Abstract
Background
Porcine respiratory and reproductive syndrome (PRRS) virus is one of the most economically significant pathogens in the Vietnamese swine industry. ORF5, which participates in many functional processes, including virion assembly, entry of the virus into the host cell, and viral adaptation to the host immune response, has been widely used in molecular evolution and phylogeny studies. Knowing of molecular evolution of PRRSV fields strains might contribute to PRRS control in Vietnam.
Results
The results showed that phylogenetic analysis indicated that all strains belonged to sub-lineages 8.7 and 5.1. The nucleotide and amino acid identities between strains were 84.5–100% and 82–100%, respectively. Furthermore, the results revealed differences in nucleotide and amino acid identities between the 2 sub-lineage groups. N-glycosylation prediction identified 7 potential N-glycosylation sites and 11 glycotypes. Analyses of the GP5 sequences, revealed 7 sites under positive selective pressure and 25 under negative selective pressure.
Conclusions
Phylogenetic analysis based on ORF5 sequence indicated the diversity of PRRSV in Vietnam. Furthermore, the variance of N-glycosylation sites and position under selective pressure were demonstrated. This study expands existing knowledge on the genetic diversity and evolution of PRRSV in Vietnam and assists the effective strategies for PRRS vaccine development in Vietnam.
Electronic supplementary material
The online version of this article (doi:10.1186/s12917-016-0885-3) contains supplementary material, which is available to authorized users.
Keywords: PRRSV, Vietnam, ORF5, Phylogeny
Background
Porcine reproductive and respiratory syndrome (PRRS) is a major infectious disease affecting pork industries worldwide. Its outbreaks were first reported in the USA and EU in the late 1980s and early 1990s, respectively [4, 5, 42]. The main clinical signs of the disease are respiratory problems in pigs of all ages and reproductive failure in pregnant sows. In Vietnam, PRRS outbreaks have continuously occurred since 2007 [8, 23, 28]. PRRS viruses, the causative agents of the disease, can be divided into two distinct genotypes, type I (EU type) and type II (American type), which present with identical disease symptoms, despite their genetic differences [24]. PRRSV is a mono-partite, linear, positive-sense single stranded RNA virus belonging to the Arterviridae family [5]. Its genome of approximately15 kb in size is organized into 10 open reading frames (ORFs) [24, 38]. Two large ORF1a and ORF1b genes encode non-structural proteins that play important roles in viral replication and virulence [13, 18]. The other ORFs encode for structural proteins that are necessary for production of infectious virions [44]. ORF5, which participates in many functional processes, including virion assembly [44], entry of the virus into the host cell [7], and viral adaptation to the host immune response [41], has been widely used in molecular evolution and phylogeny studies [30, 34, 35].
Evolutionary studies indicate that PRRSV diverged long before the first detected outbreaks of the disease. Evolutional analyses based on ORF5, as well as serological evidence, indicated that PRRSV type 2 first appeared around the 1980s [3, 35, 48]. In contrast, type 1 PRRSV originated approximately 100 years ago [30]. Further analysis of the whole PRRSV genome shows that the two types of PRRSV diverged from a common ancestor about 800 years ago [46]. Furthermore, genetic analyses indicate that the evolutionary trends, antigenic characteristics, and genetic diversity of PRRSV in different regions have distinct patterns [6, 11, 17, 32, 36, 40].
Thus far, type 2 PRRSV has been divided into 10 sub-lineages, including 9 old sub-lineages [34] occurring worldwide, and a new sub-lineage, which recently appeared in Thailand [40]. In Vietnam, several studies show that the circulating PRRSV strains belong to a highly pathogenic (HP) variant that recently emerged in China and South East Asian countries [12, 28]. However, few studies have focused on the evolutionary trends and characterization of PRRSV presenting in Vietnam. Thus, the aim of this study was to investigate the genetic diversity, selective pressure, and glycosylation patterns in GP5 of PRRSV strains that appeared in Vietnam during 2007–2015.
Methods
Sample collection
For this study, we used 40 PRRS-positive sera or tissue samples, as confirmed by RT-PCR; the samples were collected from pigs in provinces in North Vietnam during 2011–2015. Total PRRSV RNA was extracted using TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s instruction. Reverse transcription was performed using SuperScript™ III First-Strand Synthesis SuperMix (Thermo Fisher, USA). ORF5 sequences were amplified by RT-PCR using previously described primers [12]. PCR products were directly sequenced (Macrogen, Seoul, Korea). The raw sequences were assembled and aligned using BioEditv7.2.5 [14] against the corresponding ORF5 sequences from GenBank to construct the complete ORF5 sequence. Additional 104 Vietnamese ORF5 reference sequences from field isolates collected from GenBank were also used in this study (Additional file 1: Table S1).
Phylogenetic analysis and classification
In order to identify the lineage classifications for all the PRRSV strains circulating in Vietnam, an ORF5-based phylogeny was reconstructed using a restricted parameter substitution model [35] with IQ-TREE software [27]. The total data set in this study contained 144 Vietnamese ORF5 gene sequences and 612 worldwide ORF5 reference sequences for lineages 1 to 9 [35]. Bootstrap values were obtained using the ultrafast bootstrap approximation method with 1000 replicates [25] (both programs are available at http://iqtree.cibiv.univie.ac.at/).
Bayesian phylogenetic inference of ORF5 from Vietnamese strains
The coalescent-based Bayesian Markov Chain Monte Carlo (MCMC) method was used to investigate the phylogenetic relationship among Vietnamese PRRSV strains based on ORF5 sequences. The SRD06 codon-based model was used as a nucleotide substitution model [29, 31] and combined with (i) 5 molecular clock models (Strict clock, uncorrelated lognormal relaxed clock, uncorrelated exponential relaxed clock, random clock, and fixed local clock) and (ii) 7 demographic coalescent tree models (constant size, exponential growth, logistic growth, expansion growth, Bayesian skyline, extended Bayesian skyline plot, and Bayesian skygrid). In each analysis, the MCMC chain (50 million generations, sampling every 5000 stages) was performed using BEAST v1.8.2 software [9]. Five independent runs were done to verify the distribution in the MCMC run. The corresponding output log files were combined by LogCombiner before subsequently analyzing via Tracer v1.6 to select the best-fit data models for molecular clock and coalescent tree priors using Akaike’s information criterion (AICM) analysis with 1000 replicates [2]. A Bayesian phylogenetic tree was selected from combined trees files from the above chosen best-fit models using TreeAnnotator in BEAST package.
Glycosylation site prediction
Glycosylation sites in the Vietnamese PRRSV strains were predicted using the NetNGlyc server web utility (http://www.cbs.dtu.dk/services/NetNGlyc/). Adefault threshold of 0.5 was used to identify potential N-glycosylation sites, followed by additional thresholds of 0.75 and 0.9 to identify the potential N-glycosylation sites with higher confidence levels.
Selective pressure
GP5 sites undergoing positive selection were inferred using 5 algorithms: SLAC, FEL, IFEL, FUBAR, and MEME (available on the Datamonkey web server: www.datamonkey.org). Sites undergoing negative selection were predicted using 4 algorithms: SLAC, FEL, IFEL, and FUBAR. To identify other sites undergoing potential selective pressure, sites were analyzed for either diversifying or purifying selection at P-value ≤0.1 using SLAC, FEL, IFEL, and MEME methods, or for posterior probability ≥ 90% using the FUBAR method.
Results
To investigate the evolution of Vietnamese PRRSV strains, we analyzed the time scale phylogenetic tree, the genetic diversity among strains, the time of most common ancestor of PRRSV strains in Vietnam as well as the change of N-glycosylation pattern during this time.
Phylogenetic analysis of the ORF5 sequence
Based on the constructed phylogenetic tree, the major PRRSV strains (n = 138) isolated in Vietnam could be classified into sub-lineage 8.7, which is closely related to the highly pathogenic PRRSV strains recently isolated in China, including JXA1 and SX2009 [47]. The remaining strains (n = 6) were classified into sub-lineage 5.1, which contains VR-2332-related strains [35] (Additional file 2: Figure S1). Further analysis based on Bayesian inference showed that HP-PRRSVs in Vietnam can be divided into two main sub-groups (Fig. 1). Interestingly, most of the PRRSV strains collected in North Vietnam during 2013–2015 belonged to sub-group ii (Fig. 1). Under the best-fit model selected, the substitution rate in the ORF5 gene of the Vietnamese PRRSV strains was about4.459 × 10−3 (95% highest posterior density (HPD) intervals: 3.0981 × 10−3–5.8523 × 10−3). In addition, the geometric mean time to the most recent common ancestor (TMRCA) of the HP-PRRSV isolated in Vietnam was approximately 13 years ago and the TMRCA of sub-lineage 5.1 PRRSV strains was more than 16 years ago (95% HPD was 9.2708–18.8409 and 6.9872–32.5917 for the HP-PRRSV group and sub-lineage 5.1 group, respectively).
Fig. 1.
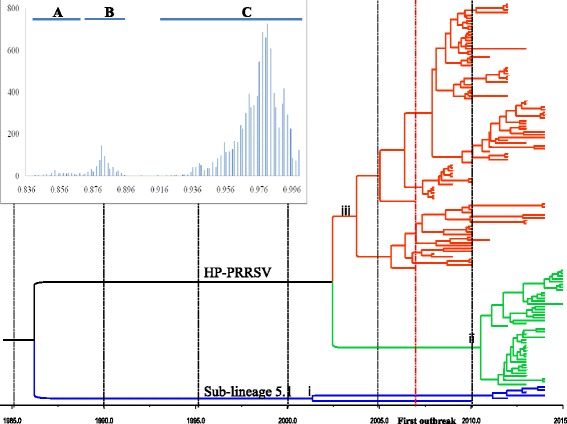
Phylogenetic tree based on nucleotide sequence of the ORF5 gene of 144 PRRSV strains isolated in Vietnam during 2007–2015. The phylogenetic tree, generated via the MCMC method using BEAST v1.8.2 software, identified three different groups. The inserted histogram illustrates pairwise sequence comparisons of Vietnamese PRRSV type 2 strains. Three distinct nucleotide identity distribution peaks are shown. The time-scale (in years) represented in the tree is indicated by the scale bar
Genetic diversity of the Vietnamese PRRSV strains during 2007–2015
Genetic comparison of the ORF5 gene of the Vietnamese PRRSV strains collected from 2007 to 2015 showed that 144Vietnamese PRRSV strains in this study shared 81–100% nucleotide identity (Table 1). Furthermore, the similarity among the ORF5 sequences presented in the same year was about 84.5–100%. Especially in 2010, 2012, 2013, and 2014, when the appearance of PRRSV sub-lineage 5.1 strains was recorded, differences among nucleotide sequences was up to 15.5% while in the remaining years, it was just about 2%. Further analysis showed that the similarity among Vietnamese HP-PRRSV strains was of 91.6–100% while the difference among sub-lineage 5.1 strains was up to 9.5%.
Table 1.
Nucleotide and deduced amino acid identities among 144 Vietnamese PRRSV strains
| Year | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2015 | nt | 99.3–100 | 88.1–99.6 | 88.1–99.5 | 89.1–99.1 | 98.1–99.1 | 86–99.1 | 98.1–99.1 | 98.1–99.1 | 98.5–99.1 |
| aa | 98–100 | 85.5–95.5 | 85.5–98.5 | 88–98.5 | 96.5–99 | 84–99 | 97–99 | 97–99 | 97.5–98.5 | |
| 2014 | nt | 84.8–100 | 85.1–100 | 86–99.5 | 87.8–99.6 | 84.1–99.1 | 87.8–99.1 | 87.8–99.1 | 88–99.1 | |
| aa | 82–100 | 82–100 | 84–99.5 | 85.5–100 | 81–99.5 | 86–99.5 | 86–99.5 | 85.5–99 | ||
| 2013 | nt | 86.5–100 | 87–99.5 | 87.8–99.3 | 83.6–99 | 87.8–99 | 87.8–99 | 88–99 | ||
| aa | 83.5–100 | 84.5–99.5 | 86–99 | 81.5–99.5 | 86–99 | 86–99 | 85.5–98.5 | |||
| 2012 | nt | 87.8–100 | 88.8–99.8 | 84.6–100 | 88.8–99.5 | 88.8–99.5 | 89–98.6 | |||
| aa | 87–100 | 88.5–99.5 | 81–100 | 88.5–99.5 | 88.5–99.5 | 88–99 | ||||
| 2011 | nt | 98.1–100 | 85.6–100 | 98.1–99.5 | 98.1–99.5 | 98.1–99.3 | ||||
| aa | 98–100 | 83–100 | 98–100 | 98–100 | 97.5–99.5 | |||||
| 2010 | nt | 84.5–100 | 85.3–100 | 85.3–100 | 85.5–99.3 | |||||
| aa | 81.5–100 | 83–100 | 83–100 | 83.5–99.5 | ||||||
| 2009 | nt | 98.1–100 | 98.1–100 | 98.1–99.3 | ||||||
| aa | 98.5–100 | 98.5–100 | 98–99.5 | |||||||
| 2008 | nt | 98.1–100 | 98.1–99.3 | |||||||
| aa | 98.5–100 | 98–99.5 | ||||||||
| 2007 | nt | 98.5–100 | ||||||||
| aa | 98.5–100 | |||||||||
The deduced amino acid sequence encoded by the ORF5 gene of 144 Vietnamese PRRSV strains shared 82–100% identity. For each sub-lineage group, the amino acid identity was 90–100% and 86.5–100% for HP-PRRSV and sub-lineage 5.1, respectively (Table 1).
Glycosylation site variants
A total of 7 potential N-glycosylation sites (amino acids 30, 32, 33, 34, 35, 44 and 51) were found for the Vietnamese PRRSV strains isolated in Vietnam during the 2007–2015 period. The identified positions and the total numbers of N-glycosylation sites were diverse. Notably, PRRSV strains isolated during the outbreaks in 2014 had the greatest variation in N-glycosylation patterns, followed by those from the outbreaks in 2013, which had 8 and 7 glycotypes (Table 2). Glycosylation site variations were located between amino acids 32 and 35, while N44 and 51 seemed to be conserved in most of the Vietnamese strains, presenting in 97.2% and 100% of strains, respectively. Furthermore, N41 was predicted as a glycosylation site with higher potential (≥0.75). An N-glycosylation pattern of N30, N35, N44, and N51 seems to be the main glycotype in Vietnamese PRRSV strains, accounting for nearly 61%. Interestingly, we observed differences in the frequencies of N-glycosylation positions between sub-lineage 5.1 strains and sub-lineage 8.7 strains. To be specific, N30, N32, N33, N34, and N35 were identified as potential N-glycosylation sites in sub-lineage 8.7, accounting for 92.09, 2.88, 12.95, 18.71 and 82.73% of strains, respectively, whereas only N30, N33, and N34 were predicted as in sub-lineage 5.1 accounting for 33.33, 66.67, and 33.33% of strains, respectively (data not shown). Furthermore, only two Vietnamese PRRSV sub-lineage 5.1 strains had similar N-glycosylation patterns as the vaccine strain VR2332, while the other strains lacked the potential N-glycosylation site at N30. On the other hand, 88 Vietnamese HP-PRRSV strains had the same N-glycosylation pattern as the JXA1 vaccine strains.
Table 2.
Glycosylation pattern of PRRSV strains in Vietnam during 2007–2015
| Year/Ref Strain | N-glycosylation site | Number of sequence | % of total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 30 | 32 | 33 | 34 | 35 | 44 | 51 | |||
| VR-2332 | x | x | xxx | x | |||||
| JXA1 | x | x | xxx | x | |||||
| 2007 | x | x | xx | x | 1 | 0.69 % | |||
| x | xx | x | 1 | 0.69 % | |||||
| 2008 | x | x | xx | x | 6 | 4.17 % | |||
| 2009 | x | x | xx | x | 7 | 4.86 % | |||
| 2010 | x | x | xx | x | 31 | 21.53 % | |||
| x | x | x | xx | x | 1 | 0.69 % | |||
| x | x | x | 1 | 0.69 % | |||||
| x | xx | x | 4 | 2.78 % | |||||
| x | x | xx | x | 3 | 2.08 % | ||||
| 2011 | x | x | xx | x | 1 | 0.69 % | |||
| x | x | xx | x | 2 | 1.39 % | ||||
| 2012 | x | x | xx | x | 11 | 7.64 % | |||
| x | x | xx | x | 2 | 1.39 % | ||||
| x | x | xx | x | 3 | 2.08 % | ||||
| x | x | x | 1 | 0.69 % | |||||
| 2013 | x | x | x | xx | x | 14 | 9.72 % | ||
| x | x | x | xx | x | 2 | 1.39 % | |||
| x | x | xx | x | 1 | 0.69 % | ||||
| x | xx | x | 2 | 1.39 % | |||||
| x | x | xx | x | 6 | 4.17 % | ||||
| x | xx | x | 1 | 0.69 % | |||||
| x | xx | x | 3 | 2.08 % | |||||
| 2014 | x | x | x | 1 | 0.69 % | ||||
| x | x | x | 1 | 0.69 % | |||||
| x | x | x | xx | x | 6 | 4.17 % | |||
| x | x | xx | x | 15 | 10.42 % | ||||
| x | x | xx | x | 5 | 3.47 % | ||||
| x | x | xx | x | 1 | 0.69 % | ||||
| x | xx | x | 2 | 1.39 % | |||||
| x | xx | x | 1 | 0.69 % | |||||
| 2015 | x | x | xx | x | 9 | 6.25 % | |||
x: indicating the potential N-glycosylation site at cut off value; xx and xxx: indicating the potential N-glycosylation site at additional value (>0.75 and >0.9, respectively)
Selective pressure in GP5
To identify positions under selective pressure, SLAC, FEL, IFEL, MEME, and FUBAR methods were implemented separately. Since each method utilizes a different algorithm for predicting sites under positive or negative selection, for our study, we considered sites to be undergoing diversifying selection if so predicted by all 5 of the methods, and to be undergoing purifying selection if predicted by 4 of the methods. Consequently, we identified 7 positions as potentially undergoing positive selection (codons 25, 33, 34, 35, 58, 59, and 104). Most of the positive selection sites were located in ecto-domain 1 (n = 5), while only 1 site undergoing diversifying selection was found in each ecto-domain 2 and signal domain (Fig. 2).
Fig. 2.
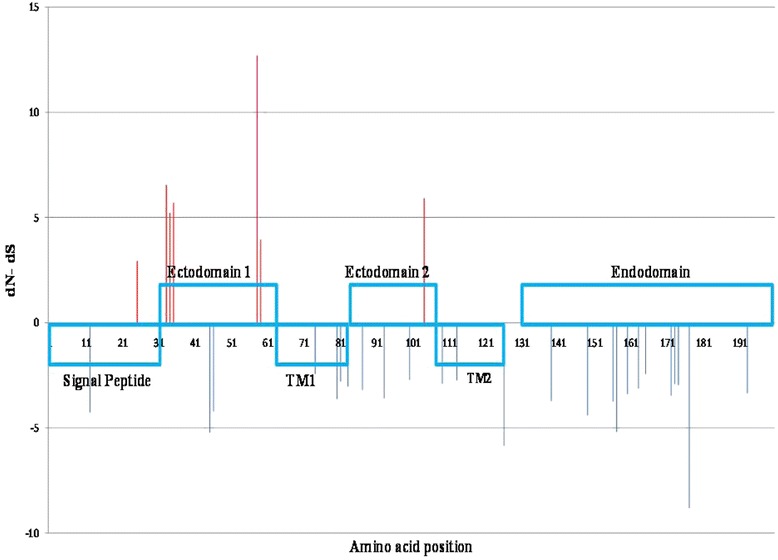
Amino acids under selective pressure. Upper rectangles indicate the ecto-domain 1, ecto-domain 2, and endo-domain, whereas lower rectangles indicate the signal peptide, trans-membrane 1, and trans-membrane 2 regions. The red lines and blue lines indicate sites under significant positive and negative selection, respectively. dN-dS represents the normalized dN–dS value according to the FEL method
A different pattern was observed for the negative selection sites. In our study, 25 sites were predicted to be undergoing negative selection (codons 12, 45, 46, 74, 80, 81, 83, 87, 93, 100, 109, 113, 126, 139, 149, 156, 157, 160, 163, 165, 172, 173, 174, 177, and 193). Purifying selection sites were mainly located in the endo-domain (n = 12). Furthermore, negative selection sites were detected in trans-membrane 1, trans-membrane 2, and ecto-domain 2 (positions 4, 3, and 3, respectively) (Fig. 2).
Discussion
Since first identified in the late 1980s, PRRSV has become the most significant porcine reproductive pathogen. The ORF5 gene is the most diverse gene not only in PRRSV, but also in other arteriviruses, and has been an important target for investigations on the genetic characterization and evolution of PRRSV worldwide [34].
In this study, most of the PRRSV strains isolated in Vietnam during 2007–2015 belonged to sub-lineage 8.1, except for 6 strains that belonged to sub-lineage 5.1 (Additional file 2: Figure S1). This result is consistent with that of previous reports, indicating that most of the PRRSV strains isolated in Vietnam are close related to JXA1 [12, 28]. In fact, attenuated vaccine strains belonging to sub-lineage 5.1, such as VR2332 and BSL-PS, have been approved for use in Vietnam. Furthermore, although HP-PRRSV strains are the main agents of PPRS in countries around Vietnam, such as China and others in South East Asia, strains from other type 2 PRRSV lineages such as lineages 1, 3, and 5 have also circulated [22, 40]. Therefore, these data suggest that the appearance of sub-lineage 5.1 in Vietnam may be due to vaccine descendants or commercial activities. In addition, our study indicates that the HP-PRRSV strains circulating during 2013–2015 were distantly related to the other HP-PRRSV strains. This may have resulted from the introduction of new PRRSV strains into Vietnam. However, the limited number of Vietnamese PRRSV ORF5 sequences used in this study may not exactly reflect the genetic diversity of PRRSVs in Vietnam.
Although, the PRRSV strains circulating in Vietnam clustered within sub-lineage 5.1 and 8.7, their percentages of intra-sub-lineage genetic diversity were 90.5–100% and 91.6–100%, respectively. The intra-sub-lineage genetic diversity in our study was higher than in a previous study [35]. This result might be due to the high substitution rate detected in the ORF5 gene. In addition, the substitution rate in the ORF5 gene of the Vietnamese strains, which was 4459 × 10−3, was slightly faster than the substitution rates observed in common type II PRRSV strains [31] This supports our hypothesis. According to our analysis, the TMRCA of sub-lineage 5.1 strains was approximately 17 years ago, which is supported by serological evidence for anti-PRRSV antibodies in Vietnam during this time. The TMRCA of sub-lineage 8.7 strains was estimated to have occurred in 2002, which is similar to the TMRCA of the HP-PRRSV from China [36].
It is reported that the N-glycosylation positions in GP5 affect the adaptation of PRRSV to the host’s immune response and infectivity [1]. In our study, 11 potential N-glycotypes were observed for the GP5 protein of the Vietnamese PRRSV strains isolated between 2007 and 2015 (Table 2). Furthermore, our investigation revealed diversity in the putative N-glycosylation site amino acid positions (7 different positions) and quantity (3 to 5 sites). However, the N-glycotype diversity identified in the Vietnamese PRRSV strains was less than that recorded for the PRRSV strains from Eastern Canada isolated between 1998 and 2009 [6]. Another report shows that the PRRSV strains isolated in China from 2006 to 2009 have the same N-glycosylation sites as the Vietnamese strains [26]. In all positions, the N44 site was predicted at a high confidence level (≥0.75) and seemed to be conserved. These results are consistent with the important role of this residue in infectivity [1]. The N51 glycosylation site has also been demonstrated to affect the growth kinetics of PRRSV [1], and is highly conserved in type 2 PRRSV from many countries [6, 17, 19].
On the other hand, N-glycosylation sites seem to vary at positions 32–35 (Table 2). These sites are located within the hyper-variable region that has been previously described [39]. It is believed that the variations in N-glycosylation in this region may influence viral neutralization [16]. However, not all potential glycosylation sites in this region are glycosylated. Li et al. [20] suggested that only 2 or 3 glycosylation sites in this region are utilized and that their exact positions are still unknown.
Another notable result of this study was the distribution of diversity and purified selection positions throughout the GP5 of Vietnamese PRRSV strains. Nguyen et al. [31] can not conclude the role of selected position in typical PRRSV and HP-PRRSV. In this study, our analysis of sites under selective pressure indicated that most of the sites undergoing positive selection (amino acids 33, 34, 35, 58, and 59) are located in ecto-domain 1, which contains the linear epitope, and an additional positive selection site is also predicted in ecto-domain 2 (amino acid 104). Our results are generally consistent with those of previous studies, with the exception of the positive selection site predicted in the signal peptide [6, 15, 26]. A previous study demonstrated that site-directed mutagenesis of the amino acid residues at 102 and 104 can enhance PRRSV evasion of neutral antibodies in vitro [10]. This supports the results from our current study. In addition, the potential N-glycosylation sites at the N33, N34, and N35 have been previously identified as undergoing positive selection in the HP-PRRSV strains recently isolated in China [26, 45]. In a previous investigation, in vitro neutralization experiments showed that mutation of the N34 Asp (wt) to Asn slightly decreases the neutralizing activity of Asp-34 sera [33].
Our study showed that the main locations under negative selection were in the endo-domain, following by trans-membrane 1, trans-membrane 2, and ecto-domain 2. This agrees with the findings of Xu et al. [45]. Negative selective pressure within the trans-membrane domains may relate to the integrity or functionality of the virion whereas the distribution of sites under purifying selection in the endo-domain could relate to the budding process of PRRSV. A similar function has been observed in alphaviruses, where E2 and the nucleocapsid protein specifically interact with each other [37]. In addition, mutations within endo-domain of glycoprotein E2 affected the biological characteristic of Sindbis virus [21, 43].
Conclusions
This study first describes the molecular evolution of ORF5 of PRRSV occurred in Vietnam since the first outbreaks. Phylogenetic analysis based on ORF5 sequence indicated the diversity of PRRSV in Vietnam. Furthermore, the variance of N-glycosylation sites and position under selective pressure were demonstrated. This study expands existing knowledge on the genetic diversity and evolution of PRRSV in Vietnam and assists the effective strategies for PRRS vaccine development in Vietnam.
Acknowledgements
None.
Funding
This work was supported by the Vietnam National Project under the Project Code No: SPQG.05b.02 and by a grant (Project Code No. 313014-03-1-HD030) from the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry & Fisheries, 2013.
Availability of data and materials
The information of Vietnamese PRRSV strains using in this study is included within the supplement data (Additional file 1: Table S1). The phylogenetic data was deposited in the TreeBase at http://purl.org/phylo/treebase/phylows/study/TB2:S20099?x-access-code=a533b9962d6c27b5884321c9668bf275&format=html.
Authors’ contributions
HQD, SC, MY, TVL conducted experiment, analyzed the data and wrote the paper. TTHV and DDT assisted sample preparation and experiment. DS and TLN shared ideas and discussed the research data. VPL, DTT, DJA contributed to supervision, had the idea for the project, and directed the project. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors. Sample collection was obtained the consent from farm owners.
Abbreviations
- HP
Highly pathogenic
- HPD
Highest posterior density
- MCMC
Markov Chain Monte Carlo
- ORF
Open reading frame
- PRRS
Porcine reproductive and respiratory syndrome
- PRRSV
Porcine reproductive and respiratory syndrome virus
- TMRCA
Time to the most recent common ancestor
Additional files
The information of Vietnamese PRRSV strains using in this study. (PDF 99 kb)
Classification of Vietnamese PRRSV strains based on the reference sequences. (PDF 158 kb)
References
- 1.Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006;80(8):3994–4004. doi: 10.1128/JVI.80.8.3994-4004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29(9):2157–67. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman S, Sanford S, Dea S. Assessment of seropositivity to porcine reproductive and respiratory syndrome (PRRS) virus in swine herds in Ontario--1978 to 1982. Can Vet J. 1995;36(12):776. [PMC free article] [PubMed] [Google Scholar]
- 4.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992;4(2):117–26. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 5.Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193(1):329–39. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delisle B, Gagnon CA, Lambert ME, D’Allaire S. Porcine reproductive and respiratory syndrome virus diversity of Eastern Canada swine herds in a large sequence dataset reveals two hypervariable regions under positive selection. Infect Genet Evol. 2012;12(5):1111–9. doi: 10.1016/j.meegid.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Delputte P, Costers S, Nauwynck H. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J Gen Virol. 2005;86(5):1441–5. doi: 10.1099/vir.0.80675-0. [DOI] [PubMed] [Google Scholar]
- 8.Do DT, Park C, Choi K, Jeong J, Nguyen TT, Le DT, Vo KM, Chae C. Nucleotide sequence analysis of Vietnamese highly pathogenic porcine reproductive and respiratory syndrome virus from 2013 to 2014 based on the NSP2 and ORF5 coding regions. Arch Virol. 2016;161(3):669–75. doi: 10.1007/s00705-015-2699-1. [DOI] [PubMed] [Google Scholar]
- 9.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7(1):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan B, Liu X, Bai J, Zhang T, Zhang Q, Jiang P. The amino acid residues at 102 and 104 in GP5 of porcine reproductive and respiratory syndrome virus regulate viral neutralization susceptibility to the porcine serum neutralizing antibody. Virus Res. 2015;204:21–30. doi: 10.1016/j.virusres.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Fang Y, Schneider P, Zhang W, Faaberg K, Nelson E, Rowland R. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch Virol. 2007;152(5):1009–17. doi: 10.1007/s00705-007-0936-y. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Zhao T, Nguyen T, Inui K, Ma Y, Nguyen TH, Nguyen VC, Liu D, Bui QA, To LT, Wang C, Tian K, Gao GF. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg Infect Dis. 2008;14(11):1774–6. doi: 10.3201/eid1411.071676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grebennikova T, Clouser D, Vorwald A, Musienko M, Mengeling W, Lager K, Wesley R, Biketov S, Zaberezhny A, Aliper T. Genomic characterization of virulent, attenuated, and revertant passages of a North American porcine reproductive and respiratory syndrome virus strain. Virology. 2004;321(2):383–90. doi: 10.1016/j.virol.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic acids symposium series. 1999.
- 15.Hanada K, Suzuki Y, Nakane T, Hirose O, Gojobori T. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol Biol Evol. 2005;22(4):1024–31. doi: 10.1093/molbev/msi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Jiang P, Wang X, Li Y, Wang X, Du Y. Influence of porcine reproductive and respiratory syndrome virus GP5 glycoprotein N-linked glycans on immune responses in mice. Virus Genes. 2007;35(3):663–71. doi: 10.1007/s11262-007-0131-y. [DOI] [PubMed] [Google Scholar]
- 17.Kvisgaard LK, Hjulsager CK, Brar MS, Leung FC, Larsen LE. Genetic dissection of complete genomes of Type 2 PRRS viruses isolated in Denmark over a period of 15 years. Vet Microbiol. 2013;167(3):334–44. doi: 10.1016/j.vetmic.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Kwon B, Ansari IH, Pattnaik AK, Osorio FA. Identification of virulence determinants of porcine reproductive and respiratory syndrome virus through construction of chimeric clones. Virology. 2008;380(2):371–8. doi: 10.1016/j.virol.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol. 2011;49(9):3175–83. doi: 10.1128/JCM.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Tao S, Orlando R, Murtaugh MP. N-glycosylation profiling of porcine reproductive and respiratory syndrome virus envelope glycoprotein 5. Virology. 2015;478:86–98. doi: 10.1016/j.virol.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu LN, Lee H, Hernandez R, Brown DT. Mutations in the endo domain of Sindbis virus glycoprotein E2 block phosphorylation, reorientation of the endo domain, and nucleocapsid binding. Virology. 1996;222(1):236–46. doi: 10.1006/viro.1996.0414. [DOI] [PubMed] [Google Scholar]
- 22.Lu WH, Tun HM, Sun BL, Mo J, Zhou QF, Deng YX, Xie QM, Bi YZ, Leung FC-C, Ma JY. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol. 2015;175(2):332–40. doi: 10.1016/j.vetmic.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Metwally S, Mohamed F, Faaberg K, Burrage T, Prarat M, Moran K, Bracht A, Mayr G, Berninger M, Koster L, To TL, Nguyen VL, Reising M, Landgraf J, Cox L, Lubroth J, Carrillo C. Pathogenicity and molecular characterization of emerging porcine reproductive and respiratory syndrome virus in Vietnam in 2007. Transbound Emerg Dis. 2010;57(5):315–29. doi: 10.1111/j.1865-1682.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- 24.Meulenberg JJ. PRRSV, the virus. Vet Res. 2000;31(1):11–21. doi: 10.1051/vetres:2000103. [DOI] [PubMed] [Google Scholar]
- 25.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30(5):1188–95. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu C, Lu X, Duan E, Chen J, Li W, Zhang F, Martin DP, Yang M, Xia P, Cui B. Molecular evolution of porcine reproductive and respiratory syndrome virus isolates from central China. Res Vet Sci. 2013;95(3):908–12. doi: 10.1016/j.rvsc.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen TDT, Thi Thu N, Son NG, Ha LTT, Hung VK, Nguyen NT, Khoa DVA. Genetic analysis of ORF5 porcine reproductive and respiratory syndrome virus isolated in Vietnam. Microbiol Immunol. 2013;57(7):518–26. doi: 10.1111/1348-0421.12067. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen VG, Kim HK, Moon HJ, Park SJ, Chung HC, Choi MK, Kim AR, Park BK. ORF5-based evolutionary and epidemiological dynamics of the type 1 porcine reproductive and respiratory syndrome virus circulating in Korea. Infect Genet Evol. 2014;21:320–8. doi: 10.1016/j.meegid.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen VG, Kim HK, Moon HJ, Park SJ, Chung HC, Choi MK, Park BK. A Bayesian phylogeographical analysis of type 1 porcine reproductive and respiratory syndrome virus (PRRSV) Transbound Emerg Dis. 2014;61(6):537–45. doi: 10.1111/tbed.12058. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen VG, Kim HK, Moon HJ, Park SJ, Chung HC, Choi MK, Park BK. Evolutionary dynamics of a highly pathogenic type 2 porcine reproductive and respiratory syndrome virus: analyses of envelope protein-coding genes. Transbound Emerg Dis. 2015;62(4):411–20. doi: 10.1111/tbed.12154. [DOI] [PubMed] [Google Scholar]
- 32.Nilubol D, Tripipat T, Hoonsuwan T, Tipsombatboon P, Piriyapongsa J. Dynamics and evolution of porcine reproductive and respiratory syndrome virus (PRRSV) ORF5 following modified live PRRSV vaccination in a PRRSV-infected herd. Arch Virol. 2014;159(1):17–27. doi: 10.1007/s00705-013-1781-9. [DOI] [PubMed] [Google Scholar]
- 33.Rowland RR, Steffen M, Ackerman T, Benfield DA. The evolution of porcine reproductive and respiratory syndrome virus: quasispecies and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology. 1999;259(2):262–6. doi: 10.1006/viro.1999.9789. [DOI] [PubMed] [Google Scholar]
- 34.Shi M, Lam TT, Hon CC, Hui RK, Faaberg KS, Wennblom T, Murtaugh MP, Stadejek T, Leung FC. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. 2010;154(1–2):7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Shi M, Lam TT, Hon CC, Murtaugh MP, Davies PR, Hui RK, Li J, Wong LT, Yip CW, Jiang JW, Leung FC. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010;84(17):8700–11. doi: 10.1128/JVI.02551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J, Shen D, Cui J, Zhao B. Accelerated evolution of PRRSV during recent outbreaks in China. Virus Genes. 2010;41(2):241–5. doi: 10.1007/s11262-010-0507-2. [DOI] [PubMed] [Google Scholar]
- 37.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Li Y, Liu R, Wang X, Gao F, Lin T, Huang T, Yao H, Tong G, Fan H. Porcine reproductive and respiratory syndrome virus ORF5a protein is essential for virus viability. Virus Res. 2013;171(1):178–85. doi: 10.1016/j.virusres.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Thaa B, Sinhadri BC, Tielesch C, Krause E, Veit M. Signal peptide cleavage from GP5 of PRRSV: a minor fraction of molecules retains the decoy epitope, a presumed molecular cause for viral persistence. PLoS One. 2013;8(6):e65548. doi: 10.1371/journal.pone.0065548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tun HM, Shi M, Wong C, Ayudhya S, Amonsin A, Thanawonguwech R, Leung F. Genetic diversity and multiple introductions of porcine reproductive and respiratory syndrome viruses in Thailand. Virol J. 2011;8:164. doi: 10.1186/1743-422X-8-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vu HL, Kwon B, Yoon K-J, Laegreid WW, Pattnaik AK, Osorio FA. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol. 2011;85(11):5555-64. [DOI] [PMC free article] [PubMed]
- 42.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, et al. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991;13(3):121–30. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 43.West J, Hernandez R, Ferreira D, Brown DT. Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly. J Virol. 2006;80(9):4458–68. doi: 10.1128/JVI.80.9.4458-4468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol. 2005;79(19):12495–506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Chang X, Xiao S, Chen H, Zhou R. Evidence for the adaptive evolution of ORF 5 gene of Porcine reproductive and respiratory syndrome virus isolated in China. Acta Virol. 2010;54(4):281–5. doi: 10.4149/av_2010_04_281. [DOI] [PubMed] [Google Scholar]
- 46.Yoon SH, Kim H, Kim J, Lee H-K, Park B, Kim H. Complete genome sequences of porcine reproductive and respiratory syndrome viruses: perspectives on their temporal and spatial dynamics. Mol Biol Rep. 2013;40(12):6843–53. doi: 10.1007/s11033-013-2802-1. [DOI] [PubMed] [Google Scholar]
- 47.Yu X, Chen N, Wang L, Wu J, Zhou Z, Ni J, Li X, Zhai X, Shi J, Tian K. New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet Microbiol. 2012;158(3):291–9. doi: 10.1016/j.vetmic.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerman J, Yoon K-J, Wills R, Swenson S. General overview of PRRSV: a perspective from the United States. Vet Microbiol. 1997;55(1):187–96. doi: 10.1016/S0378-1135(96)01330-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The information of Vietnamese PRRSV strains using in this study is included within the supplement data (Additional file 1: Table S1). The phylogenetic data was deposited in the TreeBase at http://purl.org/phylo/treebase/phylows/study/TB2:S20099?x-access-code=a533b9962d6c27b5884321c9668bf275&format=html.


