Abstract
Objectives
It has long been discussed whether fitness or fatness is a more important determinant of health status. If the same genetic factors that promote body fat percentage (body fat%) are related to cardiorespiratory fitness (CRF), part of the concurrent associations with health outcomes could reflect a common genetic origin. In this study we aimed to 1) examine genetic correlations between body fat% and CRF; 2) determine whether CRF can be attributed to a genetic risk score (GRS) based on known body fat% increasing loci; and 3) examine whether the fat mass and obesity associated (FTO) locus associates with CRF.
Methods
Genetic correlations based on pedigree information were examined in a family based cohort (n = 230 from 55 families). For the genetic association analyses, we examined two Danish population-based cohorts (ntotal = 3206). The body fat% GRS was created by summing the alleles of twelve independent risk variants known to associate with body fat%. We assessed CRF as maximal oxygen uptake expressed in millilitres of oxygen uptake per kg of body mass (VO2max), per kg fat-free mass (VO2maxFFM), or per kg fat mass (VO2maxFM). All analyses were adjusted for age and sex, and when relevant, for body composition.
Results
We found a significant negative genetic correlation between VO2max and body fat% (ρG = -0.72 (SE ±0.13)). The body fat% GRS associated with decreased VO2max (β = -0.15 mL/kg/min per allele, p = 0.0034, age and sex adjusted). The body fat%-increasing FTO allele was associated with a 0.42 mL/kg/min unit decrease in VO2max per allele (p = 0.0092, age and sex adjusted). Both associations were abolished after additional adjustment for body fat%. The fat% increasing GRS and FTO risk allele were associated with decreased VO2maxFM but not with VO2maxFFM.
Conclusions
Our findings suggest a shared genetic etiology between whole body fat% and CRF.
Introduction
It has been discussed whether fitness or fatness is a more important determinant of health status. There is evidence that low cardiorespiratory fitness (CRF) and obesity are equally important predictors of mortality [1] and other health outcomes [2]. Furthermore, physical activity and high CRF are beneficial for health at any body weight [2, 3] and each of them may attenuate overweight and obesity-induced health risks [4].
CRF is commonly estimated by VO2max, a measure of the oxygen consumption during maximal exercise. Twin studies have shown that adiposity and CRF have strong genetic components, with heritability estimates of 50–90% for body-mass index (BMI) [5], 25–30% for body fat percentage (body fat%) [6] and 40–50% for CRF (VO2max) [7]. The link between development of obesity and level of physical fitness might be caused by a common genetic origin, rather than a causal effect.
Large-scale genome-wide association studies (GWAS) have identified more than one hundred loci associated with overall adiposity [8, 9], but no genetic variants are known to robustly associate with CRF. This may be due to insufficient sample sizes with data on CRF to identify variants with modest effects at the genome-wide significant level. GWAS have thus far identified twelve loci robustly associated with body fat percentage [9]. The strongest of these, FTO, was the first GWAS-identified susceptibility gene for common obesity [10]. Ever since, many studies have examined whether single nucleotide polymorphisms (SNPs) in the FTO loci are associated with lifestyle factors such as physical activity and other mediators leading to increased body weight [10]. While FTO does not seem to play a role in the regulation of physical activity levels [10], the relationship between FTO and obesity risk is modified by physical activity [11, 12]. There are, however, only few reports on FTO and physical fitness phenotypes. In a controlled exercise intervention study of 481 individuals, it was found that exercise-induced changes in adiposity were dependent on the FTO genotype [13]. In contrast, another study examining 846 young, healthy men failed to show that aerobic fitness in the untrained state is associated with the FTO genotype nor that it modifies the effect of FTO on body composition [14].
In the present study, we aimed to (1) examine genetic correlations between measures of adiposity and CRF in a family-based study sample; (2) determine whether inter-individual differences in CRF can be attributed to a genetic risk score (GRS) of GWAS-identified body fat% variants and whether GWAS-identified body fat% loci interact with CRF to modify levels of body fat%; (3) examine whether the FTO locus, known to exert the largest genetic effect on different adiposity measures, associates with CRF in two independent population-based cohorts of Danish ancestry.
Materials and Methods
Study populations
The Family cohort consists of 533 Danish individuals from 95 families with one parent suffering from type 2 diabetes and the other parent having no known diabetes. The families were identified and all non-diabetic family members (spouses, offspring and other relatives) were recruited through the outpatient clinic at the Steno Diabetes Center (Gentofte, Denmark) or through an ongoing family study at the University of Copenhagen (Copenhagen, Denmark) [15]. All participants of the Family cohort underwent measurement of height and weight. The amount of body fat was determined by bio-impedance (Biodynamics BIA 310e, H.A.W consulting, Denmark). Maximal oxygen intake (VO2max), was estimated from the heart rate response to a submaximal cycle ergometer exercise test with the Astrand-Rhyming nomogram [16]. We excluded family members if disagreement between questionnaire information on familial relationship and the actual genotypic resemblance was observed. Of the 435 individuals from families having four or more children, 230 with data on BMI, body fat% and CRF were included in the present genetic correlation analysis. The characteristics and relationships of the 230 individuals are shown in Table 1 and S1 Table.
Table 1. Clinical characteristics of three Danish study populations included into the analysis.
| Family cohort | ADDITION-PRO | Health2006 | |
|---|---|---|---|
| n (f/m) | 230 (123/103) | 716 (329/387) | 2586 (1414/1172) |
| Age, years | 39.4 (34; 42) | 66.1 (60.9; 70.7) | 49 (40; 59) |
| BMI, kg/m2 | 26.1 (4.5) | 27.1 (4.4) | 25.6 (4.3) |
| Body fat percentage, % | 32.7 (10.3) | 32.0 (8.1) | 29.4 (8.8) |
| Lean body mass, kg | 51.8 (10.7) | 53.9 (11.3) | 53.3 (11.2) |
| VO2max, ml/kg/min | 32.6 (9.5) | 29.8 (5.4) | 31.9 (8.9) |
| VO2maxFFM, ml/kg FFM/min | 48.6 (11.2) | 44.1 (7.6) | 45.0 (10.6) |
| VO2maxFM, ml/kg FM/min | 122.6 (94.6) | 94.7 (41.0) | 126.5 (74.7) |
Data in Table 1 are given as mean (standard deviation) or median (interquartile range). n: sample size, f: female, m: male, BMI: Body mass index, VO2max: maximal oxygen uptake scaled by body weight, VO2maxFFM: maximal oxygen uptake scaled by fat free mass, VO2maxFM: maximal oxygen uptake scaled by fat mass.
The ADDITION-PRO cohort is a population-based study of Danish individuals, aged 45–80 years at medium to high risk of developing type 2 diabetes, recruited during a stepwise screening procedure during 2001–2006. The screening procedure and the assessment of anthropometric measures, including BMI and body fat% for ADDITION-PRO have been described in detail elsewhere [17]. In short, height and weight, for the calculation of BMI, was measured in light indoor clothing and without shoes. Body fat% and weight were assessed by bioelectrical impedance using a leg-to-leg Tanita Body Composition Analyser (Tokyo, Japan). A subset of participants (n = 955) underwent an 8-min submaximal step test, during and after which heart rate was measured using a combined sensor (Actiheart, CamNTech Ltd., Cambridge, UK) [18]. The test was administered using the sensor manufacturer’s software to indicate the cycles of stepping up and down a 20.5-cm step bench; stepping frequency ranged from 15 to 33 step cycles per minute over the duration of the test [18]. The submaximal heart rate response to exercise load was modeled as linear [19] and extrapolated to age-predicted maximal heart rate [20] to estimate VO2max (Study characteristics in Table 1).
The Health2006 study is a population-based cohort consisting of a random sample of Danish men and women aged 18–69 years living in the southwestern part of the greater Copenhagen area [21]. Height and weight were measured wearing light clothes and no shoes. The amount of body fat was assessed by a Tanita Body Composition Analyzer (Illinois, USA)[21]. VO2max was estimated using the Danish step test according to instructions available at (www.health-calc.com/fitness-tests/the-danish-step-test). In short, the Danish step test is simple, requires little equipment and was developed for estimation of CRF in large epidemiological studies. The test is based on workload estimation of maximal oxygen uptake. It is a progressive test that starts with a stepping frequency of 0.2 steps/second which increases gradually until a maximal frequency of 0.8 steps/seconds at 6 minutes. VO2max (ml/min) was then calculated based on the height of the stepping bench, the duration of the test procedure, and the weight of the participant using a formula that has been validated against a Wattmax test [22]. Of the 2703 individuals that had been genotyped and underwent the submaximal step-test, two were excluded due to VO2max values below 10 mL/kg/min (Study characteristics in Table 1).
Prior to participation, informed written consent was obtained prior participation from all participants of the three studies described above. The Ethical Committee of Copenhagen (KA 93033 and KA 93033gm) approved the Family cohort study. The Ethical Committee of Copenhagen County (KA-20060011) and the Danish Data Protection Agency approved the Health2006 study. The Health2006 study was registered at www.clinicaltrials.gov (ClinicalTrials.gov identification number: NCT00316667, other study ID number: KA20060011). The ADDITION-PRO study was approved by the Scientifics Ethics Committee in the Central Denmark Region (20000183). All studies were conducted in accordance with the principles of the Declaration of Helsinki.
Genotyping
ADDITION-PRO
Participants of the ADDITION-PRO cohort (n = 1657) were genotyped by the Illumina Infinium HumanCoreExome Beadchip platform (Illumina, San Diego, CA). Genotypes were called using the Genotyping module (version 1.9.4) of GenomeStudio software (version 2011.1, Illumina). We excluded 109 closely related individuals, individuals with extreme inbreeding coefficients, individuals with mislabelled sex, individuals with a call rate <95%, duplicates and individuals identified as ethnic non-European outliers, leaving 1548 individuals who passed all quality control criteria. Additional genotypes were imputed into 1000 genomes phase 1 [23] using impute2 [24]. The imputation quality was high (proper_infor > 0.95) for all imputed variants included in the current study. All variants obeyed Hardy Weinberg equilibrium (p > 0.05).
Health2006
Participants from the Health2006 (n = 2883) cohorts were genotyped by Metabochip on the Illumina HiScan platform (Illumina, San Diego, CA, USA). Genotypes were called using the GenomeStudio software (version 2011.1, Illumina). We excluded individuals with low call rate, mislabeled sex, relatedness, extreme inbreeding coefficient and with a high discordance rate to previously genotyped SNPs, leaving 2804 individuals for whom genotyping was successful accomplished. All variants obeyed Hardy Weinberg equilibrium (p > 0.05).
Genetic correlation and GRS analyses
Genetic and environmental correlations
Genetic, phenotypic and environmental correlations in the Family cohort were calculated using SOLAR (http://solar.txbiomedgenetics.org, version 4.2.0) [25]. The additive effect of shared genes was calculated as described elsewhere [26]; individuals belonging to the same family were assumed to be sharing the same household. Neither body fat% nor CRF were significantly affected by the shared environment and thus it was not included in the genetic correlation analysis. We tested whether the genetic correlation is significantly different from complete genetic correlation (Pdifferent from 1) or from no correlation (Pdifferent from 0).
GRS construction and association analyses
Genotypes were coded according to the number of body fat% increasing alleles based on 12 independent variants shown to be associated with body fat% in a large-scale GWAS meta-analysis [9]. We constructed a weighted GRS by summing the number of body fat%- increasing alleles weighted by the effect size of the variant estimated in the GWAS discovery study [9].
In the discovery cohort ADDITION-PRO, all genotypes were retrieved from the imputed dataset and genetic risk scores were calculated based on dosage information. Of the 955 individuals with information on step-test derived VO2max, n = 716 individuals had valid genotype information and were included in the subsequent analyses. For the FTO association and interaction analyses, the FTO rs1558902 variant was directly genotyped in all 716 individuals and therefore not retrieved from dosage information (S2 Table).
In Health2006, seven of the twelve GWAS identified body fat% SNPs were present on the Metabochip (rs1558902, rs6567160, rs6755502, rs693839, rs543874, rs3761445, rs757318), three SNPs (rs2943646, rs7609045, rs7187776) were captured by perfect proxies (r2 = 1), one (rs4794018) was captured by a proxy with r2 = 0.93 and one SNP (rs6857) was not covered by the Metabochip. Proxy search was performed using 1000 Genomes Pilot 1 data to estimate linkage disequilibrium using the SNP annotation proxy search tool (SNAP, http://www.broadinstitute.org/mpg/snap)[27]. Hence, a total of 11 SNPs for Health2006 are included in the GRS. Of the 2586 participants that had information on both genotypes and VO2max, 96 were excluded due to missing genotype information on ≥ 1 SNP. This allowed us to include a total of 2490 individuals into the GRS analysis. Data on FTO was available for all but two of the 2586 individuals; these were included into the FTO association and interaction analysis (S2 Table).
Statistical analyses
Analyses in ADDITION-PRO and Health2006 were performed using R software (version 3.2.0, The R Foundation for Statistical Computing, Boston, MA, USA). For our analysis we expressed CRF relative to body weight, fat-free mass (FFM) and fat mass (FM), denoted VO2max (ml/kg/min), VO2maxFFM (ml/kg FFM/min) and VO2maxFM (ml/kg FM/min). After Bonferroni correction for multiple testing for the three traits tested, p<0.017 was considered statistically significant. Associations between the GRS and CRF as well as between FTO rs1558902 and CRF were examined by linear regression using additive genetic models. Analyses were adjusted for age and sex or age, sex and body composition (BMI, body fat%, FFM, FM). Linear regression models including an interaction term were used to test gene × CRF interactions on body fat%, adjusted for age and sex. All association analyses were first performed in ADDITION-PRO and then replicated in Health2006. Subsequently, we combined the effect size estimates and standard errors (SE) derived from the linear regression analyses using fixed effects meta-analyses, including a total of 3206 individuals (nHealth2006 = 2490, nADDITION-PRO = 716) for the GRS analysis and 3300 individuals (nHealth2006 = 2584, nADDITION-PRO = 716) for the FTO analysis using the ‘meta’ package (version 4.2–0) for R. We focus the following sections of this manuscript on the results from the meta-analysis. All cohort specific results for the associations are shown in S3 Table (Genetic risk score associations) and S4 Table (FTO associations).
Results
Heritability estimates and genetic correlations
Within the Family cohort, we found that additive genetic effects (h2) explained 25% (SE 15%) of the variation in BMI, 53% (SE 12%) of the variation in body fat%, 48% (SE 12%) of the variation in fat mass (kg) and 41% (SE 11%) of the variation in fat-free body mass (kg). We found that additive genetic effects explained 49% (SE 15%) of the variation in CRF relative to body weight (VO2max), 37% (SE 15%) of the variation in CRF relative to fat-free mass (VO2maxFFM) and 57% (SE 15%) in CRF relative to fat mass (VO2maxFM). All heritability estimates were adjusted for age and sex (S5 Table).
We calculated genetic (ρG) and phenotypic (ρP) correlations between VO2max and measures of body composition such as BMI, lean body fat (kg), fat mass (kg) and body fat%. Significant genetic correlations between VO2max and body fat% (ρG = -0.72 (SE ±0.13); pdifferent from 0 < 0.01, pdifferent from 1 < 0.01) and absolute VO2 (ml/min) and fat mass (kg) (ρG = -0.70 (SE ±0.17); pdifferent from 0 < 0.01, pdifferent from 1 < 0.01) were found. No genetic correlations were found between VO2max and BMI (ρG = -0.28 (SE ±0.22); pdifferent from 0 = 0.24, pdifferent from 1 < 0.01) nor VO2max and lean body mass (kg) (ρG = 0.39 (SE ±0.26); pdifferent from 0 = 0.13, pdifferent from 1 = 0.01).
Genetic risk score association and interaction analysis
To examine whether genetic variants known to associate with body fat% contribute to the significant genetic correlation between VO2max and body fat%, we tested for associations between CRF and a GRS for body fat%, the latter constructed of the 12 known GWAS identified variants. The body fat% GRS ranged from 2 to 18 risk alleles (median = 10). The GRS was associated with body fat% (β = 0.22% per allele; p < 0.0001; age and sex adjusted) and showed an inverse association with VO2max (β = -0.15 mL/kg/min per additional risk allele, p = 0.0034). The association was abolished after additional adjustment for body fat% (β = -0.05, p = 0.27), indicating that about two thirds of the association was mediated by adiposity. We also found that the GRS was associated with VO2maxFM but not with VO2maxFFM (Table 2).
Table 2. Associations between body fat% GRS and CRF expressed in VO2max (ml/kg/min), VO2maxFFM (ml/kg FFM/min) as well as VO2maxFM (ml/kg FM/min) after meta-analysis of two Danish cohorts (ADDITION-PRO (n = 716) and Health2006 (n = 2490)).
| Trait | Covariates | Per allele effect size on CRF (95% CI) | P |
|---|---|---|---|
| VO2max (ml/kg/min) | age, sex | -0.15 (-0.26 to -0.05) | 0.0034 |
| VO2max (ml/kg/min) | age, sex, body fat% | -0.05 (-0.15 to 0.04) | 0.27 |
| VO2max (ml/kg/min) | age, sex, fat-free mass | -0.11 (-0.21 to -0.0006) | 0.04 |
| VO2maxFFM (ml/kg FFM/ min) | age, sex | -0.06 (-0.19 to 0.07) | 0.38 |
| VO2maxFFM (ml/kg FFM/min) | age, sex, body fat% | -0.07 (-0.20 to 0.06) | 0.30 |
| VO2maxFM (ml/kg FM/ min) | age, sex | -0.01 (-0.02 to -0.007) | < 0.0001 |
| VO2maxFM (ml/kg FM/min) | age, sex, fat-free mass | -0.008 (-0.01 to -0.003) | 0.0033 |
Associations between body fat% GRS and CRF were examined by linear regression using additive genetic models. Data for VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition. Cohort specific association results can be found in S3 Table.
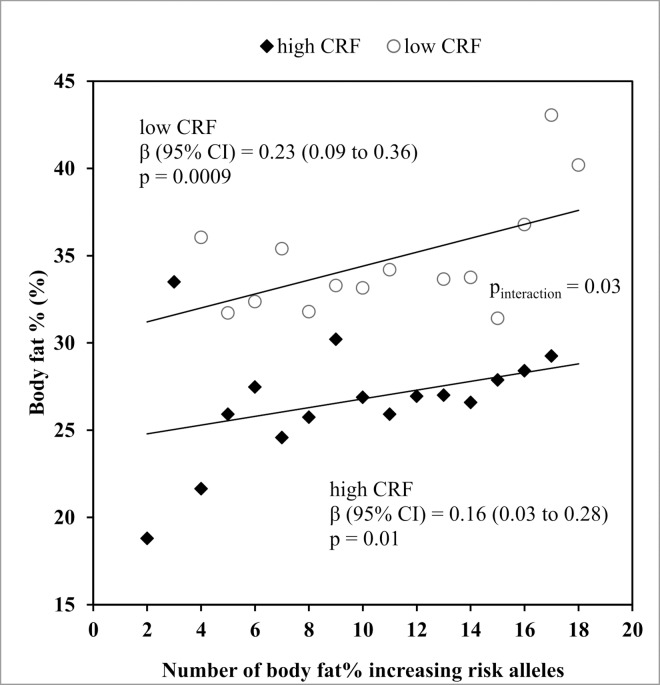
After stratifying the study participants of both cohorts by the respective median of VO2max to form “high CRF” and “low CRF” groups, we demonstrated that the magnitude of the effect of each additional body fat% increasing risk allele was 30% smaller in individuals with high CRF (β = 0.16 kg/m2) than in individuals with low CRF (β = 0.23 kg/m2). The test of interaction between CRF and body fat% GRS on body fat% showed the expected direction of effect but was not statistically significant after adjustment for multiple testing (βinteraction = -0.01, p = 0.03) (Fig 1).
Fig 1. Association between the body fat% GRS and body fat% in the low CRF and high CRF groups after meta-analysis of two Danish cohorts (ADDITION-PRO and Health2006, a total n = 3206).
Weighted mean body fat% for each number of body fat% risk alleles was calculated based on the relative weight for each cohort. CRF was objectively measured by step test and the study participants of both cohorts were stratified by the respective median of VO2max to form “high CRF” and “low CRF” groups. We tested for body fat% GRS x CRF interaction effects using linear models (Pinteraction = 0.03).
FTO association analysis
The minor FTO rs1558902 A allele was associated with higher body fat% (β = 0.62 kg/m2; p < 0.0001; age and sex adjusted) and we found that VO2max associated with body fat% (p < 0.0001). The body fat%-increasing FTO allele was associated with a 0.42 mL/kg/min unit lower VO2max per allele (p = 0.009, adjusted for age and sex). The association was abolished when additional adjustment for BMI (p = 0.58) was done. FTO was also significantly associated with VO2maxFM but not with VO2maxFFM (Table 3).
Table 3. Associations between the FTO rs1558902 A-allele and VO2max (ml/kg/min), VO2maxFM (ml/kg FFM/min) as well as VO2maxFFM (ml/kg FM/min) after meta-analysis of two Danish cohorts (ADDITION-PRO (n = 716) and Health2006 (n = 2586)), assuming a fixed effect model.
| Covariates | Effect | SE (range) | P | |
|---|---|---|---|---|
| VO2max (ml/kg/min) | mage, sex | -0.42 | (-0.74 to -0.09) | 0.0092 |
| VO2max (ml/kg/min) | age, sex, BMI | -0.11 | (-0.40 to 0.18) | 0.46 |
| VO2max (ml/kg/min) | age, sex, fat-free mass | -0.22 | (-0.54 to 0.09) | 0.17 |
| VO2maxFFM (ml/kg FFM/min) | age, sex | -0.12 | (-0.53 to 0.30) | 0.58 |
| VO2maxFFM (ml/kg FFM/min) | age, sex, BMI | -0.07 | (-0.47 to 0.34) | 0.74 |
| VO2maxFFM (ml/kg FFM/min) | age, sex, fat- mass | -0.004 | (-0.02 to 0.16) | 0.68 |
| VO2maxFM (ml/kg FM/min) | age, sex | -0.03 | (-0.05 to -0.01) | 0.00040 |
| VO2maxFM (ml/kg FM/min) | age, sex, BMI | -0.0019 | (-0.01 to 0.010) | 0.76 |
| VO2maxFM (ml/kg FM/min) | age, sex, fat-free mass | -0.01 | (-0.03 to 0.002) | 0.080 |
Associations between of the A- allele of FTO rs1558902 and CRF, expressed in VO2max (ml/kg/min), VO2maxFFM (ml/kg FFM/min) as well as VO2maxFM (ml/kg FM/min), were examined by linear regression assuming additive genetic models. Data for VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition. Study characteristics of ADDITION-PRO (n = 716) and Health2006 (n = 2586) according to FTO rs1558902 genotype can be found in S6 Table. Cohort specific association results can be found in S4 Table.
Discussion
In the present study, we found that additive genetic effects explain part of the variation in CRF and exposed a pleiotropic effect between genes contributing to VO2max and body fat, both when measured as % and in kg. A GRS based on 12 validated body fat% increasing genetic variants identified through GWAS, and the minor FTO rs1558902 allele alone, showed an association with decreased VO2max that was mediated by adiposity. Lastly, the interaction of VO2max × body fat%-associated GRS on body fat% showed the expected, direction of effect where fitness attenuated the effect of body fat%-associated loci on body fat%, but this result was non-significant after adjustment for multiple testing.
The estimated genetic component of BMI, body fat% and VO2max was within the range of previous reports [5–7]. We added another dimension to our study, namely expressing VO2max relative to fat-free mass (VO2maxFFM) and fat mass (VO2maxFM). While there are reports concluding that VO2max relative to fat-free mass is truly independent of adiposity and the best indirect estimate of metabolic capacity of the skeletal muscles [28, 29], we also calculated VO2max relative to fat mass to be able to distinguish between CRF scaled by body weight, fat-free mass and fat-mass. We found that VO2maxFM is more heritable compared to VO2maxFFM.
We found a significant negative genetic correlation between body fat (expressed in kg as well as %) and CRF but not between BMI or lean body mass and CRF. Body fat% is a more accurate measure of adiposity than is BMI. Body fat% distinguishes between fat mass and lean body mass, whereas BMI is a composite measure of fat mass and lean mass. Further, lean body mass is thought to be the body compartment explaining the major influence of body weight on VO2max[28]; yet, we did not find any pleiotropic genetic effects between VO2max (ml/kg/min) and lean body mass. The identified genetic correlation is indicative of shared genetics between the two traits (body fat% and VO2max) and may exist due to pleiotropy between both traits, meaning that the same genes are influencing both phenotypic traits or a pathway common for the phenotypes or due to gene-gene interactions, where the impact of one gene is influenced by another (set of) gene(s). We find that both scenarios are equally intriguing as these results are suggestive of a crosstalk between adipose tissue and the ability to consume oxygen in the lean body compartment, knowing that oxygen uptake in fat tissue is negligible.
In a comprehensive twin-study (n = 756 complete twin pairs) that explored genetic and environmental correlations between CRF and adiposity, it appeared that genetic, environmental as well as phenotypic influences linked greater CRF to lower levels of adiposity [30]. The authors of the same study proposed that there may be some genetic variants that contribute both to the propensity to develop adiposity and a preference for a sedentary lifestyle, resulting in reduced CRF, or resistance to formation of adiposity and preference for being physically active, resulting in an increased CRF [30]. Moreover, physical fitness and adiposity are related to inflammatory markers (such as cytokines, adipokines, C-reactive proteins, etc.) and it has been suggested that it may not be adiposity per se that mediates the association between fitness and inflammation status but instead that both, CRF and adiposity, may share the same causal pathways [31]. A further potential mechanism regulating body fat% and CRF through a common set of genes may be mediated via hypothalamic brain regions. At least three of the known body fat% increasing SNPs: FTO[8], TMEM18[8] and CRTC1[9] are thought to be expressed in hypothalamic brain regions, the brain region that regulates energy homeostasis. Studies in animals have shown that the control of voluntary movement, and therefore potentially CRF, resides in similar central neural pathways as energy intake [32, 33]. The central nervous system could therefore be an upstream region where body fat% increasing SNPs share a common biological influence on both adiposity and physical fitness phenotypes.
We then tested the effects of body fat% GRS on CRF in a meta-analysis of two Danish cohorts and demonstrated that each additional risk allele was associated with a 0.14 mL/min/kg decrease in VO2max, explaining 0.06% of the variance in VO2max in ADDITION-PRO and 0.2% in Health2006. We tested whether VO2maxFM and VO2maxFFM affect the body fat% GRS association differently and found that scaling CRF by fat-free mass abolished the association, whereas scaling CRF by fat mass seemed to signify our results (also after controlling for fat-free mass) as compared with the traditional scaling by body mass. Therefore, variants known to associate with body fat% may have no direct influence on CRF but expressing CRF relative to body mass or fat mass, both heavily influenced by adiposity, may cause the associations. Our approach to investigate the genetic overlap between body fat% and CRF in testing for associations between adiposity SNPs and CRF is inconclusive. Therefore, we suggest applying more refined techniques such as large-scale multivariate GWAS analysis in future studies, to be able to define the common set of genes that we hypothesize to be regulating oxygen uptake in muscle and body fat% simultaneously.
While we found an adiposity mediated association between CRF and body fat% GRS, it is unlikely that all of the gene variants comprising this GRS contribute to this effect. Individually, we tested whether FTO, the strongest known susceptibility locus for common obesity associates with VO2max and we found an inverse, adiposity-mediated association. The obesity risk gene FTO encodes a 2-oxoglutarate-dependent nucleic acid demethylase [34, 35], that is expressed in several peripheral tissues as well as in brain regions affecting energy balance [10, 36]. While previous studies tried to uncover a link between FTO and appetite regulation and the propensity to exercise controlled by the brain (reviewed in [10]), recent emerging data now points to a peripheral role for risk variants in the FTO locus in a pathway that is regulating adipocyte metabolism and controlling energy storage and energy dissipation [37, 38].
Participants of the ADDITION-PRO study were recruited by a stepwise screening procedure that identified Danes at a medium-to-high-risk scale of developing diabetes versus the Health2006 study population consisted of a randomly selected general population sample recruited through the Civil registration system from inhabitants in eleven municipalities in the Capital Region of Denmark. Individuals in ADDITION-PRO were overall less fit and older as compared to individuals in Health2006 and as such the two studies are not fully comparable. We speculate that the difference in age, health status and overall physical fitness across the cohorts could have diminished the effect of genetic variants known to associate with body fat% on VO2max therefore the effect may penetrate stronger in ADDITION-PRO. Our speculation of observed cohort differences due to physical fitness levels are in line with the study set up by Huuskonen et al. of healthy young males that did not show an association between the FTO obesity-linked variant and CRF. The cross-sectional study included 846 healthy young males and CRF was objectively assessed by a maximal bicycle ergometer test [14]. Even though a trend was observed, it is very likely that any effects of FTO variants on CRF was diminished by insufficient statistical power or overall fairly high baseline physical fitness levels throughout the study population. Ultimately, our findings are only generalizable to other populations with similar characteristics and therefore, our findings need replication in other independent study populations. We are aware that other factors that we could not control for in our analysis may have effects on estimates of CRF. We estimated that 49% of variation in CRF is heritable, but other determinants such as physical activity, dietary intake, disease status, biochemical measures, body composition and age are found to account for the remaining variation in CRF [39].
In ADDITION-PRO and Health2006, CRF was estimated by a submaximal step test [17, 21]. While submaximal tests do not provide accurate estimates of CRF at the individual level, the estimates are valid for studies in large populations. In a validation study, the correlation between the Danish step test and a maximal test of cardiorespiratory fitness was moderate to high, 0.77 in women and 0.69 in men, and the authors suggested the use of the Danish step test as a safe and feasible alternative to the more time-consuming watt-max test as a method for estimation of VO2max in large population-based studies [22]. Nonetheless, it is important to replicate our observations using direct measures of CRF in future studies.
In conclusion, our results suggest a shared genetic etiology between body fat% and CRF. If replicated in future studies, ideally with direct measures of maximal oxygen consumption and more accurate measures of body fat% such as DEXA, our findings suggest that fat mass may have an indirect effect on maximal oxygen uptake. However, polymorphisms known to associate with body fat% could not explain the shared genetics independent of adiposity with FTO having a potential pleiotropic effect.
Supporting Information
(XLSX)
An overview of the SNPs presented in Lu et al. and the SNPs investigated in the population-based Health2006 and ADDITION-PRO cohort and proxy (r2) when included in the analysis for Health2006. *Fat% increasing allele, EAFs (effect allele frequencies), other allele and effect sizes from the SNPs reported by Lu et al. 2015 for the sex-combined European-ancestry GWAS + ExomeChip analysis [9].
(XLSX)
Associations between BMI and body fat% GRS and CRF were examined by linear regression using additive genetic models. Data for VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition.
(XLSX)
Data are given in means ± standard error (SE) and median (interquartile ranges). Associations between FTO-rs1558902 and CRF were examined by linear regression using additive genetic models. VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition.
(XLSX)
ACE modelling is used to indicate what proportion of variance in a trait is contributed to its additive genetic effects (h2). h2 (SE) = additive genetic effect. FM = fat mass, FFM = fat-free mass.
(XLSX)
Data are given in means ± standard error (SE) and median (interquartile ranges).
(XLSX)
Acknowledgments
The authors thank A. Forman, B. Andersen and G.J. Klavsen from the Novo Nordisk Foundation Center for Basic Metabolic Research for their dedicated and careful technical assistance and G. Lademann for secretarial support. The project was supported by the Danish Diabetes Academy supported by the Novo Nordisk Foundation, the research programme "Governing Obesity" funded by the University of Copenhagen Excellence Programme for Interdisciplinary Research (www.go.ku.dk) as well as the Danish Medical Research Council. The ADDITION-PRO study was funded by an unrestricted grant from the European Foundation for the Study of Diabetes/Pfizer for Research into Cardiovascular Disease Risk Reduction in Patients with Diabetes (74550801), by the Danish Council for Strategic Research and by internal research and equipment funds from Steno Diabetes Center. The work of SB was funded by the UK Medical Research Council (MC_UU_12015/3). The Health2006 study was financially supported by grants from the Velux Foundation; the Danish Medical Research Council, Danish Agency for Science, Technology and Innovation; the Aase and Ejner Danielsens Foundation; ALK-Abello´ A/S (Hørsholm, Denmark), Timber Merchant Vilhelm Bangs Foundation, MEKOS Laboratories (Denmark) and Research Centre for Prevention and Health, the Capital Region of Denmark. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk).
Data Availability
Relevant data for the present study are within the paper and its Supporting Information files. If you wish to see additional data, the authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Data is available from the Novo Nordisk Foundation Center for Basic Metabolic Research, section of Metabolic Genetics whose authors may be contacted at torben.hansen@sund.ku.dk.
Funding Statement
The project was supported by the Danish Diabetes Academy supported by the Novo Nordisk Foundation, the research programme "Governing Obesity" funded by the University of Copenhagen Excellence Programme for Interdisciplinary Research (www.go.ku.dk) as well as the Danish Medical Research Council. The ADDITION-PRO study was funded by an unrestricted grant from the European Foundation for the Study of Diabetes/Pfizer for Research into Cardiovascular Disease Risk Reduction in Patients with Diabetes (74550801), by the Danish Council for Strategic Research and by internal research and equipment funds from Steno Diabetes Center. The work of SB was funded by the UK Medical Research Council (MC_UU_12015/3). The Health2006 study was financially supported by grants from the Velux Foundation; the Danish Medical Research Council, Danish Agency for Science, Technology and Innovation; the Aase and Ejner Danielsens Foundation; ALK-Abello´ A/S (Hørsholm, Denmark), Timber Merchant Vilhelm Bangs Foundation, MEKOS Laboratories (Denmark) and Research Centre for Prevention and Health, the Capital Region of Denmark. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk).
References
- 1.Blair SN, Brodney S. Effects of physical inactivity and obesity on morbidity and mortality: current evidence and research issues. Medicine and science in sports and exercise. 1999;31:S646–S62. [DOI] [PubMed] [Google Scholar]
- 2.Blair SN, Church TS. The fitness, obesity, and health equation: is physical activity the common denominator? Jama. 2004;292(10):1232–4. 10.1001/jama.292.10.1232 [DOI] [PubMed] [Google Scholar]
- 3.Blair SN, Cheng Y, Holder JS. Is physical activity or physical fitness more important in defining health benefits? Medicine and science in sports and exercise. 2001;33(6; SUPP):S379–S99. [DOI] [PubMed] [Google Scholar]
- 4.LaMonte MJ, Blair SN. Physical activity, cardiorespiratory fitness, and adiposity: contributions to disease risk. Current Opinion in Clinical Nutrition & Metabolic Care. 2006;9(5):540–6. [DOI] [PubMed] [Google Scholar]
- 5.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behavior genetics. 1997;27(4):325–51. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard C, Pérusse L, Leblanc C, Tremblay A, Thériault G. Inheritance of the amount and distribution of human body fat. International journal of obesity. 1987;12(3):205–15. [PubMed] [Google Scholar]
- 7.Bouchard C, Daw EW, Rice T, Pérusse L, Gagnon J, Province MA, et al. Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Medicine and science in sports and exercise. 1998;30(2):252–8. [DOI] [PubMed] [Google Scholar]
- 8.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Bishop D. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nature Communications. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loos RJ, Yeo GS. The bigger picture of FTO [mdash] the first GWAS-identified obesity gene. Nature Reviews Endocrinology. 2014;10(1):51–61. 10.1038/nrendo.2013.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilpeläinen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS medicine. 2011;8(11):1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. 10.2337/db07-0910 [DOI] [PubMed] [Google Scholar]
- 13.Rankinen T, Rice T, Teran‐Garcia M, Rao DC, Bouchard C. FTO Genotype Is Associated With Exercise Training–induced Changes in Body Composition. Obesity. 2010;18(2):322–6. 10.1038/oby.2009.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huuskonen A, Lappalainen J, Oksala N, Santtila M, Häkkinen K, Kyröläinen H, et al. Aerobic fitness does not modify the effect of FTO variation on body composition traits. 2012. [DOI] [PMC free article] [PubMed]
- 15.Gjesing AP, Hornbak M, Allin KH, Ekstrøm CT, Urhammer SA, Eiberg H, et al. High heritability and genetic correlation of intravenous glucose-and tolbutamide-induced insulin secretion among non-diabetic family members of type 2 diabetic patients. Diabetologia. 2014;57(6):1173–81. 10.1007/s00125-014-3207-y [DOI] [PubMed] [Google Scholar]
- 16.Åstrand P-O, Ryhming I. A nomogram for calculation of aerobic capacity (physical fitness) from pulse rate during submaximal work. Journal of applied physiology. 1954;7(2):218–21. [DOI] [PubMed] [Google Scholar]
- 17.Johansen NB, Hansen A-LS, Jensen TM, Philipsen A, Rasmussen SS, Jørgensen ME, et al. Protocol for ADDITION-PRO: a longitudinal cohort study of the cardiovascular experience of individuals at high risk for diabetes recruited from Danish primary care. BMC public health. 2012;12(1):1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brage S, Brage N, Franks P, Ekelund U, Wareham N. Reliability and validity of the combined heart rate and movement sensor Actiheart. European journal of clinical nutrition. 2005;59(4):561–70. 10.1038/sj.ejcn.1602118 [DOI] [PubMed] [Google Scholar]
- 19.Brage S, Ekelund U, Brage N, Hennings MA, Froberg K, Franks PW, et al. Hierarchy of individual calibration levels for heart rate and accelerometry to measure physical activity. Journal of Applied Physiology. 2007;103(2):682–92. 10.1152/japplphysiol.00092.2006 [DOI] [PubMed] [Google Scholar]
- 20.Tanaka H, Monahan KD, Seals DR. Age-predicted maximal heart rate revisited. Journal of the American College of Cardiology. 2001;37(1):153–6. [DOI] [PubMed] [Google Scholar]
- 21.Thuesen BH, Cerqueira C, Aadahl M, Ebstrup JF, Toft U, Thyssen JP, et al. Cohort Profile: The Health2006 cohort, Research Centre for Prevention and Health. International journal of epidemiology. 2014;43(2):568–75. 10.1093/ije/dyt009 [DOI] [PubMed] [Google Scholar]
- 22.Aadahl M, Zacho M, Linneberg A, Thuesen BH, Jørgensen T. Comparison of the Danish step test and the watt-max test for estimation of maximal oxygen uptake: the Health2008 study. European journal of preventive cardiology. 2013;20(6):1088–94. 10.1177/2047487312462825 [DOI] [PubMed] [Google Scholar]
- 23.Consortium GP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. The American Journal of Human Genetics. 1998;62(5):1198–211. 10.1086/301844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjesing AP, Ribel-Madsen R, Harder MN, Eiberg H, Grarup N, Jørgensen T, et al. Genetic and phenotypic correlations between surrogate measures of insulin release obtained from OGTT data. Diabetologia. 2015;58(5):1006–12. 10.1007/s00125-015-3516-9 [DOI] [PubMed] [Google Scholar]
- 27.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, De Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. 10.1093/bioinformatics/btn564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goran M, Fields D, Hunter G, Herd S, Weinsier R. Total body fat does not influence maximal aerobic capacity. International journal of obesity. 2000;24(7):841–8. [DOI] [PubMed] [Google Scholar]
- 29.Hinriksdóttir G, Tryggvadóttir Á, Ólafsdóttir AS, Arngrímsson SÁ. Fatness but not fitness relative to the fat-free mass is related to C-reactive protein in 18 year-old adolescents. PLoS One. 2015;10(6):e0130597 10.1371/journal.pone.0130597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson W, de Ruiter I, Kyvik KO, Murray AL, Sørensen TI. Genetic and environmental transactions underlying the association between physical fitness/physical exercise and body composition. Behavior genetics. 2015;45(1):84–105. 10.1007/s10519-014-9690-6 [DOI] [PubMed] [Google Scholar]
- 31.Hamer M. The relative influences of fitness and fatness on inflammatory factors. Preventive medicine. 2007;44(1):3–11. 10.1016/j.ypmed.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 32.Knab AM, Bowen RS, Hamilton AT, Gulledge AA, Lightfoot JT. Altered dopaminergic profiles: implications for the regulation of voluntary physical activity. Behavioural brain research. 2009;204(1):147–52. 10.1016/j.bbr.2009.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knab AM, Lightfoot JT. Does the difference between physically active and couch potato lie in the dopamine system? International journal of biological sciences. 2010;6(2):133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72. 10.1126/science.1151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC biochemistry. 2007;8(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stratigopoulos G, Padilla SL, LeDuc CA, Watson E, Hattersley AT, McCarthy MI, et al. Regulation of Fto/Ftm gene expression in mice and humans. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2008;294(4):R1185–R96. 10.1152/ajpregu.00839.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–5. 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. New England Journal of Medicine. 2015;373(10):895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laukkanen JA, Laaksonen D, Lakka TA, Savonen K, Rauramaa R, Mäkikallio T, et al. Determinants of cardiorespiratory fitness in men aged 42 to 60 years with and without cardiovascular disease. The American journal of cardiology. 2009;103(11):1598–604. 10.1016/j.amjcard.2009.01.371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
An overview of the SNPs presented in Lu et al. and the SNPs investigated in the population-based Health2006 and ADDITION-PRO cohort and proxy (r2) when included in the analysis for Health2006. *Fat% increasing allele, EAFs (effect allele frequencies), other allele and effect sizes from the SNPs reported by Lu et al. 2015 for the sex-combined European-ancestry GWAS + ExomeChip analysis [9].
(XLSX)
Associations between BMI and body fat% GRS and CRF were examined by linear regression using additive genetic models. Data for VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition.
(XLSX)
Data are given in means ± standard error (SE) and median (interquartile ranges). Associations between FTO-rs1558902 and CRF were examined by linear regression using additive genetic models. VO2maxFM was log-transformed. Models were adjusted for age and sex or age, sex and body composition.
(XLSX)
ACE modelling is used to indicate what proportion of variance in a trait is contributed to its additive genetic effects (h2). h2 (SE) = additive genetic effect. FM = fat mass, FFM = fat-free mass.
(XLSX)
Data are given in means ± standard error (SE) and median (interquartile ranges).
(XLSX)
Data Availability Statement
Relevant data for the present study are within the paper and its Supporting Information files. If you wish to see additional data, the authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Data is available from the Novo Nordisk Foundation Center for Basic Metabolic Research, section of Metabolic Genetics whose authors may be contacted at torben.hansen@sund.ku.dk.