Abstract
In the age of personalized medicine stratifying tumors into molecularly defined subtypes associated with distinctive clinical behaviors and predictable responses to therapies holds tremendous value. Towards this end, we developed a custom microfluidics-based bladder cancer gene expression panel for characterization of archival clinical samples. In silico analysis indicated that the content of our panel was capable of accurately segregating bladder cancers from several public datasets into the clinically relevant basal and luminal subtypes. On a technical level, our bladder cancer panel yielded robust and reproducible results when analyzing formalin-fixed, paraffin-embedded (FFPE) tissues. We applied our panel in the analysis of a novel set of 204 FFPE samples that included non-muscle invasive bladder cancers (NMIBCs), muscle invasive disease (MIBCs), and bladder cancer metastases (METs). We found NMIBCs to be mostly luminal-like, MIBCs to include both luminal- and basal-like types, and METs to be predominantly of a basal-like transcriptional profile. Mutational analysis confirmed the expected enrichment of FGFR3 mutations in luminal samples, and, consistently, FGFR3 IHC showed high protein expression levels of the receptor in these tumors. Our bladder cancer panel enables basal/luminal characterization of FFPE tissues and with further development could be used for stratification of bladder cancer samples in the clinic.
Introduction
Bladder cancer is the fifth most common malignancy worldwide, with close to 400,000 newly diagnosed cases and ~150,000 associated deaths per year [1, 2]. Approximately 75% of patients present with low grade, non-muscle-invasive bladder cancers (NMIBCs) at the time of initial diagnosis [3–5]. NMIBCs recur in ~50% of instances and can progress in ~30% of cases to more serious muscle-invasive bladder cancers (MIBCs) [3, 4]. Recurrence risk factors include tumor histopathological features such as tumor stage and grade [3, 4]. The presence of high risk histological characteristics often prompts physicians to recommend more aggressive therapeutic intervention, including radical cystectomy and adjuvant therapy [3, 4]. MIBCs (T2 and T3) constitute ~15% of new cases and they are more likely than NMIBCs to develop into metastatic bladder cancers (METs) (pT4) [3–5]. Although METs represents <10% of newly diagnosed cases, they are associated with a poor 5-year survival likelihood of ~15% and represent a major unmet medical need [5–9].
Transcriptionally defined classes of bladder cancers were identified and have been shown to be associated with distinctive clinical behaviors and responses to therapies [10–15]. Accordingly, transcriptional characterization of bladder cancers has the potential to complement histopathological assessment in guiding treatment decisions. Recently, Choi et. al. described subsets of MIBCs that were transcriptionally reminiscent of breast cancers, including a luminal subtype characterized by a high frequency of activating FGFR3 mutations and a favorable outcome, and a basal subtype that exhibited a low frequency of FGFR3 mutations and was associated with a poor prognosis [14]. Consistent with these findings, Damrauer and colleagues reported on luminal and basal transcriptionally defined subtypes of primary MIBCs that were associated with distinct clinical outcomes [10]. The significant progress in our molecular understanding of bladder cancer offers a unique opportunity to classify tumors into the clinically actionable basal and luminal subtypes. However, RNA derived from FFPE samples is of much lower quality than that obtained from frozen samples, which are typically used to derive transcriptional classifiers. Thus, a specially designed gene expression panel that is optimized for robust transcriptional characterization of archival tissues could have high clinical utility. Furthermore, basal/luminal features have only been studies in MIBCs and NMIBCs [10, 14, 16, 17], and no studies have been conducted to date on the basal/luminal characteristics of METs.
In this study, we introduce a novel microfluidics-based panel that is custom designed to measure the expression of bladder cancer-relevant genes in FFPE tissues. In silico, we demonstrate that the content of our panel is capable of distinguishing between basal and luminal bladder cancers. We employ our panel in the analysis of FFPE tissues from a novel cohort of 204 tumors and show that NMIBCs from our sample set are mostly luminal-like, carry FGFR3 mutations at high frequency, and express high protein levels of the receptor tyrosine kinase. On the other hand, MIBCs from our cohort are mixed with respect to luminal- and basal-like status, and the expression patterns seen in METs are consistent with a more basal-like transcriptional state.
Results
Development and in silico validation of a microfluidics-based panel for transcriptional characterization of archival bladder cancer tissues
Here, we introduce a microfluidics-based bladder cancer gene expression panel that is optimized for the analysis of FFPE tissues. Our panel is comprised of 90 unique genes that were selected to capture key attributes of bladder cancer biology [18–31], as well as two housekeeping genes for data normalization (Figs 1A and S1, S1 Table). The genes include components of receptor tyrosine kinase (RTK) pathways such as FGFR, ERBB, and MET, the PI3K/AKT and MAPK axes, cell cycle and genome stability genes like TP53, genes involved in the regulation of cell differentiation and development, and epithelial-mesenchymal transition (EMT) genes [18–31] (Figs 1A and S1, S1 Table).
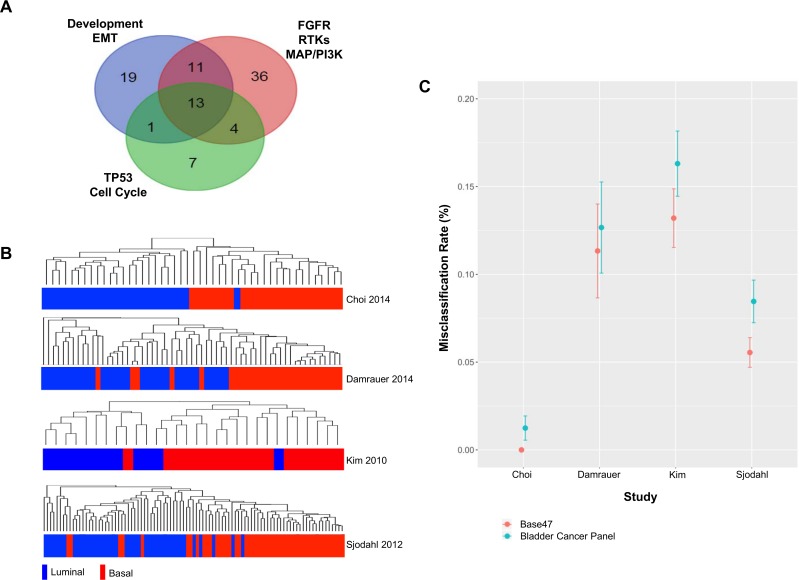
Fig 1. Development and in silico validation of a bladder cancer gene expression panel for characterization of FFPE tissues.
(A) Venn diagram of genes comprising a novel bladder cancer panel illustrating the number overlapping (and unique) genes belonging to three main pathway groups based on Ingenuity® analysis: 1) FGFR, RTK, MAPK, and PI3K pathways; 2) development and EMT axes; 3) TP53, genome stability, and cell cycle regulation networks. (B) Hierarchical clustering of samples from four public datasets based on the signals from probes corresponding to genes on the bladder panel, and corresponding published basal/luminal status. (C) Basal/luminal misclassification rates represented as % of all cases in four public datasets comparing calls made by the BASE47 gene signature from literature to assignments made based on the expression of genes on the bladder panel.
Given the biological and clinical relevance of the recently described basal and luminal subtypes of bladder cancer [10, 14], we carried out an in silico assessment of the ability of the genes on our panel to distinguish between these two molecular subtypes in public datasets. We used samples from four public data sets that were designated as basal or luminal by Choi et. al. [14] or through BASE47 scoring by Damrauer, et. al. [10] and hierarchically clustered samples based on the expression of probes corresponding to genes on our panel (Fig 1B). The content of our panel was able to correctly segregate samples based on luminal and basal status from the Choi discovery dataset [14] in 23/24 (96%) and 23/23 (100%) of instances, respectively (Fig 1B, Table 1). Furthermore, based on our genes we could accurately separate specimens from the Damrauer discovery set [10] into luminal and basal subtypes in 33/33 (100%) and 23/28 (82%) of the cases, respectively (Fig 1B, Table 1). Finally, the content of our panel correctly distinguished between 10/12 (83%) of the luminal and 17/18 (94%) of the basal tumors from the Kim cohort [12], and 39/44 (80%) of the luminal and 42/47 (89%) of the basal samples from the Sjodhal dataset [13], as determined by Damrauer using BASE47 classification [10] (Fig 1B, Table 1). Interestingly, only 1/90 and 23/90 genes from our panel overlapped with the BASE47 Damerauer [10] and Choi [14] basal/luminal signatures, respectively (S2 Table, data not shown).
Table 1. Basal/luminal misclassification rates in public datasets using bladder cancer panel and BASE47 gene sets.
| Public dataset | GEO accession | Sample # | # of samples with available basal/luminal calls | % of basal/luminal samples correctly classified by bladder cancer panel | BASE47 misclass rate +/- SD | Bladder panel misclass rate +/- SD |
|---|---|---|---|---|---|---|
| Choi et. al., 2014 (Discovery set) | GSE48075 | 73 | 23/24 | 100%/96% | 0.000+/-0.000 | 0.008+/-0.006 |
| Damrauer et. al., 2014 | GSE5287 | 30 | 18/12 | 82%/100% | 0.107+/-0.027 | 0.140+/-0.034 |
| Sjodahl et. al., 2012 | GSE32894 | 308 | 49/44 | 94%/83% | 0.057+/-0.009 | 0.082+/-0.017 |
| Kim et al., 2010 | GSE13507 | 165 | 28/33 | 89%/80% | 0.133+/-0.013 | 0.156+/-0.020 |
| Damrauer et. al., 2014 | GSE5287 | 30 | 18 / 12 | 82% / 100% | 0.107 +/- 0.027 | 0.140 +/- 0.034 |
Next, we further evaluated the ability of the genes on our panel to distinguish between basal and luminal samples by performing classifier analysis and used cross-validation to calculate the average misclassification rates in assigning the correct basal/luminal status. More specifically, for a given literature data set samples were divided into training and test sets. A diagonal linear discriminant analysis (DLDA) classifier [32] was trained to distinguish between basal and luminal samples in the training samples based on all bladder cancer panel genes. The classifier was then used to predict whether the remaining test samples were most likely to be basal or luminal. These predictions were compared to the assigned basal/luminal status by Choi et. al. [14] and Damrauer and colleagues [10], and this information was used to calculate the basal/luminal misclassification rate. Random permutation in sample assignments to test and training sets was repeated until all samples had been included as a test sample one time. Classifier analysis was performed for each public data set separately. Using this approach, we observed similar basal/luminal misclassification error rates for our bladder cancer panel compared to assignments made using the BASE47 gene set in the Choi discovery samples [14] (Fig 1C, Table 1). We also observed comparable basal/luminal misclassification rates for the Damraurer samples [10], as well as for samples from the Sjodhal [13] and Kim [12] data sets (Fig 1C, Table 1). These results indicate that the content of our panel is capable of classifying MIBCs into basal and luminal subtypes with similar accuracy to that obtained by the BASE47 gene set.
Our initial cross-validation classifier analysis demonstrated that the full content of our panel allowed for accurate classification of samples from four different public datasets into basal and luminal subtypes (Fig 1C, Table 1). However, the panel contains genes related to diverse bladder cancer pathways not all of which may be important in distinguishing between basal and luminal cancers. We investigated whether subsets of the genes on our panel could inform basal/luminal status in the public datasets. We used a gene ranking system based on Welch’s t-test and incorporated a gene selection step as part of the cross-validation process where only the top 10, 20, 30, 40, 50, 60, 70 and 80 most differentially expressed genes between basal and luminal samples were included in the classifier. We found that as few as 20 genes could classify samples with similar misclassification rates compared the full panel (S2 Fig). This observation was consistent across all four public data sets (S2 Fig). Using the entire set of samples from each of the four public datasets, we then calculated the Welch’s t-test statistic and p-value significance levels for all genes on our panel. This allowed us to rank all the genes on our panel and select a smaller subset of genes that were significantly differentially expressed between basal and luminal bladder cancer for future studies (S2 Table).
We next explored if the genes on our panel could classify cancers into disease-relevant subtypes beyond the basal and luminal groups. The cohort described by Choi et. al. [14] contained basal and luminal MIBCs as well as tumors that exhibited a p53-like signature that were resistant to frontline chemotherapy based on transcriptional data from frozen tissues [14]. Hierarchical clustering of samples from the Choi [14] cohort demonstrated that the expression of genes on our panel was able to correctly classify 20/24 (83%) luminal, 22/23 (96%) basal, and 17/26 (65%) p53-like samples (S3 Fig).
The findings from our in silico validation analysis suggest that the genes on our panel are capable of classifying tumors into basal and luminal subtypes with similar accuracy to the public classifiers, which were specifically developed to differentiate between basal and luminal samples. Furthermore, our panel was capable of differentiating between p53-like and basal/luminal bladder cancer samples, albeit without the same degree of accuracy as when distinguishing between basal and luminal samples. Thus, we focused on applying our panel in basal/luminal characterization of bladder cancers.
Technical validation of the bladder cancer panel
To assess the robustness of our panel and its reproducibility in measuring gene expression we conducted a series of quality control experiments. First, we carried out serial dilutions on high quality universal RNA (uRNA) to evaluate the performance of each of the assays, redesigning primers when necessary, to achieve linear standard dilution curves for all tests (S4 Fig). During this process, the FGF23, ERBB3, USP7, BMP2, DUSP3, SRC, CSK, MTOR, GLI1, and MET assays did not yield satisfactory standard dilution curves the first time and were re-designed for optimal performance. To assess performance of the QRT-PCR assays on our panel, we ran in triplicate high quality uRNA control samples and lower quality RNA derived from 204 FFPE clinical tissues and counted the number of times an assay failed to produce a measurable signal above background (Fig 2A). Five assays (RPS6, KDR, PTEN, AKT1, CCND2) had at least 100 failures over 664 attempted measurements (>15% failure rate) (Fig 2A). However, even for RPS6, which was the poorest performing of these five assays, we observed measurable signal above background in 319/664 attempts (48% success rate). Notably, the MYC assay had 635/664 failures (96% failure rate) and was the poorest performing assay on the panel (Fig 2A).
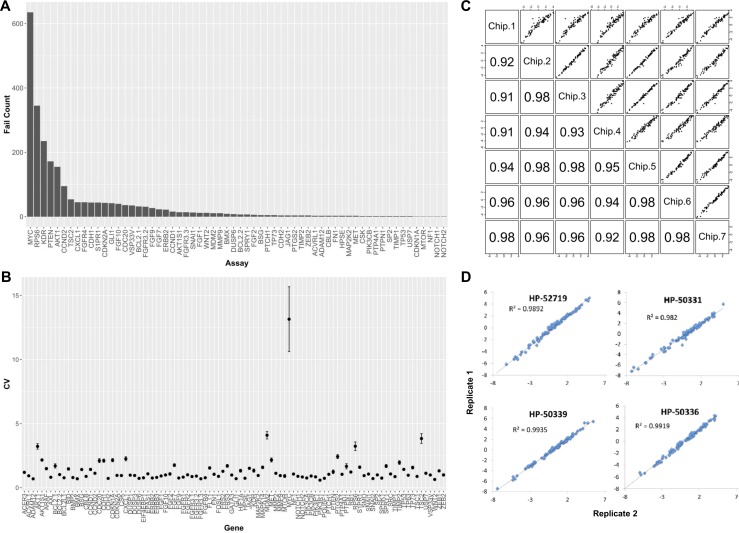
Fig 2. Technical validation of the bladder cancer panel.
(A) Bar chart reflecting the failure counts for each of the assays on the bladder cancer panel, as determined by the number of times an assay failed to produce a measurable signal above background in 664 attempted measurements. (B) CV calculations from triplicate experimental measurements using standard deviation over the mean expression values for each of the assays on the panel. (C) Chip-to-chip data reproducibility for high quality control uRNA samples from different runs. (D) Run to run data reproducibility for archival tissues, as seen for two representative FFPE-derived RNA samples run on different days.
Next, we evaluated assay variability by calculating coefficient of variation (CV) over the three replicate measurements that were obtained from our quality control experiments for each assay and sample combination. CV was calculated as the ratio of standard deviation over the mean expression measurement. The average CV for all assays on the panel was under 5% except for the MYC assay, which had an average CV of 13% when analyzing cell line- and FFPE tissue-derived RNA (Fig 2B). Notably, a CV value of 22% was observed for the MYC test when analyzing high quality uRNA samples (S5 Fig), suggesting that the poor performance was an assay- rather than a RNA quality-related issue. Because of the high failure rate and high CV values obtained with the MYC assay, this test was dropped from all subsequent analyses.
We also assessed data reproducibly when running high quality uRNA and lower quality RNA from FFPE tissues on different runs from different days. We noted a high degree of chip-to-chip data reproducibility for each gene on our panel when we ran uRNA control samples as part of seven independent runs (Fig 2C, R2 range = 0.91–0.98). Furthermore, we ran replicate FFPE-derived RNA samples on different days and observed a high degree of data concordance for each of the genes on the panel (Fig 2D, R2 range = 0.98–0.99). The results from our qualitative and quantitative assessments suggest that all assays on the panel, with the exception of the MYC test, were robust and yielded reproducible data when analyzing both high quality uRNA and lower quality RNA from FFPE tissues.
Application of the bladder cancer panel in the basal/luminal characterization of FFPE tissues from a novel clinical cohort
To begin to assess the utility of our panel in the basal/luminal characterization of FFPE tissues, we analyzed a set of 204 samples comprised of NMIBCs, MIBCs, as well as lymph node and distal METs (Table 2, S3 Table). We first examined how our samples compared to those from several public datasets with respect to basal/luminal transcriptional features. As expected and described earlier in Fig 1B, hierarchical clustering of samples from the four public datasets [10, 12–14] using median centered expression from probes corresponding to genes on our panel revealed a clear separation between basal and luminal samples (S6A Fig). Notably, luminal samples from the Kim dataset clustered with the basal samples from the three other datasets and were outliers in this analysis (S6A Fig). MIBCs from our cohort clustered alongside basal samples from all four public datasets (S6A Fig), consistent with a basal-like transcriptional profile. In our dataset, most NMIBCs clustered with luminal samples and, thus, were likely to be luminal-like in nature (S6A Fig). To our knowledge, there have been no previous reports on the basal/luminal features of METs. METs from our cohort clustered with basal samples from the public datasets (S6A Fig).
Table 2. Clinicopathologic features of a novel bladder cancer cohort.
| Number | Percentage | |
|---|---|---|
| Age (at diagnosis) | ||
| Median | 65 | |
| Range | 35–91 | |
| Standard Deviation | 11.51 | |
| Gender | ||
| Male | 164 | 79% |
| Female | 43 | 21% |
| Tissue Specimens | ||
| Primary Bladder Cancer (T0-T4) | 154 | 75.5% |
| Non-Muscle-Invasive(T0, T1, Tis) | 93 | 45.5% |
| Muscle-Invasive (T2, T3) | 61 | 30.0% |
| Metastatic Bladder Cancer (T4) | 53 | 26.0% |
| Lymph Nodes | 45 | 22.0% |
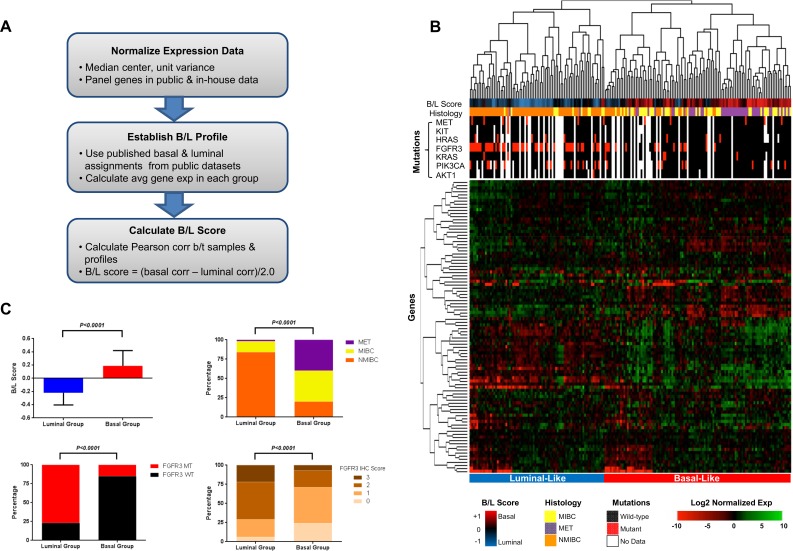
We next established a method for calculating basal/luminal similarity scores using transcriptional signatures from our FFPE samples. We began by median-centering the expression of all probes corresponding to the genes on our panel in public datasets and calculated an average gene expression value in basal and luminal groups, as assigned by Choi and Damrauer and colleagues [10, 14] (Figs 3A and S6B). This allowed us to obtain an average view of the expression of each of these genes in the luminal and basal groups from the public datasets (S6B Fig). For example, FGFR3 had the highest average expression value in luminal samples than any other gene on our panel, and the average FGFR3 expression level was one of the lowest in basal samples from the public datasets (S6B Fig). We then derived basal/luminal (B/L) similarity scores for each of the 204 FFPE samples by calculating their correlation to the public basal and luminal profiles (Fig 3A). Hierarchical clustering of our FFPE samples revealed two main branches with distinct patterns of gene expression: 1) a right branch under which the majority of luminal-like samples with low B/L similarity scores were co-clustered; and 2) a left branch that was comprised mostly of basal-like samples with high B/L scores (Fig 3B). Statistical analysis confirmed a significantly lower B/L score for samples under the left branch of the clustergram, which is consistent with luminal-like status, and a significantly higher B/L score consistent with basal-like status for specimens under the right arm of the hierarchical tree (Fig 3B and 3C, top left panel; P<0.001). We observed a significantly higher fraction of NMIBCs in the luminal-like compared to the basal-like group (Fig 3B and 3C, top right panel; P = 0.0001), consistent with the notion that NMIBCs from our cohort are mostly luminal-like in nature. MIBCs were represented in both basal- and luminal-like groups; however, a significantly higher proportion of MIBCs from our series was observed in the basal-like group (Fig 3B and 3C, top right panel; P = 0.0001). Interestingly, almost all METs clustered under the basal-like arm of the hierarchical tree, suggesting that the majority of the METs in our cohort exhibited a basal transcriptional profile (Fig 3B and 3C, top right panel; P = 0.0001).
Fig 3. Molecular characterization of a novel cohort of FFPE tissues using the bladder cancer panel.
(A) Methodology used for computing basal/luminal signatures from public data and then applying these signatures to bladder panel data to determine basal/luminal similarity of samples from a novel clinical cohort. (B) Hierarchical clustering (average-linkage, 1 –Pearson correlation distance metric) of 204 FFPE samples based on bladder cancer panel gene expression and corresponding B/L scores, histology, and mutational status of cancer-relevant genes. (C) Statistical analysis of the B/L scores (top left panel), distribution of NMIBCs, MIBCs, and METs (top right panel), prevalence of FGFR3 mutations (bottom left), and FGFR3 IHC scores (bottom right) in samples from the transcriptionally-defined luminal- and basal-like clusters in Fig 3B.
One of the characteristic features of NMIBCs is that they carry somatic activating mutations in FGFR3 in ~60–80% of the cases, and a much lower frequency of ~15% has been reported in MIBCs and METs [19, 25, 27, 33–37]. We assessed the mutational status of FGFR3 and other cancer-relevant genes in samples from cohort using a custom allele-specific PCR panel [19, 33, 34, 36–38]. We found that >75% of the tumors that co-clustered under the luminal-like label carried FGFR3 mutations, a significantly higher fraction than the <20% FGFR3 mutation rate we observed in samples from the basal-like group (Fig 3C, bottom left panel; P<0.0001). Mutations in FGFR3 have been reported to drive higher expression levels of the RTK [19]. Consistent with this finding, we noted significantly higher levels of FGFR3 protein, as measured by IHC, in samples from the luminal-like compared to the basal-like groups (Fig 3C, bottom right panel; P<0.0001). We did not observe significant associations between basal- or luminal-like status and any of the other mutations on our panel (Fig 3B). These findings suggest that our bladder cancer panel can classify FFPE samples into luminal- and basal-like groups. The enrichment of FGFR3 mutations and up-regulation of the RTK on the protein level that we observed in the luminal-like samples and the absence of these features in basal-like samples provides molecular confirmation of the basal/luminal nature of the samples in our cohort.
Discussion
Transcriptional analyses have provided valuable insights into the molecular underpinnings of bladder cancer [11–15, 31, 39, 40]. However, the majority of these studies relied on the use of frozen tissues that yield high quality nucleic acids and are not well-suited for the characterization of poor quality RNA from FFPE samples. Our microfluidics-based bladder cancer panel is custom designed for robust transcriptional characterization of FFPE tissue-derived RNA.
An intriguing finding in the bladder cancer field was that MIBCs were reminiscent of the basal and luminal classes of breast cancers, and that they defined different disease subtypes that were associated with distinct clinical outcomes [10, 14]. The genes on our panel were able to accurately segregate samples from four public datasets into basal and luminal groups [10, 12–14], pointing to the possibility of using our platform for detection of basal/luminal status in FFPE tissues from the clinic. The content of our panel performed as well as the BASE47 classifier [10] in stratifying bladder cancers into basal and luminal subtypes. Cross-validated misclassification rates were comparable between our panel and the BASE47 classifier for all four public datasets tested and ranged from 0–16%.
There was minimal overlap between the genes on our panel and those from the public basal/luminal signatures [10, 14]. This suggests that the basal/luminal features of bladder cancers include broad-ranging differences in gene expression beyond those described by Damrauer and Choi et. al. [10, 14]. The content of our panel provides an additional advantage in that it measures the expression of genes belonging to bladder-cancer relevant pathways that the public signatures might not include. As such, it may provide a preliminary assessment of these pathways in archival clinical samples. Our panel includes genes that are relevant in indications beyond bladder cancer. Future studies may be warranted to assess the ability of these genes to distinguish between basal/luminal tumors from other indications. Although our bladder panel performed well when classifying bladder cancers into basal/luminal subtypes, it was not as robust in identifying P53-like tumors. Therefore, we recommend using our panel for the basal/luminal but not p53-like characterization of bladder cancers. Given the unique intrinsic features of p53-like tumors, the stability of the p53-like subtype should be examined in additional cohorts in future studies.
On a technical level, 85/90 assays on our panel performed robustly when analyzing high quality uRNA and lower quality RNA derived from FFPE tissue, as evidenced by failure rates of <15% and a CV of <5% across all samples. Five assays (RPS6, KDR, PTEN, AKT1, CCND2) had failure rates of >15%; however, even the worst performing assay (RPS6) had a success rate of 48%, indicating that these assays were still informative in about half of the samples. We excluded the MYC assay from further analysis because it was the worst performing test on the panel with a 96% failure rate, and it also exhibited a CV of 22% when we analyzed high quality uRNA samples and a CV of 13% when using RNA from FFPE tissues. When we assessed data reproducibly we observed a high degree of data concordance for the assays on our panel for uRNA replicates from independent runs. Furthermore, a high degree of data concordance was observed for each of the assays when we ran replicate FFPE-derived RNA samples on different days. Our qualitative and quantitative assessments suggest that our panel can provide robust and reproducible results when analyzing archival clinical samples.
We utilized our panel in the characterization of a novel cohort of 204 FFPE tissues and were able to identify samples that were transcriptionally similar to the recently described basal and luminal bladder cancer subtypes [10, 14]. By deriving a basal/luminal scoring system using the genes on our panel, we confirmed previous findings using our FFPE cohort that MIBCs can exhibit basal or luminal transcriptional patterns [10, 14]. To our knowledge, this is the first report on the basal/luminal features of METs. We found the majority of METs from our cohort to have a basal-like transcriptional profile. There has been a recent controversy around the luminal/basal make-up of NMIBCs [16, 17]. McConkey and colleagues found the majority of their chemotherapy-naïve NMIBCs to be luminal [17]. On the other hand, in a recent study Hedegaard et. al. showed that NMIBCs from their cohort were mostly basal in nature [16]. The basal-/luminal-like features may vary depending on the spectrum of tumors included in different cohorts. In our sample series we found the majority of NMIBCs to be luminal-like. Mutational status of the FGFR3 gene and expression levels of the RTK by IHC provided molecular confirmation of the luminal and basal status of our samples, by virtue of the high frequency of FGFR3 mutations and high expression levels of the protein that was detected in luminal samples and the low frequency of mutations and protein expression levels that was observed in the basal samples [10, 14, 19]. Differences in the etiologies of NMIBCs might be one reason for the observed differences in the basal/luminal make up of samples from different cohorts. Additional studies into to the basal and luminal characteristics of NMIBCs will be required to resolve these discrepancies.
This study provides proof-of-concept that our microfluidics-based bladder cancer panel can robustly classify FFPE tissues into the clinically-relevant basal and luminal disease subtypes. Further validation efforts on additional archival tissues will be required before this panel can be implemented in the clinic. By making the panel publically available at this point in time we provide the field with the opportunity to apply this rapid, robust, and cost effective approach in the basal/luminal characterization of FFPE clinical samples. This approach offers additional important advantages over global transcriptional profiling for basal/luminal classification, including relatively simple data analysis and suitability for analyzing FFPE-derived RNA, which is often the only available substrate when working with clinical specimens.
Material and Methods
Selection of bladder cancer panel genes
The bladder cancer panel is built on a commercially available microfluidics platform, and this work is not intended for marketing or commercialization of any product. Selection of gene content was based on a comprehensive review of the literature and prioritization of pathways and related genes that have been shown to be involved in bladder cancer development [12, 18, 20, 23, 26, 30] (S1 Table, S1 Fig). Due to the limitation in the number of assays that could be accommodated on the panel and the need to include controls for data QC and normalization (2 housekeeping genes), only 91 unique bladder cancer-relevant genes could be selected. Due to poor technical performance, the MYC assay was dropped from all analyses, bringing the total number of genes on the panel down to 90. Panel genes were binned into three main categories (Development and EMT, FGFR, RTK, MAPK, and PI3K, and TP53, genome stability, and cell cycle genes) based on Ingenuity® pathway analysis (Fig 1A).
Analysis of public datasets
Analyses were performed using the R statistical language (R version 3.2.2) with the Bioconductor library [41] (version 3.1) to support microarray analysis, unless noted otherwise. Several public datasets were used to assess the ability of the 90 genes on our panel to capture basal/luminal status. Public data sets were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) using the GEOquery R library (version 2.34.0). Expression data was normalized using median polish [42] (medpolish command from the R stats library). The ability of the 90 genes on our bladder panel to identify basal-like and luminal-like samples based on transcriptional profiling data was assessed in four literature data sets (GSE32894, GSE5287, GSE13507, GSE48075). Basal or luminal classification was as described by Damrauer et. al. [10].
Cross-validation classifier analysis of public data sets
The genes on our panel were evaluated for their ability to distinguish between basal and luminal subtypes in public data sets. Using the CMA R library (version 1.28.1) we performed 5-fold cross-validation, training a diagonal linear discriminant analysis (DLDA) classifier [32] using all genes and 80% of randomly selected samples. The classifier performance was then evaluated on the remaining 20% of the samples for validation. This procedure was repeated 5 times until all samples had been included in the test set one time. Misclassification rates were evaluated for each fold and mean rates and standard errors were calculated. To identify the highest performing subsets of genes on our panel in distinguishing between basal and luminal subtypes, we repeated the cross-validation process but incorporated a gene sub-selection process at each iteration. Specifically, with each iteration the top n genes were selected from the panel based on their differential expression in the 80% training samples from each public dataset. Differential expression was evaluated using a Welch’s t-test. The number of selected genes was n = 10, 20, 30, 40, 50, 60, 70 and 80, and mean misclassification rates and standard errors were calculated in each case.
Calculating basal/luminal expression profiles from public datasets
The Damrauer discovery dataset [10] (N = 30, Affymetrix Human Genome U133A Array) was downloaded from the NCBI Gene Expression Omnibus (GSE5287). Affymetrix probe sets corresponding to genes on the bladder panel were used. The remaining subset of log-transformed expression data were then mean centered and normalized to unit variance. Average gene expression values were calculated for the 90 genes across the 12 samples classified as luminal-like and 18 samples classified as basal-like (classifications received through author correspondence) to produce basal and luminal expression profiles. This process was repeated for three other public datasets (GSE32894, GSE13507, GSE48075).
Tissues and histopathology
A collection of 204 FFPE bladder cancer samples was obtained from Cureline, Inc. (South San Francisco, CA) following approval of the Ethics Committee of Saint Petersburg City Clinical Oncology Hospital and appropriate confirmation of written informed consent, or from The MT Group (Van Nuys, CA) following IRB approval (http://www.sterlingirb.com). The clinical samples were of a ~80%/20% male/female ratio, and the proportions of NMIBCs (T0, T1), MIBCs (T2, T3) and metastases were 45%, 30%, and 26%, respectively (Table 2, S3 Table). Hematoxylin & Eosin-stained (H&E) sections were evaluated by two independent pathologists (DF and OF). Primary tumors were divided into NMIBCs (T0, T1, Tis) and MIBCs (T2 and T3), and bladder cancer metastases were either from lymph nodes or distal organs (Table 2, S3 Table). Tumor areas for cases with less than 70% neoplastic cellularity were marked by a pathologist for macro-dissection and subsequent tissue lysis for nucleic acid preparation. Overall, tumor content ranged from 70–90%. RNA and DNA were extracted from macro-dissected samples as previously described [38, 43].
Expression analysis of FFPE tissues
Gene expression analysis was carried out on RNA extracted from macro-dissected FFPE tissues using the High Pure FFPE RNA Micro Kit (Roche Diagnostics, Indianapolis, IN) after de-paraffinization with Envirene, as described previously [44]. RNA was quantified using NanoDrop® (Thermo Scientific, Willmington, DE), and the average 260/280 reading was 1.74. Due to the limiting amounts of available RNA it was not possible to assess the quality of the RNA electrophoretically to calculate RNA integrity number (RIN) values. Gene expression analysis was performed on patient specimens starting with 100ng total RNA that was reverse-transcribed to cDNA and pre-amplified in a single reaction using Superscript III/Platinum Taq and pre-amplification reaction mix (Invitrogen, Carlsbad, CA). All Taqman primer/probe sets were included in the pre-amplification reaction at a final dilution of 0.05x original Taqman assay concentration (Applied Biosystems, Foster City, CA). The thermocycling conditions were as follows: 1 cycle of 50°C for 15 min, 1 cycle of 70°C for 2 min, then 14 cycles of 95°C for 15 sec and 60°C for 4 min. Pre-amplified cDNA was diluted 2-fold and then amplified using Taqman Universal PCR MasterMix (Applied Biosystems, Foster City, CA) on the BioMark BMK-M-96.96 platform (Fluidigm, South San Francisco, CA) according to the manufacturer’s instructions. All samples were assayed in triplicate. Cycle threshold (Ct) values were converted to relative expression using the ΔCt method [44], where ΔCt was the mean of the target gene minus the geometric mean of reference genes calculated for the respective patient specimen. For genes assessed on the bladder cancer panel, Cycle threshold (Ct) values were normalized using median polish [42] (medpolish command from the R stats library). Hierarchical clustering was carried out on normalized data with the average-linkage method using 1 –Pearson correlation as a distance metric using the heatmap.2 function from the gplots R library (version 2.17.0) and subsequently visualized using R [45].
Mutation analysis
Mutation analyses were carried out on genomic DNA extracted from macro-dissected FFPE tissues using the QIAamp FFPE kit (Qiagen, Valencia, CA) after de-paraffinization with Envirene [15]. Mutations in PIK3CA, EGFR, KRAS, NRAS, HRAS, FGFR3, MET, BRAF, KIT, AKT1, FLT3 were detected using mutation specific qPCR as described previously [38].
Immunohistochemistry
IHC was performed on 4-μm thick FFPE tissue section using the Ventana Discovery XT Autostainer platform (Ventana Medical Systems Inc, Tucson, AZ). For the detection of FGFR3, the slides underwent pretreatment using CC1 extended antigen retrieval followed by anti-FGFR3, clone 15C3 (Genentech), primary antibody diluted to 1 μg/mL and incubated for 60 minutes at room temperature. For amplification of FGFR3 signal, an unconjugated rabbit-anti-mouse linker antibody (Jackson Immunoresearch, West Grove, PA) was applied at 1 μg/mL and incubated for 32 minutes at room temperature. This was followed by an anti-rabbit-OMNIMAP-HRP kit (Ventana Medical Systems Inc, Tucson, AZ. Catalog no. 760–4310). Sections were counter stained with hematoxylin, dehydrated, cleared and cover-slipped for viewing.
Calculation of basal/luminal similarity scores and statistical analysis
Basal/luminal similarity scores for FFPE samples were determined by first deriving basal/luminal expression profiles from the public Damrauer discovery data set, as previously described [10]. Next, basal and luminal Pearson correlations between each profile and each mean-centered, unit variance normalized FFPE sample were determined. These two correlation scores were combined to produce an overall B/L similarity score: B/L score = (basal profile correlation–luminal profile correlation)/2.0. The B/L score had a range of +1.0 to -1.0, with scores above zero indicating a basal-like sample, and scores below zero indicating a luminal-like sample. For the significance of differential mutation frequency between clusters, Fisher's exact T-tests were performed. Gene differential expression was calculated using the multi t-test (mt.maxT) from the multtest library (version 2.26) in Bioconductor [41] (S2 Table). The test statistic columns show the two-sample Welch t-statistic (unequal variances). This statistic was calculated as the fold change difference divided by the sum of the basal and luminal variances weighted by number of samples in each group. The sign of the statistic reflected the fold change direction, where a positive score implied the gene is more highly expressed in basal samples. Values were left blank in the table when a gene from the 90-gene panel was not present in the literature data set.
Supporting Information
(TIF)
Number of classifier genes ranging form 10–80 was used to train a DLDA classifier on 80% of samples from each public data set, and misclassification rates were calculated on the remaining 20% of the samples. Procedure was repeated 5 times through cross-validation to calculate average misclassification rates and to estimate standard errors.
(TIF)
Mutation data based on the Choi study is also provided.
(TIF)
(TIF)
Standard deviations over the mean expression values was calculated for each of the assays on the panel.
(TIF)
(A) Heatmap showing unsupervised clustering of genes and samples from public data sets, as well as NMIBCs, MIBCs, and METs from a novel FFPE tissue cohort. White blocks represent genes that are not found in the respective data sets. (B) Basal profile (red bars) and luminal profile (blue bars) for bladder cancer panel genes calculated from the Damrauer discovery samples. Mean-centering and unit variance normalization was applied to the log-transformed expression values, and mean log normalized expression levels were calculated independently for basal and luminal sample groups to form the final profiles.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Ling Fu, Rupal Desai, An Do, and Dr. Teiko Sumiyoshi for excellent technical support, Dr. Bob Yauch for input into the design of PCR-based FGFR3 mutation assays, and Drs. Ling Huw and Craig Cummings for assistance with figure preparation.
Data Availability
All relevant data are within the manuscript and the Supporting Information files.
Funding Statement
DK, YC, OF, RNT, RP, EBS, MM, DF, CVW, EP, LM, RR, LCA, GMH, MRL, and OK are current or former employees of Genentech, Inc. Genentech, Inc. provided support in the form of salaries for these authors but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. JI is an employee of Alternative Allele Consulting. Alternative Allele Consulting provided support in the form of salary for this author but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. 10.3322/caac.20107 . [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA: a cancer journal for clinicians. 2011;61(4):212–36. 10.3322/caac.20121 . [DOI] [PubMed] [Google Scholar]
- 3.Nargund VH, Tanabalan CK, Kabir MN. Management of non-muscle-invasive (superficial) bladder cancer. Seminars in oncology. 2012;39(5):559–72. 10.1053/j.seminoncol.2012.08.001 . [DOI] [PubMed] [Google Scholar]
- 4.Resnick MJ, Bassett JC, Clark PE. Management of superficial and muscle-invasive urothelial cancers of the bladder. Current opinion in oncology. 2013;25(3):281–8. 10.1097/CCO.0b013e32835eb583 . [DOI] [PubMed] [Google Scholar]
- 5.Heney NM. Natural history of superficial bladder cancer. Prognostic features and long-term disease course. The Urologic clinics of North America. 1992;19(3):429–33. . [PubMed] [Google Scholar]
- 6.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62(1):10–29. Epub 2012/01/13. 10.3322/caac.20138 . [DOI] [PubMed] [Google Scholar]
- 7.Hall MC, Womack S, Sagalowsky AI, Carmody T, Erickstad MD, Roehrborn CG. Prognostic factors, recurrence, and survival in transitional cell carcinoma of the upper urinary tract: a 30-year experience in 252 patients. Urology. 1998;52(4):594–601. Epub 1998/10/08. . [DOI] [PubMed] [Google Scholar]
- 8.Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, Palou J, Vicente-Rodriguez J. Multivariate analysis of the prognostic factors of primary superficial bladder cancer. The Journal of urology. 2000;163(1):73–8. . [DOI] [PubMed] [Google Scholar]
- 9.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–62. Epub 2014/11/28. 10.1038/nature13904 . [DOI] [PubMed] [Google Scholar]
- 10.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):3110–5. Epub 2014/02/13. 10.1073/pnas.1318376111 ; PubMed Central PMCID: PMCPmc3939870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nature genetics. 2003;33(1):90–6. Epub 2002/12/07. 10.1038/ng1061 . [DOI] [PubMed] [Google Scholar]
- 12.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Molecular cancer. 2010;9:3 Epub 2010/01/12. 10.1186/1476-4598-9-3 ; PubMed Central PMCID: PMCPmc2821358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(12):3377–86. Epub 2012/05/04. 10.1158/1078-0432.ccr-12-0077-t . [DOI] [PubMed] [Google Scholar]
- 14.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer cell. 2014;25(2):152–65. Epub 2014/02/15. 10.1016/j.ccr.2014.01.009 ; PubMed Central PMCID: PMCPmc4011497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindgren D, Sjodahl G, Lauss M, Staaf J, Chebil G, Lovgren K, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLOS ONE. 2012;7(6):e38863 Epub 2012/06/12. 10.1371/journal.pone.0038863 ; PubMed Central PMCID: PMCPmc3369837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedegaard J, Lamy P, Nordentoft I, Algaba F, Hoyer S, Ulhoi BP, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer cell. 2016;30(1):27–42. Epub 2016/06/21. 10.1016/j.ccell.2016.05.004 . [DOI] [PubMed] [Google Scholar]
- 17.McConkey DJ, Choi W, Shen Y, Lee IL, Porten S, Matin SF, et al. A Prognostic Gene Expression Signature in the Molecular Classification of Chemotherapy-naive Urothelial Cancer is Predictive of Clinical Outcomes from Neoadjuvant Chemotherapy: A Phase 2 Trial of Dose-dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin with Bevacizumab in Urothelial Cancer. European urology. 2016;69(5):855–62. Epub 2015/09/08. 10.1016/j.eururo.2015.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nature reviews Cancer. 2005;5(9):713–25. Epub 2005/08/20. 10.1038/nrc1697 . [DOI] [PubMed] [Google Scholar]
- 19.Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. The Journal of pathology. 2007;213(1):91–8. Epub 2007/08/02. 10.1002/path.2207 ; PubMed Central PMCID: PMCPmc2443273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Carbayo M, Cordon-Cardo C. Molecular alterations associated with bladder cancer progression. Seminars in oncology. 2007;34(2):75–84. Epub 2007/03/27. 10.1053/j.seminoncol.2006.12.013 . [DOI] [PubMed] [Google Scholar]
- 21.Ross JS, Wang K, Gay LM, Al-Rohil RN, Nazeer T, Sheehan CE, et al. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(1):68–75. Epub 2013/11/07. 10.1158/1078-0432.ccr-13-1992 . [DOI] [PubMed] [Google Scholar]
- 22.Rebouissou S, Bernard-Pierrot I, de Reynies A, Lepage ML, Krucker C, Chapeaublanc E, et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Science translational medicine. 2014;6(244):244ra91 Epub 2014/07/11. 10.1126/scitranslmed.3008970 . [DOI] [PubMed] [Google Scholar]
- 23.McConkey DJ, Lee S, Choi W, Tran M, Majewski T, Lee S, et al. Molecular genetics of bladder cancer: Emerging mechanisms of tumor initiation and progression. Urologic oncology. 2010;28(4):429–40. Epub 2010/07/09. 10.1016/j.urolonc.2010.04.008 ; PubMed Central PMCID: PMCPmc2901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Zhang W, Geng D, He J, Zhao Y, Yu L. Clinical significance of fibroblast growth factor receptor-3 mutations in bladder cancer: a systematic review and meta-analysis. Genetics and molecular research: GMR. 2014;13(1):1109–20. Epub 2014/03/19. 10.4238/2014.February.20.12 . [DOI] [PubMed] [Google Scholar]
- 25.Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLOS ONE. 2010;5(11):e13821 Epub 2010/11/13. 10.1371/journal.pone.0013821 ; PubMed Central PMCID: PMCPmc2972209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles MA. Tumor suppressor loci in bladder cancer. Frontiers in bioscience: a journal and virtual library. 2007;12:2233–51. Epub 2006/11/28. . [DOI] [PubMed] [Google Scholar]
- 27.Gust KM, McConkey DJ, Awrey S, Hegarty PK, Qing J, Bondaruk J, et al. Fibroblast growth factor receptor 3 is a rational therapeutic target in bladder cancer. Molecular cancer therapeutics. 2013;12(7):1245–54. Epub 2013/05/10. 10.1158/1535-7163.mct-12-1150 ; PubMed Central PMCID: PMCPmc3707970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George B, Datar RH, Wu L, Cai J, Patten N, Beil SJ, et al. p53 gene and protein status: the role of p53 alterations in predicting outcome in patients with bladder cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25(34):5352–8. Epub 2007/12/01. 10.1200/jco.2006.10.4125 . [DOI] [PubMed] [Google Scholar]
- 29.Choi W, Shah JB, Tran M, Svatek R, Marquis L, Lee IL, et al. p63 expression defines a lethal subset of muscle-invasive bladder cancers. PLOS ONE. 2012;7(1):e30206 Epub 2012/01/19. 10.1371/journal.pone.0030206 ; PubMed Central PMCID: PMCPmc3254658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urologic oncology. 2010;28(4):401–8. Epub 2010/07/09. 10.1016/j.urolonc.2009.04.019 . [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147(2):418–29.e8. Epub 2014/05/06. 10.1053/j.gastro.2014.04.039 ; PubMed Central PMCID: PMCPmc4107146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudoit S, Fridlyand J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome biology. 2002;3(7):Research0036 Epub 2002/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Rhijn BW, Montironi R, Zwarthoff EC, Jobsis AC, van der Kwast TH. Frequent FGFR3 mutations in urothelial papilloma. The Journal of pathology. 2002;198(2):245–51. Epub 2002/09/19. 10.1002/path.1202 . [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(17):6280–90. Epub 2005/09/08. 10.1158/1078-0432.ccr-05-0282 . [DOI] [PubMed] [Google Scholar]
- 35.Juanpere N, Agell L, Lorenzo M, de Muga S, Lopez-Vilaro L, Murillo R, et al. Mutations in FGFR3 and PIK3CA, singly or combined with RAS and AKT1, are associated with AKT but not with MAPK pathway activation in urothelial bladder cancer. Human pathology. 2012;43(10):1573–82. Epub 2012/03/16. 10.1016/j.humpath.2011.10.026 . [DOI] [PubMed] [Google Scholar]
- 36.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nature genetics. 1999;23(1):18–20. Epub 1999/09/02. 10.1038/12615 . [DOI] [PubMed] [Google Scholar]
- 37.Al-Ahmadie HA, Iyer G, Janakiraman M, Lin O, Heguy A, Tickoo SK, et al. Somatic mutation of fibroblast growth factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. The Journal of pathology. 2011;224(2):270–9. Epub 2011/05/07. 10.1002/path.2892 ; PubMed Central PMCID: PMCPmc3235805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleifman EB, Tam R, Patel R, Tsan A, Sumiyoshi T, Fu L, et al. Next generation MUT-MAP, a high-sensitivity high-throughput microfluidics chip-based mutation analysis panel. PLOS ONE. 2014;9(3):e90761 Epub 2014/03/25. 10.1371/journal.pone.0090761 ; PubMed Central PMCID: PMCPmc3962342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst CD, Platt FM, Taylor CF, Knowles MA. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(21):5865–77. Epub 2012/08/31. 10.1158/1078-0432.ccr-12-1807 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, et al. Bladder cancer outcome and subtype classification by gene expression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(11):4044–55. Epub 2005/06/03. 10.1158/1078-0432.ccr-04-2409 . [DOI] [PubMed] [Google Scholar]
- 41.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Meth. 2015;12(2):115–21. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tukey JW. Exploratory Data Analysis. Reading, Massachusetts:: Addison-Wesley; 1977. [Google Scholar]
- 43.Schleifman EB, Desai R, Spoerke JM, Xiao Y, Wong C, Abbas I, et al. Targeted biomarker profiling of matched primary and metastatic estrogen receptor positive breast cancers. PLOS ONE. 2014;9(2):e88401 Epub 2014/02/13. 10.1371/journal.pone.0088401 ; PubMed Central PMCID: PMCPmc3919784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(14):3670–83. Epub 2010/05/11. 10.1158/1078-0432.ccr-09-2828 . [DOI] [PubMed] [Google Scholar]
- 45.Team RDC. R: a language and environment for statistical computing. 2006.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Number of classifier genes ranging form 10–80 was used to train a DLDA classifier on 80% of samples from each public data set, and misclassification rates were calculated on the remaining 20% of the samples. Procedure was repeated 5 times through cross-validation to calculate average misclassification rates and to estimate standard errors.
(TIF)
Mutation data based on the Choi study is also provided.
(TIF)
(TIF)
Standard deviations over the mean expression values was calculated for each of the assays on the panel.
(TIF)
(A) Heatmap showing unsupervised clustering of genes and samples from public data sets, as well as NMIBCs, MIBCs, and METs from a novel FFPE tissue cohort. White blocks represent genes that are not found in the respective data sets. (B) Basal profile (red bars) and luminal profile (blue bars) for bladder cancer panel genes calculated from the Damrauer discovery samples. Mean-centering and unit variance normalization was applied to the log-transformed expression values, and mean log normalized expression levels were calculated independently for basal and luminal sample groups to form the final profiles.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and the Supporting Information files.