days after expressing SNAT3, prevented the rapid suppression of glutamine uptake by protein kinase C activator, phorbol-12-myristate-13-acetate [70]. These findings raise the possibility that CAV1 controls glutamine uptake and utilisation in cancer by influencing SNAT3 or other amino acid transporters reviewed in [71, 72], and could lead to novel understanding of glutamine dependency in cancer cells.
CAV1 in fatty acid metabolism
Several studies have reported that CAV1 modulates fatty acid metabolism in normal and cancer cells (Fig. 1). Fatty acid synthase (FASN) is a crucial enzyme in fatty acid biosynthesis, and catalyzes the formation of palmitate from acetyl-CoA and malonyl-CoA. Aberrant FASN function is associated with tumourigenesis in various human cancers, making it a potential cancer drug target [73–75]. CAV1 and FASN are coordinately regulated in melanoma and prostate cancers, implying that their concomitant expression may enhance tumourigenesis [76–78]. The mechanism explaining CAV1 and FASN interaction to promote tumourigenesis is still unknown, but a possible biochemical link is the dynamics in palmitoylation – a posttranslational modification process, in which long chain fatty acids (mainly palmitate) bind to cysteine residues in proteins via a thioesther bond. CAV1 has palmitoylated residues (Fig. 3a), and the effects of palmitoylation on membrane proteins include regulation of membrane interaction, stability, spatial organisation, and intracellular trafficking reviewed in [79, 80], all of which could alter how CAV1 interacts with metabolic targets. It is likely that by being co-expressed with FASN, CAV1 ensures availability of the lipids required for maintaining membrane integrity of tumour cells. Insights could be gained from study in visceral adipose tissue (VAT) of obese patients where a positive correlation between CAV1 expression and lipogenic genes, e.g. acetyl-CoA carboxylase and FASN was reported [81].
CAV1 modulates several lipid metabolic processes in hepatocytes, including hepatic lipid storage, fatty acid oxidation (FAO) and ketogenesis [57, 58, 82]. For instance, Cav1 deletion led to reduced hepatic triglyceride content and impaired lipid storage [58]. Furthermore, genomic profiling of liver, adipose tissue, and MEFs from fasted CAV1-deficient mice, all showed downregulation of lipid metabolic processes as a major consequence of loss of CAV1 [82]. These findings corroborate a previous report on reduced whole body FAO in Cav1-KO mice [57]. The reduction of FAO and ketogenesis in CAV1-deficient mice is attributed to impaired hepatic peroxisome proliferator activated receptor α (PPARα) activity [82]. Accordingly, although some PPARα target genes (e.g. Gsta2, Scarb1, Nox4 and Per2) were upregulated in the CAV1-deficient mouse liver, there was a predominant suppression of several others, including carnitine palmitoyltransferases (Cpt1α and Cpt1β), acetyl-CoA acyltransferase 2 (Acaa2), and dehydrogenase/reductase (SDR Family) – Dhrs4 and Dhrs8. In line, over 15 genes involved in peroxisomal and mitochondrial FAO, such as Acox1, Hadhb, Cpt2 and Acadm, were downregulated upon loss of CAV1. Ketogenesis was likewise suppressed as confirmed by downregulation of 3-hydroxybutyrate dehydrogenase and reduced plasma level of β-hydroxybutyrate [82]. These findings underscore a strong link between CAV1 and lipid metabolism that is yet untested in cancer settings. Further evidence reveals that loss of CAV1 causes a switch from lipid towards glucose metabolism, as observed in endothelial cells [52], and murine hepatocytes [58]. Accordingly, decreased glycolytic intermediates after CAV1 knockdown in endothelial cells was attributed to accelerated utilisation, whereas increased level of fatty acids, such as palmitate, arachidonate, myristate, and essential fatty acids (e.g. linoleic and linolenic acids) were linked to a defective lipid metabolism [52]. Similarly, analysis of liver tissue showed that CAV1-deficient mice resorted to carbohydrate – instead of fatty acid metabolism [58]. Whether CAV1 influences such a crucial switch in cancer is not yet reported. Indeed, it is worthy to recall that lipids are important for raft remodeling – a crucial function of CAV1 [23, 25, 26]. Therefore, beyond modulating lipid metabolism, CAV1 depletion can compromise the functions of other molecules involved in lipid raft function. An example of such molecule is fatty acid-binding protein 7 (FABP7), which binds to and facilitates uptake of long-chain fatty acids. Recently, it was found that CAV1 mediates lipid raft activity of FABP7, namely receptor accumulation [83]. Specifically, knockout of Fabp7 in mice suppressed CAV1 and ligand-dependent accumulation of Toll-like receptor 4 (TLR4) in astrocytes upon lipopolysaccharide stimulation, whereas overexpression of CAV1 in FABP7-deficient astrocytes was sufficient to restore TLR4 recruitment [83].
CAV1 level may determine the effect of lipid load on cancer cells. For instance, docosahexaenoic acid (DHA)– an omega-3 polyunsaturated fatty acid – is associated with cancer risk, progression and therapy [84, 85]. CAV1 participates in DHA mediated apoptosis in MDA-MB-231 cells [86]. Accordingly, DHA treatment caused cholesterol-dependent co-localisation of CAV1 and epidermal growth factor receptor (EGFR) with lysosome associated membrane protein 1 (LAMP-1), with an onward down-regulation of lipid-raft associated onco-proteins, including EGFR, HSP90, AKT and SRC [86]. Furthermore, expression of tyrosine-14 phosphorylated CAV1 accelerated palmitate-induced apoptotic cell death in pancreatic beta cells [87]. Taken together, these studies reveal that CAV1 is associated with fatty acid metabolism, offering insights for onward investigation in cancer. It will also be interesting to determine whether differential expression of CAV1 influences the expression or function of fatty acid enzymes that have recently emerged to be important mediators in cancer, e.g. acyl-CoA synthetase, carnitine palmitoyltransferase 1C, and carnitine acyltransferase [88–90].
Caveolin-1 in metabolic abnormalities – obesity and insulin resistance
Obesity and insulin resistance, the major cause of type 2 diabetes (T2DM), are two intricately connected metabolic abnormalities of significant global health burden, and are strongly linked to cancer [91, 92]. For instance, in the case of obesity, there has emerged sufficient evidence in humans that reduced body fatness also reduces the risk of most cancers, prominent among which are liver, pancreatic, ovarian cancers, and multiple myeloma [92]. Similarly, a recent consensus report concluded that although several questions remain unanswered, “diabetes (primarily type 2) is associated with increased risk for some cancers (liver, pancreas, endometrium, colon and rectum, breast, bladder)” though also the reduced risk of prostate cancer [93]. Thus, whether in obesity and insulin resistance, understanding the activity of CAV1 could shed light on its role in metabolism that may be applicable in cancer. Indeed, studies in cancer and normal cells show that CAV1 modulates insulin signaling [48, 49, 57]. The loss of CAV1 leads to insulin resistance, and several of its genetic variants are associated with type 2 diabetes and lipid disorders [94]. It is believed that CAV1 stabilizes insulin receptor and so enhances cellular response to insulin stimuli as is necessary for promoting glucose uptake and metabolism. Besides, in a differentiating murine preadipocyte model 3T3-L1, Cav1 promoter hypomethylation caused its overexpression and phosphorylation of its protein, accompanied by increased expression of insulin receptor, glucose transporter 4, and adipokines (adiponectin, interleukin 6 and leptin) [95]. CAV1 expression, therefore, apparently contribute to both insulin sensitivity and obesity. Consistent with the latter, CAV1 is overexpressed in VAT and subcutaneous adipose tissue of obese patients with normoglycaemia or T2DM (basically characterized by hyperglycemia and hyperinsulinemia) – and is positively correlated with body fat and body mass index [96]. Although still poorly understood, the mechanistic link between obesity, CAV1 and cancer may be partly due to increased adipokine activity. For instance, murine high fat diet model of obesity exposed to anti-obesity drug (orlistat) or restricted caloric intake had smaller adipocyte size, low CAV1 and suppressed adipokines (leptin and resistin) – in vivo growth of murine (B16F10) and human (A375) melanoma cell lines was also reduced in the obese mice [97]. Interestingly, the authors showed that stimulation with the adipokines enhanced proliferation and CAV1 level in A375 cells in vitro [97]. This finding supports a potential involvement of CAV1 via adipokines in obesity-driven cancer. Indeed, the involvement of CAV1 or its genetic variants in obesity and insulin resistance offers a pathophysiological clue for future understanding of its role in tumour glucose and fatty acid metabolism.
CAV1 in autophagy
Autophagy is a process in which cells breakdown their cytoplasmic components to release metabolites for meeting nutritional needs [98]. Thus, autophagy is either a routine metabolic process in itself or is a pivotal mechanism initiated for long-term cellular energy homeostasis, especially during starvation [98, 99]. Autophagy is crucial in cancer, which explains why its inhibition leads to cancer cell death [100–102]. In addition, there is evidence that autophagy in surrounding tumour cells, as recently shown in pancreatic stellate cells [11], may actually be key for tumour nutrient supply in the hypovascularized microenvironment. Several studies have found autophagy to be very context-dependent. Thus, while CAV1 is implicated in the regulation of autophagy reviewed in [33, 103], it is not surprising that the molecular mechanisms seem inconsistent – supporting both direct and inverse associations. For example, in HCC, CAV1 inhibited autophagy while promoting cell proliferation, migration, and angiogenesis [104]. Accordingly, CAV1 inversely correlated with autophagy markers ATG5 and BECLIN-1 in clinical HCC and cell lines, while its knockdown increased autophagosome formation [104]. A well-acknowledged marker of autophagic activity commonly used to indicate increased autophagosome formation or decreased turnover is the microtubule-associated protein 1 light chain 3 beta (LC3B) [105, 106]. The suppression of CAV1 markedly increased LC3B-II in endothelial cells [52]. CAV1 deficiency in breast cancer also promoted late stage autophagy by enhancing lysosomal function and autophagosome-lysosome fusion towards improved cell survival under nutrient starvation [107]. Similarly, knockdown of CAV1 in stromal fibroblast induced autophagy, and hypoxia-induced degradation of CAV1 led to upregulation of autophagy markers [108].
Studies also show that CAV1 is directly associated with or regulates autophagy. For example, CAV1 correlates with estradiol-mediated autophagy in the BT474 breast cancer cell line. These cells expressed higher CAV1 and autophagy-related proteins (BECLIN-1, light chain (LC3)-II and ATG12/5) upon estradiol treatment [109]. In lung epithelial cells, CAV1 interacts with and regulates expression of autophagy proteins, notably ATG12, ATG5, and the ATG12-ATG5 complex that is important for autophagosome formation [110]. CAV1 is also an upstream regulator of transient receptor potential melastatin 3 (TRPM3), which controls Ca2+ and Zn2+ flux, and induces oncogenic autophagy via LC3A/B-II [111]. In this setting, TRPM3 expression requires CAV1, while knockdown of their encoding genes reduce LC3A/B-II accumulation in renal carcinoma cell lines [112]. Cav1 was also among genes downregulated following bleomycin-induced pulmonary fibrosis in mice with autophagy gene Atg4b deletion [112]. The downregulation of Cav1 coincided with altered expression of several profibrotic genes, including upregulation of Tgfbr2, Smad3, Tgfb1, Tgfb3, and suppression of Smad4, 7 and Snai1 [112], suggesting a link between its expression and fibrosis via autophagy. In non-small lung adenocarcinoma cells (A549), expression of CAV1 was protective by blocking a switch from autophagy to apoptosis [113]. Taken together, these studies show a context-dependent involvement of CAV1 in autophagy that requires further clarification, especially in cancer types where CAV1 is a key player.
Conclusions
There is abundant evidence that CAV1 plays diverse roles in normal and cancer cell metabolism (Figs. 1 and 2), which appears to be spatiotemporal and dependent on cell types, microenvironmental factors, nutritional availability and probably even additional perturbations, e.g. hypoxia or drug treatment. For instance, while CAV1 overexpression may drive glycolysis and glucose dependency, its knockdown also enhances glycolytic phenotypes in cancer and normal cells. Similar discrepancies also exist in autophagy, where different studies show that both loss and overexpression of CAV1 induces the lysosomal marker LC3B. Given these conflicting findings, more studies are required to unravel the exact mechanisms by which CAV1 controls cell metabolism – especially in cancer types associated with its high expression. Based on evidences from literature, we propose that mechanisms by which CAV1 modulate cancer metabolism may include: enabling the expression of membrane-localized metabolic enzymes; coordinating a switch between metabolic pathways; relaying signals for transcriptional activation or suppression of metabolic regulators, or inhibiting anti-metabolic stimuli (Fig. 3b).
To understand the role of CAV1 in cancer metabolism, studies should employ experimental models that are as close to the human cancer situation as is possible. In addition, studies designed to extend current knowledge on CAV1 function should endeavor to validate some previous observations using the same experimental models, i.e. cell and tissue types (Table 1). This will ensure consistency and also help to detect confounding experimental variables. Diseases such as obesity and diabetes could also provide an avenue through which molecular clues on the involvement of CAV1 in cellular metabolism can be unraveled for subsequent investigation in cancer. From a technical viewpoint, complementing CAV1 gain or loss of function studies with high throughput molecular biology methods, such as metabolomics, genomics and proteomics, will help to clarify its function in cancer cell metabolism.
Table 1.
Cell, tissue types and disease conditions in which CAV1 was found to correlate with or influence metabolic processes
| CAV1 promoted or directly correlated with metabolic processes | Ref. | CAV1 suppressed or inversely correlated with metabolic processes |
Ref. |
|---|---|---|---|
| Glycolysis | |||
| •Colon cancer [HCT116, HT29, LoVo]* •Prostate cancer [LNCaP, PC-3] Smooth muscle cells, lymphocytes |
47, 48 49 51 |
BAEC CA fibroblast [hTERT–BJ1] Breast cancer cell [MCF7] Mouse liver (including gluconeogenesis) |
52 55 56 58 |
| Mitochondrial function | |||
| BAEC Hepatic cells Microglia Colon cancer [HCT116, HT29] MEFs |
52 59 60 61 57, 62 |
NIH3T3 cells oncogenically transformed with H-RAS mutant | 43 |
| Glutamine metabolism | |||
| Xenopus laevis oocyte | 70 | Stromal fibroblast; [hTERT–BJ1] | 64, 103 |
| Fatty acid metabolism | |||
| BAEC | 52 | – | – |
| Mouse liver | 57, 58, 82 | ||
| Mouse hepatocyte [AML12] | 58 | ||
| Adipocytes | 57, 82 | ||
| MEFs | 57, 82 | ||
| Melanoma, prostate cancer | 76, 77, 78 | ||
| Metabolic diseases | |||
| Obesity: | Diabetes: | ||
| Adipose tissues of patients | 96 | Insulin insensitivity, hyperglycemia | 57 |
| Murine preadipocyte model [3T3-L1] | 95 | Insulin resistance (Type 2 diabetes) | 94 |
| High fat diet mouse model | 97 | ||
| Autophagy | |||
| Breast cancer cell [BT474] treated with estradiol | 109 | BAEC | 52 |
| Lung epithelial cells [BEAS-2B] | 110 | •Hepatocellular carcinoma [HCCLM3]* | 104 |
| •Renal carcinoma cell [786-O, A498]* | 111 | •Breast cancer [MCF7, MDA-MB-231]* | 107 |
| Murine fibrotic lung tissue | 112 | MEFs | 107 |
| Lung cancer cell [A549] | 113 | CA fibroblast [hTERT–BJ1] | 108 |
MEFs mouse embryonic fibroblasts, BAEC Bovine aortic endothelial cells, CA Cancer associated; •Including in clinical samples; In squared bracket [] are cell lines used for mechanistic studies; *other cell lines are mentioned in the studies
Finally, the identification of key initiating factors – whether genomic or signaling mutations – that drive metabolic alterations are crucial steps in fine-tuning anti-cancer strategies. For instance, if low CAV1 expression in cancer also causes high dependency on glucose over lipid metabolism as reported in non-cancer cells [52, 58], then it raises an attractive prospect of using CAV1 expression levels to stratify cancer patients into those that could benefit from either inhibitors of glucose or lipid metabolism. Interestingly, we highlighted evidences that CAV1 interacts with known mediators of altered metabolism, e.g. LDHA, PKM2 and FASN, which are currently being studied as cancer drug targets [17, 114, 115]. It is therefore conceivable that understanding the expression and function of CAV1 in cancer could serve as a pointer for exposing novel metabolic genes or pathway alterations of clinical importance.
Acknowledgements
We acknowledge financial support by Deutsche Forschungsgemeinschaft and Ruprecht-Karls-Universität Heidelberg within the funding programme Open Access Publishing.
Funding
SD is supported by funds from the Deutsche Forschungsgemeinschaft (DFG) Do373/13-1, the BMBF program LiSyM (Grant PTJ-FKZ: 031 L0043), as well as from Marie Curie Actions of the European Union’s Seventh Framework Programme (FP7/2007-2013) Grant PITN-GA-2012-316549 (IT LIVER: Inhibiting TGF-beta in liver diseases). CM receives support by a grant from the Deutsche Forschungsgemeinschaft (Me4532/1-1). ZCN is a recipient of a PhD Scholarship from the Niger Delta Development Commission, Nigeria, and appreciates the generous supports from the Graduate School (HBIGS), University of Heidelberg, Germany. The funding bodies did not influence the content of this article.
Availability of data and materials
Not applicable.
Authors’ contributions
ZCN conceived the outline, carried out literature search and wrote the paper. MPE, SD, and CM supported through critical reading, comments and corrections of the manuscript. All authors read the final version.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ∆ψ
Mitochondrial membrane potential
- ALDOs
Aldolases
- ATP
Adenosine triphosphate
- CAV1
Caveolin-1
- ETC.
Electron transport chain
- FASN
Fatty acid synthase gene
- G6PD
Glucose-6-phospate dehydrogenase
- GLUTs
Glucose transporters
- HCC
Hepatocellular carcinoma
- HIF
Hypoxia inducible factor
- HK2
Hexokinase 2
- IGF-1
Insulin-like growth factor 1
- KO
Knockout
- LDHA
LDHD, Lactate dehydrogenase A, D
- MCTs
Monocarboxylates
- MEFs
Mouse embryonic fibroblasts
- OAA
Oxaloacetate
- OXPHOS
Oxidative phosphorylation
- PFK
Phosphofructokinase
- PKM2
Pyruvate kinase, muscle isoform
- PPP
Pentose phosphate pathway
- SLCs
Solute carrier families
- TCA
Tricarboxylic acid
- VAT
Visceral adipose tissue
- αKG
Alpha ketoglutarate
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–56. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014;19(2):285–92. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–6. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75(3):544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–83. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 13.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16(24):5928–35. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3(8):a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weljie AM, Jirik FR. Hypoxia-induced metabolic shifts in cancer cells: moving beyond the Warburg effect. Int J Biochem Cell Biol. 2011;43(7):981–9. doi: 10.1016/j.biocel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–70. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Outschoorn UE, Prisco M, Ertel A, Tsirigos A, Lin Z, Pavlides S, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10(8):1271–86. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowicki S, Gottlieb E. Oncometabolites: tailoring our genes. FEBS J. 2015;282(15):2796–805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–41. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, et al. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell. 2013;51(2):236–48. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta R, Toufaily C, Annabi B. Caveolin and cavin family members: dual roles in cancer. Biochimie. 2014;107(Pt B):188–202. doi: 10.1016/j.biochi.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Meyer C, Liu Y, Dooley S. Caveolin and TGF-β entanglements. J Cell Physiol. 2013;228(11):2097–102. doi: 10.1002/jcp.24380. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. J Biol Chem. 2002;277(44):41295–8. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 26.Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14(2):98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 27.Razani B, Combs TP, Wang XB, Frank PG, Park DS, Russell RG, et al. Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. J Biol Chem. 2002;277(10):8635–47. doi: 10.1074/jbc.M110970200. [DOI] [PubMed] [Google Scholar]
- 28.Navarro A, Anand-Apte B, Parat MO. A role for caveolae in cell migration. FASEB J. 2004;18(15):1801–11. doi: 10.1096/fj.04-2516rev. [DOI] [PubMed] [Google Scholar]
- 29.Burgermeister E, Liscovitch M, Röcken C, Schmid RM, Ebert MP. Caveats of caveolin-1 in cancer progression. Cancer Lett. 2008;268(2):187–201. doi: 10.1016/j.canlet.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 30.Patani N, Martin LA, Reis-Filho JS, Dowsett M. The role of caveolin-1 in human breast cancer. Breast Cancer Res Treat. 2012;131(1):1–15. doi: 10.1007/s10549-011-1751-4. [DOI] [PubMed] [Google Scholar]
- 31.Meyer C, Liu Y, Kaul A, Peipe I, Dooley S. Caveolin-1 abrogates TGF-β mediated hepatocyte apoptosis. Cell Death Dis. 2013;4:e466. doi: 10.1038/cddis.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunez-Wehinger S, Ortiz RJ, Diaz N, Diaz J, Lobos-Gonzalez L, Quest AF. Caveolin-1 in cell migration and metastasis. Curr Mol Med. 2014;14(2):255–74. doi: 10.2174/1566524014666140128112827. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Wang N, Liu P, Peng F, Tang H, Chen Q, et al. Caveolin-1, a stress-related oncotarget, in drug resistance. Oncotarget. 2015;6(35):37135–50. doi: 10.18632/oncotarget.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savage K, Lambros MB, Robertson D, Jones RL, Jones C, Mackay A, et al. Caveolin 1 is overexpressed and amplified in a subset of basal-like and metaplastic breast carcinomas: a morphologic, ultrastructural, immunohistochemical, and in situ hybridization analysis. Clin Cancer Res. 2007;13(1):90–101. doi: 10.1158/1078-0432.CCR-06-1371. [DOI] [PubMed] [Google Scholar]
- 35.Goetz JG, Lajoie P, Wiseman SM, Nabi IR. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev. 2008;27(4):715–35. doi: 10.1007/s10555-008-9160-9. [DOI] [PubMed] [Google Scholar]
- 36.Ho CC, Kuo SH, Huang PH, Huang HY, Yang CH, Yang PC. Caveolin-1 expression is significantly associated with drug resistance and poor prognosis in advanced non-small cell lung cancer patients treated with gemcitabine-based chemotherapy. Lung Cancer. 2008;59(1):105–10. doi: 10.1016/j.lungcan.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Tang Y, Zeng X, He F, Liao Y, Qian N, Toi M. Caveolin-1 is related to invasion, survival, and poor prognosis in hepatocellular cancer. Med Oncol. 2012;29(2):977–84. doi: 10.1007/s12032-011-9900-5. [DOI] [PubMed] [Google Scholar]
- 38.Tse EY, Ko FC, Tung EK, Chan LK, Lee TK, Ngan ES, Man K, Wong AS, Ng IO, Yam JW. Caveolin-1 overexpression is associated with hepatocellular carcinoma tumourigenesis and metastasis. J Pathol. 2012;226(4):645–53. doi: 10.1002/path.3957. [DOI] [PubMed] [Google Scholar]
- 39.Faggi F, Chiarelli N, Colombi M, Mitola S, Ronca R, Madaro L, et al. Cavin-1 and Caveolin-1 are both required to support cell proliferation, migration and anchorage-independent cell growth in rhabdomyosarcoma. Lab Invest. 2015;95(6):585–602. doi: 10.1038/labinvest.2015.45. [DOI] [PubMed] [Google Scholar]
- 40.Carver LA, Schnitzer JE. Caveolae: mining little caves for new cancer targets. Nat Rev Cancer. 2003;3(8):571–81. doi: 10.1038/nrc1146. [DOI] [PubMed] [Google Scholar]
- 41.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8(23):3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 42.Yang SF, Yang JY, Huang CH, Wang SN, Lu CP, Tsai CJ, et al. Increased caveolin-1 expression associated with prolonged overall survival rate in hepatocellular carcinoma. Pathology. 2010;42(5):438–45. doi: 10.3109/00313025.2010.494293. [DOI] [PubMed] [Google Scholar]
- 43.Rimessi A, Marchi S, Patergnani S, Pinton P. H-Ras-driven tumoral maintenance is sustained through caveolin-1-dependent alterations in calcium signaling. Oncogene. 2014;33(18):2329–40. doi: 10.1038/onc.2013.192. [DOI] [PubMed] [Google Scholar]
- 44.Friedrich T, Richter B, Gaiser T, Weiss C, Janssen KP, Einwächter H, Schmid RM, Ebert MP, Burgermeister E. Deficiency of caveolin-1 in Apc(min/+) mice promotes colorectal tumorigenesis. Carcinogenesis. 2013;34(9):2109–18. doi: 10.1093/carcin/bgt142. [DOI] [PubMed] [Google Scholar]
- 45.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012;7:423–67. doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]
- 46.Ng KL, Morais C, Bernard A, Saunders N, Samaratunga H, Gobe G, Wood S. A systematic review and meta-analysis of immunohistochemical biomarkers that differentiate chromophobe renal cell carcinoma from renal oncocytoma. J Clin Pathol. 2016;69:661–71. doi: 10.1136/jclinpath-2015-203585. [DOI] [PubMed] [Google Scholar]
- 47.Kawaraguchi Y, Horikawa YT, Murphy AN, Murray F, Miyanohara A, Ali SS, et al. Volatile anesthetics protect cancer cells against tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis via caveolins. Anesthesiology. 2011;115(3):499–508. doi: 10.1097/ALN.0b013e3182276d42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ha TK, Her NG, Lee MG, Ryu BK, Lee JH, Han J, et al. Caveolin-1 increases aerobic glycolysis in colorectal cancers by stimulating HMGA1-mediated GLUT3 transcription. Cancer Res. 2012;72(16):4097–109. doi: 10.1158/0008-5472.CAN-12-0448. [DOI] [PubMed] [Google Scholar]
- 49.Tahir SA, Yang G, Goltsov A, Song KD, Ren C, Wang J, et al. Caveolin-1-LRP6 signaling module stimulates aerobic glycolysis in prostate cancer. Cancer Res. 2013;73(6):1900–11. doi: 10.1158/0008-5472.CAN-12-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salani B, Maffioli S, Hamoudane M, Parodi A, Ravera S, Passalacqua M, et al. Caveolin-1 is essential for metformin inhibitory effect on IGF1 action in non-small-cell lung cancer cells. FASEB J. 2012;26(2):788–98. doi: 10.1096/fj.11-192088. [DOI] [PubMed] [Google Scholar]
- 51.Raikar LS, Vallejo J, Lloyd PG, Hardin CD. Overexpression of caveolin-1 results in increased plasma membrane targeting of glycolytic enzymes: the structural basis for a membrane associated metabolic compartment. J Cell Biochem. 2006;98(4):861–71. doi: 10.1002/jcb.20732. [DOI] [PubMed] [Google Scholar]
- 52.Shiroto T, Romero N, Sugiyama T, Sartoretto JL, Kalwa H, Yan Z, et al. Caveolin-1 is a critical determinant of autophagy, metabolic switching, and oxidative stress in vascular endothelium. PLoS One. 2014;9(2):e87871. doi: 10.1371/journal.pone.0087871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Roche O, Xu C, Moriyama EH, Heir P, Chung J, et al. Hypoxia promotes ligand-independent EGF receptor signaling via hypoxia-inducible factor-mediated upregulation of caveolin-1. Proc Natl Acad Sci U S A. 2012;109(13):4892–7. doi: 10.1073/pnas.1112129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A, et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle. 2010;9(17):3534–51. doi: 10.4161/cc.9.17.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hart PC, Ratti BA, Mao M, Ansenberger-Fricano K, Shajahan-Haq AN, Tyner AL, et al. Caveolin-1 regulates cancer cell metabolism via scavenging Nrf2 and suppressing MnSOD-driven glycolysis. Oncotarget. 2016;7(1):308–22. doi: 10.18632/oncotarget.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asterholm IW, Mundy DI, Weng J, Anderson RG, Scherer PE. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metab. 2012;15(2):171–85. doi: 10.1016/j.cmet.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández-Rojo MA, Restall C, Ferguson C, Martel N, Martin S, Bosch M, et al. Caveolin-1 orchestrates the balance between glucose and lipid-dependent energy metabolism: implications for liver regeneration. Hepatology. 2012;55(5):1574–84. doi: 10.1002/hep.24810. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y-H, Lin W-W, Liu C-S, Hsu L-S, Lin Y-M, et al. Caveolin-1 Provides Palliation for Adverse Hepatic Reactions in Hypercholesterolemic Rabbits. PLoS One. 2014;9(1):e71862. doi: 10.1371/journal.pone.0071862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niesman IR, Zemke N, Fridolfsson HN, Haushalter KJ, Levy K, Grove A, et al. Caveolin isoform switching as a molecular, structural, and metabolic regulator of microglia. Mol Cell Neurosci. 2013;56:283–97. doi: 10.1016/j.mcn.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fridolfsson HN, Kawaraguchi Y, Ali SS, Panneerselvam M, Niesman IR, Finley JC, et al. Mitochondria-localized caveolin in adaptation to cellular stress and injury. FASEB J. 2012;26(11):4637–49. doi: 10.1096/fj.12-215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bosch M, Marí M, Herms A, Fernández A, Fajardo A, Kassan A, et al. Caveolin-1 deficiency causes cholesterol-dependent mitochondrial dysfunction and apoptotic susceptibility. Curr Biol. 2011;21(8):681–6. doi: 10.1016/j.cub.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ko YH, Lin Z, Flomenberg N, Pestell RG, Howell A, Sotgia F, et al. 61: implications for preventing chemotherapy resistance. Cancer Biol Ther. 2011;12(12):1085–97. doi: 10.4161/cbt.12.12.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timme TL, Goltsov A, Tahir S, Li L, Wang J, Ren C, Johnston RN, Thompson TC. Caveolin-1 is regulated by c-myc and suppresses c-myc-induced apoptosis. Oncogene. 2000;19(29):3256–65. doi: 10.1038/sj.onc.1203654. [DOI] [PubMed] [Google Scholar]
- 67.Park DS, Razani B, Lasorella A, Schreiber-Agus N, Pestell RG, Iavarone A, Lisanti MP. Evidence that Myc isoforms transcriptionally repress caveolin-1 gene expression via an INR-dependent mechanism. Biochemistry. 2001;40(11):3354–62. doi: 10.1021/bi002787b. [DOI] [PubMed] [Google Scholar]
- 68.Roy UK, Henkhaus RS, Ignatenko NA, Mora J, Fultz KE, Gerner EW. Wild-type APC regulates caveolin-1 expression in human colon adenocarcinoma cell lines via FOXO1a and C-myc. Mol Carcinog. 2008;47(12):947–55. doi: 10.1002/mc.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang G, Goltsov AA, Ren C, Kurosaka S, Edamura K, Logothetis R, et al. Caveolin-1 upregulation contributes to c-Myc-induced high-grade prostatic intraepithelial neoplasia and prostate cancer. Mol Cancer Res. 2012;10(2):218–29. doi: 10.1158/1541-7786.MCR-11-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balkrishna S, Bröer A, Kingsland A, Bröer S. Rapid downregulation of the rat glutamine transporter SNAT3 by a caveolin-dependent trafficking mechanism in Xenopus laevis oocytes. Am J Physiol Cell Physiol. 2010;299(5):C1047–57. doi: 10.1152/ajpcell.00209.2010. [DOI] [PubMed] [Google Scholar]
- 71.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121(1):29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 72.McCracken AN, Edinger AL. Nutrient transporters: the Achilles’ heel of anabolism. Trends Endocrinol Metab. 2013;24(4):200–8. doi: 10.1016/j.tem.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–77. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 74.Swierczynski J, Hebanowska A, Sledzinski T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J Gastroenterol. 2014;20(9):2279–303. doi: 10.3748/wjg.v20.i9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaytseva YY, Elliott VA, Rychahou P, Mustain WC, Kim JT, Valentino J, et al. Cancer cell-associated fatty acid synthase activates endothelial cells and promotes angiogenesis in colorectal cancer. Carcinogenesis. 2014;35(6):1341–51. doi: 10.1093/carcin/bgu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Vizio D, Adam RM, Kim J, Kim R, Sotgia F, Williams T, et al. Caveolin-1 interacts with a lipid raft-associated population of fatty acid synthase. Cell Cycle. 2008;7(14):2257–67. doi: 10.4161/cc.7.14.6475. [DOI] [PubMed] [Google Scholar]
- 77.Witkiewicz AK, Nguyen KH, Dasgupta A, Kennedy EP, Yeo CJ, Lisanti MP, Brody JR. Co-expression of fatty acid synthase and caveolin-1 in pancreatic ductal adenocarcinoma: implications for tumor progression and clinical outcome. Cell Cycle. 2008;7(19):3021–5. doi: 10.4161/cc.7.19.6719. [DOI] [PubMed] [Google Scholar]
- 78.Pandey V, Vijayakumar MV, Ajay AK, Malvi P, Bhat MK. Diet-induced obesity increases melanoma progression: involvement of Cav-1 and FASN. Int J Cancer. 2012;130(3):497–508. doi: 10.1002/ijc.26048. [DOI] [PubMed] [Google Scholar]
- 79.Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. 2010;191(7):1229–38. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280(12):2766–74. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 81.Fernández-Real JM, Catalán V, Moreno-Navarrete JM, Gómez-Ambrosi J, Ortega FJ, Rodriguez-Hermosa JI, et al. Study of caveolin-1 gene expression in whole adipose tissue and its subfractions and during differentiation of human adipocytes. Nutr Metab (Lond) 2010;7:20. doi: 10.1186/1743-7075-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernández-Rojo MA, Gongora M, Fitzsimmons RL, Martel N, Martin SD, Nixon SJ, et al. Caveolin-1 is necessary for hepatic oxidative lipid metabolism: evidence for crosstalk between caveolin-1 and bile acid signaling. Cell Rep. 2013;4(2):238–47. doi: 10.1016/j.celrep.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 83.Kagawa Y, Yasumoto Y, Sharifi K, Ebrahimi M, Islam A, Miyazaki H, et al. Fatty acid-binding protein 7 regulates function of caveolae in astrocytes through expression of caveolin-1. Glia. 2015;63(5):780–94. doi: 10.1002/glia.22784. [DOI] [PubMed] [Google Scholar]
- 84.Azrad M, Turgeon C, Demark-Wahnefried W. Current evidence linking polyunsaturated fatty acids with cancer risk and progression. Front Oncol. 2013;3:224. doi: 10.3389/fonc.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vaughan VC, Hassing MR, Lewandowski PA. Marine polyunsaturated fatty acids and cancer therapy. Br J Cancer. 2013;108(3):486–92. doi: 10.1038/bjc.2012.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee EJ, Yun UJ, Koo KH, Sung JY, Shim J, Ye SK, et al. Down-regulation of lipid raft-associated onco-proteins via cholesterol-dependent lipid raft internalization in docosahexaenoic acid-induced apoptosis. Biochim Biophys Acta. 2014;1841(1):190–203. doi: 10.1016/j.bbalip.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 87.Wehinger S, Ortiz R, Díaz MI, Aguirre A, Valenzuela M, Llanos P, et al. Phosphorylation of caveolin-1 on tyrosine-14 induced by ROS enhances palmitate-induced death of beta-pancreatic cells. Biochim Biophys Acta. 2015;1852(5):693–708. doi: 10.1016/j.bbadis.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 88.Pei Z, Sun P, Huang P, Lal B, Laterra J, Watkins PA. Acyl-CoA synthetase VL3 knockdown inhibits human glioma cell proliferation and tumorigenicity. Cancer Res. 2009;69(24):9175–82. doi: 10.1158/0008-5472.CAN-08-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25(10):1041–51. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pacilli A, Calienni M, Margarucci S, D’Apolito M, Petillo O, Rocchi L, et al. Carnitine-acyltransferase system inhibition, cancer cell death, and prevention of myc-induced lymphomagenesis. J Natl Cancer Inst. 2013;105(7):489–98. doi: 10.1093/jnci/djt030. [DOI] [PubMed] [Google Scholar]
- 91.van Dieren S, Beulens JW, van der Schouw YT, Grobbee DE, Neal B. The global burden of diabetes and its complications: an emerging pandemic. Eur J Cardiovasc Prev Rehabil. 2010;17(Suppl 1):S3–8. doi: 10.1097/01.hjr.0000368191.86614.5a. [DOI] [PubMed] [Google Scholar]
- 92.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, International Agency for Research on Cancer Handbook Working Group Body Fatness and Cancer--Viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794–8. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, et al. Diabetes and Cancer. Diabetes Care. 2010;33(7):1674–85. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Méndez-Giménez L, Rodríguez A, Balaguer I, Frühbeck G. Role of aquaglyceroporins and caveolins in energy and metabolic homeostasis. Mol Cell Endocrinol. 2014;397(1–2):78–92. doi: 10.1016/j.mce.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 95.Palacios-Ortega S, Varela-Guruceaga M, Milagro FI, Martínez JA, de Miguel C. Expression of Caveolin 1 is enhanced by DNA demethylation during adipocyte differentiation. status of insulin signaling. PLoS One. 2014;9(4):e95100. doi: 10.1371/journal.pone.0095100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Catalán V, Gómez-Ambrosi J, Rodríguez A, Silva C, Rotellar F, Gil MJ, et al. Expression of caveolin-1 in human adipose tissue is upregulated in obesity and obesity-associated type 2 diabetes mellitus and related to inflammation. Clin Endocrinol (Oxf) 2008;68(2):213–9. doi: 10.1111/j.1365-2265.2007.03021.x. [DOI] [PubMed] [Google Scholar]
- 97.Malvi P, Chaube B, Pandey V, Vijayakumar MV, Boreddy PR, Mohammad N, et al. Obesity induced rapid melanoma progression is reversed by orlistat treatment and dietary intervention: role of adipokines. Mol Oncol. 2015;9(3):689–703. doi: 10.1016/j.molonc.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 2012;4(9):a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5(9):726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 101.Coker-Gurkan A, Arisan ED, Obakan P, Guvenir E, Unsal NP. Inhibition of autophagy by 3-MA potentiates purvalanol-induced apoptosis in Bax deficient HCT 116 colon cancer cells. Exp Cell Res. 2014;328(1):87–98. doi: 10.1016/j.yexcr.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 102.Yang A, Kimmelman AC. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy. 2014;10(9):1683–4. doi: 10.4161/auto.29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer. 2015;15:225–37. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- 104.Liu WR, Jin L, Tian MX, Jiang XF, Yang LX, Ding ZB, et al. Caveolin-1 promotes tumor growth and metastasis via autophagy inhibition in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40(2):169–78. doi: 10.1016/j.clinre.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 105.Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol. 2010;221(2):117–24. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi Y, Tan SH, Ng S, Zhou J, Yang ND, Koo GB, et al. Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy. 2015;11(5):769–84. doi: 10.1080/15548627.2015.1034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFκB activation in the tumor stromal microenvironment. Cell Cycle. 2010;9(17):3515–33. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang R, He W, Li Z, Chang W, Xin Y, Huang T. Caveolin-1 functions as a key regulator of 17β-estradiol-mediated autophagy and apoptosis in BT474 breast cancer cells. Int J Mol Med. 2014;34(3):822–7. doi: 10.3892/ijmm.2014.1836. [DOI] [PubMed] [Google Scholar]
- 110.Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, et al. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2014;306(11):L1016–25. doi: 10.1152/ajplung.00268.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hall DP, Cost NG, Hegde S, Kellner E, Mikhaylova O, Stratton Y, et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26(5):738–53. doi: 10.1016/j.ccell.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M, López-Otín C, Selman M, Pardo A. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11(4):670–84. doi: 10.1080/15548627.2015.1034409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J, Ma K, Qi T, Wei X, Zhang Q, Li G, Chiu JF. P62 regulates resveratrol-mediated Fas/Cav-1 complex formation and transition from autophagy to apoptosis. Oncotarget. 2015;6(2):789–801. doi: 10.18632/oncotarget.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12(11):829–46. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 115.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, et al. Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19(5):795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


