Abstract
Modern antiretroviral therapies have provided HIV-1 infected patients longer lifespans and better quality of life. However, several neurological complications are now being seen in these patients due to HIV-1 associated injury of neurons by infected microglia and astrocytes. In addition, these effects can be further exacerbated with opiate use and abuse. One possible mechanism for such potentiation effects of opiates is the interaction of the mu opioid receptor (MOR) with the chemokine receptor CCR5 (CCR5), a known HIV-1 co-receptor, to form MOR-CCR5 heterodimer. In an attempt to understand this putative interaction and its relevance to neuroAIDS, we designed and synthesized a series of bivalent ligands targeting the putative CCR5-MOR heterodimer. To understand how these bivalent ligands may interact with the heterodimer, biological studies including calcium mobilization inhibition, binding affinity, HIV-1 invasion, and cell fusion assays were applied. In particular, HIV-1 infection assays using human peripheral blood mononuclear cells, macrophages, and astrocytes revealed a notable synergy in activity for one particular bivalent ligand. Further, a molecular model of the putative CCR5-MOR heterodimer was constructed, docked with the bivalent ligand, and molecular dynamics simulations of the complex was performed in a membrane-water system to help understand the biological observation.
Keywords: NeuroAIDS, bivalent ligands, GPCR dimerization, structure-activity relationship
Introduction
The progression of human immunodeficiency virus (HIV)-1/acquired immunodeficiency syndrome (AIDS) has been shown to be accelerated by abused substances such as opioids, cocaine, and alcohol.1–5 While both abusive and addictive behavior of opioids are mainly associated with the mu opioid receptor (MOR), opioids can also negatively impact the immune system via immunomodulation regulated through the MOR.5,6 These deleterious results on the immune system may affect the progression of HIV/AIDS.7
The major co-receptor that regulates the invasion of monotropic (or R5-tropic) HIV-1 is the chemokine receptor CCR5, which is expressed in immune and non-immune cells.8–11 In 2007, maraviroc, a CCR5 antagonist, was approved by the FDA. In combination with other antiretroviral therapies (ART), maraviroc has improved the overall health of HIV-1 infected indidivuals.12 However, despite the use of ART and reductions in plasma viral loads to near undetectable levels, there remain significant CNS complications. In particular, HIV-associated neurocognitive disorders (HAND) are evident in nearly half of AIDS patients and lead to abnormalities in neurocognition, behavior, and motor control.13 The neurological complications of neuro-acquired immunodeficiency syndrome (neuroAIDS) are largely due to the injury of neurons caused by inflammation and the release of viral products from infected microglia and astrocytes.14
The progression of neuroAIDS has been linked to opioid abuse and addiction. A key site of HIV and opioid convergence may be caused by interactions between the CCR5 and the MOR.4,6,15–18 For example, MOR agonists can upregulate the expression of CCR5 and promote HIV-1 infection, while MOR antagonists may block these effects.19 Opioids can also exacerbate the amount of indirect neuronal injury in neurons and glia through HIV-1 induced CNS inflammation.15,17 There is additional evidence that the specific opioid-dependent neuronal injury may be primarily induced by MOR expressing glia in the CNS.6
Moreover, the MOR and the CCR5 have been shown to heterodimerize with each other and undergo crosstalk.20–22 This interaction affects immune cell function and may produce the synergistic effects of HIV and opioid drug co-exposure seen in neuroAIDS progression.19,21 Recently, a bivalent compound containing both a MOR and a CCR5 antagonist pharmacophore was designed and synthesized in our lab in an attempt to study the pharmacological implication of MOR–CCR5 heterodimerization in neuroAIDS (Figure 1).23–25 This new ligand (compound 1b) showed higher potency in antiviral activity in human astrocytes than maraviroc or a mixture of naltrexone (a MOR antagonist) and maraviroc. Presumably, by targeting the putative MOR–CCR5 heterodimer, the bivalent ligand was able to block viral invasion more effectively.25 More significantly, when morphine was present, the inhibition of infectivity by maraviroc was abolished in astrocytes, but the bivalent ligand retained its antiviral activity under the same conditions.25 To understand the pharmacological profile and structure-activity relationship (SAR) of this bivalent ligand, we further designed and studied several analogues of compound 1b along with their monovalent control compounds. Herein, we report the SAR study of this series of bivalent compounds as chemical probes targeting the putative MOR-CCR5 heterodimer through binding assays, functional assays, HIV-1 infection assays, and molecular modeling simulations.
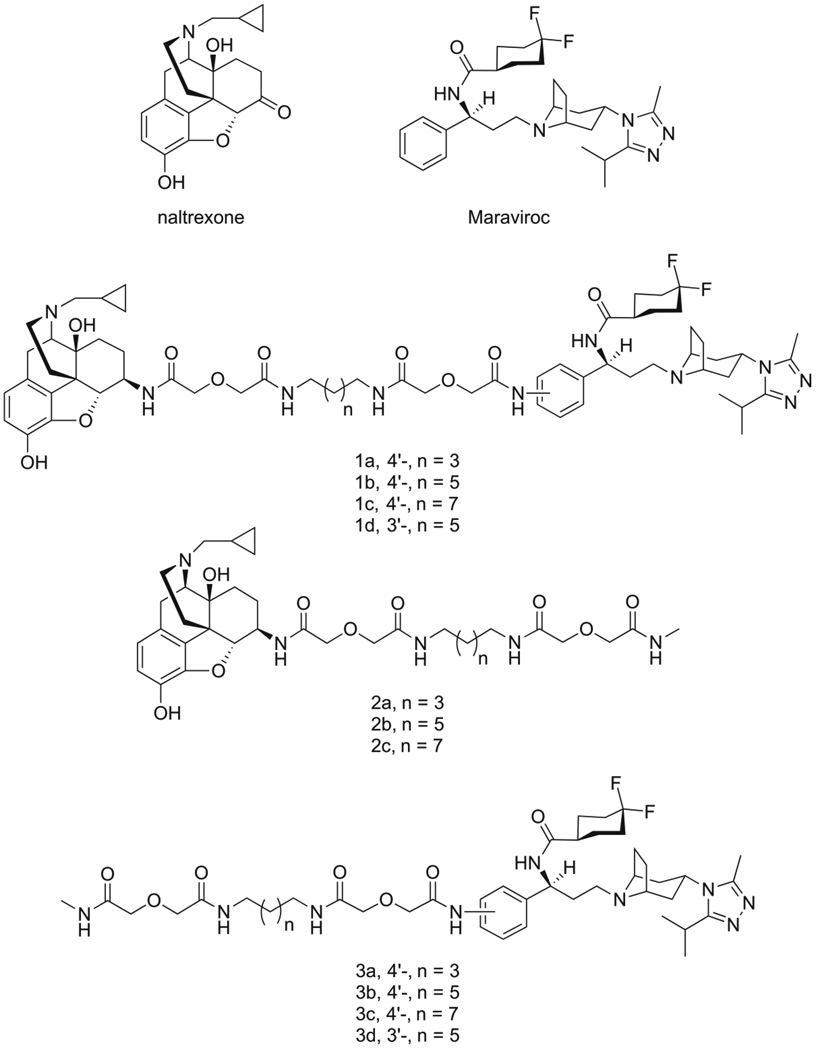
Figure 1.
Chemical structures of naltrexone, maraviroc, bivalent ligands (1a–d), and monovalent ligands (2a–c), and 3a–d).
Results and Discussion
Compound Design and Synthesis
It has been postulated that the linker length between the two pharmacophores of bivalent ligands is critical for their activity and may serve as an indicator for the distance between the two binding pockets of a GPCR dimer.26 The first bivalent ligand (designated as compound 1b, Figure 1) was designed with an overall length of 21 atoms based upon previous bivalent ligand reports involving the MOR,23 along with two control compounds with the same length of spacer (compound 2b for monovalent control attaching maraviroc, compound 3b for monovalent control attaching naltrexone). In order to study how linker length affects activity, the overall length was decreased or increased by two atoms in compounds 1a or 1c respectively. Concurrently, four new monovalent control compounds were synthesized with either a 19 atom linker (2a and 3a) or a 23 atom linker (2c and 3c) for both maraviroc and naltrexone pharmacophores.
To study linker attachment position influence, another ligand (1d) was designed by switching the linker attachment position from the 4’-position (para, as in 1b) of phenyl ring in maraviroc to its 3’-position (meta). The corresponding control compound 3d was then prepared.
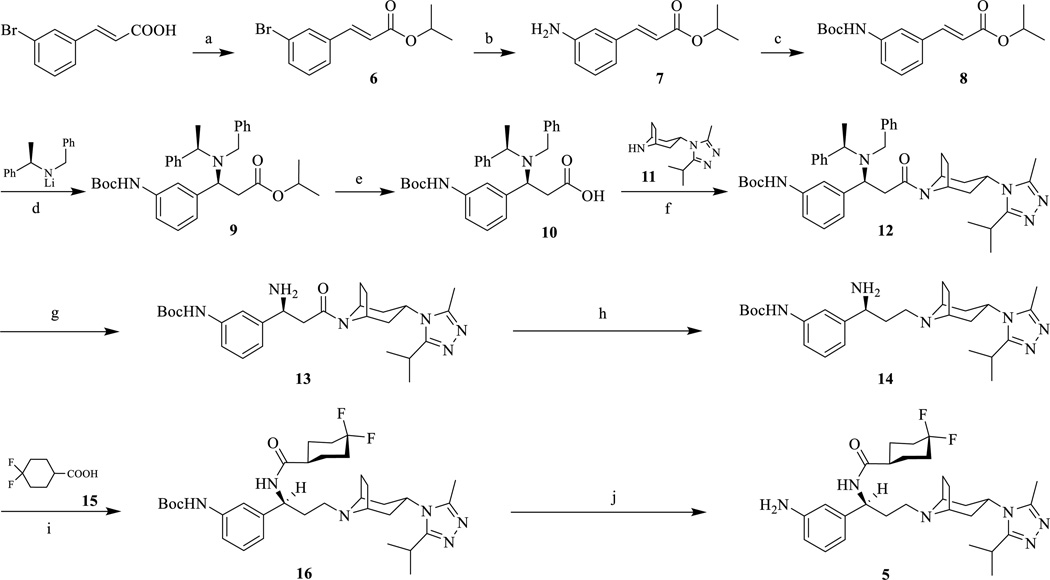
The synthetic route for 3’-amino maraviroc precursor (5) that was needed to prepare ligand 1d is shown in Scheme 1. 3-bromocinnamic acid was first protected via an esterification reaction using isopropanol (i-PrOH) and a catalytic amount of H2SO4 while being refluxed. The overall yield of 6 was 79%. The bromide was then converted to the amine (7) using lithium hexamethyldisilazide (LHMDS), Pd2(dba)3, and P(t-Bu)3 with yields ranging from 50% to 70%. Immediately after purification, the amine of 7 was protected with a Boc group using di-tert-butyl dicarbonate stirred in a 1:1 mixture of H2O/Dioxane with NaHCO3 at room temperature which gave 8 at yields up to 76%. The stereoselective Michael addition to form 9 was achieved by using lithium (R)-(+)-N-benzyl-α-methylbenzylamide. This reaction has been used previously in multiple synthetic routes to selectively form enantiomerically pure adducts.23,27–29 Both column chromatography and recrystallization were used to purify the product with yields up to 50%. Saponification of the isopropyl ester (9) to form the carboxylic acid 10 was accomplished by refluxing in MeOH/H2O with LiOH. After reaction workup, a yield of 88% was achieved.
Scheme 1.
Synthetic route of 3’-aminomaraviroc (5).Reagents and conditions: (a) i-PrOH, H2SO4 (conc.), reflux, 79%; (b) i) LHMDS, Pd2(dba)3, P(t-Bu)3, toluene, rt; ii) 1N HCl, rt, 69%; (c) Boc2O, THF, reflux, 76%; (d) THF, −78 °C, 41%; (e) LiOH, MeOH-H2O (2/1), reflux, 88%, two steps; (f) EDCI, HOBt, TEA, 11, 4Å MS, DCM, 0°C to rt, 74%; (g) 10% Pd-C, 60 psi, MeOH, 91%; (h) i) LiAlH4, THF, 0°C to rt; ii) H2O, NaOH, 79%; (i) EDCI, HOBt, TEA, 15, 4Å MS, DCM, 0°C to rt, 60%; (j) CF3COOH, DCM, 0°C to rt, 79%.
Next, an amide coupling between 10 and 11 to form 12 was done by using EDCI with a yield of 74%. The reduction of the (R)-(+)-N-benzyl-α-methylbenzylamide to form the amine 13 proved to be more difficult compared to the same reaction for the 4-amino maraviroc derivative.23 The hydrogenation of 12 was first attempted using 10% Pd/C and 60 psi H2 in MeOH, but very little product was formed even after 7 days. Therefore, new conditions were tried using 10% Pd/C, 60 psi H2, and 2 equivalents of AcOH in MeOH. A yield of 91% for 13 was achieved. Therefore, acid was essential for facilitating the reduction of 12 to 13. Reduction of the amide, 13, to form 14 was accomplished by using lithium aluminum hydride. Another EDCI mediated amide coupling was performed between 14 and 15 to form 16. The 4,4-difluorocyclohexanecarboxylic acid (15) had previously been synthesized from ethyl-4-oxocyclohexanecarboxylate.23,30 The Boc-deprotection of 16 was accomplished using 10% trifluoroacetic acid to afford 5 quantitatively.
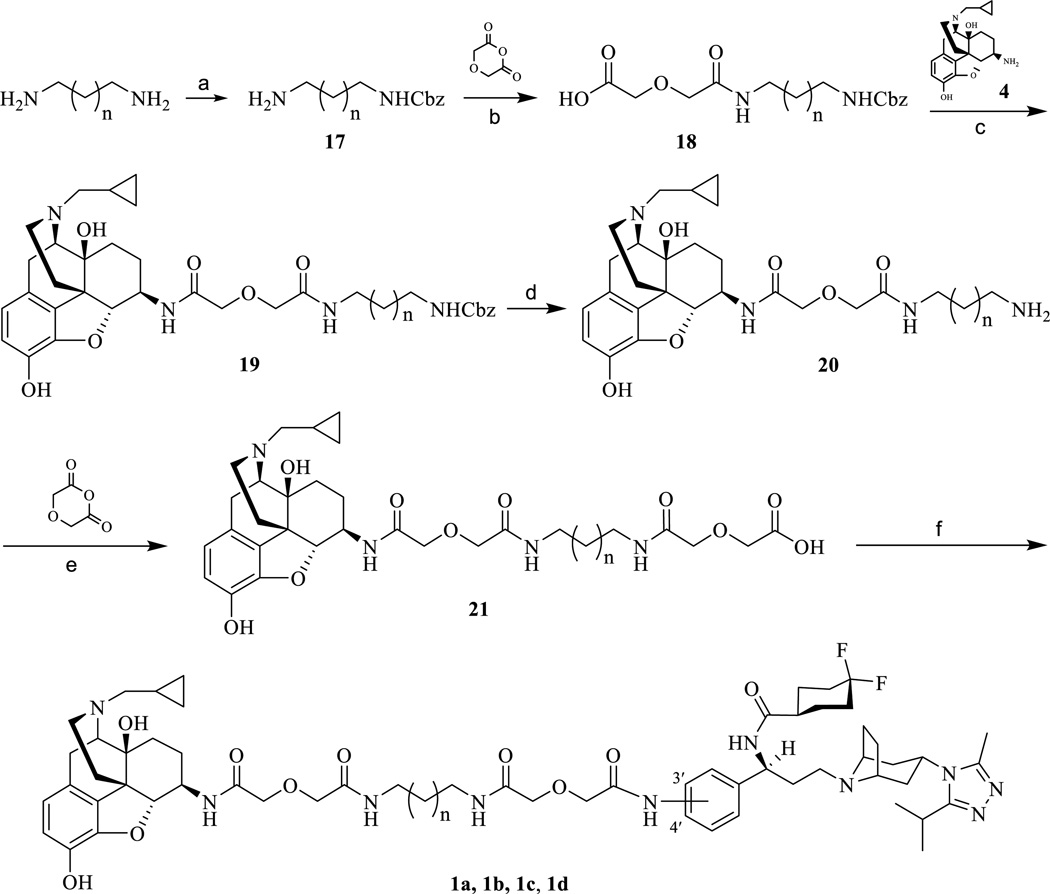
The synthesis of compound 1d (Scheme 2) was facilitated by coupling 5 with 21 using EDCI with a final yield of 26%. The synthetic route for 4’-amino maraviroc precursor has been reported and the chemical synthesis for all the bivalent compounds is depicted in Scheme 2 and it was similar to the one reported previously.24
Scheme 2.
Synthetic route of bivalent ligands (1a, 1b, 1c, and 1d). (a) CbzCl, DCM, MeOH, 5 °C, 60%; (b) THF, diglycolic anhydride, rt, 64%; (c) EDCI, HOBt, TEA, 4·2HCl, 4Å MS, DMF, 0 °C to rt, 70%; (d) 10% Pd–C, 60 psi, MeOH, 89%; (e) DMF, diglycolic anhydride, rt, 95%; (f) EDCI, HOBt, TEA, 6, 4Å MS, DMF, 0 °C to rt, 26%.
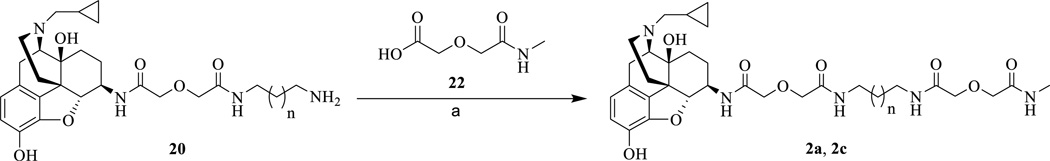
The monovalent control compounds 2a, 3a, 2c, and 3c were synthesized in the same manner as previously described (Schemes 3 and 4).23 The 3’-amino maraviroc monovalent control compound, 3d, was synthesized using the synthetic route in Scheme 4. The monoprotected diamine 17 was coupled with methylcarbamoylmethoxy-acetic acid (22) to form 23. Next, 23 was deprotected using Pd/C mediated hydrogenation (24) and a diglycolic anhydride group was then added to bring the total linker length to 21 atoms (25). Finally, 25 and 5 were coupled using EDCI to form the monovalent compound 3d (33% yield).
Scheme 3.
Synthetic route of monovalent ligands (2a, and 2c). Regents and conditions: (a) EDCI, HOBt, TEA, 22, 4Å MS, DMF, 0 °C to rt, 30%.
Scheme 4.
Synthetic route of monovalent ligands (3a, 3c, and 3d). Regents and conditions: (a) EDCI, HOBt, TEA, 22, 4Å MS, DMF, 0 °C to rt, 37%; (b) 10% Pd–C, 60 psi, MeOH, 30%; (c) DMF, diglycolic anhydride, rt, 44%; (d) EDCI, HOBt, TEA, 6, 4Å MS, DMF, 0 °C to rt, 33%.
Biological Evaluation
MOR calcium mobilization assays and radioligand binding assays
The compounds were first subjected to their agonism and antagonism property tests in the MOR monocloned CHO cells. The results from MOR calcium mobilization and competitive binding assays indicated that all the compounds maintained their recognition to the receptor MOR (Table 1), which supported our original molecular design pertaining to the MOR part. Compared to the parent pharmacophore naltrexone, all of the bivalent compounds showed relatively higher IC50 values for both calcium mobilization antagonism and receptor binding affinity, while the linker length did not drastically affect the bivalent compounds’ affinity on the MOR. The 3’-position bivalent compound, 1d, showed the highest Ki value for MOR, but conversely had the lowest IC50 values for the calcium reflux functional activities, indicating that the linkage position on the CCR5 pharmacophore did not influence the recognition on the MOR, which seemed to be reasonable. Furthermore, the monovalent control compounds, compared to the corresponding bivalent compounds, did not show any significant difference regarding their affinity and function on the receptor, which indicated that the linkers were well tolerated in the recognition of the MOR.
Table 1.
MOR Ca2+ inhibition and [3H]NLX competitive binding assays results.a
| Compd | n | Linkage | Ca2+ assay IC50 (nM) |
[3H]NLX binding Ki (nM) |
|---|---|---|---|---|
| NTX | NA | NA | 2.87 ± 0.27 | 0.39 ± 0.04 |
| 1a | 3 | 4′ | 21.8 ± 5.6 | 3.80 ± 0.55 |
| 2a | 3 | NA | 92.1 ± 20.1 | 0.78 ± 0.12 |
| 1b | 5 | 4′ | 40.0 ± 4.8b | 3.24 ± 0.34 |
| 2bb | 5 | NA | 37.8 ± 4.4 | 9.2 ± 3.4 |
| 1c | 7 | 4′ | 21.9 ± 3.7 | 6.49 ± 0.16 |
| 2c | 7 | NA | 41.4 ± 24.6 | 1.11 ± 0.08 |
| 1d | 5 | 3′ | 17.1 ± 4.9 | 10.0 ± 0.6 |
The values are the means ± S.E.M. of at least three independent experiments. Ca2+ mobilization assay was performed on hMOR-CHO cells. Membranes for radioligand binding assays were prepared from mMOR-CHO cells. NLX, Naloxone. NA, not applicable.
Data taken from Ref. 24.
CCR5 Calcium mobilization assays and radioligand binding assays
Compounds were then tested for both their agonism and antagonism for the CCR5 in the MOLT-4 cells. We intended to use the calcium mobilization assay as the preliminary screening tool for its high throughput property and much lower cost. Prior to use, CCR5-MOLT-4 cells were transiently transfected with a chimeric G protein, Gqi5, in order to boost their calcium signaling levels.31 Over a range of concentrations, compounds showed no apparent agonism of the CCR5 (data not shown).
The CCR5 antagonism assay was then tested through the inhibition of RANTES (CCL5) stimulated calcium mobilization and indicated that modification of maraviroc at its phenyl ring was not very well tolerated (Table 2). The bivalent ligands 1a–d all showed significantly lower potency compared to maraviroc, the parent pharmacophore, which was consistent with previously described data for similar modifications on the compound.24 In particular, bivalent compound 1b with the 21 atom linker showed a 60-fold decrease of antagonist potency as compared to maraviroc.23 The decrease in CCR5 antagonism was not improved by either increasing or decreasing linker length as seen for bivalent compounds 1c or 1a. As a matter of fact, the optimal linker length for CCR5 antagonism was still 21 atoms. However, the linker lengths of the monovalent control compounds 3a, 3b, and 3c were directly correlated to their CCR5 antagonism: increasing linker length led to an increase of CCR5 antagonism.
Table 2.
CCR5 Ca2+ mobilization and [125I]MIP-1α competitive binding assays results.a
| Compd | n | Linkage | Ca2+ assay IC50 (nM) |
Radioligand bindingb Ki (nM) |
|---|---|---|---|---|
| maraviroc | - | - | 2.1 ± 0.4 | 0.24 ± 0.06 |
| 1a | 3 | 4′ | 2413 ± 617 | ND |
| 3a | 3 | 4′ | 948 ± 34 | ND |
| 1bb | 5 | 4′ | 126 ± 28 | 239 ± 56 |
| 3bb | 5 | 4′ | 622 ± 36 | 151 ± 44 |
| 1c | 7 | 4′ | 543 ± 79 | ND |
| 3c | 7 | 4′ | 392 ± 51 | ND |
| 1d | 5 | 3′ | 1340 ± 110 | ND |
| 3d | 5 | 3′ | 129 ± 42 | ND |
The values are the means ± S.E.M. of at least three independent experiments. Membranes for radioligand binding assay were prepared from mMOR-CHO cells. Ca2+ mobilization assay was performed on MOLT4-CCR5 cells. NA, not applicable. ND, not determined.
Data taken from Ref. 24.
When the linker attachment on maraviroc was switched from the 4’- to 3’- position of the phenyl ring, a significant difference in CCR5 antagonism was observed, that was, ligand 1b was ten times more potent than 1d as a CCR5 antagonist. Therefore, for the bivalent compounds, a 4’-position attachment was more favored over the 3’-position attachment.
To further verify the direct binding affinity of compound 1b, it was submitted for radioligand binding affinity study at the CCR5 along with the parent pharmacophore, maraviroc, and its monovalent control (compound 3b) at EMD Millipore, a division of Merck KGaA, Darmstadt, Germany. Apparently introduction of a spacer at the phenyl ring of maraviroc did dramatically influence the recognition to the receptor CCR5. It was decided not to apply such test for other less potent compounds.
HIV-1 p24 production in PMBC and macrophage cultures
PBMCs, macrophages, and microglia expressing CCR5 are preferentially susceptible to infection by R5-tropic strains of HIV.33–36 Accordingly, the use of PBMCs and macrophages is highly appropriate for testing the effects of these bivalent ligands on blocking infection by R5-tropic strains of HIV.
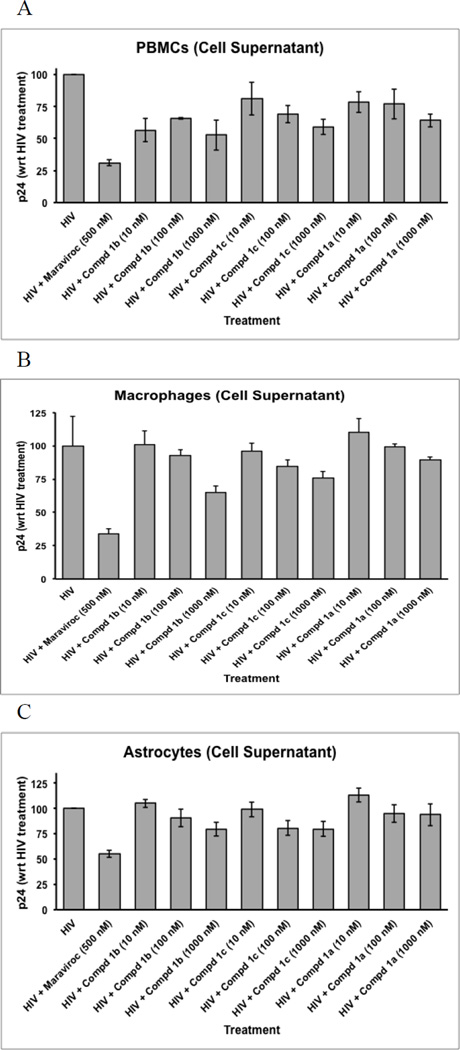
Viral production in HIV-infected PBMCs was assessed by measuring p24 levels following 5 days of exposure to maraviroc and/or the bivalent ligands (Figure 2A). Overall, there was a significant effect of the treatments on viral production in PBMCs (p < 0.005; ANOVA). As anticipated, exposure to 500 nM maraviroc (MVC) significantly decreased viral production (p < 0.00025 vs. HIV-1 infection alone; Dunnett’s test, two-tailed). Exposure to bivalent ligand 1b also significantly reduced p24 levels in PBMCs. Interestingly, viral levels were reduced following exposure to all three concentrations of bivalent ligand 1b (p < 0.02; Dunnett’s test) without apparent concentration response effect. Bivalent ligand 1c markedly reduced p24 levels only at the highest concentration (1 µM) (p < 0.05; Dunnett’s test), while bivalent ligand 1a did not significantly reduce viral production in PBMC cultures at any of the concentrations tested.
Figure 2.
Preliminary screening results of HIV-1 p24 production in PMBC, macrophage, and astrocyte cultures. PMBC, macrophage, and astrocyte cultures were exposed to R5-tropic HIV virus and varying concentrations of compounds 1a–c. Inhibition of p24 production was measured using ELISA and compared to the control (maraviroc).
Not unexpectedly, the bivalent ligands that inhibited HIV-1 production in PBMCs also reduced p24 levels in macrophages resulting in a significant overall treatment effect in macrophages (p < 0.00025; ANOVA) (Figure 2B). Maraviroc alone (MVC) significantly restricted viral production (p < 0.00005 vs. HIV-1 infection alone; Dunnett’s test, two-tailed). Exposure to bivalent ligands 1b and 1c significantly reduced p24 levels in macrophages, but only at the highest 1 µM concentration (p < 0.005 and p < 0.025, respectively; Dunnett’s test). Similar to the findings in PBMCs, bivalent ligand 1a failed to diminish p24 levels in macrophages even at the highest 1 µM concentration.
HIV-1 p24 production in astrocyte cultures
Primary human astrocytes were chosen because they are a major cellular site of infection in the CNS and are a key cellular site where opioids act to potentiate the pathophysiological effects of HIV-1 infection.5–6,16,18,25,37–39 Concentration response effects of viral infection inhibition were observed for all three bivalent compounds (Figure 2C). By virtue of the fact that astrocytes display lower rates of HIV-1 infectivity than PBMCs or macrophages and because of reduced rates of relative infectivity using the p24 assay in the present study, we additionally assayed infectivity by examining HIV-1 Tat expression in astrocytes.40–43
HIV-1 Tat expression in astrocytes
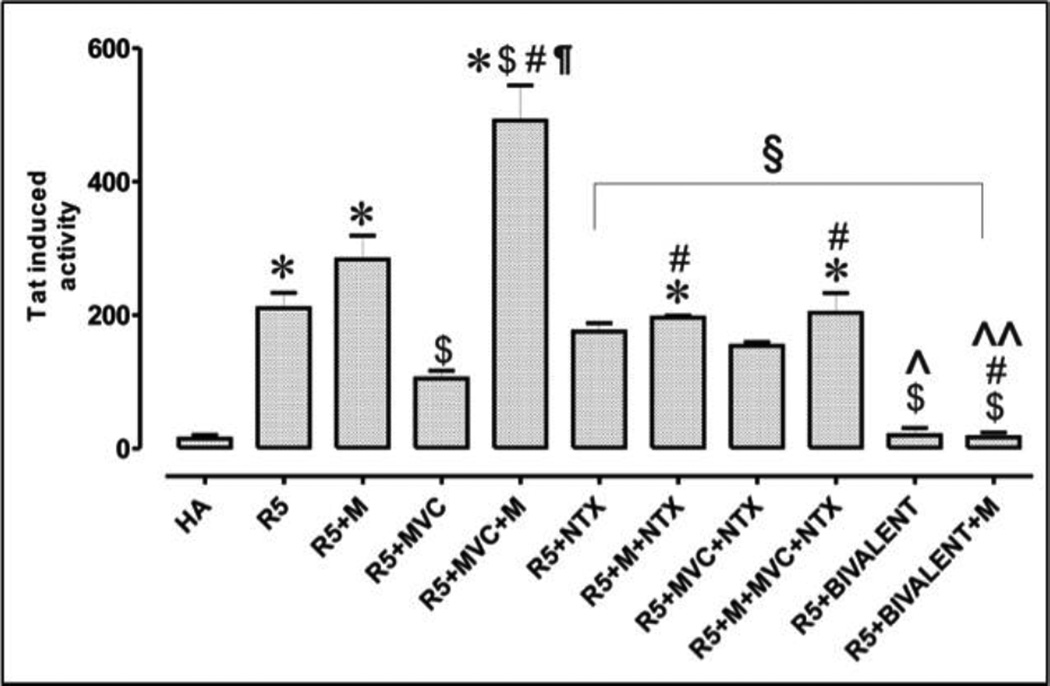
Among three bivalent compounds, compound 1b was picked to be evaluated in this assay because of its favorable binding affinities at both receptors as well as its more significant effect in all three HIV-1 p24 production assay results. Figure 3 shows the effect that 1b and maraviroc have on the infection of astrocytes by HIV-1 with and without the presence of morphine stimulation. Relative expression of Tat was significantly increased in astrocytes after infection with R5 HIV-1SF162 alone or in combination with morphine (M). As expected, exposure to maraviroc (MVC) significantly decreased viral entry, while co-exposure with morphine completely abolished the antiviral effect of maraviroc causing a <4-fold increase in Tat expression in astrocytes. The addition of the bivalent ligand 1 was extremely effective in inhibiting viral entry in astrocytes, causing a 3.3-fold decrease in viral entry when compared to maraviroc alone and a 7-fold decrease when compared to MVC + NTX. Co-exposure with morphine had no significant effect on the viral inhibition effect of the bivalent ligand 1b. The results showed that morphine impairs the antiviral function of maraviroc in human astrocytes while the bivalent ligand 1b can function as a potent inhibitory entity in astrocytes even under exposure from morphine interactions. These results supported our hypothesis that a properly designed bivalent ligand may synergistically block the HIV-1 invasion to host cells.
Figure 3.
HIV-1 invasion assay in human astrocytes. HIV-1SF162 infectivity in human glia was determined based on the relative amount of Tat protein expressed by the virus using a luciferase based assay. HA: human astrocytes, uninfected cells; R5: R5-tropic HIV-1; M: morphine; MVC: maraviroc; NTX: naltrexone; BIVALENT: compound 1b. Values are absorbance ± SEM of 3 independent experiments at 18 h post-infection (*p < 0.005 vs. un-infected cells; $p < 0.05 vs. R5 HIV-1; #p <0.05 vs. M; ¶p < 0.05 vs. MVC; §p < 0.05 vs. M + MVC;^p <0.05 vs. MVC + NTX; ^^p <0.05 vs. M + MVC + NTX; Ωp <0.05 vs. bivalent).
Cell fusion assays
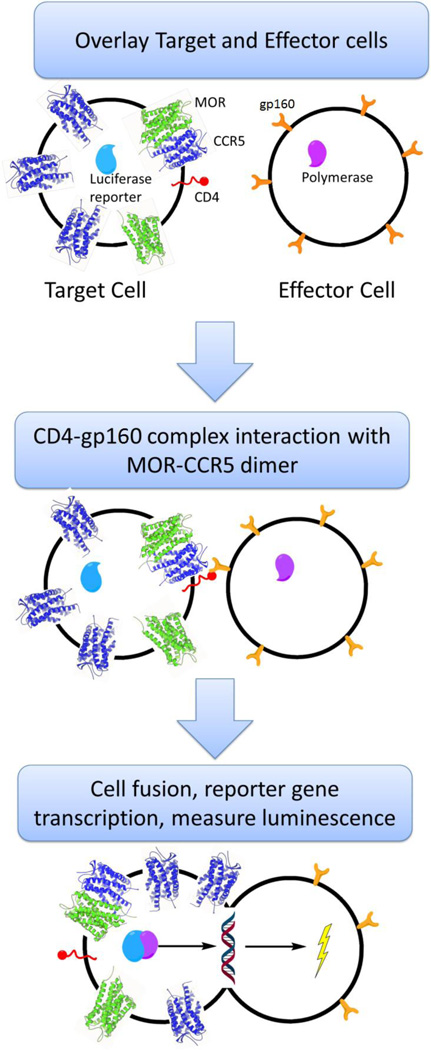
While the calcium mobilization assays can measure the activity of the compounds at the receptor level, it does not show the anti-HIV invasion activity of the compounds. Cell fusion assays are an alternative to working with live virus and have been shown to mimic the HIV invasion process.44 Figure 4 illustrates the general process for the cell fusion assay in which two cell populations, called the target and effector cells, are used. Fundamentally, the target cells act as the host cells that are infected by HIV and the effector cells act as the virus. A CCR5-MOR CHO cell line was used as the basis for the target cells and was transiently transfected with CD4 and a luciferase reporter. Human embryonic kidney (HEK) cells were used as the effector cells and were transiently transfected with HIV-1 gp120 and T7 polymerase. Once overlaid, CD4 and gp120 form a complex and interact with the CCR5-MOR heterodimer and initiate the fusion process. Upon cell fusion, the luciferase gene reporter is transcribed, and after 18 hours luminescence is measured. Adding a CCR5 antagonist, such as maraviroc, during the overlay process inhibits the fusion process and leads to a decrease in luminescence. Therefore, addition of the bivalent compounds should also inhibit the fusion process.
Figure 4.
Cell fusion assay scheme adopted to mimic HIV invasion without live virus.
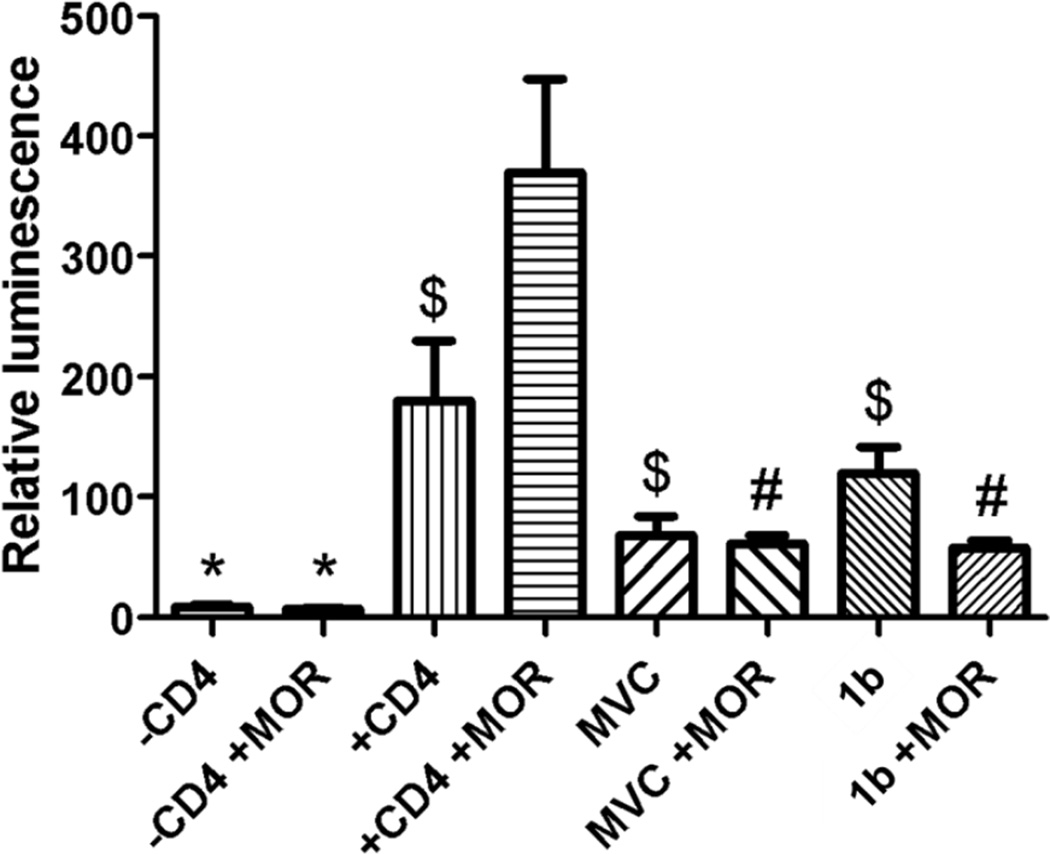
Figure 5 is the results from a cell fusion assay with and without morphine stimulation during the fusion process. Upon the addition of morphine and +CD4 to effector cells, there was a significant increase (p < 0.05) in fusion compared to the +CD4 effector cells alone. Addition of maraviroc significantly lowered cell fusion, while surprisingly; addition of morphine did not influence the cell fusion inhibition effect of maraviroc. On the other hand, the inhibitory effect of 1b alone on the cell fusion was not as significant as maraviroc alone while its inhibitory effect was amplified by 2-fold when morphine was present. These results were partially agreeable with what was observed in the virus invasion assay consisting of astrocytes and HIV-1 in a native system that the bivalent ligand 1b showed more significant inhibitory effect upon the addition of morphine than without morphine.
Figure 5.
Cell fusion assay based upon luminescence from expressed luciferase reporter gene. For morphine stimulation, 300 nM morphine was added. 100 nM Maraviroc, and 3000 nM 1b were used. Values are representative of 4 assays run. (* p < 0.001 vs. +CD4 +morphine; $ p < 0.05 vs. +CD4 +morphine; # p < 0.01 vs. +CD4 +morphine) This trend was seen in an additional three assays.
mRNA levels of CCR5 and MOR
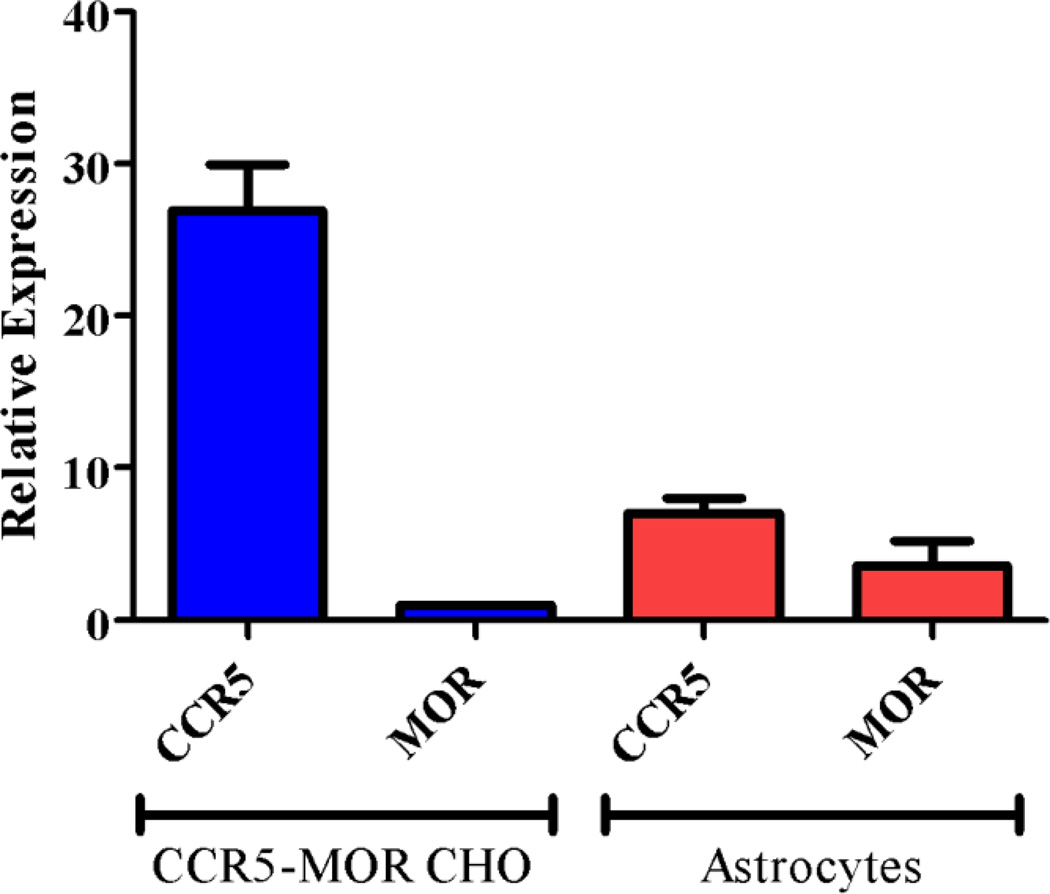
To understand the disconnection between the cell fusion assay results and the astrocyte HIV-1 invasion assay results, we tried to find out the relative protein expression levels in the cells between the two assays. Using RT-PCR, the mRNA expression levels of CCR5 and MOR were analyzed for both astrocytes and the CCR5-MOR CHO cells (Figure 6). From two batches of primary human astrocytes we adopted in the assays, the CCR5 mRNA level was about 11-fold higher than the MOR while in the CCR5-MOR CHO cell line the mRNA level of the CCR5 was at least 24-fold higher than the MOR. That is, there was at least 2-fold difference in the ratio of MOR and CCR5 between the two cell lines with the CCR5-MOR CHO cell line having a much higher expression of the CCR5 than the MOR. Therefore we postulated that with much higher amounts of CCR5 than MOR, there would be a lower level of formation of CCR5-MOR heterodimers in the CCR5-MOR CHO cell line than in the astrocytes. Since the bivalent compounds would preferentially bind to the putative CCR5-MOR heterodimers, fewer available heterodimers for bivalent ligand binding in the CCR5-MOR CHO cells than in the astrocytes would lead to less significant inhibitory effect of the ligand.
Figure 6.
mRNA levels of MOR and CCR5 in the CCR5-MOR CHO cell line and in primary human astrocytes.25
Molecular Modeling Studies
To understand potential interaction of the ligand 1b with the putative receptor dimers, as well as its relatively low affinity to the CCR5 receptor, we adopted computational modeling to explore the relationship between the CCR5-MOR heterodimer and bivalent compound 1b. Several methods have been used to model GPCR homodimers and heterodimers. Until recently, the most prominent way to model dimerization was to use protein-protein docking programs such as ZDOCK, GRAMM, or Rosetta.45 Recently, several GPCR homodimer crystal structures have been characterized and offered a new route to model GPCR dimerization.46–48 These structures have either a TM4-TM5 or a TM5-TM6 interface, which both represent feasible GPCR dimer model.45,47,49 Current knowledge also suggests that GPCRs do not undergo significant conformational changes upon dimerization.45 Therefore, GPCR dimers can now be modeled by using the experimentally observed dimers and overlaying the receptors being studied onto it and aligning them based upon sequence homology. This technique has been successfully applied to model 5-HT1A homodimers and has been experimentally verified.49
Upon the time of our initial molecular design effort, neither the crystal structure of the MOR or the CCR5 was available for structure-based design. Thus compound 1b and its analogues were designed based on the homology models of the CCR5 and the MOR from our own efforts.50,51 Later on the crystal structure of the MOR homodimer was made available47 and then adopted by us as the template for the CCR5-MOR heterodimer. Their heterodimer interface was assumed to be between TM5 and TM6. For the CCR5 counterpart, we had to adopt the then available CXCR4 crystal structure48 as a template to rebuild our CCR5 homology model in order to construct the heterodimer model, as elaborated below. More recently the crystal structures of the CCR5 and its dimers were reported,52 thus, providing a meaningful comparison for our future exploration.
In detail, first, a CCR5 homology model was constructed using the CXCR4 crystal structure as the template structure.48 Second, one MOR receptor in the MOR dimer crystal structure was kept in place while the other was overlaid with the CCR5 homology model, aligned based upon their homology, and replaced. Before ligand docking, preliminary heterodimer model refinement was carried out through energy minimization using the MMFF94 force field. An alternative way to build this dimer model would be to construct CCR5 homology model based on one of the MOR monomers in its homodimer structure. Though it would provide a more conserved heterodimer model, the lower homology between the MOR and the CCR5 compared to that between the CXCR4 and the CCR5 could be an intrinsic shortcoming, and therefore was not adopted for this method.
Docking compound 1b into both binding pockets of the heterodimer model simultaneously proved to be difficult. Therefore, a new method had to be devised so that the two different portions of the bivalent compound could be docked individually, in their respective receptor, and then connected to each other with the (19-atom) linker. Since the MOR homodimer was co-crystalized with its irreversible antagonist β-FNA,47 the naltrexone portion of the bivalent compound was well aligned with the morphinan structure of β-FNA.47 Once aligned, the 6-β position of the naltrexone portion points upward toward the TM-5/TM-6 heterodimer interface, which provided an appropriate orientation to allow for the linker to connect to maraviroc and reach for the CCR5 binding pocket, with the assumed TM5/TM6 heterodimer interface.
Next maraviroc was docked into the CCR5 portion of the heterodimer using GOLD.53 The docking poses subsequently obtained were manually evaluated to select those that allow geometrically reasonable attachment between the phenyl group of maraviroc and the linker portion of 1b near the TM5 and TM6 heterodimer interface. Of those poses, the one with the highest GOLD docking score was used. After attaching maraviroc to the 19-stom linker portion of compound 1b, the system was energy minimized.
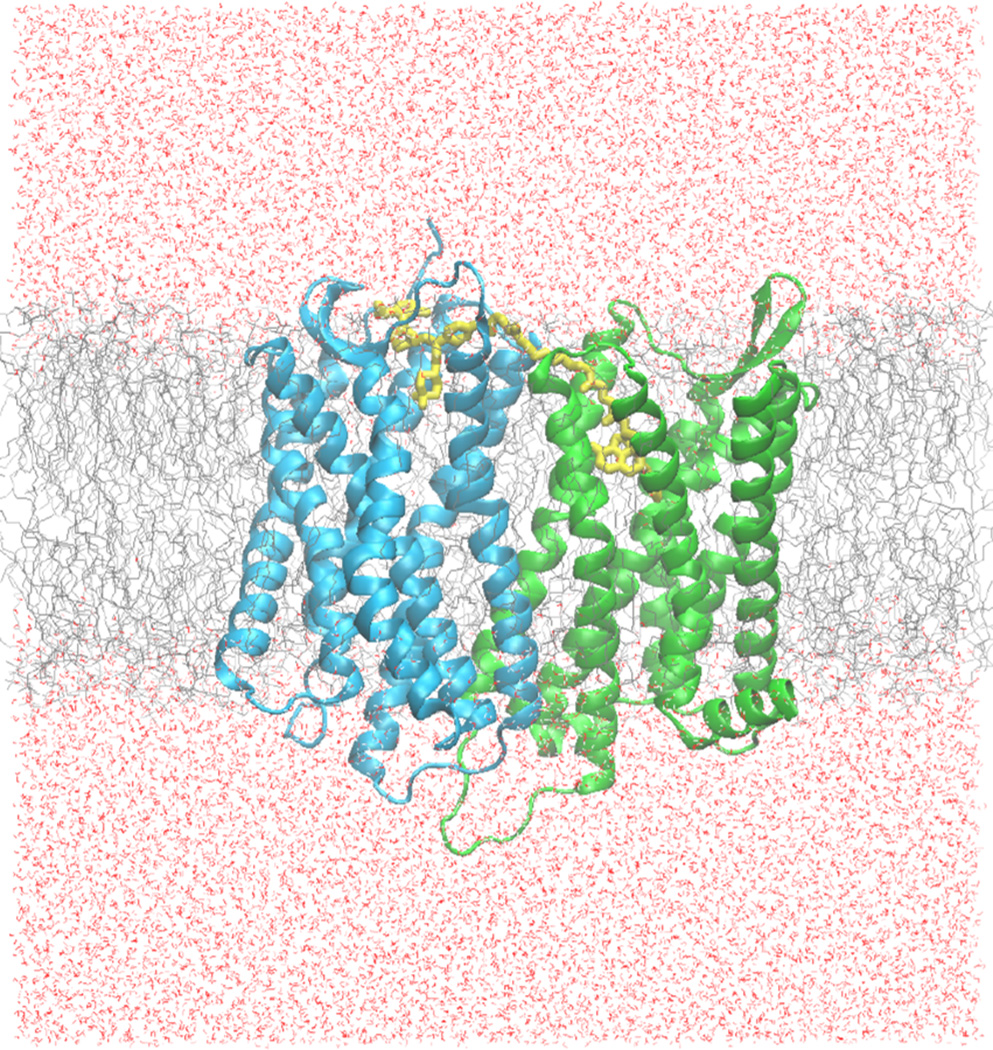
Molecular dynamics, NAMD, was then used to interpret the stability of the heterodimer-bivalent compound complex.54 Several steps were taken in order to prepare the heterodimer-ligand complex for dynamics simulation: the complex was first added to a lipid bilayer and then solvated with a pre-defined water box with appropriate types and concentrations of ions to accurately simulate its native membrane environment (Figure 7).49,55,56 Altogether, the dimer-ligand-lipid-water-ion system had 162,385 atoms. A series of minimizations was then done in a step-wise manner to slowly equilibrate and energy minimize the components of the complex. After 10 ns of dynamics simulation the system was equilibrated as indicated by the RMSD (Figure S1, Supporting Information) and total energy (Figure S2, Supporting Information) of the system. A longer period of simulation (another 10 ns) did not change the outcome significantly and therefore, was not included in the discussion.
Figure 7.
Molecular simulation system for the CCR5-MOR heterodimer in a membrane (gray), and water box (red) system. The green protein represents the MOR and the blue protein is the CCR5, while compound 1b is colored in yellow.
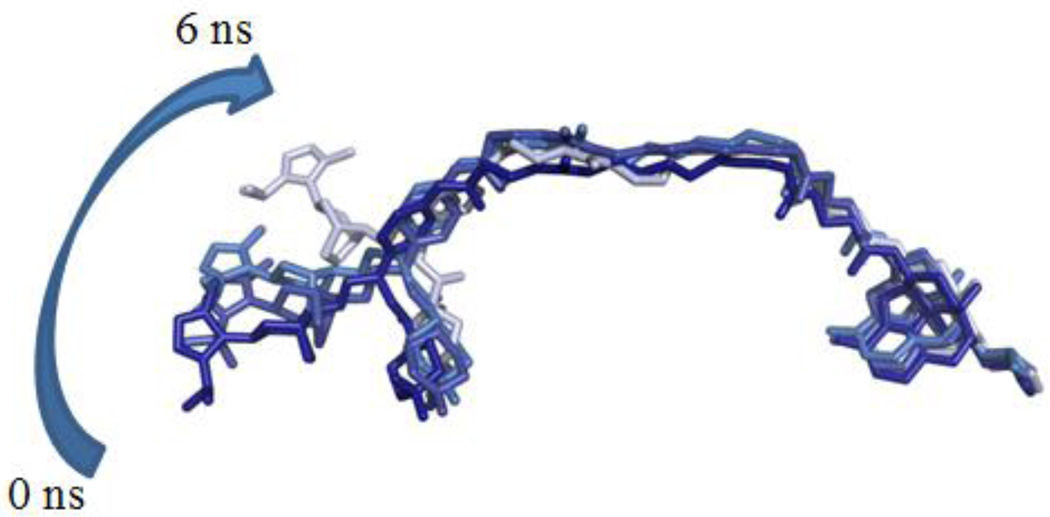
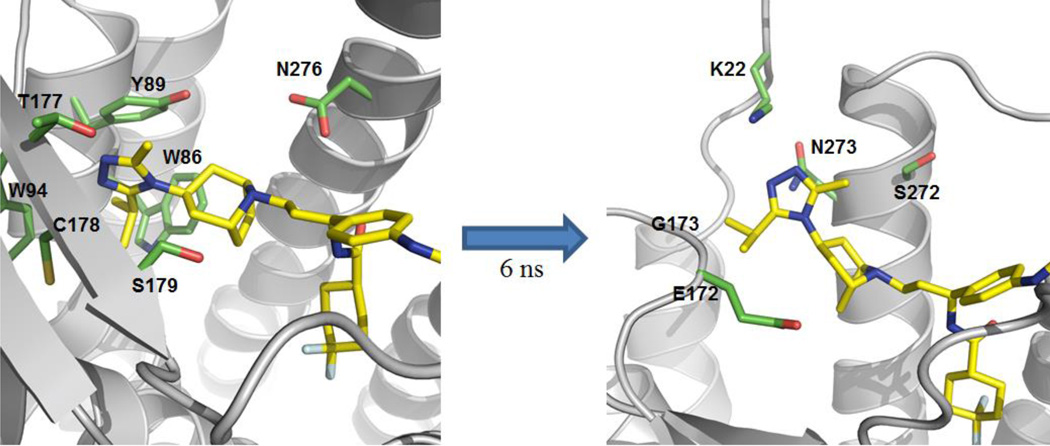
During the 10 ns of dynamics simulation, the maraviroc portion of compound 1b partially dislodged from the CCR5 binding pocket, whereas the naltrexone portion maintained its binding pose largely within the MOR binding pocket observed in the MOR crystal structure (Figure 8).47 This result indicated that for the heterodimer model, the initial binding mode for compound 1b in CCR5 was not energetically favored. However, it is also important to note that only the triazole moiety of maraviroc moved out of the original CCR5 binding pocket. Figure 8 illustrates that after 6.0 ns of dynamic simulation the triazole ring rotated upward out of its initial binding pocket. This shift upward was reflected in the changes in the RMSD of 1b (Figure S3, Supporting Information). Such an observation actually was partially in agreement with the later available maraviroc bound CCR5 crystal structure.52
Figure 8.
Trajectory of 1b in the CCR5-MOR heterodimer at 0, 2.4, 4.4, and 6.0 ns, with dark blue representing 1b at 0 ns and subsequently becoming a light blue at 6.0 ns.
The change in interactions between compound 1b and the CCR5-MOR heterodimer at 0 ns and at 6.0 ns was shown in Table 3. The opioid portion of 1b retained the majority of its interactions with the MOR binding pocket from its initial binding pose. This binding pose matched well with that of β-FNA within the MOR crystal structure.28 However, there were significant changes in the CCR5 interactions of 1b between these two time points (Figure 9). After an additional 4 ns of stimulation, i.e., until the end of the simulation, 1b maintained the binding pocket pose observed at 6.0 ns. These results helped explain the lower CCR5 binding affinity of the bivalent compound as compared to maraviroc.
Table 3.
Major amino acids in the CCR5 and MOR binding pockets in the heterodimer interacting with compound 1b.
| Time Frame | CCR5 Binding Pocketa | MOR Binding Pocket |
|---|---|---|
| 0 ns |
W86, Y89, W94, T177, C178, S179, I198, L255, N258, Q261, D276, M279 |
D147, Y148, N150, M151, I293, H294, V297, W315, I319, Y323 |
| 6 ns | K22, E172, G173, Y184, K191, I198, L255, N258, Q261, S272, N273, D276, M279 |
D147, Y148, N150, M151, I293, V236, H294, W315, I319, Y323 |
Figure 9.
Different binding pocket (green) for the triazole moiety of 1b (yellow) at 0 ns and 6.0 ns.
The dynamics simulation study could also help explain the changes in functional activities observed between maraviroc and 1b. As suggested by the simulations, addition of the linker to the p-phenyl portion of maraviroc led to 1b being able to adopt only one general binding mode that may represent a lower affinity mode. Within this binding mode there was an unstable binding pocket for the triazole portion of the molecule that led to it adopting two different conformations during the simulation. These observations were in agreement with the experimental data observed. For the CCR5 calcium antagonism assays, the loss in activity between maraviroc and compounds 1b and 3b can be explained by the unstable triazole binding pocket. In comparison, MOR calcium antagonism between naltrexone and compounds 1b and 2b was affected to a much lesser extent. During the simulation, the naltrexone portion of 1b did not move from its original binding pocket, which suggests that the 6β-attachment did not notably affect MOR binding. To be noticed, during the dynamics simulation, the maraviroc portion of 1b interacted with I198, L255, N258, Q261, and M279. Among them I198 and L255 have been deemed essential for maraviroc binding and N258 has been implicated in HIV-1 gp120 binding.57,58 This could partially explain the anti-HIV invasion effect of ligand 1b in the astrocytes.
Overall, the dynamics simulations indicated that 1b may recognize the putative CCR5-MOR heterodimer in an acceptable manner. Furthermore, though the CCR5 binding mode might not be optimal for 1b due to the limitation of our initial molecular design, it still blocked gp120 mediated invasion/fusion as seen in both the cell fusion assay and the HIV-1 invasion assay. Within the HIV-1 invasion assay, 1b showed even higher potency for inhibiting invasion than maraviroc or a simple combination of maraviroc and naltrexone. Thus, the binding mode observed from our dynamics simulations suggested an explanation for the observed enhancement in inhibitory effects, as ligand 1b might simultaneously bound to both the MOR and the CCR5 to contribute its inhibitory effect on HIV invasion.
Conclusion
Bivalent ligands provide a new tool to study heterodimerization of GPCRs. Targeting the putative CCR5-MOR heterodimer may be a novel and potentially efficacious antiviral strategy to treat neuroAIDS. Among the ligands we designed, synthesized, and tested, compound 1b has proven to be a potent inhibitor in both an artificial cell fusion assay mimicking HIV invasion and a native HIV invasion assay using live virus. Importantly, in the native cell HIV invasion assay, maraviroc was unable to inhibit HIV infection effectively in the presence of morphine in primary human astrocytes. However, compound 1b was a more potent inhibitor than maraviroc in primary human astrocytes with and without morphine (3.3-fold higher virus inhibition than maraviroc without morphine, and 7-fold higher virus inhibition than maraviroc with morphine). Utilizing molecular modeling and dynamics simulations, a possible binding mode of 1b was postulated and helped explain the possible mechanism of invasion inhibition by 1b. The intrinsic shortcoming of our initial molecular design based on the homology model of CCR5 at least partially rendered a less compelling affinity of compound 1b to both receptors and their putative dimers. On the other hand, as a proof-of-concept, this compound seems to be a promising lead to further develop more potent chemical probes to study neuroAIDS by targeting such a dimer. In all, these results encourage us to pursue another wave of molecular design based on this lead compound and the recently available crystal structure of CCR5 bound to maraviroc.
Experimental
Chemical syntheses
Chemical reagents were purchased from Sigma-Aldrich, Alfa Aesar, Combi-blocks, or AK Scientific and used without further purification. TLC analyses were carried out on Analtech Uniplate F254 plates. Chromatographic purification was accomplished on silica gel columns (230–400 mesh, Bodman). IR spectra were recorded on a Nicolet Avatar 360 FT-IR Instrument with ATR attachment. 1H (400 MHz) and 13C (100 MHz) nuclear magnetic resonance (NMR) spectra were acquired on a Bruker Ultrashield 400 Plus spectrometer. HREIMS analysis was performed on a Quattro II triple quadrupole mass spectrometer, a Waters Micromass QTOF-II instrument (ESI source), or an Applied Bio Systems 3200 Q trap with a turbo V source for TurbolonSpray.
(E)-Isopropyl 3-(3-bromophenyl)acrylate (6)
3-bromocinnamic acid (5 g, 22.02 mmol) was dissolved in 100 mL isopropyl alcohol in a round bottom flask. Several drops of concentrated H2SO4 (~100 µL) were added to the solution. The mixture was refluxed at 120 °C in an oil bath and monitored with TLC. After 24 h the reaction mixture was cooled down to ambient temperature and the solvent was evaporated. Ethyl acetate was then added to dissolve the residue and washed with NaHCO3 (aq.) and dried over Na2SO4, filtered and purified using column chromatography. A total of 4.71 g 6 was received with a yield of 79%. 1H NMR (400 MHz, CDCl3) δ 7.65 (t, J = 1.72 Hz, 1H), 7.56 (d, J = 16.00 Hz, 1H), 7.47 (m, 1H), 7.41 (d, J = 7.76 Hz, 1H), 7.23 (t, J = 7.86 Hz, 1H), 6.40 (d, J = 15.96 Hz, 1H), 5.14 (d, J = 6.28 Hz, 1H), 1.31 (d, J = 6.28 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 165.95, 142.48, 136.67, 132.86, 130.69, 130.32, 126.57, 122.99, 120.37, 68.00, 21.91 (× 2). IR (ATR, cm−1) νmax 3061, 2979, 2936, 2874, 1706, 1638, 1144, 1105.
(E)-Isopropyl 3-(3-aminophenyl)acrylate (7)
Compound 6 (4.71 g, 17.5 mmol) was dissolved in 60 mL anhydrous toluene. To it, in a stepwise manner, was added Pd2(dba)3 (0.801 g, 5%) and P(t-Bu)3 (0.142 g, 4%), and the mixture was allowed to stir for 15 min under N2 protection. To the suspension, LHMDS in toluene (19.25 mL, 1 M in toluene, 19.25 mmol) was added dropwise and the reaction mixture was allowed to stir overnight under N2 protection. An additional 2.5% Pd2(dba)3, 2% P(t-Bu)3, and 0.5 equiv LHMDS was added subsequently to the reaction mixture and stirred overnight under N2 protection. The resulting reaction mixture was then quenched using 1 N HCl very slowly over ice. The mixture was stirred for an additional 2 h and filtered through celite and diluted with dichloromethane. The organic layer was extracted and washed with saturated aqueous NaHCO3, then brine and dried over Na2SO4. The crude product was then purified using column chromatography to give 2.461 g of title compound at a yield of 69%. 1H NMR (400 MHz, CDCl3) δ 7.57 (d, J = 16.00 Hz, 1H), 7.14 (t, J = 7.80 Hz, 1H), 6.90 (d, J = 7.60 Hz, 1H), 6.80 (m, 1H), 6.68 (m, 1H), 6.35 (d, J = 16.00 Hz, 1H), 5.12 (sep, J = 6.28, 1H), 3.74 (brs, 2H), 1.29 (d, J = 6.28 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 166.63, 146.85, 144.63, 135.55, 129.73, 118.58, 118.56, 117.00, 114.12, 67.73, 21.95 (× 2). IR (ATR, cm−1) νmax 3457, 3420, 3369, 2979, 2935, 1694, 1633, 1458, 1271, 1173, 1103.
(E)-Isopropyl 3-(3-(tert-butoxycarbonylamino)phenyl)acrylate (8)
Compound 7 (2.54 g, 12.4 mmol) was added to 30 mL H2O and to it NaHCO3 (3.12 g, 24.8 mmol) was added and allowed to stir for 10 min. The solution was cooled to 5 °C and di-tert-butyl dicarbonate (4.06 g, 18.6 mmol) in 20 mL dioxane was added dropwise. The resultant solution was cooled to 0 °C for 1 h and allowed to stir at ambient temperature overnight. The aqueous solution was then washed with 50 mL of ethyl acetate and the organic layer was then extracted with saturated NaHCO3 (aq.). The aqueous layers were then combined and acidified with 10% HCl to a final pH of 1. The aqueous solution was then extracted with ethyl acetate, and the organic layer was dried over Na2SO4 and concentrated. The crude product was then purified with column chromatography and a total of 2.884 g title compound was obtained with a yield of 76%. 1H NMR (400 MHz, CDCl3) δ 7.65 (m, 1H), 7.62 (d, J = 16.08 Hz, 1H), 7.28 (m, 2H), 7.19 (m, 1H), 6.55 (brs, 1H), 6.42 (d, J = 16.00 Hz, 1H), 5.13 (sep, J = 6.28 Hz, 1H), 1.53 (s, 9H), 1.30 (d, J = 6.28 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 166.43, 152.61, 144.03, 138.97, 135.46, 129.39, 122.73, 120.05, 119.34, 117.64, 80.81, 67.79, 28.33 (× 3), 21.93 (× 2). IR (ATR, cm−1) νmax 3308, 3058, 2979, 2936, 1702, 1547, 1485, 1440, 1229, 1169, 1104.
(S)-Isopropyl 3-(benzyl((R)-1-phenylethyl)amino)-3-(3 (tertbutoxycarbonylamino)phenyl) propanoate (9)
R-(+)-N-benzyl-α-methylbenzylamine (4.6 g, 21.8 mmol) was dissolved in 30 mL anhydrous THF and stirred at 0 °C under N2 protection. To it, n-butyl-lithium (8.76 mL, 2.5 M in hexane, 21.8 mmol) was added dropwise and stirred for 30 min. During the addition, the reaction mixture went from being clear to a deep purple color. The reaction mixture was then cooled down to −78 °C and 8 (2.68 g, 8.78 mmol) in 15 mL anhydrous THF was added dropwise and allowed to stir for 2 h. Saturated NH4Cl (50 mL) was then added to the reaction mixture and it was allowed to warm up to ambient temperature over 1.5 h. Ethyl acetate was then added to the reaction mixture and extracted. The organic layer was then washed twice with 1 N HCl, dried over Na2SO4, filtered, and concentrated. MeOH was then added to the residue and then concentrated to get rid of any residual ethyl acetate. The title compound was then recrystallized from hot MeOH and a total of 1.827 g was received with a 41% yield as the first crop. 1H NMR (400 MHz, CDCl3) δ 7.41 (d, J = 7.36 Hz, 2H), 7.38–7.27 (m, 6H), 7.25–7.20 (m, 4H), 7.17 (m, 1H), 7.08 (d, J = 7.56 Hz, 1H), 6.43 (brs, 1H), 4.79 (sep, J = 6.28 Hz, 1H), 4.39 (dd, J = 5.28, 9.68 Hz, 1H), 4.00 (q, J = 6.88 Hz, 1H), 3.68 (s, 2H), 2.65–2.45 (m, 2H), 1.53 (s, 9H), 1.26 (d, J = 6.84 Hz, 3H), 1.06 (d, J = 6.24 Hz, 3H), 1.00 (d, J = 6.24 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 171.30, 152.60, 144.07, 142.84, 141.55, 138.30, 137.50, 128.73, 128.12, 128.06, 127.88, 126.82, 126.54, 122.86, 118.26, 117.30, 80.40, 67.54, 59.55, 57.09, 50.91, 46.25, 37.82, 28.37 (× 3), 21.59, 21.58, 16.27. IR (ATR, cm−1) νmax 3380, 2977, 2933, 2162, 1723, 1614, 1540, 1155. MS (ESI) m/z calcd 517.3061, found 517.113 (M + H)+.
(S)-3-(Benzyl((R)-1-phenylethyl)amino)-3-(3-(tert-butoxycarbonylamino)phenyl) propanoic acid (10)
Compound 9 (1.4 g, 2.71 mmol) was dissolved in a 2:1 mixture of MeOH/H2O (30 mL). To it LiOH (0.32 g, 13.55 mmol) was added while the reaction was stirring. The suspension was then refluxed (~85 °C) using a preheated oil bath under N2 protection overnight. The reaction mixture was allowed to cool to ambient temperature and was adjusted to pH = 1 using 10% HCl. The solution was then extracted with dichloromethane three times and the resulting organic layers were dried over Na2SO4, filtered, and concentrated. No additional purification was required and a total of 1.12 g title compound was received with 88% yield. 1H NMR (400 MHz, CDCl3) δ 7.47 (brs, 1H), 7.35–7.32 (m, 6H), 7.32–7.26 (m, 5H), 7.23 (m, 1H), 7.04 (m, 1H), 6.64 (brs, 1H), 4.42 (dd, J = 5.2, 10.28 Hz, 1H), 4.18 (q, J = 6.88 Hz, 1H), 4.00 (d, J = 13.8 Hz, 1H), 3.71 (d, J = 13.8 Hz, 1H), 2.88 (dd, J = 10.3, 16.76 Hz, 1H), 2.45 (dd, J = 5.2, 16.76 Hz, 1H), 1.54 (s, 9H), 1.28 (d, J = 6.92 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 172.06, 161.25, 152.75, 143.24, 140.60, 139.01, 128.47, 128.10 (× 2), 128.04 (× 2), 127.93 (× 2), 127.74 (× 2), 126.92, 126.61, 122.12, 118.65, 117.40, 79.32, 58.44, 57.20, 49.89, 34.60, 27.27 (× 3), 15.89. IR (ATR, cm−1) νmax 3293, 2978, 2931, 2520, 1713, 1594, 1495, 1154.
tert-Butyl (3-((1S)-1-(benzyl((R)-1-phenylethyl)amino)-3-(-3-(3-isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl)-3-oxopropyl)phenylcarbamate (12)
In a 25 mL flask, acid 10 (600 mg, 1.264 mmol) was dissolved in 6 mL anhydrous dichloromethane. To the solution N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (364 mg, 1.8965 mmol), 1-hydroxybenzotriazole hydrate (256 mg, 1.8965 mmol), triethylamine (0.54 mL, 3.793 mmol), and 4 Å molecular sieves were added and stirred under nitrogen protection at 0 °C for 0.5 h. 3-(3-Isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octane 111(314 mg, 1.5172 mmol) was then added to the reaction mixture and allowed to proceed to ambient temperature over the period of 96 h, and monitored via TLC. Once completed, the reaction mixture was filtered and the solvent was evaporated under reduced pressure. The reaction mixture was then washed once with brine. The organic layer was dried over anhydrous sodium sulfate, filtered and evaporated under reduced pressure. Column chromatography was then conducted and a total of 645 mg of the title compound as a mixture of atropisomers (with a ratio of 2:3 based on 1H NMR) was received in 74% yield. 1H NMR (400 MHz, CDCl3) δ 7.61 (brs, 0.4H), 7.49 (d, J = 7.92 Hz, 1H), 7.48 (brs, 0.6H), 7.44 (d, J = 7.56 Hz, 1H), 7.35–7.10 (m, 11H), 6.67 (s, 0.6H), 6.65 (s, 0.4H), 4.71 (m, 0.4H), 4.67–4.55 (m, 1H), 4.45 (dd, J = 4.12, 10.52 Hz, 0.6H), 4.36 (m, 1H), 4.02 (qu, J = 6.74 Hz, 1H), 3.90–3.65 (m, 3H), 2.83 (sep, J = 6.60 Hz, 1H), 2.68–2.52 (m, 1.4H), 2.48 (dd, J = 4.16, 13.92 Hz, 0.6H), 2.27 (s, 1.8H), 2.19 (brs, 0.4H), 2.12 (s, 1.2H), 2.10–2.05 (m, 1.6H), 1.89 (m, 1H), 1.80–1.60 (m, 4H), 1.53–1.45 (m, 10H), 1.37–1.25 (m, 9H); 13C NMR (100 MHz, CDCl3) δ 167.21 (166.98), 158.88 (158.76), 152.64 (152.60), 150.41 (150.56), 144.25 (144.36), 143.02 (143.38), 142.09, 138.59 (138.65), 129.03 (128.94), 128.19 (128.15), 128.09 (128.04), 127.92 (× 2), 126.79 (126.70), 126.61 (126.54), 122.92, 117.99 (118.11), 117.39 (117.30), 80.41, 61.33, 59.78, 56.43 (56.72), 53.87, 51.16 (50.77), 50.53 (50.62), 46.67, 38.46 (37.88), 37.51 (37.68), 35.75 (35.63), 28.37 (× 3), 28.26 (28.43), 26.64 (26.89), 25.83 (25.77), 21.65 (21.51), 21.57 (× 2), 13.97 (14.79), 13.10 (13.01). IR (ATR, cm−1) νmax 3247, 2973, 2932, 2185, 2050, 1716, 1632, 1529, 1436, 1158.
tert-Butyl (3-((1S)-1-amino-3-(3-(3-isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl)-3-oxopropyl)phenylcarbamate (13)
In a 250 mL hydrogenation flask, acetic acid (0.166 mL, 2.9 mmol) was added to 60 mL MeOH. To that, 1-(trifluoromethyl)-4-(4-isopropoxy-3-nitrophenyl)piperazine (1.0 g, 1.45 mmol) was added to the solution along with 10 % w/w palladium on carbon (0.2 g). The flask was placed on a hydrogenator at 60 psi H2 gas for 48 h, and monitored via TLC. Once completed, the reaction mixture was vacuum filtered through celite, and then evaporated under reduced pressure. Column chromatography was then conducted and a total of 0.66 g of the title compounds as a mixture of atropisomers (with a ratio of 1:1) was received with a final yield of 91%. 1H NMR (400 MHz, CDCl3) δ 7.55 (s, 0.5 H), 7.48 (s, 0.5H), 7.25–7.15 (m, 2H), 7.08 (m, 1H), 6.70 (brs, 1H), 4.88 (m, 1H), 4.53 (m, 2H), 4.28 (m, 1H), 2.93 (sep, J = 6.84 Hz, 1H), 2.76–2.54 (m, 2H), 2.45 (s, 1.5H), 2.36 (s, 1.5H), 2.44–1.62 (m, 10H), 1.51 (s, 9H), 1.39 (m, 6H); 13C NMR (100 MHz, CDCl3) δ 166.64 (166.60), 158.90, 153.03 (152.96), 150.96, 150.53, 139.17, 129.45, 121.16, 118.45 (118.35), 117.10 (117.06), 80.47 (80.38), 53.89 (53.83), 52.68, 52.57, 51.01, 50.97, 50.47, 50.42, 46.88 (46.78), 37.58 (37.45), 35.88 (35.74), 28.32 (× 3), 26.86, 25.80 (25.72), 21.59 (21.54), 21.49, 13.29 (13.08). IR (ATR, cm−1) νmax 3256, 2973, 2933, 2879, 2162, 1715, 1610, 1440, 1158. MS (ESI) m/z calcd 497.3235, found 497.162 (M + H)+.
tert-Butyl (3-((1S)-1-amino-3-(3-(3-isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl)propyl)phenylcarbamate (14)
Lithium aluminum hydride (191 mg, 5.035 mmol) was added to 15 mL anhydrous THF at 0 °C under N2 protection. Compound 13 (500 mg, 1.007 mmol) was dissolved in 15 mL anhydrous THF and then added dropwise to the above suspension. The resultant mixture was stirred at 0 °C for 15 min and then allowed to reach ambient temperature over a 3 h period. The reaction mixture was then cooled to 0 °C in an ice bath and quenched with the sequential addition of 0.2 mL H2O, 0.2 mL 4 N NaOH, and then 0.6 mL H2O and stirred at ambient temperature for 1 h. The suspension was filtered and the filtrate was washed with THF and diethyl ether. The organic filtrates were combined, dried over Na2SO4, filtered, and then evaporated to dryness. After column chromatography, a total of 0.38 g title compound was obtained with a yield of 79%. 1H NMR (400 MHz, CDCl3) δ 7.51 (s, 1H), 7.25 (t, J = 7.84 Hz, 1H), 7.11 (dd, J = 1.22, 7.98 Hz, 1H), 7.00 (d, J = 7.60 Hz, 1H), 6.50 (brs, 1H), 4.29 (m, 1H), 4.06 (t, J = 6.64 Hz, 1H), 3.39 (m, 2H), 2.99 (sep, J = 6.80 Hz, 1H), 2.48 (s, 3H), 2.43 (t, J = 6.94 Hz, 1H), 2.21 (m, 2H), 2.05 (m, 2H), 1.90–1.55 (m, 9H), 1.51 (s, 9H), 1.38 (d, J = 6.84 Hz, 1H). IR (ATR, cm−1) νmax 3362, 2931, 2875, 1159, 1016, 915.
4,4-Difluorocyclohexanecarboxylic acid (15)
Ethyl 4-oxycyclohexanecarboxylate (1.13 g, 6.67 mmol) was dissolved in anhydrous dichloromethane (10 mL) in a high density polyethylene (HDPE) container. To it, 4-tert-butyl-2,6-dimethylphenylsulfur trifluoride (2.5 g, 9.99 mol) was added and stirred under N2 at 0 °C. HF-pyridine (0.64 mL, 2.64 mmol) was added to the vessel and the reaction was allowed to reach ambient temperature. After 5 h, the reaction mixture was quenched with saturated aqueous NaHCO3. The organic layer was allowed to stir at ambient temperature in 2 N NaOH for 1 h. The aqueous layer was washed with dichloromethane and then acidified to pH 1 and extracted with dichloromethane. A total of 0.435 g title compound was obtained at 27% yield. 1H NMR (400 MHz, CDCl3) δ 2.498–2.278 (m, 1H), 1.984–1.881 (m, 4H), 1.872–1.753 (m, 4H).
tert-Butyl (3-((1S)-1-(4,4-difluorocyclohexanecarboxamido)-3-(3-(3-isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl)propyl)phenylcarbamate (16)
In a 10 mL flask, acid 15 (132 mg, 0.801 mmol) was dissolved in 2 mL anhydrous dichloromethane. To the solution N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (177 mg, 0.924 mmol), 1-hydroxybenzotriazole hydrate (125 mg, 0.924 mmol), triethylamine (0.26 mL, 1.848 mmol), and 4 Å molecular sieves were added and stirred under nitrogen protection at 0 °C for 0.5 h. Compound 14 (300 g, 0.616 mmol) was then added to the reaction mixture and allowed to proceed to ambient temperature over the period of 48 h, and monitored via TLC. Once completed, the reaction mixture was filtered, washed with brine, dried over Na2SO4. The solvent was evaporated under reduced pressure. Column chromatography was then conducted and a total of 0.234 g of a yellow oil title compound was received with a final yield of 60%. 1H NMR (400 MHz, CDCl3) δ 7.55 (s, 1H), 7.24 (t, J = 8.00 Hz, 1H), 7.08 (dd, J = 1.26, 8.02 Hz, 1H), 6.93 (d, J = 7.64 Hz, 1H), 6.60 (m, 2H), 5.10 (q, J = 6.80 Hz, 1H), 4.30 (sep, J = 5.68 Hz, 1H), 3.39 (m, 2H), 2.98 (sep, J = 6.84 Hz, 1H), 2.51 (s, 3H), 2.43 (t, J = 6.48 Hz, 2H), 2.30–2.10 (m, 5H), 2.06 (m, 2H), 2.03–1.88 (m, 4H), 1.88–1.55 (m, 8H), 1.51 (s, 9H), 1.39 (d, J = 6.80 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 173.24, 159.14, 152.67, 150.63, 142.98, 138.94, 129.29, 121.16, 117.41, 117.41, 116.23, 80.63, 58.79, 58.16, 53.43, 52.20, 47.66, 47.26, 42.89, 35.31, 35.12, 34.69, 33.04, 32.79, 32.55, 28.34 (× 3), 26.84, 26.01, 25.92, 25.87, 21.66 (× 2), 13.22. IR (ATR, cm−1) νmax 3257, 2968, 2936, 2875, 2162, 1980, 1717, 1656, 1527, 1444, 1367, 1237, 1159. MS (ESI) m/z calcd 629.3985, found 629.292 (M + H)+.
N-((1S)-1-(3-Aminophenyl)-3-(3-(3-isopropyl-5-methyl-4H-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl)propyl)-4,4-difluorocyclohexanecarboxamide (5)
Compound 16 (200 mg, 0.3181 mmol) was dissolved in 5 mL anhydrous dichloromethane and stirred at 0 °C. To the solution, trifluoroacetic acid (0.5 mL) was added dropwise and the solution was allowed to reach ambient temperature and stirred for 2 h. The solution was then cooled to 0 °C in an ice bath and saturated aqueous Na2CO3 was added and the aqueous layer was adjusted to pH 12 and extracted three times with dichloromethane. The combined organic layers were then washed with brine, dried over Na2SO4, filtered, and evaporated to dryness. A total of 0.215 g title compound of yellow oil was received with quantitative yield. 1H NMR (400 MHz, CDCl3) δ 7.70 (m, 1H), 7.53 (m, 1H), 7.13 (t, J = 7.80 Hz, 1H), 6.66 (d, J = 7.68 Hz, 1H), 6.38 (d, J = 6.56 Hz, 1H, exchangeable), 5.01 (qu, J = 6.96 Hz, 1H), 4.29 (m, 1H), 3.70 (brs, 2H, exchangeable), 3.39 (m, 2H), 2.98 (seq, J = 6.88 Hz, 1H), 2.50 (s, 3H), 2.43 (m, 2H), 2.30–2.02 (m, 7H), 2.02–1.72 (m, 8H), 1.72–1.52 (m, 4H), 1.38 (d, J = 6.84 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 173.15, 159.13, 150.62, 146.86, 143.04, 132.47, 122.57 (JCF 239.9 Hz), 116.26, 114.32, 113.37, 58.76, 58.23, 52.13, 47.81, 47.25, 42.95, 35.31, 35.16, 34.71, 32.83 (2JCF 24.4 Hz), 26.83, 26.00 (3JCF 9.6 Hz), 25.86, 21.66, 13.19. IR (ATR, cm−1) νmax 3318, 3224, 2957, 2932, 2873, 2177, 1724, 1651, 1520, 1456, 1345, 1106.
Benzyl 5-aminopentylcarbamate (17a)
On an ice-water bath, to the solution of cadaverine (1.022 g, 10 mmol) in dichloromethane (250 mL) was added the solution of benzyl chloroformate (853 mg, 5 mmol) in dichloromethane (50 mL) dropwise within 12 h while keeping the temperature below 5°C. The reaction mixture was then concentrated to 50 mL. Water (50 mL) was added, and the aqueous layer was adjusted to pH = 2 using 6 N HCl. The layers were separated. The aqueous layer was washed with dichloromethane (50 mL × 3), then adjusted to pH = 12 with 10 N NaOH and extracted with dichloromethane (50 mL × 3). The combined organic layers were dried over Na2SO4, concentrated and purified by flash column using dichloromethane/MeOH to give 710 mg white semi-solid in 60% yield. 1H NMR (400 MHz, CDCl3) δ 7.35 (m, 5H), 5.10 (s, 2H), 4.79 (brs, 1 H), 3.19 (m, 2H), 2.69 (m, 2H), 1.50–1.47 (m, 4H), 1.36 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 156.49, 136.74, 128.45 (× 2), 128.00 (× 2), 66.51, 41.73, 40.95, 32.77, 29.74, 23.92. IR (ATR, cm−1) νmax 3328, 2922, 2852, 1686, 1537, 1266, 1248, 697.
Benzyl 7-aminononylcarbamate (17b)
Previously reported.23
Benzyl 9-aminononylcarbamate (17c)
The title compound was prepared in a similar way as 17a in 62% yield. 1H NMR (400 MHz, CDCl3) δ 7.34 (m, 5H), 5.09 (s, 2H), 4.82 (brs, 1H), 3.18 (q, J = 6.16 Hz, 2H), 2.67 (m, 2H), 1.49 (m, 2H), 1.42 (m, 2H), 1.29 (m, 10H); 13C NMR (100 MHz, CDCl3) δ 156.41, 136.76, 128.47 (× 3), 128.02 (× 2), 66.54, 42.24, 41.11, 33.84, 29.96, 29.46, 29.37, 29.17, 26.83, 26.69. mp 41–43 °C. IR (ATR, cm−1) νmax 3342, 2922, 2850, 1683, 1527, 1255, 1235, 1023, 726, 694.
[(9-Benzyloxycarbonylamino-pentylcarbamoyl)-methoxy]acetic acid (18a)
To the solution of 17a (520 mg, 2.20 mmol) in THF (20 mL) was added diglycolic anhydride (268 mg, 2.31 mmol) in one portion. The resultant mixture was stirred at ambient temperature for 12 h. After removing THF under reduced pressure, the residue was crystallized with EtOAc/hexane to give 493 mg white solid as first crop, in 64% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.74 (brs, 1H), 7.79 (m, 1H), 7.38–7.30 (m, 5 H), 7.18 (m, 1H), 5.00 (s, 2H), 4.09 (s, 2H), 3.94 (s, 2H), 3.08 (q, J = 6.39 Hz, 2H), 2.97 (q, J = 6.31 Hz, 2H), 1.42–1.38 (m, 4H), 1.23 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 171.37, 168.54, 156.06, 137.30, 128.29 (× 3), 127.67 (× 2), 70.21, 67.93, 65.06, 38.05, 29.02, 28.74 (× 2), 23.57. mp 71–71.5 °C. IR (ATR, cm−1) νmax 3358, 3311, 2930, 1686, 1552, 1266, 1216, 1131, 692.
3,13-dioxo-1-phenyl-2,15-dioxa-4,12-diazaheptadecan-17-oic acid (18b)
Previously reported.23
3,15-Dioxo-1-phenyl-2,17-dioxa-4,14-diazanonadecan-19-oic acid (18c)
The title compound was prepared in a similar way as 18a in 73% yield. 1H NMR (400 MHz, DMSO-d6) δ 7.86 (m, 0.5H), 7.73 (m, 0.5H), 7.38–7.28 (m, 5 H), 7.18 (m, 1H), 5.00 (s, 2H), 4.20 (s, 1H), 4.08 (s, 1H), 3.94 (s, 1H), 3.93 (s, 1H), 3.66 (s, 1H), 3.08 (q, J = 6.60 Hz, 2H), 2.97 (q, J = 6.66 Hz, 2H), 1.39 (m, 4H), 1.24 (m, 10H); 13C NMR (100 MHz, DMSO-d6) δ 171.40, 168.56, 168.20, 156.05, 137.33, 128.28, 127.66, 127.62, 70.28, 70.14, 68.11, 67.75, 65.03, 51.45, 38.10, 29.04, 28.86, 28.61, 26.30, 26.16. mp 37–38 °C. IR (ATR, cm−1) νmax 3338, 2924, 1682, 1645, 1528, 1235, 1138, 696.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(3′,11′-Dioxo-1′-phenyl-2′,13′-dioxa-4′,10′-diazapentadecanamido)morphinan (19a)
The title compound was prepared according to the general amide coupling procedure by reacting acid 18a with 6β-naltrexamine hydrochloride14·2HCl in DMF overnight. The crude product was purified with chromatography using CH2Cl2/MeOH as eluent to give 667 mg white solid, in 70% yield. 1H NMR (400 MHz, CD3OD) δ 7.30–7.25 (m, 5H), 6.65 (d, J = 8.12 Hz, 1H), 6.59 (d, J = 8.16 Hz, 1H), 5.06 (s, 2H), 4.54 (d, J = 7.56 Hz, 1H), 4.05 (s, 2H), 4.04 (s, 2H), 3.79–3.72 (m, 1H), 3.28–3.24 (m, 3H), 3.14–3.09 (m, 3H), 2.73–2.69 (m, 2H), 2.54–2.43 (m, 2H), 2.33–2.21 (m, 2H), 1.92 (m, 1H), 1.60–1.33 (m, 10H), 0.91 (m, 1H), 0.56 (m, 2H), 0.20 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 171.62, 171.48, 158.95, 143.82, 142.02, 138.52, 132.28, 129.46, 128.94, 128.74, 124.97, 120.21, 118.80, 92.84, 71.68, 71.62, 67.34, 63.94, 60.12, 52.47, 45.71, 41.70, 40.00, 31.60, 31.20, 30.53, 30.09, 25.31, 25.10, 23.73, 9.86, 4.68, 4.10. mp 80–83 °C. IR (ATR, cm−1) νmax 3296, 2934, 1655, 1538, 1242, 1128, 1035, 697.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(3′,13′-dioxo-1′-phenyl-2′,15′-dioxa-4′,12′-diazaheptadecanamido)morphinan (19b)
Previously reported.23
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(3′,15′-dioxo-1′-phenyl-2′,17′-dioxa-4′,14′-diazanonadecanamido)morphinan (19c)
The title compound was prepared in a similar way as 19a in 63% yield. 1H NMR (400 MHz, CD3OD) δ 7.33–7.23 (m, 5H), 6.65 (d, J = 8.12 Hz, 1H), 6.58 (d, J = 8.12 Hz, 1H), 5.05 (s, 2H), 4.54 (d, J = 7.52 Hz, 1H), 4.06 (s, 2H), 4.05 (s, 2H), 3.76 (m, 1H), 3.26 (t, J = 7.14 Hz, 2H), 3.20 (m, 1H), 3.13–3.07 (m, 3H), 2.73–2.66 (m, 2H), 2.52 (m, 1H), 2.43 (m, 1H), 2.32–2.18 (m, 2H), 1.91 (m, 1H), 1.62–1.41 (m, 8H), 1.31 (m, 10H), 0.90 (m, 1H), 0.60–0.51 (m, 2H), 0.23–0.17 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 171.54, 171.45, 158.91, 143.82, 142.00, 138.55, 132.30, 129.47 (× 2), 128.94, 128.75 (× 2), 125.07, 120.22, 118.80, 92.87, 71.68 (× 2), 71.62, 67.31, 63.91, 60.14, 52.45, 48.75, 45.66, 41.88, 40.16, 31.68, 31.21, 30.91, 30.55, 30.48, 30.32 (× 2), 27.99, 27.79, 25.33, 23.73, 9.89, 4.70, 4.17. mp 70–72 °C. IR (ATR, cm−1) νmax 3304, 2929, 2857, 1652, 1557, 1453, 1247, 1128, 1034, 742.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(2’-(2’-(5’-aminopentylamino)-2’-oxoethoxy)acetamido)morphinan (20a)
A solution of 19a (494 mg, 0.73 mmol) in methanol (20 mL) was hydrogenated in the presence of 10% Pd/C (50 mg) under a H2 atmosphere (60 psi) at room temperature for 6 h. The mixture was filtered, and the filtrate was concentrated and crystallized with MeOH/Et2O to give 20a as white solid (352 mg, 89% yield). 1H NMR (400 MHz, CD3OD) δ 6.63 (d, J = 8.12 Hz, 1H), 6.57 (d, J = 8.16 Hz, 1H), 4.51 (d, J = 7.64 Hz, 1H), 4.07 (s, 2H), 4.06 (s, 2H), 3.75 (m, 1H), 3.29 (m, 2H), 3.12 (d, J = 5.76 Hz, 1H), 3.08 (d, J = 18.64 Hz, 1H), 2.78 (t, J = 7.34 Hz, 2H), 2.70 –2.61 (m, 2H), 2.48–2.36 (m, 2H), 2.29–2.21 (m, 1H), 2.19–2.10 (m, 1H), 1.91 (m, 1H), 1.63–1.37 (m, 10H), 0.88 (m, 1H), 0.54 (m, 2H), 0.16 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 171.75, 171.47, 143.79, 142.08, 132.50, 125.41, 120.12, 118.70, 92.98, 71.74, 71.62, 63.82, 60.30, 52.57, 49.84, 45.30, 41.60, 39.80, 31.98, 31.27, 31.06, 30.10, 25.48, 24.98, 23.59, 10.28, 4.44, 4.19. mp 145–148 °C. IR (ATR, cm−1) νmax 3275, 2929, 1652, 1552, 1323, 1130, 1036.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-2′-[2′′-(7′′-aminoheptylamino)-2′-oxoethoxy]acetamidomorphinan (20b)
Previously reported.23
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-2′-[2′′-(9′′-aminononylamino)-2′-oxoethoxy]acetamidomorphinan (20c)
The title compound was prepared in a similar way as 20a in 64% yield (first crop). 1H NMR (400 MHz, CD3OD) δ 6.63 (d, J = 8.12 Hz, 1H), 6.56 (d, J = 8.16 Hz, 1H), 4.51 (d, J = 7.56 Hz, 1H), 4.06 (s, 2H), 4.05 (s, 2H), 3.76 (m, 1H), 3.26 (t, J = 7.12 Hz, 2H), 3.12–3.05 (m, 2H), 2.72–2.60 (m, 4H), 2.40 (m, 2H), 2.25 (m, 1H), 2.15 (m, 1H), 1.90 (m, 1H), 1.59–1.43 (m, 8H), 1.30 (m, 10H), 0.87 (m, 1H), 0.54–0.50 (m, 2H), 0.18–0.14 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 171.57, 171.43, 143.85, 142.17, 132.48, 125.34, 120.10, 118.74, 92.96, 71.74, 71.62, 71.57, 63.82, 60.33, 52.50, 48.93, 45.28, 42.00, 40.10, 32.14, 32.06, 31.25, 30.48, 30.45, 30.36, 30.27, 27.94, 27.79, 25.46, 23.58, 10.32, 4.46, 4.23. mp 125–128 °C. IR (ATR, cm−1) νmax 3274, 2928, 1652, 1557, 1323, 1129, 1035, 743.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(1′-carboxy-4′,12′-dioxo-2′,14′-dioxa-5′,11′-diazahexaadecanamido)morphinan (21a)
To the solution of 20a (311 mg, 0.61 mmol) in THF (2 mL) was added diglycolic anhydride (71 mg, 0.61 mmol) in one portion. The resultant mixture was stirred at ambient temperature for overnight. After removal of THF under reduced pressure, the residue was crystallized by MeOH/Et2O to give 381 mg light yellow solid, in 95% yield. 1H NMR (400 MHz, DMSO-d6) δ 8.28 (d, J = 8.44 Hz, 1H, exchangeable), 8.20 (m, 1H, exchangeable), 8.07 (t, J = 5.80 Hz, 1H, exchangeable), 6.62 (d, J = 8.04 Hz, 1H), 6.55 (d, J = 8.12 Hz, 1H), 4.64 (d, J = 7.76 Hz, 1H), 4.03 (s, 2H), 3.95 (s, 2H), 3.935 (s, 2H), 3.928 (s, 2H), 3.51 (m, 1H), 3.26 (m, 2H), 3.17–3.04 (m, 5H), 2.75–2.61 (m, 3H), 2.24 (m, 1H), 2.11 (m, 1H), 1.81 (m, 1H), 1.54–1.41 (m, 6H), 1.34–1.20 (m, 4H), 0.90 (m, 1H), 0.51 (m, 2H), 0.20 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 171.80, 168.85, 168.47, 168.28, 142.11, 140.68, 130.84, 122.64, 118.58, 117.30, 90.31, 70.59, 70.44, 69.58, 68.87, 61.82, 57.97, 50.56, 48.56, 46.63, 44.37, 38.09, 38.06, 29.91, 29.50, 28.84, 28.69, 24.32, 23.75, 22.43, 8.24, 3.97, 3.33. mp 156–157 °C. IR (ATR, cm−1) νmax 3395, 2935, 1655, 1559, 1123, 1035.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(1′-carboxy-4′,14′-dioxo-2’,16’-dioxa-5′,13′-diazaicosanamido)morphinan (21b)
Previously reported.23
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(1′-carboxy-4′,16′-dioxo-2’,18’-dioxa-5′,15′-diazaicosanamido)morphinan (21c)
The title compound was prepared in a similar way as 21a in 99% yield. 1H NMR (400 MHz, DMSO-d6) δ 9.14 (brs, 1H, exchangeable), 8.26 (d, J = 8.36 Hz, 1H, exchangeable), 8.10 (m, 1H, exchangeable), 8.04 (t, J = 5.78 Hz, 1H, exchangeable), 6.61 (d, J = 8.08 Hz, 1H), 6.55 (d, J = 8.08 Hz, 1H), 4.63 (d, J = 7.76 Hz, 1H), 4.03 (s, 2H), 3.94 (s, 2H), 3.93 (m, 4H), 3.52 (m, 1H), 3.21 (m, 2H), 3.18–3.00 (m, 6H), 2.73–2.64 (m, 2H), 2.22 (m, 1H), 2.09 (m, 1H), 1.81 (m, 1H), 1.52–1.30 (m, 6H), 1.25 (m, 12H), 0.90 (m, 1H), 0.50 (m, 2H), 0.19 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 171.72, 168.78, 168.39, 168.25, 142.09, 140.65, 130.89, 122.77, 118.56, 117.25, 90.33, 70.42, 70.38, 69.58, 68.72, 61.79, 58.03, 56.00, 50.57, 46.69, 44.27, 38.15, 38.11, 29.95, 29.60, 29.17, 29.01, 28.84, 28.62, 26.34, 26.29, 24.35, 22.40, 18.52, 8.39, 3.92, 3.35. mp 150–151 °C. IR (ATR, cm−1) νmax 3236, 2929, 1645, 1604, 1545, 1318, 1127.
Bivalent ligand 1a
The title compound was prepared following the general procedure by reacting 4-aminomaraviroc1 (60 mg, 0.1135 mmol) with 21a (112 mg, 0.170 mmol) in 49% yield. Hydrochloride salt: 1H NMR (400 MHz, CD3OD) δ 7.64 (d, J = 8.52 Hz, 2H), 7.42 (d, J = 8.52 Hz, 2H), 6.76 (d, J = 8.2 Hz, 1H), 6.72 (d, J = 8.28 Hz, 1H), 4.98 (m, 1H), 4.69 (m, 2H), 4.32 (m, 1H), 4.24 (m, 3H), 4.16 (s, 2H), 4.10 (m, 4H), 3.94 (d, J = 5.72 Hz, 1H), 3.71 (m, 2H), 3.40–3.32 (m, 3H), 3.25–3.00 (m, 9H), 2.95–2.55 (m, 8H), 2.52–2.38 (m, 4H), 2.36–2.15 (m, 4H), 2.15–1.73 (m, 9H), 1.68–1.49 (m, 7H), 1.47–1.30 (m, 7H), 1.14–1.08 (m, 2H), 0.82 (m, 1H), 0.74 (m, 1H), 0.55–0.49 (m, 2H), 0.29 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 177.86, 171.87, 171.76, 171.74, 170.32, 143.78, 143.10, 138.59, 138.49, 130.77, 128.40 (× 2), 122.02 (× 2), 121.90, 121.01, 119.68, 91.90, 71.84, 71.63, 71.45, 71.39 (× 2), 64.84, 64.32, 63.58, 62.69, 58.82, 52.44, 51.73, 50.39, 47.62, 43.46, 39.98, 39.93, 35.01, 33.82 (2JCF 24.3 Hz, × 2), 32.00, 31.14, 29.88 (× 2), 28.91, 27.22, 27.16, 27.12, 26.67, 25.12, 24.87, 24.68, 24.49, 21.80 (3JCF 4.92 Hz, × 2), 15.43, 12.19, 6.88, 6.18, 3.47. IR (ATR, cm−1) νmax 3251, 2941, 1645, 1452, 1127, 1109, 1035. mp 214–217 °C. HRMS (ESI) m/z calcd 1169.6569, found 1169.6567 (M + H)+, 585.3308 (M + 2H)2+.
Bivalent ligand 1b
Previously reported.23
Bivalent ligand 1c
The title compound was prepared following the general procedure by reacting 4-aminomaraviroc (40 mg, 0.0666 mmol) with 21c (71 mg, 0.0999 mmol) in 81% yield. Free base: 1H NMR (400 MHz, CD3OD) δ 7.64 (d, J = 8.52 Hz, 2H), 7.40 (d, J = 8.56 Hz, 2H), 6.74 (AB, J = 8.24 Hz, 2H), 4.99 (m, 1H), 4.69 (d, J = 7.92 Hz, 1H), 4.57 (m, 1H), 4.22 (s, 2H), 4.14 (s, 2H), 4.07 (m, 4H), 3.94 (d, J = 5.72 Hz, 1H), 3.71 (m, 1H), 3.39–3.34 (m, 3H), 3.28–3.23 (m, 4H), 3.21–3.08 (m, 4H), 2.88 (s, 2H), 2.80–2.50 (m, 7H), 2.50–2.28 (m, 5H), 2.28–2.05 (m, 6H), 2.05–1.71 (m, 9H), 1.70–1.45 (m, 8H), 1.42–1.29 (m, 15H), 1.12 (m, 1H), 0.82 (m, 1H), 0.74 (m, 1H), 0.51 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 171.72 (× 2), 171.60, 170.25, 161.64, 160.48, 143.81, 143.11, 138.59, 130.78, 128.40 (× 2), 122.05 (× 2), 121.91, 120.99, 119.73, 91.94, 71.90, 71.70, 71.56, 71.48, 71.42 (× 2), 64.42, 58.88, 56.54, 52.44, 51.78, 47.64, 43.48, 43.39, 40.11, 40.10, 37.29, 35.95, 35.49, 33.82 (2JCF 24.4 Hz, × 2), 31.15, 30.35 (× 2), 30.32, 30.12 (× 2), 28.96, 27.82, 27.80, 27.19, 27.10, 26.64, 25.32, 24.65, 24.48, 22.24 (3JCF 6.11 Hz, × 2), 15.66, 12.36, 6.90, 6.13, 3.48. IR (ATR, cm−1) νmax 3246, 2933, 1648, 1541, 1127, 1110, 1034. Hydrochloride salt: mp 209–212 °C. HRMS (ESI) m/z calcd 1225.7195, found 1225.7203 (M + H)+, 613.3623 (M + 2H)2+.
Bivalent ligand 1d
The title compound was prepared following the general procedure by reacting 3-aminomaraviroc 5 (56 mg, 0.1059 mmol) with 21b (73 mg, 0.1059 mmol) in 26% yield. Free base: 1H NMR (400 MHz, DMSO-d6) δ 10.00 (s, 1H), 9.03 (s, 1H), 8.27 (d, J = 8.68 Hz, 1H), 8.21 (d, J = 8.32 Hz, 1H), 8.08 (t, J = 5.82 Hz, 1H), 8.02 (t, J = 5.72 Hz, 1H), 7.61 (s, 1H), 7.44 (d, J = 9.04 Hz, 1H), 7.27 (t, J = 7.88 Hz, 1H), 7.03 (d, J = 7.52 Hz, 1H), 6.58 (d, J = 7.88 Hz, 1H), 6.52 (d, J = 8.64 Hz, 1H), 4.93 (q, J = 7.44 Hz, 1H), 4.88 (s, 1H), 4.59 (d, J = 8.08 Hz, 1H), 4.22 (m, 1H), 4.13 (s, 2H), 4.03 (s, 2H), 3.94 (s, 2H), 3.93 (s, 2H), 3.50 (m, 1H), 3.30–3.20 (m, 2H), 3.20–3.08 (m, 6H), 3.00 (m, 1H), 2.65–2.60 (m, 3H), 2.40–2.25 (m, 10H), 2.10–1.95 (m, 3H), 1.93 (m, 1H), 1.85–1.70 (m, 7H), 1.70–1.50 (m, 7H), 1.43 (m, 7H), 1.30–1.20 (m, 12H), 0.83 (1H), 0.46 (m, 2H), 0.10 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 173.88, 169.11, 168.86, 168.62, 167.75, 159.16, 150.79, 143.11, 137.87, 130.68, 129.23, 122.82, 119.31, 119.19, 118.24, 118.09, 91.65, 71.63, 71.38, 70.95, 70.07, 62.34, 59.26, 58.90, 58.46, 58.33, 51.89, 50.31, 47.84, 47.25, 42.66, 38.99, 38.77, 35.14, 35.00, 32.82 (2JCF 24.4 Hz, × 2), 29.31, 28.99, 28.25, 26.66, 26.28, 26.00, 25.92, 25.79, 23.53, 22.69, 21.60, 13.04, 9.26, 4.08, 3.74. IR (ATR, cm−1) νmax 3271, 3078, 2931, 2858, 2162, 2036, 1980, 1655, 1537, 1448, 1323, 1252, 1107, 1035. HRMS (ESI) m/z calcd 1197.6882, found 599.3652 (M + 2H)2+.
Monovalent ligand 2a
The title compound was prepared following the general procedure by reacting methylcarbamoylmethoxy-acetic acid 221 (24 mg, 0.163 mmol) with 20a (86 mg, 0.158 mmol) in 30% yield. Free base: 1H NMR (400 MHz, CD3OD) δ 6.75 (m, 2H), 4.70 (d, J = 7.68 Hz, 1H), 4.080 (s, 2H), 4.076 (s, 2H), 4.04 (s, 4H), 3.72 (m, 1H), 3.28–3.20 (m, 4H), 3.12 (m, 2H), 2.88 (m, 2H), 2.80 (m, 4H), 2.75–2.55 (m, 2H), 2.02 (m, 1H), 1.76 (d, J = 13.04 Hz, 1H), 1.70–1.50 (m, 8H), 1.39 (m, 2H), 1.12 (m, 1H), 0.84 (m, 1H), 0.75 (m, 1H), 0.52 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 171.75, 143.80, 143.15, 130.75, 121.85, 121.02, 119.72, 91.89, 71.64, 71.50, 71.39, 64.40, 58.87, 52.41, 47.62, 39.97, 39.92, 31.18, 30.05, 28.94, 25.95, 25.21, 24.67, 24.55, 6.90, 6.25, 3.48. IR (ATR, cm−1) νmax 3274, 2934, 1645, 1549, 1323, 1125, 1034, 857. mp 115–118 °C. HRMS (ESI) m/z calcd 672.3603, found 672.3627 (M + H)+.
Monovalent ligand 2b
Previously reported.23
Monovalent ligand 2c
The title compound was prepared following the general procedure by reacting methylcarbamoylmethoxy-acetic acid 22 (23 mg, 0.156 mmol) with 20c (91 mg, 0.152 mmol) in 25% yield. Free base: 1H NMR (400 MHz, CD3OD) δ 6.65 (d, J = 8.12 Hz, 1H), 6.60 (d, J = 8.16 Hz, 1H), 4.55 (d, J = 7.64 Hz, 1H), 4.07 (s, 2H), 4.06 (s, 2H), 4.03 (s, 4H), 3.75 (m, 1H), 3.28–3.22 (m, 4H), 3.20–3.08 (m, 2H), 2.82–2.64 (m, 5H), 2.49 (m, 2H), 2.29 (m, 2H), 1.94 (m, 1H), 1.65–1.42 (m, 8H), 1.40–1.25 (m, 10H), 0.91 (m, 1H), 0.57 (m, 2H), 0.23 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 172.17, 171.57, 171.51, 171.49, 143.75, 142.15, 132.06, 124.89, 120.27, 118.78, 92.73, 71.66, 71.58, 71.56, 71.53, 71.50, 71.44, 71.40, 63.89, 60.04, 60.01, 52.49, 40.11, 40.08, 31.23, 30.48, 30.44, 30.42, 30.26, 27.96, 27.92, 27.90, 25.89, 25.30, 23.74, 9.70, 4.80, 4.06. IR (ATR, cm−1) νmax 3273, 2926, 1651, 1549, 1323, 1126, 1035, 747. mp 112–115 °C. HRMS (ESI) m/z calcd 728.4229, found 728.4217 (M + H)+.
Benzyl 5-(2-(2-(methylamino)-2-oxoethoxy)acetamido)pentylcarbamate (23a)
The title compound was prepared according to the general amide coupling procedure by reacting acid 22 (474 mg, 3.225 mmol) with amine 17a (508 mg, 2.15 mmol) in 37% yield. 1H NMR (400 MHz, CD3OD) δ7.34–7.27 (m, 5 H), 5.06 (s, 2H), 4.01 (s, 4H), 3.24 (m, 2H), 3.11 (t, J = 6.96 Hz, 2H), 2.79 (s, 3H), 1.58–1.48 (m, 4H), 1.34 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 172.14, 171.52, 158.94, 138.53, 129.48, 128.96, 128.76, 71.54, 71.51, 67.34, 41.69, 39.98, 30.55, 30.09, 25.90, 25.10. mp 85–86.5 °C. IR (ATR, cm−1) νmax 3308, 2936, 1655, 1250, 1124, 732, 697.
Benzyl (7-(2-(2-(methylamino)-2-oxoethoxy)acetamido)heptyl)carbamate (23b)
The title compound was prepared according to the general amide coupling procedure by reacting acid 22 (179 mg, 1.22 mmol) with amine 17b (268 mg, 1.01 mmol) in 67% yield. 1H NMR (400 MHz, CDCl3) δ 7.40–7.28 (m, 5H), 6.57–6.35 (m, 2H), 5.09 (s, 2H), 4.80 (brs, 1H), 4.04 (s, 2H), 4.03 (s, 2H), 3.29 (q, J = 6.80 Hz, 2H), 3.18 (q, J = 6.66 Hz, 2H), 2.87 (d, J = 4.92 Hz, 3H), 1.51 (m, 4H), 1.32 (m, 6H). IR (ATR, cm−1) νmax 3312, 3093, 2935, 1665, 1361, 1278, 1264, 1239, 1081, 984, 953.
Benzyl 9-(2-(2-(methylamino)-2-oxoethoxy)acetamido)nonylcarbamate (23c)
The title compound was prepared according to the general amide coupling procedure by reacting acid 22 (327 mg, 2.22 mmol) with amine 17c (432 mg, 1.48 mmol) in 58% yield. 1H NMR (400 MHz, CD3OD) δ7.33–7.29 (m, 5 H), 5.06 (s, 2H), 4.02 (s, 4H), 3.25–3.22 (m, 2H), 3.10 (t, J = 7.02 Hz, 2H), 2.79 (s, 3H), 1.54–1.46 (m, 4H), 1.31 (m, 10H); 13C NMR (100 MHz, CD3OD) δ 172.13, 171.45, 158.92, 138.55, 129.48, 128.95, 128.76, 71.55, 71.51, 67.32, 41.88, 40.26, 40.14, 30.92, 30.54, 30.47, 30.31, 27.97, 27.80, 25.93. mp 98–100 °C. IR (ATR, cm−1) νmax 3323, 2924, 2854, 2473, 1683, 1640, 1529, 1236, 1123, 722.
N-(5-aminopentyl)-2-(2-(methylamino)-2-oxoethoxy)acetamide (24a)
The title compound was prepared in a similar way as 20a. The obtained product was used for next step without further purification. 1H NMR (400 MHz, CD3OD) δ4.05 (s, 4H), 3.26 (t, J = 7.08 Hz, 2H), 2.79 (s, 3H), 2.72 (t, J = 7.28 Hz, 2H), 1.60–1.51 (m, 4H), 1.38 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 172.20, 171.64, 71.44, 71.38, 41.81, 39.88, 31.66, 30.15, 25.89, 25.06. IR (ATR, cm−1) νmax 3293, 2937, 1651, 1564, 1125, 746.
N-(7-aminoheptyl)-2-(2-(methylamino)-2-oxoethoxy)acetamide (24b)
The title compound was prepared in a similar way as 20a. The obtained product was used for next step without further purification.
N-(9-aminononyl)-2-(2-(methylamino)-2-oxoethoxy)acetamide (24c)
The title compound was prepared in a similar way as 20a. The obtained product was further recrystallized with MeOH/EtOAc to give 74 mg white solid in 30% yield. 1H NMR (400 MHz, CD3OD) δ4.02 (s, 4H), 3.24 (t, J = 7.18 Hz, 2H), 2.79 (s, 3H), 2.77 (m, 2H), 1.54 (m, 4H), 1.34 (m, 10H); 13C NMR (100 MHz, CD3OD) δ 172.15, 171.49, 71.53, 71.48, 41.53, 40.06, 30.95, 30.73, 30.43, 30.25, 30.23, 27.91, 27.64, 25.89. mp 90–94 °C. IR (ATR, cm−1) νmax 3310, 2923, 2852, 1652, 1546, 1125.
17-Methylamino-5,13,17-trioxo-3,15-dioxa-6,12-diazaheptadecan-1-oic acid (25a)
The title compound was prepared in a similar way as 18a. The obtained product was used for next step without further purification. 1H NMR (400 MHz, CD3OD) δ4.18 (s, 2H), 4.04 (s, 2H), 4.03 (s, 4H), 3.26 (m, 4H), 2.79 (s, 3H), 1.61–1.54 (m, 4H), 1.38 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 173.77, 172.18, 171.97, 171.55, 71.59, 71.52, 71.50, 69.32, 39.95, 39.84, 30.05, 30.03, 25.91, 25.19.
19-Methylamino-5,15,19-trioxo-3,17-dioxa-6,14-diazanonadecan-1-oic acid (25b)
The title compound was prepared in a similar way as 18a. The obtained product was used for next step without further purification. 1H NMR (400 MHz, DMSO-d6) δ7.93 (m, 1H), 7.48 (t, J = 7.52 Hz, 1H), 7.37 (t, J = 7.58 Hz, 1H), 4.20 (s, 2H), 3.94 (s, 2H), 3.91 (s, 4H), 2.65 (d, J = 4.48 Hz, 1H), 1.41 (m, 4H), 1.26 (m, 6H). IR (ATR, cm−1) νmax 3307, 3091, 2930, 2857, 2532, 1736, 1633, 1552, 1437, 1220, 1130, 1048.
21-Methylamino-5,17,21-trioxo-3,19-dioxa-6,16-diazahenicosan-1-oic acid (25c)
The title compound was prepared in a similar way as 18a. The obtained product was further recrystallized with MeOH/EtOAc to give 46 mg white solid in 44% yield. 1H NMR (400 MHz, CD3OD) δ4.14 (m, 2H), 4.10 (m, 1H), 4.04–4.03 (m, 5H), 3.24 (m, 4H), 2.79 (s, 3H), 1.54 (m, 4H), 1.34 (m, 10H); 13C NMR (100 MHz, CD3OD) δ 174.27, 172.18, 171.99, 171.49, 71.50 (× 2), 71.47, 69.72, 40.10, 40.02, 30.48, 30.43, 30.35, 30.26 (× 2), 27.92, 27.89, 25.86. mp 67–70 °C. IR (ATR, cm−1) νmax 3244, 2927, 1640, 1549, 1156, 681.
Monovalent ligand 3a
The title compound was prepared following the general procedure by reacting acid 25a (40 mg, 0.0832 mmol) with 4-aminomaraviroc (50 mg, 0.0832 mmol) in 66% yield. Hydrochloride salt: 1H NMR (400 MHz, CD3OD) δ7.64 (d, J = 7.84 Hz, 2H), 7.42 (d, J = 8.00 Hz, 2H), 4.98 (m, 1H), 4.34 (m, 1H), 4.24–4.21 (m, 3H), 4.13 (s, 2H), 4.03 (s, 4H), 3.80 (m, 1H), 3.28–3.22 (m, 6H), 2.88 (m, 4H), 2.79 (s, 3H), 2.70 (m, 1H), 2.55–2.34 (m, 7H), 2.34–2.18 (m, 3H), 2.18–2.03 (m, 2H), 2.03–1.65 (m, 6H), 1.65–1.50 (m, 4H), 1.50–1.30 (m, 8H); 13C NMR (100 MHz, CD3OD) δ 162.43, 154.33, 138.66, 138.43, 128.42, 122.08, 98.08, 72.01, 71.79, 71.51, 71.48, 63.43, 62.80, 56.57, 51.74, 43.50, 43.44, 39.98, 39.93, 37.38, 36.00, 34.84, 34.78, 33.96 (2JCF 24.8 Hz, × 2), 31.99, 30.06, 30.02, 27.19, 27.10, 27.05, 26.75, 25.93, 25.22, 24.92, 21.62 (3JCF 3.49 Hz, × 2), 15.65. IR (ATR, cm−1) νmax 3270, 1644, 1549, 1402, 1126, 1108. mp 157–159 °C. HRMS (ESI) m/z calcd 858.5048, found 858.5054 (M + H)+.
Monovalent ligand 3b
Previously reported.23
Monovalent ligand 3c
The title compound was prepared following the general procedure by reacting acid 25c (32 mg, 0.0799 mmol) with 4-aminomaraviroc (40 mg, 0.0666 mmol) in 62% yield. Hydrochloride salt: 1H NMR (400 MHz, CD3OD) δ7.64 (d, J = 8.12 Hz, 2H), 7.42 (d, J = 8.28 Hz, 2H), 4.98 (m, 1H), 4.34 (m, 1H), 4.24 (m, 3H), 4.15 (s, 2H), 4.061 (s, 2H), 4.058 (s, 2H), 3.79 (m, 1H), 3.27–3.22 (m, 6H), 3.17–3.11 (m, 2H), 2.88 (m, 4H), 2.80 (s, 3H), 2.45 (m, 7H), 2.28 (m, 2H), 2.09 (m, 2H), 1.92–1.74 (m, 6H), 1.55 (m, 4H), 1.44 (d, J = 6.48 Hz, 6H), 1.33 (m, 10H); 13C NMR (100 MHz, CD3OD) δ 171.76, 171.60, 170.32, 162.42, 161.77, 154.23, 138.57, 138.52, 128.41, 122.05, 71.83, 71.60, 71.34, 71.31, 63.43, 62.80, 56.57, 51.74, 50.49, 50.01, 43.49, 43.42, 40.11, 40.07, 37.36, 35.98, 34.82, 34.77, 33.83 (2JCF 24.3 Hz, × 2), 31.97, 30.30, 30.08, 30.06, 27.77, 27.06, 26.74, 25.96, 24.91, 21.60 (3JCF 3.3 Hz, × 2), 15.65, 12.13. IR (ATR, cm−1) νmax 3244, 2943, 1647, 1540, 1407, 1132, 1110, 1038. mp 204–207 °C. HRMS (ESI) m/z calcd 914.5674, found 914.5600 (M + H)+.
Monovalent ligand 3d
The title compound was prepared following the general procedure by reacting acid 25b (40 mg, 0.1065 mmol) with 3-aminomaraviroc 5 (70 mg, 0.1165 mmol) in 33% yield. 1H NMR (400 MHz, MeOD) δ 7.79 (s, 1H), 7.48 (d, J = 8.04 Hz, 1H), 7.38 (t, J = 7.86 Hz, 1H),7.23 (d, J = 7.44 Hz, 1H), 5.00 (m, 1H), 4.71 (m, 1H), 4.34 (m, 1H), 4.23 (m, 3H), 4.14 (s, 2H), 4.03 (s, 4H), 3.68 (seq, J = 6.66 Hz, 1H), 3.28–3.20 (m, 5H), 3.13 (m, 1H), 2.80 (m, 8H), 2.55–2.20 (m, 9H), 2.10 (m, 2H), 1.96 (m, 1H), 1.95–1.67 (m, 5H), 1.56 (m, 4H), 1.42 (d, J = 6.56 Hz, 6H), 1.37 (m, 6H); 13C NMR (400 MHz, MeOD) δ171.96, 168.83, 165.97, 158.67, 139.58, 130.51, 121.41, 120.36, 71.98, 71.70, 71.43, 66.89, 63.68, 43.49, 40.07, 40.01, 36.45, 34.94, 33.87, 30.32, 29.92, 27.80, 26.70, 21.78, 21.72, 15.43. IR (ATR, cm−1) νmax 3256, 3055, 2933, 2857, 2531, 2162, 1652, 1545, 1445, 1108. HRMS (ESI) m/z calcd 886.5361, found 886.5404 (M + H)+, 908.5211 (M + Na)+.
Radioligand binding assay
MOR radioligand binding
MOR–CHO cell culture and membrane homogenate preparation followed the literature report.50 Saturation binding was performed by incubating membranes for 90 min at 30 °C with 0.5–15 nM [3H] naloxone in assay buffer in a 0.5 mL volume. Non-specific binding was determined with 5 µM naltrexone. For competition assays, membranes were incubated as above with 2 nM [3H] naloxone and various concentrations of unlabeled ligand, to determine competitor IC50 for MOR. The reaction was terminated by rapid filtration through Whatman GF–B glass fiber filters, followed by 3 washes with 3 mL ice-cold Tris buffer. Bound radioactivity was determined by liquid scintillation spectrophotometry at 45% efficiency for [3H].
CCR5 radioligand binding
The CCR5 competitive radioligand binding assay was conducted by EMD Millipore, St. Charles, MO. Compounds were mixed 1:1 with [125I]-MIP-1α (concentrations of sample compounds were 25µM with subsequent three-fold dilutions; fixed concentration of [125I]- MIP-1β were 0.25nM). The reaction was initiated by the addition of CCR5, rhesus macaque membranes prepared in assay buffer at (2×) with a final 1 unit/well. After all sample additions, the assay plate was allowed to incubate at room temperature in a non-binding plate for 120 minutes. Prior to harvesting, the FC filter plate was pre-coated with 0.3% PEI for one hour. Samples were collected and the filter plate was washed three times in wash buffer (50mM HEPES, pH 7.4, 500mM NaCl, and 0.1% BSA). The filtration plate was allowed to dry, followed by the addition of scintillation fluid at 50µL per well. Radiolabeled samples were measured on a Perkin Elmer (Wallac) 1450 Microbeta TriLux liquid scintillation counter to determine assay counts per minute.
Calcium mobilization assays
MOR calcium mobilization
hMOR-CHO cells4 were first transfected with Gqi5 pcDNA16 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommended procedure. Then cells were incubated for 6 hours at 37 °C and 5% CO2 and then trypsinized and transferred to a clear bottom, black 96-well plate (Greiner Bio-one) at 20,000 cells per well in DMEM/F- 12 supplemented with 5% fetal bovine serum, 100 u/mL penicillin, 100 µg/mL streptomycin, and 0.25 mg/mL hygromycin B. Forty eight hours after transfection the growth media was decanted and wells were washed with 100 µL of 50:1 HBSS:HEPES assay buffer. Cells were then incubated with 55 µL of Fluo4 loading buffer [30 µL 2 µM Fluo4-AM (Invitrogen), 84 µL 2.5 mM probenacid, in 5.5 mL assay buffer] for 30 minutes. Varying concentrations of ligands and controls were added to the wells to bring the total volume up to 80 µL in each well and the plates were subsequently incubated for 15 minutes. Plates were then read on a FlexStation3 microplate reader (Molecular Devices) at 494/516 ex/em for a total of 90 seconds. After 15 seconds of reading, 20 µL of 1.25 µM DAMGO in assay buffer, or assay buffer alone, was added to the wells to bring the total volume up to 100 µL. The changes in Ca2+ mobilization were monitored and peak height values were obtained using SoftMaxPro software (Molecular Devices). Non-linear regression curves and IC50s were generated using GraphPad Prism 3.0 (San Diego, CA). All experiments were repeated a total of three times.
CCR5 calcium mobilization
CCR5-MOLT-4 cells were transfected with Gqi5 pcDNA16 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommended procedure and maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 u/mL penicillin, 100 µg/mL streptomycin, and 1 mg/mL G418 at 37 °C and 5% CO2. Forty eight hours after transfection, a total of 2,500,000 cells were spun down and brought back up in 8 mL of 50:1 HBSS:HEPES assay buffer. Cells were then plated at 25,000 cells per well into a clear bottom, black 96-well plate (Greiner Bio-one) and 50 µL of Fluo4 loading buffer [40 µL 2 µM Fluo4-AM (Invitrogen), 100 µL 2.5 mM probenacid, in 5 mL assay buffer] was added to bring the volume up to 130 µL. After incubating for 45 minutes, 50 µL of varying concentrations of ligands and controls were added and the plate was incubated for an additional 15 minutes. Plates were then read on a FlexStation3 microplate reader (Molecular Devices) at 494/516 ex/em for a total of 120 seconds. After 16 seconds of reading, 20 µL of 200 nM RANTES (Biosource) in assay buffer, or assay buffer alone, was added to the wells to bring the total volume up to 200 µL. The changes in Ca2+ mobilization were monitored and peak height values were obtained using SoftMaxPro software (Molecular Devices). Non-linear regression curves and IC50s were generated using GraphPad Prism 3.0 (San Diego, CA). All experiments were repeated a total of three times.
Cell fusion assay
For the cell fusion assay two cell populations were constructed: target cells containing CCR5, MOR, CD4, and pT7EMCLuc; and effector cells containing pCAGGS-SF162gp160 and pCAGT7pol. The established CCR5-MOR cells (target cells) were transfected with the plasmids pcDNA3.1 CD4 (PMID: 17722977) and pT7EMCLuc (PMIDs: 9770428, 9349488, and 14625051) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommended procedure. HEK-293T (GenHunter Corporation; Nashville, TN, USA; catalog number Q401) cells (effector cells) were also transfected with plasmids pCAGGS-SF162gp160 (PMIDs: 10890360, 9737584, and 8995695) and pCAGT7pol using polyethylenimine (Polysciences, Inc.; Warrington, PA, USA; catalog number 23966). Prior to being overlaid, compound dilutions were added to a 96-well, white, clear bottom plate at 25 µL of 5 times concentration stock. For morphine stimulation assays, morphine stock was added to the 5 times concentrated stocks to give a final concentration of 500 nM in test wells. 24 h post transfection, the target and effectors cells were detached and overlaid onto each other at a 1:1 mixture in the 96-well white, clear bottom plate at a final concentration of 15,000 cells/well and incubated at 37 °C and 5% CO2. After an additional 24 h, 96 well plates are allowed to reach room temperature in darkness. Once equilibrated, 100 µL of a luciferin-lysis buffer solution was added (Bright-Glo Luciferase Assay System, Promega). Plates were allowed to incubate for 2 min and read luminescence for each well with a FlexStation3 plate reader (Molecular Devices). IC50s were obtained using GraphPad Prism. All experiments repeated a total of three independent times.
HIV-1 invasion assay
In a 24-well plate, primary human astroglia (ScienCell catalog #1901) were infected by incubation with the CCR5- (R5-) tropic HIV-1 strain SF162 obtained through the NIH AIDS Research and Reference Reagent Program. A concentration of HIV-1 p24 50 pg /106 cells was used and uninfected cells served as a negative control. Cells were treated with and without morphine (500 nM) along with naltrexone (1.5 µM), maraviroc (increasing concentrations of 10, 50, 100, 500 nM), and bivalent compound 1b (increasing concentrations of 10, 50, 100, 500 nM) 60 minutes before HIV-1 infection. After approximately 18 to 20 hours, the supernatant was removed and stored at −80 °C, cells were rinsed twice with PBS and lysed. The lysate was subsequently tested for the relative Tat protein expression by using a luciferase assay system (Promega). Luciferase activity was measured using a PHERAstar FS plate reader (BMG Labtech).
Interactions between opioids and HIV-1 entry inhibitors in human glial: compare maraviroc and the bivalent ligand 1. Primary human astrocytes (HA) (ScienCell catalog # 1901) were cultured in 24-well plates and transfected with the plasmid pBlue3'LTR-luc (NIH AIDS Research and Reference Reagent Program) using Lipofectamine 2000 (Invitrogen) followed by treatment with the CCR5 antagonist, maraviroc (MVC, 100 nM), morphine (M, 500 nM), naltrexone (NTX, 1.5 µM), bivalent ligand 1 (100 nM) as indicated and infected with HIV-1SF162 (R5) at a concentration of HIV-1 p24 50 pg /106 cells (Figure 5). Eighteen hours later, relative Tat protein expression was determined by measuring luciferase using the Luciferase Assay System (Promega) according to manufacturer’s protocol.
PCR study of CCR5-HMOR CHO and Astrocytes
Total RNA was isolated from the CCR5-MOR CHO cell line and two lots of primary human astrocytes from two different individuals (ScienCell Research Laboratories; Carlsbad, CA, USA; catalog number 1800) using the miRNeasy Mini Kit (Qiagen, Inc.; Valencia, CA, USA) and used to generate cDNA templates by reverse transcription using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Carlsbad, CA, USA) according to the manufacturer's instructions. PCR reactions were performed in a total volume of 20 µL containing SensiMix SYBR qPCR reagents (Bioline USA, Inc.; Tauton, MA, USA) using a Corbett Rotor-Gene 6000 real-time PCR system (Qiagen, Inc.). PCR conditions consisted of an initial hold step at 95 °C for 10 min followed by 35 amplification cycles of 95 °C for 5 s, 55 °C for 10 s, and 72 °C for 20 s. Sequences of the primer sets used were forward: 5'- CCCAACCTCTTCCAACATTGAGCAA -3' and reverse: 5'- AACGGAGCAGTTTCTGCTTCCAGAT -3' for MOR-1; forward: 5'- CTGCTCAACCTGGCCATCTCT -3' and reverse: 5'- CTTTTAAAGCAAACACAGCAT GGAC -3' for CCR5; forward: 5'- CATGGCACCGTCAAGGCTGAGAA -3' and reverse: 5'- CAGTGGACTCCACGACGTACTCA -3' for human GAPDH; and forward: 5'- CTGGAGAAACCTGCCAAGTA -3' and reverse: 5'- ACCACTCTGTTGCTGTAGCC -3' for hamster GAPDH. The specificity of the amplified products was verified by melting curve analysis and agarose gel electrophoresis. qRT-PCR data were calculated as relative expression levels by normalization against GAPDH mRNA using the 2−ΔΔCt method (reference PMID: 11846609).
HIV-1 infection assay on peripheral blood mononuclear cells (PBMCs), macrophages, and astrocytes
PBMCs were isolated from blood (Leukopak; New York Blood Center) by Ficoll density gradient centrifugation. Briefly, blood was diluted with 1× phosphate-buffered saline (PBS) at a ratio of blood:PBS (1:2). Diluted blood (30 mL), loaded very slowly onto 10 mL of Ficoll-containing lymphocyte separation medium (LSM™; MP BioMedicals), and centrifuged at 1,200 rpm for 30 min at room temperature. PBMCs from the buffy coat layer were collected and washed twice with ice cold PBS, and cultured in RPMI-1640 medium (Invitrogen) supplemented with 10% FBS (Gibco). To obtain macrophages, PBMCs were differentiated by treating with 100 ng/mL of macrophage-colony stimulating factor (M-CSF; Peprotech) for 6 days. Cell culture medium was aspirated and the adherent monocyte-derived macrophages were washed twice with Hank's balanced salt solution (Gibco) to provide macrophages for subsequent experiments. Human astrocytes were obtained from and cultured as recommended by ScienCell Research Laboratories.
PBMCs, macrophages, and astrocytes were plated at 1 × 105 cells/mL in their respective types of cell culture medium, and stimulated with interleukin-2 (IL-2; 100 ng/mL; Sigma-Aldrich) and phytohaemagglutinin (PHA; 5 µg/mL; Sigma-Aldrich) for 48 hours. Stimulated cells were treated with polybrene (2 µg/mL; Sigma-Aldrich) for 30 min and re-suspended in fresh medium. Cells were treated with maraviroc (500 nM; Sigma-Aldrich), or bivalent compounds (1b, 1c, and 1a) at 10, 100, and 1000 nM concentrations, for 1 hour prior to infection with HIVBal (HIV-p24 = 1000 pg/mL; Advanced Biotechnologies Inc., Columbia, MD) an R5-tropic HIV-1 strain
Cell Supernatant and Lysate Protein Collections and HIV-p24 Quantifications
At 5 days post-infection, cell culture supernatants were collected and cell debris was removed by spinning at 1,200 rpm for 5 min. Cells were lysed using radioimmunoprecipitation assay buffer (RIPA buffer; Sigma-Aldrich) supplemented with protease and phosphatase inhibitors (Roche Applied Sciences), and cell lysate proteins were collected by spinning at 14,000 rpm for 15 min at 4°C. Cell culture supernatants and cell lysate proteins were stored at −80°C. Cell supernatant and lysate samples were quantified for HIV-p24 levels using ELISA (HIV-1 p24 Antigen Capture Assay; Advanced Bioscience Laboratories).
Dynamics simulation studies
All ligands used in the docking studies were built with standard bond lengths and angles using the molecular modeling package SYBYL-X 2.0. The small molecules were assigned Gasteiger-Hückel charges and energy minimized with the Tripos Force Field.
All molecular modeling was collected using the SYBYL-X 2.0 molecular modeling package (Tripos LP, St. Louis, MO) on dual-core AMD Opteron(tm) 2.4 GHz processors. The amino acid sequence of chemokine receptor CCR5 was obtained from UniProtKB/Swiss-Prot (P51681). Within ClustalX a multiple alignment was performed with a gap opening penalty of 15 using the BLOSUM protein weight matrix series.47,59 Sequence alignment between CCR5 and CXCR4 was further optimized based on the most conserved residues among most GPCRs and used for model construction for both the inactive and active models. The comparative modeling software, MODELLER 9v8, was used to generate 100 homology models for each state using the default parameters.60
Model screening was performed by using the genetic-algorithm docking program GOLD 5.1 (Cambridge Crystallographic Data Centre, Cambridge, UK) to dock maraviroc into the CCR5 homology models using GOLD score as the fitness function.53 Once receptor model was chosen based upon the discrete optimized protein energy (DOPE) scores, fitness function values, and the electronic and steric interactions between the ligands and receptor. Further model refinement was done using molecular mechanics based energy minimization in Sybyl-X 2.0. Briefly, the model was minimized using a Tripos Force Field with Gasteiger-Hückel charges, a non-bonded interaction cutoff of 8 Å with a distance-dependent dielectric constant of ε = 4 being terminated at 0.05 kcal/(mol Å). The minimized models were then analyzed using PROCHECK and ProTable within SYBYL-X 2.0 to ensure the overall quality of the models (i.e. acceptable torsion angles, steric clashes, bond lengths, etc.).
The heterodimer was built within SYBYL-X 2.0 using the above described CCR5 homology model and the mu opioid receptor crystal structure functional dimer (PDB code: 4DKL).48 MOR was crystallized as both a dimer and both a TM5/TM6 and a TM1/TM2 dimer interface were observed.47 The TM5/TM6 has more extensive packing and network of interactions which make it a more plausible dimer interface. In order to construct the heterodimer, one of the MOR units was aligned with the CCR5 homology model according to their homology levels. The subsequent MOR was removed and a MOR-CCR5 heterodimer was left. Initial heterodimer refinement was done using molecular mechanics based energy minimization in Sybyl-X 2.0. Briefly, the model was minimized using a MMFF94 force field with Gasteiger-Hückel charges, a non-bonded interaction cutoff of 8 Å with a distance-dependent dielectric constant of ε = 4 being terminated at 0.05 kcal/(mol Å). The minimized heterodimer was then analyzed using PROCHECK and ProTable within SYBYL-X 2.0 to ensure the overall quality of the models (i.e. acceptable torsion angles, steric clashes, bond lengths, etc.).
The heterodimer interface had extensive hydrophobic and polar interactions similar to the ones seen in the MOR homodimer.48 Using APBS, the electrostatic interfaces between MOR and CCR5 were mapped.61,62
The optimized heterodimer model was then subjected to another round of docking of the agonists and antagonists. Using GOLD 5.1 the ligands were docked into both the heterodimer. The putative binding area was restricted to a 15 Å radius around E283 and maraviroc was docked into the receptor a total of 100 iterations using the generic GOLD docking parameters.57,58 Concurrently, naltrexone was aligned/overlapped with the morphanin antagonist β-FNA within the MOR binding pocket of the heterodimer model. The attachment site of the linker to naltrexone allows for the linker to span into the CCR5 binding pocket through the TM5/TM6 interface. Therefore, of the 100 docked poses of maraviroc, the poses with the linker portion pointed towards the TM5/TM6 interface were sorted out for further analysis. The pose with the highest GOLD score and that was within the proper 21-atom distance to naltrexone (linker length: 21 atoms long) was chosen. Once both the naltrexone and maraviroc modes were chosen, they were connected to each other using SYBYL X 2.0 with the 21-atom linker to yield compound 1b. The subsequent bivalent compound was then merged with the heterodimer and the whole system was energy minimized using a MMFF94 force field.
All molecular dynamics simulations were run using the Teal cluster housed at the Virginia Commonwealth University Center for High Performance Computing. The cluster consists of ~2480 64 bit AMD computer cores, each with 2–4 GB RAM/core.
The heterodimer-1b complex was further analyzed using molecular dynamics with the CHARMM force field using nanoscale molecular dynamics, NAMD.54,63,64 Using the program VMD (Visual Molecular Dynamics), a solvated 150 Å × 150 Å phosphatidylcholine (POPC) was constructed on the x-y plane.65 The CCR5-MOR bound 1b complex was then properly orientated for insertion into the lipid bilayer using the orientations of proteins in membranes (OPM) database.66 After inserting the protein into the middle of the membrane, lipids within 0.8 Å of the protein were removed. Next the system was solvated with TIP3 water and equilibrated with 0.15 M NaCl ions. In the completed system there were a total of 162385 atoms. A modified CHARMM27 force field was constructed with the parameters for compound 1b; the online server SwissParam was used to calculate the CHARMM force field for the ligand.67
Using NAMD, the system was equilibrated in a three-step process. First, 500 ps of molecular dynamic simulation was run (with a time step of 2 fs) on only the lipid tails of the POPC bilayer while keeping the protein, water, ions, ligand, and lipid-head groups fixed. During the second round of equilibration, the protein and ligand were harmonically constrained while the rest of the system was allowed to move. The simulation was run for 500 ps (2 fs time step) while keeping water out of the lipid bilayer. The third step was run completely without constraints for 500 ps while keeping a constant area for the water box.
Molecular dynamics stimulation was then run on the equilibrated system for 10 ns with a time step of 2 fs with the area of the membrane kept constant. Langevin dynamics helped maintained a constant temperature of 310 K and a hybrid Nosé-Hoover Langevin piston method was used to keep a constant pressure of 1 atm with an oscillation period of 200 fs. Electrostatics were maintained using periodic boundary conditions and the particle mesh ewalds method. A 12 Å non-bonded cutoff and a grid spacing of 1 Å per point in each dimension while calculating van der Waals energies using a switching radius of 10 Å and a cutoff radius of 12 Å. Trajectory analyses were carried out using VMD focusing on the heterodimer and 1b interactions.
Supplementary Material
Acknowledgments
The authors thank NIH/NIDA for financial support. This work was partially supported by NIH/NIDA DA 037096 (Y.Z./K.F.H.) and DA024022 (Y.Z.).
References
- 1.Nath A, Hauser KF, Wojna V, Booze RM, Maragos W, Prendergast M, Cass W, Turchan JT. Molecular Basis for Interactions of HIV and Drugs of Abuse. JAIDS, J. Acquir. Immune Defic. Syndr. 2002;31:S62–S69. doi: 10.1097/00126334-200210012-00006. [DOI] [PubMed] [Google Scholar]
- 2.Hauser KF, El-Hage N, Buch S, Berger JR, Tyor WR, Nath A, Bruce-Keller AJ, Knapp PE. Molecular Targets of Opiate Drug Abuse in neuroAIDS. Neurotoxic. Res. 2005;8:63–80. doi: 10.1007/BF03033820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norman KF, Basso M, Kurmar A, Malow R. Neuropsychological Consequences of HIV and Substance Abuse: A Literature Review and Implications for Treatment and Future Research. Curr. Drug Abus. Rev. 2009;2:143–156. doi: 10.2174/1874473710902020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony IC, Arango JC, Stephens B, Simmonds P, Bell JE. The Effects of Illicit Drugs on the HIV Infected Brain. Front. Biosci. 2008;13:1294–1307. doi: 10.2741/2762. [DOI] [PubMed] [Google Scholar]
- 5.Hauser KF, Fitting S, Dever SM, Podhaizer EM, Knapp PE. Opiate Drug Use and the Pathophysiology of neuroAIDS. Curr. HIV Res. 2012;10:435–452. doi: 10.2174/157016212802138779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou S, Fitting S, Hahn YK, Welch SP, El-Hage N, Hauser KF, Knapp PE. Morphine Potentiates Neurodegenerative Effects of HIV-1 Tat through Actions at M-Opioid Receptor-Expressing Glia. Brain. 2011;134:3613–3628. doi: 10.1093/brain/awr281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noel RJJ, Rivera-Amill V, Buch S, Kurmar A. Opiates, Immune System, Acquired Immunodeficiency Syndrome, and Nonhuman Primate Model. J. Neurovirol. 2008;14:279–285. doi: 10.1080/13550280802078209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alkhatib G, Comadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: A RANTES, MIP-1alpha, MIP-1beta Receptor as a Fusion Cofactor for Macrophage-Tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Shall TJ, Littman DR, Landau NR. Identification of a Major Co-Receptor for Primary Isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Gabuzda D, Wang J. Chemokine Receptors and Virus Entry in the Central Nervous System. J. Neurovirol. 1999;5:643–658. doi: 10.3109/13550289909021293. [DOI] [PubMed] [Google Scholar]
- 11.Luster AD. Chemokines - Chemotactic Cytokines That Mediate Inflammation. N. Engl. J. Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 12.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori G, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a Potent, Orally Bioavailable, and Selective Small-Molecule Inhibitor of Chemokine Receptor CCR5 with Broad-Spectrum Anti-Human Immunodeficiency Virus Type 1 Activity. Antimicrob. Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lind KA, Marks DR, Kolson DL, Jordan-Sciutto KL. HIV-Associated Neurocognitive Disorder: Pathogenesis and Therapeutic Opportunities. J. Neuroimmune. Pharmacol. 2010;5:294–309. doi: 10.1007/s11481-010-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minagar A, Commins D, Alexander JS, Hoque R, Chiappelli F, Signer EJ, Nikbin B, Shapshak P. NeuroAIDS: Characteristics and Diagnosis of the Neurological Complications of AIDS. Mol. Diagn. Ther. 2008;12:25–43. doi: 10.1007/BF03256266. [DOI] [PubMed] [Google Scholar]
- 15.Turchan-Cholewo J, Liu Y, Gartner S, Reid R, Jie C, Peng X, Chen KC, Chauhan A, Haughey N, Cutler R, Mattson MP, Pardo C, Conant K, Sacktor N, McArthur JC, Hauser KF, Gairola C, Nath A. Increased Vulnerability of ApoE4 Neurons to HIV Proteins and Opiates: Protection by Diosgenin and L-Deprenyl. Neurobiol. Dis. 2006;23:109–119. doi: 10.1016/j.nbd.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 16.El-Hage N, Wu G, Wang J, Ambati J, Knapp PE, Reed JL, Bruce-Keller AJ, Hauser KF. HIV-1 Tat and Opiate-Induced Changes in Astrocytes Promote Chemotaxis of Microglia through the Expression of MCP-1 and Alternative Chemokines. Glia. 2006;53:132–146. doi: 10.1002/glia.20262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurwell JA, Nath A, Sun Q, Zhang J, Martin KM, Chen Y, Hauser KF. Synergistic Neurotoxicity of Opioids and Human Immunodeficiency Virus-1 Tat Protein in Striatal Neurons in Vitro. Neuroscience. 2001;102:555–563. doi: 10.1016/s0306-4522(00)00461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser KF, El-Hage N, Steine-Martin A, Maragos WF, Nath A, Peridsky Y, Volsky DJ, Knapp PE. HIV-1 Neuropathogenesis: Glial Mechanisms Revealed through Substance Abuse. J. Neurochem. 2007;100:567–586. doi: 10.1111/j.1471-4159.2006.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S, Chuang LF, Yau P, Doi RH, Chuang RY. Interactions of Opioid and Chemokine Receptors: Oligomerization of Mu, Kappa, and Delta with CCR5 on Immune Cells. Exp. Cell Res. 2002;280:192–200. doi: 10.1006/excr.2002.5638. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan SD, Schwartz SA, Shanahan TC, Chawda RP, Nair MPN. Morphine Regulates Gene Expression of A- and B-Chemokines and Their Receptors on Astroglial Cells via the Opioid Receptor. J. Immunol. 2002;169:3589–3599. doi: 10.4049/jimmunol.169.7.3589. [DOI] [PubMed] [Google Scholar]
- 21.Szabo I, Chen XH, Xin L, Adler MW, Howard OM, Oppenheim JJ, Rogers TJ. Heterologous Desensitization of Opioid Receptors by Chemokines Inhibits Chemotaxis and Enhances the Perception of Pain. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10276–10281. doi: 10.1073/pnas.102327699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Li J, Bot G, Szabo I, Rogers TJ, Liu-Chen LY. Heterodimerization and Cross-Desensitization between the Mu-Opioid Receptor and the Chemokine CCR5 Receptor. Eur. J. Pharmacol. 2004;483:175–186. doi: 10.1016/j.ejphar.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Yuan Y, Arnatt CK, Li G, Haney KM, Ding D, Jacob JC, Selley DE, Zhang Y. Design and Synthesis of a Bivalent Ligand to Explore the Putative Heterodimerization of the Mu Opioid Receptor and the Chemokine Receptor CCR5. Org. Biomol. Chem. 2012;10:2633–2646. doi: 10.1039/c2ob06801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y, Arnatt CK, El-Hage N, Dever SM, Jacob JC, Selley DE, Hauser KF, Zhang Y. A Bivalent Ligand Targeting the Putative Mu Opioid Receptor and Chemokine Receptor CCR5 Heterodimer: Binding Affinity versus Functional Activities. Med. Chem. Comm. 2013;4:847–851. doi: 10.1039/C3MD00080J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Hage N, Dever SM, Podhaizer EM, Arnatt CK, Zhang Y, Hauser KF. A Novel Bivalent HIV-1 Entry Inhibitor Reveals Fundamental Differences in CCR5 - [mu]-Opioid Receptor Interactions in Human Glia. AIDS. 2013;27:2181–2190. doi: 10.1097/QAD.0b013e3283639804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnatt CK, Zhang Y. Bivalent Ligands Targeting Chemokine Receptor Dimerization: Molecular Design and Functional Studies. Curr. Top. Med. Chem. 2014;14:1606–1618. doi: 10.2174/1568026614666140827144752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams PG, Moore RE, Paul VJ. Isolation and Structure Determination of Lyngbyastatin 3, a Lyngbyastatin 1 Homologue from the Marine Cyanobacterium Lyngbya Majuscula. Dertermination of the Configuration of the 4-Amino-2,2-Dimethyl-3-Oxopentanoic Acid Unit in Majusculamide C, Dolastatin. J. Nat. Pod. 2003;66:1356–1363. doi: 10.1021/np0302145. [DOI] [PubMed] [Google Scholar]
- 28.Pu X, Ma D. Asymmetric Total Synthesis of (−)-Alkaloid 223A and its 6-Epimer. J. Org. Chem. 2003;68:4400–4405. doi: 10.1021/jo0341261. [DOI] [PubMed] [Google Scholar]
- 29.Davies SG, Mulvaney AW, Russell AJ, Smith AD. Parallel Synthesis of Homochiral Beta-Amino Acids. Tetrahedron:Asymmetry. 2007;18:1554–1566. [Google Scholar]
- 30.Haycock-Lewandowski SJ, Wilder A, Ahman J. Development of a Bulk Enabling Route to Maraviroc (UK-427,857), a CCR-5 Receptor Antagonist. Org. Process Res. Dev. 2008;12:1094–1103. [Google Scholar]
- 31.Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. Substitution of Three Amino Acids Switches Receptor Specificity of Gq to that of Gi Alpha. Nature. 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 32.Cisneros IE, Ghorpade A. HIV-1, Methamphetamine and Astrocyte Glutamate Regulation: Combined Excitotoxic Implications for Neuro-AIDS. Curr. HIV Res. 2012;10:392–406. doi: 10.2174/157016212802138832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, Mackay CR. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hoffman W, Newman W, Mackay CR, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 36.Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage succeptibility to infection by human immunodeficiency virus type 1. J. Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito Y, Sharer LR, Epstein LG, Michaels J, Mintz M, Louder M, Golding K, Cvetkovich TA, Blumberg BM. Overexpression of nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology. 1994;44:474–481. doi: 10.1212/wnl.44.3_part_1.474. [DOI] [PubMed] [Google Scholar]
- 38.Gorry PR, Howard JL, Churchill MJ, Anderson JL, Cunningham A, Adrian D, McPhee DA, Purcell FJ. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J. Virol. 1999;73:352–361. doi: 10.1128/jvi.73.1.352-361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Churchill MJ, Wesselingh SL, Cowley D, Pardo CA, McArthur JC, Brew BJ, Gorry PR. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann. Neurol. 2009;66:253–258. doi: 10.1002/ana.21697. [DOI] [PubMed] [Google Scholar]
- 40.Conant K, Tornatore C, Atwood W, Meyers K, Traub R, Major EO. In vivo and in vitro infection of the astrocyte by HIV-1. Adv. Neuroimmunol. 1994;4:287–289. doi: 10.1016/s0960-5428(06)80269-x. [DOI] [PubMed] [Google Scholar]
- 41.Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- 42.Canki M, Thai JN, Chao W, Ghorpade A, Potash MJ, Volsky DJ. Highly productive infection with pseudotyped human immunodeficiency virus type 1 (HIV-1) indicates no cellular restrictions to HIV-1 replication in primary human astrocytes. J. Virol. 2001;75:7925–7933. doi: 10.1128/JVI.75.17.7925-7933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer-Hammerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto T, Ushijima H, Okitsu S, Suzuki E, Sakai K, Morikawa S, Muller WE. Establishment of an HIV Cell-Cell Fusion Assay by Using Two Genetically Modified HeLa Cell Lines and a Reporter Gene. J. Virol. Methods. 2003;114:159–166. doi: 10.1016/j.jviromet.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Simpson LM, Taddese B, Wall ID, Reyonds CA. Bioinformatics and Molecular Modelling Approaches to GPCR Oligomerization. Curr. Opin. Pharmacol. 2010;10:30–37. doi: 10.1016/j.coph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-Force Microscopy: Rhodopsin Dimers in Native Disc Membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 47.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal Structure of the M-Opioid Receptor Bound to a Morphinan Antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorinski N, Kowalsman N, Renner U, Wirth A, Reinartz MT, Seifert R, Zeug A, Ponimaskin E, Niv MY. Computational and Experimental Analysis of the Transmembrane Domain 4/5 Dimerization Interface of the Serotonin 5-HT1a Receptor. Mol. Pharmacol. 2012;82:448–463. doi: 10.1124/mol.112.079137. [DOI] [PubMed] [Google Scholar]
- 50.Li G, Aschenbach LC, Chen J, Cassidy MP, Stevens DL, Gabra BH, Selley DE, Dewey WL, Westkaemper RB, Zhang Y. Design, Synthesis, and biological Evaluation of 6alpha- and 6beta-N-Heterocyclic Substituted Naltrexamine Derivatives as Mu Opioid Receptor Selective Antagonists. J. Med. Chem. 2009;52:1416–1427. doi: 10.1021/jm801272c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Haney KM, Richardson AC, Wilson E, Gerwitz DA, Ware JL, Zehner ZE, Zhang Y. Anibamine, a Natural Product CCR5 Antagonist, as a Novel Lead for the Development of Anti Prostate Cancer Agents. Bioorg. Med. Chem. Lett. 2010;20:4627–4630. doi: 10.1016/j.bmcl.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved Protein-Ligand Docking Using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 54.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnston JM, Filizola M. Showcasing Modern Molecular Dynamics Simulations of Membrane Proteins through G Protein-Coupled Receptors. Curr. Opin. Struct. Biol. 2011;21:552–558. doi: 10.1016/j.sbi.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spijker P, Vaidehi N, Freddolino PL, Hilbers PAJ, Goddard WAI. Dynamic Behavior of Fully Solvated 2-Adrenergic Receptor, Embedded in the Membrane with Bound Agonist or antagonistNo Title. Proc. Natl. Acad. Sci. USA. 2006;103:4882–4887. doi: 10.1073/pnas.0511329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Perez J, Rueda P, Alcami J, Rognan D, Arenzana-Seisdedos F, Lagane B, Kellenberger E. Allosteric Model of Maraviroc Binding to CC Chemokine Receptor 5 (CCR5) J. Biol. Chem. 2011;286:33409–33421. doi: 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M. Molecular Interations of CCR5 with Major Classes of Small-Molecule Anti-HIV CCR5 Antagonists. Mol. Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- 59.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 61.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holst MJ, Saied F. Multigrid Solution of the Poisson-Boltzmann Equation. J. Comp. Chem. 1993;14:105–113. [Google Scholar]
- 63.Mackerell ADJ, Feig M, Brooks CLI. Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 64.Brooks BR, Brooks CLI, Mackerell ADJ, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J. Comp. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 66.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes Database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 67.Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam, a Fast Force Field Generation Tool for Small Organic Molecules. J. Comp. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.